Abstract
Phospholipid flippase (type 4 P-type ATPase) plays a major role in the generation of phospholipid asymmetry in eukaryotic cell membranes. Loss of Lem3p-Dnf1/2p flippases leads to the exposure of phosphatidylserine (PS) and phosphatidylethanolamine (PE) on the cell surface in yeast, resulting in sensitivity to PS- or PE-binding peptides. We isolated Sfk1p, a conserved membrane protein in the TMEM150/FRAG1/DRAM family, as a multicopy suppressor of this sensitivity. Overexpression of SFK1 decreased PS/PE exposure in lem3Δ mutant cells. Consistent with this, lem3Δ sfk1Δ double mutant cells exposed more PS/PE than the lem3Δ mutant. Sfk1p was previously implicated in the regulation of the phosphatidylinositol-4 kinase Stt4p, but the effect of Sfk1p on PS/PE exposure in lem3Δ was independent of Stt4p. Surprisingly, Sfk1p did not facilitate phospholipid flipping but instead repressed it, even under ATP-depleted conditions. We propose that Sfk1p negatively regulates transbilayer movement of phospholipids irrespective of directions. In addition, we showed that the permeability of the plasma membrane was dramatically elevated in the lem3Δ sfk1Δ double mutant in comparison with the corresponding single mutants. Interestingly, total ergosterol was decreased in the lem3Δ sfk1Δ mutant. Our results suggest that phospholipid asymmetry is required for the maintenance of low plasma membrane permeability.
INTRODUCTION
In eukaryotic cells, the composition of phospholipids is quite different between the exoplasmic (outer) and the cytosolic (inner) membrane leaflets of the plasma membrane. The bulk of phosphatidylcholine (PC) and sphingolipids are contained in the exoplasmic leaflet, whereas phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphoinositides are predominantly distributed in the cytosolic leaflet (Devaux, 1991; Zachowski, 1993; Murate et al., 2015). In the model membranes, phospholipid transfer between the bilayer hardly occurs spontaneously, because polar heads of phospholipids are hydrophilic, whereas the center of the bilayer is hydrophobic region. Therefore, transbilayer movement of phospholipids in cell membranes is catalyzed by three types of phospholipid translocases: flippase, floppase, and scramblase. Flippases are type 4 P-type ATPases that translocate phospholipids from the exoplasmic to cytosolic leaflet (flip) (Tanaka et al., 2011; Sebastian et al., 2012; Panatala et al., 2015). Some of the ABC transporters are reported to act as floppases that translocate phospholipids from the cytosolic to exoplasmic leaflet (flop) (Coleman et al., 2013). Scramblase catalyzes bidirectional transbilayer movement in an energy-independent manner (Montigny et al., 2016).
Flippases are implicated in the fundamental body functions including vision, hearing, and learning and their diseases (Levano et al., 2012; van der Mark et al., 2013; Coleman et al., 2014). At the cellular level, flippases in endomembranes are important for the membrane trafficking processes: endosomal trafficking (Hua et al., 2002; Pomorski et al., 2003; Furuta et al., 2007; Lee et al., 2015; Tanaka et al., 2016), biogenesis of secretory vesicles (Gall et al., 2002; Poulsen et al., 2008; Mioka et al., 2014), and protein sorting in the trans-Golgi network (TGN) (Hachiro et al., 2013; Hankins et al., 2015). On the other hand, flippases located in the plasma membrane are not only involved in the membrane trafficking (Hua et al., 2002; Pomorski et al., 2003; Furuta et al., 2007) but also participate in other various functions: apoptosis signaling (Segawa et al., 2014), mating signaling (Sartorel et al., 2015), apical membrane barrier (Paulusma et al., 2006), cell polarity (Iwamoto et al., 2004; Saito et al., 2007; Das et al., 2012), and cell migration (Kato et al., 2013).
The relevance of flippases to cellular functions has been mostly found in studies using a model organism, budding yeast. Budding yeast has five flippases: Drs2p, Dnf1p, Dnf2p, Dnf3p, and Neo1p. Drs2p, Dnf1/2p, and Dnf3p form complexes with Cdc50p, Lem3p, and Crf1p noncatalytic subunits, respectively (Saito et al., 2004; Furuta et al., 2007). These interactions are required for ER exit, proper localization, function, and translocase activity (Saito et al., 2004; Furuta et al., 2007; Lenoir et al., 2009; Takahashi et al., 2011; Puts et al., 2012). Therefore, defects in drs2Δ, dnf1Δ dnf2Δ, and dnf3Δ mutants are phenocopied by cdc50Δ, lem3Δ, and crf1Δ mutants, respectively (Saito et al., 2004; Furuta et al., 2007). Of the five flippases, only Neo1p is essential, and does not bind to any of Cdc50 family proteins (Saito et al., 2004; Furuta et al., 2007). Cdc50p–Drs2p, Crf1p–Dnf3p, and Neo1p are located primarily in the TGN and endosomes (Chen et al., 1999; Saito et al., 2004; Wicky et al., 2004; Furuta et al., 2007), while Lem3p–Dnf1/2p is mainly localized to the plasma membrane (Kato et al., 2002; Pomorski et al., 2003). Loss of the Lem3p–Dnf1/2p causes the decrease in uptake of exogenous phospholipid analogues, and the exposure of PE and PS (Kato et al., 2002; Hanson et al., 2003; Parsons et al., 2006). Lem3p–Dnf1/2p are also required for the uptake of exogenous lysophospholipids (Riekhof and Voelker, 2006; Riekhof et al., 2007).
In addition to Lem3p–Dnf1/2p, some factors affecting the phospholipid asymmetry in the plasma membrane have been identified. Serine/threonine kinases Fpk1/2p phosphorylate Dnf1/2p and up-regulate their flippase activity (Nakano et al., 2008). Two other kinases, Ypk1p and Gin4p, and sphingolipids indirectly modulate the phospholipid asymmetry through the regulation of Fpk1/2p (Roelants et al., 2010, 2015). Pdr5p and Yor1p, two multidrug ABC transporters, contribute to PE flop in the plasma membrane (Decottignies et al., 1998; Pomorski et al., 2003). Opt2p, a member of the oligopeptide transporter family, is implicated in phospholipid flop and plays a role in maintenance of phospholipid asymmetry mediated by the RIM101 pathway (Yamauchi et al., 2015).
In this study, to further understand the regulatory mechanisms and the physiological role of the phospholipid asymmetry in the plasma membrane, we searched for novel regulators of the phospholipid asymmetry and identified Sfk1p, a conserved transmembrane protein belonging to the TMEM150/FRAG1/DRAM family (Chung et al., 2015).Our results suggest an unprecedented function of Sfk1p; Sfk1p may negatively regulate the rate of spontaneous flip and flop of phospholipids in the plasma membrane. Moreover, we found that the order of phospholipid asymmetry correlated with passive permeability of the plasma membrane.
RESULTS
Sfk1p, in conjunction with Lem3p-Dnf1/2p, regulates PS and PE asymmetry in the plasma membrane
PC, PE, and PS are thought to be potential substrates for Lem3p-Dnf1/2p, but unlike nitrobenzoxadiazole (NBD)-PC and -PE, the internalization of NBD-PS occurs even in lem3Δ or dnf1Δ dnf2Δ cells (Kato et al., 2002; Saito et al., 2004; Stevens et al., 2008). To examine the contribution of Lem3p-Dnf1/2p to the flip of endogenous PS, we tested growth sensitivity to papuamide B (pap B), a depsipeptide, that binds to PS in biological membranes (Parsons et al., 2006). lem3Δ and dnf1Δ dnf2Δ exhibited high sensitivity to pap B and duramycin, a small tetracyclic peptide which binds to PE (Iwamoto et al., 2007) (Figure 1A). These results strongly suggest that Lem3p-Dnf1/2p actively translocates endogenous PS, as well as PE, in the plasma membrane. Lem3p-Dnf1/2p is recycled through the endocytic recycling pathway from the plasma membrane to the TGN via early endosomes (Saito et al., 2004; Takagi et al., 2012), and this pathway is impaired in cdc50Δ and drs2Δ cells (Saito et al., 2004; Liu et al., 2007). Therefore, the milder sensitivity of cdc50Δ and drs2Δ to pap B and duramycin may be due to reduced levels of Lem3p-Dnf1/2p in the plasma membrane due to the recycling defect (Figure 1A). Consistent with this idea, mutants defective in the endocytic recycling pathway (e.g., vps51Δ and ypt6Δ) exhibited a sensitivity to a PE-binding peptide and intracellularly accumulated Dnf2p-green fluorescent protein (GFP) (Takagi et al., 2012).
FIGURE 1:
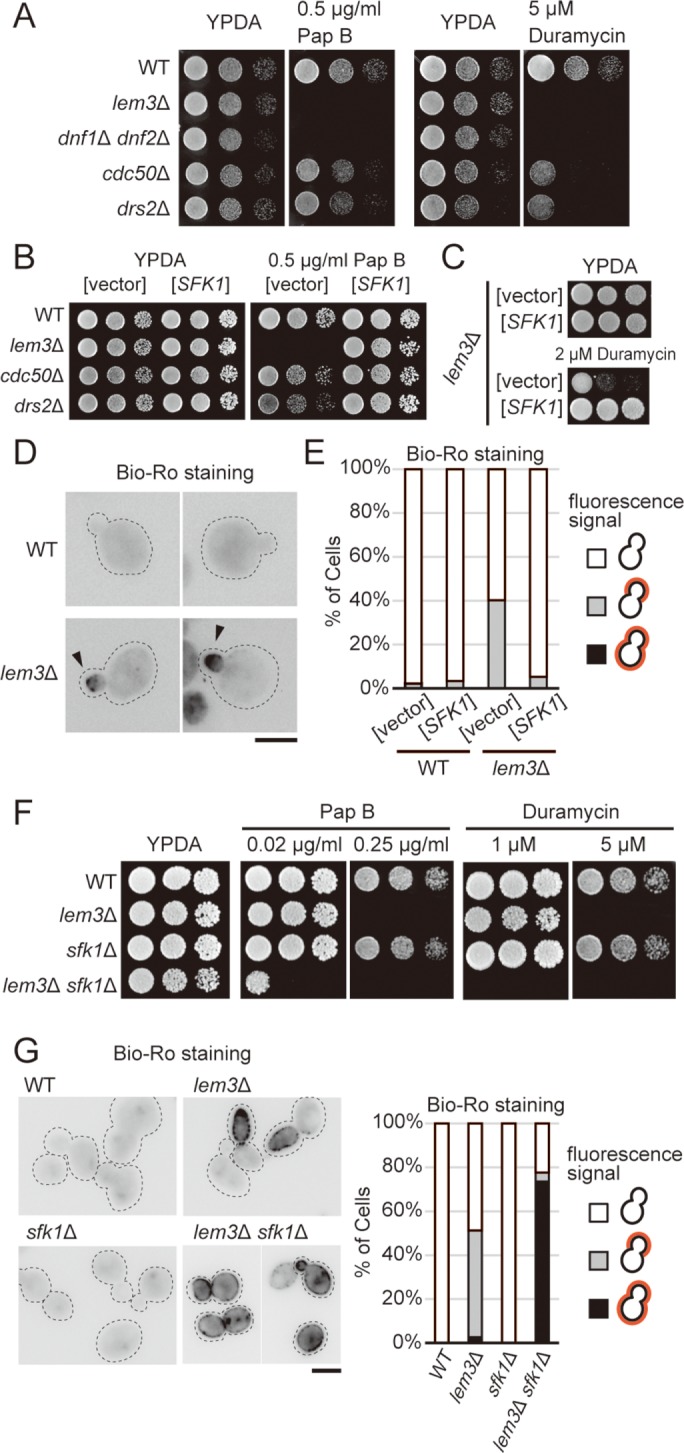
Identification of SFK1 as a multicopy suppressor of pap B sensitivity in lem3Δ mutant cells. (A) High sensitivity to pap B and duramycin in lem3Δ and dnf1Δ dnf2Δ cells. Cells were cultured in YPDA medium at 30°C, serially diluted, and spotted onto YPDA plates containing 0.5 μg/ml pap B or 5 μM duramycin, followed by incubation for 1.5 d at 30°C. For the pap B assay including (B), YKT2112 (cdc50Δ) and YKT2000 (drs2Δ) strains were used, whereas YKT249 (cdc50Δ) and YKT745 (drs2Δ) were used for the duramycin assay. (B) Suppression of pap B sensitivity in flippase mutants by overexpression of SFK1. Cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in A for 2 d at 30°C. (C) Suppression of duramycin sensitivity in lem3Δ by overexpression of SFK1. lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in A for 2 d at 30°C. (D) Staining of exposed PE in the lem3Δ mutant with biotinylated Ro-peptide (Bio-Ro) and Alexa Fluor 488–labeled streptavidin. Arrowheads indicate Alexa Fluor 488 signal on the bud tip. Dashed lines indicate cell edges. Scale bar: 5 μm. (E) PE exposure in lem3Δ is decreased by overexpression of SFK1. Wild-type and lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C and then treated with Bio-Ro. Cells were categorized into three patterns: 1) cells with no signal, 2) those with signal at the bud, and 3) those with signal all over the cell surface. Percentages for each pattern are shown (n > 100 cells). (F) Deletion of SFK1 increases sensitivity to pap B and duramycin in lem3Δ. Cell growth was examined as in A for 2-d at 30°C. (G) Deletion of SFK1 increases exposure of PE in lem3Δ. Cells were cultured in YPDA medium at 30°C, and exposed PE was visualized as in D. Dashed lines indicate cell edges. Scale bar: 5 μm. Cells were categorized as in E (n > 100 cells).
