1. Introduction
A critical step in the assembly of many dsDNA viruses inside infected host cells is the packaging of newly synthesized viral genomes into procapsid shells. The procapsids have a small, ~3 nm diameter “portal” through which the ~2 nm diameter DNA must enter. A molecular motor complex transiently assembles at the portal and converts chemical energy from ATP hydrolysis into mechanical work needed to translocate the DNA into the procapsid. This is a remarkable process from a biophysical point of view because the fully packed DNA attains very tight confinement, reaching crystalline density against huge resisting forces arising from electrostatic self-repulsion, bending rigidity, and entropic penalty.
This chapter reviews “single-molecule” studies of viral DNA packaging, which have been an exciting and powerful development during the last decade. In these approaches the packaging of a single DNA molecule into a single viral procapsid can be measured in real time. Using “optical tweezers”, in particular, one can directly measure binding of the procapsid-motor-DNA complex, initiation of packaging, and measure the DNA translocation dynamics with better than 1 nanometer displacement resolution and 0.1 second time resolution. One can also directly measure the forces exerted by the motor on the DNA with piconewton-level resolution. The optical tweezers method has now been applied to the study of three systems—bacteriophages ϕ29, λ, and T4—and this approach has provided a much more detailed picture of viral DNA packaging than traditional biochemical assays.
New findings have provided insight on motor force generation, velocity and processivity, internal forces resisting packaging, procapsid expansion and rupture, details of the mechanochemical kinetic cycle and motor stepping dynamics, functional roles of structural motifs, and conformational dynamics of motor units. These advances will be reviewed in detail below. Though other single-molecule techniques such as fluorescence imaging and spectroscopy have been applied to the study of viral DNA packaging, we limit the scope of this chapter predominantly to the advances made using optical tweezers due to space limitations. We also emphasize that single-molecule techniques will likely continue to have high applicability in the field of viral DNA packaging. Potential future directions, such as further extension of high-resolution and mutational studies in the ϕ29, λ, and T4 systems, and studies of packaging initiation and termination mechanisms, will be discussed in the conclusion.
2. Single-molecule approaches
Viral DNA packaging has long been studied by traditional methods from biochemistry and structural biology. Structural methods such as cryo-electron microscopy and X-ray crystallography can provide detailed information on structures of viral proteins and assemblies with atomic or near-atomic resolution, but do not provide information on the kinetics of packaging or conformational dynamics in protein subunits. Ensemble biochemical techniques, on the other hand, can provide information on molecular dynamics. However, to measure kinetics quantitatively and accurately requires synchronizing populations of complexes, which is often difficult or impossible due to heterogeneity in the ensemble. This heterogeneity stems both from the inherent stochasticity (i.e., randomness due to thermal fluctuations) in the molecular processes, and from potential structural and conformational variability in individual complexes. Traditionally, viral DNA packaging has been assessed in bulk biochemical reactions by quantifying the amount of DNA protected by the capsid from degradation (Grimes and Anderson 1989). This is achieved by adding a nuclease (DNase or sometimes a restriction enzyme) to a packaging reaction to digest any unpackaged DNA, releasing any packed DNA by treatment with proteinase K, and quantifying it by gel electrophoresis. This assay can be used, for example, to determine the efficiency of packaging (the fraction of DNA successfully packaged) or estimate the total time to complete packaging. It is quite difficult, however, to go beyond these basic measurements and extract quantitative information on rates of packaging, how they vary in time, and how they vary among different complexes.
Single-molecule techniques provide a powerful complementary method for studying packaging kinetics in detail. Unlike traditional bulk methods, these techniques do not rely on temporal and population averaging, instead collecting statistics from individual packaging complexes. Moreover, conformational dynamics and force generation can be detected directly in real-time with single-molecule techniques, providing a clear advantage over ensemble methods. Techniques such as single-molecule fluorescence, optical traps, and magnetic tweezers (reviewed in (Neuman and Block 2004, Myong, et al 2006, Greenleaf, et al 2007, Moffitt, et al 2008, Neuman and Nagy 2008, Joo, et al 2008, Rickgauer and Smith 2008)) have been instrumental in deciphering the mechanism of a wide range of biological phenomena, and have been applied to the study of viral DNA packaging in recent years. While much of the work has been with optical tweezers, single-molecule fluorescence microscopy has also recently been employed. Single fluorophore imaging methods were used to search for conformational changes and subunit stochiometries in the ϕ29 motor (Hugel, et al 2007, Shu, et al 2007), and fluorescence correlation spectroscopy and single-pair fluorescence resonance energy transfer were recently used to detect packaging and conformational changes in DNA substrates in T4 packaging (Sabanayagam, et al 2007, Oram, et al 2008, Ray, et al 2010b, Ray, et al 2010a). This chapter, however, will focus primarily on reviewing findings in our main area of expertise: single DNA molecule packaging experiments using optical tweezers.
2.1. Optical traps
Light carries momentum and thus can be used to exert forces on microscopic objects. Optical traps or “tweezers” use a tightly focused laser beam to trap a dielectric object such as a polystyrene microsphere in all three dimensions near the focal point (Ashkin 1986). The trap exerts a restoring force proportional to the displacement of the microsphere. Typically, the position of the microsphere in its trap can be detected to great precision by imaging the trap light scattered by the microspheres onto position-sensitive photodetectors (Neuman and Block 2004). This method can provide sub-nanometer (nm) resolution of displacements and sub-piconewton resolution of forces (pN = 10−12 N).
Optical traps have been used to study the dynamics of a variety of different biomolecular processes, ranging from how individual macromolecules such as proteins, DNA, and RNA unfold under force (Bustamante, et al 2000, Cecconi, et al 2005, Tinoco, et al 2006) to how molecular motors translocate and exert forces (Ross, et al 2008, Michaelis, et al 2009). Of particular interest, single-molecule optical trap experiments have provided novel insights into the mechanism of nucleic acid translocases. In a typical measurement, an individual DNA molecule is tethered between a microsphere held in an optical trap and a second attachment point. This attachment point can be the surface of a sample chamber, or a second microsphere suctioned onto the end of a micropipette or held in a second trap (reviewed in (Moffitt, et al 2008, Chemla 2010, Fuller, et al 2006, Smith, et al 2003)). The tethered DNA molecule may be stretched by an applied force and displacements of the trapped microsphere report on the actions of the biological system under study. Alternately, the instrument can be controlled by a feedback loop in order to maintain a constant force on the trapped microsphere. In this force feedback mode, the position of the trap or attachment point is actively controlled to apply a steady tension to the tethering molecule (Neuman and Block 2004, Smith, et al 2001). Instead of detecting the displacement of the microsphere, which is constant, the readout is the change in separation between one trap and the second attachment point.
2.2. Single-molecule measurement of ϕ29 packaging
Bacteriophage ϕ29 was the first viral DNA packaging system successfully studied using single-molecule optical tweezers measurements (Smith, et al 2001). The initial approach for carrying out this type of measurement was to create a “stalled complex” consisting of a prohead with partially packaged DNA hanging out. Prior to preparing the stalled complex, one end of the ϕ29 DNA was labeled with biotin by cutting the molecule with a restriction endonuclease and using DNA polymerase I to incorporate biotinylated nucleotides. Packaging was initiated in a bulk in vitro reaction and allowed to proceed for ~1 minute until roughly 30–50% of the DNA was packaged. An excess of non-hydrolyzable ATP analog (γS-ATP) was then added, which caused the motors to stall with the unpackaged biotin-labeled end of the DNA dangling out of the prohead.
The single-molecule measurements were performed by carrying out the following steps. Stalled complexes were first attached to streptavidin-coated polystyrene microspheres via the biotin tag. Then, a second batch of microspheres coated with anti-ϕ29 antibodies was prepared. Both types of microspheres were injected into a microfluidic flow chamber in the optical tweezers instrument. In the initial work of Smith et al., the anti-ϕ29 microsphere was held by suction onto the end of a glass micropipette and the microsphere carrying the ϕ29 packaging complex was trapped with an optical trap. The fluid chamber was filled with a solution containing ATP, such that the non-hydrolyzable ATP was eluted away as soon as the microspheres were injected, whereupon stalled complexes resumed packaging. When the two microspheres were brought into proximity by moving the pipette (attached to the chamber) with a piezo-actuated stage, the prohead bound the anti-ϕ29 microsphere, tethering the DNA between the two microspheres (Figure 1A). Successful packaging events were observed from progressive shortening of the DNA tether as the motor reeled in its DNA against the force exerted by the trap, pulling the two microspheres together. Packaging was monitored either by allowing the force to increase as the microspheres were pulled out of their traps by the phage (Figure 1B), or by using force feedback and measuring the decreasing tether length as DNA was packaged (Figure 1C).
Figure 1.
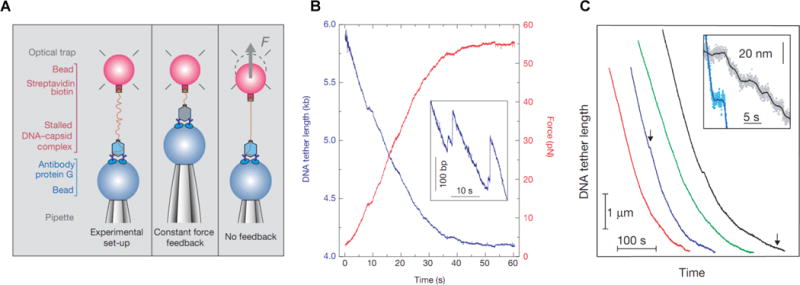
Single-molecule viral DNA packaging assay. A) Schematic of the experimental setup used in the earliest work. A single DNA molecule hanging out of a stalled φ29 packaging complex was tethered at one end to a microsphere held in an optical trap while the procapsid was bound to a second microsphere held by a micropipette. After initiating packaging with ATP two measurement modes were used: “Constant force feedback”, where the separation between the microspheres was adjusted to keep the DNA stretching force constant, or “No feedback” where the separation was fixed and the DNA stretching force was allowed to rise as packaging proceeded. B) Force vs. time (red line) for a packaging event measured without feedback, reaching ~55 pN before the motor paused or stalled, and corresponding tether length vs. time (blue line). Inset is a zoomed view illustrating occasional slipping events where the DNA moved backwards out of the capsid. C) DNA tether length (i.e., unpackaged DNA length) vs. time during packaging with 5 pN force feedback (the four different colored lines indicate four different single packaging events, shifted arbitrarily along the time axis for clarity). D) Inset is a zoomed view of the regions marked with arrows, illustrating occasional pauses in translocation.
This single-molecule approach has been further improved and also successfully extended to study the packaging motors of different viruses (bacteriophages λ and T4). In the following sections, we highlight the contributions made by this assay to our understanding of viral DNA packaging through its initiation (Section 3), its early stages (Section 4), and completion (Section 5).
3. Initiation of DNA packaging
During their life cycles, viruses co-opt the host cell’s own machinery to replicate their genomes and to express the proteins essential to their proliferation. In the case of many dsDNA viruses, including many tailed bacteriophages, expression of structural proteins coded by the viral genome lead to self-assembly of empty procapsid shells into which viral genomes must be packaged. Procapsids contain a portal ring through which DNA is imported. The first step in viral DNA packaging is the assembly of a molecular motor complex on the portal that can translocate the DNA into the procapsid (Catalano 2005). Packaging motors are multi-component, multi-subunit complexes. Unlike many cellular molecular motors such as myosins or kinesins, viral packaging motors assemble only transiently for the purpose of packaging the viral DNA and then disassemble prior to the formation of mature infectious viruses. The transient nature of these motor complexes is one feature that makes them challenging to study. Here, we review features of the initiation of packaging in the phage ϕ29, T4, and λ systems, focusing on recent findings from single-molecule studies.
3.1. Initiation in ϕ29
The B. subtilis phage ϕ29 is one of the smallest tailed dsDNA phages, having a 19.3-kbp genome, and has been extensively characterized using genetic, biochemical, and structural methods (Grimes, et al 2002). The packaging motor is situated at a unique five-fold portal vertex in the 40-nm by 50-nm prolate (elongated) icosahedral prohead (Morais, et al 2005). The motor complex consists of a dodecameric head-tail connector (“portal ring”) comprised of gene product 10 (gp10), a ring of RNA molecules (“prohead RNA” or “pRNA”) attached to the narrow end of the connector, and five copies of the gp16 ATPase (Simpson, et al 2000, Morais, et al 2008). The ϕ29 genome, like that of human adenovirus, has a terminal protein covalently bound to each 5′ end (DNA-gp3) that primes DNA replication and enhances DNA packaging efficiency and selectivity (Grimes and Anderson 1989).
DNA can be efficiently packaged in vitro by mixing together ϕ29 DNA, gp16 motor protein, and empty ϕ29 proheads (procapsids) in a buffer containing ATP (Grimes and Anderson 1989), facilitating detailed biochemical and biophysical studies. Empty proheads can be produced with a mutant that lacks gp16 but contain the gp10 portal ring and pRNA. The original single-molecule experiments with stalled complexes were useful for initial studies (discussed in Section 4), but did not permit study of the initiation and early stages of packaging. Thus, an improved method facilitating measurement of packaging from initiation to completion was developed (Rickgauer, et al 2008).
3.1.1. Method for tracking single DNA packaging from initiation to completion
In the improved method of Rickgauer et al., complexes consisting of proheads and the packaging ATPase gp16 were assembled with γS-ATP in the absence of DNA. An optical tweezers instrument consisting of two traps was also used, facilitating more accurate measurements of packaged DNA length (Fuller, et al 2006, Rickgauer, et al 2006). A microsphere carrying DNA was injected into the chamber and captured in one optical trap and a microsphere carrying prohead-gp16 complexes was injected and captured in a second trap. Packaging was initiated by bringing the two microspheres into near contact, allowing a DNA molecule to bind to a prohead-motor complex (Figure 2A). Binding was observed to occur within seconds and DNA translocation within one second of binding.
Figure 2.
