Abstract
Histone post-translational modifications (PTMs) generate a complex combinatorial code that regulates gene expression and nuclear functions, and whose deregulation has been documented in different types of cancers. Therefore, the availability of relevant culture models that can be manipulated and that retain the epigenetic features of the tissue of origin is absolutely crucial for studying the epigenetic mechanisms underlying cancer and testing epigenetic drugs. In this study, we took advantage of quantitative mass spectrometry to comprehensively profile histone PTMs in patient tumor tissues, primary cultures and cell lines from three representative tumor models, breast cancer, glioblastoma and ovarian cancer, revealing an extensive and systematic rewiring of histone marks in cell culture conditions, which includes a decrease of H3K27me2/me3, H3K79me1/me2 and H3K9ac/K14ac, and an increase of H3K36me1/me2. While some changes occur in short-term primary cultures, most of them are instead time-dependent and appear only in long-term cultures. Remarkably, such changes mostly revert in cell line- and primary cell-derived in vivo xenograft models. Taken together, these results support the use of xenografts as the most representative models of in vivo epigenetic processes, suggesting caution when using cultured cells, in particular cell lines and long-term primary cultures, for epigenetic investigations.
INTRODUCTION
Histones, which represent the protein component of chromatin, are site of many dynamic and reversible post-translational modifications that play a fundamental role in the regulation of the underlying genes (1,2), influencing gene expression and cell fate. Aberrations in the levels of histone PTMs, which is usually a consequence of the deregulation of the enzymes responsible for the deposition and removal of the modifications, known as histone modifying enzymes (HMEs), have been linked with different types of cancer (3). Indeed, anomalous expression, mislocalization and mutations of HMEs have been reported in many different tumors (4–6); likewise, the disruption of normal histone PTMs patterns was identified as a general hallmark of cancer (7) and linked with patient prognosis in various tumor types (8–10). Therefore, studying epigenetic processes -and particularly histone PTMs- in cancer holds great potential for the discovery of biomarkers for patient stratification, as well as of possible epigenetic mechanisms underlying cancer onset and development. Furthermore, because epigenetic changes -unlike genetic ones- are reversible, epigenetic therapies aimed at correcting epigenetic aberrations are emerging as a promising avenue in translational research. A few drugs targeting HMEs are now in clinical use for hematological malignancies, and several more are in clinical trials for the treatment of solid tumors (11). In this scenario, the availability of relevant culture models that can be manipulated and that retain the epigenetic features of the tissue from which they were derived is absolutely crucial for studying epigenetic mechanisms underlying different pathologies, as well as for testing epigenetic drugs and uncovering possible epigenetic biomarkers.
Models to study cancer include cancer cell lines, primary cells and xenografts. Because of their accessibility, ease of growth and manipulation, cell lines are the most widely used model system. However, although they have been extensively used for research purposes, there is still a debate on whether cancer cell lines are truly representative of primary tumors. Many studies suggest that they mirror many, but not all, molecular features of primary tumors (12). Typically, cancer cell lines exhibit oncogene mutations, chromosomal rearrangements, allelic loss and gene amplifications. For instance, in breast cancer, one of the tissue types where culture models have been most extensively characterized, the comparison of genomic features and transcriptional profiles showed high similarity between primary tumors and cell lines, which carried most of the recurrent genomic abnormalities associated with clinical outcome in primary tumors (13). Breast cancer cell lines also displayed similar patterns of DNA copy number alterations, and retained expression patterns that allow distinguishing luminal and basal subtypes, although with some differences compared with primary tumors (12–15). Furthermore, comparison of RNA-sequencing transcriptomes and DNA methylation profiles showed that breast cancer cell lines overall resemble primary tumors, but with some discrepancies (16,17). Important drug targets in breast cancer, such as HER2, ESR1, PGR, EGFR showed a high correlation in tumors and cell lines, while a low correlation was observed in phosphorylated proteins (12). In glioblastoma, cell lines show drastically altered gene expression patterns compared to the original tumor, and they usually do not fully mirror the characteristic invasive growth phenotype of glioblastomas when returned in vivo in xenografts models (18). Another important issue related to cell lines is that they fail to recapitulate the heterogeneity found in tumors (19). Finally, the experimental results obtained with cancer cell lines are relevant in most case only for rapidly proliferating high-grade tumors, from which most cell lines are derived, but not for the lower grade ones.
Primary cell cultures, which are derived directly from patient tumors, can be used as an alternative to cancer cell lines, with two main advantages: they maintain some of the heterogeneity of the original tumor and they are usually kept in culture conditions for shorter times. The gene expression comparison of tumor tissue and early-passage primary cultures showed that primary cultures resemble the malignant tissue much more closely than cell lines in breast cancer and gliomas (20,21). Nevertheless, a recent study based on DNA methylation profiling showed that mouse embryonic fibroblasts undergo epigenetic and transcriptional re-programming much earlier than previously expected, with a global 5-hydroxymethylcytosine (5hmC) erasure detectable as soon as three days after culture initiation (22).
Finally, human xenografts are one of the most widely used models to study human cancer in vivo, in the context of a complex microenvironment similar to that found in the original tumor. They involve transplantation of human tumor cells or tumor fragments into immunocompromised mice, and allow studying cancer mechanisms and potential cancer treatments in much more physiological conditions compared with culture models, although the lack of the immune component generates a less realistic tumor microenvironment and must be taken into consideration.
Although many studies have addressed the comparison of different culture systems to primary tumors under different molecular aspects, no information is available regarding histone PTMs in the transition from cancer tissue to culture conditions. In this study, we investigated the effects of the adaptation to culture conditions on histone PTM patterns by profiling them in different culture models through mass spectrometry (MS)-based approaches, which allow the comprehensive and quantitative investigation of up to 38 differentially modified peptides from histone H3 and H4. By profiling histone PTM patterns in a large collection of primary cultures, cell lines, and mouse xenograft models belonging to three representative tumor types and two non-cancerous models, we highlighted many common changes that are determined by culture conditions and are time-dependent, but, remarkably, are reverted in cell-derived xenograft models.
MATERIALS AND METHODS
Cell lines
Breast cancer cell lines were grown in the media indicated in Supplementary Table S1. Glioblastoma (GBM) cell lines U118, T98G and LN405 and ovarian cancer cell lines A2780, Caov-3 and HEY were grown in Dulbecco's modified Eagle's medium (DMEM) (Lonza) supplemented with 10% South American (SA) FBS (Lonza). U138 were grown in Ham's F10 (Lonza) with 10% SA-FBS, while U87 were grown in MEM (Lonza) supplemented with 0.1 mM non-essential amino acids (Gibco), sodium pyruvate and 10% SA-FBS. The ovarian cancer cell line OVCAR3 was grown in RPMI medium (Lonza) supplemented with 10% North American (NA) FBS and Insulin 0.01 mg/ml. MCF10A cells were grown in MEBM complemented with bovine pituitary extract 3 mg/ml, hydrocortisone 0.5 mg/ml, hEGF 10 ug/ml, insulin 5 mg/ml and 100 ng/ml cholera toxin. DU4475 were grown in RPMI 1640 supplemented with 20% NA-FBS, 1mM Sodium Pyruvate, 10mM HEPES and 2.5 g/l d-glucose. HBL-100 were grown in McCoy'S 5A medium supplemented with 10% NA-FBS and 2 mM l-glutamine. All growth media were supplemented with antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) (Gibco) and 2 mM l-glutamine (Gibco). Cell lines were grown in a humidified 37°C incubator with 5% CO2.
Tissue specimens and primary cultures
Human breast cancer specimens or normal breast tissues surrounding the tumors were obtained from patients undergoing surgery for the removal of clinically confirmed neoplasia (Supplementary Table S2). The patients provided informed consent and this study was approved by the Ethical Committee of the European Institute of Oncology. For breast cancer samples, the levels of hormone receptors, Her-2 and Ki-67 were ascertained by immunohistochemistry (23). Luminal A-like and Triple Negative subtypes were defined as follows: Luminal A-like: ER and/or PgR(+), HER2(–), Ki67 < 20%; Triple Negative: ER, PgR and HER2(–), irrespective of Ki67 score. To obtain breast cancer primary cells, tumor pieces were minced and incubated for 4–6 h in the enzyme digestion mixture (DMEM with penicillin/streptomycin, 2 mM l-glutamine, 5 mg/ml insulin, 0.25 mM hydrocortisone, 200 U/ml collagenase (Sigma-Aldrich), 100 U/ml hyaluronidase (Sigma-Aldrich), 10 ng/ml epidermal growth factor (EGF)). The suspension was filtered through a 70 μm strainer, centrifuged, and resuspended in F-12/DMEM (1:1) supplemented with 1% fetal calf serum, 20 μg/ml gentamicin (Lonza), 10 mM HEPES pH 7.5 (Lonza), 10 nM triiodothyronine (Sigma-Aldrich), 35 μg/ml bovine pituitary extraction (Gibco), 50 μM l-ascorbic acid (Sigma-Aldrich), 15 nM sodium selenite (Sigma-Aldrich), 10 nM β-estradiol (Sigma-Aldrich), 0.1 M ethanolamine (Sigma-Aldrich), 10 ng/ml EGF, 1 μg/ml insulin, 1 μg/ml hydrocortisone, 10 μg/ml transferrin (Lonza), 50 ng/ml cholera toxin, 1 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 250 ng/ml amphotericin B (Lonza). Primary breast cancer cells were passaged in conditions that favor attachment of tumoral epithelial cells compared with normal cells through differential trypsinization, eliminating both fibroblast and normal cells, which attach less (24). Primary breast cancer cells were cultured in average for approximately 3 passages before they underwent senescence and died; therefore, they are all short-term cultures.