To investigate the regulatory mechanism and physiological role of PS asymmetry, we screened for multicopy suppressor genes that rescue the pap B sensitivity of the lem3Δ mutant. The screen identified SFK1 (YKL051W), which encodes a conserved plasma membrane protein of the TMEM150/FRAG1/DRAM family with six membrane-spanning domains (Audhya and Emr, 2002; Chung et al., 2015). Overexpression of SFK1 suppressed growth sensitivity to pap B in lem3Δ and cdc50Δ/drs2Δ mutants (Figure 1B) and also suppressed the duramycin sensitivity in the lem3Δ mutant (Figure 1C). Overexpression of SFK1 did not suppress growth sensitivity of wild-type cells to a high concentration of pap B or duramycin (Suppemental Figure S1A). To confirm the effect of overexpression of SFK1 to the exposure of PE in the plasma membrane, we visualized exposed PE using biotinylated Ro 09-0198 peptide (Bio-Ro), an analogue of duramycin, and Alexa Fluor 488–conjugated streptavidin (Emoto et al., 1996; Iwamoto et al., 2004; Figure 1D). We observed fluorescence signals in the bud tip of lem3Δ cells (40.3%, n > 100 cells), but these signals were significantly reduced in SFK1-overexpressing lem3Δ cells (5.2%, n > 100 cells) (Figure 1E).
Consistent with the effects of overexpression, deletion of SFK1 aggravated the growth sensitivities to pap B and duramycin in lem3Δ cells at lower concentrations of these drugs (Figure 1F). The increase in exposed PE in lem3Δ sfk1Δ cells was confirmed by Bio-Ro staining: fluorescence signals were detected all over the plasma membrane in 73.6% of lem3Δ sfk1Δ cells (n > 100 cells) (Figure 1G). Quantitative analysis of fluorescence signals suggested that ∼80% of lem3Δ sfk1Δ cells exhibited Alexa Fluor 488 signal in more than 70% of cell surface area (Supplemental Figure S1B). sfk1Δ single-mutant cells were not sensitive to either pap B or duramycin at concentrations at which lem3Δ mutant cells were sensitive (Figure 1F) and did not expose PE (Figure 1G). These results suggest that Sfk1p, in conjunction with Lem3p-Dnf1/2p, regulates the asymmetric distribution of PS and PE.
Reduced exposure of PS and PE by SFK1 overexpression is independent of phosphoinositides, ABC transporters, and phospholipase B
SFK1 was originally isolated as a multicopy suppressor of a temperature-sensitive mutation of STT4, which encodes a type III phosphatidylinositol 4-kinase. Sfk1p was implicated in the proper localization of Stt4p to the plasma membrane and Stt4p-mediated PI4P production. However, Sfk1p was not essential for either localization to the plasma membrane or kinase activity of Stt4p (Audhya and Emr, 2002). On the other hand, Ypp1p and Efr3p play essential roles in the assembly of the functional Stt4p complex at the plasma membrane (Baird et al., 2008). Thus, Sfk1p may have another function not related to Stt4p. Overexpression of neither STT4 nor MSS4, which encodes a phosphatidylinositol-4-phosphate 5-kinase (Desrivieres et al., 1998; Homma et al., 1998), rescued growth sensitivity to pap B or duramycin in the lem3Δ mutant (Figure 2A). We further examined the involvement of Stt4p in the effect of overexpression of SFK1. TMEM150A, a mammalian homologue of Sfk1p, binds to PI4KIIIα via its C-terminal cytoplasmic region (Chung et al., 2015). We constructed the SFK1ΔC mutant that lacks the C-terminal cytoplasmic region (Figure 2B), which was properly expressed and localized to the plasma membrane (Figure 2, C and D). Consistent with the results in mammalian cells, overexpression of SFK1ΔC hardly suppressed the temperature sensitivity of stt4-1 (Figure 2E). In contrast, overexpression of SFK1ΔC suppressed the pap B sensitivity of lem3Δ (Figure 2F). Furthermore, the stt4-1 mutation did not aggravate sensitivity to pap B in the lem3Δ mutant (Figure 2G). These results indicate that the effects of overexpression or deletion of SFK1 on the PS and PE exposure in the lem3Δ mutant are not mediated by Stt4p.
FIGURE 2:
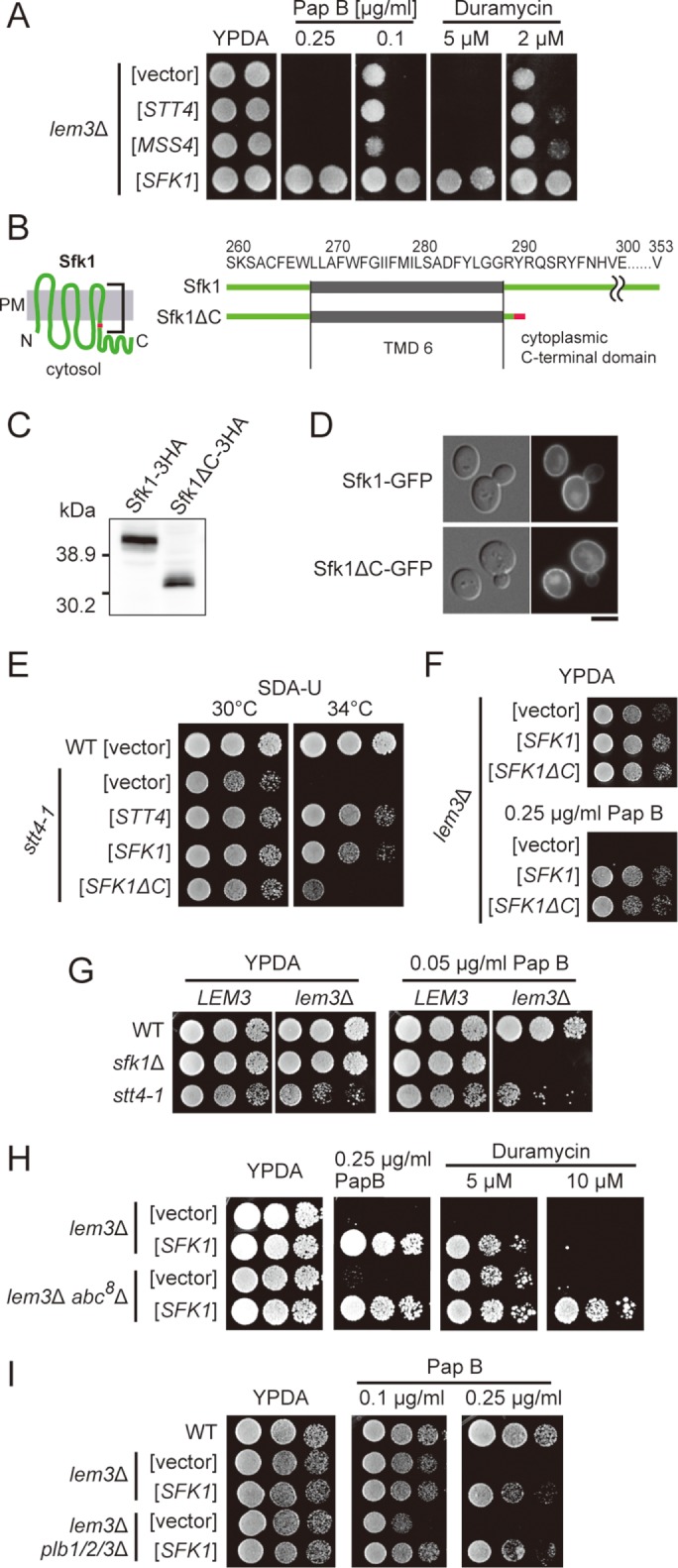
The effect of SFK1 overexpression is independent of phosphoinositides, ABC transporters, and phospholipase B. (A) Pap B and duramycin sensitivity of lem3Δ are not suppressed by overexpression of either PI 4-kinase Stt4p or PI(4)P 5-kinase Mss4p. lem3Δ cells transformed with YEp24, YEp352-STT4, YEp352-MSS4, or YEp24-SFK1 were cultured in SDA-U medium at 30°C, serially diluted, and spotted onto YPDA plates containing 0.25 or 0.1 μg/ml pap B, or 5 or 2 μM duramycin, followed by incubation for 2 d at 30°C. (B) Structures of Sfk1p and Sfk1ΔCp. Sfk1ΔCp was generated by deleting the C-terminal 64 amino acids of Sfk1p. The C-terminal amino acid of Sfk1ΔCp is shown in red for comparison. (C) Expression of Sfk1ΔCp. Wild-type cells expressing SFK1-3HA or SFK1ΔC-3HA under the control of the GAL1 promoter were cultured in YPGA for 6 h. Total lysates were prepared and separated by SDS–PAGE, followed by immunoblotting with antibody against HA. (D) Sfk1ΔCp is properly localized to the plasma membrane. Wild-type cells expressing Sfk1p-EGFP or Sfk1ΔCp-EGFP under the control of the GAL1 promoter were cultured in YPGA for 12 h, followed by fluorescence microscopic observation. Scale bar: 5 μm. (E) Overexpression of SFK1ΔC hardly suppresses the temperature-sensitive growth of stt4-1. Wild-type and stt4-1 cells transformed with YEplac195, YEp352-STT4, YEplac195-SFK1, or YEplac195-SFK1ΔC were cultured in SDA-U medium at 30°C, serially diluted, and spotted onto SDA-U plates, followed by incubation for 2 d at 30°C or 34°C. (F) Overexpression of SFK1ΔC suppresses pap B sensitivity of lem3Δ. lem3Δ cells transformed with YEplac195, YEplac195-SFK1, or YEplac195-SFK1ΔC were cultured in SDA-U medium at 30°C, and cell growth was examined as in A for 1 day at 30°C. (G) The stt4-1 mutation does not affect the sensitivity to pap B in lem3Δ cells. Cells were cultured in YPDA medium at 30°C, and cell growth was examined as in A. (H) The effect of SFK1 overexpression is independent of ABC transporters. lem3Δ and lem3Δ abc8Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in A. (I) The effect of SFK1 overexpression is independent of phospholipase Bs. lem3Δ and lem3Δ plb1/2/3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in A.
Some ABC transporters act as floppases in both mammalian cells (Aye et al., 2009) and yeast (Pomorski et al., 2003). To investigate the possibility that Sfk1p inhibits flop of PS and PE by down-regulating an ABC transporter, we created a lem3Δ abc8Δ strain deleted for all eight known plasma membrane-localized ABC transporters, YOR1, PDR5, SNQ2, PDR10, PDR11, PDR12, PDR15, and AUS1 (Jungwirth and Kuchler, 2006) (see Materials and Methods). The deletion of the ABC transporters did not suppress the pap B sensitivity in lem3Δ but did partially suppress the duramycin sensitivity, suggesting that these ABC transporters are not responsible for PS flop but may make some contribution to PE flop (Pomorski et al., 2003). Overexpression of SFK1 suppressed both pap B and duramycin sensitivity in the lem3Δ abc8Δ mutant (Figure 2H). These results suggest that ABC transporters are not involved in the reductions in PS and PE exposure on overexpression of SFK1.