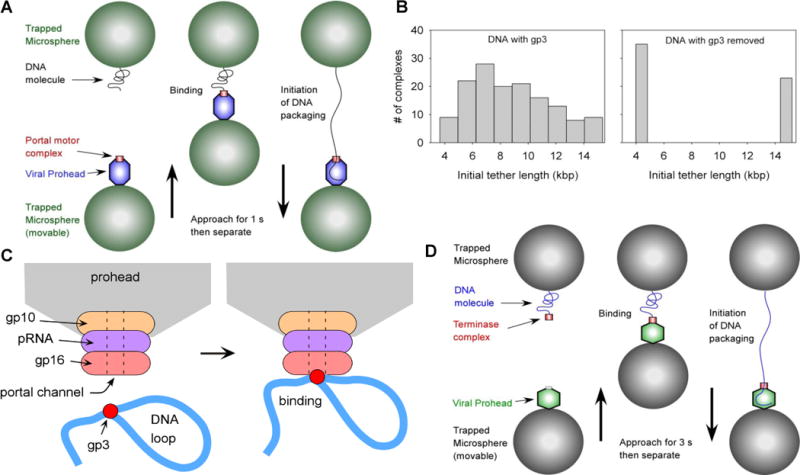
Measurements of the initiation of viral DNA packaging A) Schematic of the experimental setup used to initiate φ29 packaging in real time. Instead of using a stalled, partially packaged complex as in Fig. 1, a pre-assembled prohead-motor complex is “fed” DNA. Each microsphere was held in a separate optical trap. The same approach was used for initiating phage T4 packaging. B) Measured distribution of initial tether lengths for packaging native φ29 DNA with its gp3 terminal protein, or with gp3 removed by digestion with proteinase K. C) Proposed model for gp3-mediated DNA looping at the initiation of φ29 packaging. D) Modified approach used to initiate single-molecule phage λ packaging. A motor-DNA complex is preassembled and brought into proximity of a λ procapsid (instead of preassembling a motor-procapsid complex).
While the order of assembly of the components was unclear in prior bulk packaging assays, these single-molecule studies demonstrate an assembly pathway wherein a motor-prohead complex forms first and then engages the DNA. In these studies, γS-ATP appeared to stabilize a packaging-competent conformation of the prohead-motor complex and ATP appeared to destabilize it, but following initiation the translocating complex was highly stable in the presence of ATP. As with many DNA processing enzymes, Mg2+ was found to be a necessary cofactor; 1 mM Mg2+, beyond that complexed with ATP, was necessary for initiation of packaging (Fuller, et al 2007b).
3.1.2. Effect on initiation of packaging of the ϕ29 gp3 terminal protein
Following initiation of packaging as described above, it was unexpectedly found that the initial end-to-end extension of stretched DNA tethers was highly variable, ranging from 30 to 100% of the full-length of the DNA substrate (Figure 2B) (Rickgauer, et al 2008). This suggests ϕ29 DNA has a complex structure that does not bind the motor by a free end. The variability was shown to be due to the gp3 terminal protein, since it disappeared when the ϕ29 DNA was pretreated with proteinase K. Both left end (15 kb) and right end (4 kb) restriction fragments with proteolysed gp3 were readily packaged with full tether lengths at initiation (Figure 2B). Arbitrary non-native DNA molecules generated by PCR could also be packaged with no tether length variability. While non-gp3 DNA is not packaged in vivo and packaged inefficiently in bulk in vitro reactions compared with gp3-DNA, non-gp3 DNA was efficiently packaged in the optical tweezers assay (Rickgauer, et al 2008). The observed variability in DNA extension immediately following initiation of DNA packaging in the optical tweezers is most likely due to gp3-mediated DNA looping. Electron microscopy shows that ϕ29 DNA can form loops at its termini and that purified gp16 motor protein can bind at DNA-gp3 loop junctions (Grimes and Anderson 1997). The observed extension variability in the optical tweezers measurements further suggests that packaging initiates with binding of prohead-gp16 complexes to these loop junctions (Figure 2C). A recent study of T4 packaging suggests, based on co-localization of the packaged DNA ends detected by single-pair FRET, that the T4 motor might also be capable of packaging looped DNA (Ray, et al 2010a). On the other hand, it is unclear how such looped DNA can fit through the portal channel since X-ray crystal structures suggest that it could not easily thread more than one segment of dsDNA at a time (Simpson, et al 2000).
After initiation, no abrupt increase in DNA extension was observed, indicating that the putative loop does not open prior to DNA translocation. One possibility is that the section of looped DNA is cleaved prior to DNA translocation. Although the ϕ29 genome is replicated as a monomer, related viral motors including λ and T4 have endonuclease activities that are needed to excise their genomes from concatemeric substrates (Rao and Feiss 2008). Alternatively, it has been suggested that gp16 may have gyrase activity, based on the observation that it appears to be capable of supercoiling DNA-gp3 loops (Grimes and Anderson 1997). It is therefore possible that cleavage of the DNA loop may result from stretching the DNA during gyrase action. A second possibility is that a DNA loop is present throughout packaging and the motor is capable of translocating DNA in one side of a loop while packaging the DNA from the other side of the loop.
3.2. Initiation in phage T4 by procapsid-motor complex assembly
T4 is an important model for large tailed phages that exhibits distinct differences from ϕ29. Notably, T4 has a much larger capsid size (120 × 86 nm), and must package a 9x longer genome (Rao and Feiss 2008, Fokine, et al 2004). T4 is also a prototype for viruses that package DNA by a “headful” mechanism, in which unit length DNA segments that fill the procapsid must be excised by the packaging motor from a concatenated string of multiple T4 genomes produced by rolling circle replication. The T4 packaging motor consists of a terminase complex comprised of a small subunit, gp16, and large subunit, gp17 (containing the packaging ATPase), which appears from cryo-EM studies to form a pentameric ring (V. Rao, personal communication) connected to a dodecameric portal ring (gp20). Notably, T4 is the only virus for which an atomic structure of the ATPase subunit responsible for powering DNA packaging (gp17) has been determined by X-ray crystallography (Sun, et al 2008).
The single-molecule T4 packaging assay employed a similar strategy as used with ϕ29. An efficient defined in vitro T4 packaging system consisting of only three components—empty proheads (containing a portal connector ring), the large terminase subunit (gp17, the packaging ATPase) and DNA—was developed by V. Rao and coworkers (Kondabagil, et al 2006), also building on work by L. Black and coworkers (Baumann and Black 2003). It was found that a packaging-competent prohead-motor complex could be prepared by incubating the T4 gp17 with empty procapsids in the presence of non-hydrolyzable γS-ATP (Fuller, et al 2007). These complexes were then bound to microspheres, and DNA molecules were attached to separate microspheres. As in the method of initiation developed by Rickgauer et al. for ϕ29, DNA was “fed” to the T4 packaging motor by rapidly bringing both types microspheres into close proximity inside a flow chamber containing ATP (Figure 2A). Although a small terminase subunit (gp16) is needed for packaging in vivo, inclusion of only the large subunit (gp17) was found to be sufficient for efficient in vitro packaging.
3.3. Initiation in phage λ by motor-DNA complex assembly
Phage λ has been one of the most important model systems in molecular biology for over half a century (Gottesman and Weisberg 2004). This E. coli virus has a 62-nm diameter icosahedral capsid containing a 48.5-kbp genome. Packaging is carried out by λ terminase, a hetero-oligomer composed of the viral gene products gpA (large terminase subunit) and gpNu1 (small subunit) (Catalano 2005). Like T4, the viral DNA is copied by rolling circle replication, yielding concatenated genomes that must be excised. However, unlike T4, λ terminase binds to and cleaves the DNA at a specific site (cos site) where packaging initiates. A stable terminase complex can be assembled onto DNA by adding recombinant gpA and gpNu1 proteins to form a stable intermediate referred to as Complex I. The gpNu1 subunit mediates the assembly of terminase at this site while gpA possesses endonuclease, strand separation, and ATPase/DNA translocation activities. Biochemical studies indicate that the proteins assemble into a stable gpA1/gpNu12 heterotrimer and these trimers can assemble into a homogeneous tetrameric ring of sufficient size to encircle dsDNA (Maluf, et al 2006). Presumably the terminase ring assembles at a cos site, then binds to the portal of a procapsid (to form “Complex II”), and then initiates DNA translocation. At least two accessory proteins aid packaging: E. coli IHF appears to aid the formation of Complex I, and λ gpFI aids the formation of Complex II (Sippy and Feiss 2004, Gaussier, et al 2006).
The strategy used by Fuller et al. to develop a single-molecule packaging assay was opposite of that used with the ϕ29 and T4 systems. Instead of assembling a procapsid-motor complex and feeding it DNA, the terminase complex was assembled on a DNA substrate containing a cos site (required for λ packaging) near one terminus and biotin-labeled nucleotides at the opposite terminus (Fuller, et al 2007a). This DNA was tethered to a streptavidin-coated microsphere and attached λ procapsids to a separate batch of microspheres. Packaging was initiated by injecting the two microspheres into a microfluidic chamber containing ATP, trapping them in separate optical traps, and bringing the two microspheres into proximity such that the terminase-DNA complex could bind the procapsid (Figure 2D). As in the ϕ29 and T4 systems DNA binding and initiation of translocation occurred rapidly within seconds. Although different pathways for initiation, via either a motor-DNA complex (λ) or motor-prohead complex (ϕ29 and T4) have been observed in the single molecule studies, it is unclear whether multiple pathways may be followed in vivo.
4. Early stages of DNA packaging
Upon successful assembly of the packaging motor complex, the viral genome is internalized into the procapsid. This is accomplished in one continuous process in which the portal motor translocates the viral DNA using the energy of ATP hydrolysis. It is useful for our purposes to distinguish between “early” and “late” stages of packaging. During the initial stages, DNA encounters relatively little resistance to encapsidation. However, as the capsid is filled, internal forces resisting DNA confinement build, and the motor operates under a significant load. In this section we will focus on motor function in the early stage of packaging, where there is little load on the motor, and how single-molecule studies have revealed key aspects of its mechanism. The late stages of packaging will be discussed in Section 5.
4.1. Insights on packaging from structural and biochemical studies
Prior to the single-molecule measurements highlighted in this chapter, years of extensive structural and biochemical studies provided important insights into the mechanism of viral DNA packaging. An early structural model for the mechanism of the packaging motor was provided by the observation of a mismatch between the six-fold symmetry of the portal connector and five-fold symmetry of the capsid vertex in which it resides. This observation led Hendrix (Hendrix 1978) to propose that packaging might be driven by a “nut-and-bolt” mechanism, in which rotation of the connector (the nut) causes linear motion of the helical DNA (the bolt). More recently, the first X-ray structures of the ϕ29 connector by Simpson et al. (Simpson, et al 2000) verified this symmetry mismatch, though the connector was found to have 12-fold symmetry, rather than 6-fold. The portal structure is remarkably conserved in other phages, hinting at a common mechanism. Based on this new structural information, several rotary motor models were proposed, involving such mechanisms as cyclic compression and relaxation of the connector (Simpson, et al 2000), electrostatic interaction with lysine rings inside the portal channel (Guasch, et al 2002), and sequential movement of loops (molecular levers) in the connector tunnel in kind of a “Mexican wave” (Lebedev, et al 2007) (reviewed in (Rao and Feiss 2008)). All of these proposed mechanisms necessitate connector rotation to align the proper structural motifs of the motor with the helical pitch of the DNA during translocation, and also require coordinating this rotation with ATP hydrolysis in the ATPases.
Despite the attractiveness of these models, recent experiments have challenged these “connector-centric” mechanisms of packaging. Portal rotation was tested directly in T4 by cross-linking the connector to the capsid using the Hoc protein (Baumann, et al 2006). If rotation was essential for packaging, tethering of the connector would have abolished packaging. However, packaging efficiency was comparable to that in wildtype phages. Single-molecule fluorescence experiments in ϕ29 led to the same conclusion. Here, portals were labeled with a single fluorescent Cy3 dye whose polarized emission was used as a reporter for the orientation of the protein. In measurements of labeled complexes that were actively packaging, assessed by observing the translocation of tethered microspheres, no evidence was found for portal rotation (Hugel, et al 2007). Lastly, in ϕ29, mutations to the connector loops predicted in one model to interact with DNA did not lead to any observable decrease in packaging efficiency (R. Atz, S. Grimes, D.L. Anderson, personal communication). These studies together strongly suggest that the portal does not play an active role in DNA translocation. Interestingly, the ϕ29 loop mutation studies indicate that mutants are more prone to release packaged DNA than wildtype phages, implying that the connector may be important in retention of packaged DNA, rather than in its translocation.
The alternative to a portal-driven packaging mechanism is that the ATPase motor component directly translocates DNA (Black 1989, Fujisawa and Morita 1997). Compelling evidence supporting this mechanism has come from the recently solved structures of the T4 large terminase, gp17 (Sun, et al 2007). These studies, along with sequence homology analyses (Draper and Rao 2007), indicate a similar architecture in gp17 to monomeric SF1 and SF2 DNA (and RNA) helicases. Comparisons of X-ray crystal and cryo-electron microscopy structures of T4 gp17 monomers revealed two globular sub-domains separated by a “hinge”. These can adopt “relaxed” and “tensed” conformations separated by approximately 2 bp that correspond to the apo- and nucleotide-bound states, respectively (Sun, et al 2008). These ATP-driven conformational rearrangements in the sub-domains support an inchworm-type mechanism for DNA translocation, in which a C-terminal DNA binding domain ratchets the DNA into the prohead. Further supporting this “terminase-centric” model of packaging, the observed conformational switch in gp17 is consistent with estimations of the motor step size by biochemical assays. Measurements of ATP consumption in bulk, in vitro packaging with phage ϕ29 (Guo, et al 1987, Chemla, et al 2005) and T3 (Morita, et al 1993) have been used to determine the “coupling ratio”, the number of ATPs hydrolyzed per length of DNA packaged. These measurements revealed that on average ~2 bp of DNA is packaged per 1 ATP hydrolyzed. These have been interpreted as indicating that the motor steps in increments of 2 bp, consistent with the structural model of T4 gp17.
Despite the many insights provided by these structural and biochemical studies, many questions on viral DNA packaging remained open. Below we discuss recent single-molecule optical trap experiments and how they have shaped our understanding of key aspects of this process: the kinetics of packaging, the mechanism of force generation, how ATP hydrolysis is coupled to translocation, how the ATPases are coordinated, the precise step size, and the interaction of the motor to DNA.