Glioblastomas (GBMs, World Health Organization grade IV) were collected from consenting patients at the Department of Neurosurgery of the Istituto Neurologico Carlo Besta, Milan, Italy (Supplementary Table S3). GBM tissues were either snap frozen or mechanically dissociated immediately after surgery and enzymatically digested with papain (2 mg/ml; Worthington Biochemical) at 37°C to obtain a single cell suspension. Human GBM cells were grown either as spheroid aggregates in order to establish neurosphere cultures (3D cells) or were maintained in adherence (2D cells). The formers were grown in F-12/DMEM (1:1) supplemented with B-27 (Life Technologies), 20 ng/ml EGF, 10 ng/ml basic fibroblast growth factor (bFGF) (PeproTech), and 0.0002% heparin (Sigma-Aldrich), while the latters were grown in 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin. GBM neurospheres were maintained by weekly passaging through mechanical dissociation and seeding at 50 000 cells/ml in proliferation media (Thermo Scientific). GBM adherent cells were plated at a density sufficient to reach confluence in 3–4 days (50 000 cells/cm2). 3D GBM cells were cultured in a medium specifically designed for the enrichment of the self-renewing cells, while 2D GBM cells were established by depleting normal cells (deriving either from the brain or the stroma) through differential trypsinization. Notably, all the 3D GBM cells used in this study are able to form orthotopic xenograft tumors (25), a functional hallmark that distinguishes cancer from normal cells. Supplementary Table S3 summarizes all the glioblastoma samples tested. Non tumoral neural stem and progenitor cells were isolated from the sub-ventricular zone of adult C57-BL6 mice and expanded as floating neurospheres, as described above for GBM neurosphere cultures. Primary cultures were grown in a humidified 37°C incubator with 5% CO2.
Ovarian cancer samples (Supplementary Table S4) were obtained upon informed consent from patients undergoing surgery at the Gynecology Division of the European Institute of Oncology (Milan). High-grade serous epithelial ovarian cancer and mucinous borderline cystadenoma cells were derived from tumor biopsies of patients who had received chemotherapy. Tissues were minced into small fragments and enzymatically digested as described (26). Tissue dissociation was monitored by microscope observation and, when single cells or small cell clusters were visible, the cell suspension was centrifuged for 5 min at 1200 rpm and the cell pellet was treated with an ammonium–chloride–potassium (ACK) buffer at room temperature for 2 min to lyse red blood cells. Primary epithelial tumor cells were plated on collagen I-coated flasks (Corning Biocoat) and cultured in epithelial growth medium (26), at 37°C in a humidified atmosphere with 5% CO2. All primary cell batches were cultured for a maximum of two passages after tissue digestion in order to preserve the heterogeneity of original tissue. The purity of primary cell cultures, monitored by immunostaining for the epithelial cytokeratins 5, 7 and 8, pan-cytokeratins or for MUC-16, was consistently over 95%.
Samples were collected and snap frozen or fixed overnight in 4% formalin and embedded in paraffin. All frozen breast cancer samples had a tumor cellularity >50%, with the exception of frozen #3 and 4, which had a tumor cellularity of 20 and 30%, respectively, as assessed by hematoxylin and eosin (H&E) staining. Glioblastoma samples had tumor cellularity >70% and <50% of necrosis, and specimens were derived only from the tumor core excluding the invasive rim, in order to minimize the presence of normal brain tissue. Ovarian cancer samples had a tumor cellularity >50%, except for frozen #1 which was 35%. The cellularity of the two mucinous cystadenoma samples was 20–25%.
Xenografts
For the establishment of patient derived xenografts (PDXs), GBM-derived neurospheres were mechanically dissociated, and cells were resuspended in 2 μl of phosphate-buffered saline and stereotaxically injected into the nucleus caudatus (1 mm posterior, 3 mm left lateral, 3.5 mm in depth from bregma) of 5-week-old female nu/nu CD1 mice (Charles River). Tumorigenic human glioblastoma cell lines were also transplanted intracerebrally into CD1 mice. In parallel, an immunocompetent model of murine GBM was established through intracranial implantation of murine GL261 neurospheres into C57/BL6 mice (27). In all cases, GBM xenografts were maintained until the development of neurological signs. Then, the brain was removed, fixed overnight in 4% formalin and embedded in paraffin. To obtain xenografts-derived primary cells, the tumour was enucleated and processed for cell isolation as described before. Experimental procedures involving animals complied with the Guidelines of the Italian National Institute of Health, and were approved by the Institutional Ethical Committee.
Histone enrichment
Histones were purified from breast cancer cell lines through nuclei isolation on a sucrose cushion followed by acidic extraction, as previously described (28), while they were enriched from primary cultures and GBM, ovarian cancer, brain or breast cell lines by resuspending 0.5–2 × 106 cells in 1 ml of PBS buffer containing 0.1% Triton X-100 and protease inhibitors (PBS-Triton buffer). Nuclei were isolated through a 10-min centrifugation at 2300 × g, resuspended in 100 μl of the same buffer containing 0.1% SDS and incubated for few minutes at 37°C in the presence of 250 U of benzonase (Merk Millipore) to digest nucleic acids. A comparison of enrichment methods with and without acidic extraction was performed using three cell lines, to verify that the purification strategy does not affect histone PTM quantification (Supplementary Figure S1). To enrich histones from frozen tissues, at least 20 mg of tissue were thawed on ice, cut with scissors and homogenized in 1 ml of PBS-Triton using a Dounce homogenizer. The homogenate was filtered through a 100 μm cell strainer and nuclei were isolated and lysed as described for primary cells. Histones were isolated from FFPE tissues using the PAT-H-MS protocol, as recently described (28). Briefly, four 10-μm tissue sections were deparaffinized and rehydrated using standard procedures. Tissue samples were homogenized by sonication in 200 μl of 20 mM Tris pH 7.4 containing 2% SDS and proteins were extracted and de-crosslinked at 95°C for 45 min and 65°C for 4 h. Histones were obtained from paraffin-embedded whole mouse brains harboring either orthotopic GBM patient-derived xenografts or an orthotopic model for murine GBM from 7–10 μM-thick H&E stained sections that were subjected to manual macrodissection to isolate either the tumor or the normal brain tissue prior to PAT-H-MS (29). PAT-H-MS coupled with laser microdissection (LMD) was instead used to isolate breast tumor cells from non-tumoral cells in human breast cancer samples, starting from 7–16 10-μM-thick H&E stained sections, as recently described (29). Of note, few modified histone peptides, including H3K18me1 and methylations on H3K79, cannot be profiled in FFPE tissues, due to the insurgence of artifacts likely caused by formalin fixation (28). The yield of histones deriving from the different purification protocols was estimated by SDS-PAGE gel by comparison with known amounts of recombinant histone H3.1 (New England Biolabs), following protein detection with colloidal Comassie staining (Expedeon).
Super-SILAC
MDA-MB-231, MDA-MB-468, MDA-MB-453 and MDA-MB-361 breast cancer cells lines were grown in SILAC-DMEM (Euroclone) supplemented with 2 mM l-glutamine, 146 mg/l of lysine (Sigma-Aldrich), 84 mg/l l-13C615N4-arginine (Arg-10, Sigma-Aldrich), 10% dialyzed serum (Life Technologies) and penicillin/streptomycin for at least eight doublings to obtain complete labeling with heavy-labeled aminoacids. Histones were isolated as previously described (30), mixed in equal amounts, lyophilized, and stored at −80°C until use. Because the most abundant histone PTMs found in GBM and ovarian cancer samples are present in the breast cancer super-SILAC mix, the same standard was used to analyze all samples.
