Abstract
HomeoboxA10 (HOXA10) is a transcription factor that is crucial for the development and patterning of the uterus during embryogenesis. In the adult endometrium, HOXA10 expression is regulated by steroid hormones and embryonic signals. Expression of sufficient HOXA10 messenger RNA is essential to endometrial receptivity and embryo implantation. Aberrant methylation is believed to alter the expression of HOXA10. Methylation of this gene may be associated with decreased fertility, implantation defects, and/or reproductive wastage seen in certain disease states that affect the female reproductive tract. This study describes the differences in methylation patterns of HOXA10 gene in uterine myomas, endometriosis, uterine septum, Asherman syndrome, or uterine polyps of women undergoing hysteroscopic surgery. In the endometrium of uteri with polyps, submucosal myomas, and intramural myomas, there were CpG sites within the HOXA10 gene that were highly methylated compared to controls. The HOXA10 gene in women with endometriosis was hypomethylated compared to controls. DNA methylation may be a common molecular mechanism that results in reproductive dysfunction seen in gynecologic disease.
Keywords: HOXA10, DNA methylation, endometriosis, endometrium
Introduction
Many common gynecologic conditions such as endometriosis or uterine myomas are associated with infertility.1–3 Embryo implantation defects are at least partly responsible for the decreased fertility seen in women with these conditions.2,4–8 The HOX/Hox (human/rodent) family of genes encode transcription factors that have a role in embryo development and particularly in female reproductive tract.9–11 HomeoboxA10 (HOXA10) is expressed in the uterus of the developing embryo.12 In adult humans, HOXA10 is expressed in endometrial glands and stroma. Its expression peaks during the midluteal phase of the cycle, which coincides with the time of endometrial receptivity to embryo implantation.13–15 Expression of sufficient HOXA10 is known to be essential to endometrial receptivity and embryo implantation.16–18 This is clearly demonstrated in the HOXA10 knockout mouse, which is sterile secondary to impaired endometrial receptivity. The HOXA10−/− mouse produces viable embryos, yet neither HOXA10−/− nor wild-type embryos implant. However, when the HOXA10−/− embryos are transferred to the wild-type mouse, they implant and develop normally.19–21 Likewise, in humans, cyclic expression of HOXA10 in the adult endometrium9,13 is required for endometrial receptivity. In several common gynecologic disease states that are associated with implantation defects, such as endometriosis, hydrosalpinges, polyps, and submucosal myomas, defective HOXA10 expression has been demonstrated.22–29
DNA methylation regulates gene expression30–32 and has been associated with several inflammatory diseases and cancers.33–36 Aberrant methylation is known to alter the expression of HOXA10.37 Methylation of this gene may be associated with decreased fertility, implantation defects, and/or reproductive wastage seen in certain disease states that affect the female reproductive tract.38–41 Our objective was to examine methylation of the HOXA10 gene in the endometrium in disease states that affect the female reproductive tract and embryo implantation, specifically uterine myoma, endometriosis, uterine septum, hydrosalpinges, uterine polyps, and Asherman syndrome. We hypothesized that methylation of the HOXA10 gene may be a common mechanism explaining the aberrant expression of the gene and reproductive dysfunction observed in these disorders.
Materials and Methods
Sample Collection
We collected endometrial samples from a total 84 women with submucosal uterine myomas (n = 13), intramural uterine myomas (n = 13), endometriosis (n = 27), uterine septum (n = 6), Asherman syndrome (n = 8), hydrosalpinx (n = 4), or uterine polyps (n = 11) undergoing hysteroscopic surgery as well as endometrial samples from controls (n = 7) who were anonymous egg donors free from gynecologic disorders undergoing oocyte retrieval. Some women had more than 1 condition (ie, endometriosis and hydrosalpinx) and were included separately in each group (Table 1). This study was approved by the Yale University Human Investigation Committee. At the time of biopsy, each endometrial tissue sample was stored in 1 mL of RNAlater (Qiagen, Valencia, California) at −80°C until RNA and DNA isolation.
Table 1.