4.1. Single-molecule kinetics of packaging
4.1.1. Motor velocities with saturating ATP at low force and low capsid filling
The single-molecule optical trap assay allowed, for the first time, extensive and accurate characterizations of packaging kinetics in phages ϕ29, T4 and λ (Smith, et al 2001, Rickgauer, et al 2008, Fuller, et al 2007, Fuller, et al 2007a, Chemla, et al 2005). In these three systems, the observed kinetics proved to be far more complex than anticipated. Packaging is interrupted by pauses and slips, where the motor transiently disengages and reengages its DNA substrate, and occurs at a rate that depends on how much DNA is encapsidated, on load force applied to the motor, and can vary for individual motor complexes.
Measurements of motor velocities in the early stages of packaging (near-zero capsid filling, where there is little resistance to packaging) and at a saturating ATP concentration were carried out in all three systems. Force feedback was used to apply a small constant force, typically ~5 pN, to keep the DNA stretched and allow continuous tracking of packaging in real time (Smith, et al 2001). Under these conditions, and in standard packaging buffer (25 mM Tris-HCl (pH 7.8), 50 mM NaCl, 5 mM MgCl2), the ϕ29 motor translocates at an average rate of ~145 bp/s (Rickgauer, et al 2008). Solution conditions can affect motor velocity (Fuller, et al 2007b); average speeds were found to vary between 170 bp/s (at 5 pN load) with 100 mM NaCl and 114 bp/s with zero NaCl. As discussed further below, velocities are also dependent on the load force applied by the optical trap. Using a ramped DNA stretching method, Rickgauer et al. were able to extract packaging rates at near-zero applied load, revealing a slightly higher average motor velocity of 165 bp/s (Rickgauer, et al 2008). Studies employing a temperature-controlled sample chamber also showed that increasing the temperature from 20 to 35°C (a more physiologically relevant temperature) increased the motor velocity two- to three-fold (unpublished data, M. White and D. Smith). Phage λ has a 2.5× longer genome than ϕ29 and its average motor velocity was measured to be ~4.2× higher under similar conditions, 590 bp/s at a 5 pN load. T4, one of the largest phages, has an enormous 171 kbp genome and its average motor velocity was measured to be ~5.4× higher than ϕ29, 770 bp/s at a 5 pN load, (Fuller, et al 2007). This observed trend of faster packaging rates for longer genome lengths is consistent with the need for each virus to package its complete genome in a limited time window of ~2–5 minutes to complete the infection cycle within 20–30 minutes. Kinetic parameters for ϕ29, T4, and λ from optical trap measurements are summarized in Table 1.
Table 1.
Summary of kinetic parameters from phage φ29, λ, and T4
| Phage | Genome length (kbp) | Maximal velocitya (bp/s) | Std/mean velocity (%) | Hydrolysis rate (ATP/s−1) | Processivityb (kbp) |
|---|---|---|---|---|---|
| φ29 | 19.3 | 145 | 10 | >6 | ~20 |
| λ | 48.5 | 590 | 20 | >25 | >48.5 |
| T4 | 171 | 770 | 40 | >40 | 13 |
Measured at room temperature, in standard packaging buffer containing saturating ATP, at low capsid filling, and under an externally applied load of 5 pN.
Defined as the distance between slipping events (scored as >50 bp backward movement of the DNA out of the capsid at a load of 5 pN).
Another intriguing finding revealed by the single-molecule studies is that the velocity of individual motors can vary significantly in time and the average velocity of different individual motors can also vary significantly. Such variability was most striking in T4 (Figure 3A) (Fuller, et al 2007). The average velocities of different individual motors ranged from as low as 70 bp/s to as high as 1840 bp/s with a standard deviation of 320 bp/s, or 40% of the mean. By comparison the average velocities of ϕ29 and λ motors were also variable, though by smaller amounts (~10% and 20% of the mean velocities, respectively). (Rickgauer, et al 2008, Fuller, et al 2007a) (Figure 3B). Some individual T4 motors exhibited velocity changes in time ranging as wide as 500 to 1500 bp/s (Figure 3C). The variations in T4 motor velocities are too large to be reconciled in standard kinetic models as being due to inherent stochastic thermal fluctuations alone, suggesting that individual motors can adopt different active conformational states gearing different DNA translocation velocities and can switch between these states in time (Fuller, et al 2007). Static and dynamic variability (also termed “disorder”) in enzyme kinetics has been reported in single-molecule studies of several other simpler enzyme complexes including lactate dehydrogenase, cholesterol oxidase, λ exonuclease, and RecBCD helicase (Perkins, et al 2004, Xue and Yeung 1995, Lu, et al 1998, van Oijen, et al 2003). An understanding of the structural origin of these heterogeneities is currently lacking. They may be due, in part, to changes in conformation or chemical structure of individual motor subunits, variation in the number of active subunits in individual complexes, or conformational changes in the arrangement of subunits in the whole motor complex.
Figure 3.
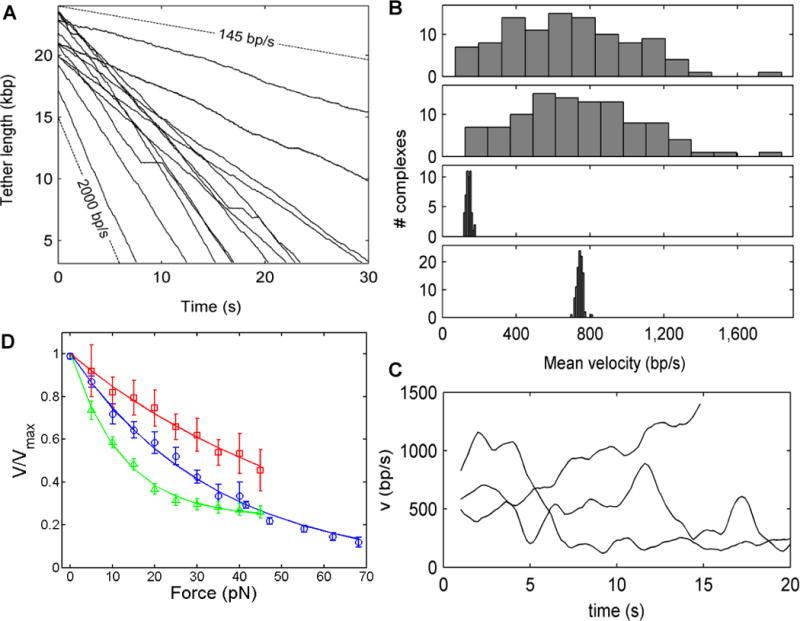
Phage T4 DNA packaging dynamics. A) Repeated measurements of DNA tether length vs. time with 5 pN force feedback, showing variable packaging rates for individual complexes ranging from ~145–2000 bp/s, and showing occasional pauses in translocation (plateaus). B) Histogram of average packaging rates measured for individual T4 motors (top panel); same histogram but with rates calculated not including pauses (2nd panel); histogram of average φ29 packaging rates for comparison (3rd panel); histogram showing stochastic variation in T4 packaging rates predicted by a simple Poisson-stepper model if individual complexes are assumed to have uniform kinetics (4th panel). C) Examples of three packaging events where large variations in instantaneous motor velocity vs. time were observed. D) Dependence of motor velocity on applied load force for T4 (red squares), λ (green triangles), and φ29 (blue circles). Velocities are normalized to unity at zero load.
4.1.2. Pauses and slips
Optical tweezers measurements revealed that packaging motors are remarkably processive, meaning that many basepairs of DNA are continuously packaged with only occasional pausing or backward slipping of the DNA out of the capsid (Smith, et al 2001, Rickgauer, et al 2008, Fuller, et al 2007, Fuller, et al 2007a) (Figure 1B&C, 3A). While the appearance of pauses and slips is intriguing and not fully understood, reversible pauses and slips only reduce the overall average motor velocity at low capsid filling by ~10% (Fuller, et al 2007) (although the λ motors were occasionally observed to abruptly pause and never resumed packaging even after >1–2 minutes, after which data collection was stopped). The ϕ29 and λ motors exhibited less than one significant slip (>50 bp backwards movement of the DNA out of the capsid) per genome length packaged. The T4 motor, which has the longest genome to package, exhibited the most slipping (one slip every 13 kbp), but is still a highly processive motor. The majority of slips were relatively small, less than a few hundred bp, meaning that once packaging initiates the probability that DNA slips out of the capsid completely appears to be quite small.
The mechanisms of pausing and slipping are not known, but the finding that their frequencies increase with applied load and that neither type of event correlates with particular positions along the DNA suggests that they may be off-pathway events that stochastically occur during normal DNA translocation. The dependence on load may indicate that force is able to disrupt the motor-DNA interactions made during translocation, leading to temporarily arrests in packaging (pauses) or disengagements with the molecule (slips). The capacity of these motors to reversibly pause, slip, and change velocity may be biologically relevant since packaging in vivo must be coordinated with other biochemical processes potentially ongoing on the same DNA substrate, including DNA transcription, recombination, and repair.
4.1.3. Dependence of motor velocity and power on applied load
In all three systems studied, ϕ29, λ, and T4, the motor velocity with saturating ATP was found to decrease with applied force (Smith, et al 2001, Rickgauer, et al 2008, Fuller, et al 2007, Fuller, et al 2007a). A general implication of this finding is that the rate-limiting step in packaging must involve DNA translocation (Wang, et al 1998). Displacement against an opposing force implies that mechanical work must be done by the motor, leading to an increased free energy barrier for motor stepping and slowed reaction rate. In the simplest single-reaction energy barrier model, the extra work required to translocate DNA against the optical trap force is F∆x1, where ∆x1 is the distance to the “transition state” (∆x1 ≤ d, the step size), defined along a reaction coordinate corresponding to the amount of DNA packaged. The translocation rate thus decreases as , where kB is the Boltzmann constant and T is the absolute temperature. As shown in Figure 3D, the dependence of velocity on force for each system displayed quantitative differences. The T4 motor velocity dropped by ~40% as the force was increased from 5 to 40 pN, whereas the ϕ29 and λ motor velocities dropped ~60%. Neither the ϕ29 nor λ velocity vs. force datasets could be well fit by a single force-dependent rate suggested by the simple single-reaction energy barrier model, indicating that different kinetic transitions become rate limiting at high force. A notable feature of the λ motor was that the motor velocity appeared to plateau at ~200 bp/s above ~50 pN, rather than approaching zero, suggesting that a purely chemical transition (i.e., one not involving DNA translocation) becomes rate-limiting at high force. In T4, the velocity decreased approximately linearly with force, over the range studied, possibly indicating a very small transition state distance for that system. Despite commonalities in behavior, these differences in force dependence may point to different mechanisms of translocation. Future work will be necessary to resolve these differences further.
By multiplying the average applied force by the corresponding average velocity, the mechanical power generated by the motor can be calculated. The maximum power observed to be generated by the λ motor occurred with a load of 45 pN, where the motor velocity was 208 bp/s, implying an average power of 9400 pN·bp/s = 3200 pN·nm/s. This is ~4× higher than the maximum power detected for the ϕ29 motor. Assuming a free-energy release of 130 pN·nm per ATP (note that we express this free energy in units of pN·nm (force × distance) relevant in the single-molecule trap studies and ~130 pN·nm = 73 kJ/mol), this implies an ATP hydrolysis rate of at least 3200 pN·nm/s ÷ 130 pN·nm per ATP = 25 ATP/s for the λ motor. This figure is higher than the figure of ~10 ATP/s previously estimated in bulk biochemical assays, suggesting that those assays underreport the rate due to difficulties in accounting for “futile” ATP hydrolysis not associated with packaging. The maximum T4 mechanical power was observed with 40 pN load, where the velocity was 380 bp/s, yielding a power of 15200 pN·bp/s = 5200 pN·nm/s, about 7× higher than that detected for ϕ29, and implying an ATP hydrolysis rate of at least 5200 pN·nm/s ÷ 130 pN·nm per ATP ≅ 40 ATP/s. While these power figures may seem small, it must be kept in mind that the motor is a nanoscale device occupying a volume of only ~(10 nm)3, implying a power density on the order of 5000 kiloWatts/m3, which is roughly twice that generated by a high performance automobile engine.
4.1.4. Motor force generation
One of the most striking features revealed by the single-molecule measurements was that viral DNA packaging motors generate very high forces, among the highest known for biomolecular motors (Ross, et al 2008, Michaelis, et al 2009, Oster and Wang 2003). Optical tweezers measurements allow one to measure directly the packaging force exerted by the motor on the DNA since that force is transmitted directly to the trapped microspheres. Measurements at low capsid filling, where internal forces resisting DNA confinement are small (discussed in Section 5), revealed that the ϕ29, λ, and T4 motors are all able to translocate DNA against externally applied load forces of >50–60 pN (Smith, et al 2001, Fuller, et al 2007, Fuller, et al 2007a). These figures are lower bounds because most measurements ended with the DNA tether detaching from the microspheres, likely due to rupture of the prohead-antibody-microsphere linkage. While some motors did stall at forces <50 pN others were still translocating when rupture occurred at >50 pN. Extrapolation of measurements made at high capsid filling, where forces resisting DNA confinement (discussed in Section 5) contribute a large additional load on the motor suggest that the ϕ29 motor can exert total forces as high as 110 pN (Rickgauer, et al 2008), strikingly large compared with many cellular molecular motors. For example, skeletal muscle myosin II, which powers skeletal muscle contraction, only generates 2–3 pN of force (Finer, et al 1994). It is likely that large forces are necessary to package DNA against the enormous internal forces generated from compacting the viral genome into the capsid. This issue will be discussed in depth in Section 5 on completion of packaging.
Current understanding dictates that molecular motors translocate in discrete-sized steps tightly coupled to the hydrolysis of ATP molecules. Energetic considerations then impose a tradeoff between a motor’s mechanical step size and force generation. Each ATP hydrolysis releases a free energy on the order of 130 pN·nm, depending on solution conditions (Lehninger, et al 1993). A motor, depending on its efficiency, can then convert up to ~130 pN·nm of chemical energy into mechanical work to translocate one step. If the motor translocates a step size d against force F, the total mechanical work performed is Fd, which must be less than ~130 pN·nm. Thus a force of ~60 pN generated by the packaging motor places an upper bound of ~2 nm = 6 bp on its step size. For ϕ29, for which a maximum total force of 110 pN was reported, the step size must be smaller than ~1.2 nm, or 3.5 bp. (Precise direct measurements of motor step size are discussed below.) It is tempting to speculate that different types of motors may have evolved to generate more or less force depending on the physical bounds on their step size. Thus, cytoskeletal motors like myosin must take large steps, as dictated by the periodicity of the actin tracks on which they translocate, and thus generate small forces. On the other hand, viral DNA packaging motors, which are built to exert large forces to counteract the internal forces generated by compaction of the DNA in the capsid, must take relatively small steps.