LC–MS/MS analysis of histone PTMs
About 1–5 μg of histones per run per sample were mixed with an approximately equal amount of super-SILAC mix and separated on a 17% SDS-PAGE gel. Bands corresponding to histones H3 and H4 were excised, chemically alkylated with D6-acetic anhydride or propionic anhydride, in-gel digested with trypsin (the combination of chemical alkylation and trypsin digestion generates an ‘Arg-C-like’ digestion) and desalted on handmade nanocolumns (StageTips) as previously described (28). Peptide mixtures were separated by reversed-phase chromatography on an in-house-made 25-cm column (inner diameter 75 μm, outer diameter 350 μm outer diameter, 1.9 μm ReproSil, Pur C18AQ medium), using a ultra nanoflow high-performance liquid chromatography (HPLC) system (EASY-nLC™ 1000, Thermo Fisher Scientic), or an EASY-Spray column (Thermo Fisher Scientific), 50-cm long (inner diameter 75 μm, PepMap C18, 2 μm particles), which were connected online to a Q Exactive HF instrument (Thermo Fisher Scientific) through a Nanospray Flex™ or an EASY-Spray™ Ion Sources (Thermo Fisher Scientific), respectively. Solvent A was 0.1% formic acid (FA) in ddH2O and solvent B was 80% ACN plus 0.1% FA. Peptides were injected in an aqueous 1% TFA solution at a flow rate of 500 nl/min and were separated with a 100-min linear gradient of 0–40% solvent B, followed by a 5-min gradient of 40–60% and a 5-min gradient of 60–95% at a flow rate of 250 nl/min. For ovarian cancer and normal mouse brain samples, a shorter linear gradient of 60 min was used. The Q Exactive HF instrument was operated in the data-dependent acquisition (DDA) mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra (m/z 300–1650) were analyzed in the Orbitrap detector with a resolution of 35 000 at m/z 400. The 10 most intense peptide ions with charge states ≥2 were sequentially isolated to a target value for MS1 of 3 × 106 and fragmented by HCD with a normalized collision energy setting of 25%. The maximum allowed ion accumulation times were 20 ms for full scans and 50 ms for MS/MS and the target value for MS/MS was set to 1 × 106. The dynamic exclusion time was set to 20 s, and the standard mass spectrometric conditions for all experiments were as follows: spray voltage of 2.4 kV, no sheath and auxiliary gas flow.
Histone PTM data analysis
Acquired RAW data were analyzed using the integrated MaxQuant software v.1.5.2.8, which performed peak list generation and protein identification using the Andromeda search engine. The Uniprot HUMAN_histones 1502 database was used for histone peptide identification. Enzyme specificity was set to Arg-C. The estimated false discovery rate (FDR) of all peptide identifications was set at a maximum of 1%. The mass tolerance was set to 6 ppm for precursor and fragment ions. One missed cleavage was allowed, and the minimum peptide length was set to 6 amino acids. Variable modifications include lysine D3-acetylation (+45.0294 Da)/propionylation (+56.0262 Da), lysine monomethylation (+59.0454 or +70.0422, for the D3-acetylation and propionylation protocols respectively, which corresponds to the sum of monomethylation (+14.016 Da) and the chemical alkylating agent), dimethylation (+28.031 Da), trimethylation (+42.046 Da), and lysine acetylation (+42.010 Da). To reduce the search time and the rate of false positives, which increase with increasing the number of variable modifications included in the database search (31), the raw data were analyzed through multiple parallel MaxQuant jobs (32), setting different combinations of variable modifications: (i) D3-acetylation/propionylation, lysine monomethylation with D3-acetylation/propionylation, dimethylation and lysine acetylation, (ii) D3-acetylation/propionylation, lysine monomethylation with D3-acetylation/propionylation, dimethylation and trimethylation, (iii) D3-acetylation/propionylation, lysine monomethylation with D3-acetylation/propionylation, trimethylation and lysine acetylation. Peptides with Andromeda score <60 (corresponding to a Mascot score of 15 (33), which has been previously used as a cut-off value (34)) and localization probability score <0.75, were removed. Identifications and retention times were used to guide the manual quantification of each modified peptide using QualBrowser version 2.0.7 (ThermoFisher Scientific). Site assignment was evaluated using QualBrowser and MaxQuant Viewer 1.3.0.5. Extracted ion chromatograms (XIC) were constructed for each doubly charged precursor based on its m/z value, using a mass tolerance of 10 ppm and a mass precision up to four decimals. For each histone modified peptide, the relative abundance (RA) was estimated by dividing the area under the curve (AUC) of each modified peptide for the sum of the areas corresponding to all the observed forms of that peptide (35). For SILAC experiments, Arg10 was selected as heavy label (multiplicity = 2) in MaxQuant. The heavy form of each modified peptide was quantified from its XIC and the relative abundance quantified. Of note, because the histone H3 peptide 3–8 is particularly short and hydrophilic, and as such particularly challenging to analyze by MS, and the H3K4me3 mark is low abundance, we could not be reliably quantify it in the complete set of samples analyzed using our current approach, therefore we did not include it in our results. The AUC values for all the samples analyzed are reported in Datasets S1(tumoral samples) and S2 (normal samples).
Statistical analysis
Normalized L/H ratios, defined as L/H ratios of relative abundances normalized over the average value across the samples, were visualized and clustered using Perseus, with correlation distance and average linkage as parameters (36). The optimal number of clusters ( = 3) for the breast cancer samples shown in Figure 1A was determined by using the XSTAT package. The 3 main clusters explain 41% of the variance among samples (7.2 out of 12.3 total variance). Within class variances were 6.1, 5.2 and 10 for the cluster composed by frozen, primaries (plus T47D) and cell line samples. Principal component analyses (PCA) were performed using Perseus. Changes in single histone modifications among two or more groups were evaluated by t-test or one-way ANOVA followed by Bonferroni's post-hoc test, respectively, on log2 transformed L/H ratios, using GraphPad Prism. When only two sample measurements were available, a t-test was performed and samples with a P-value <0.05 were indicated as ‘trends’ (Figure 3B and Supplementary Figure S4).
Figure 1.
Epigenetic profiling of breast cancer primary tumors, primary cells and cell lines. (A) Heatmap display and hierarchical clustering of the log2 of ratios obtained for the indicated histone PTMs for breast cancer frozen biopsies (black), primary cells (pink) and cell lines (blue). L/H relative abundances ratios obtained with the super-SILAC strategy (light channel: breast cancer biopsy, heavy channel: spike-in super-SILAC standard), normalized over the average value across the samples are shown. All frozen samples had a tumor cellularity greater than 50%, with the exception of frozen #3 and 4, which had a tumor cellularity of 20 and 30%, respectively. (B) Modified peptides significantly changing by one-way ANOVA followed by Bonferroni's post-hoc test (P< 0.05) in primary cells or cell lines compared with frozen breast cancer samples. (C) Modified peptides significantly changing by t-test (P< 0.05) in Luminal-A like and Triple Negative breast cancer cell lines compared with FFPE Luminal-A-like and Triple Negative breast cancer samples where tumor cells were isolated by laser microdissection (n = 3 for each subtype). The lighter color arrows indicate non-significant trends (P≤ 0.1). The grey color indicates those peptides that cannot be quantified in FFPE tissues. (D) Principal component analysis (normalized and centered) of the samples shown in A. (E) L/H ratios obtained for the H3K9ac/K14ac, H3K18me1 and H3K27me1/K36me3 peptides in matching frozen and primary cells. (F) Line plot showing trends of modification changes in primary cells and cell lines compared to frozen tissues. Log2 of the L/H ratios of the samples divided by the L/H average ratio for frozen samples are shown (averages from 8 frozen tumors, 9 primary cells and 12 cell lines). Peptides significantly changing (by one-way ANOVA and Bonferroni's post-hoc test, P< 0.05) in cell lines and primary cells, cell lines only, or primary cells only are shown in the left, middle and right panel, respectively. If not otherwise specified, the peptides are from histone H3. (G) L/H Ratios obtained for the H3K27me3/K36me1 and H3K9me3/K14ac peptides in Luminal-A-like (LuA) and Triple Negative (TN) FFPE tumors and cell lines. FFPE microdissected samples and cell lines belonging to the same subtype were compared by t-test (P< 0.05). Error bars represent standard error of the mean (SEM) from 3–5 samples.
Figure 3.
