Abstract
Endometriosis is a common gynecological disease found in approximately 10% of reproductive-age women. Gene expression analysis has been performed to explore alterations in gene expression associated with endometriosis; however, the underlying transcription factors (TFs) governing such expression changes have not been investigated in a systematic way. In this study, we propose a method to integrate gene expression with TF binding data and protein–protein interactions to construct an integrated regulatory network (IRN) for endometriosis. The IRN has shown that the most regulated gene in endometriosis is RUNX1, which is targeted by 14 of 26 TFs also involved in endometriosis. Using 2 published cohorts, GSE7305 (Hover, n = 20) and GSE7307 (Roth, n = 36) from the Gene Expression Omnibus database, we identified a network of TFs, which bind to target genes that are differentially expressed in endometriosis. Enrichment analysis based on the hypergeometric distribution allowed us to predict the TFs involved in endometriosis (n = 40). This included known TFs such as androgen receptor (AR) and critical factors in the pathology of endometriosis, estrogen receptor α, and estrogen receptor β. We also identified several new ones from which we selected FOXA2 and TFAP2C, and their regulation was confirmed by quantitative real-time polymerase chain reaction and immunohistochemistry (IHC). Further, our analysis revealed that the function of AR and p53 in endometriosis is regulated by posttranscriptional changes and not by differential gene expression. Our integrative analysis provides new insights into the regulatory programs involved in endometriosis.
Keywords: regulatory programs, transcription factor, endometriosis, gene expression, ChIP-seq
Introduction
Endometriosis is a very common gynecological disease. The main clinical manifestations of endometriosis include dysmenorrhea, pelvic pain, and infertility. More than 50% of infertility cases and 60% of pelvic pain diagnoses are associated with endometriosis.1,2 It typically occurs due to migration of endometrial cells from uterine cavity to the peritoneum.3 Many theories have been proposed to explain the reasons for the formation of ectopic endometrium4 such as retrograde menstruation,5 Müllerianosis,6 coelomic metaplasia,7 and transplantation.8 Based on experimental and clinical evidence, we have proposed that ectopic differentiation of endometrial stem cells may provide a novel mechanism of endometriosis.9–12 Although there are likely multiple causes of endometriosis, microarray studies have consistently revealed the altered expression of many genes in this disease compared to normal endometrium.13–16
Gene expression is under precise regulation by specific transcription factors (TFs). To understand the molecular mechanisms of endometriosis, it is important to identify the regulatory programs underlying this disease. Several TFs have been found to be involved in endometriosis. Increased expression of nuclear factor κB (NF-κB), RHOC, and FOXL2 have been reported in endometriosis.17–19 Octamer-binding transcription factor 4 (OCT4) is significantly upregulated in human ectopic endometriotic tissues, resulting in ectopic endometrial growth by stimulating the migration of endometrial cells.20 Estrogen receptor α (Esr1) and estrogen receptor β (Esr2) control the proliferation and apoptosis of endometrium cells through transcriptional regulation of multiple genes, including hepatocyte nuclear factor 1b (HNF-1b), cyclin D1, and growth regulation by estrogen in breast cancer (GREB).21,22 Gonzalez-Ramos et al showed that in endometriotic cells NF-κB stimulates inflammation and cell proliferation and inhibits apoptosis to promote the development and maintenance of endometriosis.23 Our laboratory previously reported the function of the TF Homeobox A10 (HOXA10) in endometriosis and showed the regulatory functions of TFs that regulate its expression.24–28 Also, Lu et al reported the downregulation of HOXA10 in endometriosis.29 However, the underlying regulatory network for endometriosis has never been investigated. Gene expression profiling from microarray or ribonuclease-sequencing (RNA-Seq) experiments can be used to identify differentially expressed genes (DEGs), including TFs. However, in many cases, the deregulation of TFs in disease might not be reflected by expression changes at the messenger RNA (mRNA) level. A nonsynonymous mutation in p53 gene can completely abolish its transcriptional activity without detectable expression change at mRNA and/or protein level. Posttranslational modifications frequently alter TF activity. Furthermore, many TFs are expressed at very low levels, and consequently their changes in expression are not detected in gene expression array data. As TFs are regulatory molecules, small changes in TFs can have an amplified biologic effect due to their profound regulation of gene expression. Thus, to identify the role of TFs in disease either a candidate gene approach or computational studies are often required.
Computational methods have been proposed to identify the changes in the expression of target genes due to TFs to unearth the regulatory programs in disease.30–36 Rhodes et al studied the regulatory programs in cancer and identified gene expression signatures in cancers compared to normal tissues.35 Further, they predicted that the differential gene expression is based on the existence of TF binding sites in the promoters of putative target genes that act as regulatory signatures. This method is very effective in unveiling the deregulation of TFs, but its application has been limited by the high positive rate of the predicted TF target genes. Recently, the development of chromatin immunoprecipitation with microarray (ChIP-chip) or massively parallel DNA sequencing (ChIP-Seq) has revolutionized the study of TF target genes,37,38 and thus provide efficient methods to identify accurate lists of binding sites for TFs in the regulatory signatures. Thus, a combination of gene expression data with TF target genes from the ChIP-chip or ChIP-Seq experiments provides us with the opportunity to understand transcriptional regulation underlying in endometriosis. Here, we report an integrated regulatory network (IRN) that elucidates molecular mechanisms driving endometriosis.
Materials and Methods
Gene Expression, TF Binding, and Protein–Protein Interactions
We searched the Gene Expression Omnibus (GEO) database to collect gene expression data sets associated with endometriosis.39 To ensure an adequate statistical power in the subsequent analysis, we selected 2 data sets, GSE7305 and GSE7307, which contained at least 10 samples for both endometriosis and eutopic endometrium controls. In both data sets, the disease samples are ovarian endometriomas, representing the most frequent lesion location for endometriosis. The first data set (GSE7305) adopted a paired experiment design, namely, the eutopic endometrium and the ectopic endometrium were collected from the same patients.16 In contrast, the samples in the second data set (GSE7307) were unpaired with ectopic endometrium from women with endometriosis and normal endometrium from women without endometriosis and analyzed using normal t test (groupwise). For comparison, a microarray data set of peritoneal endometriosis (GSE11691) was also investigated, which contained paired eutopic and ectopic endometrial samples from 9 women with peritoneal endometriosis.40 The target gene data set was obtained from the ChIP Enrichment Analysis (ChEA) database,41 which provided 83 target gene sets for 32 sequence-specific human TFs based on their genomic occupation profiles from the ChIP-Chip or ChIP-Seq experiments. A TF may associate with multiple target gene sets from independent experiments, usually being performed in different tissue or cell lines. The protein–protein interactions (PPIs) were available from the Interologous Interaction Database (I2D)42 or from a more specific study by Ravasi et al.43 The I2D integrates known experimental and predicted PPIs. The latter data set contains PPIs of human TFs from a systematic screening experiment.