4.2. Mechanochemistry of packaging
The viral packaging machine requires ATP as a cofactor to drive processive encapsidation of DNA. A fundamental question is how the energy contained in a molecule of ATP is utilized by the motor to generate the large forces exerted during DNA translocation. The conversion of chemical energy into mechanical action—termed “mechanochemistry”—is a process carried out by all molecular motors, and provides an important clue to their mechanism. In all nucleic acid translocases like DNA packaging motors, a mechanism couples ATP hydrolysis to DNA movement. ATP hydrolysis can be viewed as a multi-step process involving: docking of ATP into the catalytic cleft, accommodation of the nucleotide into the proper orientation, nucleophilic attack of the γ-phosphate, and release of the hydrolysis products phosphate and ADP, to name a few examples. These steps must somehow connect to the translocation cycle, during which the motor must engage with the DNA, translocate it by the motor step size, disengage the DNA, and reset the machinery for the subsequent cycle. Thus, one key goal in understanding this process is identifying which step or steps in the ATP hydrolysis cycle lead to DNA translocation. A second key goal is to understand the structural specifics governing these steps.
Recent structural studies of T4 gp17 terminase have provided important clues on the mechanochemical coupling process in packaging motors. The observation of a “C-motif”, in which a network of hydrogen bonds connects the γ-phosphate of ATP to DNA (Draper and Rao 2007), provides the first structural basis for the communication between chemical and the mechanical activities of the protein. While instructive, molecular structures only provide static pictures of the mechanochemical conversion process. Single-molecule measurements provide complementary information in the form of real-time measurements of the DNA translocation dynamics. In particular, optical traps provide an ideal platform for investigating the mechanochemistry of molecular motors because force can be utilized as a probe for mechanical motion. In the single-molecule packaging assays described above, the phage translocates DNA against the force exerted by the optical trap; increasing this tension thus decreases the rate at which the translocation step occurs while leaving the other (force-independent) steps unperturbed. This feature of optical trap measurements is unique but loosely analogous to classical enzymology, where how an enzyme binds a substrate can be measured by varying the substrate concentration or by adding inhibitors that compete with the substrate. Here, rather than vary concentrations of a chemical species, force is used to modulate the translocation rate in the mechanochemical cycle of the protein.
4.2.1. Single-molecule measurements of mechanochemistry in ϕ29
Chemla et al. (Chemla, et al 2005) investigated mechanochemical coupling in the DNA packaging motor of phage ϕ29 exploiting this unique ability of optical traps. Combined with methods of classical enzymology to understand the chemical aspects of packaging and force dependence to decipher the mechanical aspects, they were able to provided key insights on the coupling between the two. To achieve this understanding, Chemla et al. performed an extensive set of experiments measuring the generation of force by the ϕ29 motor as a function of ATP, non- (or slowly-) hydrolyzable ATP analogs (AMP-PNP and γS-ATP), and product (ADP and Pi) concentrations. In the first set of measurements, ATP and force were varied and their effect on the packaging rate was investigated. The packaging rate V was found to depend on ATP concentration according to the classical Michaelis-Menten equation: . At low forces (~5 pN) a Michaelis-Menten constant KM ~ 30 μM was observed. More illuminating were measurements of the force dependence of the packaging rate at different ATP concentrations, as shown in Figure 4A. At saturating ATP levels ( ), the velocity depended strongly on force, indicating that the DNA translocation step was rate-limiting. In contrast, at low ATP levels ( ), where ATP binding was now rate-limiting, the packaging rate was largely independent of force. This observation revealed that the force-generating DNA translocation step must not occur during ATP binding.
Figure 4.
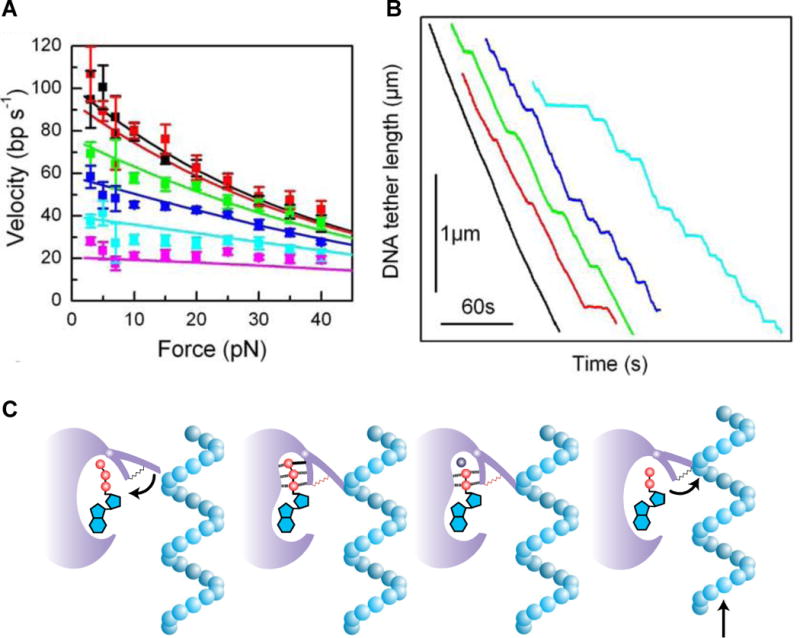
Mechanochemistry of φ29. A) Force-velocity behavior. As ATP was decreased from 500 μM (black data) to 10 μM (magenta), the packaging velocity displayed a decreasing dependence on force, indicating that ATP binding is not a force-generating kinetic event. B) Analog-induced stalls. As the non-hydrolyzable ATP analog AMP-PNP was added to the packaging buffer (containing 100 μM ATP), packaging was interrupted by stalls. The frequency of the stalls increased as AMP-PNP was increased from 0 (black data) to 5 μM (cyan). C) Model of mechanochemical coupling in φ29. ATP binding in the catalytic cleft of gp16 induces elastic strain that is stored as a compressed spring. This binding step occurs while the ATPase is disengaged from the DNA, such that no translocation occurs and binding is insensitive to tension. Upon hydrolysis and release of the cleaved phosphate, this elastic strain is relieved in a “recoil” step. Here the gp16 is engaged to the DNA, and recoil drives the force-generating translocation.
Poisoning the ATP packaging buffer with non-hydrolyzable analogs was found to induce stalls in packaging lasting several seconds (Figure 4B) and at a frequency (measured in number of induced stalls per length packaged) proportional to the analog concentration. Neither the frequency nor the duration of these stalls—kinetic parameters corresponding to the binding and unbinding of the analog to the motor, respectively—displayed any dependence on force. These results again indicated that the nucleotide binding step in the mechanochemical cycle of the motor does not generate force. Measurements in the presence of hydrolysis products (ADP and Pi) illuminated the role of product release in the motor cycle. Though no stalls were detected, the packaging rate did decrease significantly with increasing ADP concentration, in a manner consistent with it acting as a competitive inhibitor to ATP. (Presumably, ADP binding and release were too rapid to observe stalls of significant duration). Moreover, similarly to the force dependence of the packaging velocity at varying ATP concentration described above, the pattern of ADP inhibition at various forces was inconsistent with ADP release being a force-generating step. Finally, increasing the inorganic phosphate level a thousand-fold had no discernable effect on the packaging rate, indicating that the release of phosphate was highly irreversible. Taken together, these results led Chemla et al. to conclude that DNA translocation must occur at some step between hydrolysis (i.e. after ATP binding) and phosphate release (i.e. before ADP release and resetting of the cycle).
4.2.2. Model of ATP coupling in ϕ29
Theoretical models of mechanochemistry in other molecular machines, particularly in other ringed ATPases such as the members of the AAA superfamily, provide a useful context for interpreting these optical trap measurements. In F1-ATPase, part of the rotary ATP synthase motor, it has been proposed that the energy driving the mechanical rotation is ultimately derived from ATP binding (Oster and Wang 2000). In this model, ATP binds to each catalytic site in a zippering of hydrogen bonds that induces elastic strain and produces the first of two force-generating “power strokes” driving the motor. A second power stroke occurs when that elastic strain is relieved in a “recoil” step triggered by phosphate release and immediately preceding ADP release. The hydrolysis step itself is not believed to drive any mechanical motion, because it involves only small rotation of the terminal phosphate, which is almost iso-energetic in this protein (Oster and Wang 2000).
The measurements of Chemla et al. on ϕ29 suggest a model for the packaging motor partly consistent with this mechanism. One prediction of the F1-ATPase mechanochemical model is that both ATP binding and recoil steps should involve large changes in free energy in order to drive mechanical motion. This is consistent with two observations in ϕ29: (1) long stalls with ATP analogs, indicating that nucleotides are tightly bound to the motor and release at very slow rates; and (2) the evidence for irreversible phosphate release. On the other hand, the F1-ATPase model proposes that ATP binding should drive mechanical motion, yet this step was shown to be force-independent in the trap experiments. This suggests that in ϕ29, ATP binding occurs at a gp16 subunit that is disengaged with the DNA, and thus insensitive to the forces applied by the optical trap. This is supported by the observation that the motor is more likely to slip, or disengage from DNA, at low ATP concentrations (Chemla, et al 2005) favoring the apo state. This suggests that elastic strain induced by ATP binding may cock the spring that drives translocation. After ATP binds, the gp16 engages the DNA and relieves this elastic strain, releasing the spring and translocating the DNA right before ADP is released. The data is thus consistent with a recoil step generating force, with phosphate release acting as an irreversible trigger committing the motor to the rest of the reaction cycle. This mechanism proposed by Chemla et al. is depicted schematically in Figure 4C.
4.2.3. Coupling mechanism and motor structure/function relationships in T4 and λ
Studies of λ and T4 phage are so far consistent with this mechanism of mechanochemical coupling. In T4, structures of gp17 in its apo and nucleotide-bound states by Sun et al. (Sun, et al 2008) qualitatively fit with the model proposed by Chemla et al. In the apo state, gp17 is in a relaxed conformation, and putatively disengaged from DNA, whereas the ATP-bound state is tensed and putatively engaged with the DNA. This is consistent with the ATPase being “cocked” by nucleotide binding in the Chemla model. Upon hydrolysis, the C-motif of gp17 is proposed to unlock the terminal phosphate of ATP and the catalytic cleft recoils to its open state, driving translocation of the DNA, consistent with the proposed ϕ29 mechanism.
In the phage λ system, optical tweezers studies of the effects of amino acid point mutations in the motor’s large terminase subunit (gpA) on packaging dynamics recently shed light on structure-function relationships (summarized in Table 2) (Tsay, et al 2009, Tsay, et al 2010). Photo-crosslinking with 8-azido-ATP showed that residues Y46 and K84 of gpA interact with ATP (Hang, et al 2000). On the basis of sequence homology with AAA+ superfamily ATPases, gpA was predicted to have a “Walker A-like” phosphate binding motif at 76-KSARVGYS-83 (Mitchell and Rao 2004). In support of this hypothesis, optical tweezers measurements indicated that change K84A, immediately adjacent to this region, decreased motor velocity by ~40% but did not alter processivity or the steepness of the velocity-force dependence (Figure 5A) (Tsay, et al 2009). This finding is consistent with the notion that the Walker A motif is involved in ATP binding but not coupling, and support a model in which ATP binding and hydrolysis are not the force-generating steps in λ as in the ϕ29 system discussed above.
Table 2.
Summary of point mutations in phage λ gpA.
| Residue change | Putative Location | Maximal velocity a | Force-dependence a | Processivity a |
|---|---|---|---|---|
| Y46F | Q-motif | Reduced 40% | Steep | Reduced >10× |
| K84A | Walker A | Reduced 40% | Wildtype | Wildtype |
| T194M | Loop-helix-loopb | Reduced 8× | Wildtype | Wildtype |
| T194V | Loop-helix-loopb | Wildtype | Wildtype | Wildtype |
| G212S | C-motif | Reduced 3× | not yet measured | Reduced 6× |
Measured relative to wildtype gpA.
Modeling suggests this Loop-helix-loop may position the Walker B and C-motif in the ATPase center.
Figure 5.
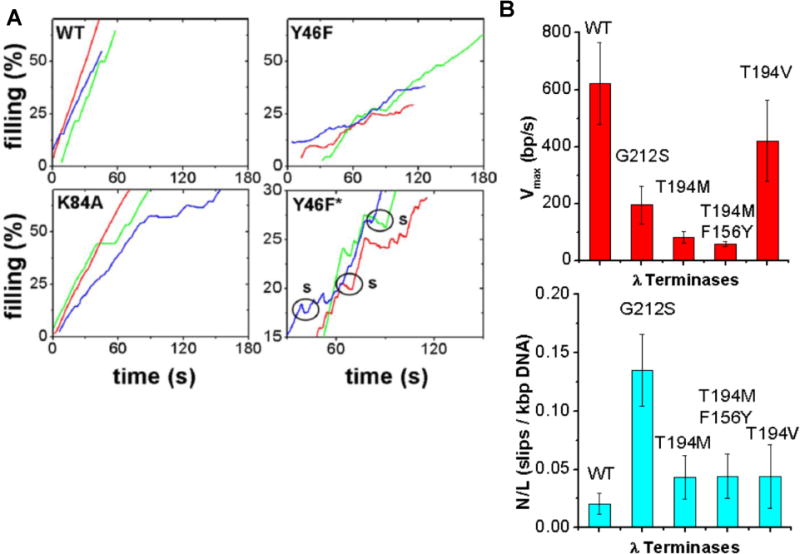
Effects on phage λ packaging dynamics of mutations altering the gpA large terminase subunit of the λ packaging motor. A) Representative recordings of length of DNA packaged (expressed as % of wildtype genome length) vs. time for wildtype (WT), Y46F mutants (residue change in the putative Q motif), and K84A mutants (residue change adjacent the putative Walker A motif). The panel labeled “Y46F*” is a zoomed view of the Y46F data illustrating frequent motor slipping events (labeled “s”) observed with this mutant. B) Bar charts comparing average motor velocity (top chart) and slipping frequency (bottom chart) for WT, G212S mutants (change in the putative C-motif), T194M mutants (change in a structurally conserved loop-helix-loop), T194M/F156Y double mutants, and T194V pseudo-revertants.
