Abstract
The vertebrate hedgehog pathway is organized in primary cilia, and hedgehog components relocate into or out of cilia during signaling. Defects in intraflagellar transport (IFT) typically disrupt ciliary assembly and attenuate hedgehog signaling. Determining whether IFT drives the movement of hedgehog components is difficult due to the requirement of IFT for building cilia. Unlike most IFT proteins, IFT27 is dispensable for cilia formation but affects hedgehog signaling similarly to other IFTs, allowing us to examine its role in the dynamics of signaling. Activating signaling at points along the pathway in Ift27 mutant cells showed that IFT is extensively involved in the pathway. Similar analysis of Bbs mutant cells showed that BBS proteins participate at many levels of signaling but are not needed to concentrate Gli transcription factors at the ciliary tip. Our analysis showed that smoothened delivery to cilia does not require IFT27, but the role of other IFTs is not known. Using a rapamycin-induced dimerization system to sequester IFT-B proteins at the mitochondria in cells with fully formed cilia did not affect the delivery of Smo to cilia, suggesting that this membrane protein may not require IFT-B for delivery.
INTRODUCTION
Primary cilia are microtubule-based hairlike sensory organelles that are crucial regulators of cell signaling. The receptors and downstream effectors of many signaling proteins are enriched and sequestered in the cilium, and this compartmentalization allows for fine temporal and spatial regulation of pathway activation as well as downstream signal propagation. In vertebrates, the best-studied cilia-linked pathway is sonic hedgehog, which plays fundamental roles during development and in adult tissue homeostasis. All the key components of the pathway are enriched in the cilium (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi and Scott, 2007; Ocbina and Anderson, 2008), and their localization changes dynamically in response to the activation of the pathway. In its off-state, patched-1 (Ptch1), the hedgehog ligand receptor, accumulates in the primary cilium and prevents ciliary accumulation and the activation of smoothened (Smo). On binding of ligand, Ptch1 is removed from the cilium and Smo is derepressed and accumulates in the cilium. Smo subsequently activates downstream signaling, which results in the accumulation of the Gli family of transcription factors at the tip of the cilium before their modification and translocation to the nucleus where they modulate target genes.
The movement of hedgehog pathway components has been well studied. Ptch1 and Smo come and go from the cilium depending on the activation status of the pathway (Corbit et al., 2005; Rohatgi and Scott, 2007; Kim et al., 2009). Gli2 and Gli3 localize to the tip of primary cilia where they accumulate to high levels on pathway activation (Haycraft et al., 2005; Kim et al., 2009; Keady et al., 2012; Santos and Reiter, 2014). SuFu localizes to the primary cilium in a Gli-dependent manner (Haycraft et al., 2005; Tukachinsky et al., 2010; Zeng et al., 2010). Kif7, like the Gli factors, localizes at the ciliary tip and accumulates when the pathway is activated (He et al., 2014). However, mechanisms underlying their trafficking to and within the cilium remain elusive. Part of these movements is facilitated by intraflagellar transport (IFT) (Keady et al., 2012; Eguether et al., 2014) and perturbing IFT disrupts hedgehog signaling (Huangfu et al., 2003; Liem et al., 2012; Duran et al., 2016). However, it is difficult to know whether IFT is participating directly in the transport of many of the components as perturbing IFT typically disrupts ciliary assembly, which could cause indirect effects. Hence, it is fundamental to experimentally disconnect IFT and primary cilia formation to better understand the function of the IFT system in hedgehog signaling. Previously, we showed that the complex B proteins (IFT25 and IFT27) are dispensable for cilia formation but are fundamental for the regulation of hedgehog signaling (Keady et al., 2012; Eguether et al., 2014), thus describing a system in which IFT and primary cilia formation are naturally uncoupled. In this study, we further examined the role of IFT27 in hedgehog signaling and compared it to the BBSome. We find that IFT27 is involved in most of the major steps of hedgehog signal transduction and unexpectedly plays roles independent of the BBSome.
Our work indicates that IFT25 and IFT27 are extensively involved in hedgehog signaling, but the question of whether other IFT proteins have direct roles in signaling remains. To test this, we designed a system to perturb IFT in fully formed cilia and asked how this affects hedgehog signaling. Our approach uses an in-cell dimerization system based on rapamycin proprieties. Rapamycin can simultaneously bind the FK506 binding protein 12 (FKBP) and the FKBP12-rapamycin binding (FRB) domain of mTOR with high affinity (Chen et al., 1995). Proteins that would not normally dimerize can be tagged with the FKBP and FRB domains and be brought together on addition of rapamycin (Bayle et al., 2006). The system is highly efficient and has been used to study receptor-mediated endocytosis (Varnai et al., 2006), clathrin-coated vesicles (Robinson et al., 2010), actin network (Castellano et al., 1999), and the gating properties of the transition zone of primary cilia (Lin et al., 2013). In our system, an IFT-B protein is tagged with the FKBP domain while the FRB domain is sequestered at the mitochondria. In the absence of rapamycin, the IFT-FKBP fusion protein participates in IFT and assembles cilia. On addition of rapamycin, IFT-B components are sequestered at the mitochondria to prevent their participation in IFT.
RESULTS
IFT27 is involved in signal propagation downstream of Smo
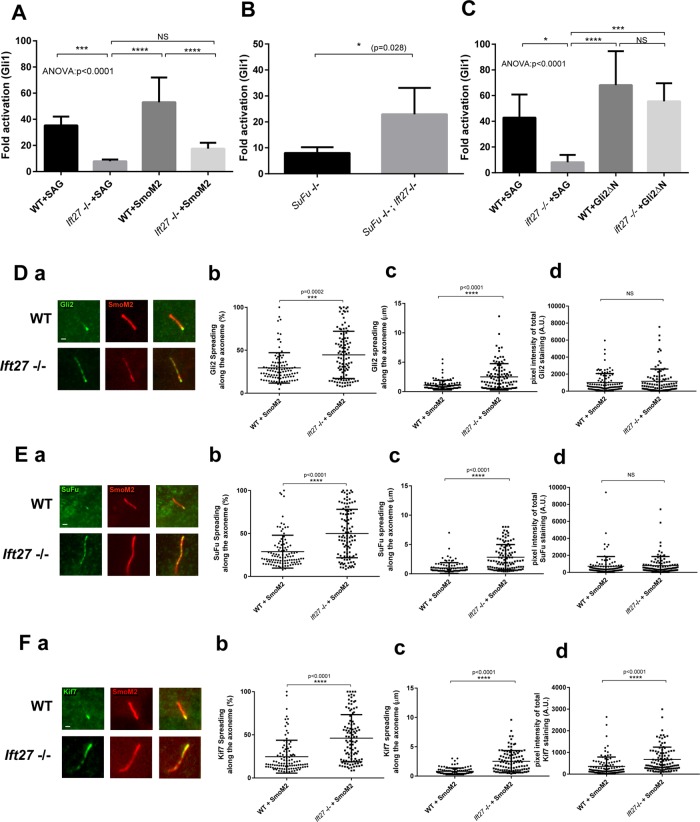
We previously showed (Eguether et al., 2014) that the loss of IFT27 attenuated hedgehog signaling and reduced the amount of Gli2 at the ciliary tip after pathway stimulation. At the molecular level, IFT27 is required to remove Smo from cilia when the pathway is inactive, suggesting that IFT27 functions in the signaling cascade at or prior to the steps regulated by Smo. To determine whether IFT27 also functions downstream of Smo, we expressed the SmoM2 constitutively active form of Smo in control and Ift27 mutant cells. As measured by Gli1 gene expression, SmoM2 is sufficient to activate the pathway in wild-type cells but is not sufficient in Ift27 mutant cells (Figure 1A). In spite of not being sufficient to activate the pathway in Ift27 mutant cells, SmoM2 rescued the ciliary levels of Gli2. However, instead of being concentrated at the ciliary tip, Gli2 distributes along the cilium when IFT27 is missing (Figure 1D and Eguether et al., 2014). Like Gli2, Kif7 and SuFu also localize to the ciliary tip, and the amount at the tip increases on pathway activation (Haycraft et al., 2005; He et al., 2014). Similarly to what we observed with Gli2, the loss of IFT27 caused both Kif7 and SuFu to spread out along the cilium. Ciliary SuFu levels are not significantly different between control and mutant cells expressing SmoM2, but ciliary Kif7 is increased in Ift27 mutant cilia (Figure 1, E and F). These observations suggest that IFT27 plays a role in transport of Gli2, SuFu, and Kif7 to the ciliary tip or in assembly of the ciliary tip compartment. Alternatively, if Gli2, Kif7 and SuFu bind BBSome components, then the spreading from the tip could be a secondary consequence of the elevated level of BBSome proteins in cilia.