Identification of DEGs in Endometriosis
To identify genes that are differentially expressed, we compared the expression values of all probe sets in endometriosis samples with those in normal endometrium samples using the t test. For the paired data set (GSE7305), a paired t test was applied. The genes with at least 1 significant probe set (P < .001) were identified as DEGs. We identified upregulated and downregulated genes for each of the 2 data sets, and the genes that are significant in both were selected for subsequent analysis.
Inferring Regulatory Activity Changes of TFs in Endometriosis
The modification of TF regulatory activity can often be reflected by changes in the expression of its target genes. If a TF is deregulated in endometriosis with respect to normal endometrium, we would expect that the target genes of this TF are more likely to be differentially expressed, either upregulated or downregulated. Thus, we identified the TFs that were potentially involved in endometriosis by examining the enrichment of their target genes in the DEGs. We applied Fisher exact test to specifically examine the overlapping of 2 gene sets: the DEG set GD and the target gene set of a TF.44 Given the 2 gene sets, the number of overlapping genes k follows a hypergeometric distribution:
where m is the number of genes,39 n is the number of genes in GD, and N is the total number of genes. The significance for enrichment of genes in GD can be calculated as P(x ≥ k), the probability of observing an equal or larger intersection between the 2 gene sets. We examined the enrichment of all TF target gene sets in the DEGs GD. To correct for multiple testing, we calculated the adjusted P values for all target gene sets using the Benjamini & Hochberg method. The TFs with an adjusted P < .001 were reported as endometriosis-associated TFs.
Construction of Integrative Regulatory Network
To understand the changes in regulatory programs underlying endometriosis, we constructed an integrative regulatory network by combining gene expression data, TF–gene regulatory relationships, and PPIs. First, the regulatory programs that were associated with endometriosis were identified as described in the previous section. Each regulatory program was consistent of a TF and its target genes that were differentially expressed. These regulatory programs were interconnected by TF→TF regulatory interactions identified by the ChIP-Seq or ChIP-chip experiments (available from ChEA database). Second, genes with PPIs were connected. The integrative regulatory network contains 2 types of interactions: TF→TF–gene regulatory interactions and PPIs. The majority of genes in the network are differentially expressed in endometriosis versus normal endometrium except for a few TFs, which are not differentially expressed but are inferred to be endometriosis associated according to target gene enrichment analysis.
Enrichment of PPIs in Target Genes of TFs
To investigate the hypothesis that target genes of the same TF are more likely to interact, we calculated the enrichment of PPIs in the target genes of each of the TFs. First, we calculated the probability of a pair of random selected genes to interact: , where s is the total number of interactions in the PPI data and N is the total number of genes. Second, the number of PPI interactions i among the m target genes of a TF follows a binomial distribution: , where b = m (m − 1)/2, the number of possible interactions. Finally, the P value for enrichment of PPI interactions in the target gene set was calculated as P(x ≥ i), the probability of observing equal or larger number of interactions. Again the Benjamini & Hochberg method was used for correcting multiple testing. Note that self-interactions in the PPI data were excluded from the analysis.
Gene Ontology Analysis
Gene ontology analysis was performed using the online bioinformatics resource the Database for Annotation, Visualization and Integrated Discovery (DAVID).45
Sample Collection and Preparation
Normal endometrial tissue was collected from 40 women without endometriosis who underwent a curettage or hysterectomy for nonendometrial disease including fibroid uterus and prolapse (samples collected from 2012 to 2013 in Department of Gynecology, Minially Invasive Gynecology Center, Beijing Obstetrics and Gynecology Hospital Capital Medical University). Only samples with benign histology were included in the study. The study was approved by the Institutional Review Boards of both the institutions. Eutopic and ectopic endometrial tissues were collected from 40 women with ovarian endometriosis who underwent laparoscopy. All women (controls and participants with endometriosis) were between 23 and 43 years of age with normal menstrual cycles and did not use hormonal therapies for at least 3 months prior to surgery. Samples were not used when menstrual cycle dating as determined by last menstrual period was not concordant with histologic dating. Participants with endometrial hyperplasia, cancer, untreated thyroid disease, elevated prolactin, or infection were excluded. Dissected tissues were immediately frozen in liquid nitrogen for RNA and protein extraction or fixed in 10% formalin and embedded in paraffin for histological analysis.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated using TRIzol (Invitrogen, Carlsbad, California) and purified in RNeasy minicolumns (QIAGEN, Valencia, California) with on-column deoxyribonuclease digestion, per the manufacturers’ instructions. First-strand complementary DNA (cDNA) was reverse transcribed using iScript cDNA Synthesis Kit and iQ SYBR Green Supermix-based assays (Bio-Rad, Hercules, California) were performed. Total FOXA2 mRNA levels were measured by quantitative polymerase chain reaction (PCR) with specific gene primers as follows:
FOXA2-1: 5′-GCCCCAACAAGATGCTGAC-3′
FOXA2-2: 5′-CACCTTCAGGAAACAGTCGTTG-3′
TFAP2C-1: 5′-GACCAAGAACCCTCTGAACCTC-3′
TFAP2C-2: 5′-GGGACAGTCGCCTCTGTACTTC-3′
β-actin-1: 5′-TGACGTGGACATCCGCAAAG-3′
β-actin-2: 5′-CTGGAAGGTGGACAGCGAGG-3′
iCycler iQ real-time PCR detection system and SYBR green PCR mix were used to carry out the quantitative real-time PCR (qRT-PCR). For each experimental sample, a control without reverse transcriptase was run to verify that the amplification product arose from cDNA and not from genomic DNA. The 3-step optimized thermal cyclic conditions used were: initial denaturation and enzyme activation for 3 minutes at 95°C followed by 45 cycles (denaturation for 15 seconds at 95°C, annealing for 20 seconds at 60°C, and extension for 25 seconds at 72°C), followed by melt curve for 1 minute at 95°C and for 1 minute at 55°C, and holding temperature is at 4°C. The relative expression levels, normalized to β-actin, were determined using the comparative cycle threshold (Ct) method (known as the 2ΔΔ CT method).46,47
The relative abundance of FOXA2 and TFAP2C transcripts was quantified using the comparative Ct method with β-actin as an internal control. The data were analyzed from 3 independent experiments, and statistical significance was validated by Student t test.
Immunohistochemistry
Slides with tissue sections from endometriosis and normal endometrial tissue were deparaffinized in a degraded concentration of ethanol. After blocking with 3% hydrogen peroxide for 15 minutes, the sections were heated in 0.01 mol/L sodium citrate for 3 minutes, followed by washing in phosphate-buffered saline (PBS) with 0.1% Tween 20 and then treated with 3% hydrogen peroxide solution for 5 minutes. Immunohistochemical staining was performed on slides with rabbit polyclonal antibody directed against FOXA2 (ab2363; Abcam, Cambridge, MA, USA) overnight at 4°C. Goat immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, California) was used as the negative control. Biotinylated secondary antibody was purchased from Beyotime Institute of Biotechnology (Jiangsu, China) and applied for 1 hour at 4°C. The samples were finally washed in PBS and diaminobenizidine tetrahydrochloride solution was applied, which was followed by washing under running tap water as well as nuclear counterstaining with hematoxylin (Sigma, St. Louis, MO, USA). The stained slides were viewed with microscope (Olympus Optical Co, Ltd, Miami, FL, USA) under 400× magnification. Positive cells were characterized by the brown staining of FOXA2 antibody. Data were analyzed by 2 dependent investigators with Image Pro Plus 6.0.