Based on sequence comparisons with T4 gp17 and RNA helicases, gpA was predicted to have an adenine-binding “Q-motif” at 46-YQ-47 involved in mechanochemical coupling (Draper and Rao 2007, Mitchell, et al 2002). This motif, located 17 residues upstream of the Walker A motif, was recently discovered in SF2 RNA helicases and observed to contain highly conserved aromatic residues proposed to aid in hydrophobic stacking interactions with adenine. Experiments with helicase mutants indicate that the Q-motif plays a role in regulating nucleic acid affinity and conformational changes driven by nucleotide and ATP hydrolysis (Cordin, et al 2004). Consistent with those findings, optical tweezers studies of λ terminase showed that mutation Y46F in its putative Q-motif decreased motor velocity 40% and increased the frequency of motor slipping during DNA translocation by >10-fold (Figure 5A) (Tsay, et al 2009). This alteration in function is in sharp contrast to that observed with Y46, which also exhibited a 40% reduction in velocity but no significant change in motor slipping. These findings support the hypothesis that viral DNA packaging motors contain an adenine-binding Q-motif motif that regulates substrate affinity, analogous to that found in RNA helicases. Such a feature qualitatively fits with the proposed translocation models for ϕ29 and T4. In addition, optical tweezers measurements revealed that change Y46F caused motor velocity to decrease more steeply with increasing load force relative to wildtype, suggesting that the Q-motif can also regulate motor power.
gpA is also hypothesized to have a “C-motif” involved in coupling at 212-GST-214 (Draper and Rao 2007). In helicases, C-motif residues hydrogen bond with the γ-PO4 of ATP and the DNA. Mutants generally retain ATPase and DNA binding activities, but fail to translocate, due to loss of coupling between the two activities (Banroques, et al 2010). In T4 certain mutants with changes in the putative C-motif were shown to be able to hydrolyze one ATP but not turn over (Draper and Rao 2007). In λ, optical tweezers measurements found that change G212S in this motif caused a 3-fold decrease in velocity and a 6-fold reduction in processivity, again consistent with a coupling defect (Figure 5B).
Genetic screening experiments also revealed several packaging defective mutants in a region of λ gpA outside any known functional motifs (Duffy and Feiss 2002). Change G191S resulted in no detectable packaging activity while change T194M caused an 8-fold reduction in motor velocity without substantially changing motor processivity or force dependence (Figure 5B) (Tsay, et al 2010). Structural modeling of gpA based on homology with T4 gp17 indicates that T194 is part of a loop-helix-loop region that connects the β4 (Walker B) and β5 (C-motif) strands of the nucleotide-binding domain. This region has high structural similarity with analogous regions in T4 gp17, chromosome transport motor FtsK, and MjDEAD RNA helicase. Underscoring the importance of this region, change D584A in the proximal (predicted) loop segment of the prokaryotic SpoIIIE chromosome segregation motor (a close homolog of FtsK) was reported to reduce DNA translocation rate significantly (~3-fold) (Burton, et al 2007). Together, these findings suggest the presence of a conserved structural region between the Walker B motif and C-motif that may be part of a mechanism that governs motor velocity and processivity in several different types of nucleic acid translocases. Variations in this region may explain how the ϕ29, λ, and T4 motors evolved different packaging motor velocities that scale with viral genome size.
4.3. Subunit coordination and step size
Though the optical trap measurements of Chemla et al. provided a framework for understanding the coupling of the chemical and mechanical reactions in each gp16 subunit of the ϕ29 motor, they did not fully address how the subunits coordinate their actions to package DNA. On one hand, the observation of Michaelis-Menten kinetics in this work suggests that there is no cooperativity in ATP binding in the pentameric ring. At the same time, the measurements of stalls with non-hydrolyzable ATP analogs indicate that the whole machinery is locked when one catalytic site is unable to complete its reaction, and the subunits cannot act independently. According to the model proposed by Sun et al. based on structural data on the T4 large terminase, coordination between ATPases may be imposed by the DNA itself. Geometrical constraints based on the helical structure of B-DNA (10.5 bp per turn), the pentameric ring conformation of the packaging ATPases, and the putative 2-bp step size would dictate that each subunit must pass off DNA to its neighbor in ordinal sequence in order to remain in register with the molecule (Sun, et al 2008, Chemla, et al 2005). The alternative would be that coordination results from an underlying inherent communication between the subunits, independent of DNA geometry. Recent optical trap measurements have begun to address these issues in the ϕ29 system.
4.3.1. High-resolution optical trap measurements of ϕ29 packaging
The development of high-resolution optical tweezers recently enabled the direct observation of viral DNA packaging step by step. Whereas the optical traps used in the packaging experiments described above were not sufficiently sensitive to monitor the stepwise encapsidation of DNA, these new instruments are capable of detecting motion at the scale of a single base pair (Abbondanzieri, et al 2005, Moffitt, et al 2006, Carter, et al 2009) (reviewed in (Moffitt, et al 2008, Chemla 2010)). With this technical breakthrough, Moffitt et al. (Moffitt, et al 2009) were able to detect directly, for the first time, the fundamental stepping motion of the ϕ29 packaging motor, revealing not only its step size but also its inter-subunit coordination.
In a first set of experiments, packaging was monitored with the high-resolution optical traps at low force and at varying ATP concentrations. Contrary to expectations, Moffitt et al. observed that packaging occurred in large 10-bp “bursts” in which the DNA was translocated rapidly, separated by “dwells”, regions in which the motor remained at one position on the DNA (Figure 6A). Further insight into this surprising result came from an analysis of the kinetics of the two classes of events. As ATP concentration was increased, the same 10-bp burst-dwell structure was observed. The duration of the dwells became shorter, while that of the bursts remained constant, indicating that ATP binding occurred solely during the dwells. Moreover, the dwell durations varied with ATP in a manner consistent with the Michaelis-Menten kinetics observed in the previous optical trap work. By measuring the durations of hundreds of dwells at each ATP concentration, accurate distributions of dwell times under each condition were compiled. If a single rate-limiting kinetic step governed the dwells, then an exponential distribution would have been expected. However, Moffitt et al. instead observed peaked distributions (Figure 6B), better described by a convolution of multiple exponentials, consistent with multiple rate-limiting kinetic events during each dwell period. This key observation, in tandem with the ATP dependence of the dwell durations, led to the conclusion that several ATP molecules must bind to the motor during each dwell prior to translocation in 10-bp bursts.
Figure 6.
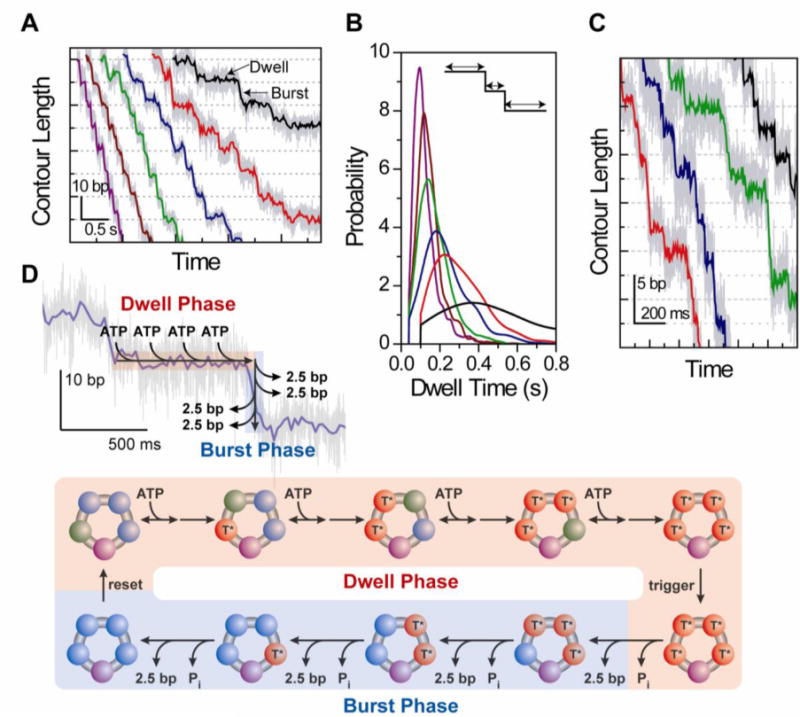
High-resolution measurement of φ29 step size. A) Burst-dwell behavior. At low tensions (~8 pN) packaging was observed to occur in large 10-bp “bursts” separated by flat “dwells”. The behavior was seen across the full range of ATP, from 10 μM (black data) to 500 μM (purple). B) Dwell time distributions. The duration of each dwell in the stepping traces was measured and used to compile a probability distribution at each ATP concentration (same color code as A). The distributions were peaked rather than exponential, indicating that multiple rate-limiting kinetic steps occurred during the dwells. C). Sub-step size. At higher tensions (~40 pN) and at 250 μM ATP, conditions under which DNA translocation is rate-limiting, the 10-bp bursts were observed to consist of four 2.5-bp substeps. D) Model of intersubunit coordination in φ29. Packaging occurs via a biphasic mechanism in which the gp16 ring loads multiple (most likely 4) ATPs during “dwells”, and translocates the DNA in 4 rapid and successive 2.5-bp sub-steps during the 10-bp “bursts”.
These studies indicate a wholly unexpected mechanism of coordination in the pentameric gp16 ring. Rather than each subunit binding ATP, hydrolyzing it, and translocating DNA in succession, the high-resolution kinetics showed that the motor must wait to load multiple ATPs before translocation can occur. A question in this model is how the 10-bp bursts are generated by the motor. In the first experiment of Moffitt et al., the finite duration of the bursts already provided a hint that these were themselves composite events made up of multiple sub-steps. At the low forces in these measurements, however, most bursts were too rapid to observe such sub-structure. Thus, the authors performed a second set of experiments exerting higher forces with the optical traps to slow down each DNA translocation step. These measurements indeed revealed sub-steps within the 10-bp bursts. Unexpectedly, however, the sub-steps were a non-integer number of base pairs in size, 2.5 bp, as shown in Figure 6C. An analysis of the durations of the “micro-dwells” preceding each 2.5-bp sub-step revealed that three in four were fast events and one in four was slow, corresponding to the long dwells taken every 10-bp as observed at low forces.
The surprising picture emerging from these experiments is that packaging in ϕ29 occurs via a biphasic mechanism in which ATP binding and DNA translocation are temporally segregated, as depicted in Figure 6D. During a dwell, multiple ATPs load to the gp16 ring. Based on the observation of multiple rate-limiting kinetic events during the dwell at low forces and the four 2.5-bp sub-steps at high forces, the likeliest scenario is that four ATPs bind the motor during this phase. Then, following a dwell, the motor translocates DNA in a burst of four rapid and successive 2.5-bp sub-steps totaling 10 bp. At first glance, the requirement that multiple ATPs bind the motor prior to translocation may appear at odds with the observation of Michaelis-Menten kinetics, which would indicate a lack of cooperativity between subunits. However, as argued by Moffitt et al., the two results can be reconciled if and only if the binding of each ATP involves an irreversible step that commits it to the remainder of the cycle. This “commitment” step is consistent with the large free energy change upon ATP binding in the mechanochemical model of Chemla et al., as discussed above. Thus, each gp16 subunit must bind ATP in succession. Since DNA is not translocated during the dwell, this time-ordered binding of ATP around the ring must indicate some level of communication between subunits, rather than a coordination imposed by the helical geometry of the DNA. The simplest model is that ATP binding at one subunit allosterically activates the catalytic site of its neighboring subunit, leading to successive binding around the ring.
4.3.2. Implications of high-resolution optical trap studies
Two key questions remain regarding this mechanism. The first is how to explain the measurement of a 2.5-bp step size. Assuming 4 ATPs are consumed during each burst-dwell cycle, a coupling ratio of 2.5 bp/ATP is obtained, which differs slightly from the value of ~2 bp/ATP measured in bulk. However, the bulk measurements of ATP hydrolysis are often difficult to make precise because of basal ATPase activity of nonfunctional complexes in the ensemble, or because of futile hydrolysis by active proteins. If hydrolysis of ATP does not necessarily lead to translocation, then the coupling ratio need not equal the step size of the motor. As discussed above single-molecule experiments show that packaging is interrupted by slips in which DNA spills out of the capsid and is repackaged by the motor. These events lead to futile ATP hydrolysis and an underestimate of the coupling stoichiometry. Initiation of packaging may also requires ATP hydrolysis. This overhead of ATP consumption not directly tied to translocation could also lead to an underestimate of the coupling ratio. Thus the discrepancy between 2.5 and 2 bp estimated step sizes is likely consistent given these systematic errors expected for the ensemble measurements. An interesting implication of a 2.5-bp step size is that it indicates that the motor is highly efficient. Assuming this step size persists at the highest force believed to be exerted by the motor, 110 pN (a caveat being that Moffitt et al. measured this step size out to only ~45 pN), then over 70% of the energy available from ATP hydrolysis can be converted into mechanical work driving DNA translocation. This would make ϕ29 one of the most efficient molecular machines known (Bustamante, et al 2004). Whether the remaining energy available from ATP hydrolysis is simply dissipated as heat, or utilized to drive conformational changes unrelated to DNA translocation remains unclear.
A second question is why the motor must wait to bind the requisite number of ATPs (likely 4) before translocation can occur. A related issue is why only four of the five ATPases in the ring participate in each mechanochemical cycle. It is currently not understood why there is a “special” subunit in the pentameric ring that does not translocate DNA and whether it is the same subunit every 10-bp cycle or if its position rotates around the ring after each cycle. One attractive model is that after every cycle, the last subunit to step remains engaged with the DNA to keep it from slipping out of the capsid while the others begin loading with ATP. This subunit would most likely have to be bound to a nucleotide since the apo state is known to have a weak affinity for DNA, leaving only four available sites to bind ATP. A consequence of this model is that the identity of the special subunit would rotate around the ring after every each 10-bp burst. What event triggers the switch from ATP-loading dwell phase to burst phase is not explained by this mechanism. One possibility is that once all available sites are occupied, ATP hydrolysis can occur spontaneously in one subunit, triggering a concerted wave of hydrolysis and translocation through the ring. Another is that the binding of each ATP induces strain in the ring, and that a critical threshold is attained upon binding four molecules that triggers a conformational switch in the motor, leading to the translocation burst. An alternative mechanism proposed by Moffitt et al. to explain four translocation steps in a pentameric ring is that the ATPase ring could be broken at one subunit-subunit interface. Though “open” ATPase ring structures have been observed in ringed ATPase motors notably in ϕ8 bacteriophage (Lisal, et al 2005), there is no structural evidence to date for an active open conformation. Thus, we do not discuss this class of model presently.