We next examined the possibility that Sfk1p contributes to the degradation of exposed PS and PE. Budding yeast has three phospholipase Bs (PLBs), PLB1, PLB2, and PLB3. Yeast PLBs are localized in the periplasmic space or the cell wall and have hydrolase activity toward PC, PE, PS, and PI (Lee et al., 1994; Merkel et al., 1999). To test the possibility that Sfk1p accelerates the PLB-mediated hydrolysis of exposed PS and PE, we generated the lem3Δ plb1Δ plb2Δ plb3Δ mutant and checked its sensitivity to pap B. Deletion of PLBs slightly increased the pap B sensitivity of lem3Δ, suggesting that PLBs contribute to the hydrolysis of exposed PS to a minor extent (Figure 2I). Overexpression of SFK1 suppressed the pap B sensitivity of lem3Δ plb1Δ plb2Δ plb3Δ, suggesting that the suppression was not mediated by up-regulation of PLBs.
Phospholipid flipping is decreased by the overexpression of SFK1
In the next series of experiments, we investigated whether overexpression of SFK1 would promote PS and PE flipping. We first examined the localization of other flippases, as SFK1 overexpression might recruit another flippase to the plasma membrane in lem3Δ. Enhanced GFP (EGFP)-tagged Dnf3p, Drs2p, and Neo1p were distributed normally in punctate structures corresponding to the TGN/endosomes (Hua et al., 2002; Wicky et al., 2004), and they were not localized to the plasma membrane in SFK1-overexpressing lem3Δ cells, suggesting that the effect of SFK1 overexpression on lem3Δ cells is not mediated by these flippases (Figure 3A). We also confirmed that Dnf1p-EGFP and Dnf2p-EGFP were trapped in the ER in lem3Δ (Saito et al., 2004) on overexpression of SFK1 (Figure 3A).
FIGURE 3:
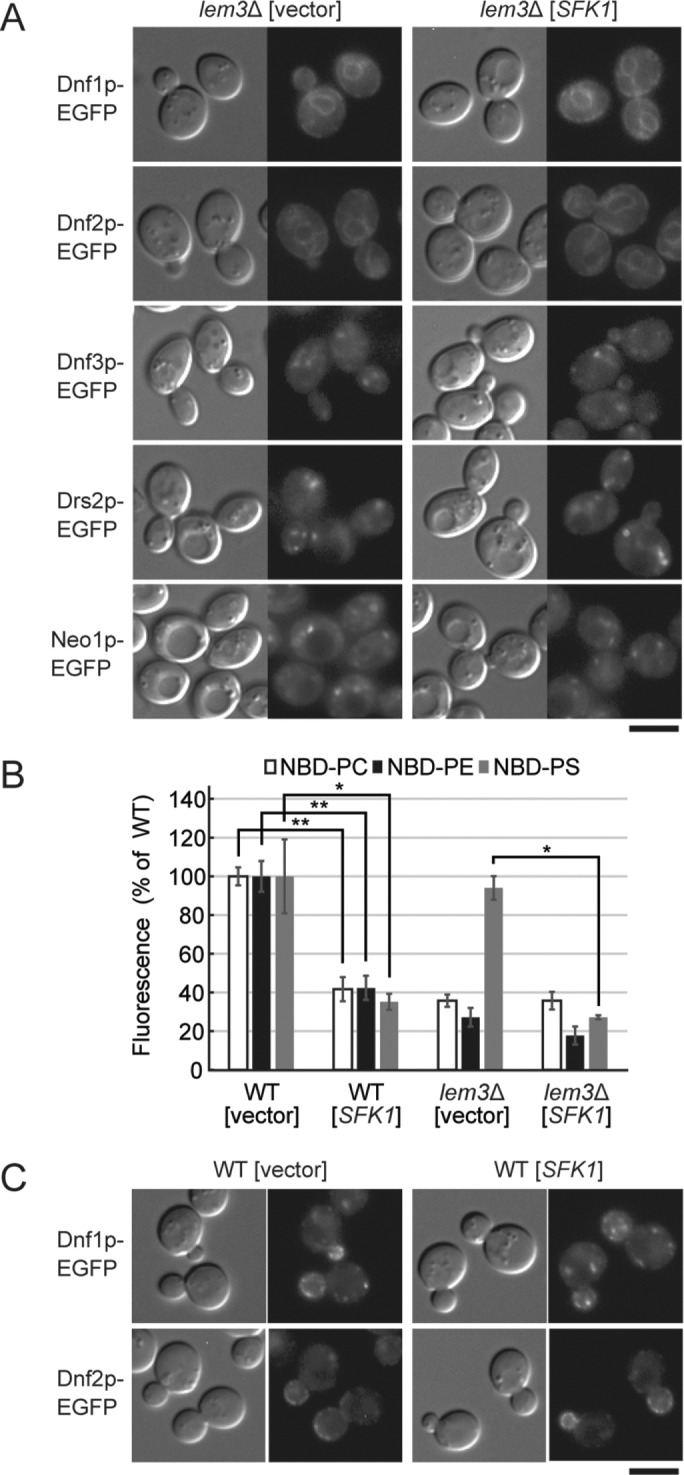
Phospholipid flipping is repressed by overexpression of SFK1. (A) Overexpression of SFK1 does not alter the localization of flippases Dnf1p, Dnf2p, Dnf3p, Drs2p, and Neo1p in lem3Δ. EGFP-tagged flippase-expressing lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C and subjected to fluorescence microscopy. Scale bar: 5 μm. (B) Overexpression of SFK1 decreases uptake of NBD-PC, -PE, and -PS. Wild-type and lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C and labeled with either NBD-PC, -PE, or -PS for 30 min at 30°C. The percentage of average accumulation of NBD-lipids relative to wild-type carrying YEp24 vector is presented with SD of three independent experiments (*p < 0.05, **p < 0.01; Tukey’s test). (C) Overexpression of SFK1 does not alter the localization of flippases Dnf1/2p. Dnf1p-EGFP- or Dnf2p-EGFP-expressing wild-type cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C and subjected to fluorescence microscopy. Scale bar: 5 μm.
We next determined the internalization of NBD-labeled phospholipids. As previously reported, the bulk of internalization of NBD-PC and -PE, but not -PS, depended on Lem3p-Dnf1/2p (Kato et al., 2002; Saito et al., 2004; Stevens et al., 2008) (Figure 3B). Considering that lem3Δ and dnf1Δ dnf2Δ were hypersensitive to pap B (Figure 1A), the internalization of NBD-PS may be achieved by an NBD-PS-specific ATP-dependent translocase other than Dnf1/2p (Stevens et al., 2008). Surprisingly, overexpression of SFK1 in wild-type cells decreased uptake of all three NBD-phospholipids by ∼60% (Figure 3B). This was not due to the internalization of Dnf1p or Dnf2p: EGFP-tagged versions of these proteins were localized normally to the plasma membrane at polarized sites in SFK1-overexpressing cells (Figure 3C). In lem3Δ cells, overexpression of SFK1 significantly decreased the NBD-PS uptake. These results rule out the possibility that Sfk1p enhances flip of PS and PE and instead suggest that Sfk1p represses phospholipid flipping.
Sfk1p may negatively regulate transbilayer movement of phospholipids in both directions
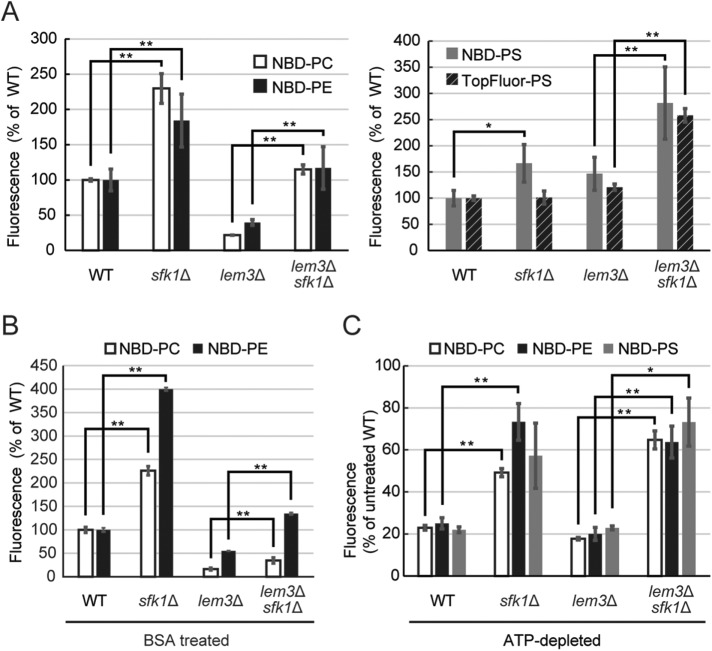
If Sfk1p represses phospholipid flip, then this repression would be released by deletion of SFK1. Consistent with this, deletion of SFK1 increased uptake of NBD-PC and -PE in wild-type and lem3Δ cells (Figure 4A, left panel). In an assay for flip of a fluorescently labeled PS, we employed TopFluor-PS, with properties more similar to PS than NBD-PS (Kay et al., 2012). TopFluor-PS, like NBD-PS, was internalized into cells in a LEM3-independent manner (Figure 4A, right panel). Deletion of SFK1 increased uptake of NBD-PS, but not of TopFluor-PS, in wild-type cells. However, deletion of SFK1 increased uptake of both NBD-PS and TopFluor-PS in the lem3Δ mutant (Figure 4A, right panel). These effects by deletion of SFK1 were also observed when cells were washed at the last step of the assay with bovine serum albumin (BSA) that extracts NBD-lipids from the outer leaflet of the plasma membrane, suggesting that Sfk1p affects flipping but not the insertion of NBD-lipids into the outer leaflet of the plasma membrane (Figure 4B). Interestingly, the increase in internalization of NBD-PC, -PE, and -PS by sfk1Δ occurred even under ATP-depleted conditions (Figure 4C). These results suggest that Sfk1p represses NBD-phospholipid uptake in a manner independent of flippases and other ATP-dependent phospholipid translocases.
FIGURE 4:
Phospholipid flipping is increased by deletion of SFK1. Increased uptake of NBD- or TopFluor-labeled phospholipids by deletion of SFK1. (A–C) Fluorescent phospholipid uptake assay was performed as described under Materials and Methods. Cells were cultured in SD medium at 30°C and labeled with each fluorescent lipid analogue for 30 min at 30°C. For assays with BSA treatment (B), cells were washed with PBS containing 2.5% BSA before flow cytometry. For assays under ATP-depleted conditions (C), cells were preincubated with 1 mM sodium azide for 30 min at 30°C. Percentage average accumulation of fluorescent lipid analogues relative to wild type in the presence of ATP is presented, with SD of three independent experiments (*p < 0.05, **p < 0.01; Tukey’s test).
To summarize the effect of SFK1, PS/PE exposure in lem3Δ cells was decreased by overexpression of SFK1, but increased by deletion of SFK1. These results seemed to be consistent with the hypothesis that Sfk1p, like flippases, enhances phospholipid flipping. However, we obtained the opposite results: internalization of NBD-phospholipids was decreased by overexpression of SFK1 but increased by deletion of SFK1. On the basis of these seemingly contradictory results, we propose an unprecedented function for SFK1, namely that Sfk1p restricts transbilayer movement of phospholipids independently of direction. That is, Sfk1p may negatively regulate both flip and flop in the plasma membrane (see Discussion). We attempted to determine the effect of Sfk1p on the flop of preinternalized NBD-phospholipids, as described previously (Hanson and Nichols, 2001). However, because overexpression or deletion of SFK1 interfered with equivalent loading of NBD-phospholipids between the tested strains, we were unable to accurately measure the flop rate.