Results
Overview of the Integrative Network Analysis
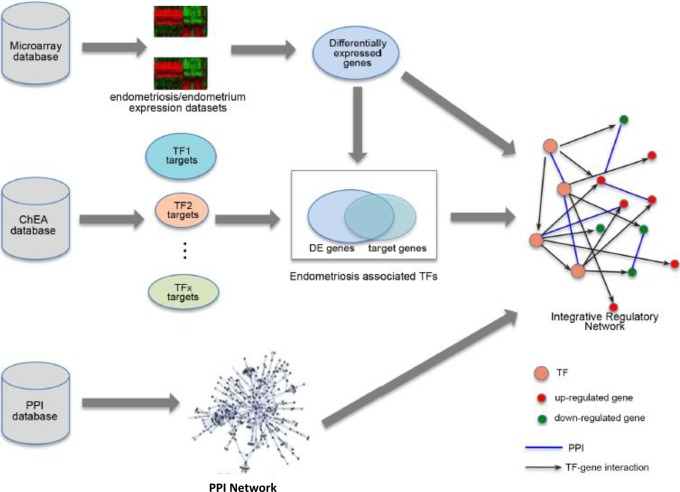
To understand the underlying regulatory programs, we constructed an IRN for endometriosis as shown in Figure 1. The network was based on an integrative analysis of 3 sources: gene expression profiles, TF binding information, and PPIs. Gene expression was quantified from microarray data from samples of endometriosis and normal endometrium. Differentially expressed genes in endometriosis and normal endometrium were identified. Then, we obtained the target gene sets for ∼70 TFs based on ChIP-chip or ChIP-Seq experiments from ChEA database. We examined the enrichment of target genes of TFs in the DEGs and identified regulatory programs in endometriosis. Each regulatory program consists of a TF, and its target genes that are significantly up- or downregulated in endometriosis. These regulatory programs are connected via TF→TF interactions and form a regulatory network. The PPIs data are also incorporated in the network to form an integrative regulatory network.
Figure 1.
The schematic diagram of our integrative analysis. PPI indicates protein–protein interaction.
Differentially Expressed Genes in Endometriosis Versus Normal Endometrium
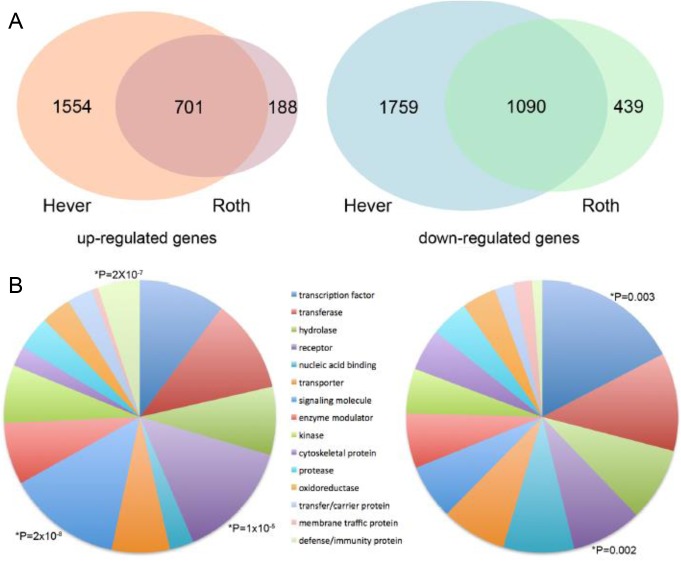
Microarray data on endometriosis were collected from GEO database39 to obtain 2 data sets (Hever and Roth) and identify genes in each data set that were significantly up- or downregulated in endometriosis versus normal endometrium using t test. We compared the t scores of all genes in the 2 data sets and found that the 2 t score profiles were highly correlated (ρ = .77), suggesting a good consistency between the 2 data sets. We identified 2255 upregulated and 2849 downregulated genes (adjusted P < .001) in the Hever data set and 889 upregulated and 1529 downregulated genes with same significance in the Roth data set. The combination of results from both data sets produced 701 upregulated and 1090 downregulated genes as shown in Figure 2A, and the distribution of these genes was examined in 29 gene categories from the Protein Analysis Through Evolutionary Relationships (PANTHER) Classification System.48 As shown in Figure 2B, the 701 upregulated genes were prominent in the receptor signaling molecule and defense/immunity protein categories, while 1090 downregulated genes were classified in TF and cytoskeletal categories. These results are consistent with pathway analysis results reported by Hever et al16 and Zhao et al22 who found that many immune-related pathways were involved in endometriosis.
Figure 2.
Differentially expressed genes between endometriosis and normal endometrium. (A) Comparison of upregulated genes and downregulated genes identified from the Hever data and the Roth data. (B) Gene Ontology analysis of shared upregulated and downregulated genes.
Regulatory Programs in Endometriosis
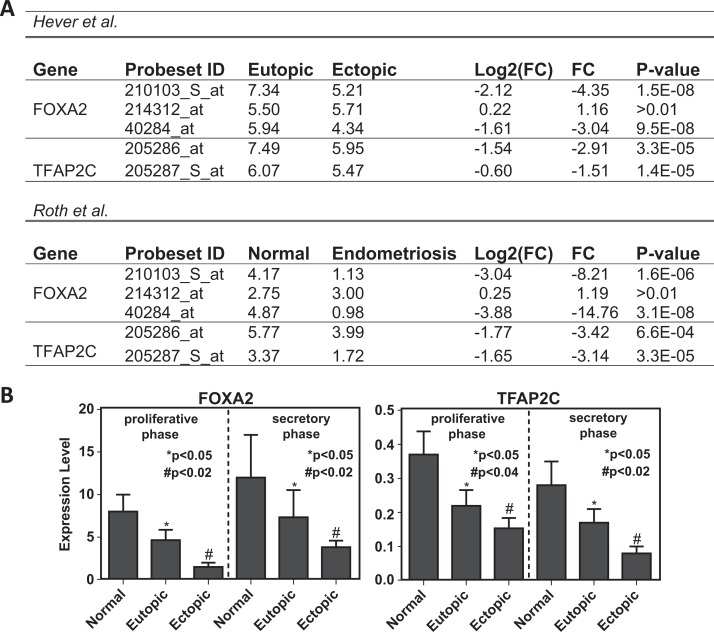
To identify regulatory programs in endometriosis, we used Rhodes et al method35 with a slight modification. This method identified the enrichment of TF target genes in the DEGs based on ChIP-Seq data. A total of 83 target gene sets for 63 TFs were collected from ChEA database. Note that the target gene number is more than the number of TFs because some TFs had multiple target gene sets. Among these target gene sets, we found 30 targets for 26 TFs which are significantly enriched (adjusted P < .001) in the DEGs. Then, we defined 26 transcriptional regulatory programs each with a TF and target genes that were up- or downregulated in endometriosis. These regulatory programs connected into a regulatory network, since a TF in one regulatory program can be the target of the TF from another program. Strikingly, most of the identified TFs do not show significant differential expression between endometriosis and normal endometrium based on microarray analysis. Among these 26 TFs, only 5 are significant at the mRNA level in both the Hever and the Roth microarray data sets. Esr2 is upregulated, whereas FOXA2, TFAP2C1 (Figure 3B), Esr1, and POU5F1 are downregulated in endometriosis.