FIGURE 1:
IFT27 is fully integrated in hedgehog signaling pathway. (A) qRT–PCR quantification of Gli1 expression in wild-type and Ift27 knockout cells activated by SAG or by the transfection of SmoM2. Five independent analyses were performed. (B) qRT–PCR quantification of Gli1 expression in SuFu knockout cells or SuFu, Ift27 double knockout cells. Five independent analyses were performed. (C) qRT–PCR quantification of Gli1 expression in wild-type and Ift27 knockout cells activated by SAG or by the transfection of constitutively active form of Gli2 (Gli2ΔN). Five independent analyses were performed. (D–F) Localization of hedgehog effectors in wild-type and Ift27 knockout cells. Immunofluorescence staining of Gli2 (Da), SuFu (Ea), and Kif7 (Fa). Quantification of the spreading pattern presented as percentage of the axoneme covered by the staining (Db, Eb, Fb) or length (Dc, Ec, Fc). Quantification of total fluorescence intensity for each of the cilia (Dd, Ed, Fd). In each case, ∼120 cilia were measured. Data were analyzed using one-way Kruskal–Wallis test (analysis of variance [ANOVA]) with Tukey’s multiple comparison test (A, C), two-tailed Kolmogorov–Smirnov test (B), or Mann–Whitney test (C, Db, Dc, Dd, Eb, Ec, Ed, Fb, Fc, Fd). Error bars are SD. Bars, 1 micron.
SuFu, through sequestration of the Gli transcription factors, is a major repressor of the mammalian hedgehog pathway, and its loss leads to the activation of hedgehog signaling (Svärd et al., 2006; Tukachinsky et al., 2010). As the activation of the pathway by loss of SuFu is independent of the primary cilium (Jia et al., 2009), we reasoned that a double knockout of SuFu and Ift27 would allow us to test the involvement of IFT27 in the postciliary steps of hedgehog transduction. To create double-mutant cells, we used CRISPR/CAS9 to knock out Ift27 in SuFu null cells (Supplemental Figure S1A). If the pathway downstream of SuFu is independent of IF27, then the loss of IFT27 should have no effect on the increase in Gli1 expression seen in SuFu null cells. If IFT27 acts to propagate the signal, then Gli1 expression should be reduced in the double mutants. Surprisingly, the double knockout shows a greater activation of the pathway, indicating that IFT27 is a repressor of the pathway at the level of SuFu or downstream (Figure 1B).
To test the involvement of IFT27 in the activation of target genes in the nucleus, we examined how expression of Gli2ΔN affects gene expression in Ift27 mutants. This version of Gli2 lacks the N-terminal repressor domain and constitutively activates the pathway (Tanimura et al., 1998). No differences in Gli1 expression were seen between wild-type and Ift27 mutant cells, indicating that IFT27 is dispensable for Gli2 entry in the nucleus and activation of target genes (Figure 1C).
BBSome and LZTFL1 have different roles in hedgehog signaling
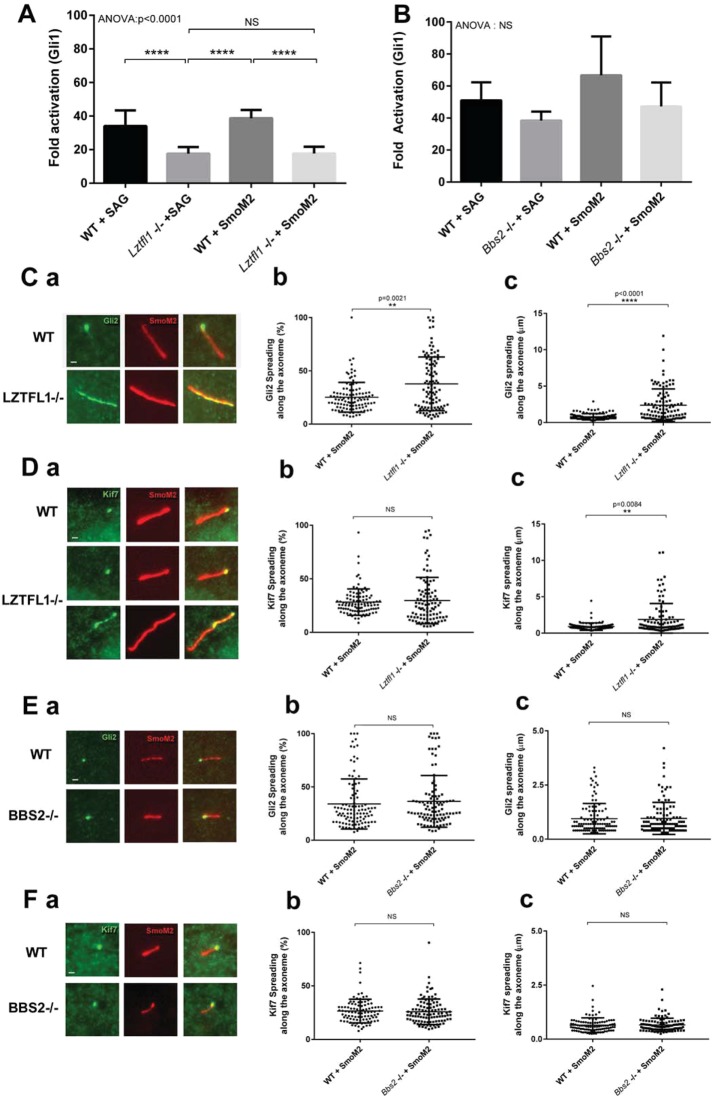
Defects in the BBSome and the BBSome regulator LZTFL1 (Seo et al., 2011; Zhang et al., 2012) cause retention of Smo in cilia of nonactivated cells like what we observe in Ift27 mutant cells. Since defects in IFT27 also disrupt the ciliary localization of the BBSome and LZTFL1, we hypothesized that IFT27 works through the BBSome and LZTFL1 to remove Smo from cilia of nonactivated cells (Eguether et al., 2014). To understand whether all the hedgehog defects seen in Ift27 mutants are due to interactions with the BBSome and LZTFL1, we directly compared the phenotypes resulting from mutations in each. Activation of the pathway by smoothened agonist (SAG) or by expression of SmoM2 showed that Gli1 expression is attenuated in Lztfl1 mutant cells, but the effect is not as large as seen in the Ift27 mutants. On the other hand, the BBSome mutant Bbs2 (Figure 2B) was less affected than either the Ift27 or Lztfl1 mutant. While the differences seen in our Bbs cells were similar to published work (75% of normal) (Zhang et al., 2012), they did not reach significance. These findings suggest that IFT27 is more integrally involved in hedgehog signaling than LZTFL1 or the BBSome.