LEM3 and SFK1 are required for maintenance of membrane impermeability
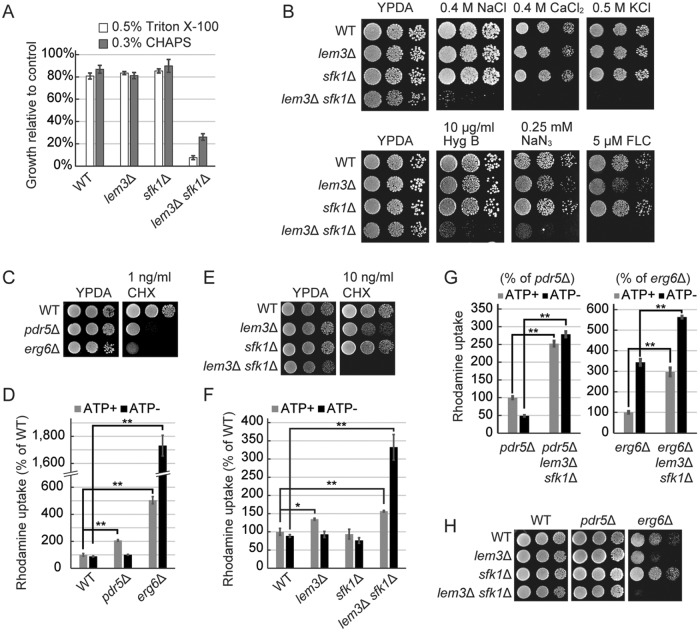
Deletion of SFK1 exacerbated the disruption of phospholipid asymmetry in lem3Δ cells. To understand the physiological significance of phospholipid asymmetry in the plasma membrane, we investigated the phenotypes of lem3Δ sfk1Δ cells. We found that lem3Δ sfk1Δ cells were highly sensitive to detergents (Figure 5A). This is consistent with a previous report that the dysfunction of ATP8B1, a P4-ATPase in mammalian cells, leads to high sensitivity to hydrophobic bile salts in liver cells (Paulusuma et al., 2006). Thus, the asymmetric distribution of phospholipids in the plasma membrane may act as a barrier to protect cells against membrane-destabilizing agents. In addition, lem3Δ sfk1Δ cells were also highly sensitive to various metal cations and cytotoxic compounds, including hygromycin B, sodium azide, and fluconazole (FLC), which have different intracellular targets (Figure 5B). To exclude the possibility that this drug sensitivity was due to weakened cell wall, we examined effects of mutations in CWP1 and CWP2, which encode cell wall mannoproteins. The cwp1Δ cwp2Δ mutant shows a decrease in cell wall thickness and an increase in the permeability to genotoxic agents (Zhang et al., 2008). The cwp1Δ cwp2Δ mutations did not confer sensitivity to 5 μM FLC and 10 ng/ml cycloheximide (CHX) on wild-type, lem3Δ, and sfk1Δ cells, whereas lem3Δ sfk1Δ cwp1Δ cwp2Δ cells were sensitive to these drugs (Supplemental Figure S2A). These results raise the possibility that the permeability of the plasma membrane is elevated in lem3Δ sfk1Δ cells.
FIGURE 5:
Plasma membrane permeability is increased in the lem3Δ sfk1Δ double mutant. (A) Sensitivity of lem3Δ sfk1Δ to detergents. Cells in exponential phase were grown in YPDA medium with or without detergents for 10 h at 30°C, and optical density at 600 nm was measured. Growth relative to detergent-free control is presented with mean ± SD from three independent experiments. (B) Sensitivity of lem3Δ sfk1Δ to metal cations and cytotoxic compounds. Cells were cultured in YPDA medium at 30°C, serially diluted, and spotted onto YPDA plates containing the indicated concentrations of divalent cations and compounds, followed by incubation for 2 d at 30°C. FLC; fluconazole. (C–F) Membrane permeability is increased in erg6Δ and lem3Δ sfk1Δ. (C, E) Sensitivity to cycloheximide. Cell growth was examined as in B. (D, F) Intracellular accumulation of rhodamine. Cells were cultured in YPDA medium at 30°C, preincubated in SD medium with or without 1 mM sodium azide for 30 min at 30°C, and incubated with rhodamine 6G for 60 min at 30°C. Rhodamine accumulation was measured as described under Materials and Methods. Percentage average accumulation of rhodamine relative to wild type in the presence of ATP is presented, with SD from three independent experiments (*p < 0.05, **p < 0.01; Tukey’s test). (G) Elevated membrane permeability in lem3Δ sfk1Δ is independent of Pdr5p and Erg6p. Intracellular accumulation of rhodamine was measured as in D and F. Percentage average accumulation of rhodamine relative to pdr5Δ or erg6Δ in the presence of ATP is presented, with SD from three independent experiments (**p < 0.01; Student’s t test). (H) lem3Δ sfk1Δ exhibits a synthetic growth defect with erg6Δ. Cell growth was examined as in B.
Emter et al. (2002) suggested that ergosterol, a major sterol in budding yeast, restricts the rate of passive diffusion across the plasma membrane. In support of this hypothesis, deletion of ERG6, an enzyme in the ergosterol biosynthetic pathway, resulted in high sensitivity to CHX and significant accumulation of rhodamine dye. This effect of ERG6 on the membrane permeability was independent of PDR5, a multidrug ABC transporter acting as an efflux pump. Consistent with the results of Emter et al. (2002), pdr5Δ and erg6Δ cells were highly sensitive to CHX (Figure 5C) but only erg6Δ cells exhibited rhodamine accumulation in the absence of ATP, confirming that energy-dependent efflux by Pdr5p was unrelated to membrane permeability (Figure 5D). We observed that lem3Δ sfk1Δ cells exhibited high sensitivity to CHX and an ATP-independent accumulation of rhodamine (Figure 5, E and F). Further, the effect of lem3Δ sfk1Δ on rhodamine accumulation was additive to the effects of pdr5Δ and erg6Δ (Figure 5G). These results suggest that membrane permeability is increased by simultaneous deletion of LEM3 and SFK1.
Rhodamine accumulation and sensitivity to cytotoxic compounds, including CHX, did not differ significantly between wild-type and sfk1Δ cells but were slightly higher in lem3Δ cells and highest in lem3Δ sfk1Δ cells (Figure 5, B, E, and F). The pattern of these phenotypes is similar to that of pap B/duramycin sensitivity (Figure 1F), which led us to hypothesize that disruption of phospholipid asymmetry causes an increase in membrane permeability. Interestingly, lem3Δ sfk1Δ exhibited a synthetic growth defect with erg6Δ, but not pdr5Δ (Figure 5H), reflecting the functional relationship between phospholipid asymmetry and ergosterol in the plasma membrane.
Overexpression of SFK1 suppresses the increase in membrane permeability in lem3Δ cells
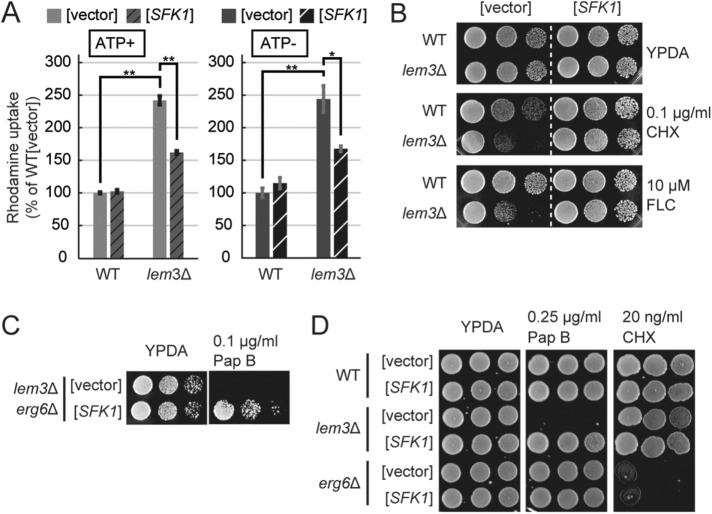
We found that the lem3Δ cells grown in synthetic medium accumulated higher levels of rhodamine than those grown in rich medium (Figure 5F), even under ATP-depleted conditions (Figure 6A). Next, we tested the effect of overexpression of SFK1 on membrane permeability in the lem3Δ mutant. Overexpression of SFK1 did not change rhodamine uptake in wild-type cells but suppressed the increase in rhodamine accumulation in lem3Δ cells, even under ATP-depleted conditions (Figure 6A). Furthermore, the sensitivities of lem3Δ cells to CHX and FLC were also suppressed by overexpression of SFK1 (Figure 6B). This suppression occurred in lem3Δ cwp1Δ cwp2Δ triple mutant cells (Supplemental Figure S2B), whereas overexpression of SFK1 did not suppress sensitivity to calcofluor white (CFW), which binds to cell wall chitin, in the lem3Δ cells (Supplemental Figure S2C). These results are consistent with our notion that the increase in membrane permeability is caused by perturbed phospholipid asymmetry in the lem3Δ mutant.
FIGURE 6:
Suppression of elevated membrane permeability in lem3Δ by overexpression of SFK1. (A) Overexpression of SFK1 reduces the influx of rhodamine into lem3Δ cells in an ATP-independent manner. Wild-type and lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, preincubated in SDA-U medium with or without 1 mM sodium azide for 30 min at 30°C, and incubated with rhodamine 6G for 60 min at 30°C. Percentage average accumulation of rhodamine relative to wild-type carrying YEp24 vector is presented, with SD of three independent experiments (*p < 0.05, **p < 0.01; Tukey’s test). (B) Suppression of sensitivity to CHX and FLC in lem3Δ by overexpression of SFK1. Wild-type and lem3Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, serially diluted, and spotted onto YPDA plates containing 0.1 μg/ml CHX or 10 μM FLC, followed by incubation for 2 d at 30°C. (C) Sfk1p is functional in the erg6Δ background. lem3Δ erg6Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in B. (D) Overexpression of SFK1 does not suppress the CHX sensitivity in erg6Δ. Wild-type, lem3Δ, and erg6Δ cells transformed with YEp24 or YEp24-SFK1 were cultured in SDA-U medium at 30°C, and cell growth was examined as in B.
We next examined the effect of SFK1 in erg6Δ cells. Overexpression of SFK1 suppressed the pap B sensitivity of lem3Δ erg6Δ, suggesting that Sfk1p was functional in the plasma membrane with accumulated intermediates in ergosterol biosynthesis (Figure 6C). However, the sensitivity of erg6Δ to CHX was not suppressed by overexpression of SFK1, and, consistent with this, erg6Δ did not exhibit high sensitivity to pap B (Figure 6D). These results suggest that the increase in membrane permeability in erg6Δ is not associated with perturbation of phospholipid asymmetry.
Fluidity of the plasma membrane is increased in the lem3Δ sfk1Δ mutant
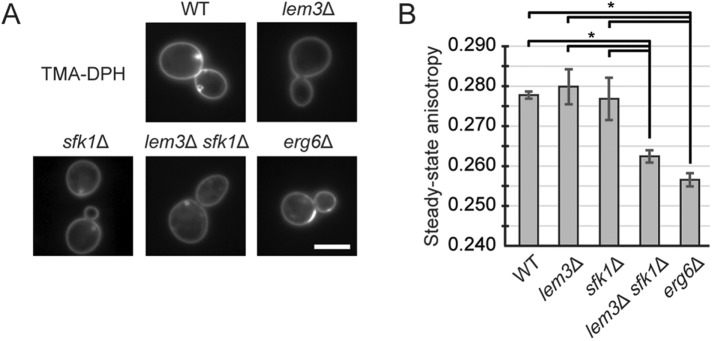
Membrane permeability to most nonionic substances is correlated to membrane fluidity (Lande et al., 1995). Membrane fluidity can be assayed by fluorescence anisotropy using a fluorescent probe, trimethylammonium diphenylhexatriene (TMA-DPH). TMA-DPH is anchored at the plasma membrane and does not enter into the cell due to its trimethylammonium group (Abe and Hiraki, 2009; Figure 7A). To assess the fluidity of the plasma membrane, we assayed steady-state anisotropy of intact cells with TMA-DPH. Deletion of ERG6 causes an increase in membrane permeability and membrane fluidity (Emter et al., 2002; Abe and Hiraki, 2009), which is reflected in a decrease in steady-state anisotropy (Abe and Hiraki, 2009; Figure 7B). Steady-state anisotropy was not changed in lem3Δ and sfk1Δ cells but decreased in lem3Δ sfk1Δ as well as in erg6Δ cells compared with wild-type cells (Figure 7B). These results suggest that membrane fluidity is also increased in the lem3Δ sfk1Δ mutant.
FIGURE 7:
Membrane fluidity is increased in the lem3Δ sfk1Δ double mutant. (A) TMA-DPH is retained in the plasma membrane. Cells were cultured in SD medium at 25°C and treated with 0.5 μM TMA-DPH. Stained cell suspensions were kept on ice for 45 min and subjected to fluorescence microscopy. Scale bar: 5 μm. (B) Steady-state anisotropy is decreased in the lem3Δ sfk1Δ mutant. Cells were cultured in SD medium at 25°C, treated with 0.5 μM TMA-DPH, and subjected to an analysis by spectrophotometry. Measurement of anisotropy was performed as described under Materials and Methods. Average of steady-state anisotropy is presented, with SD from three independent experiments (*p < 0.05; Tukey’s test).
