To the Editor
An aim of systems biology is to understand complex interactions between genes, proteins and metabolites by integrating and modeling multiple data sources. We report an ‘integrated-omics’ approach within XCMS Online1 that automatically superimposes raw metabolomic data onto metabolic pathways and integrates it with transcriptomic and proteomic data (http://XCMSOnline.scripps.edu/).
Mapping downstream metabolite changes onto metabolic pathways and biological networks can provide considerable mechanistic insight that can be confirmed by association to multi-omic data. However, pathway analysis using untargeted metabolomics requires intense data curation, including feature filtering, statistical analysis and metabolite identification. Subjectively defined values such as fold change, P value and signal intensity cut-off are needed to identify significantly dysregulated metabolite features within enormous data sets. Confirming metabolite identities for pathway analysis typically requires performing additional tandem mass spectrometry (MS/MS) experiments and matching the spectra to standards or MS/MS spectral databases. The magnitude of these data sets makes it impractical to manually interpret, and therefore the use of bioinformatic tools at each step is essential. Multiple analysis platforms are often needed to complete the entire workflow, which can take several weeks, depending on the size of the sample cohort and the experience of the analyst.
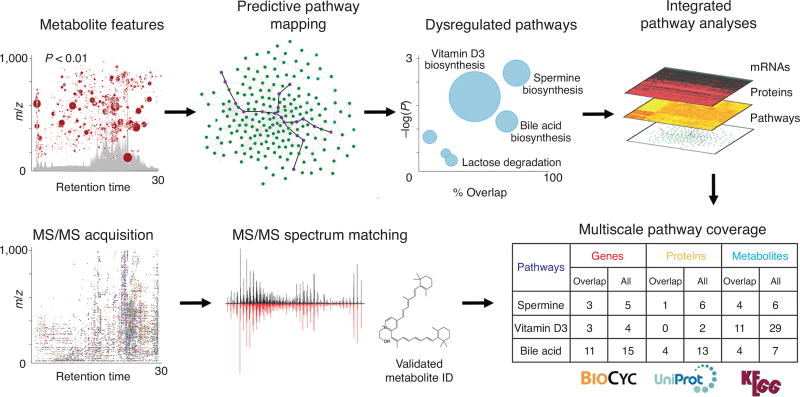
XCMS was originally developed as a metabolomics data processing algorithm to extract metabolic features from raw MS data and perform statistical analysis. The evolution of XCMS from a command line tool2 to an intuitive cloud-based online platform1 facilitated its use by a broader community. However, the community is still in need of user-friendly tools to take metabolomic output and associate it with metabolic pathways to identify aberrant biological processes. To address this demand, we implemented automated predictive pathway analysis3, operating directly on the entire metabolic feature table, into the XCMS Online workflow (Fig. 1), removing the need to transfer data to another application and enabling quick and efficient pathway analysis. This process involves uploading raw MS data to XCMS Online, where the statistically significant features are identified; then, using Fisher’s exact test, dysregulated metabolic pathways are identified from the processed accurate mass data3. If gene and protein data are available, they are uploaded and overlaid with the results of the metabolomic analysis. Currently there are over 7,600 metabolic models available for pathway analysis from BioCyc4 v19.5–20.0, with contents being updated regularly. Further confirmation of dysregulated pathways can be performed by comparing metabolite spectra, obtained via targeted or autonomous MS/MS, with standard fragmentation spectra from METLIN, which contains MS/MS data on over 14,000 molecules5. To address instances in which a standard spectrum is not available, we have also recently added machine learning in silico fragmentation data to METLIN, generating MS/MS spectra on over 220,000 more molecules. Our workflow enables (i) evaluation of biochemical relevance by mapping high resolution MS data directly onto pathways, (ii) cross-integration of genomic and proteomic data and (iii) metabolite identity verification via data-dependent MS/MS analysis, either separately or as part of the autonomous workflow5.
Figure 1.
Workflow for metabolomic data and pathway analysis using XCMS Online. A metabolite feature table of statistically significant features is generated from standard XCMS processing; these features automatically undergo predictive pathway mapping using a specified biological model. The pathway cloud plot shows dysregulated pathways (blue circles) with increasing statistical significance on the y axis, metabolite overlap on the x axis and total number of metabolites in the pathway represented by the circle radius. The multiscale pathway coverage table presents enriched metabolic pathways with overlapped and total metabolites, genes and proteins. MS/MS data confirm dysregulated pathways by matching metabolite MS/MS spectra with the METLIN database.
