Abstract
Oncogene activation disturbs cellular processes and accommodates a complex landscape of changes in the genome that contribute to genomic instability, which accelerates mutation rates and promotes tumourigenesis. Part of this cellular turmoil, involves deregulation of physiological DNA replication, widely described as replication stress. Oncogene-induced replication stress is an early driver of genomic instability and is attributed to a plethora of factors, most notably aberrant origin firing, replication-transcription collisions, reactive oxygen species and defective nucleotide metabolism.
Keywords: DNA replication stress, oncogenes, genomic instability, cancer
Introduction
Genomic instability (GIN) has been highlighted as a driving force of tumourigenesis by Hanahan and Weinberg in their celebrated ‘Hallmarks of cancer’ paper (1). GIN can result from changes in the number or structure of chromosomes (chromosomal instability), changes in the number of oligonucleotide repeats in microsatellite sequences (microsatellite instability), or base pair mutations, all of which are associated with activated oncogenes. Deregulation of DNA replication, known as replication stress (RS), is linked to GIN and is increased during the early steps of carcinogenesis (2–4). In particular, RS has been associated with chromosomal instability (5) as well as activation of the APOBEC3 family of deaminases (6) which increase the mutagenic load that fuels tumourigeneseis. In this review, we cover the latest findings on the RS response and discuss in detail the various mechanisms through which oncogenes induce RS.
DNA Replication
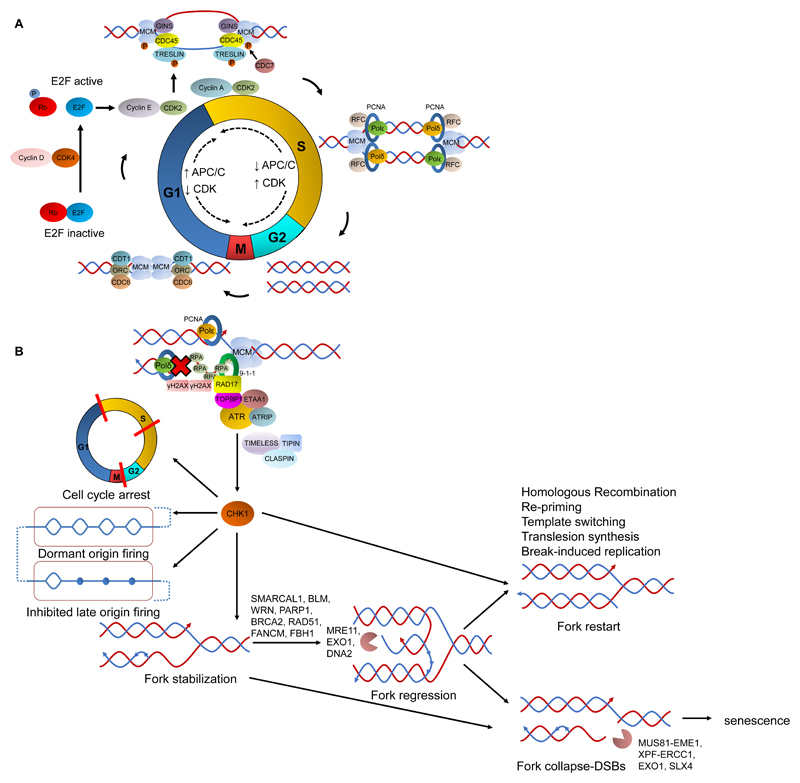
DNA replication ensures the precise duplication of DNA during each cell cycle. It is a tightly regulated process that consists of two stages: licensing and initiation (reviewed in ref. 7). In eukaryotic cells, the licensing stage is restricted during late mitosis and G1-phase when thousands of replication origins are established along the genome and ensures that DNA replication occurs only once per cell cycle. For an origin to form, the origin recognition complex (ORC) binds at the origin site and recruits CDT1 and CDC6, which in turn facilitate loading of the mini-chromosome maintenance 2-7 (MCM2-7) helicases to form the pre-replicative complex (pre-RC) (Fig. 1A).