Ergosterol is decreased in the lem3Δ sfk1Δ mutant
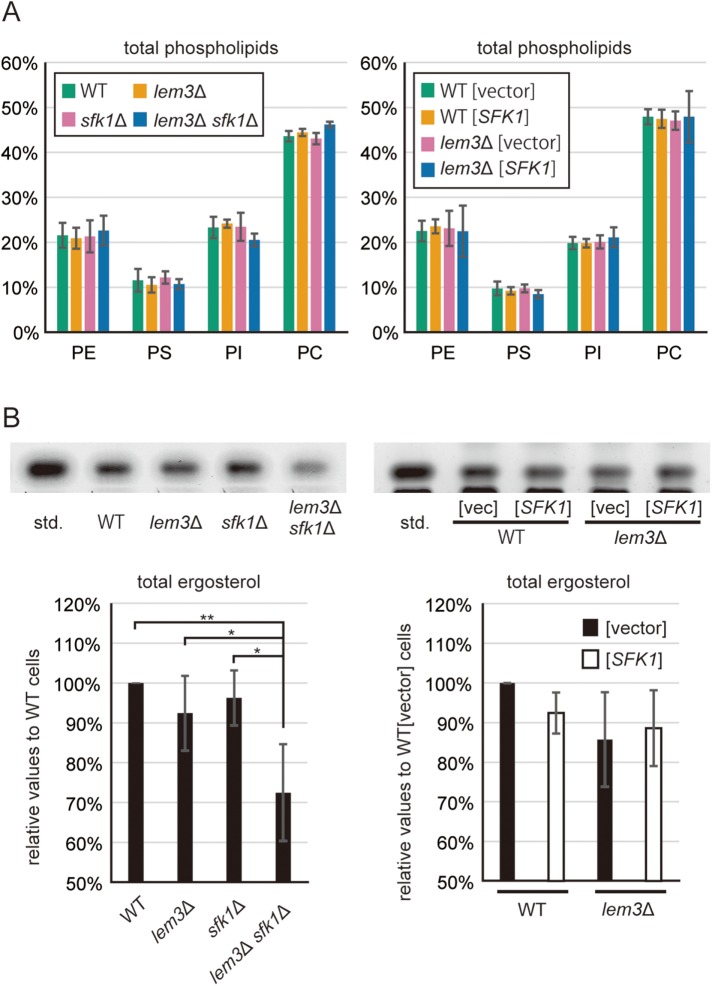
To examine whether Sfk1p affects membrane lipid composition, we measured phospholipid content in total cellular lipids of mutant cells. Neither deletion nor overexpression of SFK1 altered the phospholipid composition in wild-type and lem3Δ cells (Figure 8A). We next examined ergosterol content in mutant cells. Surprisingly, free ergosterol was decreased in lem3Δ sfk1Δ cells compared with wild-type cells (Figure 8B). Because most of the free ergosterol is found in the plasma membrane (Zinser et al., 1993), our results suggest that ergosterol is decreased in the plasma membrane of the lem3Δ sfk1Δ mutant, which is consistent with the increased permeability (Figure 5) and increased fluidity (Figure 7) of the plasma membrane in lem3Δ sfk1Δ cells. Overexpression of SFK1 did not change ergosterol levels in wild-type and lem3Δ cells (Figure 8B). These results suggest that the decreased level of ergosterol was due to severe defects in phospholipid asymmetry caused by lem3Δ sfk1Δ double mutations rather than to a specific effect of Sfk1p on ergosterol levels.
FIGURE 8:
Ergosterol is decreased in the lem3Δ sfk1Δ mutant. (A) Total phospholipid composition was not altered by either deletion or overexpression of SFK1. Cells were grown in YPDA or SDA-U medium, followed by extraction of total cellular lipids. Phospholipid contents were analyzed by TLC as described under Materials and Methods. The data present percentages of total phospholipids with SD of three independent experiments. (B) Ergosterol is decreased in lem3Δ sfk1Δ cells but not altered in SFK1-overexpressing cells. Cells were grown in YPDA or SDA-U medium, followed by extraction of total cellular lipids. Ergosterol contents were analyzed by TLC as described under Materials and Methods. Percentage average ergosterol levels relative to wild-type or wild-type carrying YEp24 vector are presented, with SD of six independent experiments (*p < 0.05, **p < 0.01; Tukey’s test). Top panel shows the scanned images of HPTLC plates.
Expression of SFK1 is up-regulated by some stresses
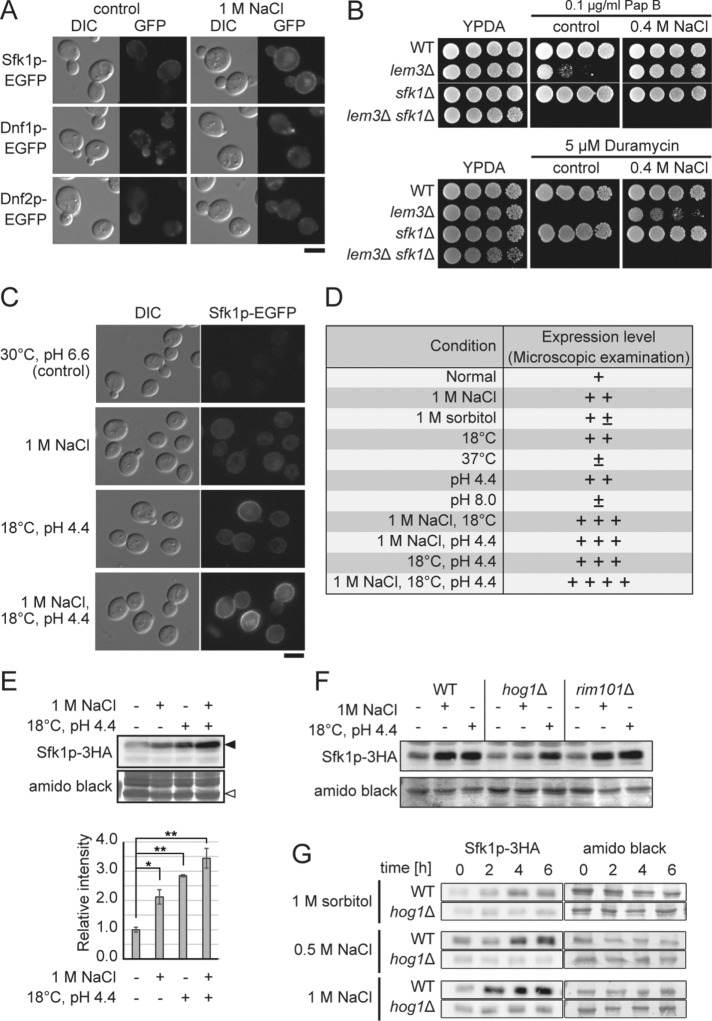
Growth of cells lacking Lem3p and Sfk1p was sensitive to NaCl (Figure 5B). We checked expressions of Dnf1p, Dnf2p, and Sfk1p under conditions of NaCl stress by fluorescent microscopy and found that expression of Sfk1p-EGFP, but not that of Dnf1/2p-EGFP, was up-regulated by 1 M NaCl (Figure 9A), which was previously reported in a genomewide transcriptome analysis (Yale and Bohnert, 2001). Consistent with this, growth sensitivity to pap B and duramycin in lem3Δ cells was suppressed by 0.4 M NaCl, as well as overexpression of SFK1 (Figure 9B). We confirmed that C-terminally tagged versions of Sfk1p were functional (Supplemental Figure S3).
FIGURE 9:
Expression of SFK1 is up-regulated by several stresses. (A) Up-regulation of Sfk1p-EGFP by 1M NaCl. Sfk1p-EGFP-, Dnf1p-EGFP-, or Dnf2p-EGFP-expressing wild-type cells were cultured in YPDA medium with or without 1 M NaCl at 30°C for 6 h in the exponential phase and subjected to fluorescence microscopy. Scale bar: 5 μm. (B) Suppression of sensitivity to pap B and duramycin in lem3Δ by NaCl. Cells were cultured in YPDA medium at 30°C, serially diluted, and spotted onto YPDA plates containing 0.1 μg/ml pap B or 5 μM duramycin with or without 0.4 M NaCl, followed by incubation for 2 d at 30°C. (C–E) Up-regulation of Sfk1p by cold temperature and low pH. (C) Sfk1p-EGFP was induced by low pH and cold temperature. Wild-type cells expressing SFK1-EGFP were cultured in YPDA medium under the indicated conditions for >12 h in the exponential phase and then subjected to fluorescence microscopy. Scale bar: 5 μm. (D) Expression of Sfk1p-EGFP was microscopically examined under various conditions, and relative intensity of GFP fluorescence is shown. (E) Wild-type cells expressing SFK1-3HA were cultured in YPDA as in C. Total lysates were prepared and separated by SDS–PAGE, followed by immunoblotting with antibody against HA. Amido black-stained membrane is shown as a loading control. Expression levels were determined by the ratio of Sfk1p-3HA (closed arrowhead) to a band in amido black control (open arrowhead). The band intensity was quantified with ImageJ. The data present average expression levels relative to control with SD of three independent experiments (*p < 0.05, **p < 0.01; Tukey’s test). (F, G) Up-regulation of Sfk1p by high osmolarity is dependent on the Hog1p pathway. (F) Cells expressing SFK1-3HA were cultured in YPDA under the indicated conditions for 8 h in the exponential phase. (G) Cells expressing SFK1-3HA were cultured in YPDA under the indicated conditions and collected at the indicated time points. SDS–PAGE and immunoblotting were performed as in E, and the band in amido black control (open arrowhead) was used as a loading control.
Up-regulation of Sfk1p-EGFP by NaCl prompted us to investigate whether Sfk1p was induced by other stresses, including high or low pH of medium (pH 8.0 and 4.4), high or low growth temperature (37° and 18°C), and high osmolarity (1 M sorbitol). We found that Sfk1p-EGFP was highly induced when low pH medium was combined with a low growth temperature (Figure 9, C and D). This induction was additive with the effect of 1 M NaCl, as confirmed by Western blotting (Figure 9, C–E). We examined whether the up-regulation of Sfk1p by stresses is mediated by the Hog1p and/or Rim101p pathways, which are involved in the response to high osmolarity and high salinity, respectively (Brewster et al., 1993; Marques et al., 2015). Although 1 M sorbitol had little effect on the expression of Sfk1p-EGFP in our microscopic examination (Figure 9D), the response to 1 M NaCl stress, but not to low pH and low temperature, was dependent on the Hog1p pathway (Figure 9F). Then we tested expression of Sfk1p in 1 M sorbitol and 0.5 M NaCl, which is equivalent to 1 M sorbitol in osmolarity. Sfk1p was induced in 1 M sorbitol as well as in 0.5 M NaCl in a Hog1p-dependent manner, but the induction level was lower compared with 1 M NaCl (Figure 9G).
We monitored growth of the lem3Δ sfk1Δ cells at 18°C and pH 4.4, but they grew normally, implying that SFK1 might be redundant with a gene other than LEM3 that is required for growth under this condition. Although we do not know how harmful NaCl, low pH, and low temperature are to the cell membrane, Sfk1p may counteract the effects of these stresses by down-regulating phospholipid translocation.
DISCUSSION
Sfk1p is a novel regulator of phospholipid asymmetry in the plasma membrane
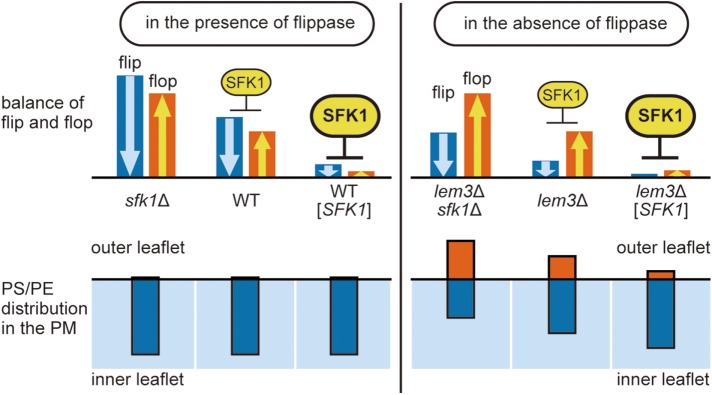
We identified Sfk1p as a novel regulator of phospholipid asymmetry in the plasma membrane. Overexpression of SFK1 repressed phospholipid flipping as well as PS/PE exposure in lem3Δ, but deletion of SFK1 exhibited opposite effects. Although we were unable to measure the flop rate due to a technical difficulty, our results are most consistent with the notion that Sfk1p represses both flip and flop. A model for Sfk1p action is depicted in Figure 10. In the presence of Lem3p-Dnf1/2p, neither deletion nor overexpression of SFK1 influences phospholipid asymmetry, because the rate of flip by Lem3p-Dnf1/2p always exceeds the rate of flop, resulting in maintenance of normal phospholipid asymmetry. On the other hand, in the absence of Lem3p-Dnf1/2p, PS and PE are exposed on the cell surface due to the reversal of flip/flop rates. This PS/PE exposure is suppressed by overexpression of SFK1 and enhanced by deletion of SFK1 through repression and disinhibition of flip and flop, respectively.