Figure 3.
Downregulation of FOXA2 and TFAP2C in endometriosis. (A) Expression levels of FOXA2 and TFAP2C measured by microarray. FOXA2 and TFAP2C are represented by 3 and 2 probed sets, respectively. In the Hever data set, expression levels are measured and compared between eutopic and ectopic endometrium tissues. In the Roth data set, expression levels are measured and compared between normal endometrium and endometriosis cells. The P value is calculated based on Student t test. (B) qRT-PCR. FOXA2 and TFAP2C downregulation in 40 participants with endometriosis compared to controls using qRT-PCR. The expression levels of FOXA2 and TFAP2C in normal, eutopic, and ectopic endometrium cells were measured in proliferative and secretory phases. Each bar represents the mean + SD for data from 3 individual experiments, and each experiment was performed in triplicate.
*denotes statistical significance (P < .05) between normal and eutopic.
#denotes statistical significance (P < 0.02) between normal and ectopic.
The P value for TFAP2C in proliferative phase is <.04 comparing normal with ectopic. qRT-PCR indicates quantitative real-time polymerase chain reaction; SD, standard deviation.
The endometriosis-associated TFs are showed in Table 1. The table shows that there were 373 target genes for androgen receptor (AR) which showed significant differential expression in endometriosis versus normal endometrium. These target genes were enriched in both upregulated (146 genes, P = 6 × 10−6) and downregulated (227 genes, P = 3 × 10−9). Some of the TFs may act predominantly as activators or repressors; for example, E2F4 targets were enriched in the upregulated genes (P = 3 × 10−13) but depleted in the downregulated ones (P = 2 × 10−4). More specifically, in the 95 differentially expressed target genes of E2F4, 88 are downregulated and only 7 are upregulated.
Table 1.
Regulatory Programs Associated With Differential Gene Expression in Endometriosis.a
| TF | Cell Type | #Targets | Differential Genes | Upregulated Genes | Downregulated Genes | |||
|---|---|---|---|---|---|---|---|---|
| Number | P Value | Number | P Value | Number | P Value | |||
| AR | PC3 | 2941 | 373 | 2.2E-15 | 146 | 5.6E-06 | 227 | 2.5E-09 |
| CDX2 | CACO-2 | 389 | 54 | 0.00067 | 13 | .69 | 41 | 7.1E-05 |
| CTNNB1 | HCT116 | 897 | 112 | 8.2E-05 | 37 | .21 | 75 | .00014 |
| E2F4 | JURKAT | 697 | 95 | 1.3E-05 | 7 | 1 | 88 | 2.7E-13 |
| EOMES | HESC | 865 | 113 | 1.3E-05 | 45 | .0084 | 68 | .0015 |
| ERG | JURKAT | 294 | 43 | .00089 | 19 | .017 | 24 | .041 |
| ESR1 | MCF7 | 216 | 37 | .00011 | 11 | .21 | 26 | .00025 |
| ESR1 | T-47D | 176 | 31 | .00025 | 12 | .046 | 19 | .0065 |
| ESR2 | MCF7 | 396 | 71 | 1.1E-08 | 25 | .0084 | 46 | 2.7E-06 |
| EWS-FLI1 | SK-N-MC | 517 | 77 | 4.8E-06 | 38 | 4.4E-05 | 39 | .027 |
| FOXA2 | HepG2 | 2823 | 355 | 2.9E-14 | 139 | 1.5E-05 | 216 | 1.4E-08 |
| HNF4A | HepG2 | 5778 | 602 | 9.8E-09 | 232 | .0026 | 370 | 5.6E-06 |
| MITF | Melanoma | 5067 | 533 | 3.3E-08 | 211 | .00078 | 322 | 6.7E-05 |
| MYC | AK7 | 1783 | 195 | .00033 | 80 | .014 | 115 | .020 |
| MYC | Medulloblastoma | 1162 | 162 | 1.9E-09 | 70 | 1.5E-05 | 92 | .00014 |
| MYCN | Shep-21N | 287 | 46 | 8.2E-05 | 8 | .92 | 38 | 1.2E-06 |
| NANOG | HESC | 1152 | 141 | 2.3E-05 | 45 | .26 | 96 | 1.5E-05 |
| PAX3-FKHR | Rhabdomyosarcoma | 969 | 135 | 4.0E-08 | 61 | 1.6E-05 | 74 | .0019 |
| POU3F2 | 501MEL | 1310 | 157 | 2.3E-05 | 63 | .0084 | 94 | .0026 |
| POU5F1 | HESC | 468 | 77 | 9.2E-08 | 26 | .027 | 51 | 4.0E-06 |
| PPARD | Myofibroblast | 3038 | 324 | 1.8E-05 | 137 | .00072 | 187 | .015 |
| SCL | HPC-7 | 207 | 36 | .00010 | 16 | .0084 | 20 | .015 |
| SMAD2 | HaCaT | 1698 | 204 | 8.9E-07 | 73 | .045 | 131 | 1.4E-05 |
| SMAD3 | HaCaT | 1698 | 204 | 8.9E-07 | 73 | .045 | 131 | 1.4E-05 |
| SOX2 | HESC | 878 | 109 | .00012 | 37 | .19 | 72 | .00032 |
| SOX2 | LN229-GBM | 2824 | 356 | 2.4E-14 | 147 | 5.1E-07 | 209 | 3.7E-07 |
| SOX2 | SW620 | 2104 | 350 | 8.8E-36 | 126 | 3.0E-09 | 224 | 5.4E-25 |
| TFAP2C | MCF7 | 1115 | 159 | 5.9E-10 | 50 | .059 | 109 | 2.3E-09 |
| TP53 | U2OS | 728 | 111 | 9.8E-09 | 53 | 2.4E-06 | 58 | .0026 |
| TRIM28 | NTERA2 | 677 | 86 | .00033 | 44 | .00016 | 42 | .20 |
Abbreviations: TF, Transcription Factor; CDX2, Caudal-type homeobox transcription factor 2; CTNNB1, Catenin beta-1; E2F4, E2F transcription factor 4; EOMES, Eomesodermin; ERG, ETS-related gene; EWS-FLI1, Ewings sarcoma/Friend leukemia integration factor; FOXA2, Forkhead box A2; HNF4A, Hepatocyte nuclear factor 4-alpha; MITF, Microphthalmia-associated transcription factor; MYC, c-Myc; MYCN, v-Myc; NANOG, Nanog; PAX3-FKHR, Paired box3 forkhead box; POU3F2, POU class 3 homeobox 2; POU5F1, POU class 5 homeobox 1; PPARD, Peroxisome proliferator-activated receptor delta; SCL, Stem cell leukemia; SMAD2, Mothers against decapentaplegic homolog 2; SMAD3, Mothers against decapentaplegic homolog 3; SOX2, SRY (sex determining region Y) box 2; TFAP2C, Transcription factor AP2 (activating enhancer binding protein 2); TP53, Tumor protein p53; TRIM28, Tripartite Motif containing 28; AR, androgen receptor; ESR1, estrogen receptor α; ESR2, estrogen receptor β.