Figure 1.
DNA replication and replication stress response. A, in late mitosis and throughout G1-phase, the pre-replicative complex comprised by the ORC complex, CDC6 and CDT1 is recruited to replication origins to facilitate loading of the MCM2-7 complex. During G1-phase, retinoblastoma (Rb) is bound to E2F rendering it inactive. Phosphorylation of Rb by the Cyclin D-CDK4 complex alleviates its inhibitory effect on E2F. APC/C activity is high from late M- to late G1-phase regulating CDK activity. Upon entry in S-phase, APC/C is inhibited and CDKs are activated throughout S, G2 and early M-phase. CDKs form complexes with E2F-regulated cyclins that collaborate with CDC7 to phosphorylate TRESLIN and MCM2-7 complex, activating the CMG (CDC45-MCM2-7-GINS) helicase complex. Simultaneously, clamp loader RFC and sliding clamp PCNA are recruited and enable polymerases δ and ε to initiate replication in the lagging and leading strands respectively. B, when a replication fork is stalled, ssDNA is generated as the CMG complex unwinds DNA. ssDNA binds RPA that recruits ATR (through ATRIP), RAD17-RFC and 9-1-1 complexes at the stalled fork. ATR is then activated by TOPBP1/RAD17/9-1-1 and ETAA1 and phosphorylates H2AX, while through TIMELESS/TIPIN/CLASPIN phoshoprylates CHK1. CHK1 then organises the RS response by arresting the cell cycle, inhibiting new origin firing, enabling dormant origin firing and stabilizing the fork, which can then be reversed by various proteins in a chicken foot structure. An unprotected reversed fork is susceptible to nucleolytic degradation by MRE11, EXO1 and DNA2. A stalled fork can restart through homologous recombination, re-priming, template switching, translesion synthesis or break-induced replication. Alternatively, it will collapse into DSBs by the combined activity of MUS81-EME1, XPF-ERCC1, EXO1 and SLX4 that will drive the cell to senescence.
Cell cycle progression is controlled by cyclin dependent kinases (CDK) and the retinoblastoma/E2F pathway. CDK activity depends on binding to their regulatory subunits, cyclins, whose levels are regulated throughout the cell cycle by the anaphase promoting complex/cyclosome (APC/C). APC/C activity is high from late M to late G1-phase, hence CDK activity oscillates accordingly, being low during G1 and high during S/G2-phases. Mitogenic signalling by RAS triggers CYCLIN D/CDK4 to phosphorylate retinoblastoma, which renders it inactive, thus alleviating its inhibitory effect on the E2F family of transcription activators. E2F proteins promote expression of CYCLIN A and E, amongst other key S-phase genes, which upon binding to CDK2 during G1/S-phase partake in promoting entry into S-phase.
Replication initiation occurs as the cell proceeds into S-phase and requires the concerted action of CDK2 and DBF4/DRF1-dependent CDC7 kinases, which phosphorylates the pre-RC allowing recruitment of CDC45 and the GINS complex, and leads to the activation of the replicative helicase (CMG complex). Once this occurs, a replication bubble is formed and replication forks proceed bi-directionally from the origin (Fig. 1A). Under normal conditions, an excess of origins is licensed but only a small number of them becomes activated, with the remaining dormant origins reserved as a backup.
The DNA Replication Stress Response
Replication is susceptible to impediments in DNA caused by both exogenous and endogenous DNA damaging agents and by the intrinsic properties of certain DNA sequences to adopt secondary structures. In particular, fork progression can be hindered due to interference with the transcription machinery, torsional stress or non-B DNA structures (cruciforms, hairpins, trinucleotide repeats, R-loops, G-quadruplexes).