FIGURE 2:
The BBSome and LZTFL1 play different roles in hedgehog signaling. (A) qRT–PCR quantification of Gli1 expression in wild-type and Lztfl1 knockout cells activated by SAG or by the transfection of SmoM2. (B) qRT–PCR quantification of Gli1 expression in wild-type and Bbs2 knockout cells activated by SAG or by the transfection of SmoM2. (C, D) Localization of Hedgehog effectors in wild-type and Lztfl1 knockout cells. Staining of Gli2 (Ca) and Kif7 (Da). Quantification of the spreading pattern in percentage of the axoneme covered by the staining (Cb, Db) and length (Cc, Dc). In each case, ∼120 cilia were measured. (E, F) Localization of Hedgehog effectors in wild-type and Bbs2 knockout cells. Staining of Gli2 (Ea) and Kif7 (Fa). Quantification of the spreading pattern in percentage of the axoneme covered by the staining (Eb, Fb) and length (Ec, Fc). In each case, ∼120 cilia were measured. Data were analyzed using one-way Kruskal–Wallis test (ANOVA) with Tukey’s multiple comparison test (A, B) or Mann-Whitney test (Cb, Cc, Db, Dc, Eb, Ec, Fb, Fc). Error bars are SD. Bars, 1 micron.
To more carefully examine the functions of LZTFL1 and the BBSome in hedgehog signal propagation, we expressed SmoM2 in Lztfl1 and Bbs2 mutants and quantitated the effects on ciliary Kif7 and Gli2. As observed in Ift27 mutants, Lztfl1 mutants showed expansion of the zone of Gli2 and Kif7 label down from the tip of the cilium (Figure 2, C and D). Analysis of the results is complicated by the fact that expression of SmoM2 in Lztfl1 mutant cells increased ciliary length. This caused the percentage of the cilium labeled by Kif7 to not be significantly different between mutants and controls even though the zone was physically larger in the mutants (Figure 2D, b and c). In Bbs2 cells, no difference was found between wild-type and mutant cells for either Gli2 or Kif7 staining (Figure 2, E and F).
Overall, our quantitative reverse transcription-PCR (qRT-PCR) and immunofluorescence experiments show minimal involvement of BBS2 in hedgehog signaling. LZTFL1 is important for hedgehog signaling but is less involved than IFT27. This suggests that IFT27 has BBSome-independent functions and further suggests that LZTFL1 also plays roles independent of the BBSome.
IFT27 and LZTFL1 cells are defective in Gli2/3 expression and processing
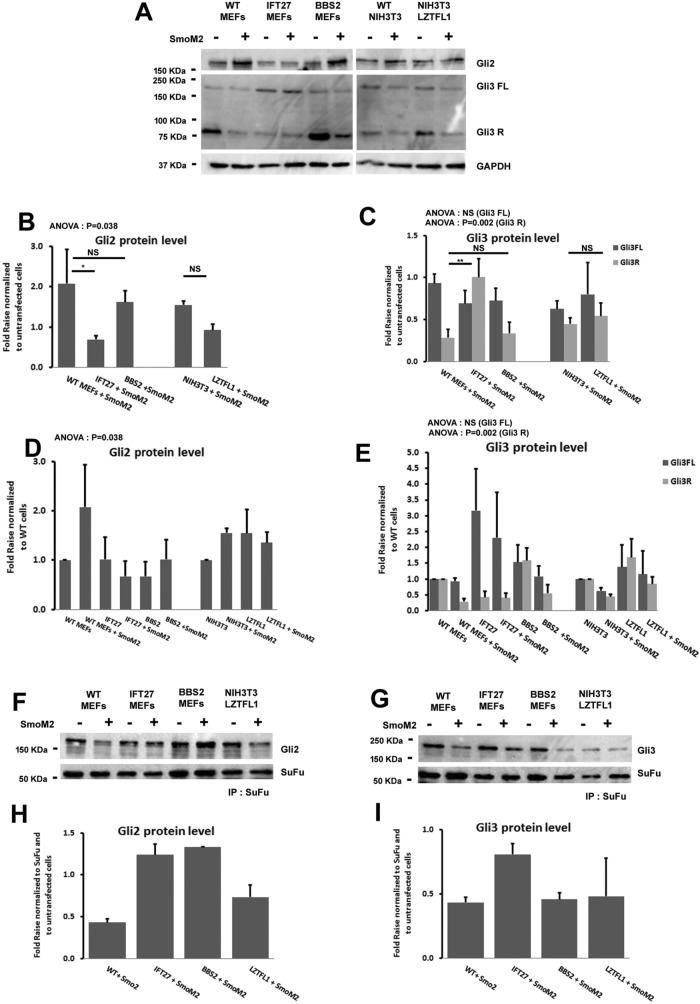
Our experiments expressing SmoM2 in Ift27 mutants suggests that IFT27 has critical functions downstream of Smo but upstream of events in the nucleus. Possible roles for IFT27 in these steps include regulating the levels of the Gli factors or regulating the conversion of the transcription factors from active to inactive forms. To test these ideas, we examined the levels of Gli2 and Gli3 in wild-type, Ift27–/–, Lztfl1–/–, and Bbs2–/– cell lines expressing SmoM2 compared with untransfected cells (Figure 3A). All values were normalized to GAPDH and then to either the SmoM2-untransfected counterparts (Figure 3, B and C) or to each of the wild-type untreated cell lines (Figure 3, D and E).
FIGURE 3:
Ift27 and Lztfl1 mutant cells are defective in Gli2/3 expression and processing. (A–C) Immunoblots of Gli2 and Gli3 in wild-type, Ift27, Bbs2, and Lztfl1 knockout cells transfected or not with SmoM2. GAPDH is the loading control. Quantification of Gli2 (B), Gli3 full length (Gli3FL), and repressor form (Gli3R) (C) expression from three experiments normalized to untransfected cells expression. (D, E) Quantification of Gli2 (D), Gli3 full length (Gli3FL), and repressor form (Gli3R) (E) expression from three experiments normalized to wild-type cells expression. (F) Wild type, Ift27. (F) Pairs of control and SmoM2-transfected cells were lysed, immunoprecipitated with SuFu antibody, and analyzed by immunoblotting with Gli2 antibody to evaluate SuFu/Gli2 binding. (G) Pairs of control and SmoM2-transfected cells were lysed, immunoprecipitated with SuFu antibody and analyzed by immunoblotting with Gli3 antibody to evaluate SuFu/Gli3 binding. (H, I) Quantification of Gli2 (H) and Gli3 full-length (I) expression from three experiments normalized to untransfected cells expression. Graphs represent three technical replicates. Error bars are SD. Data were analyzed using one-way ANOVA followed with Tukey’s multiple comparison test.