Patients’ Characteristics.a
| Condition | Age (Mean ± SD), years | Gravity (Mean) | Parity (Mean) |
|---|---|---|---|
| Controls (n = 7) | 34.9 ± 3.9 | 1.7 | 1.1 |
| Asherman (n = 8) | 33.5 ± 3.3 | 1.4 | 0.5 |
| Hydrosalpinx (n = 4) | 37.5 ± 5.8 | 1.5 | 1 |
| Endometriosis (n = 27) | 32.3 ± 6.7 | 1.1 | 0.7 |
| Intramural myoma (n = 13) | 39.2 ± 7.6 | 1.5 | 1 |
| Submucosal myoma (n = 13) | 36.6 ± 6.2 | 0.6 | 0.6 |
| Polyp (n = 11) | 33.8 ± 3.7 | 0.5 | 0.5 |
| Septum (n = 6) | 32.7 ± 5.1 | 1.2 | 1.2 |
Abbreviation: SD, standard deviation.
a N sums to greater than 84 as there was an overlap of patients’ diagnosis. There were 6 patients with more than 1 diagnosis: 2 with submucosal myomas and septum, 1 with history of endometriosis and polyp, 1 with endometriosis and hydrosalpinx, and 1 with endometriosis and submucosal myoma.
Quantitative Real-Time Polymerase Chain Reaction Analysis
The RNA was extracted using the RNeasy Mini kit (Qiagen), according to the manufacturer’s protocol. Messenger RNA (mRNA) levels were analyzed by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) using the Bio-Rad iCycler iQ system (Bio-Rad Laboratories, Hercules, California). For each sample, 500 ng of total RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories). Real-time RT-PCR was performed using a MyiQ Single-Color Real-Time PCR Detection System and iQ SYBR Green Supermix (both from Bio-Rad Laboratories). The sequences of all primers and the real-time RT-PCR reaction conditions have been described previously.42 Each assay was run in duplicate with each set of primers, and samples without mRNA were included as negative controls. HomeoboxA10 gene expression was normalized to the expression of β-actin for each run. Relative mRNA expression for each gene was calculated using the comparative cycle threshold (Ct) method (known as 2ΔΔ CT method).43–44 Results are presented as the mean ± standard error (SE). Statistical significance was determined and analyzed by Student t test. P values less than .05 were considered statistically significant.
DNA Isolation and Methylation
DNA was isolated using the DNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol. Quantitative DNA methylation analysis was performed on the collected tissue, using Sequenom MassARRAY quantitative methylation analysis and EpiTyper technology, in 3 CpG-rich fragments in 2 regions of the HOXA10 gene (total 39 distinct CpG sites/fragments), to detect differences in methylation. The first CpG island is located 50 base pair upstream of exon 1, and 2 additional islands are located in the intron flanked by exons 1 and 2. Briefly, genomic DNA underwent bisulfite treatment followed by PCR amplification using a T7-promotor tag. In vitro RNA transcription was performed followed by uracil-specific cleavage. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry was used to analyze the cleavage products and produced distinct signal pattern pairs indicating nonmethylated and methylated DNA. Due to limitations of this technique, there were a few cleavage products that were indistinguishable as they have the same mass. In fragment 1, sites CpG37 and CpG4, sites CpG38 and CpG21, and sites CpG17 and CpG30 could not be distinguished from each other. Similarly, sites CpG8 and CpG9 in fragment 2 were indistinguishable. Differences in endometrial HOXA10 expression and mean percentage methylation at different CpG areas between the disease groups and controls were determined using Student t test. P < .05 was considered statistically significant. To determine whether there was an association between the level of methylation and gene expression, a Pearson product-moment correlation coefficient was calculated.
Results
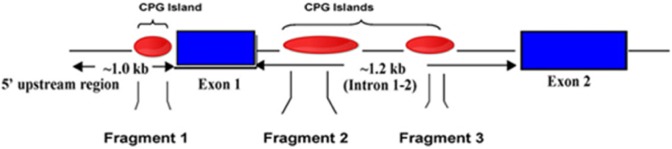
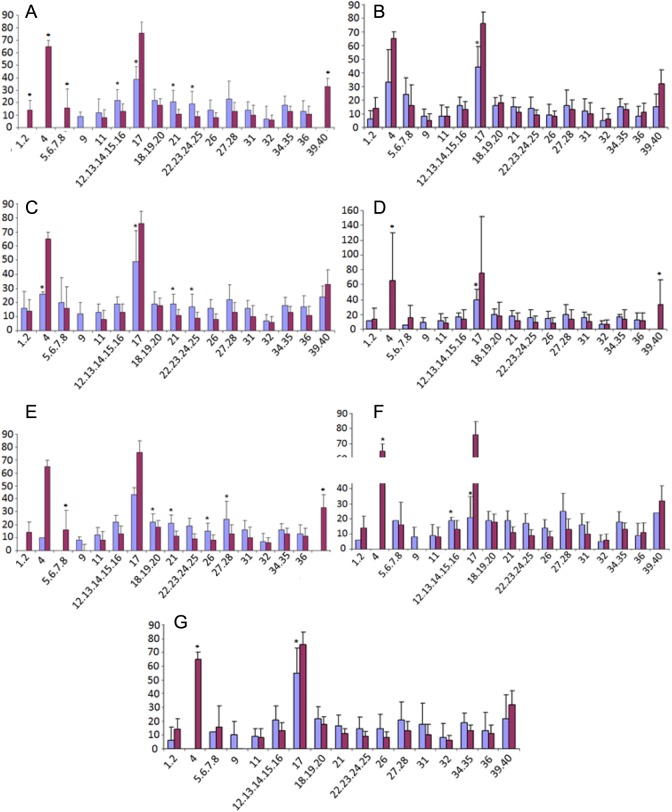
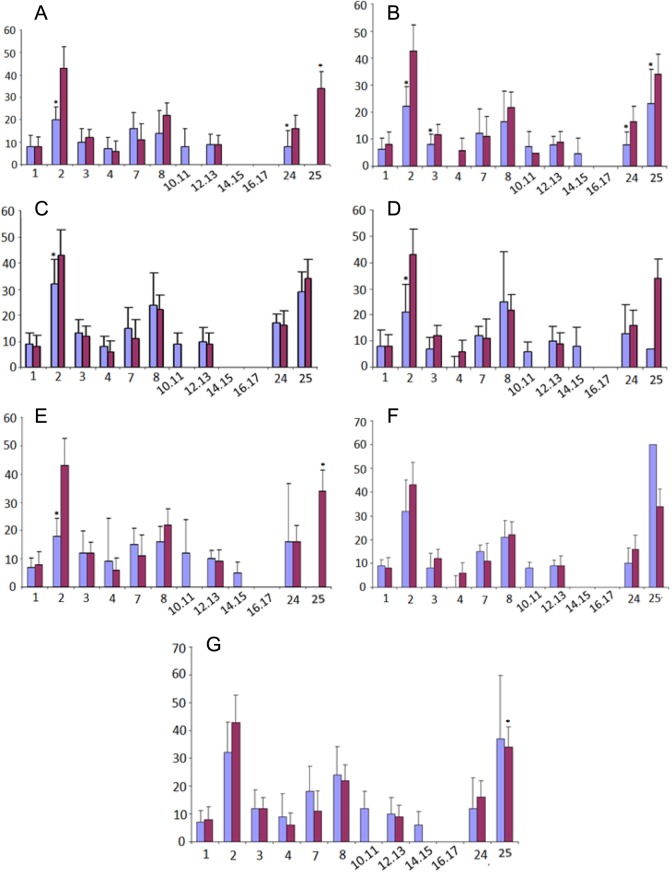
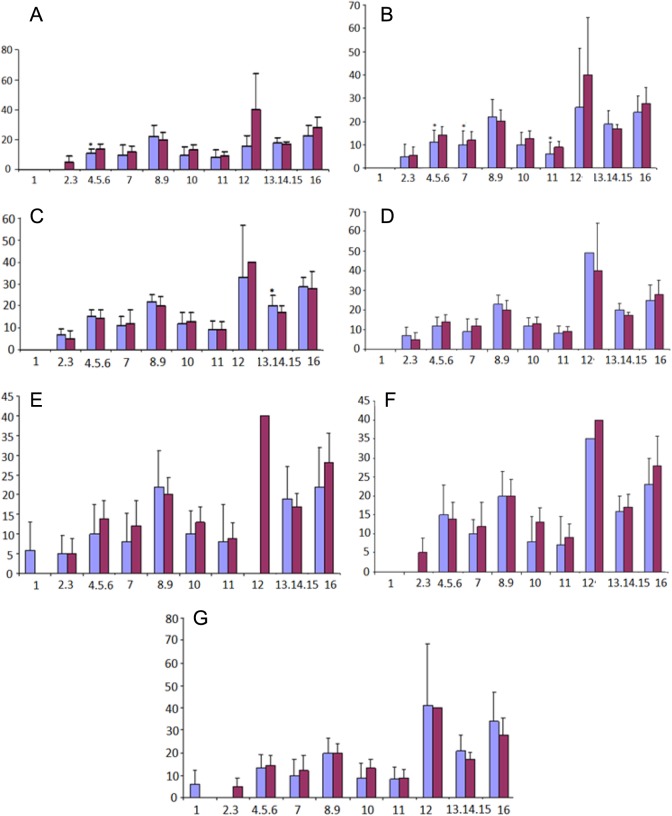
The CpG islands present in HOXA10 gene are shown in Figure 1. The CpG sites or clusters present at different regions in HOXA10 gene are designated as fragment 1 if in the promoter region, while fragments 2 and 3 are located in the intron regions. The summary of the Sequenom MassARRAY quantitative methylation analysis of the CPG areas located in the 5′ promoter region (fragment 1) is shown in Figure 2, while in the intron region (fragment 2 and 3) is shown in Figures 3 and 4. The average percentage methylation value at each numbered CpG site or cluster of CpG sites for the 84 endometrial samples is shown. The results show the overall low levels of methylation. There were very few CpG sites with greater than 50% methylation. However, some individual sites were highly and specifically methylated. The number of CpG sites with significantly altered methylation status compared to controls by disease is shown in Table 2. Specifically, CpG17 and CpG4 in the promoter region and CpG12 in the second intronic region of HOXA10 were highly methylated in the control samples and had significantly greater percentage methylation than many of the disease states. At other CpG sites, the disease state endometrium had CpG sites with significantly greater percentage methylation compared to controls. Specifically, in polyp endometrium, 4 of the 6 statistically different CpG clusters in the promoter region had higher percentage methylation compared to controls (Figure 2; CpG18-CpG20, CpG21, CpG26, and CpG27-28). In endometrium of uteri with intramural myomas, 2 of the 4 statistically different CpG clusters in the promoter region (Figure 2: CpG 21 and 22-25), as well as 1 CpG site in the second intronic region (Figure 4: CpG13- CpG15), had higher methylation compared to controls. In endometrium of uteri with submucosal myomas, 3 of the 4 statistically different CpG clusters in the promoter region had higher degrees of methylation compared to controls. Interestingly, endometrium of submucous and intramural myomas as well as polyp