The ATR/CHK1 pathway
In response to RS, the cell initiates a DNA damage response (DDR) with the aim of resolving the damage or DNA secondary structures and restore fork progression (reviewed in ref. 8). During replication fork stalling, uncoupling between the replicative helicase and polymerase leads to the accumulation of single strand DNA (ssDNA) which is bound by RPA. ssDNA-RPA in turn allows the recruitment of the ATR kinase through ATRIP, as well as RAD17-RFC and RAD9-RAD1-HUS1 (9-1-1). The 9-1-1 complex also interacts with TOPBP1, which triggers ATR-ATRIP kinase activity, leading to the phosphorylation of numerous down-stream factors that collectively respond to RS. Recently, ETAA1 was identified as a novel RPA binding protein that activates ATR in response to DNA damage in parallel to TOPBP1/RAD17/9-1-1, and is involved in fork restart (9, 10). The synergistic action of the TIMELESS/TIPIN complex promotes binding of CLASPIN to RPA, which allows ATR to phosphorylate its primary substrate kinase, CHK1, at Ser-317 and Ser-345. Additionally, ATR phosphorylates histone H2AX at Ser-319 (γH2AX) early in the response. This modification then spreads away from the stalled fork, and is further sustained by two other DDR kinases ATM and DNA-PKcs.
CHK1 organises the cellular DDR by inducing cell cycle arrest, inhibiting late origin firing, activating dormant origin firing, and promoting fork stabilization and fork restart (Fig. 1B). Cell cycle arrest allows sufficient time for the cell to effect lesions repair and also prevent premature entry into mitosis with under-replicated DNA. In response to stress, CHK1 phosphorylates CDK activators CDC25A/C, which leads to their degradation or nuclear export, thus triggering arrest at S, G2 or G2/M-phases. At the same time, CHK1 phosphorylates and activates the CDK antagonist WEE1 causing G2 delay.
During unperturbed early S-phase, ATR protects the genome from ssDNA formation by inhibiting origin firing and promoting nucleotide synthesis at the same time (11). In general, ATR regulates origin firing by phosphorylating MLL at Ser-516, stabilizing it on chromatin where it methylates histone H3K4, inhibits CDC45 loading and blocks origin activation (12). Additionally, CHK1 regulates replication initiation by binding and phosphorylating TRESLIN, which inhibits CDC45 loading onto origins (13). Recently, a backup pathway of CHK1 activation was identified, when upon ATR inhibition, accumulation of ssDNA produces aberrant DNA structures, that after processing by SLX4-MUS81 induce a DNA-PK-dependent CHK1 activation that inhibits origin firing (11).
In order to complete DNA replication in response to any disturbance, CHK1 inhibits origin firing at new replication factories (late origins), while at the same time allows the firing of dormant origins within active replication factories that experience stress (14, 15) (Fig. 1B).
Replication fork stabilization and reversal
Stabilization of the replication fork has long been considered a CHK1 response to RS, protecting it from deleterious nucleolytic processing. This view has been challenged recently by evidence from yeast, showing that fork stability is retained in the absence of checkpoint kinases (16). In addition, a SILAC-iPOND study in human cells, revealed ATR to be responsible for fork but not replisome stability in response to RS and that ATR protects the fork from various forms of collapse (17). Notably, RS at an active fork triggers accumulation of homologous recombination proteins, whereas RS arising in the context of an origin that fired due to a checkpoint deficiency triggers accumulation of non-homologous end joining proteins (17).
A common mechanism of fork stabilisation involves the annealing of the parental DNA strands, followed by binding of the newly synthesized strands, thus forming a reversed fork structure, also known as regressed fork or a ‘chicken foot’ (Fig. 1B). Early studies in S. cerevisiae, showed that reversed forks accumulate in response to checkpoint defects (18), which led to the view that fork reversal is a pathological response. However, an expanding body of evidence has shown that reversed forks are also important for fork stability and protection from collapse. Reversed forks are formed through the action of many proteins, including RAD51 (19), PARP1 (20), BLM (21), WRN (22), SMARCAL1 (22), FANCM (23), FBH1 (24), HLTF (25) and ZRANB3 (26) and are favoured by positive supercoiling. Reversed forks are protected by BRCA1/2 (27), RAD51 (27), TOP1 (20), FANCA/B (28), FANCD2 (28), REV1 (29), WRN (30), BOD1L (31), RECQL5 (32) and WRNIP1 (33), in whose absence they are susceptible to the activity of nucleases MRE11 (27), DNA2 (34) and EXO1 (35) or resolvase Yen1 (36). Despite its positive role in resolving stalled forks, if unrestrained, fork reversal can cause fork collapse (37), highlighting the need for a regulated balance between reversal and restart.