Our multi-omic analysis tool uses embedded BioCyc4 and Uniprot6 databases to map user-uploaded gene and protein data onto the predicted metabolic pathways (Supplementary Fig. 1). Results can be viewed in table form or using the interactive Pathway Cloud plot (Fig. 1). Dysregulated pathways with greater percent overlap and statistical significance appear in the upper right of the cloud plot. Graph features can be clicked to view more information on overlapping gene, protein and metabolite data, with links to BioCyc, KEGG and METLIN. Important features can be readily identified, helping to decipher underlying biological mechanisms. Details on the pathway analysis and integrated omics workflow can be found in the Supplementary Methods. Data sharing is possible between collaborators and the public, and we encourage users to share their data in the XCMS Online community.
To demonstrate metabolic pathway analysis and multi-omic integration, we describe representative sample sets in the Supplementary Note, including metabolic pathway analysis using progenitor cell proliferation data and a bacterially induced corrosion study (Supplementary Fig. 2); proteomic integration with an aging study (Supplementary Fig. 3); transcriptomic and proteomic integration using a human colon cancer study (Supplementary Fig. 4 and Supplementary Table 1); a nitrate stress response study in sulfate-reducing bacteria (Supplementary Fig. 5) and a media stress response study in Escherichia coli (Supplementary Fig. 6 and Supplementary Table 2); and a cohort of 1,600 diabetes plasma samples (Supplementary Fig. 7), which helps illustrate the scalability of the cloud-based XCMS Online.
Other notable tools providing pathway analysis and multi-omic integration include Galaxy-M7, Open MS from KNIME8 and MetaboAnalyst9. However, many of these tools still require separate preprocessing of tandem liquid chromatography—mass spectrometry data and are not fully integrated into a single program. Our workflow automatically maps metabolomic data directly onto pathways and integrates transcriptomics and proteomics for systems-wide interpretation in one cohesive platform. Additionally, metabolic network mapping is available based on the predictive activity network algorithm3 for analysis of metabolomic data only, with multi-omics networking in development. In the future, we will incorporate unique metabolic pathways and networks from other sources to provide more comprehensive biological resources.
Data availability
To assist users with the workflow, we have provided a sample data set entitled “Ecoli_glucose-vs-adenosine” (Job ID #1133019) that can be found on XCMS Online under XCMS Public (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=listPublicShares), as well as two instructional videos available on the XCMS Institute website (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=institute) under the Omics tab and by clicking Integrated Omics or Pathway Cloud Plot.
Supplementary Material
Acknowledgments
The authors thank J. Nazroo, G. Tampubolon, N. Pendleton and F.C.W. Wu from the University of Manchester for constructive discussions alongside Medical Research Council grant MRC G1001375/1 (R.G.) for generous funding. The authors thank the following for funding assistance: Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research under contract number DE-AC02-05CH11231 (G.S.); and National Institutes of Health grants R01 GMH4368 (G.S.) and PO1 A1043376-02S1 (G.S.).
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
T.H., E.M.F., D.R., H.P.B., A.A., B.H., T.I., M.W.W.A., P.D.R., L.J.N., M.W.F. and G.S. contributed to multi-omic platform design and development; T.H., E.M.F., C.H.J., M.F., G.K., M.P.T., L.L.L., F.L.P. E.L.M., J.D.W., N.J.W.R. and R.G. contributed to data collection and analysis. T.H. and E.M.F. share first authorship. C.H.J., J.I., T.I., and G.S. also contributed to manuscript writing.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Gowda H, et al. Anal. Chem. 2014;86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 3.Li SZ, et al. PLoS Comput. Biol. 2013;9:7. doi: 10.1371/journal.pcbi.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caspi R, et al. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benton HP, et al. Anal. Chem. 2015;87:884–891. doi: 10.1021/ac5025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The UniProt Consortium. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson RL, Weber RJM, Liu HY, Sharma-Oates A, Viant MR. Gigascience. 2016;5:10. doi: 10.1186/s13742-016-0115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aiche S, et al. Proteomics. 2015;15:1443–1447. doi: 10.1002/pmic.201400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia J, Sinelnikov IV, Han B, Wishart DS. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To assist users with the workflow, we have provided a sample data set entitled “Ecoli_glucose-vs-adenosine” (Job ID #1133019) that can be found on XCMS Online under XCMS Public (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=listPublicShares), as well as two instructional videos available on the XCMS Institute website (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=institute) under the Omics tab and by clicking Integrated Omics or Pathway Cloud Plot.