In wild-type cells, the level of Gli2 protein (the main activator at the transcription level) increases about twofold on pathway activation in mouse embryonic fibroblasts (MEFs) and ∼1.5 fold in NIH 3T3 cells (Figure 3B). Bbs2 mutants behave like wild type, consistent with our failure to detect significant defects in signaling downstream of Smo in the cells. Interestingly, Ift27–/– and Lztfl1–/– cells showed decreases in the level of basal Gli2 and did not show an increase after pathway activation (Figure 3, B and D).
In wild-type cells, Gli3 exists as a 190-kDa full-length isoform that is thought to have limited ability to activate the pathway and an 87-kDa isoform with only repressive activity (Figure 3A) (Wang and Li, 2006). In control unstimulated cells, Gli3 is mostly converted to the repressor form, which is destroyed on pathway activation. Bbs2–/– and Lztfl1–/– cells behave much like wild type. However, Ift27-/– cells have increased levels of full-length Gli3 and show little processing to the repressor form (Figure 3, C and E). If full-length Gli3 has activator function as has been suggested (Wang et al., 2007), then the increased level of Gli3 full length seen in Ift27 mutants may explain the increased activation seen in SuFu–/–, Ift27–/– cells as compared with SuFu–/– cells; however, other unknown functions could also be at play.
At the basal state, SuFu binds the full-length forms of the Gli transcription factors to stabilize and sequester to prevent activation of gene expression. On pathway activation, SuFu releases the Gli transcription factors to allow their activation and participation in transcription (Tukachinsky et al., 2010). To understand whether the loss of IFT27 altered the interactions between SuFu and Gli2 or Gli3, we precipitated SuFu from controls and cells expressing SmoM2 and quantified the amount of Gli2 and Gli3 that coprecipitates (Figure 3, F–I). As expected, activation of wild-type cells reduced the amount of Gli2 and Gli3 that bound SuFu. Lztfl1 mutants behaved like wild type for both Gli2 and Gli3. Bbs2 mutants showed normal behavior regarding Gli3 but had abnormally high levels of binding to Gli2 after pathway activation. Ift27 mutants had abnormally high binding of SuFu to both Gli2 and Gli3 after pathway activation, suggesting that IFT27 participates in the release of the Gli transcription factors from SuFu.
Development of a system to stop IFT after ciliary assembly
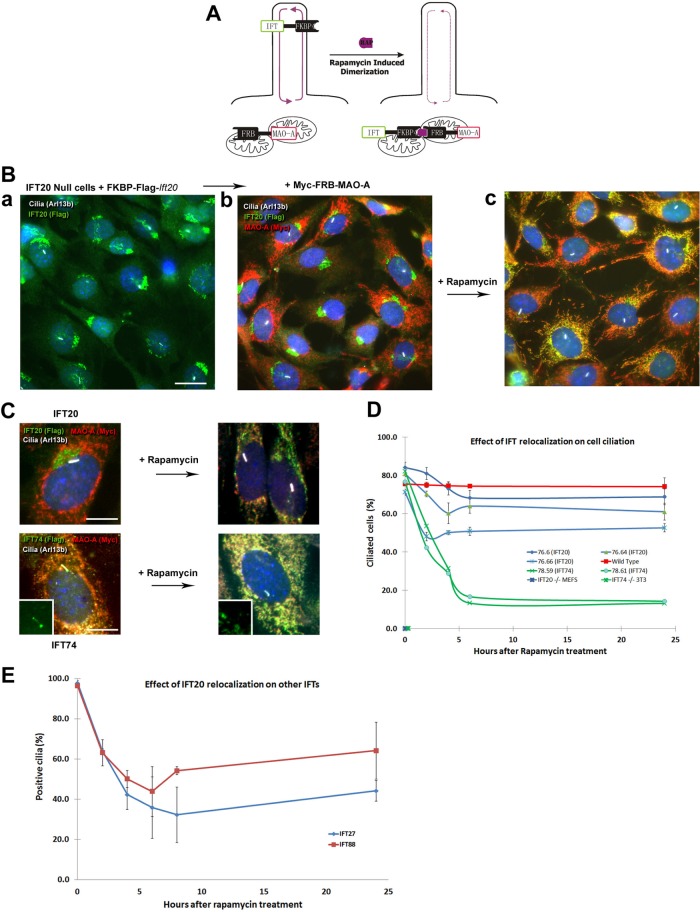
To study the role of IFT in ciliary signaling without the complication that IFT is required to build the organelle, we developed a rapamycin-dimerization approach to temporally perturb IFT. To do this, we created lines where Ift null cells were transfected with IFT-FKBP and FRB-MAO-A expression constructs. In the absence of rapamycin, the FRB-MAO-A fusion protein is sequestered at the mitochondria and the IFT-FKBP protein participates in IFT to assembly cilia. The addition of rapamycin dimerizes the FKBP and FRB domains, sequestering the IFT protein at the mitochondria and preventing its participation in IFT (Figure 4A).
FIGURE 4:
Sequestering IFTs at the mitochondria. (A) Schematic of the rapamycin-induced sequestration process. The FKBP fragment is fused to mitochondrial membrane protein monoamine oxidase A (MAO-A). The FRB fragment is fused to an IFT. Addition of rapamycin traps the IFT fusion protein in the mitochondrial compartment. (B) Ift20-null cells were rescued with an FKBP-Flag-Ift20 construct (a) and then transfected with an Myc-FRB-MAO-A construct (b). Addition of rapamycin relocates IFT20 from the Golgi complex to the mitochondria (c). Bar is 10 microns. (C) Higher-magnification images showing the relocalization of IFT20 and IFT74 by rapamycin treatment. Bar is 5 microns. (D) Quantification of percentage of ciliated cells after treatment with rapamycin in control cells, three clones expressing the IFT20-Flag-FKBP construct, and two clones expressing the IFT74-Flag-FKBP construct. In each case, ∼120 cells were counted. Parental Ift20 and Ift74 mutant fibroblasts are not ciliated. (E) Quantification of IFT27- and IFT88-positive cilia after treatment with rapamycin in IFT20 cells (clone TE79.6). In each case, ∼120 cells were counted. Error bars are SD.
For these experiments, we transfected Ift20 mutant MEFs and NIH-3T3 Ift74 mutant CRISPR cells (Supplementa Figure S1B) with TE79 (4xFKBP-3xFlag-MmIft20) and TE78 (MmIft74-3xFlag-4xFKBP), respectively. Like wild-type IFT20, the IFT20-FKBP fusion is localized at the Golgi complex while IFT74-FKBP is localized to the cilium and around the base of the cilium. Both constructs rescue the ciliation defects present in the parental lines (Figure 4, B and C). Uniformly expressing cell lines were obtained by dilution cloning and transfected with TE76 (3xMyc-FRB-MAO-A), which expresses a mitochondrial targeted FRB domain (Figure 4, B and C). After another round of dilution cloning, cell lines with functional cilia were identified by their ability to activate the hedgehog pathway and accumulate smoothened in cilia after SAG stimulation. Rapamycin treatment relocalized the IFT74-FKBP and the IFT20-FKBP to the mitochondria in both lines, which resulted in a marked depletion of the cilia pool of IFT74 and the Golgi pool of IFT20 (Figure 4C). We were unable to examine the loss of the ciliary pool of IFT20 as the epitope recognized by our antibody is blocked when IFT20 is in the cilium (Follit et al., 2006).