The model described above implies a symmetry mismatch between the DNA helical pitch and the translocation by the ATPase ring. While B-form DNA makes one full turn per 10.5 bp, the motor translocates DNA by 10 bp using only 4 of 5 subunits, i.e. four-fifths of a complete turn around the ring. Thus, the motor subunits and DNA would have to rotate relative to each other to remain in register. Preliminary single-molecule measurements (Craig Hetherington, personal communication) indicate that DNA indeed rotates during packaging in an under-winding direction. Encouragingly, this is so far consistent with the mechanism described above, in which one special subunit remains engaged with the DNA after the translocation burst and its identity rotates in the ring after every mechanochemical cycle (in a counterclockwise direction around the ring, as viewed outward from inside the capsid). Further measurements will be needed to confirm this aspect of the model.
4.4. Motor-DNA interaction
The apparent 2.5-bp step size of the ϕ29 motor has important implications regarding the interaction between the motor and DNA. Prior to the high-resolution studies, models of packaging have explicitly or implicitly assumed the motor translocates DNA through interactions with phosphates located every base pair along the DNA backbone. The observed 2.5-bp step size, however, suggests that a different type of motor-DNA contact must be made. To better understand the nature of the motor-DNA interaction in ϕ29, Aathavan et al. (Aathavan, et al 2009) performed extensive optical trap measurements challenging the ability of the motor to package modified substrates. To test the electrostatic nature of the motor-DNA contact, the authors incorporated methylphosphonate (i.e. neutral) DNA of varying lengths on each or both DNA strands; to reveal the structural requirements of this interaction they utilized substrates such as abasic DNA, single-stranded DNA, DNA bulges, and even unstructured non-DNA linkers. These modified substrates were integrated within an 8-kb ordinary double-stranded DNA molecule to ensure normal initiation and start to packaging.
Faced with these modified inserts, the ϕ29 motors displayed a uniform pattern of pausing at the modification and either traversing it or dissociating completely (Figure 7A). The probability of traversal depended not only on the type of modification, but also on its length, which strand was modified, and the tension in the substrate. In general, inserts residing in the 5′-3′ strand (as measured along the direction of packaging) decreased traversal, whereas those on the 3′–5′ strand had little effect on packaging progress, indicating that the motor preferentially tracked the 5′-3′ strand. However, surprisingly, the motor displayed a remarkable ability to accommodate most modifications. Provided they were short (10 bp or less), neutral or abasic DNA, bulges, and even unstructured non-DNA linkers were traversed with over 80% probability. How the ϕ29 motor could package a substrate with no resemblance to canonical dsDNA is an intriguing question. One possibility is that the motor is able to “jump” over these barriers by diffusion, while another is that it can actively package through a variety of inserts. To test these possibilities, Aathavan et al. studied the effect of substrate tension on traversal and pause duration. For a diffusive model, they predicted force would stretch the modified substrate, creating a larger barrier for the motor to jump over, leading to a lower traversal probability and longer pauses. While increased tension on the substrate did decrease the traversal probability and increase pause duration, the effect was much less dramatic than that predicted by diffusion alone. The authors also decreased the ATP concentration and observed a decrease in traversal probability and increase in pause duration, favoring an active—as opposed to purely diffusive—mechanism of traversal. Thus, the surprising conclusion is that over short length scales (≤ 10 bp) the motor is able to actively package through many types of substrates and neither phosphate charge nor DNA structure appears essential.
Figure 7.
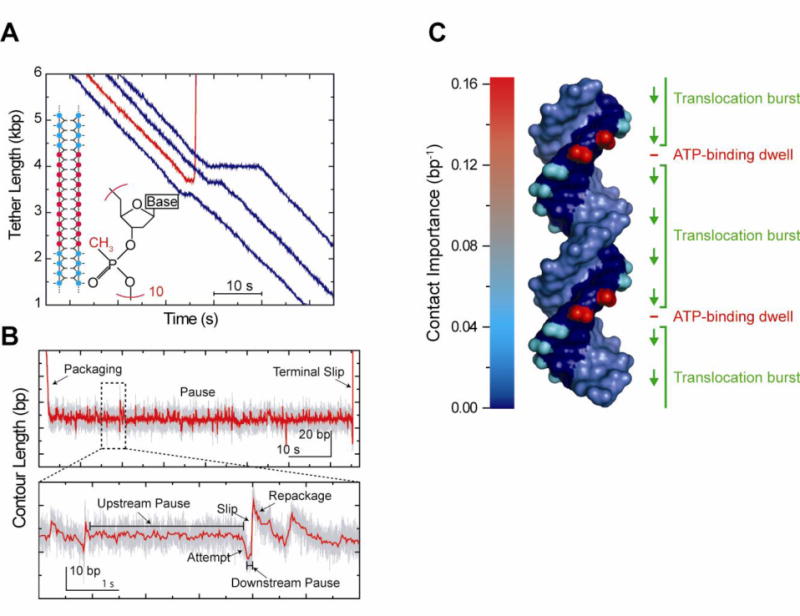
Motor-DNA interactions in φ29. A) Motor behavior at the site of modified DNA. The packaging complexes were challenged with modified DNA inserts of various lengths and compositions (inset; here showing a 10-bp methylphosphonated dsDNA insert), at various tensions (here ~5 pN) and ATP (1 mM). Upon reaching the modified inserts in the course of normal packaging, motors were observed to pause and either traverse the insert (blue traces) or dissociate completely from the DNA (red). Pause durations and the probabilities of traversal were dependent on the insert type and on tension. B) High-resolution measurement of pause and dissociation. Pauses were composite events consisting of long “upstream” pauses, followed by either packaging attempts with short “downstream” pauses, small slips, or terminal dissociation. The two pause types are believed to occur during the dwell and burst phases of the motor, respectively. C) “Heat map” of motor-DNA interactions in φ29. The biphasic coordination mechanism of φ29 dictates that two different types of contacts are made to DNA. During packaging, the motor tracks the 5′-3′strand, making strong ionic contacts to adjacent phosphates (red) during the dwell phase, and making transient, promiscuous contacts along the backbone (cyan) during the translocation burst. (The color map indicates the “contact importance” scale with red highest, blue lowest).
As the insert length is increased, however, this behavior changed remarkably. While double-stranded methyphosphonate DNA stretches 10 bp or shorter were packaged with 80% or better probability, increasing the length by a single base pair to 11 bp dramatically decreased the traversal probability to less than 50%. The shift in behavior between 10 and 11 bp is highly suggestive, because this corresponds to the burst size of the motor, as determined by the high-resolution studies. Thus, Aathavan et al. argued the shift may indicate that contacts made within a burst (i.e. on length scales < 10 bp) may be different than those made during dwells (which occur every 10 bp) in the mechanochemical phases of the motor. To probe this idea further, the authors carried out high-resolution optical trap experiments with a 10-bp methylphosphonate dsDNA insert. These measurements revealed that the pauses at the inserts were composite events with multiple packaging attempts, sub-pauses, and temporary disengagements from DNA or slips (Figure 7B). They were able to identify two distinct types of sub-pauses termed “upstream” and “downstream” depending on whether they occurred at longer or shorter tether lengths, respectively. Upstream pauses were long-lasting (1s), occurred exclusively before slips or packaging attempts, and took place at the same position (within 1 bp) on the substrate, suggesting that they occurred at the boundary between normal and neutral DNA. Downstream pauses, on the other hand, were shorter in duration (80 ms), and occurred after packaging attempts and at a broader set of positions. Based on this observation and other evidence, Aathavan et al. proposed that upstream pauses may correspond to dwells, and downstream pauses to the micro-dwells during bursts. According to this identification, dwells were more affected by neutral DNA, lasting much longer than their counterparts in normal DNA, compared to the bursts. The authors also predicted that, if this model were correct, phages would be able to bypass neutral inserts shorter than one 10-bp burst as long as no dwells occurred within the modified DNA. Indeed, when phages were challenged with 5-bp methylphosphonate DNA insert, 50% of the phages traversed without exhibiting long pauses. This fraction is consistent with the phage arriving at the insert with an arbitrary “phase”, such that half are able to burst over the modified 5 bp and half are required to dwell within this region, leading to a pause.
The data of Aathavan et al. paint a picture of motor-DNA interaction that is in concert with the biphasic mechanism proposed in Moffitt et al. Two types of contacts are made with the 5′–3′ DNA strand during packaging, corresponding to the two mechanochemical phases of the motor. During dwells, phosphate contacts are made every 10 bp. Since the traversal probability decreases at 11 bp, not 10 bp, the authors argued that motor may be interacting with adjacent phosphates on the DNA. These contacts are important; their absence stalls the motor, suggesting that they play a sensory role in the mechanochemical coordination of the motor. During the bursts, phosphate contacts are not essential, pointing to weaker “promiscuous” interactions with the substrate. These non-specific contacts may explain why a non-integer base pair step size is observed on double-stranded DNA, and how the motor can actively traverse short substrates structurally dissimilar to canonical B-form DNA. This model of motor-DNA interaction is summarized in Figure 7C. Based on current understanding of the coordination of the motor subunits, strong ionic contacts might be required during dwells, when the motor must wait for the requisite number of ATP molecules to load. On the other hand weak, transient contacts may be preferable during bursts to encapsidate DNA quickly. At present there is no model for two different motor-DNA contacts in the available T4 gp17 structures, though it is possible that a single molecular lever could be responsible for both electrostatic and non-ionic contacts. Crystal structures of ϕ29 gp16 and λ gpA would likely provide additional insight into this mechanism.
5. Completion of packaging
The previous sections have reviewed studies probing the function of the packaging motor at low capsid filling where the forces resisting DNA confinement are negligible. To complete packaging, however, the whole viral genome must be translocated into the capsid, resulting in very high densities of DNA. Remarkably, the DNA is confined at ultra-high densities of ~0.5 g/ml, meaning that roughly half the volume of the capsid is filled by DNA and half by closely associated water molecules and ions. This tight packing of DNA is a highly unfavorable conformation from the point of view of DNA bending energy, electrostatic self-repulsion, and conformational entropy loss. In addition to characterizing motor function, single-molecule studies have enabled the characterization of the forces resisting the tight confinement of the DNA in the capsid (Smith, et al 2001, Rickgauer, et al 2008, Fuller, et al 2007b). While many single-molecule studies involve isolating motor complexes away from their natural contexts (e.g., isolation of single myosin molecules from muscle fibers), the packaging motor dynamics can be measured during the natural biological task of translocating the viral DNA into the viral procapsid.
5.1. Internal force resisting DNA confinement in phage ϕ29
In solution, ϕ29 DNA forms a random coil of radius ~250 nm (Robertson, et al 2006). This DNA must be compressed by approximately 1000-fold in volume to fit inside a ϕ29 capsid of average radius ~24 nm (Morais, et al 2005). The persistence length of DNA is 50 nm, meaning that significant energy is required to bend the DNA to fit in the capsid. DNA in solution is also highly charged, carrying one negative charge per phosphate group. While this charge is predicted to be 80–90% screened by mono- and divalent cations such as Na+, Mg2+, the residual charge would still create large electrostatic repulsion forces when the DNA is packaged (Riemer and Bloomfield 1978).
Thus, the motor must do mechanical work to translocate DNA against “internal forces” resisting DNA confinement. An approach for inferring these internal forces is based on the observation that the motor velocity steadily decreases with increasing amount of DNA packaged, slowing to nearly a stop when the full genome length is packaged (Figure 8A–C) (Smith, et al 2001, Rickgauer, et al 2008). Since measurements at low capsid filling showed that the motor velocity decreases with an increasing applied load force, the progressive slowing with increasing capsid filling can be interpreted as arising from increasing load due to the internal force resisting DNA packaging. (Incidentally, accounting for this decrease in velocity with capsid filling means that the total time to package the full ϕ29 genome length is ~2–3 minutes on average). Careful measurements of the velocity vs. force relationship at low capsid filling and the velocity vs. filling relationship with low applied load allow one to deduce the internal force vs. filling. Measurements with low (<10%) capsid filling were also made using the initiation method described above and measurements with low force (<0.15 pN on average) were made using a ramped DNA stretching technique (Rickgauer, et al 2008) (not shown, see reference). Measurements made in this manner were more accurate than, while revealing similar trends as, earlier measurements (Smith, et al 2001). The earlier work used complexes with higher initial filling (>30%), a 5 pN force clamp, a less-accurate micropipette-trap system rather than a dual trap system, and DNA with the gp3 terminal protein (which appears to cause DNA looping, as described in the initiation section).
Figure 8.
Motor velocity and internal forces resisting DNA packaging. A) φ29 motor velocity vs. % of genome packaged (measured with 5 pN force feedback) in various ionic conditions (see legend). B) φ29 motor velocity vs. applied load force (measured at low capsid filling with no feedback). C) φ29 internal force vs. % of genome packaged deduced from the measurements in panels A&B. D) Internal force vs. % of genome packaged measured for phage λ in the absence of gpD (a putative capsid stabilizing protein). Proposed capsid expansion and capsid rupture events are marked by the dashed vertical lines.
In the standard ϕ29 in vitro packaging buffer (25 mM Tris-HCl buffer (pH 7.8), 50 mM NaCl, 5 mM MgCl2, 0.5 mM ATP) the internal force was found to rise to ~7 pN at one-third filling (~6% of the maximum), ~14 pN at half filling (~12% of the maximum), and increase sharply beyond 70 pN during the final stages of filling (Rickgauer, et al 2008). Extrapolation of the velocity vs. load data to the low velocities observed in the final stages of packaging yielded an estimate of 110 ± 9 pN for the maximum internal force. The presence of such high internal forces resisting DNA confinement rationalizes the observed capability of the viral packaging motor to exert very high forces, among the highest reported any molecular motors. The forces that build during packaging also likely play a critical biological role to drive DNA ejection when the virus infects a host cell (Smith, et al 2001, Kindt, et al 2001, Evilevitch, et al 2003).