FIGURE 10:
Model for the Sfk1p action. Top panel, Blue and orange bars represent the rate of flip and flop, respectively. In this model, Sfk1p represses both flip and flop in a dose-dependent manner. Bottom panel, Blue and orange bars represent the distribution of PS/PE in the inner and outer leaflet of the plasma membrane, respectively. Deletion or overexpression of SFK1 affects phospholipid asymmetry in the absence of flippase (right) but not in the presence of flippase (left).
It remains to be elucidated how Sfk1p represses flip and flop. Importantly, the increase in NBD-lipid uptake on deletion of SFK1 occurred in an ATP-independent manner (Figure 4C). Sfk1p could affect flip/flop by inhibiting a phospholipid scramblase, but no scramblase has yet been identified in yeast. One possibility is that Sfk1p is a novel regulatory factor of spontaneous flip and flop, which are thought to occur rarely in the plasma membrane due to the tight packing of lipid bilayer and the amphipathic nature of phospholipids. In model membranes, packing defects at the phase-transition temperature were implicated in rapid flip and flop (John et al., 2002). Sterols, sphingolipids, and saturated acyl chains, all of which are abundant in the plasma membrane, are key components of tightly packed membranes (Bigay and Antonny, 2012). Sfk1p might be involved in the maintenance of packing order via regulation of the distribution, content, or interactions of these key lipids. The decrease of ergosterol in the lem3Δ sfk1Δ mutant may suggest that Sfk1p is involved in the regulation of ergosterol levels, but neither the sfk1Δ single mutation nor overexpression of SFK1 affected ergosterol contents, suggesting that the decrease in ergosterol is rather caused by the defects in phospholipid asymmetry.
Our results suggested that the effect of SFK1 on the phospholipid asymmetry is STT4 independent (Figure 2, A and E–G), but the interaction between these two proteins implies that the regulation of phospholipid asymmetry by Sfk1p is functionally relevant to PI4P signaling. Recent work showed that the C-terminal region of TMEM150A, a mammalian homologue of Sfk1p, also binds to PI4-kinase type IIIα (Chung et al., 2015). The conserved motif in the TMEM150/FRAG1/DRAM family is located in the region including six transmembrane domains except for the C-tail (Chung et al., 2015), suggesting that the interactions with PI4 kinases are specific to Sfk1p/TMEM150A.
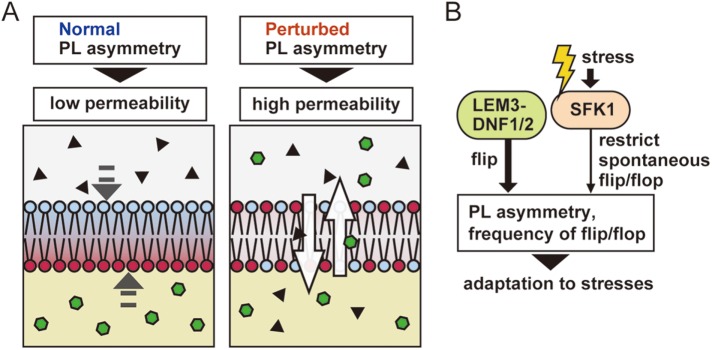
The role of phospholipid asymmetry in passive membrane permeability of the plasma membrane
Low membrane permeability is a very important property for the barrier function of the plasma membrane, specifically, the entry of exogenous toxic molecules. Our results revealed that normal phospholipid asymmetry is required for the impermeability of the plasma membrane; this is a novel finding regarding the physiological significance of the phospholipid asymmetry (Figure 11A). Loss of phospholipid asymmetry leads to a decreased level of ergosterol, which may account for the increased permeability of the plasma membrane in the lem3Δ sfk1Δ mutant.
FIGURE 11:
Physiological role and regulation of phospholipid asymmetry. (A) Contribution of phospholipid asymmetry to the barrier function of the plasma membrane. Perturbation of phospholipid asymmetry causes high permeability in the plasma membrane. (B) Sfk1p may be involved in the adaptation to environmental stresses by regulating spontaneous flip and flop.
Phospholipid asymmetry of the plasma membrane seems to play an important role for ergosterol homeostasis. In the lem3Δ sfk1Δ mutant, free ergosterol may be decreased due to increased esterification for storage or decreased synthesis. A recent study reported an association between PS and cholesterol: PS is required for retaining cholesterol in the cytosolic leaflet of the plasma membrane (Maekawa and Fairn, 2015). Although a similar interaction of PS with ergosterol has yet to be demonstrated, the abnormal distribution of ergosterol caused by the perturbation of PS asymmetry may trigger imbalance of ergosterol homeostasis in lem3Δ sfk1Δ cells. On the other hand, PC is also a potential substrate of Lem3p-Dnf1/2p (Kato et al., 2002; Pomorski et al., 2003). ATP8B1, a mammalian P4-ATPase located in the plasma membrane, preferentially flips NBD-PC (Takatsu et al., 2014). Mutations in ATP8B1 cause progressive familial intrahepatic cholestasis type 1 (PFIC1), a severe liver disease (Bull et al., 1998; Klomp et al., 2004). Interestingly, a PFIC1-type mutation in ATP8B1 leads to high sensitivity to the detergent action of hydrophobic bile salts in mouse (Paulusma et al., 2006), as well as a reduction in NBD-PC flipping (Takatsu et al., 2014). Thus, the perturbation of PC asymmetry may also bring about imbalance of ergosterol homeostasis, resulting in high membrane permeability and high detergent sensitivity in lem3Δ sfk1Δ cells.
Sfk1p is up-regulated by various stresses
We found that the protein level of Sfk1p was up-regulated by high osmolarity, low temperature, and low pH (Figure 9). The additive effects of these stresses on SFK1 expression suggest multiple regulatory pathways for SFK1, including the Hog1p pathway. Meanwhile, the protein level of Dnf1/2p was not altered by these stresses (Figure 9A and Supplemental Figure S4). The flippase activity of Dnf1/2p is regulated by serine/threonine kinases Fpk1/2p (Nakano et al., 2008), and Fpk1/2p are regulated in turn by Ypk1p and Gin4p kinases and sphingolipids (Roelants et al., 2010, 2015). Therefore, Dnf1/2p may be regulated at the level of flippase activity in response to environmental stresses.
Changes in pH and salinity of medium alter the lipid composition of the plasma membrane (Turk et al., 2007). Growth temperature and carbon source also influence the composition of membrane lipid and fatty acid (Klose et al., 2012). Our findings suggest that the inhibition of phospholipid transbilayer movement, and thus regulation of phospholipid asymmetry, plays a role in adaptation to stress conditions (Figure 11B). Investigation of the mechanisms the cell uses to adapt to these stresses will reveal novel function of phospholipid asymmetry, beyond the maintenance of low membrane permeability.
MATERIALS AND METHODS
Chemicals, media, and genetic techniques
Chemicals were purchased from Wako Pure Chemicals Industries (Osaka, Japan) unless otherwise indicated. Papuamide B was from the collection of R. Andersen. Duramycin was purchased from Sigma-Aldrich (St. Louis, MO). Standard genetic manipulations and plasmid transformation of yeast were performed as described previously (Elble, 1992; Guthrie and Fink, 2002). Yeast strains were cultured in rich yeast extract peptone dextrose adenine (YPDA) medium containing 1% yeast extract (Difco Laboratories, Detroit, MI), 2% Bacto peptone (Difco), 2% glucose, and 0.01% adenine, or synthetic dextrose (SD) medium containing 0.17% yeast nitrogen base without amino acids and ammonium sulfate (Difco), 0.5% ammonium sulfate, 2% glucose, and the required amino acids or nucleic acid bases. Strains carrying URA3-harboring plasmids were cultured in SD medium containing 0.5% casamino acids (Difco), 0.03% tryptophan, and 0.01% adenine (SDA-U). For serial dilution spot assays, cells were grown to early log phase in appropriate medium and adjusted to a concentration of 1.0 × 107 cells/ml. From 10-fold dilutions, 4-μl drops were spotted onto appropriate plates.
Strains and plasmids
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Standard molecular biological techniques (Sambrook and Russell, 2001) were used for the construction of plasmids, PCR amplification, and DNA sequencing. PCR-based procedures were used to construct gene deletions and gene fusions with EGFP, 3HA, and the GAL1 promoter (Longtine et al., 1998). All constructs produced by the PCR-based procedure were verified by colony PCR to confirm that the replacement occurred at the expected locus. Sequences of PCR primers are available on request.
TABLE 1:
Saccharomyces cerevisiae strains used in this study.
| Straina | Mating type | Genotype | Reference or source |
|---|---|---|---|
| YKT38 | a | Wild-type (lys2-801 ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1) | Misu et al. (2003) |
| YKT249 | a | cdc50Δ::HIS3MX6 | Misu et al. (2003) |
| YKT715 | a | lem3Δ::TRP1 | Saito et al. (2004) |
| YKT745 | α | drs2Δ::KanMX6 | Saito et al. (2004) |
| YKT770 | α | lem3Δ::TRP1 DRS2-EGFP::KanMX6 | Saito et al. (2004) |
| YKT771 | α | DNF1-EGFP::KanMX6 | Saito et al. (2004) |
| YKT773 | α | lem3Δ::TRP1 DNF1-EGFP::KanMX6 | Saito et al. (2004) |
| YKT918 | a | dnf1Δ::HphMX6 dnf2Δ::KanMX6 | This study |
| YKT921 | α | DNF2-EGFP::KanMX6 | Furuta et al. (2007) |
| YKT923 | α | lem3Δ::TRP1 DNF2-EGFP::KanMX6 | Furuta et al. (2007) |
| YKT927 | α | lem3Δ::TRP1 DNF3-EGFP::KanMX6 | This study |
| YKT930 | α | lem3Δ::TRP1 NEO1-EGFP::KanMX6 | This study |
| YKT1161 | a | lem3Δ::TRP1 pdr5Δ::KanMX6 | This study |
| YKT1162 | a | pdr5Δ::KanMX6 | This study |
| YKT1709 | a | stt4-1 ade8 TRP1 | Yoshida et al. (1994) |
| YKT2000 | α | drs2Δ::HphMX4 TRP1 | This study |
| YKT2112 | a | cdc50Δ::HphMX4 TRP1 | This study |
| YKT2117 | a | sfk1Δ::KanMX6 | This study |
| YKT2118 | a | lem3Δ::HIS3MX6 sfk1Δ::KanMX6 | This study |
| YKT2119 | a | erg6Δ::KanMX6 TRP1 | This study |
| YKT2120 | α | SFK1-EGFP::KanMX6 | This study |
| YKT2121 | α | lem3Δ::CaLEU2 plb1Δ::HIS3MX6 plb2Δ::KanMX6 plb3Δ::HphMX6 | This study |
| YKT2122 | a | lem3Δ::HIS3MX6 stt4-1 TRP1 | This study |
| YKT2123 | α | lem3Δ::HIS3MX6 yor1Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 pdr10Δ::TRP1 pdr11Δ::TRP1 pdr12Δ::TRP1 pdr15Δ::TRP1 aus1Δ::TRP1 | This study |
| YKT2124 | a | yor1Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 | This study |
| YKT2125 | a | lem3Δ::HIS3MX6 yor1Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 | This study |
| YKT2126 | a | lem3Δ::TRP1 sfk1Δ::HphMX6 | This study |
| YKT2129 | a | lem3Δ::TRP1 pdr5Δ::KanMX6 sfk1Δ::HphMX6 | This study |
| YKT2130 | α | lem3Δ::HISMX6 erg6Δ::TRP1 | This study |
| YKT2131 | a | sfk1Δ::HphMX6 erg6Δ::TRP1 | This study |
| YKT2132 | α | lem3Δ::HIS3MX6 sfk1Δ::HphMX6 erg6Δ::TRP1 | This study |
| YKT2133 | a | SFK1-3HA::KanMX6 | This study |
| YKT2134 | a | hog1Δ::KanMX6 SFK1-3HA::HIS3MX6 | This study |
| YKT2135 | a | rim101Δ::KanMX6 SFK1-3HA::HIS3MX6 | This study |
| YKT2136 | a | HIS3MX6::PGAL1-SFK1-3HA:: KanMX6 | This study |
| YKT2137 | a | HIS3MX6::PGAL1-SFK1ΔC-3HA:: KanMX6 | This study |
| YKT2138 | a | HIS3MX6::PGAL1-SFK1-GFP:: KanMX6 | This study |
| YKT2139 | a | HIS3MX6::PGAL1-SFK1ΔC-GFP:: KanMX6 | This study |
| YKT2140 | a | cwp1Δ::HIS3MX6 cwp2Δ::KanMX6 | This study |
| YKT2141 | α | lem3Δ::TRP1 cwp1Δ::HIS3MX6 cwp2Δ::KanMX6 | This study |
| YKT2142 | a | sfk1Δ::hphMX6 cwp1Δ::HIS3MX6 cwp2Δ::KanMX6 | This study |
| YKT2143 | a | lem3Δ::TRP1 sfk1Δ::hphMX6 cwp1Δ::HIS3MX6 cwp2Δ::KanMX6 | This study |
| YKT2144 | α | lem3Δ::TRP1 SFK1-EGFP::KanMX6 | This study |
| YKT2145 | a | lem3Δ::TRP1 SFK1-3HA::KanMX6 | This study |
aYKT strains are isogenic derivatives of YEF473 (Longtine et al., 1998). Only relevant genotypes are described.