Sequestration of IFT20 or IFT74 leads to cilia resorption
In Chlamydomonas, temperature-sensitive IFT mutants gradually shorten their cilia starting about 2 h after shift to nonpermissive temperatures, with cilia disappearing 4–10 h after shift (Huang et al., 1977). To understand if mammalian cilia behave similarly, we followed percentage of ciliation after treatment with rapamycin. Three IFT20 and two IFT74 cell lines were chosen for analysis (Figure 4D). The three IFT20 lines showed a moderate but robust diminution of ciliation ranging from 23% for line 76.66 to 15% for line 76.6. Curiously, ciliary loss plateaued after a few hours and did not continue to decrease. Rapamycin has no effect on the percentage of ciliated wild-type cells. For IFT74, deciliation was more extensive, and both lines lost approximatively 60% of their cilia by 6 h of treatment. The difference in efficiency of deciliation between IFT74 and IFT20 may be related to the abundance of the two proteins. The plateau is not fully understood but may be caused by saturation of the FRB binding sites on the mitochondria since rapamycin binding to targets is irreversible. Line 76.6 was chosen for more study as it had robust removal of IFT20 from the Golgi complex but retained cilia the longest, giving more time for analysis before cilia loss.
If the sequestration of IFT20 or IFT74 at the mitochondria disrupts IFT, then we would expect that other IFT proteins would be lost from the cilium. To test this, we quantified the percentage of cilia positive for IFT88 and IFT27 after rapamycin treatment. At the start of the experiment, nearly all ciliated cells showed the expected staining pattern with IFT27 and IFT88 antibodies. Addition of rapamycin caused a gradual decrease in the percentage of positive cilia to ∼40% of the population before the percentage plateaued and started to rebound (Figure 4E).
Ciliary entry of Smo is independent of IFT
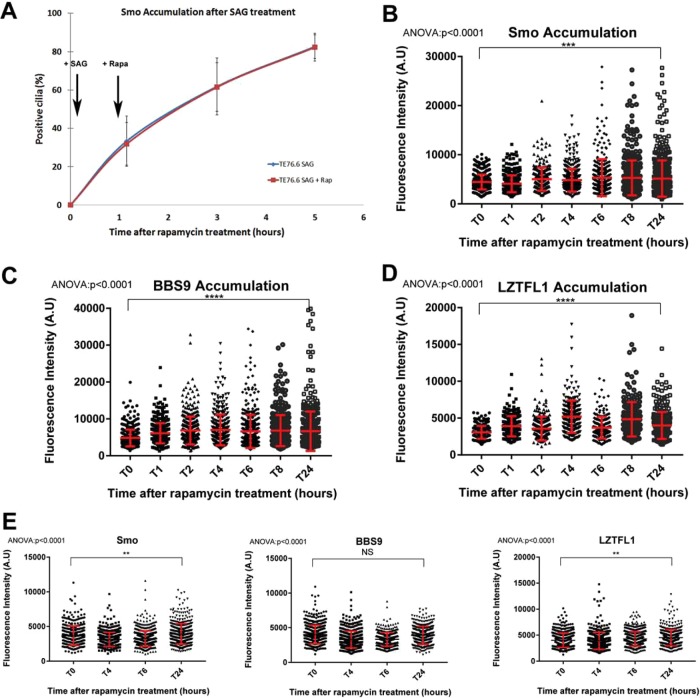
Our previous work on IFT25 and IFT27 implicated IFT in the removal of Smo from cilia. This work clearly showed that IFT25 and IFT27 were not needed for the entry of Smo but other parts of the IFT system could still be involved in Smo transport to cilia. To test this possibility, we asked how perturbing IFT would affect the delivery of Smo to cilia in response to pathway activation. We reasoned that if the IFT system was involved in Smo transport, accumulation of Smo in the cilium on SAG activation should be blocked or attenuated by addition of rapamycin to our cell system. Cells were treated with SAG for 1 h and then treated with rapamycin. Cells were fixed at intervals and ciliary Smo quantified. If IFT is required for Smo delivery, then the level of ciliary Smo should not increase as fast in the rapamycin-treated cells compared with nonrapamycin-treated cells. Quantification showed the same accumulation profile regardless of rapamycin treatment, suggesting that Smo entry into the cilium may be independent of IFT-B (Figure 5A).
FIGURE 5:
Independent entry of hedgehog effectors in the cilium. (A) Quantification of Smo-positive cilia after activation of the pathway with SAG (added at 5 min) and treatment with rapamycin (added at 1 h to half of the samples). (B) Quantification of Smo fluorescence intensity over time in rapamycin-treated unstimulated 76.6 cells. Rapamycin was added at time zero, no SAG was added. (C) Quantification of BBS9 fluorescence intensity after activation of the pathway under the same experimental paradigm as in B. (D) Quantification of LZTFL1 fluorescence intensity after activation of the pathway under the same experimental paradigm as in B. (E) Quantification of Smo, BBS9, and LZTFL1 fluorescence intensity over time in rapamycin-treated unstimulated wild-type cells. Data were analyzed using one-way ANOVA with Dunett’s multiple comparison test. In each panel, ∼300 cells were counted. Error bars are SD.
To further examine the question of IFT involvement in Smo entry, we quantified Smo accumulation in cells with IFT impaired by rapamycin but not stimulated with ligand. If Smo delivery is independent of IFT, then we expect these cells to accumulate Smo like we observed in cells missing IFT25 or IFT27 (Keady et al., 2012; Eguether et al., 2014). Over time, Smo increased in cilia on IFT-impaired cells while treatment of wild-type cells with rapamycin had no effect on ciliary Smo levels (Figure 5, B and E), thus supporting the possibility that Smo entry into the primary cilium is independent of IFT-B.
In addition to accumulating Smo, Ift27-mutant cells also accumulated BBS9 and LZTFL1. We assumed that these BBS-related components were delivered by other IFT proteins and removed by IFT25/IFT27. If this is true, then BBS components should not accumulate in cells with IFT impaired by rapamycin. Surprisingly, the treatment of our cells with rapamycin caused an accumulation of both proteins (Figure 5, C and D). Wild-type cells treated with rapamycin showed no accumulation of either BBS9 or LZTFL1 in the cilium (Figure 5E). These findings suggest that BBS9 and LZTFL1 entry in the cilium is at least partially independent of the IFT system, whereas their removal depends on IFT proteins.