had two of the same CpG sites or clusters, CpG21 and CpG cluster (CpG_22.23.24.25) located in the promoter region, hypermethylated compared to controls. Two of the diseases (submucosal myoma and polyp) shared a second CpG cluster (CpG_12.13.14.15.16) also located in the promoter, which had greater methylation as compared to controls in both. Submucous and intramural leiomyoma also showed distinct methylation differences as well. Overall, the percentage of CpG sites or clusters within each fragment with statistically significant different methylation levels between diseases and controls was low. However, all diseases studied had at least 1 CpG cluster in the promoter region of HOXA10, which was differently methylated when compared to controls. Many of the diseases also demonstrated at least 1 CpG cluster with methylation significantly different from controls in intronic fragment 2 and few diseases were associated with differences in methylation in intronic fragment 3 as shown in Figures 1, 3, and 4.
Figure 1.
HomeoboxA10 (HOXA10) gene structure. The genomic region of HOXA10 showing the locations of the CpG islands. The CpG sites or clusters present on the gene are designated as fragments 1, 2, and 3. Intron region containing 2 clusters of CpG islands (fragment 1 and 2), while promoter region having 1 cluster (fragment 1).
Figure 2.
Summation of Sequenom MassARRAY methylation analysis of HomeoboxA10 (HOXA10) CpG islands in the promoter region in each disease state. The percentage DNA methylation of individual CpG sites or clusters, averaged for each disease state, from controls is indicated by the bars (blue is disease and red is controls). Each graph represents a different disease, submucosal myoma (A), endometriosis (B), intramural myoma (C), septum (D), polyp (E), hydrosalpinx (F), and Asherman syndrome (G). The CpG positions from each genomic region analyzed are numbered in ascending order. Some CpG sites are grouped because of the limits of the technique, which do not allow them to be resolved individually. *Particular CpG sites or clusters with significantly different methylation levels compared to controls. An absent bar indicates that insufficient methylation data were obtained due to very low levels of methylation. (The color version of this figure is available in the online version at http://rs.sagepub.com/)
Figure 3.
Summation of Sequenom MassARRAY methylation analysis of HomeoboxA10 (HOXA10) CPG island intronic region 1 (fragment 2) in each disease state, submucosal myoma (A), endometriosis (B), intramural myoma (C), septum (D), polyp (E), hydrosalpinx (F), and Asherman syndrome (G). *Particular CpG sites or clusters with significantly different methylation levels compared to controls. Percentage DNA methylation is represented by the bars, with disease state showed in blue and controls in red. (The color version of this figure is available in the online version at http://rs.sagepub.com/)
Figure 4.