Table 2:
Plasmids used in this study.
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| YEp24 | URA3 2μm | Carlson and Botstein (1982) |
| pKT1724 [YEp24-SFK1] | SFK1 URA3 2μm | This study |
| pKT1721 [YEp352-STT4] | STT4 URA3 2μm | A gift from Yoshikazu Ohya (The University of Tokyo) |
| pKT1722 [YEp352-MSS4] | MSS4 URA3 2μm | A gift from Yoshikazu Ohya |
| pKT2193 [YEplac195-SFK1] | SFK1 URA3 2μm | This study |
| pKT2194 [YEplac195-SFK1ΔC] | SFK1ΔC URA3 2μm | This study |
A multicopy plasmid carrying SFK1ΔC, in which the C-terminal 64 amino acids of Sfk1p were deleted, was constructed as follows. Two DNA fragments, which contained the SFK1 promoter region and SFK1ΔC coding region (1374 base pairs) and the SFK1 terminator region (478 base pairs), respectively, were PCR amplified with the genomic DNA of wild-type cells (YKT38) as a template. These DNA fragments were ligated into the YEplac195 vector plasmid. Schemes detailing the construction of plasmids and DNA sequences of nucleotide primers are available on request.
To create the lem3Δ abc8Δ strain in which all eight known plasma membrane-localized ABC transporters were deleted, the lem3Δ::HIS3MX6 yor1Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 strain (lem3Δ abc3Δ) was first generated by crossing the lem3Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 strain with the yor1Δ::HIS3MX6 pdr5Δ::HphMX6 snq2Δ::KanMX6 strain. lem3Δ::HIS3MX6 and yor1Δ::HIS3MX6 were confirmed by colony PCR. Next, PDR10, PDR11, PDR12, PDR15, and AUS1 were individually disrupted in the lem3Δ abc3Δ strain using the TRP1 disruption cassette. Crosses among these five strains, followed by sporulation and tetrad dissection, yielded strains containing combinations of the disrupted alleles, which were confirmed by colony PCR. This procedure was performed repeatedly, ultimately yielding the lem3Δ abc8Δ strain.
Isolation of multicopy suppressors of papuamide B sensitivity in the lem3Δ mutant
The lem3Δ strain was transformed with a yeast genomic DNA library constructed in the multicopy plasmid YEp24 (Carlson and Botstein, 1982). A total of 12,000 transformants were selected on SDA-U plates at 30°C and then replica-plated onto YPDA plates containing 0.25 μg/ml pap B. The transformants that contained LEM3 on the plasmid were identified by colony PCR and eliminated. Plasmids were recovered from the remaining 51 transformants. Restriction enzyme digestion of the plasmids indicated that 21 different clones had been isolated, and they were grouped into seven different genomic regions by DNA sequencing. Fragment subcloning revealed that SFK1, PDR3, PDR1, PLB3, YLR278C, KIC1, and HLR1 were responsible for the suppression. Among these genes, SFK1 was the strongest suppressor of pap B sensitivity in the lem3Δ mutant. PDR1, YLR278C, KIC1, and HLR1 exhibited modest suppression, whereas PDR3 and PLB3 exhibited weak suppression (Supplemental Figure S5).
Microscopic observations
Cells were observed using a Nikon ECLIPSE E800 microscope (Nikon Instec, Tokyo, Japan) equipped with an HB-10103AF super-high-pressure mercury lamp and a 1.4 numerical aperture 100× Plan Apo oil immersion objective lens (Nikon Instec) with appropriate fluorescence filter sets (Nikon Instec) or differential interference contrast optics. Images were acquired using a cooled digital charge-coupled devicecamera (C4742-95-12NR; Hamamatsu Photonics, Hamamatsu, Japan) and AQUACOSMOS software (Hamamatsu Photonics). GFP-tagged proteins were observed in living cells, which were grown to early to midlogarithmic phase, harvested, and resuspended in SD medium. For detection of GFP fluorescence, cells were immediately observed using a GFP bandpass filter set. Observations were compiled from the examination of at least 100 cells.
Staining of phosphatidylethanolamine with biotinylated Ro09-0198 peptide
Staining of PE in the outer leaflet of the plasma membrane was performed using the biotinylated Ro09-0198 peptide (Bio-Ro) as described (Iwamoto et al., 2004; Saito et al., 2007), with the following modifications. Bio-Ro, which was prepared essentially as described (Aoki et al., 1994), was a gift from M. Umeda (Kyoto University, Japan). Cells were cultured in YPDA or SDA-U medium at 18°C for 12 h, until reaching midlogarithmic phase. A 1-ml culture of cells at a cell density of OD600/ml = 0.4–0.6 was harvested, resuspended in 20 μl of YPDA containing 40 μM Bio-Ro, and incubated for 3 h on ice. The cells were washed once with phosphate-buffered saline (PBS) and fixed with 5% formaldehyde in PBS for 1 h at room temperature. After two washes with spheroplast buffer (1.2 M sorbitol, 0.1 M potassium phosphate, pH 7.4), cells were resuspended in 100 μl of spheroplast buffer containing 1 mg/ml Zymolyase 100T (Seikagaku Kogyo, Tokyo, Japan) and 0.2 μl of β-mercaptoethanol (from 14.2 M stock solution), followed by incubation for 30 min at 30°C. To visualize Bio-Ro, the cells were washed three times with PBS and then incubated in PBS containing 5 μg/ml Alexa Fluor 488–streptavidin (Invitrogen, Carlsbad, CA) for 1 h at room temperature. After five washes with PBS, cells were observed using a fluorescein isothiocyanate (FITC) filter set.
Lipid preparation
1-Palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine (NBD-PC), 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PE), 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine (NBD-PS), 1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-phospho-l-serine (TopFluor PS), and dioleoylphosphatidylcholine (DOPC) were obtained from Avanti Polar Lipids (Alabaster, AL). To prepare large unilamellar vesicles, lipids were mixed at 40 mol% NBD- or TopFluor-phospholipid and 60 mol% DOPC. Chloroform was removed by evaporation followed by vacuum desiccation. Desiccated phospholipids were solubilized in SD medium; the mixture was passed seven times through a LiposoFast-Basic stabilizer (Avestin, Ottawa, Canada), equipped with 0.1-μm filters to produce evenly sized vesicles, containing a 1 mM total concentration of lipids.
Internalization of fluorescence-labeled phospholipids into yeast cells
Fluorescently labeled phospholipid internalization experiments were performed as described (Kato et al., 2002). Briefly, cells were grown to early logarithmic phase in SD or SDA-U medium at 30°C. After dilution to 0.35 OD600/ml, cells were preincubated in SD with or without 1 mM sodium azide for 30 min at 30°C and incubated for 30 min at 30°C with vesicles containing 40% NBD- or TopFluor-phospholipid and 60% DOPC at a final concentration of 50 μM. Cells were then suspended in SD containing 20 mM sodium azide. Flow cytometry of fluorescently labeled cells was performed on a FACSCalibur cytometer using the CellQuest software (BD Biosciences, San Jose, CA).
Labeling with TMA-DPH and measurement of steady-state anisotropy
Labeling with TMA-DPH was performed as described (Abe et al., 2009). Briefly, cells were grown in SD medium at 25°C at 0.5–1.0 OD600. The cells (2 OD600 units) were collected, washed twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.0), and labeled with TMA-DPH (Invitrogen, Carlsbad, CA) at 25°C for 10 min in the dark. After washing twice, the cells were subjected to fluorescence microscopy or analysis by spectrophotometry. Before the microscopic observation, TMA-DPH-stained cells were kept on ice for 45 min, and the cells were observed using a UV-1A filter set.
For measurement of the steady-state anisotropy of TMA-DPH, an RF-5300PC spectrofluorometer (Shimadzu Co., Kyoto, Japan) was used. TMA-DPH-stained cells were resuspended in TE buffer at 0.25 OD600 and placed on ice until use. The cell suspensions were warmed at 30°C for 5 min prior to measurements. The excitation and emission wavelengths were 361 and 432 nm, respectively. Calculation of the steady-state anisotropy was performed as described (Abe et al., 2009).
Rhodamine uptake assay
Cells were grown to early logarithmic phase in YPDA or SDA-U medium at 30°C. After dilution to 0.5 OD600/ml, cells were preincubated in SD with or without 1 mM sodium azide for 30 min at 30°C and incubated for 1 h at 30°C with rhodamine 6G (Sigma-Aldrich) at a final concentration of 50 μM. Cells were then subjected to flow cytometry analysis on a FACSCalibur cytometer using the CellQuest software.
Lipid analysis
Total lipids were extracted basically by the Bligh and Dyer method (Bligh and Dyer, 1959). Cells were grown in 100–200 ml of YPDA or SDA-U medium to 0.8–1.0 OD600/ml at 30°C. The cells were collected and resuspended in 3.8 ml of chloroform-methanol-0.1 M HCl/0.1 M KCl (1:2:0.8) and lysed by vortexing with glass beads for 1 min. Then 1.0 ml each of chloroform and 0.1 M HCl/0.1 M KCl were added, followed by centrifugation, isolation of the lipid containing phase, and evaporation of the solvent. The extracted lipids were dissolved in appropriate volume of chloroform. Total phospholipids were determined by phosphorus assay (Rouser et al., 1970).
For phospholipid analysis, samples were subjected to thin-layer chromatography (TLC) on a TLC plate (Silica gel 60; Merck Millipore), with the solvent system chloroform/methanol/acetic acid/formic acid (50:30:4.5:6.5). After migration, plates were dried and sprayed with primuline solution (Sigma-Aldrich). Fluorescent spots of PE, PS, PI, and PC were scraped into glass tubes and subjected to lipid extraction and phosphorus assay as described above.
For ergosterol analysis, the samples containing 20 nmol phosphate were subjected to TLC on a HPTLC plate (Silica gel 60; Merck Millipore), with the solvent system hexane/diethyl ether/acetic acid (80:20:1) (Dodge and Phillips, 1967). After migration, plates were dried and sprayed with a 10% (wt/vol) cupric sulfate solution in 8% (wt/vol) orthophosphoric acid. Plates were heated in an oven at 180°C for 20 min. Plates were scanned with a CanoScan 8800F image scanner (Canon, Tokyo, Japan), and the acquired images were quantified with ImageJ (https://imagej.nih.gov/ij/).
Immunoblot analysis
Immunoblot analysis was performed as described previously (Misu et al., 2003). For SDS–PAGE of Sfk1p-3HA, samples were heated at 95°C for 5 min before loading. The mouse anti-HA (HA.11) monoclonal antibody (Babco, Richmond, CA) was used at 1:1000 dilution.
Statistical analysis and data reproducibility
Significance for Figure 5G was determined using a two-sided Student’s t test. Significance for all other figures was determined by a one-way analysis of variance with a Tukey’s test. All data are representatives of at least three independent experiments of biological replicates that gave reproducible results.
Supplementary Material
Acknowledgments
We thank Yoshikazu Ohya for providing yeast strains and plasmids. We thank Masato Umeda for providing Bio-Ro. We thank Masataka Kinjo for the RF-5300PC spectrofluorometer. We thank Yukifumi Uesono and our colleagues in the Tanaka laboratory for valuable discussions and Eriko Itoh for technical assistance. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grants 20570174 (K.F.-K.), 25840064 (T.S.), and 21370085 (K.T.).
Abbreviations used:
- 3HA
three tandem repeats of the influenza virus hemagglutinin epitope
- DIC
differential interference contrast
- GFP
green fluorescent protein
- NBD
7-nitrobens-2-oxa-1,3-diazol-4-yl
- NBD-PC
1-palmitoyl-2-(6-NBD-aminocaproyl)-PC
- NBD-PE
1-palmitoyl-2-(6-NBD-aminocaproyl)-PE
- NBD-PS
1-palmitoyl-2-(6-NBD-aminocaproyl)-PS
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- TGN
trans-Golgi network
- TMA-DPH
1-[4-(trimethylamino)pheny]-6-phenyl-1,3,5-hexatriene
- TopFluor-PS
1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-PS.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0217) on March 22, 2018.