DISCUSSION
IFT27 is deeply integrated into the hedgehog pathway
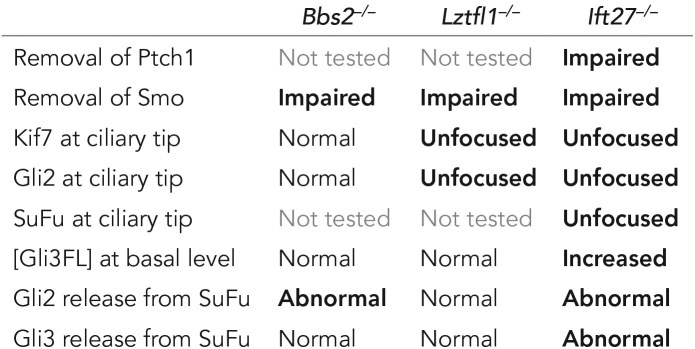
In this work, we show that Ift27 mutant cells have defects in almost every step of the hedgehog signal transduction cascade (Table 1). The defects start at the beginning with abnormal accumulation of Smo in nonactivated cilia. Farther down the pathway, we find defects in the localization of Gli2, Kif7, and SuFu at the ciliary tip. In addition, Ift27 mutants have increased levels of the full-length Gli3 transcription factor, which is not correctly processed to the repressor form in the absence of pathway stimulation. Unexpectedly, double-mutant SuFu–/–, Ift27–/– cells showed higher levels of basal Gli1 expression than single-mutant SuFu–/– cells, suggesting that this increased level of full-length Gli3 in Ift27 mutants has the ability to activate Gli1 expression if it is not repressed by SuFu.
TABLE 1:
Summary of hedgehog pathway defects.
The only step where we do not see an IFT27 effect is the activation of gene expression by truncated Gli2, suggesting that nuclear entry and transcriptional stimulation does not require IFT27. Interestingly IFT25, the obligate binding partner of IFT27, was originally identified as a nuclear protein upregulated in neuroectodermal tumors (Pozsgai et al., 2007). These tumors included medulloblastomas, which are known to be driven by upregulated hedgehog signaling (Wu et al., 2017), suggesting that there may be undiscovered roles for IFT25 and IFT27 in the nucleus.
IFT27 has BBSome-dependent and BBSome-independent roles in hedgehog signaling
As Lztfl1 and Bbs mutants share the Smo accumulation phenotype with Ift27 mutants and LZTFL1 and BBS proteins accumulate in Ift27–/– cilia, we suggested that IFT27 functions through LZTFL1 and the BBSome. Our current studies show that Ift27 and Lztfl1 but not Bbs2 mutants have defects in the localization of Kif7 and Gli2 at the ciliary tip and Ift27-mutants have increased levels of full length Gli3 not seen in Bbs or Lztfl1 mutants (Table 1). The extent of the protein’s involvement in the pathway correlates with severity of the corresponding mouse phenotypes. Ift27 mutants are most strongly affected with no animals living past the day of birth. About half of Lztfl1 mutants die in utero. The remaining survivors become obese and develop retinal degeneration (Datta et al., 2015; Jiang et al., 2016). Bbs2 mutant mice are initially smaller than littermates and develop obesity, retinal degeneration, and kidney cysts with age (Nishimura et al., 2004). While numerous hedgehog-related phenotypes are observed in Ift27 mutants, no hedgehog phenotypes such as polydactyly, cleft palate, holoprosencephaly, cardiac malformations, or lung isomerisms were reported in the Lztfl1 or Bbs2 mutant mice. These data indicate that both IFT27 and LZTFL1 have BBSome-independent functions in hedgehog signaling.
Development of a system to uncouple IFT role in ciliogenesis from signaling
Temperature-sensitive alleles of IFT components are powerful tools for uncovering the role of IFT in fully formed cilia. The difficulty in obtaining temperature-sensitive alleles in mice drove us to develop an alternative approach based on rapamycin-induced in-cell dimerization to perturb IFT in mature cilia. Previously, we had shown that Smo entry was independent of IFT25 and IFT27, and our work suggests that it may be independent of IFT-B in general. These data are consistent with published work showing that Smo accumulates in cilia on cells with defective IFT dynein without pathway activation (Ocbina and Anderson, 2008). Smo may be an unusual membrane protein, as it is thought to enter the ciliary compartment from a plasma membrane pool rather than from an internal membrane pool (Milenkovic et al., 2009). Our newly developed tool will allow us to address the role of IFT in the delivery of other membrane proteins.
Entry of the BBSome may be IFT independent
In Caenorhabditis elegans and Chlamydomonas, the BBSome is transported through cilia by IFT (Ou et al., 2005; Lechtreck et al., 2013), and it is expected to be carried into cilia by the IFT kinesin motors as part of a large complex with IFT-A and IFT-B. Unexpectedly, we found that LZTFL1 and BBS9 accumulate in cilia when we perturb IFT. Similar observations were previously reported in Chlamydomonas, where BBS4 levels initially dropped after IFT was stopped in a temperature-sensitive allele. However, once the IFT proteins were depleted from the cilia, BBS4 started to accumulate. The authors attributed the increase in ciliary BBS4 to increased numbers of cilia being loaded at later time points due to ciliary shortening after temperature shift (Lechtreck et al., 2013), but in light of our observations, this interpretation should be reexamined. The experimental approaches in the two experiments were different, but the similar observations indicate that BBSome entry into cilia is at least partially independent of IFT. One possibility is that the BBSome can independently engage kinesin-2 for entry. However, BBS4 accumulated in Chlamydomonas cilia with kinesin-2 mutations suggesting that a different motor is responsible for initial delivery.
MATERIALS AND METHODS
DNA Constructs
SmoM2-mCherry.
SmoM2 in pHAGE_DN_CMV vector (gift from D. Nedelcu and A. Salic, Harvard Medical School).
TE72 (mNeon-Gli2ΔN).
MmGli2 amplified from AA325 to the end and inserted in TE44 vector (mNeonGreen_pHAGE_DN_CMV_Nuc::eGFP) mimicking human Gli2ΔN truncation (Roessler et al., 2005).
TE85 (IFT27 CRIPSR gRNA).
IFT27 guide RNAs oligos designed in Liew et al. (2014) were annealed together and inserted in LentiCRISPRv2 plasmid (gift from Fen Zhang) (Sanjana et al., 2014) (Addgene Plasmid #52961).
TE32 (CRISPR pBSKgRNA with IFT74 oligo #5).
Candidate sgRNAs were identified by searching for G(N)20GG motifs 300 bases upstream and 100 bases downstream of the targeting sequence that conform with the nucleotide requirements for U6 Pol III transcription and the spCas9 PAM recognition element (NGG) (Jinek et al., 2012; Mali et al., 2013) using Web-based software ZiFiTtargeter 4.2 (Sander et al., 2010). Sequences generated were aligned to mouse genome using nBLAST to search for potential off-target sites. Pairs of oligonucleotides were subsequently annealed together and cloned into pBSK-gRNA (gift of R. Maehr, University of Massachusetts Medical School). IFT74 target sequence is GTTCTCGTGGTGGTCCCTTA.
TE76 (3xMyc-FRB-MAO-A in pHAGE-DN-CMV-nuc::eGFP).
The FKBP-rapamycin binding domain (FRB) fused to Mm MAO-A was PCR amplified from a CFP-FRB-MAO-A construct (gift of T. Inoue, Johns Hopkins University) with the 3xMyc tag in the 5’ primer and cloned into the lentiviral vector pHAGE-DN-CMV-nuc::eGFP.