aTFs are identified if their target genes are significantly enriched in upregulated or downregulated genes in endometriosis versus normal endometrium.
Integrative Regulatory Network and PPIs in Endometriosis
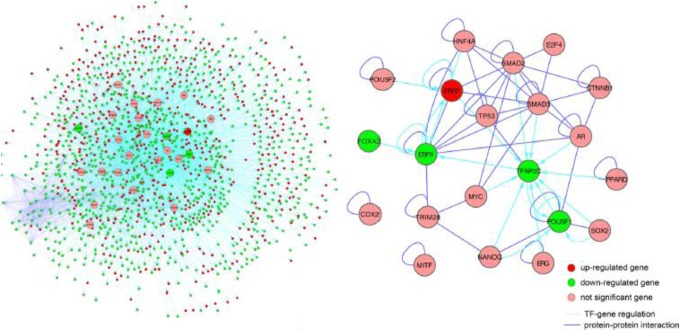
We further incorporated the PPIs into the regulatory program network made in an IRN for endometriosis (Figure 4A). The IRN is formed of 5172 TF→gene interactions and 1490 PPIs, involving 26 regulatory TFs and 1445 DEGs in endometriosis (538 upregulated and 907 downregulated). The number of target genes regulated by the 26 TFs varies dramatically, ranging from 36 to 671 with an average of 199. The number of regulatory TFs for each gene also varies significantly, roughly following an exponential distribution. The most regulated gene is RUNX1, which is targeted by 14 of the 26 endometriosis-associated TFs. Interestingly, RUNX1 itself is a TF known to be involved in the development of normal hematopoiesis.49 It is upregulated significantly in endometriosis by 3-fold in the Hever data and by 5-fold in the Roth data, consistent with a study performed in a rat endometriosis model.50 On average, each gene interacted with 1.6 partners (PPIs). The numbers of partners range from 0 to 43 and follow a power law distribution. The core of the IRN is a subnetwork consisting of regulatory relationships and PPIs among the 26 endometriosis-associated TFs (Figure 4B). Interestingly, 15 (58%) of the 26 TFs are self-interacting, with a significant increased frequency above the average (21%). Another interesting observation for the core subnetwork is that the majority of PPIs were among 9 TFs: AR, Esr1, Esr2, TP53, SMAD2, SMAD3, E2F4, HNF4A, and CTNNB1. It is also notable that TFAP2C is highly regulated by 10 TFs. As mentioned earlier, among 26 TFs, only Esr1, TFAP2C, and POU5F1 are significantly downregulated, and Esr2 is significantly upregulated, in both the Hever and the Roth data sets. The gene enrichment data analysis of TF target genes that were either differentially expressed or not showed that inner set PPIs are significantly enriched in 48 of the 83 TF target gene sets (adjusted P < .01). There are 1051 interactions between the 573 target genes of E2F4 in Jurkat cells, which is about 4-fold more than expected (P = 10−320).
Figure 4.
Integrated regulatory network for endometriosis. (A) A global view of the integrated regulatory network that contains transcription factors and their target genes (Left image). (B) A subnetwork that only includes transcription factors (Right image).
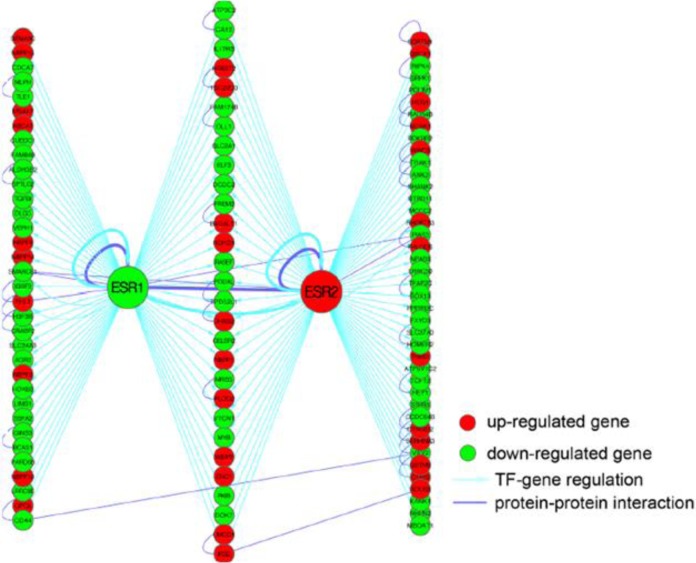
Involvement of Esr1 and Esr2 in Endometriosis
Our analysis indicates that Esr1 and Esr2 target genes are enriched in DEGs, especially in downregulated genes in endometriosis relative to normal endometrium (Figure 5). Specifically, 63 Esr1 target genes are differentially expressed, in which 21 (33%) are upregulated and 42 (67%) are downregulated. Seventy Esr2 target genes are differentially expressed, in which 24 (34%) are upregulated and 46 (66%) are downregulated in endometriosis. These results suggest that, as expected, Esr1 and Esr2 are 2 critical factors in the pathology of endometriosis. Further, our analysis observed that AR and p53 target genes are also significantly enriched in both upregulated and downregulated genes in endometriosis, implicating a function for these genes and TFs in endometriosis, even though they were not differentially expressed between endometriosis and normal endometrium. This suggests that their function is regulated at the posttranscriptional level (eg, phosphorylation).
Figure 5.
The regulatory program of Esr1 and Esr2. Esr1 and Esr2 specific targets and shared targets are shown as the left, the middle, and the right columns, respectively. Only differentially expressed Esr1 or Esr2 target genes are displayed. Esr1 indicates estrogen receptor α; Esr2 estro- gen receptor β.