A common histone PTM rearrangement in different culture models. (A) Heatmap display of the log2 of ratios obtained for the indicated histone PTMs for ovarian cancer frozen primary tumors, primary cells and cell lines. L/H relative abundances ratios obtained with the super-SILAC strategy normalized over the average value across the samples are shown. Modified peptides significantly changing in primary cultures or cell lines compared with frozen tumors are indicated by arrows on the right (P<0.05, lighter color corresponds to P< 0.1). (B) Summary of the modified peptides significantly changing in breast cancer (BC), GBM and ovarian cancer (OC) cultures. The majority of these changes were also detected in breast and brain cultures (the detailed heatmap display for these experiments can be found in Supplementary Figure S4). Grey: not quantified. The asterisk indicates that the modified peptides was not quantified in brain tissue, but the change was observed in long- vs short-term cultures. The lighter color corresponds to P<0.1 for ovarian cancer samples, and trends for normal breast samples (see Materials and Methods). (C) Principal component analysis (normalized and centered) of tumor tissue and cell lines for the cancer models analyzed. The breast samples also include FFPE laser microdissected tumor populations, which are indicated by empty circles and cluster with the other breast cancer samples. (D) Examples of two modified peptides showing significant differences in the frozen samples belonging to distinct tumor models. The differences were lost/reduced in cell lines. Error bars represent standard error from 3–8 samples. *P< 0.05 by one way-ANOVA and Bonferroni's post-hoc test.
Global proteome analysis
The proteomes of matching GBM FFPE tumor tissue and primary GBM 3D cultures (short- and long-term) deriving from patient #4 (Supplementary Table S3) were profiled by quantitative label-free analysis, as described in detail in the Supplementary information. Identified Protein groups and corresponding quantification are reported in Dataset S3.
GO analysis of the proteins significantly changing in the comparison between short-term cultures and tissue, or long-term cultures and short-term cultures was carried out using the Gorilla algorithm (37), on two unranked lists of proteins, with a P-value threshold of 0.001. The list of proteins quantified in six out of six samples in the two experiments (3090 for the tissue vs. short-term culture comparison, and 5262 for the short- vs. long-term culture comparison) was used as a background set, while the significantly up- and down- regulated proteins in short-term cultures compared with tissue (439 up and 256 down) or long-term cultures compared with short-term cultures (219 up and 244 down) were used as target sets. Significantly enriched GO terms are reported in Dataset S4, and were summarized by reducing functional redundancies and visualized by treemap using the REVIGO web server (38), with allowed similarity 0.5 (small), database Homo Sapiens, and semantic similarity measure SimRel.
RESULTS
Profiling of breast cancer frozen tissues, primary cultures and cell lines reveals extensive rewiring of histone PTMs in culture conditions
To investigate the consequences of the transition from tumor tissue to culture, we comprehensively profiled by mass spectrometry 38 differentially modified peptides from histone H3 and H4 in 8 breast cancer human frozen tumors (Supplementary Table S2), 10 breast cancer primary cells (8 of which matching with the frozen tissues (Supplementary Table S2), and 12 commonly used cell lines (Supplementary Table S1), using the histone focused super-SILAC approach that we have recently described (28,39). Non-supervised clustering based on relative changes of histone PTM levels defined three main clusters containing either frozen tissues, primary cells and cell lines, with the exception of the T47D cell line, which clustered with primary cells (Figure 1A). Both the heatmap display and the PCA analysis (Figure 1D) showed that primary cells are closer to primary tumors than cell lines. Some changes were detected already in primary cells (e.g. H3K18me1, H3K9acK14ac, H3K27me1K36me3, Figure 1A, B and F, left panels), and comparison with their matching frozen samples showed that they occur very systematically, namely in all the samples tested and in the same direction (Figure 1E). With only few exceptions (Figure 1B and F, right panel), the significant changes observed in primary cells were also present in cell lines, where many additional significantly changing marks could be detected (Figure 1B and F, middle panels). Interestingly, acetylation marks generally decrease in both primary and cell line cultures, while the trend of methylations appeared more composite.
We confirmed most of the above-mentioned changes when comparing cell lines with pure tumor populations that were laser microdissected from FFPE sections, for a small set of patient samples (29); this suggests that reduction of tissue heterogeneity overall does not account for the histone PTM changes that we observed in culture (Figure 1C and G, Supplementary Figure S2). In this experiment, we analyzed samples belonging to the Luminal-A-like and Triple Negative subtypes, which are characterized by different molecular features, prognosis and cell of origin, to account for subtype heterogeneity, and compared them with cell lines belonging to the same subtypes (Supplementary Table S1). While most of the changes were confirmed in both subtypes, H3K9me3 containing peptides were increased in Luminal-A-like only samples, a result in accordance with our initial frozen dataset being composed only by Luminal samples, and suggest a possible subtype-specific change occurring during culturing. Another interesting finding that we achieved from this experiment came from the comparison of Luminal-A-like and Triple Negative patterns in tissues and cell lines. We have previously identified several marks, including H3K27me3- and H3K9me3-containing peptides, H3K27me2/K36me1 and H3K36me2 (28,29) that distinguish these two subtypes. We confirmed most of them in this small FFPE tissue dataset, but found no differences, or much less marked ones, by comparing Luminal-A-like and Triple Negative cell lines, as shown in Figure 1G for H3K9me3/K14ac and H3K27me3/K36me1.
Overall, these results demonstrate that substantial alterations are generated at the histone PTM level upon adaptation of breast cancer tumor cells to culture, and that these changes are more marked and numerous in cell lines compared with primary cells originated from patient tumors and kept in culture for a few weeks.
Histone modification profiling in GBM culture models
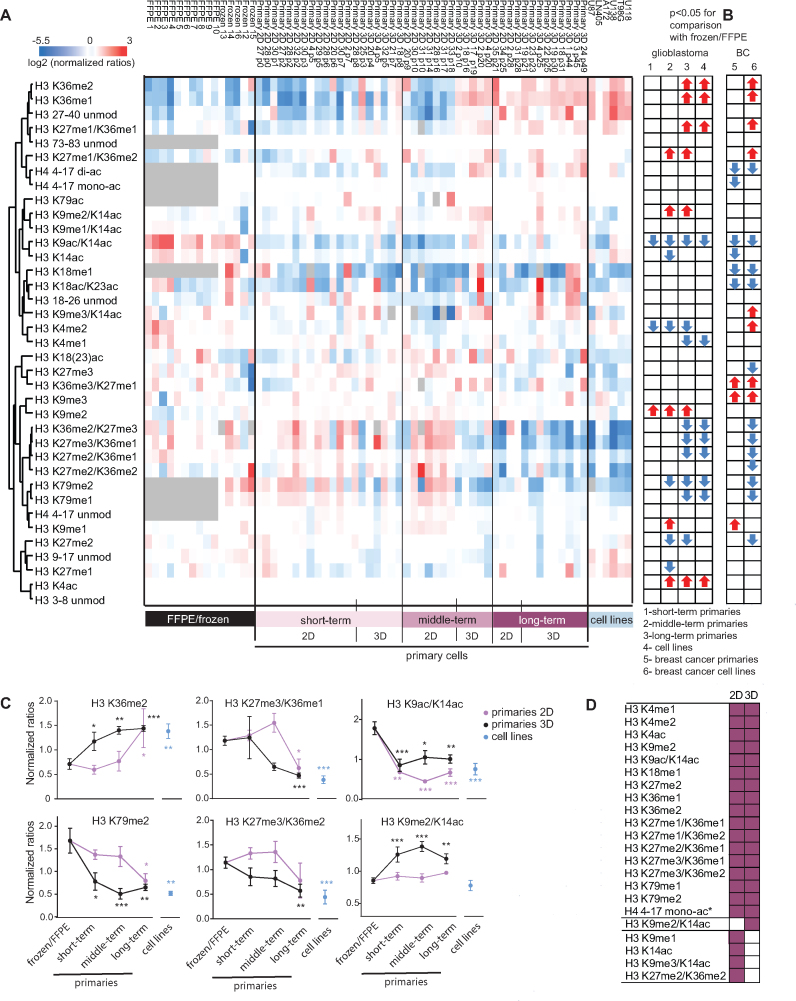
Next, we analyzed a set of GBM samples composed of 11 primary tissue samples (stored as frozen or FFPE tissue), 45 primary cultures, and 6 cell lines (Supplementary Table S3). Because GBM primary cultures can be maintained in culture for much longer compared to breast cancer primary cells, we divided them in short-term (cells kept in culture for up to 8 passages), middle-term (9–20 passages) and long-term (>20 passages). Furthermore, in addition to 2D adherent cultures similar to the breast cancer primary cells, we tested 3D neurospheres, which are expected to better mimic the GBM pathophysiology upon orthotopic transplantation (21,25). The histone PTM heatmap display that summarizes these results shows a progressive shift away from the pattern of the tumor tissue in cultured cells, which is more marked in long-term primary cultures (either 2D and 3D models) and cell lines (Figure 2A). Examples of this behavior are histone H3 peptides bearing methylations on K36, K27 and K79, which show significant differences starting in late cultures and cell lines, but not in earlier primary cultures. Some modifications, mostly on the histone H3 K9-17 peptide, appear in early primary cultures and are maintained in later passages cultures (e.g. H3K9ac/K14ac), while a few others are instead detectable early but are then lost (e.g. H3K9me2). When analyzed separately, 2D and 3D primary cultures displayed overall similar histone PTM changes and similar trends, with long-term cultures showing more/more pronounced changes compared with short-term cultures. Most of the changes were common to 2D and 3D cultures, but they generally appeared earlier in 3D cultures (Figure 2C and D, Supplementary Figure S3).