TE78 (MmIFT74-3xFlag–4xFKBP in pHAGE-puro).
Four tandem repeats (4xFKBP) of the FK506 binding protein were PCR amplified from a vector containing IFT20-4xFKBP12 (gift of T. Inoue). IFT74-3xFlag was amplified from TE60. Both fragments were inserted in the lentiviral vector pHAGE-puro with the Gibson Assembly system (New England Biolabs).
TE79 (4xFKBP-3xFlag-MmIFT20 in pHAGE-puro).
Four tandem repeats (4xFKBP) of the FK506 binding protein and IFT20-3xFlag fragments were PCR amplified from a vector containing IFT20-4xFKBP12 (gift of T. Inoue). Both fragments were inserted in the lentiviral vector pHAGE-puro with the Gibson Assembly system (New England Biolabs).
Cell culture
Ift27 and Lztfl1 mutant cell lines are described elsewhere (Eguether et al., 2014). Bbs2 mutant MEFs were isolated from E13.5 Bbs2 mutant embryos (Nishimura et al., 2004) and immortalized with the large T antigen from SV40 virus. The SuFu mutant cell line was a gift from A. Salic. All DNA constructs were transfected into the cells via lentiviral infection followed by drug selection to create stable cell lines.
NIH3T3 fibroblasts, MEFs, and their derivatives were grown at 37°C in 5% CO2 in DMEM (Life Technologies) with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (Life Technologies). For SAG experiments, cells were plated at near-confluent densities and serum starved (same culture medium described above but with 0.25% FBS) for 48 h prior to treatment to allow ciliation. SAG (Calbiochem) was used at 400 nM.
To generate IFT74 rapamycin dimerization cells (NIH3T3 + TE32cl.33 + TE78 + TE76), NIH 3T3 were electroporated with TE32 and the Cas9-puro vector to knock out IFT74, selected with puromycin, and dilution cloned. After Western blot screening for loss of IFT74, clone 33 was selected and transfected with TE78. After blasticidin selection, cells were dilution cloned and screened for rescue of primary cilium formation and ability to activate the hedgehog pathway under SAG stimulation. Clone 71 was selected and transfected with TE76.
To generate IFT20 rapamycin dimerization cells (14176.1T MEFs + TE79 + TE76), MEFs from Ift20 mutant embryos (E9.5) were immortalized with the Large T antigen from SV40. Immortalized cells were transfected withTE79, blasticidin selected, dilution cloned, and screened for rescue of primary cilium formation and ability to activate the hedgehog pathway on SAG stimulation. Clone 16 was selected and transfected with TE76.
Cell lines were generated from mouse models or edited by CRISPR and were authenticated by genotyping loci of interest to the study. The presence of mycoplasma is monitored by 4’,6-diamidine-2’-phenylindole dihydrochloride stain.
Lentivirus production
Lentiviral packaged pHAGE-derived plasmids (Wilson et al., 2008) were used for transfection. These vectors are packaged by a third-generation system comprising four distinct packaging vectors (Tat, Rev, Gag/Pol, VSV-g) using HEK 293T cells as the host. DNA (Backbone: 5 μg; Tat: 0.5 μg; Rev: 0.5 μg; Gag/Pol: 0.5 μg; VSV-g: 1 μg) was delivered to the HEK cells using Effectene (Qiagen) or as calcium phosphate precipitates. After 48 h, supernatant was harvested, filtered through a 0.45-mm filter, and precipitated with Lenti-x concentrator (Clontech). Precipitated viral particles were resuspended in DMEM with polybrene (5 mg/ml) and added to 10–20% confluent cells. After 24 h, cells were selected with antibiotic.
Protein and mRNA analysis
For Western blots, MEFs were lysed directly into denaturing gel loading buffer (Tris-HCl, 125 mM, pH 6.8, glycerol 20% vol/vol, SDS 4% vol/vol, β-mercaptoethanol 10% vol/vol, bromophenol blue). Western blots were developed by chemiluminescence (Super Signal West Dura, Pierce Thermo) and imaged using a LAS-3000 imaging system (Fujifilm).
For immunoprecipitations, cells were serum starved for 48 h and proteins were extracted with lysis buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 1 mM MgCl2) with 0.5% digitonin and protease inhibitor (Complete EDTA-Free, Roche). Insoluble components were removed by centrifugation at 20,000 × g. Primary antibodies preadsorbed to protein G-sepharose beads (GE Healthcare) were added to the cell extract and the mixture incubated for 2 h at 4°C. After centrifugation, beads were washed with lysis buffer supplemented with 0.1% digitonin before elution in denaturing gel loading buffer for SDS–PAGE electrophoresis and Western blotting analysis.
Isolation of mRNA and quantitative mRNA analysis was performed as previously described (Jonassen et al., 2008) using the described primers (Table 2).
TABLE 2:
Quantitative RT–PCR primers.
In-cell rapamycin dimerization
IFT20 and IFT74 engineered cells were plated on glass coverslips, grown to confluence, and serum starved for 48 h with DMEM containing 0.25% FBS to allow cilia growth. To trigger dimerization of the TE76 and TE78/79 constructs, cells were treated with rapamycin (100 nM in low-serum medium) and coverslips were then fixed at desired time points.
To study hedgehog signaling in the context of IFT20 removal, cells were plated on glass coverslips, grown to confluence, and serum starved for 48 h with DMEM containing 0.25% FBS in order to allow cilia growth. Cells were treated with SAG for 1 h to activate the hedgehog pathway. To trigger dimerization of the two constructs, rapamycin was added at a final concentration of 100 nM. Coverslips were fixed at various time points.
Immunofluorescence
Cells were fixed with 2% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, and treated with 0.05% SDS for 5 min to retrieve antigens. The primary antibodies are described in Table 3.
TABLE 3:
Antibodies.
| Primary antibody | Clone name | Concentration | Supplier |
|---|---|---|---|
| Acetylated Tubulin | 6-11B-1 | 1:10,000 | Sigma |
| Flag | F1804 | 1:1000 | Sigma |
| Arl13b | 1:1000 | Davis/NIH NeuroMab | |
| Kif7 | 1:1000 | Gift from K.V. Anderson, Memorial Sloan Kettering Cancer Center | |
| SuFu | 1:500 (IF) | Gift from J. Eggenschwiler, University of Georgia, Athens | |
| SuFu | C81H7 | 1:50 (IP) | Cell Signaling |
| Gli2 | 1:1000 | Gift from J. Eggenschwiler | |
| Gli3 | AF3690 | 1:200 | RD Systems |
| Smoothened | E5 | 1:100 | Santa Cruz |
| IFT27 | 1:250 | Lab made | |
| IFT88 | 1:250 | Lab made |
Antibody verification was done by Western blot and immunofluorescence. Kif7, SuFu, Gli2, Gli3, and Smo antibodies recognized expected size proteins on Western blot, and their staining of cilia matched previously published patterns in unstimulated and hedgehog-activated cells. IFT27 and IFT88 antibodies recognized appropriate sized bands on Western blots from wild-type cells but not from corresponding mutants. By immunofluorescence IFT88 and IFT27 antibodies labeled the mother centriole or basal body in wild-type cells but not in corresponding mutants.