Validation of FOXA2 and TFAP2C downregulation in Endometriosis
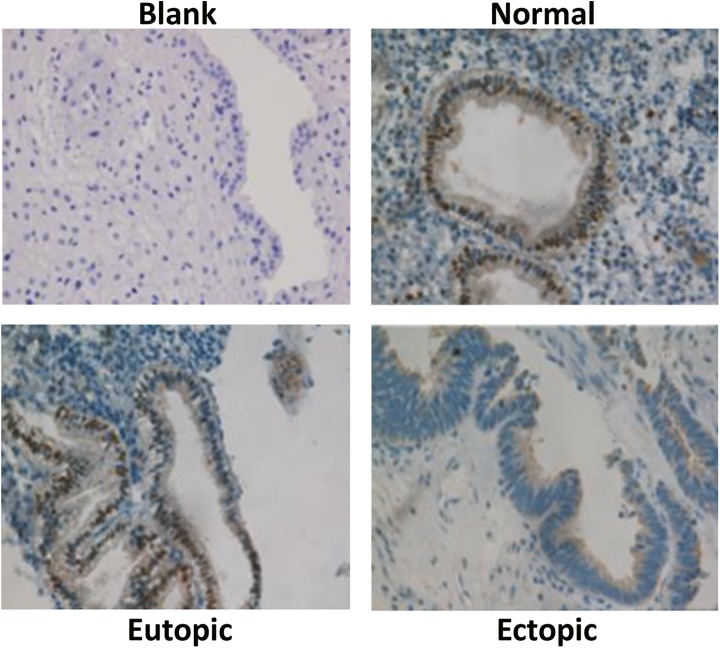
The target genes of these 2 TFs were highly enriched in the genes that were identified as upregulated or downregulated in endometriosis (Table 1). Microarray data from both the Hever and the Roth data sets showed the downregulation of FOXA2 and TFAP2C in endometriosis (Figure 3A).16 To confirm the downregulation of these 2 TFs in endometriosis, we carried out qRT-PCR with the RNA extracted from eutopic and ectopic endometrium of 40 patients with endometriosis as well as of 40 normal endometrium. As shown in Figure 3B, FOXA2 and TFAP2C are significantly downregulated in eutopic and ectopic endometrium relative to normal endometrium (P < .01), in both proliferative and secretory phases. The reduction in FOXA2 protein levels in endometriosis is further validated by immunohistochemical staining (Figure 6). The level of FOXA2 protein decreases substantially in the ectopic tissue from patients with endometriosis relative to normal endometrium, which corresponds with the results reported for placenta protein (PP-14) from serum of patients with endometriosis.51 The staining also reveals that FOXA2 is mainly present in the nucleus of both endometrial glandular and luminal epithelial cells but not in stroma cells. These results confirmed our integrative analysis results, suggesting that FOXA2 and TFAP2C might be implicated in endometriosis.
Figure 6.
Immunohistochemistry of FOXA2. Protein distribution in normal endometrium compared to the eutopic and ectopic tissues from participants with endometriosis. Original magnification ×60.
Discussion
In this study, we describe a method based on gene expression, TF-gene pathways/connections, and PPIs that can identify an IRN for understanding molecular mechanisms involved in endometriosis. We predicted several TFs that might play critical roles in the pathology of endometriosis. The study revealed that 1 TF might act as a key regulator in multiple regulatory programs, since it can regulate a different set of target genes under different conditions or in different tissues. E2F4 binds to 1002 genes in Jurkat cells, while in lymphoblastoid cells it binds to 2,998 genes. Additionally, 259 genes are targeted by E2F4 in both cell types. Rhodes et al applied a similar analysis to identify regulatory programs in cancer transcriptome, where TF target genes were predicted by searching TF binding sites in the promoter regions of genes.35 However, this method is valid only for the TFs with known high affinity binding motifs, and an extremely large fraction of these predicted targets are false positives. Thus, target genes determined by ChIP-seq or ChIP-chip experiments are more highly accurate. The microarray data analysis from endometriosis samples revealed that the expression of Esr1 and Esr2 is downregulated and upregulated, respectively, suggesting that both genes have a function in endometriosis. This is in consistent with previous report that Esr1 and Esr2 have opposite effects on gene transcription,52 and upon activation by binding to estrogens, Esr1 and Esr2 can form homodimers (1,1 and 2,2) or heterodimers (1,2) that interact with specific DNA sequences to activate gene transcription.53 Esr2 has been shown to have antiproliferative effects and oppose the actions of Esr1 in reproductive tissue.54 Regulatory interactions identified by ChIP-seq showed that Esr1 regulates the transcription of Esr2 gene and both autoregulate their own transcription. These regulatory and physical interactions between Esr1 and Esr2 might be involved in the pathology of endometriosis, as we found enrichment of their target genes in DEGs in endometriosis.
We also identified an AR regulatory program in endometriosis, where its function is not regulated by change in expression but rather by posttranscriptional changes such as phosphorylation. This would not have been detected by traditional microarray or other sequencing-based studies. Androgens and AR are not only required for male reproductive function but also essential for female reproductive physiology.55 The AR signaling plays a critical role in the differentiation of human endometrial stromal cells into decidual cells.56 AR perturbation is associated with multiple gynecological disorders, including polycystic ovary syndrome,57 premature ovarian failure, and adverse pregnancy outcome. Association studies have shown that polymorphic CAG repeats of the AR gene were associated with endometriosis in different populations.58–60 The AR-CAG repeat length does not constitute an important factor for the genetic predisposition to endometriosis, but 19 AR-CAG repeats can be regarded as high-risk marker.58 The involvement of AR in endometriosis maybe related to its regulatory function in human endometrial apoptosis.61,62
It is also interesting to see the potential involvement of p53 in endometriosis. A number of association studies have reported the link between p53 polymorphism and the risk of endometriosis in several populations.63–67 In addition, decreased expression of p53 was observed more often in the endometrium of patients with endometriosis than in controls.68 Gylfason et al investigated p53 copy numbers in eutopic endometrial and endometriotic tissue and observed a link between somatic p53 locus alterations and the pathogenesis of late- or severe-stage endometriosis.69 Ovarian endometriosis can progress to epithelial cytological atypia and malignant transformation.70 Thus, our integrative analysis suggests that p53 regulatory program might be implicated in the pathology of endometriosis and implies a possible link between endometriosis and ovarian cancer.
We validated the downregulation of FOXA2 and TFAP2C in endometriosis. Our results confirm that their expression is downregulated at both mRNA and protein levels. Similar results were reported for progesterone receptor membrane components 1 and 2 in eutopic endometrium.71 However, it should be noted that the activity changes in transcriptional regulatory programs inferred from this analysis might not be reflected by expression changes in the corresponding TFs because TFs are typically subject to intensive posttranscriptional regulation and posttranslational modifications. Although the functional relevance of several TFs remains unclear, our analysis provides a useful tool to generate new biological insights about transcriptional regulation that may not be acquired directly from gene expression data. The method we proposed in this study could also be applied to determine regulatory mechanisms that are involved in other diseases.