Figure 2.
Epigenetic profiling of GBM culture models. (A) Heatmap display of the log2 of ratios obtained for the indicated histone PTMs for GBM frozen/FFPE primary tumors, primary cells (divided in early-, middle- and long-term cultures; or 2D and 3D cultures) and cell lines. L/H relative abundances ratios obtained with the super-SILAC strategy normalized over the average value across the samples are shown. Modified peptides significantly changing in primary cultures (divided in early-, middle- and long-term) or cell lines compared with FFPE/frozen GBM samples are indicated by arrows on the right. Grey: not quantified. (B) Modified peptides significantly changing in primary breast cancer (BC) primary cultures or cell lines compared with frozen breast cancer biopsies. (C) Normalized ratios for GBM FFPE/frozen, 2D and 3D primary cultures followed over time and cell lines. (D) Summary of histone PTM changes in GBM 2D and 3D cultures. Significance in A, C and D was defined by comparing the samples to frozen/FFPE by one-way ANOVA and Bonferroni's post-hoc test, P< 0.05. Error bars in C represent SEM from 3–13 samples. *P< 0.05, **P< 0.01, ***P< 0.001.
Strikingly, most of the histone PTM changes occurring in GBM cultures were also found in breast cancer cultures (Figure 2B), suggesting common processes underlying the culture-dependent epigenetic rewiring. A few exceptions could be observed, which may underlie some tissue-specific mechanisms. For instance, some acetylations (mono- and di-acetylation on the H4 peptide 4–17 and H3K18ac/K23ac) decreased only in breast cancer, and the previously mentioned H3K9me3-containing peptides increase only in Luminal-A-like cultures. A few changes were instead specific to GBM cultures, such as the decrease of H3K4me1/me2, while a few others appeared to be 2D- or 3D-specific (Figure 2D).
A core of histone modifications changes common to different culture models
To further investigate the existence of a common program of histone mark changes occurring in culture conditions, we also profiled a panel of ovarian cancer samples (four frozen tumor samples, four primary cells and four cell lines, Supplementary Table S4) confirming, in spite of the limited number of samples, most of the histone PTM changes observed in the other two culture models (Figure 3A, Supplementary Figure S4A–C). The ‘core’ of changes common to breast cancer, GBM and ovarian cancer is summarized in Figure 3B. According to a hypergeometric distribution, the expected occurrence of a significant change in the same direction in the three models tested is ∼2, against the 12 observed, a difference that corresponds to a P-value of 0.0048 by Fisher exact test. Strikingly, most of these changes were also found in two small panels of non cancerous models: (i) frozen ‘normal’ breast samples derived from the normal tissue surrounding breast tumors compared with two breast cell lines, and (ii) mouse brain tissue obtained from FFPE sections compared with short- and middle/long-term normal primary brain cells (Figure 3B, right panel, and Supplementary Figure S4B–C). Therefore, the epigenetic rewiring that we observed underlies a general phenomenon associated with transition to cell culture conditions.
Taken together, these results show that the culture conditions cause an extensive and systematic rearrangement that involves a decrease of acetylation on H3K9 and H3K14, of methylation on H3K27 and H3K79, and an increase of methylation on H3K36, which is time-dependent and very conserved across tissue types and culturing conditions. Such rearrangement is relevant to the point that differences between tissues and cell lines are more marked than those among cancer models (Figure 3C), as shown by the PCA analysis of breast cancer, GBM and ovarian cancer samples, causing the reduction or disappearance of histone PTM differences among cancer types (Figure 3D, Supplementary Figure S4D).
Histone modifier gene expression and proteomic profiling show widespread changes in GBM 3D cultures
To evaluate whether the core of histone PTMs that systematically change in all the tested culture conditions correlates with alterations in the levels of the HMEs responsible for their deposition/removal, we profiled their gene expression levels in frozen GBM human tissue and short- and long-term primary GBM cultures. Because no information is available regarding the HMEs mediating the H3K18me1 mark, and histone acetyl-transferases and deacetylases act non- specifically on many acetylation sites, we focused on the enzymes linked with H3K36, H3K27 and H3K79 methylation. The H3K36-specific methyltransferases SETD2 and SETMAR were significantly upregulated in long-term cultures (SETMAR also in long- vs. short-term primaries) compared with the tumor tissue, and others (NSD2 and NSD3) showed an increasing trend (Figure 4A and B) that correlates with the observed increase of H3K36 methylation. A decrease of the demethylase KDM4B was also consistent with the H3K36 methylation increase, while the increase of the demethylase KDM4A pointed in the opposite direction. This apparent discrepancy can be explained by the fact that we quantify global histone PTM relative variations, which do not exclude that changes in the opposite direction may occur in specific genomic regions. No enzyme related to H3K27me3 or H3K79 methylation (whose demethylase is not yet known) showed any significant expression changes (Supplementary Figure S5A).
Figure 4.
Gene expression and global proteomic analysis of GBM cultures. (A) Gene expression analysis of the indicated HMEs in frozen tumors and short-term and long-term GBM primary cells. Samples were compared by one way ANOVA and Bonferroni's post-hoc test. *P< 0.05. Error bars represent SEM from 4–5 biological replicates. (B) Summary of results obtained by gene expression analysis. Red and blue arrows indicate significant changes (as indicated in (A)), lighter color arrows indicate non-significant trends (P< 0.1). (C) NSD2, NSD3 and SETMAR mRNA levels measured by RT-qPCR analysis in glioblastoma primary cells from patient #4 expressing target-specific shRNAs, normalized over the enzyme levels in the shScramble cells. Error bars represent SEM from 3 technical replicates. (D) NSD2, NSD3 and SETMAR protein levels in the same samples as in (C), measured by MS analysis of gel bands corresponding to the molecular weights of NSD2 and NSD3 (152.5 and 162 KDa) or SETMAR (76 kDa). IBAQ values from two technical replicates, normalized over shScramble cells, are shown. Error bars indicate SEM. (E) Analysis of histone PTM levels in glioblastoma primary cells expressing NSD2, NSD3 and SETMAR shRNAs. Histone H3 peptides containing K36me1, K36me2 and K27me3 were quantified by MS and normalized over shScramble cells. (F) Immunoblot analysis and densitometric quantification (normalized to the tissue sample) of the matching tumor tissue and primary short- and long-term cultures analyzed by global proteomics. Non-relevant lanes were removed. Three biological replicates for short- and long-term cells were compared to the tissue by Student's t-test, *P< 0.05. The complete blots corresponding to the replicates can be found in Supplementary Figure S6. (G) Volcano plots showing significantly up- and down-regulated proteins by comparing short-term primary 3D glioblastoma cultures and tumor tissue (left), and long- and short- term cultures (right). Differential protein expression cutoff: FDR < 0.01, S0 = 5 (left) and S0 = 1 (right). (H) Enrichment of GO biological process (BP) terms in the up- and down-regulated proteins from (G). Each tree-map includes all biological process terms mapped to a high hierarchical level. The size of the boxes corresponds to the number of proteins in that category. The grey color in the top-left plot include: regulation of heterotypic cell-cell adhesion, single organism cellular process, signaling response to inorganic substance, cell communication, regulation of biological quality, acute inflammatory response, immune system process, acute inflammatory response, neural nucleus development, multicellular organism process, developmental process, response to stimulus, signaling, locomotion, localization, glycosaminoglycan synthesis, immune system process, cell activation, intermediate filament-based process. In the top-right plot: VEGFR signaling pathway and single-organism cellular process. In the bottom-right: developmental process, positive regulation of protein autophosphorylation, cellular response to abiotic stimulus, collagen-activated signaling pathway.
To test whether in our model the changes in the expression of the HMEs observed in culture would be sufficient to alter H3K36 methylation, we downregulated NSD2, NSD3 and SETMAR, the methyltransferases specific for the deposition of H3K36 mono- and di-methylation, through lentivirus-mediated short hairpin RNA interference (shRNA) in 3D glioblastoma long-term primary cells (Figure 4C and D and Supplementary Figure S5B) and quantified the differentially modified forms of the H3 27–40 peptide by accurate SILAC-based MS analysis. We observed a decrease of the H3K36me1 and me2 marks in NSD2 and NSD3 depleted cells, but not as clearly in SETMAR knock-down cells (Figure 4E), a trend that was confirmed by immunoblot analysis (Supplementary Figure S5C). Although both shRNA clones for NSD2 also partially decreased SETMAR levels, the effect on H3K36me1/me2 is likely mediated by NSD2 downregulation, since SETMAR knockdown alone had a much milder effect. Interestingly, the decrease of H3K36 methylation in NSD2 and NSD3 depleted cells was paralleled by a concomitant increase of H3K27me3 (Figure 4E, bottom histogram). This last finding is consistent with the notion that H3K36 methylation antagonizes H3K27 methylation, and vice-versa (40–42) and with a previous study showing that increased levels of NSD2 induced a global reduction of H3K27me3 (43), providing a potential mechanism for the decrease in the H3K27me3 mark observed in culture, in the absence of detectable changes in the enzymes that depose/remove this mark.