Summation of Sequenom MassARRAY methylation analysis of HomeoboxA10 (HOXA10) CPG island intronic region 2 (fragment 3) in each disease state, submucosal myoma (A), endometriosis (B), intramural myoma (C), septum (D), polyp (E), hydrosalpinx (F), and Asherman syndrome (G). *Particular CpG sites or clusters with significantly different methylation levels compared to controls (blue is disease and red is controls). Percentage methylation is on Y axis, and CpG site is on X axis. (The color version of this figure is available in the online version at http://rs.sagepub.com/)
Table 2.
Methylation Status Compared to Controls by Disease.a
| Condition | Increased Methylation | Decreased Methylation | Total |
|---|---|---|---|
| Asherman | 1 | 2 | 3 |
| Hydrosalpinx | 1 | 2 | 3 |
| Endometriosis | 0 | 8 | 8 |
| Intramural myoma | 3 | 3 | 6 |
| Submucosal myoma | 3 | 9 | 12 |
| Polyp | 4 | 4 | 8 |
| Septum | 0 | 4 | 4 |
a Number of CpG sites with significantly altered methylation.
When percentage methylation was averaged for each CpG island or fragment in disease states and compared to controls, 3 diseases (submucosal myoma, polyp, and endometriosis) were found to have significantly different CpG island wide percentage methylation compared to controls. In submucosal myoma, fragment 2 had 10% methylation compared to 14% in controls (P = .008) and fragment 3 had 12% methylation compared to 16% in controls (P = .014). In polyp, fragment 3 had 12% methylation compared to 16% in controls (P = .049). In endometriosis, all fragments were significantly less methylated compared to controls—fragment 1: 16% versus 21% (P = .004); fragment 2: 8% versus 14% (P = .002); fragment 3: 13% versus 16% (P = .006). In all cases, the disease states were found to have less overall methylation compared to controls. Percentage methylation was also averaged over the whole HOXA10 gene. Gene-wide methylation was found to be significantly different only in 1 disease, endometriosis, compared to controls (14% vs 17%, P = .001).
Quantitative real-time reverse transcriptase-polymerase chain reaction was performed to assess the expression levels of HOXA10 in disease and normal endometrium. RNA was available from 43 disease state endometrial samples. The RT-PCR results for the disease samples were normalized to the controls, and these data were correlated with percentage DNA methylation at significantly different CpG sites or clusters. There was a correlation between DNA methylation and gene expression in endometrium from submucous myoma at 1 CpG cluster in the second intronic region (Figure 4; CpG 4.5.6; r = .72, P = .02). There was also a correlation in endometriosis between CpG site 11 methylation in the second intronic region and gene expression (r = −.9, P = .04). We also observed that fragment-wide DNA methylation for fragment 3 in endometriosis was also correlated with gene expression (r = −.9, P = .04).
Discussion
Our study reports the differential methylation of HOXA10 CpG sites in human female reproductive diseases such as submucosal myoma, endometriosis, intramural myoma, septum, polyps, hydrosalpinx, and Asherman syndrome compared to normal female samples. These data suggest that the HOXA10 gene may be differently methylated in multiple diseases and that DNA methylation may be a common means by which these gynecologic diseases affect the HOXA10 gene and its expression in female reproductive track. DNA methylation has not been well studied in most of the gynecological diseases mentioned previously, except for endometriosis and uterine polyps.45–48 Overall, we noted low levels of methylation of the HOXA10 gene in both disease and control endometrium. A few highly specific sites had high levels of methylation in controls. Methylation is associated with the absence of disease at these sites, and loss of methylation can signal disease. The net absence or presence of methylation is less important than the specific pattern of methylation measured at the level of individual C/G base pairs.
All diseases had at least 1 CpG site that was significantly differently methylated as compared to normal. Yet each disease had a unique pattern of methylation of the HOXA10 gene, and methylation of some CpG sites appears to be disease specific. This suggests that there may be disease-specific mechanisms of HOXA10 methylation and gene regulation. Further, aberrant methylation of the HOXA10 gene is likely related to its aberrant expression, and this aberrant expression is thought to have a role in the reproductive dysfunction such as implantation failure seen in these diseases, for example, in endometriosis as reported previously.37
CpG sites located in the promoter region of HOXA10 had significantly increased methylation compared to normal in 3 diseases: polyp, intramural myoma, and submucosal myoma. CpG21 was consistently methylated. CpG methylation, especially in the promoter region of genes, typically results in decreased gene expression.48 The diseases studied are known to result in decreased expression of HOXA10 in the endometrium.49 Our findings of increased methylation at certain CpG sites in the promoter of HOXA10 suggest that this methylation correlates with previous reports of decreased HOXA10 gene expression in these diseases. Interestingly, the same sites were hypermethylated in all 3 diseases, suggesting that there may be a common mechanism regulating HOXA10 gene expression in uteri with endometrial polyps as well as intramural and submucosal myomas. Expression of HOXA10 correlated with methylation at only a few of the CpG sites tested. It is likely that a combination of methylation/demethylation of multiple sites determines the net change in gene expression. Promoter methylation or demethylation may be a common mechanism by which multiple pathologic processes affect endometrial receptivity.