Acknowledgments
We thank Zongzhi Liu for bioinformatics support and Ysabel Ilagan for manuscript editing.
Footnotes
Authors’ Note: Huan Yang and Kai Kang contributed equally to the article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH R01 HD076422 and the National Natural Science Foundation of China Grant No. 81300467.
References
- 1. Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362(25):2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rogers PA, D’Hooghe TM, Fazleabas A, et al. Defining future directions for endometriosis research: workshop report from the 2011 World Congress of Endometriosis in Montpellier, France. Reprod Sci. 2013;20(5):483–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. [DOI] [PubMed] [Google Scholar]
- 4. Vinatier D, Orazi G, Cosson M, Dufour P. Theories of endometriosis. Eur J Obstet Gynecol Reprod Biol. 2001;96(1):21–34. [DOI] [PubMed] [Google Scholar]
- 5. Fauser BC, Diedrich K, Bouchard P, et al. Contemporary genetic technologies and female reproduction. Evian Annual Reproduction. Hum Reprod Update. 2011;17(6):829–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Signorile PG, Baldi F, Bussani R, D’Armiento M, De Falco M, Baldi A. Ectopic endometrium in human foetuses is a common event and sustains the theory of mullerianosis in the pathogenesis of endometriosis, a disease that predisposes to cancer. J Exp Clin Canc Res. 2009;28:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsuura K, Ohtake H, Katabuchi H, Okamura H. Coelomic metaplasia theory of endometriosis: evidence from in vivo studies and an in vitro experimental model. Gynecol Obstet Invest. 1999;47(suppl 1):18–20; discussion 20–12. [DOI] [PubMed] [Google Scholar]
- 8. Laschke MW, Giebels C, Menger MD. Vasculogenesis: a new piece of the endometriosis puzzle. Hum Reprod Update. 2011;17(5):628–636. [DOI] [PubMed] [Google Scholar]
- 9. Taylor HS. Endometrial cells derived from donor stem cells in bone marrow transplant recipients. JAMA. 2004;292(1):81–85. [DOI] [PubMed] [Google Scholar]
- 10. Du H, Taylor HS. Contribution of bone marrow-derived stem cells to endometrium and endometriosis. Stem cells. 2007;25(8):2082–2086. [DOI] [PubMed] [Google Scholar]
- 11. Sasson IE, Taylor HS. Stem cells and the pathogenesis of endometriosis. Ann N Y Acad Sci. 2008;1127:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Figueira PG, Abrao MS, Krikun G, Taylor HS. Stem cells in endometrium and their role in the pathogenesis of endometriosis. Ann N Y Acad Sci. 2011;1221:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rogers PA, D’Hooghe TM, Fazleabas A, et al. Priorities for endometriosis research: recommendations from an international consensus workshop. Reprod Sci. 2009;16(4):335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shinohara A, Kutsukake M, Takahashi M, Kyo S, Tachikawa E, Tamura K. Protease-activated receptor-stimulated interleukin-6 expression in endometriosis-like lesions in an experimental mouse model of endometriosis. J Pharmacol Sci. 2012;119(1):40–51. [DOI] [PubMed] [Google Scholar]
- 15. Khoufache K, Bazin S, Girard K, et al. Macrophage migration inhibitory factor antagonist blocks the development of endometriosis in vivo. PloS One. 2012;7(5):e37264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hever A, Roth RB, Hevezi P, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc Natl Acad Sci U S A. 2007;104(30):12451–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Celik O, Celik E, Turkcuoglu I, et al. Surgical removal of endometrioma decreases the NF-kB1 (p50/105) and NF-kB p65 (Rel A) expression in the eutopic endometrium during the implantation window. Reprod Sci. 2013;20(7):762–770. [DOI] [PubMed] [Google Scholar]
- 18. Meola J, Dentillo DB, Rosa e Silva JC, Hidalgo Gdos S, Paz CC, Ferriani RA. RHOC: a key gene for endometriosis. Reprod Sci. 2013;20(8):998–1002. [DOI] [PubMed] [Google Scholar]
- 19. Governini L, Carrarelli P, Rocha AL, et al. FOXL2 in human endometrium: hyperexpressed in endometriosis. Reprod Sci. 2014;21(10):1249–1255. [DOI] [PubMed] [Google Scholar]
- 20. Chang JH, Au HK, Lee WC, et al. Expression of the pluripotent transcription factor OCT4 promotes cell migration in endometriosis. Fertil Steril. 2013;99(5):1332–1339. e5. [DOI] [PubMed] [Google Scholar]
- 21. Xiao W, Awadallah A, Xin W. Loss of ARID1A/BAF250a expression in ovarian endometriosis and clear cell carcinoma. Int J Clin Exp Pathol. 2012;5(7):642–650. [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao H, Wang Q, Bai C, He K, Pan Y. A cross-study gene set enrichment analysis identifies critical pathways in endometriosis. Reprod Biol Endocrinol. 2009;7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzalez-Ramos R, Defrere S, Devoto L. Nuclear factor-kappaB: a main regulator of inflammation and cell survival in endometriosis pathophysiology. Fertil Steril. 2012;98(3):520–528. [DOI] [PubMed] [Google Scholar]
- 24. Yang H, Zhou Y, Edelshain B, Schatz F, Lockwood CJ, Taylor HS. FKBP4 is regulated by HOXA10 during decidualization and in endometriosis. Reproduction. 2012;143(4):531–538. [DOI] [PubMed] [Google Scholar]
- 25. Fischer CP, Kayisili U, Taylor HS. HOXA10 expression is decreased in endometrium of women with adenomyosis. Fertil Steril. 2011;95(3):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Penna I, Du H, Ferriani R, Taylor HS. Calpain5 expression is decreased in endometriosis and regulated by HOXA10 in human endometrial cells. Mol Hum Reprod. 2008;14(10):613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim JJ, Taylor HS, Lu Z, et al. Altered expression of HOXA10 in endometriosis: potential role in decidualization. Mol Hum Reprod. 2007;13(5):323–332. [DOI] [PubMed] [Google Scholar]
- 28. Browne H, Taylor H. HOXA10 expression in ectopic endometrial tissue. Fertil Steril. 2006;85(5):1386–1390. [DOI] [PubMed] [Google Scholar]
- 29. Lu H, Yang X, Zhang Y, Lu R, Wang X. Epigenetic disorder may cause downregulation of HOXA10 in the eutopic endometrium of fertile women with endometriosis. Reprod Sci. 2013;20(1):78–84. [DOI] [PubMed] [Google Scholar]
- 30. Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27(2):167–171. [DOI] [PubMed] [Google Scholar]