Next, in order to analyze global changes, both epigenetic and not, that characterize the transition to culture conditions, we performed a label-free quantitative proteomic analysis of matched FFPE human GBM tissue and short- and long-term 3D GBM neurospheres, where we verified by both MS and WB analysis the above-mentioned histone PTM changes (Figure 4F and Supplementary Figure S6A–C). We assessed early and late changes due to the transition to culture conditions by comparing the original tissue and short-term primary cells (Figure 4G, left panels), and short- and long-term cultures (Figure 4G, right panels), respectively. We identified and quantified with three valid values approximately 5600 proteins in cells and 3100 in FFPE tissue (Supplementary Figure S6D), with the lower number in tissue likely due to technical differences in the protein extraction methods. Because of this, while in cells we were able to profile 50 HMEs, only 11 were identified and quantified in the FFPE tissue (Supplementary Table S6). The comparison between short- and long-term cultures confirmed some of the findings obtained with gene expression, with a significant increase of the H3K36me- mediating methyltransferase SETMAR, and an increasing trend for NSD3 in long-term cultures compared with short-term ones (Figure 4G, right panel). Interestingly, in this dataset we also found a general down-regulation of various linker histone H1 variants (particularly H1.0 and H1.2), which are required to stabilize higher‐order chromatin structures and generally correlate with repression of transcription, although more recent evidence showed that they can regulate specific sets of genes (44). More globally, the comparison of short-term cultures and tissue showed a dramatic rearrangement of proteins levels (Figure 4G, left panel), with 1894 significantly different proteins identified (t-test, P < 0.01, fold change (S0) = 1). Using very strict criteria to define differential protein expression (S0 > 5), we found that the 256 proteins up-regulated in tissue and the 439 up-regulated in short-term cultures were enriched for different functional annotations (Figure 4H, left panels, Dataset S4). The GO biological processes up-regulated in tissue included terms related with interaction with the microenvironment (e.g. extracellular matrix (ECM) organization, immune response and adhesion) -which is expected in a complex setting such as a tissue- as well as nervous system-specific terms. Upregulated biological processes in culture were mostly related to mRNA processing and transport, chromosome organization, response to DNA damage, metabolism and cell division, suggesting an increased rate of transcription and cell replication. Short- and long-term cultures were much more similar to each other, with 463 significantly changing proteins (t test, P < 0.01, S0 = 1), which highlighted changes in metabolic pathways and secretion (Figure 4H, right panels, Dataset S4). Only few categories were up-regulated in long-term cells (Dataset S4), and the most represented term was epithelial differentiation. This could be explained by the high phenotypic plasticity typical of stem/progenitor cell, and it is plausible that their adaptation to culture conditions might cause their transition between epithelial- and mesenchymal-like states.
Histone modifications revert in xenograft models
Next, we sought to determine whether the changes observed in culture conditions are irreversible or can be reverted by returning the cells in vivo. To this aim, we analyzed the histone PTM patterns of 10 mouse FFPE xenografts, six of which derived from GBM long-term neurospheres (Supplementary Table S3) and four from GBM cell lines that are tumorigenic in vivo, and compared them to their corresponding cells of origin and to eight FFPE human tumor samples (Supplementary Table S3). Unsupervised clustering defined two main clusters, one comprising most of the FFPE primary tumors and xenografts deriving from both primary cells and cell lines, and the other comprising mostly primary cells and cell lines (Figure 5A). Accordingly, the PCA analysis showed that xenografts are indeed much closer to the primary tumors than the cultured cells, and that the xenografts derived from primary cells are even more similar than those from cell lines (Figure 5B). Of note, the two cell lines LN405 and A172 appear to be closer to the pattern of primary samples than the others. Remarkably, most of the significant changes in modified peptides that we observed in cultures were no longer detected or much reduced in either type of xenograft (Figure 5A–C, Supplementary Figure S7). These include the H3K36me1/me2, H3K27me1/K36me1, H3K27me3/K36me2, H3K9ac/K14ac peptides. Other H3K27me2/me3-containing peptides reverted in primary cells, but not, or only partially, in cell lines, consistent with their lower similarity to primary tumors. The overall reversibility of histone PTM changes was further confirmed by the observation that 3D primary cells derived from xenografts undergo the same epigenetic changes that were observed in the transition from primary tumor to culture (Supplementary Figure S8), re-acquiring a ‘cell culture specific’ histone PTM pattern. These results show that most of the histone PTM rewiring observed in culture are reversible and suggest that the cell environment plays a crucial role in their appearance/disappearance.
Figure 5.
Histone modifications revert in xenograft models. (A) Heatmap display of the log2 of ratios obtained for the indicated histone PTMs for FFPE primary tumors, primary cells, cell lines and xenografts deriving from primary cells or cell lines. L/H relative abundances ratios obtained with the super-SILAC strategy normalized over the average value across the samples are shown. Modified peptides significantly changing in primary cultures, cell lines or xenografts compared with FFPE/frozen GBM samples are indicated with arrows. Grey: not quantified. (B) Principal component analysis (normalized and centered) of the samples shown in A. (C) Normalized ratios for specific peptides from (A). Significance in A–C was defined by one-way ANOVA and Bonferroni's post-hoc test, P< 0.05. Error bars in C represent SEM from 4–9 samples. *P< 0.05, **P< 0.01, ***P< 0.001.
DISCUSSION
The high failure rate of new agents in oncology is often attributed to the lack of pre-clinical models that closely represent the patient tumors and their heterogeneity (45); hence, a lot of effort has been devoted to the identification of the best model for cancer studies. In this study, we approached the suitability of two important in vitro models—cell lines and tumor-derived primary cells—from the epigenetic point of view, providing the first systematic profiling of histone PTMs in cancer culture systems. We showed that both cell lines and primary cells differ significantly from the original tumor in a number of histone marks. Many PTM changes can be observed already in primary cells, they increase with increasing culture time and are generally maintained in cell lines, where more modifications can be found. Histone PTM changes occur with different dynamics, with some appearing within a few weeks and others only in long-term cultures. Interestingly, all the acetylation changes observed in culture occur early, somewhat consistently with the long-standing notion that this modification is more dynamic compared with methylation.
Cell lines appear to be more different from primary tumors compared with short-term, but not with long-term primary cells, highlighting the importance of considering the amount of time spent in culture when using primary cells. In addition, among the cell lines some appear to have a modification pattern more representative of the original tumor. The importance of choosing the right cell line model for the specific study being performed has been highlighted in a recent study where comparison of copy-number changes, mutations and mRNA expression profiles revealed pronounced differences in molecular profiles between commonly used ovarian cancer cell lines and tumor samples, while other cell lines resembled much more the primary tumors (46). Many efforts have been devoted towards defining the best cell line model for tumor studies, which include the development of general computational methods to systematically define similarities between cell lines and the corresponding tumors of origin when genetic and molecular profiles are available (47,48). In our study, we identified cell lines that among those tested better recapitulate the epigenetic landscape of primary breast cancer tumors (T47D and BT474) and glioblastomas (LN405 and A172) and that should then be preferred for epigenetic studies. Interestingly, the T47D breast cancer cell line had already been reported to be representative of tumors in studies addressing gene expression, copy number variations, mutations and mRNA and protein expression (12,47).
Although 3D cultures are generally believed to better mimic the in vivo situation, here we showed that in culture they undergo the same epigenetic changes as 2D cultures do, with some exceptions that may be 2D- or 3D-specific. This result show that, considering the bulk amount of histone PTMs, no culture approach is preferable. The 2D and 3D cultures that we analyzed not only differ because the cells grow in adhesion or in suspension, but also in their medium composition and cell representation. 2D cultures are grown in the presence of serum and contain a mix of differentiated cells, including glia, microglia, neurons and immature cells, which in the presence of serum differentiate. On the contrary, the medium used for the 3D cultures (without serum, EGF, FGF and B27 supplements) is designed to enrich immature cells, such as progenitor and stem cells. As such, the 2D and 3D primary cells that we analyzed are biologically different models, that, strikingly, undergo the same epigenetic changes in culture.