In our study, we found hypomethylation of HOXA10 in the endometrium of women with endometriosis. This was noted at specific CpG sites both within the promoter as well as the CpG islands found in the intronic regions of the gene. Further, CpG island-wide as well as gene-wide decreased methylation was found in the HOXA10 gene in endometriosis compared to controls. This is in contrast to other studies, which have shown increased methylation of the HOXA10 gene, especially in the promoter region, in endometriosis.50–53 The sequenom platform allows for precise measurement and quantification of DNA methylation. This may explain some of the differences seen in methylation as compared to older studies using cruder techniques. However, this would not explain the overall low levels of methylation compared to fertile controls found in our study. Perhaps, we found a different pattern of methylation of the HOXA10 gene because our patient population differs from those reported previously. The patients with endometriosis in this study were all treated. Some patients had only a documented history of endometriosis and possibly did not have active endometriosis at the time of surgery. All patients and animals in prior studies had active disease. Perhaps, this variation in the spectrum of endometriosis and its treatment state affected the average methylation of the HOXA10 gene. Epigenetic changes such as DNA methylation, although stable through cell divisions, are believed to be reversible or modifiable by lifestyle factors.54,55 It is likely that the use of medications, such as the hormonal medications taken by some patients in this study, altered methylation patterns of this gene in agreement with the results reported for other genes.56,57 This opens up the possibility that prolonged therapy may reverse the epigenetic alterations seen in this disease. Further, expression of HOXA10 is known to be decreased in the endometrium of women with endometriosis.22,27,46,49 Perhaps, the decrease in methylation of the HOXA10 gene observed in our cohort58 would allow for the binding of inhibitors of transcription, thus explaining the lower expression of HOXA10 observed in this disease. Of all the gynecologic diseases that we studied, endometriosis had the greatest number of significant differences in HOXA10 methylation compared to normal. Therefore, we demonstrate that endometriosis is an epigenetic disease associated with aberrant methylation of HOXA10.
Limitations of this study include the fact that all patients were undergoing infertility evaluation and treatment. Different changes in methylation may be seen in women who are fertile; however, we believe that the significant differences seen with each disease make it likely that these changes are disease specific rather than all related to infertility. Further, many of the patients received previous medical therapies for their disease. We did not have sufficient numbers of patients using each individual therapy to enable each to be analyzed separately. Although several sites of epigenetic regulatory control were identified, most individual changes in methylation did not alter gene expression under the conditions tested. Changes in methylation are likely not all functionally relevant, however, the precise combination of methylation changes that regulate gene expression are likely more complex than the net level of methylation. Complex combinatorial patterns of increased and decreased methylation contribute to the control of gene expression.
We show that the endometrium of women affected by common gynecologic diseases appears to have a unique disease-specific pattern of methylation of the HOXA10 gene. We anticipate that a “methylation signature” could be developed for each disease. Analyzing DNA methylation patterns of specific genes may allow for a noninvasive method to identify patients with a specific disease or at risk for certain adverse clinical outcomes such as embryo implantation failure. Additionally, if therapies to treat disease affect DNA methylation patterns in the endometrium, analyzing these patterns may be a way to assess response to treatment. In conclusion, DNA methylation, particularly HOXA10 gene methylation, may be a common molecular mechanism that results in reproductive dysfunction seen in gynecologic disease.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH U54 HD052668 and R01 HD076422.