5.2. Effect of Ionic Screening on DNA packaging forces
The effect of varying ionic conditions on ϕ29 DNA packaging has also been studied by carrying out sets of packaging measurements with buffers containing different concentrations of Na+, Mg2+, and CoHex3+ (Fuller, et al 2007b) (Figure 8A–C). When Na+ is the dominant ion screening the DNA, the effective DNA charge is predicted to be reduced by ~80%. Mg2+ is predicted to screen more strongly, reducing DNA charge by ~90% (Manning 1978). A smaller amount of a trivalent cation such as CoHex3+ can screen more effectively than Mg2+, and beyond a critical concentration trivalent cations or ions with higher valence can induce DNA condensation (Baumann, et al 2000). In the work of Fuller et al., measurements were carried out using a 5 pN force clamp to measure velocity vs. filling, and a fixed trap position mode to measure velocity vs. force (Fuller, et al 2007b). While this is a slightly less accurate method than the lower-force, lower-filling method described in the previous section, resulting in slightly lower estimates for internal force, it was adequate for discerning the trends of how packaging forces vary with ionic condition.
Under each ionic condition the motor velocity decreased monotonically with capsid filling, and in all conditions a sharp drop in velocity was observed above ~50% filling (Figure 8A). The velocity converged towards zero at ~100% genome packaging in all cases except with a high Na+ buffer, where the velocity approached zero at ~90% filling, indicating that packaging would not proceed to completion in this condition (of highest net DNA charge). The internal forces resisting DNA confinement, deduced as described in the previous section, followed the expected trend of decreasing with increasing ionic screening (Figure 8C). Reduced forces similar to those with high Mg2+ were also observed with 1 mM CoHex3+ added to the standard packaging buffer (less than that needed to induce DNA condensation), showing that trivalent cations can screen the DNA very effectively, even when in competition with higher concentrations of mono- and divalent cations. A plateau of near-zero internal force was observed during the first one-third of packaging with the high Mg2+ and CoHex3+ buffers, but not with the lower-screening buffers. These measurements confirm the prediction that electrostatic self-repulsion is the dominant force resisting packaging (Riemer and Bloomfield 1978, Tzlil, et al 2003, Purohit, et al 2005), although agreement with theoretical predictions is not perfect, as discussed below.
5.3. Theoretical models for DNA packaging and ejection forces
Spurred by the experimental studies described above and complementary measurements of viral DNA ejection forces (Evilevitch, et al 2004) many investigators recently worked on theoretical models of the forces governing DNA confinement. Both analytical continuum-elastic models and dynamic simulations approaches were used. Analytical models predict specific packed DNA conformations that would minimize the free energy change, while the simulations predict DNA conformations resulting from stochastic dynamics. Building on earlier work (Riemer and Bloomfield 1978, Odijk 1998), Gelbart and coworkers used Brownian dynamics simulations and analytical theory to predict the forces resisting DNA compaction (Kindt, et al 2001, Tzlil, et al 2003). Their calculations suggest that the DNA conformation would transition through toroidal and spool-like geometries with increasing length packaged, resulting in forces rising to tens of piconewtons, in the same range as inferred experimentally for ϕ29 with optical tweezers measurements (and for λ as well, as discussed further below). The Brownian dynamics simulations assumed a harmonic DNA bending potential and an empirical interaction potential deduced from X-ray diffraction data on the condensation of DNA by osmotic pressure, which reveal short-range interaxial DNA separations of 2–3 nm (Rau and Parsegian 1992). Assuming a purely repulsive potential (as with modest screening by Na+ and Mg2+), a disordered structure was observed in simulations, but it was speculated that this might equilibrate to a spool-like structure, as seen in simulations with an attractive-repulsive potential on longer time scales (Kindt, et al 2001). Phillips and coworkers further developed coaxial inverse spool models and made internal force predictions for various ionic conditions and various capsid shapes and sizes (Purohit, et al 2005, Purohit, et al 2003). These models agreed well with measurements of phage λ DNA ejection forces (Evilevitch, et al 2003, Evilevitch, et al 2005, Grayson, et al 2006), and the experimentally inferred packaging force in ϕ29 with Na+ screening were within a factor of two of those predicted (2x higher than predicted). However, the experimentally inferred packaging forces with Mg2+ as the dominant screening were ~6× higher than predicted (Fuller, et al 2007b), a result that so far remains unexplained.
Equilibrium thermodynamic simulations also predicted a spool structure (Marenduzzo and Micheletti 2003), but stochastic rotation dynamics simulations predicted a more random conformation in the latter stages of packaging (Ali, et al 2006). Harvey and coworkers carried out an extensive series of molecular dynamics (MD) simulations (Harvey, et al 2009). They predicted concentric spooling with spherical capsids and folded toroidal conformations with prolate capsids, rather than coaxial spools. They also predicted, contrary to other investigators, that entropy plays a major role in the free energy of packaging. Other dynamic simulations also found evidence for folded toroids (Spakowitz and Wang 2005, Forrey and Muthukumar 2006) and evidence for non-equilibrium effects and molecular heterogeneity (i.e. different stochastic conformational dynamics of different individual packaging complexes, an effect observed in the optical tweezers experiments as well).
Several factors may potentially contribute to the discrepancy between the experimentally inferred ϕ29 packaging forces and theoretical predictions. First, ϕ29 DNA may not be uniformly packaged as an axially-symmetric coaxial spool, as assumed in the analytical theoretical models. Notably, disordered structures are predicted to result in higher internal forces. Cryo-EM reconstructions of T7 phages suggest that the fully packed DNA is at least partially organized in a coaxial spool, although the degree of order towards the interior of the capsid remains unclear (Cerritelli, et al 1997). EM images of partially filled ϕ29 capsids, on the other hand, surprisingly revealed DNA mass density uniformly distributed throughout the capsid rather than progressive outside-in layering (Comolli, et al 2008). Second, the assumption that internal forces have the same effect on motor velocity as externally applied forces may not hold exactly, leading to inaccuracy in determining the former from measurements of motor velocity vs. capsid filling. Third, energy dissipation due to friction may occur during packaging, such that the measured work done is higher than the theoretically predicted gain in potential energy of the packed DNA. However, observations of rapid slipping during ϕ29 and λ packaging and of rapid DNA ejection in the T5 and λ phages suggest that friction is negligible (Smith, et al 2001, Mangenot, et al 2005, Grayson, et al 2007). Fourth, as suggested by some simulations, the dynamic conformation adopted by the DNA during rapid packaging may not be exactly equal to the equilibrated free-energy minimum conformation. Fifth, uncertainty in the exact interior volumes of procapsids accessible to packed DNA assessed from cryo-EM data could lead to uncertainty in the theoretical predictions. Finally, DNA-DNA interaction potentials assumed in theoretical calculations may not be universally applicable in describing the DNA packaged in all types of phages in all ionic environments. The potentials used in many models were derived empirically from experiments in which straight DNA segments were condensed into hexagonally packed bundles in solution by applied osmotic pressure and probed by X-ray diffraction (Rau and Parsegian 1992).
5.4. Internal force, procapsid expansion, and procapsid rupture in phage λ
Using the approaches developed for ϕ29, Fuller et al. measured internal forces in phage λ. The average λ packaging rate decreased substantially with packaging, going from 580 bp/s to 240 bp/s as the procapsid filled from 20% to 90% of the genome length (Fuller, et al 2007a). Since the motor velocity decreases with increasing load, this decrease in velocity with capsid filling is indicative of a building internal force resisting DNA confinement in the procapsid, analogous to that observed with ϕ29. Extrapolation of the decreasing packaging rate trend versus length of DNA packaged to 100% filling suggests that the total time required to package the λ genome is 2–3 minutes, about the same as for ϕ29. In the λ system, however, the force reached 25 ± 6 pN with 90% of the genome length packaged (Figure 8D), which is notably two to three-fold lower than that found for ϕ29 under a similar ionic condition. It is especially noteworthy, however, that this internal force figure is in excellent agreement with phage λ DNA ejection forces inferred by osmotic pressure experiments (Evilevitch, et al 2004) and DNA confinement forces predicted by theoretical calculations (Tzlil, et al 2003, Purohit, et al 2003). That multiple independent approaches to inferring internal force quantitatively agree strongly support the validity of these force determinations and the proposal that the internal force that builds during packaging is the primary force driving DNA ejection when the virus infects a host cell.
An intriguing behavior observed in the λ packaging dynamics, not seen in ϕ29, was a temporary dip in the packaging rate when ~30% of the λ genome length was packaged (Fuller, et al 2007a). Electron microscopy studies showed that λ procapsids undergo a dramatic conformational change that roughly doubles the internal volume of the capsid at some point during packaging (Dokland and Murialdo 1993). The dip observed in motor velocity corresponds to a 4 pN increase then decrease in the internal force (Figure 8D). The presence of this dip strongly supports the long-standing hypothesis that internal force builds in the unexpanded procapsid and triggers expansion, which temporarily reduces the internal force due to a reduction in DNA confinement. As packaging proceeds the motor velocity again slows significantly, consistent with subsequent re-buildup of internal force in the expanded procapsid. Notably, the location and magnitude of the dip was found to be variable in individual datasets, suggesting that stochastic dynamics are involved and individual procapsids expand at different internal force and filling levels.
In measurements with phage λ a number of events were unexpectedly observed where greater than 100% of the genome length of DNA was translocated (Fuller, et al 2007a). In all of these events, after slowing dramatically, the motor abruptly accelerated to full speed, and this acceleration occurred at a distinct point between 90 and 100% packaging (Figure 8D). Translocation then continued up to ~105–146% of the genome length, significantly more than expected for the assembly of a viable phage. These events strongly suggest that the building internal force at 90–100% of genome packaging causes rupture of the expanded procapsid, which releases the confined DNA, presumably through cracks in the capsid shell, and relieves the opposing load on the motor. Such rupture likely occurred because an accessory capsid protein, gpD, which binds the procapsid during packaging in vivo, was not included in the in vitro optical tweezers experiments. These findings provide strong support for the long-held notion (Sternberg and Weisberg 1977, Perucchetti, et al 1988, Yang, et al 2008) that gpD binding stabilizes the procapsid against a building internal force that accompanies DNA packaging. Recent higher resolution cryo-EM structures of λ capsids reveal the structural mechanism for this stabilization (Lander, et al 2008).
6. Conclusions and future prospects
As described above, single-molecule approaches developed over the last decade, particularly the optical tweezers method, have led to many advances in our understanding of the detailed biophysics of DNA packaging. These single-molecule studies strongly complement traditional biochemical studies, in vivo studies, and structural studies. Experiments have informed us both about the function and mechanisms of viral molecular motor complexes, and on the nature of the motor’s task, which is to overcome the large forces resisting dense DNA confinement. Studies of three different virus systems, bacteriophage ϕ29, λ, and T4, have revealed some universal properties, such as very high force generation compared with other known motors, and some differences, such as in motor velocities, capsid expansion effects, and internal forces. High-resolution optical tweezers measurements have revealed a surprising burst-dwell stepping behavior for the ϕ29 motor, a novel mode of operation that has never before been seen in any other molecular motor system. In the λ system progress has been made in establishing motor structure-function relationships by identifying several mutants having specific alterations in packaging dynamics. Some progress has also been achieved in using single-molecule fluorescence measurements to characterize packaging and conformational dynamics of the motor and DNA substrate.
While much has been learned from single-molecule studies over the last several years, the findings also raise many new questions that will be the target of continuing future investigation. There is much more work to be done on each of the three virus systems already studied (ϕ29, λ, and T4), and it is also hoped that eventually these methods will be applicable to other virus systems that utilize analogous motor complexes for DNA packaging, such as the medically relevant herpes- and adeno-viruses. Let us review some potential directions to be pursued:
Mechanisms of initiation of packaging
Optical tweezers experiments showed that packaging can initiate within seconds when DNA is brought into close proximity with procapsid-motor complexes (in the ϕ29 and T4 systems) or when DNA-motor complexes are brought into close proximity with procapsids (in the λ system) (Rickgauer, et al 2008, Fuller, et al 2007, Fuller, et al 2007a). Studies suggest that this latter pathway of assembly is the prevalent one in λ in vivo, so it is of interest to investigate whether packaging in ϕ29 and T4 could also initiate via the formation of motor-DNA complexes. In ϕ29 initiation, optical tweezers measurements revealed an unusual DNA structure at the initiation of packaging, with a DNA loop apparently mediated by the gp3 terminal protein (Rickgauer, et al 2006). It is unclear how this looped structure is resolved during packaging, and combined optical tweezers and fluorescence measurements with labeled DNA could shed light on this. Optical tweezers can also be used to directly probe the formation of DNA loops (Gemmen, et al 2006). In the λ system, an accessory protein, gpFI, is essential for efficient viral assembly in vivo and has been implicated in influencing initiation of DNA packaging in vitro (Feiss and Catalano 2005). A possible role for gpFI in post-cleavage steps is suggested by single-molecule packaging experiments, which thus far have lacked gpFI, and demonstrate that many motor-procapsid binding events do not proceed to active translocation while others show a significant delay before translocation starts (Tsay, et al 2009, Tsay, et al 2010). It has been proposed that gpFI may facilitate diffusion of the DNA motor complex over the procapsid surface to the portal, or may coat the procapsid surface to block non-productive DNA-motor-procapsid binding, or may alter the interaction between the motor and DNA binding site (cos site) to initiate translocation (Murialdo 1991). These models can potentially be discerned by comparing initiation dynamics and efficiency with and without gpFI.
High-resolution measurements of motor stepping in the λ and T4 systems
Having uncovered the unusual coordinated 2.5-bp burst-dwell stepping behavior in the ϕ29 system (Moffitt, et al 2009), an area of immediate interest for single-molecule experiments is to investigate whether such behavior is universal among other virus systems. It will hopefully be possible to extend high-resolution optical tweezers measurements to the λ and T4 systems, which do not have significant global sequence homology with the ϕ29 system (although they may have high structural similarity). Such measurements are challenging, however, due to the 4–10× higher packaging rate of these systems (Fuller, et al 2007, Fuller, et al 2007a). Measurement resolution is ultimately limited by inherent Brownian motion of the trapped microspheres, and can only be averaged down by reducing measurement bandwidth. However, the finding that the motors can be slowed by decreasing ATP, increasing load force, decreasing temperature, and/or by appropriate mutations (Smith, et al 2001, Chemla, et al 2005, Tsay, et al 2010) should make it possible to resolve the stepping dynamics of the λ and T4 motors.