Furthermore, not only most of the histone PTM changes observed in culture were common to 2D or 3D cultures, but also to different tissue types, as shown by the similar results obtained with breast cancer, GBM and ovarian cancer samples, as well as non-cancerous models, suggesting the presence of common epigenetic processes of cell rewiring in culturing condition. Most interestingly, these processes appear to be reversible and highly dependent on the cell environment, since the majority of the modification changes revert when the cells are returned in vivo. Comprehensive gene-expression analysis studies showed that xenografts derived from the direct engrafting of tumor portions in mice (PDTX) maintain the majority of global pathway activity and the key genes found in primary tumors (49,50), and represent a better model compared with cell-derived xenografts. In addition, it has been reported that a set of tumor-specific genes expressed in small cell lung cancer primary tumors and xenografts was lost in culture conditions, and was not recovered when the tumors were re-established as secondary xenografts (49), supporting the notion that some of the changes imposed by culture conditions are irreversible. Overall, this does not seem to apply to bulk histone modifications, as in our study we tested xenografts derived from cell lines or patient-derived long-term cultures, both of which were kept in culture for a significant amount of time, and found that all the histone marks changing in culture (and measurable in FFPE) reverted when moved back to an in vivo microenvironment, both in the case of primary- and cell line-derived xenografts. An exception is represented by the H3K27me2/K36me1 peptide and the H3K27me3-containing peptides, which reverted in xenografts derived from primary cultures but not (or only slightly) in those deriving from cell lines, suggesting that a few marks become irreversibly altered after a certain amount of time of culturing.
Histone PTM changes occur in culture in a very systematic manner, as exemplified by the comparison between matching frozen and primary cells samples, and are reversed when switching between in vitro/in vivo situations, suggesting that such epigenetic rewiring is likely due to adaptation to cell culture conditions, rather than to the selection of a specific clone. Corroborating this idea, differential protein expression analysis in cultured primary GBM cells compared with the primary tumor revealed changes in many biological processes involved with the interaction with the microenvironment. For instance, we found upregulation in tissue of terms such as extracellular matrix (ECM) organization, immune response, regulation of external stimulus and cell adhesion. On the other hand, culture conditions cause upregulation of processes that include mRNA processing and transport, chromosome organization and cell division, indicative of increased transcription and cell replication. These findings are in perfect agreement with previous gene expression results comparing cell lines and tumor tissues for six different tumor types (51).
Because histone PTMs change their abundance in different phases of the cell cycle, one possibility is that differing proportions of cycling cells in the different sample types could explain the epigenetic rearrangement in culture. Different studies investigated histone PTMs during the cell cycle, with somewhat discordant results. Acetylation and H3K27/K36 methylation were reported to be decreased in G2/M in one study (52) and slightly higher in another (53), while H3K79me2 was found increased in dividing cells (54). To address this question in our models, we correlated histone PTM levels and proliferation rates in different types of samples (breast cancer Luminal A and B-like samples (28), glioblastoma tissues and breast cancer cell lines). We found a significant/close to significant correlation between the proliferation index Ki67 and H3K36me1/me2 H3K27me2/me3 in breast cancer and glioblastoma tissues (Supplementary Figure S9A–B), but this did not apply to other modifications. In addition, the correlation of these histone marks with breast cancer cell line doubling time was less marked or absent (Supplementary Figure S9C), and no differences could be observed between MCF7 breast cancer cells in starving and not-starving conditions (Supplementary Figure S9D). These results suggest that different proliferation rates may contribute to a few of the modification changes occurring in culture, but do not represent their main cause of the global epigenetic rewiring that we have described.
On the other hand, increasing evidence shows that the environmental conditions can cause changes in HME levels, which in turn can mediate metabolic shifts. For instance, KDM4B is a hypoxia inducible gene (55,56), which could be downregulated in culture compared with primary tumors due to increased oxygen concentrations. NSD2 expression and activity has also been reported to depend on oxygen levels (57), while NSD3 has been shown to cause a metabolic shift from aerobic to anaerobic metabolism (58). Therefore, it is likely that histone PTM rewiring in culture is the result of a composite response, where histone mark changes can both be the drivers and the result of the cellular response to the dramatically different conditions found in culture compared with the tumor environment.
The core of modifications that we have consistently found in all the cell culture model tested include an increase of the active transcription mark H3K36me2 (paralleled by an increase of several methyltransferases that depose this mark, and by a decrease of the demethylase KDM4B) and a decrease of the repressive mark H3K27me3. Concomitantly, we also observed a decrease of H3/H4 acetylation, a typical activation mark, and of H3K79 methylation, which is also usually associated with active transcription (59). These results suggest that the histone PTMs changes occurring in culture may exert a composite effect on gene transcription, which however results in a net increase in RNA processing and transport, as suggested both by previous studies (51) and by our proteomics results. Consistent with increased transcription, in the same experiment we also found a decrease of histone H1 isoforms, also reported in cultured MEFs (22), whose presence is usually associated with a more compact and less transcription-prone chromatin (44). In particular, histone H1.0, the most markedly changing isoform in our experiment, has been associated with differentiated cells (60), and its decrease could suggest a de-differentiation process occurring in culture. It will be interesting to investigate to what extent the epigenetic changes that we observed in culture, particularly those that are involved with transcriptional regulation, correlate with target genes and impact on global gene expression.
Taken together, our findings show that transition to culture conditions causes a rather dramatic rearrangement of histone PTM patterns in culture and reinforce the growing awareness that cell culture models, and particularly cell lines, may poorly recapitulate some aspects of in vivo biology, such as epigenetics. In particular, our results indicate that short-term primary cultures (either 2D or 3D) are a better model compared to long-term cultures, and that, not surprisingly, tumor xenografts are the most representative model for epigenetic studies, even when cell-derived. However, primary cells are not often available, they are much more difficult to grow, and their limited lifespan, together with the necessity to use them within a certain time-frame, significantly hinders their applications, while xenografts models cannot reasonably be employed in the initial phases of discovery projects. Therefore, cell lines in some cases remain the only tool for studying epigenetic mechanisms underlying cancer and to investigate novel therapeutic options. In this regard, we have identified few breast cancer and GBM cell lines that are more similar to the original tissue and should be therefore preferred for epigenetic studies in order to increase the likelihood that the conclusions reached in vitro are transferrable to the clinic. Nevertheless, our finding strongly supports the notion that results obtained in vitro should be ultimately validated in vivo.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (61) via the PRIDE partner repository with the dataset identifier PXD007235.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Cuomo for technical support, M. Soldi for critical reading of the manuscript, F. Pisati, G. Jodice and C. Luise for preparing the samples for histological analysis, G. Bertalot for histopathological assessment of tissue samples, N. Colombo and the IEO Biobank staff for providing primary ovarian tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC) (to T.B. [15741], S.C., G. Pe, U.C.); Italian Ministry of Health [GR-2011-02347880] and the CNR-EPIGEN flagship project (to T.B.); Fondazione Umberto Veronesi (FUV) supported C.R.; Fondazione IEO-CCM supported M.L. and U.C. Funding for open access charge: AIRC—ASSOCIAZIONE ITALIANA PER LA RICERCA SUL CANCRO [15741].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 2. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Portela A., Esteller M.. Epigenetic modifications and human disease. Nat. Biotechnol. 2010; 28:1057–1068. [DOI] [PubMed] [Google Scholar]
- 4. Suva M.L., Riggi N., Bernstein B.E.. Epigenetic reprogramming in cancer. Science. 2013; 339:1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albert M., Helin K.. Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 2010; 21:209–220. [DOI] [PubMed] [Google Scholar]
- 6. D’Oto A., Tian Q.W., Davidoff A.M., Yang J.. Histone demethylases and their roles in cancer epigenetics. J. Med. Oncol. Ther. 2016; 1:34–40. [PMC free article] [PubMed] [Google Scholar]
- 7. Fraga M.F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K. et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005; 37:391–400. [DOI] [PubMed] [Google Scholar]
- 8. Elsheikh S.E., Green A.R., Rakha E.A., Powe D.G., Ahmed R.A., Collins H.M., Soria D., Garibaldi J.M., Paish C.E., Ammar A.A. et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009; 69:3802–3809. [DOI] [PubMed] [Google Scholar]