References
- 1. Angioni S, Cela V, Sedda F, et al. Focusing on surgery results in infertile patients with deep endometriosis. Gynecol Endocrinol. 2015;31(8):595–598. [DOI] [PubMed] [Google Scholar]
- 2. Macer ML, Taylor HS. Endometriosis and infertility: a review of the pathogenesis and treatment of endometriosis-associated infertility. Obstet Gynecol Clin North Am. 2012;39(4):535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghahiry AA, Refaei Aliabadi E, Taherian AA, et al. Effectiveness of hysteroscopic repair of uterine lesions in reproductive outcome. Int J Fertil Steril. 2014;8(2):129–134. [PMC free article] [PubMed] [Google Scholar]
- 4. Matson PL, Yovich JL. The treatment of infertility associated with endometriosis by in vitro fertilization. Fertil Steril. 1986;46(3):432–434. [DOI] [PubMed] [Google Scholar]
- 5. Barnhart K, Dunsmoor-Su R, Coutifaris C. Effect of endometriosis on in vitro fertilization. Fertil Steril. 2002;77(6):1148–1155. [DOI] [PubMed] [Google Scholar]
- 6. Kuivasaari P, Hippelainen M, Anttila M, Heinonen S. Effect of endometriosis on IVF/ICSI outcome: stage III/IV endometriosis worsens cumulative pregnancy and live-born rates. Hum Reprod. 2005;20(11):3130–3135. [DOI] [PubMed] [Google Scholar]
- 7. Matsuzaki S, Canis M, Darcha C, Pouly JL, Mage G. HOXA-10 expression in the mid-secretory endometrium of infertile patients with either endometriosis, uterine fibromas or unexplained infertility. Hum Reprod. 2009;24(12):3180–3187. [DOI] [PubMed] [Google Scholar]
- 8. Rackow BW, Taylor HS. Submucosal uterine leiomyomas have a global effect on molecular determinants of endometrial receptivity. Fertil Steril. 93(6):2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daftary GS, Taylor HS. Endocrine regulation of HOX genes. Endocr Rev. 2006;27(4):331–355. [DOI] [PubMed] [Google Scholar]
- 10. Du H, Taylor HS. Molecular regulation of mullerian development by Hox genes. Ann N Y Acad Sci. 2004;1034:152–165. [DOI] [PubMed] [Google Scholar]
- 11. Taylor HS. The role of HOX genes in the development and function of the female reproductive tract. Semin Reprod Med. 2000;18(1):81–89. [DOI] [PubMed] [Google Scholar]
- 12. Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol Reprod. 1997;57(6):1338–1345. [DOI] [PubMed] [Google Scholar]
- 13. Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J Clin Invest. 1998;101(7):1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor HS, Igarashi P, Olive DL, Arici A. Sex steroids mediate HOXA11 expression in the human peri-implantation endometrium. J Clin Endocrinol Metab. 1999;84(3):1129–1135. [DOI] [PubMed] [Google Scholar]
- 15. Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96(12):E1925–E1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eun Kwon H, Taylor HS. The role of HOX genes in human implantation. Ann N Y Acad Sci. 2004;1034:1–18. [DOI] [PubMed] [Google Scholar]
- 17. Daftary GS, Taylor HS. Pleiotropic effects of Hoxa10 on the functional development of peri-implantation endometrium. Mol Reprod Dev. 2004;67(1):8–14. [DOI] [PubMed] [Google Scholar]
- 18. Bagot CN, Troy PJ, Taylor HS. Alteration of maternal Hoxa10 expression by in vivo gene transfection affects implantation. Gene Ther. 2000;7(16):1378–1384. [DOI] [PubMed] [Google Scholar]
- 19. Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374(6521):460–463. [DOI] [PubMed] [Google Scholar]
- 20. Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122(9):2687–2696. [DOI] [PubMed] [Google Scholar]
- 21. Taylor HS. The role of HOX genes in human implantation. Hum Reprod. 2000;6(1):75–79. [DOI] [PubMed] [Google Scholar]
- 22. Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14(5):1328–1331. [DOI] [PubMed] [Google Scholar]
- 23. Daftary GS, Taylor HS. Hydrosalpinx fluid diminishes endometrial cell HOXA10 expression. Fertil Steril. 2002;78(3):577–580. [DOI] [PubMed] [Google Scholar]
- 24. Zanatta A, Pereira RM, Rocha AM, et al. The relationship among HOXA10, estrogen receptor α, progesterone receptor, and progesterone receptor B proteins in rectosigmoid endometriosis: a tissue microarray study. Reprod Sci. 2015;22(1):31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daftary GS, Kayisli U, Seli E, Bukulmez O, Arici A, Taylor HS. Salpingectomy increases peri-implantation endometrial HOXA10 expression in women with hydrosalpinx. Fertil Steril. 2007;87(2):367–372. [DOI] [PubMed] [Google Scholar]
- 26. Rackow BW, Jorgensen E, Taylor HS. Endometrial polyps affect uterine receptivity. Fertil Steril. 2011;95(8):2690–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim JJ, Taylor HS, Lu Z, et al. Altered expression of HOXA10 in endometriosis: potential role in decidualization. Mol Hum Reprod. 2007;13(5):323–332. [DOI] [PubMed] [Google Scholar]
- 28. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. [DOI] [PubMed] [Google Scholar]