- 31. Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100(26):15522–15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alter O, Golub GH. Integrative analysis of genome-scale data by using pseudoinverse projection predicts novel correlation between DNA replication and RNA transcription. Proc Natl Acad Sci U S A. 2004;101(47):16577–16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai HK, Lu HH, Li WH. Statistical methods for identifying yeast cell cycle transcription factors. Proc Natl Acad Sci U S A. 20 2005;102(38):13532–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics. 2007;8:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37(6):579–583. [DOI] [PubMed] [Google Scholar]
- 36. Cheng C, Li LM, Alves P, Gerstein M. Systematic identification of transcription factors associated with patient survival in cancers. BMC Genomics. 2009;10:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. [DOI] [PubMed] [Google Scholar]
- 38. Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004;17: Unit 17. 17. [DOI] [PubMed] [Google Scholar]
- 39. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(database issue):D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hull ML, Escareno CR, Godsland JM, et al. Endometrial-peritoneal interactions during endometriotic lesion establishment. Am J Pathol. 2008;173(3):700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8(5):R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ravasi T, Suzuki H, Cannistraci CV, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140(5):744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gerstein MB, Lu ZJ, Van Nostrand EL, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 46. Applied Biosystems. ABI Prism 7700, User Bulletin Foster City, CA: 1997. [Google Scholar]
- 47. Barr A, Manning D. G Proteins Techniques of Analysis, Manning DR, ed. Boca Raton, FL: CRC Press, Inc.; 1999:227–245. [Google Scholar]
- 48. Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38(Database issue):D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ichikawa M, Asai T, Chiba S, Kurokawa M, Ogawa S. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle. 2004;3(6):722–724. [PubMed] [Google Scholar]
- 50. Konno R, Fujiwara H, Netsu S, et al. Gene expression profiling of the rat endometriosis model. Am J Reprod Immunol. 2007;58(4):330–343. [DOI] [PubMed] [Google Scholar]
- 51. Wang P, Zhu L, Zhang X. The role of placental protein 14 in the pathogenesis of endometriosis. Reprod Sci. 2013;20(12):1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paech K, Webb P, Kuiper GG, et al. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science.1997;277(5331):1508–1510. [DOI] [PubMed] [Google Scholar]
- 53. Li X, Huang J, Yi P, Bambara RA, Hilf R, Muyan M. Single-chain estrogen receptors (ERs) reveal that the ERalpha/beta heterodimer emulates functions of the ERalpha dimer in genomic estrogen signaling pathways. Mol Cell Biol. 2004;24(17):7681–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weihua Z, Saji S, Makinen S, et al. Estrogen receptor (ER) beta, a modulator of ERalpha in the uterus. Proc Natl Acad Sci U S A. 2000;97(11):5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cloke B, Christian M. The role of androgens and the androgen receptor in cycling endometrium. Mol Cell Endocrinol. 2012;358(2):166–175. [DOI] [PubMed] [Google Scholar]
- 56. Cloke B, Huhtinen K, Fusi L, et al. The androgen and progesterone receptors regulate distinct gene networks and cellular functions in decidualizing endometrium. Endocrinology. 2008;149(9):4462–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Apparao KB, Lovely LP, Gui Y, Lininger RA, Lessey BA. Elevated endometrial androgen receptor expression in women with polycystic ovarian syndrome. Biol Reprod. 2002;66(2):297–304. [DOI] [PubMed] [Google Scholar]
- 58. Shaik NA, Govindan S, Kodati V, Rao KP, Hasan Q. Polymorphic (CAG)n repeats in the androgen receptor gene: a risk marker for endometriosis and uterine leiomyomas. Hematol Oncol Stem Cell Ther. 2009;2(1):289–293. [DOI] [PubMed] [Google Scholar]
- 59. Lattuada D, Vigano P, Somigliana E, Odorizzi MP, Vignali M, Di Blasio AM. Androgen receptor gene cytosine, adenine, and guanine trinucleotide repeats in patients with endometriosis. J Soc Gynecol Investig. 2004;11(4):237–240. [DOI] [PubMed] [Google Scholar]
- 60. Hsieh YY, Chang CC, Tsai FJ, Wu JY, Tsai CH, Tsai HD. Androgen receptor trinucleotide polymorphism in endometriosis. Fertil Steril. 2001;76(2):412–413. [DOI] [PubMed] [Google Scholar]
- 61. Fujimoto J, Hirose R, Sakaguchi H, Tamaya T. Expression of size-polymorphic androgen receptor (AR) gene in ovarian endometriosis according to the number of cytosine, adenine, and guanine (CAG) repeats in AR alleles. Steroids. 1999;64(8):526–529. [DOI] [PubMed] [Google Scholar]
- 62. Carneiro MM, Morsch DM, Camargos AF, Reis FM, Spritzer PM. Androgen receptor and 5alpha-reductase are expressed in pelvic endometriosis. BJOG. 2008;115(1):113–117. [DOI] [PubMed] [Google Scholar]
- 63. Marshall E, Lowrey J, MacPherson S, et al. In silico analysis identifies a novel role for androgens in the regulation of human endometrial apoptosis. J Clin Endocrinol Metab. 2011;96(11):E1746–E1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ammendola M, Gloria-Bottini F, Sesti F, Piccione E, Bottini E. Association of p53 codon 72 polymorphism with endometriosis. Fertil Steril. 2008;90(2):406–408. [DOI] [PubMed] [Google Scholar]
- 65. Govatati S, Chakravarty B, Deenadayal M, et al. p53 and risk of endometriosis in Indian women. Genet Test Mol Biomarkers. 2012;16(8):865–873. [DOI] [PubMed] [Google Scholar]
- 66. Jia S, Xu L, Chan Y, et al. p53 codon 72 polymorphism and endometriosis: a meta-analysis. Arch Gynecol Obstet. 2012;285(6):1657–1661. [DOI] [PubMed] [Google Scholar]
- 67. Ribeiro Junior CL, Arruda JT, Silva CT, Moura KK. Analysis of p53 codon 72 gene polymorphism in Brazilian patients with endometriosis. Genet Mol Res. 2009;8(2):494–499. [DOI] [PubMed] [Google Scholar]
- 68. Ying TH, Tseng CJ, Tsai SJ, et al. Association of p53 and CDKN1A genotypes with endometriosis. Anticancer Res. 2011;31(12):4301–4306. [PubMed] [Google Scholar]
- 69. Gylfason JT, Dang D, Petursdottir V, et al. Quantitative DNA perturbations of p53 in endometriosis: analysis of American and Icelandic cases. Fertil Steril. 2005;84(5):1388–1394. [DOI] [PubMed] [Google Scholar]
- 70. Bayramoglu H, Duzcan E. Atypical epithelial changes and mutant p53 gene expression in ovarian endometriosis. Pathol Oncol Res. 2001;7(1):33–38. [DOI] [PubMed] [Google Scholar]
- 71. Bunch K, Tinnemore D, Huff S, Hoffer ZS, Burney RO, Stallings JD. Expression patterns of progesterone receptor membrane components 1 and 2 in endometria from women with and without endometriosis. Reprod Sci. 2014;21(2):190–197. [DOI] [PubMed] [Google Scholar]