- 9. Seligson D.B., Horvath S., McBrian M.A., Mah V., Yu H., Tze S., Wang Q., Chia D., Goodglick L., Kurdistani S.K.. Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 2009; 174:1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seligson D.B., Horvath S., Shi T., Yu H., Tze S., Grunstein M., Kurdistani S.K.. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005; 435:1262–1266. [DOI] [PubMed] [Google Scholar]
- 11. Nervi C., De Marinis E., Codacci-Pisanelli G.. Epigenetic treatment of solid tumours: a review of clinical trials. Clin. Epigenet. 2015; 7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang G., Zhang S., Yazdanparast A., Li M., Pawar A.V., Liu Y., Inavolu S.M., Cheng L.. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genomics. 2016; 17(Suppl. 7):525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006; 10:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiaradonna F., Barozzi I., Miccolo C., Bucci G., Palorini R., Fornasari L., Botrugno O.A., Pruneri G., Masullo M., Passafaro A. et al. Redox-mediated suberoylanilide hydroxamic acid sensitivity in breast cancer. Antioxid. Redox. Signal. 2015; 23:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kao J., Salari K., Bocanegra M., Choi Y.L., Girard L., Gandhi J., Kwei K.A., Hernandez-Boussard T., Wang P., Gazdar A.F. et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE. 2009; 4:e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cope L.M., Fackler M.J., Lopez-Bujanda Z., Wolff A.C., Visvanathan K., Gray J.W., Sukumar S., Umbricht C.B.. Do breast cancer cell lines provide a relevant model of the patient tumor methylome. PLoS ONE. 2014; 9:e105545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vincent K.M., Findlay S.D., Postovit L.M.. Assessing breast cancer cell lines as tumour models by comparison of mRNA expression profiles. Breast Cancer Res. 2015; 17:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seidel S., Garvalov B.K., Acker T.. Isolation and culture of primary glioblastoma cells from human tumor specimens. Methods Mol. Biol. 2015; 1235:263–275. [DOI] [PubMed] [Google Scholar]
- 19. Vargo-Gogola T., Rosen J.M.. Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer. 2007; 7:659–672. [DOI] [PubMed] [Google Scholar]
- 20. Dairkee S.H., Ji Y., Ben Y., Moore D.H., Meng Z., Jeffrey S.S.. A molecular ‘signature’ of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004; 5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006; 9:391–403. [DOI] [PubMed] [Google Scholar]
- 22. Nestor C.E., Ottaviano R., Reinhardt D., Cruickshanks H.A., Mjoseng H.K., McPherson R.C., Lentini A., Thomson J.P., Dunican D.S., Pennings S. et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015; 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ortensi B., Osti D., Pellegatta S., Pisati F., Brescia P., Fornasari L., Levi D., Gaetani P., Colombo P., Ferri A. et al. Rai is a new regulator of neural progenitor migration and glioblastoma invasion. Stem Cells. 2012; 30:817–832. [DOI] [PubMed] [Google Scholar]
- 24. Speirs V., Green A.R., Walton D.S., Kerin M.J., Fox J.N., Carleton P.J., Desai S.B., Atkin S.L.. Short-term primary culture of epithelial cells derived from human breast tumours. Br. J. Cancer. 1998; 78:1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richichi C., Osti D., Del Bene M., Fornasari L., Patane M., Pollo B., DiMeco F., Pelicci G.. Tumor-initiating cell frequency is relevant for glioblastoma aggressiveness. Oncotarget. 2016; 7:71491–71503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francavilla C., Lupia M., Tsafou K., Villa A., Kowalczyk K., Rakownikow Jersie-Christensen R., Bertalot G., Confalonieri S., Brunak S., Jensen L.J. et al. Phosphoproteomics of primary cells reveals druggable kinase signatures in ovarian cancer. Cell Rep. 2017; 18:3242–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ausman J.I., Shapiro W.R., Rall D.P.. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970; 30:2394–2400. [PubMed] [Google Scholar]
- 28. Noberini R., Uggetti A., Pruneri G., Minucci S., Bonaldi T.. Pathology Tissue-quantitative mass spectrometry analysis to profile histone Post-translational modification patterns in patient samples. Mol. Cell. Proteomics. 2016; 15:866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noberini R., Longuespee R., Richichi C., Pruneri G., Kriegsmann M., Pelicci G., Bonaldi T.. PAT-H-MS coupled with laser microdissection to study histone post-translational modifications in selected cell populations from pathology samples. Clin. Epigenet. 2017; 9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cuomo A., Moretti S., Minucci S., Bonaldi T.. SILAC-based proteomic analysis to dissect the “histone modification signature” of human breast cancer cells. Amino Acids. 2011; 41:387–399. [DOI] [PubMed] [Google Scholar]
- 31. Ong S.E., Mittler G., Mann M.. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004; 1:119–126. [DOI] [PubMed] [Google Scholar]
- 32. Bremang M., Cuomo A., Agresta A.M., Stugiewicz M., Spadotto V., Bonaldi T.. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 2013; 9:2231–2247. [DOI] [PubMed] [Google Scholar]
- 33. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 34. Jung H.R., Pasini D., Helin K., Jensen O.N.. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol. Cell. Proteomics. 2010; 9:838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pesavento J.J., Mizzen C.A., Kelleher N.L.. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem. 2006; 78:4271–4280. [DOI] [PubMed] [Google Scholar]
- 36. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 37. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Supek F., Bosnjak M., Skunca N., Smuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noberini R., Bonaldi T.. A Super-SILAC strategy for the accurate and multiplexed profiling of histone posttranslational modifications. Methods Enzymol. 2017; 586:311–332. [DOI] [PubMed] [Google Scholar]
- 40. Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B.. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011; 286:7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmitges F.W., Prusty A.B., Faty M., Stutzer A., Lingaraju G.M., Aiwazian J., Sack R., Hess D., Li L., Zhou S. et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011; 42:330–341. [DOI] [PubMed] [Google Scholar]
- 42. Zheng Y., Sweet S.M., Popovic R., Martinez-Garcia E., Tipton J.D., Thomas P.M., Licht J.D., Kelleher N.L.. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13549–13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popovic R., Martinez-Garcia E., Giannopoulou E.G., Zhang Q., Ezponda T., Shah M.Y., Zheng Y., Will C.M., Small E.C., Hua Y. et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014; 10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hergeth S.P., Schneider R.. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson J.I., Decker S., Zaharevitz D., Rubinstein L.V., Venditti J.M., Schepartz S., Kalyandrug S., Christian M., Arbuck S., Hollingshead M. et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer. 2001; 84:1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Domcke S., Sinha R., Levine D.A., Sander C., Schultz N.. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013; 4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sandberg R., Ernberg I.. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc. Natl. Acad. Sci. U.S.A. 2005; 102:2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sinha R., Winer A.G., Chevinsky M., Jakubowski C., Chen Y.B., Dong Y., Tickoo S.K., Reuter V.E., Russo P., Coleman J.A. et al. Analysis of renal cancer cell lines from two major resources enables genomics-guided cell line selection. Nat. Commun. 2017; 8:15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Daniel V.C., Marchionni L., Hierman J.S., Rhodes J.T., Devereux W.L., Rudin C.M., Yung R., Parmigiani G., Dorsch M., Peacock C.D. et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009; 69:3364–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fichtner I., Rolff J., Soong R., Hoffmann J., Hammer S., Sommer A., Becker M., Merk J.. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin. Cancer Res. 2008; 14:6456–6468. [DOI] [PubMed] [Google Scholar]
- 51. Ertel A., Verghese A., Byers S.W., Ochs M., Tozeren A.. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol. Cancer. 2006; 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bonenfant D., Towbin H., Coulot M., Schindler P., Mueller D.R., van Oostrum J.. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics. 2007; 6:1917–1932. [DOI] [PubMed] [Google Scholar]
- 53. Schulze J.M., Jackson J., Nakanishi S., Gardner J.M., Hentrich T., Haug J., Johnston M., Jaspersen S.L., Kobor M.S., Shilatifard A.. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell. 2009; 35:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mellor J. Linking the cell cycle to histone modifications: Dot1, G1/S, and cycling K79me2. Mol. Cell. 2009; 35:729–730. [DOI] [PubMed] [Google Scholar]
- 55. Pollard P.J., Loenarz C., Mole D.R., McDonough M.A., Gleadle J.M., Schofield C.J., Ratcliffe P.J.. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem. J. 2008; 416:387–394. [DOI] [PubMed] [Google Scholar]
- 56. Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., Staller P.. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 2008; 283:36542–36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hancock R.L., Masson N., Dunne K., Flashman E., Kawamura A.. The activity of JmjC histone lysine demethylase KDM4A is highly sensitive to oxygen concentrations. ACS Chem. Biol. 2017; 12:1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rona G.B., Almeida D.S.G., Pinheiro A.S., Eleutherio E.C.A.. The PWWP domain of the human oncogene WHSC1L1/NSD3 induces a metabolic shift toward fermentation. Oncotarget. 2017; 8:54068–54081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 60. Doenecke D., Alonso A.. Organization and expression of the developmentally regulated H1(o) histone gene in vertebrates. Int. J. Dev. Biol. 1996; 40:395–401. [PubMed] [Google Scholar]
- 61. Vizcaino J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J.A., Sun Z., Farrah T., Bandeira N. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014; 32:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (61) via the PRIDE partner repository with the dataset identifier PXD007235.