- 29. Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–1799. [DOI] [PubMed] [Google Scholar]
- 30. Rupp RA, Becker PB. Becker gene regulation by histone H1: new links to DNA methylation. Cell. 2005;123(7):1178–1179. [DOI] [PubMed] [Google Scholar]
- 31. Hoffman AR, Hu JF. Directing DNA methylation to inhibit gene expression. Cell Mol Neurobiol. 2006;26(4-6):425–438. [DOI] [PubMed] [Google Scholar]
- 32. Popiela A, Keith G, Borzecki A, et al. The meaning of the methylation of genomic DNA in the regulation of gene expression levels. Eur J Gynaecol Oncol. 2004;25(2):145–149. [PubMed] [Google Scholar]
- 33. Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. [DOI] [PubMed] [Google Scholar]
- 34. Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb). 2015;25(2):161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Győrffy B, Bottai G, Fleischer T, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer. 2016;138(1):87–97. [DOI] [PubMed] [Google Scholar]
- 36. Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13(7):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2009;80(1):79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cakmak H, Taylor HS. Implantation failure: molecular mechanisms and clinical treatment. Hum Reprod Update. 2011;17(2):242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24(7):2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bromer JG, Wu J, Zhou Y, Taylor HS. Hypermethylation of homeobox A10 by in utero diethylstilbestrol exposure: an epigenetic mechanism for altered developmental programming. Endocrinology. 2009;150(7):3376–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cakmak H, Taylor HS. Molecular mechanisms of treatment resistance in endometriosis: the role of progesterone-hox gene interactions. Semin Reprod Med. 2010;28(1):69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith CC, Taylor HS. Xenoestrogen exposure imprints expression of genes (Hoxa10) required for normal uterine development. FASEB J. 2007;21(1):239–246. [DOI] [PubMed] [Google Scholar]
- 43. Barr AJ, Manning DR. Agonist promoted (35S)-GTPgs binding as a probe of receptor G-protein communication in reconstituted sf9 cells In: Manning DR, ed. G Proteins Techniques of Analysis. Boca Raton, FL: CRC Press, Inc; 1999:227–245. [Google Scholar]
- 44. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)]. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 45. Ren F, Wang DB, Li T, Chen YH, Li Y. Identification of differentially methylated genes in the malignant transformation of ovarian endometriosis. J Ovarian Res. 2014;7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andersson KL, Bussani C, Fambrini M, et al. DNA methylation of HOXA10 in eutopic and ectopic endometrium. Hum Reprod. 2014;29(9):1906–1911. [DOI] [PubMed] [Google Scholar]
- 47. Guida M, Sanguedolce F, Bufo P, et al. Aberrant DNA hypermethylation of hMLH-1 and CDKN2A/p16 genes in benign, premalignant and malignant endometrial lesions. Eur J Gynaecol Oncol. 2009;30(3):267–270. [PubMed] [Google Scholar]
- 48. Keller S, Sarchiapone M, Zarrilli F, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67(3):258–267. [DOI] [PubMed] [Google Scholar]
- 49. Fischer CP, Kayisili U, Taylor HS. HOXA10 expression is decreased in endometrium of women with adenomyosis. Fertil Steril. 2010;95(3):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu Y, Halverson G, Basir Z, Strawn E, Yan P, Guo SW. Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. Am J Obstet Gynecol. 2005;193(2):371–380. [DOI] [PubMed] [Google Scholar]
- 51. Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2009;80(1):79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Szczepanska M, Wirstlein P, Luczak M, Jagodzinski PP, Skrzypczak J. Reduced expression of HOXA10 in the midluteal endometrium from infertile women with minimal endometriosis. Biomed Pharmacother. 2010;64(10):697–705. [DOI] [PubMed] [Google Scholar]
- 53. Lu Y, Nie J, Liu X, Guo SW. Reduced expression and concomitant promoter hypermethylation of HOXA10 in endometrium from women wearing intrauterine devices. Fertil Steril. 2010;95(4):1583–1588. [DOI] [PubMed] [Google Scholar]
- 54. Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–376. [DOI] [PubMed] [Google Scholar]
- 55. Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams KE, Anderton DL, Lee MP, Pentecost BT, Arcaro KF. High-density array analysis of DNA methylation in Tamoxifen-resistant breast cancer cell lines. Epigenetics. 2014;9(2):297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan M, Yan PS, Hartman-Frey C, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66(24):11954–11966. [DOI] [PubMed] [Google Scholar]
- 58. Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2008;80(1):79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]