Structure-function relationships
Studies of λ motor mutants exhibiting altered packaging dynamics have begun to inform us on the presence of various motifs and regions of the motor responsible for hydrolyzing ATP, converting the released energy into mechanical work, and/or gripping the DNA substrate (Tsay, et al 2009, Tsay, et al 2010). Future studies using site-directed mutagenesis could more systematically map out the specific critical regions and residues. Of particular interest is to further explore the region between the Walker B motif and C-motif, wherein change T194M was found to cause a sharp reduction in motor velocity without affecting processivity, pausing, or force generation (Tsay, et al 2010). Analogous mutational studies could also be extended to the ϕ29 and T4 systems and high-resolution motor stepping measurements with mutants may be very informative. Mutant studies in ϕ29 revealed that deletion of residues of the portal ring, proposed in earlier models to drive DNA translocation, did not strongly affect packaging dynamics (R. Atz, S. Grimes, D.L. Anderson, personal communication). It is thus of interest to study the effects of changes in the ϕ29 gp16 ATPase, which is analogous to the λ gpA large terminase subunit that drives DNA translocation. The T4 system is also particularly attractive for studies of structure-function relationships because the X-ray crystal structure of the large terminase subunit has been determined and the first specific structural model for motor function has been proposed (Sun, et al 2008). Specifically, electrostatic interactions between ion-pairs within two globular sub-domains separated by a flexible hinge are proposed produce force and DNA ratcheting motion. Studies with optical tweezers of the effect of site-directed mutations in these regions on packaging dynamics are underway. An ambitious future direction is to combine optical tweezers measurements of DNA translocation with single-molecule fluorescence measurements to probe conformational changes within the motor and the effects of site-directed mutations. One unsolved question is to understand why motor velocity can dramatically vary in time and for different individual complexes (Fuller, et al 2007). One explanation could be that these motor complexes may have multiple different active conformational states gearing different translocation rates.
Further understanding of forces resisting DNA packaging
Studies of ϕ29 and λ revealed that large internal force resist packaging and depend on ionic screening of the DNA charge (Smith, et al 2001, Rickgauer, et al 2008, Fuller, et al 2007b, Fuller, et al 2007a). Such studies have also not yet been done with bacteriophage T4 because current optical tweezers instruments would have to be reconfigured to attain sufficient separation range to fully stretch the extremely long T4 genome (171 kbp). While studies in ϕ29 were able to follow packaging to completion, λ packaging could not be tracked beyond 90% of the genome length without capsid rupture, presumably due to the lack of the gpD protein putatively needed to stabilize the capsid (Fuller, et al 2007a). Optical tweezers measurements with added gpD would shed further light on the role of this protein. While inferred internal forces were in good agreement with theoretical predictions for λ packaging and measurements of ejection forces (Tzlil, et al 2003, Purohit, et al 2005, Evilevitch, et al 2004), they were not as in good agreement for ϕ29, particularly with Mg2+ as the dominant screening ion. Several issues that could explain these discrepancies remain to be investigated, such as the effect of prohead shape (ϕ29 and T4 have prolate capsids), and whether packaging involve non-equilibrium dynamics and/or dissipative effects that could make the transient force resisting packaging higher than the equilibrated force later driving DNA ejection. Another unexplored issue is the effect on packaging of polyamine ions such as spermidine3+ and spermine4+, which are present in host cells and have higher screening capacity than Mg2+. At high enough concentrations such polyamines are capable of completely neutralizing the DNA charge and inducing spontaneous DNA condensation (Baumann, et al 2000). While such an effect could reduce the internal packing forces, condensation of DNA outside the capsid could interfere with packaging and/or present an “external” load on the motor. In vivo there are also DNA binding proteins of interest to study, both viral and host gene products, as they would likely have to be stripped from the DNA during packaging and likely present another load on the motor that could affect packaging dynamics.
Mechanisms of termination of packaging
While ϕ29 packages a genome that is replicated as a monomer, λ and T4 and many other dsDNA virus motors utilize an endonuclease function to excise a unit length genome from a string of catenated genomes produced by rolling circle replication (Rao and Feiss 2008). The λ motor initiates packaging at a specific cos site in the concatemer, translocates DNA into the capsid, and finally recognizes the downstream cos site, which signals the end of the genome. Arrest of the translocating terminase complex at the terminal cos site requires not only interaction with the cosQ and cosN elements but also activation of a sensor that somehow detects the extent of DNA packaging in the capsid. The existence of such a “headful” sensor mechanism is indicated by experiments that show that λ terminase fails to arrest at an ectopic cos site placed earlier in the DNA substrate such that the site is reached with <80% of the wildtype genome length packaged (Cue and Feiss 1997). Bacteriophage T4 packaging also relies on a headful mechanism independent of a specific termination site.
Several models have been proposed for how an extent-of-packaging sensor may work (Casjens, et al 1992, Lander, et al 2006). One possibility is that the density of packaged DNA is monitored by the motor, perhaps via a domain of the portal that senses internal pressure of DNA confinement inside the capsid. When a critical point is reached, a conformational change of the portal protein activates terminase’s endonuclease activity. A second possibility is that efficient binding of cosQ requires that motor velocity has slowed substantially, as occurs in the latter stages of packaging due to buildup of internal pressure resisting DNA confinement. A third possibility is that the packaging motor can directly measure the energy (or work = resisting force × distance translocated per motor step) required to package DNA and cleavage activity is enabled when a critical energy or resisting force has built up.
Single-molecule measurements should permit these models to be distinguished in λ packaging termination experiments. Specifically, Feiss, Catalano, and Smith have proposed experiments to probe for termination in a DNA substrate with an ectopic cos termination site placed at an early position that would be reached by the motor when only a small amount of DNA has been packaged. Measurements with reduced ATP, reduced temperature, and/or using the slow T194M mutant would examine whether reducing the motor velocity triggers early termination. Measurements with increasing load force applied with optical tweezers would test whether an “energy sensor” triggers early termination. If neither velocity reduction nor force increases triggers termination, this would suggest that the motor has a separate packaging sensor domain which functions independently of the DNA translocating motor domain.
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I, Marenduzzo D, Yeomans JM. Polymer packaging and ejection in viral capsids: Shape matters. Phys Rev Lett. 2006;96:208102. doi: 10.1103/PhysRevLett.96.208102. [DOI] [PubMed] [Google Scholar]
- Ashkin A. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- Banroques J, Doere M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Baumann CG, Bloomfield VA, Smith SB, Bustamante C, Wang MD, Block SM. Stretching of single collapsed DNA molecules. Biophys J. 2000;78:1965–1978. doi: 10.1016/S0006-3495(00)76744-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007;131:1301–1312. doi: 10.1016/j.cell.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr Opin Struct Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- Carter AR, Seol Y, Perkins TT. Precision surface-coupled optical-trapping assay with one-basepair resolution. Biophys J. 2009;96:2926–34. doi: 10.1016/j.bpj.2008.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. New York: Kluwer Academic Plenum Press; 2005. [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng NQ, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Chemla YR. Revealing the base pair stepping dynamics of nucleic acid motor proteins with optical traps. Phys Chem Chem Phys. 2010;12:3080–95. doi: 10.1039/b920234j. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D, Feiss M. Genetic evidence that recognition of cosQ the signal for termination of phage lambda DNA packaging, depends on the extent of head filling. Genetics. 1997;147:7–17. doi: 10.1093/genetics/147.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Duffy C, Feiss M. The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- Evilevitch A, Gober JW, Phillips M, Knobler CM, Gelbart WM. Measurements of DNA lengths remaining in a viral capsid after osmotically suppressed partial ejection. Biophys J. 2005;88:751–756. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evilevitch A, Castelnovo M, Knobler CM, Gelbart WM. Measuring the force ejecting DNA from phage. J Phys Chem B. 2004;108:6838–6843. [Google Scholar]
- Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci U S A. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M, Catalano C. Bacteriophage lambda terminase and the mechanism of viral DNA packaging 2005 [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007a;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci U S A. 2007b;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Gemmen GJ, Rickgauer JP, Dupont A, Millin R, Recouvreux P, Smith DE. A general method for manipulating DNA sequences from any organism with optical tweezers. Nucleic Acids Res. 2006;34:e15. doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit barge force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaussier H, Yang O, Catalano CE. Building a virus from scratch: Assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J Mol Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Gemmen GJ, Millin R, Smith DE. DNA looping by two-site restriction endonucleases: heterogeneous probability distributions for loop size and unbinding force. Nucleic Acids Res. 2006;34:2864–2877. doi: 10.1093/nar/gkl382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman ME, Weisberg RA. Little lambda, who made thee? Microbiol Mol Biol Rev. 2004;68:796–813. doi: 10.1128/MMBR.68.4.796-813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. The effect of genome length on ejection forces in bacteriophage lambda. Virology. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc Natl Acad Sci U S A. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–90. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes S, Jardine PJ, Anderson D. Bacteriophage phi 29 DNA packaging. Adv Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. In vitro packaging of bacteriophage phi 29 DNA restriction fragments and the role of the terminal protein gp3. J Mol Biol. 1989;209:91–100. doi: 10.1016/0022-2836(89)90172-1. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–76. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi 29. J Mol Biol. 1987;197:229–36. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Hang JQ, Tack BF, Feiss M. ATPase center of bacteriophage lambda terminase involved in post-cleavage stages of DNA packaging: Identification of ATP-interactive amino acids. J Mol Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- Harvey SC, Petrov AS, Devkota B, Boz MB. Viral assembly: a molecular modeling perspective. Physical Chemistry Chemical Physics. 2009;11:10553–10564. doi: 10.1039/b912884k. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A. 1978;75:4779–83. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci U S A. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:486–499. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry 1993 [Google Scholar]
- Lisal J, Lam TT, Kainov DE, Emmett MR, Marshall AG, Tuma R. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat Struct Mol Biol. 2005;12:460–466. doi: 10.1038/nsmb927. [DOI] [PubMed] [Google Scholar]
- Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry (N Y) 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- Mangenot S, Hochrein M, Radler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Current Biology. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Marenduzzo D, Micheletti C. Thermodynamics of DNA packaging inside a viral capsid: The role of DNA intrinsic thickness. J Mol Biol. 2003;330:485–492. doi: 10.1016/s0022-2836(03)00584-9. [DOI] [PubMed] [Google Scholar]
- Michaelis J, Muschielok A, Andrecka J, Kugel W, Moffitt JR. DNA based molecular motors. Phys Life Rev. 2009 doi: 10.1016/j.plrev.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Mitchell MS, Rao VB. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology. 2004;321:217–221. doi: 10.1016/j.virol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–52. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- Murialdo H. Bacteriophage lambda DNA maturation and packaging. Annu Rev Biochem. 1991;60:125–153. doi: 10.1146/annurev.bi.60.070191.001013. [DOI] [PubMed] [Google Scholar]
- Myong S, Stevens BC, Ha T. Bridging conformational dynamics and function using single-molecule spectroscopy. Structure. 2006;14:633–43. doi: 10.1016/j.str.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odijk T. Hexagonally packed DNA within bacteriophage T7 stabilized by curvature stress. Biophys J. 1998;75:1223–1227. doi: 10.1016/S0006-3495(98)74041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster G, Wang H. Rotary protein motors. Trends Cell Biol. 2003;13:114–121. doi: 10.1016/s0962-8924(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Oster G, Wang H. Reverse engineering a protein: the mechanochemistry of ATP synthase. Biochim Biophys Acta. 2000;1458:482–510. doi: 10.1016/s0005-2728(00)00096-7. [DOI] [PubMed] [Google Scholar]
- Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Biophys J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucchetti R, Parris W, Becker A, Gold M. Late stages in bacteriophage lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology. 1988;165:103–114. doi: 10.1016/0042-6822(88)90663-0. [DOI] [PubMed] [Google Scholar]
- Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit PK, Kondev J, Phillips R. Mechanics of DNA packaging in viruses. Proc Natl Acad Sci U S A. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The Bacteriophage DNA Packaging Motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Rau DC, Parsegian VA. Direct Measurement of the Intermolecular Forces between Counterion-Condensed Dna Double Helices - Evidence for Long-Range Attractive Hydration Forces. Biophys J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J Mol Biol. 2010a;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010b;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Smith DE. Single-Molecule Studies of DNA Visualization and Manipulation of Individual DNA Molecules with Fluorescence Microscopy and Optical Tweezers. 2008;4 [Google Scholar]
- Rickgauer JP, Fuller DN, Smith DE. DNA as a metrology standard for length and force measurements with optical tweezers. Biophys J. 2006;91:4253–4257. doi: 10.1529/biophysj.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer SC, Bloomfield VA. Packaging of Dna in Bacteriophage Heads - some Considerations on Energetics. Biopolymers. 1978;17:785–794. doi: 10.1002/bip.1978.360170317. [DOI] [PubMed] [Google Scholar]
- Robertson RM, Laib S, Smith DE. Diffusion of isolated DNA molecules: dependence on length and topology. Proc Natl Acad Sci U S A. 2006;103:7310–7314. doi: 10.1073/pnas.0601903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JL, Ali MY, Warshaw DM. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–9. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy J, Feiss M. Initial cos cleavage of bacteriophage lambda concatemers requires proheads and gpFI in vivo. Mol Microbiol. 2004;52:501–513. doi: 10.1111/j.1365-2958.2004.03990.x. [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Optical-Trap Force Transducer that Operates by Direct Measurement of Light Momentum. Methods in Enzymology. 2003;361:134. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Spakowitz AJ, Wang ZG. DNA packaging in bacteriophage: Is twist important? Biophys J. 2005;88:3912–3923. doi: 10.1529/biophysj.104.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J Mol Biol. 1977;117:733–759. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The Structure of the Phage T4 DNA Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage t4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Tinoco I, Jr, Li PT, Bustamante C. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys. 2006;39:325–360. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, del Toro D, Andrews B, Draper B, Rao V, Catalano C, Feiss M, Smith DE. Mutations altering a structurally conserved Loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. Journal of Biological Chemistry in press. 2010 doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc Natl Acad Sci U S A. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Xue Q, Yeung ES. Differences in the chemical reactivity of individual molecules of an enzyme. Nature. 1995;373:681–683. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- Yang Q, Maluf NK, Catalano CE. Packaging of a Unit-Length Viral Genome: The Role of Nucleotides and the gpD Decoration Protein in Stable Nucleocapsid Assembly in Bacteriophage lambda. J Mol Biol. 2008;383:1037–1048. doi: 10.1016/j.jmb.2008.08.063. [DOI] [PubMed] [Google Scholar]