Quantification of fluorescence in cilia
Cilia analysis was performed using an in-house developed ImageJ macro-toolset (W.S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij/, 1997–2016). Briefly, sets of images are imported into the software, and cilia are preprocessed (median filtering and background subtraction) and then detected using the “Analyze particle function.” Each cilium is extracted using a 64 × 64 pixels bounding box. Deconvolution is performed on both the structural marker channel and the signal channel with the DeconvolutionLab plug-in (Sage et al., 2016), using a synthetic point spread function (PSF) generated by the Diffraction PSF 3D plug-in (www.optinav.info/Diffraction-PSF-3D.htm). For each structure, the following parameters are extracted: total volume, average intensity, Feret length, length of the skeleton, and ratio of both the Feret and the skeleton length (an indicator of cilium bending).
Statistics
Data groups were compared using nonparametric Mann–Whitney or Kolmogorov–Smirnov tests (two groups) or a one-way Kruskall–Wallis test (more than two groups) followed by Tukey’s or Dunett’s multiple comparison tests using GraphPad Prism 7 software. Differences between groups were considered statistically significant if p < 0.05. Statistical significance is denoted with asterisks (*p = 0.01–0.05; **p = 0.001–0.01; ***p < 0.001). Error bars are all SD. Center values are all averages.
Supplementary Material
Acknowledgments
We thank our colleagues for sharing reagents described under Materials and Methods. This work was supported by funding from National Institutes of Health GM060992 and DK103632 to G.J.P. The Bordeaux Imaging Center is a service unit of the CNRS-INSERM and Bordeaux University and member of the national infrastructure France BioImaging supported by the French National Research Agency (ANR-10-INBS-04).
Abbreviations used:
- FKBP
FK506 binding protein 12
- FRB
FKBP12-rapamycin binding
- IFT
intraflagellar transport
- MEFs
mouse embryonic fibroblasts
- SAG
smoothened agonist.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0600) on March 22, 2018.
REFERENCES
- Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. (2006). Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol , 99–107. [DOI] [PubMed] [Google Scholar]
- Castellano F, Montcourrier P, Guillemot JC, Gouin E, Machesky L, Cossart P, Chavrier P. (1999). Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr Biol , 351–360. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. (1995). Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA , 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, Reiter JF. (2005). Vertebrate smoothened functions at the primary cilium. Nature , 1018–1021. [DOI] [PubMed] [Google Scholar]
- Datta P, Allamargot C, Hudson JS, Andersen EK, Bhattarai S, Drack AV, Sheffield VC, Seo S. (2015). Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc Natl Acad Sci USA , E4400–E4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran I, Taylor SP, Zhang W, Martin J, Forlenza KN, Spiro RP, Nickerson DA, Bamshad M, Cohn DH, Krakow D. (2016). Destabilization of the IFT-B cilia core complex due to mutations in IFT81 causes a spectrum of short-rib polydactyly syndrome. Sci Rep , 34232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ. (2014). IFT27 links the bbsome to ift for maintenance of the ciliary signaling compartment. Dev Cell , 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell , 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-son Y, Zhang Q, Michaud EJ, Yoder BK. (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet , e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Kapoor TM, Anderson KV. (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol , 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Rifkin MR, Luck DJ. (1977). Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J Cell Biol , 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander LA, Anderson KV. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature , 83–87. [DOI] [PubMed] [Google Scholar]
- Jia J, Kolterud Å, Zeng H, Hoover A, Teglund S, Toftgård R, Liu A. (2009). Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol , 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Promchan K, Jiang H, Awasthi P, Marshall H, Harned A, Natarajan V. (2016). Depletion of BBS protein LZTFL1 affects growth and causes retinal degeneration in mice. J Genet Genomics , 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science , 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J, San Agustin J, Follit JA, Pazour GJ. (2008). Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol , 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ. (2012). IFT25 links the signal-dependent movement of hedgehog components to intraflagellar transport. Dev Cell , 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. (2009). Gli2 trafficking links hedgehog-dependent activation of smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA , 21666–21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, Witman GB. (2013). Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol , 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV. (2012). The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol , 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew GM, Ye F, Nager AR, Murphy JP, Lee JS, Aguiar M, Breslow DK, Gygi SP, Nachury MV. (2014). The intraflagellar transport protein ift27 promotes bbsome exit from cilia through the gtpase ARL6/BBS3. Dev Cell , 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. (2013). Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol , 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. (2013). Cas9 as a versatile tool for engineering biology. Nat Methods , 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R. (2009). Lateral transport of smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol , 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura DY, Fath M, Mullins RF, Searby C, Andrews M, Davis R, Andorf JL, Mykytyn K, Swiderski RE, Yang B, et al (2004). Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci USA , 16588–16593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina PJR, Anderson KV. (2008). Intraflagellar transport, cilia, and mammalian hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn 2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. (2005). Functional coordination of intraflagellar transport motors. Nature , 583–587. [DOI] [PubMed] [Google Scholar]
- Pozsgai E, Gomori E, Szigeti A, Boronkai A, Gallyas F, Sumegi B, Bellyei S. (2007). Correlation between the progressive cytoplasmic expression of a novel small heat shock protein (Hsp16.2) and malignancy in brain tumors. BMC Cancer , 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Sahlender DA, Foster SD. (2010). Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell , 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. (2005). A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet , 2181–2188. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Scott MP. (2007). Patching the gaps in hedgehog signalling. Nat Cell Biol , 1005–1009. [DOI] [PubMed] [Google Scholar]
- Sage D, Donati L, Soulez F, Fortun D, Schmit G, Seitz A, Guiet R, Vonesch C, Unser M. (2016). DeconvolutionLab2: an open-source software for deconvolution microscopy. Methods , 28–41. [DOI] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. (2010). ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res , W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N, Reiter JF. (2014). A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J Cell Sci , 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Zhang Q, Bugge K, Breslow DK, Searby CC, Nachury MV, Sheffield VC. (2011). A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and smoothened. PLoS Genet , e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd J, Henricson KH, Persson-Lek M, Rozell B, Lauth M, Bergström Å, Ericson J, Toftgård R, Teglund S. (2006). Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian hedgehog signaling pathway. Dev Cell , 187–197. [DOI] [PubMed] [Google Scholar]
- Tanimura A, Dan S, Yoshida M. (1998). Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome. J Virol , 3958–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A. (2010). A mechanism for vertebrate hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol , 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Thyagarajan B, Rohacs T, Balla T. (2006). Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol , 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li Y. (2006). Evidence for the direct involvement of bTrCP in Gli3 protein processing. Proc Natl Acad Sci USA , 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Rüther U, Wang B. (2007). The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev Biol , 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AA, Kwok LW, Hovav AH, Ohle SJ, Little FF, Fine A, Kotton DN. (2008). Sustained expression of alpha1-antitrypsin after transplantation of manipulated hematopoietic stem cells. Am J Respir Cell Mol Biol , 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. (2017). Hedgehog signaling: from basic biology to cancer therapy. Cell Chem Biol , 252–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Jia J, Liu A. (2010). Coordinated translocation of mammalian gli proteins and suppressor of fused to the primary cilium. PLoS One , e15900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Seo S, Bugge K, Stone EM, Sheffield VC. (2012). BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum Mol Genet , 1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.