REFERENCES
- Abe F, Hiraki T. (2009). Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim Biophys Acta , 743–752. [DOI] [PubMed] [Google Scholar]
- Abe F, Usui K, Hiraki T. (2009). Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry , 8494–8504. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Uenaka T, Aoki J, Umeda M, Inoue K. (1994). A novel peptide probe for studying the transbilayer movement of phosphatidylethanolamine. J Biochem , 291–297. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD. (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell , 593–605. [DOI] [PubMed] [Google Scholar]
- Aye IL, Singh AT, Keelan JA. (2009). Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem Biol Interact , 327–339. [DOI] [PubMed] [Google Scholar]
- Baird D, Stefan C, Audhya A, Weys S, Emr SD. (2008). Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol , 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell , 886–895. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Physiol , 911–917. [DOI] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. (1993). An osmosensing signal transduction pathway in yeast. Science , 1760–1763. [DOI] [PubMed] [Google Scholar]
- Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, Klomp LW, Lomri N, Berger R, Scharschmidt BF, et al (1998). A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet , 219–224. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. (1982). Two differentially regulated mRNAs with different 5’ ends encode secreted with intracellular forms of yeast invertase. Cell , 145–154. [DOI] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. (1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol , 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Nakatsu F, Baskin JM, De Camilli P. (2015). Plasticity of PI4KIIIalpha interactions at the plasma membrane. EMBO Rep , 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Quazi F, Molday RS. (2013). Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim Biophys Acta , 555–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Zhu X, Djajadi HR, Molday LL, Smith RS, Libby RT, John SW, Molday RS. (2014). Phospholipid flippase ATP8A2 is required for normal visual and auditory function and photoreceptor and spiral ganglion cell survival. J Cell Sci , 1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Slaughter BD, Unruh JR, Bradford WD, Alexander R, Rubinstein B, Li R. (2012). Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat Cell Biol , 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A. (1998). ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem , 12612–12622. [DOI] [PubMed] [Google Scholar]
- Desrivieres S, Cooke FT, Parker PJ, Hall MN. (1998). MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem , 15787–15793. [DOI] [PubMed] [Google Scholar]
- Devaux PF. (1991). Static and dynamic lipid asymmetry in cell membranes. Biochemistry , 1163–1173. [DOI] [PubMed] [Google Scholar]
- Dodge JT, Phillips GB. (1967). Composition of phospholipids and of phospholipid fatty acids and aldehydes in human red cells. J Lipid Res , 667–675. [PubMed] [Google Scholar]
- Elble R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques , 18–20. [PubMed] [Google Scholar]
- Emoto K, Kobayashi T, Yamaji A, Aizawa H, Yahara I, Inoue K, Umeda M. (1996). Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc Natl Acad Sci USA , 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emter R, Heese-Peck A, Kralli A. (2002). ERG6 and PDR5 regulate small lipophilic drug accumulation in yeast cells via distinct mechanisms. FEBS Lett , 57–61. [DOI] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. (2007). Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell , 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. (2002). Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol , 1623–1627. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. (2002). Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press. [Google Scholar]
- Hachiro T, Yamamoto T, Nakano K, Tanaka K. (2013). Phospholipid flippases Lem3p-Dnf1p and Lem3p-Dnf2p are involved in the sorting of the tryptophan permease Tat2p in yeast. J Biol Chem , 3594–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. (2015). Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell , 4674–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PK, Malone L, Birchmore JL, Nichols JW. (2003). Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem , 36041–36050. [DOI] [PubMed] [Google Scholar]
- Hanson PK, Nichols JW. (2001). Energy-dependent flip of fluorescence-labeled phospholipids is regulated by nutrient starvation and transcription factors, PDR1 and PDR3. J Biol Chem , 9861–9867. [DOI] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. (1998). Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem , 15779–15786. [DOI] [PubMed] [Google Scholar]
- Hua Z, Fatheddin P, Graham TR. (2002). An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell , 3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Hayakawa T, Murate M, Makino A, Ito K, Fujisawa T, Kobayashi T. (2007). Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys J , 1608–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Kobayashi S, Fukuda R, Umeda M, Kobayashi T, Ohta A. (2004). Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells , 891–903. [DOI] [PubMed] [Google Scholar]
- John K, Schreiber S, Kubelt J, Herrmann A, Muller P. (2002). Transbilayer movement of phospholipids at the main phase transition of lipid membranes: implications for rapid flip-flop in biological membranes. Biophys J , 3315–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth H, Kuchler K. (2006). Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett , 1131–1138. [DOI] [PubMed] [Google Scholar]
- Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, Murakami-Murofushi K, Umeda M. (2002). A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem , 37855–37862. [DOI] [PubMed] [Google Scholar]
- Kato U, Inadome H, Yamamoto M, Emoto K, Kobayashi T, Umeda M. (2013). Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. J Biol Chem , 4922–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. (2012). Phosphatidylserine dynamics in cellular membranes. Mol Biol Cell , 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp LW, Vargas JC, van Mil SW, Pawlikowska L, Strautnieks SS, van Eijk MJ, Juijn JA, Pabon-Pena C, Smith LB, DeYoung JA, et al (2004). Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology , 27–38. [DOI] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. (2012). Flexibility of a eukaryotic lipidome—insights from yeast lipidomics. PLoS One , e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande MB, Donovan JM, Zeidel ML. (1995). The relationship between membrane fluidity and permeabilities to water, solutes, ammonia, and protons. J Gen Physiol , 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Patton JL, Fido M, Hines LK, Kohlwein SD, Paltauf F, Henry SA, Levin DE. (1994). The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem , 19725–19730. [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al (2015). Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J , 669–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir G, Williamson P, Puts CF, Holthuis JC. (2009). Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem , 17956–17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levano K, Punia V, Raghunath M, Debata PR, Curcio GM, Mogha A, Purkayastha S, McCloskey D, Fata J, Banerjee P. (2012). Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J Neurochem , 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Hua Z, Nepute JA, Graham TR. (2007). Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol Biol Cell , 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Fairn GD. (2015). Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J Cell Sci , 1422–1433. [DOI] [PubMed] [Google Scholar]
- Marques MC, Zamarbide-Fores S, Pedelini L, Llopis-Torregrosa V, Yenush L. (2015). A functional Rim101 complex is required for proper accumulation of the Ena1 Na+-ATPase protein in response to salt stress in Saccharomyces cerevisiae. FEMS Yeast Res , fov017. [DOI] [PubMed] [Google Scholar]
- Merkel O, Fido M, Mayr JA, Pruger H, Raab F, Zandonella G, Kohlwein SD, Paltauf F. (1999). Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J Biol Chem , 28121–28127. [DOI] [PubMed] [Google Scholar]
- Mioka T, Fujimura-Kamada K, Tanaka K. (2014). Asymmetric distribution of phosphatidylserine is generated in the absence of phospholipid flippases in Saccharomyces cerevisiae. Microbiologyopen , 803–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu K, Fujimura-Kamada K, Ueda T, Nakano A, Katoh H, Tanaka K. (2003). Cdc50p, a conserved endosomal membrane protein, controls polarized growth in Saccharomyces cerevisiae. Mol Biol Cell , 730–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montigny C, Lyons J, Champeil P, Nissen P, Lenoir G. (2016). On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim Biophys Acta , 767–783. [DOI] [PubMed] [Google Scholar]
- Murate M, Abe M, Kasahara K, Iwabuchi K, Umeda M, Kobayashi T. (2015). Transbilayer distribution of lipids at nano scale. J Cell Sci , 1627–1638. [DOI] [PubMed] [Google Scholar]
- Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K. (2008). Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell , 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatala R, Hennrich H, Holthuis JC. (2015). Inner workings and biological impact of phospholipid flippases. J Cell Sci , 2021–2032. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al (2006). Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell , 611–625. [DOI] [PubMed] [Google Scholar]
- Paulusma CC, Groen A, Kunne C, Ho-Mok KS, Spijkerboer AL, Rudi de Waart D, Hoek FJ, Vreeling H, Hoeben KA, van Marle J, et al (2006). Atp8b1 deficiency in mice reduces resistance of the canalicular membrane to hydrophobic bile salts and impairs bile salt transport. Hepatology , 195–204. [DOI] [PubMed] [Google Scholar]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC. (2003). Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell , 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LR, Lopez-Marques RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG. (2008). The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell , 658–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts CF, Panatala R, Hennrich H, Tsareva A, Williamson P, Holthuis JC. (2012). Mapping functional interactions in a heterodimeric phospholipid pump. J Biol Chem , 30529–30540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Voelker DR. (2006). Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem , 36588–36596. [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Wu J, Gijón MA, Zarini S, Murphy RC, Voelker DR. (2007). Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem , 36853–36861. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. (2010). A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci USA , 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Su BM, von Wulffen J, Ramachandran S, Sartorel E, Trott AE, Thorner J. (2015). Protein kinase Gin4 negatively regulates flippase function and controls plasma membrane asymmetry. J Cell Biol , 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouser G, Fkeischer S, Yamamoto A. (1970). Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids , 494–496. [DOI] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. (2004). Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell , 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Hanamatsu H, Kato U, Umeda M, Kozminski KG, Tanaka K. (2007). Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev Cell , 743–751. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sartorel E, Barrey E, Lau RK, Thorner J. (2015). Plasma membrane aminoglycerolipid flippase function is required for signaling competence in the yeast mating pheromone response pathway. Mol Biol Cell , 134–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian TT, Baldridge RD, Xu P, Graham TR. (2012). Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim Biophys Acta , 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. (2014). Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science , 1164–1168. [DOI] [PubMed] [Google Scholar]
- Stevens HC, Malone L, Nichols JW. (2008). The putative aminophospholipid translocases, DNF1 and DNF2, are not required for 7-nitrobenz-2-oxa-1,3-diazol-4-yl-phosphatidylserine flip across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem , 35060–35069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Iwamoto K, Kobayashi S, Horiuchi H, Fukuda R, Ohta A. (2012). Involvement of Golgi-associated retrograde protein complex in the recycling of the putative Dnf aminophospholipid flippases in yeast. Biochem Biophys Res Commun , 490–494. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Fujimura-Kamada K, Kondo S, Tanaka K. (2011). Isolation and characterization of novel mutations in CDC50, the non-catalytic subunit of the Drs2p phospholipid flippase. J Biochem , 423–432. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Tanaka G, Segawa K, Suzuki J, Nagata S, Nakayama K, Shin HW. (2014). Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J Biol Chem , 33543–33556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Fujimura-Kamada K, Yamamoto T. (2011). Functions of phospholipid flippases. J Biochem , 131–143. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Ono N, Shima T, Tanaka G, Katoh Y, Nakayama K, Takatsu H, Shin HW. (2016). The phospholipid flippase ATP9A is required for the recycling pathway from the endosomes to the plasma membrane. Mol Biol Cell , 3883–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk M, Montiel V, Zigon D, Plemenitas A, Ramos J. (2007). Plasma membrane composition of Debaryomyces hansenii adapts to changes in pH and external salinity. Microbiology , 3586–3592. [DOI] [PubMed] [Google Scholar]
- van der Mark VA, Elferink RP, Paulusma CC. (2013). P4 ATPases: flippases in health and disease. Int J Mol Sci , 7897–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S, Schwarz H, Singer-Kruger B. (2004). Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol , 7402–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale J, Bohnert HJ. (2001). Transcript expression in Saccharomyces cerevisiae at high salinity. J Biol Chem , 15996–16007. [DOI] [PubMed] [Google Scholar]
- Yamauchi S, Obara K, Uchibori K, Kamimura A, Azumi K, Kihara A. (2015). Opt2 mediates the exposure of phospholipids during cellular adaptation to altered lipid asymmetry. J Cell Sci , 61–69. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohya Y, Nakano A, Anraku Y. (1994). Genetic interactions among genes involved in the STT4-PKC1 pathway of Saccharomyces cerevisiae. Mol Gen Genet , 631–640. [DOI] [PubMed] [Google Scholar]
- Zachowski A. (1993). Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem J (Pt 1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liang Y, Zhang X, Xu Y, Dai H, Xiao W. (2008). Deletion of yeast CWP genes enhances cell permeability to genotoxic agents. Toxicol Sci , 68–76. [DOI] [PubMed] [Google Scholar]
- Zinser E, Paltauf F, Daum G. (1993). Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol , 2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.