Replication fork restart
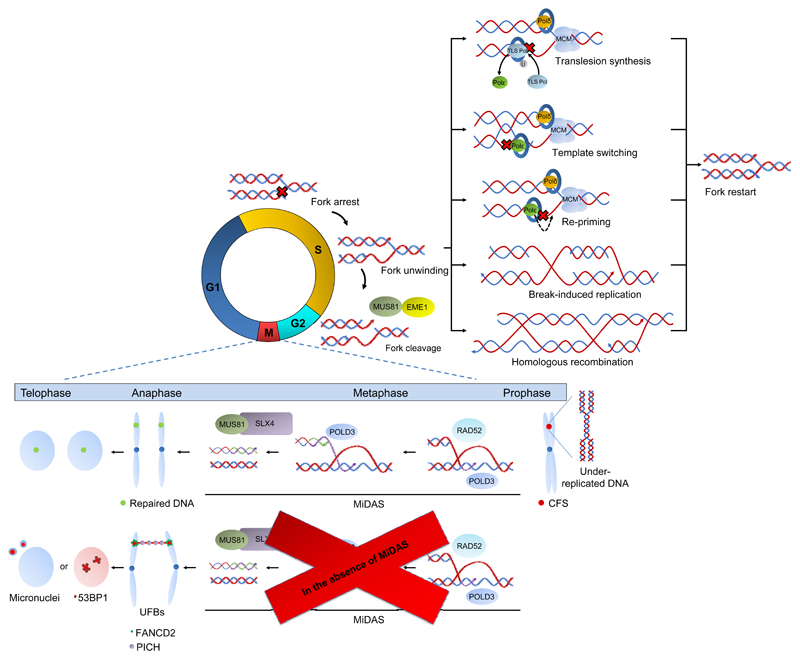
Once a fork is stalled, different pathways, including homologous recombination, re-priming, template switching, translesion synthesis and break-induced replication may occur to allow replication restart (Fig. 2). The details of these mechanisms have been described elsewhere (38) and are not the focus of this review. Alternatively, stalled forks may be resolved by an incoming fork from an adjacent origin. Evidence in yeast has shown that terminally arrested forks that are unprotected by Rad52/Rad51, cannot merge with a converging fork and appear in the ensuing mitosis as anaphase bridges (39). If fork stalling is sustained the forks often collapse into double strand breaks (DSBs) by the combined nucleolytic activities of SLX4 and nucleases MUS81-EME1, XPF-ERCC1 and EXO1 (reviewed in ref. 40).
Figure 2.
Repair of a stalled fork. Upon encountering an obstacle, replication machine arrests and the CMG complex unwinds DNA ahead of the stalled fork, leaving ssDNA behind. Replication can resume through various mechanisms, such as translesion synthesis, template switching, re-priming, break-induced replication or homologous recombination. If this fails, the stalled fork is cleaved by MUS81-EME1 during G2-phase and upon entrance in metaphase RAD52, POLD3 and MUS81-SLX4 collaborate to facilitate replication of the under-replicated DNA through MiDAS. In the absence of MiDAS and during chromosomal segregation in anaphase, UFBs are formed at the CFSs, which will appear as micronuclei or 53BP1 bodies in the daughter cells.
Recent insights from the Hickson and Halazonetis labs discovered how under-replicated DNA is managed if it escapes replication/repair during S-phase. According to their data, regions of under-replicated DNA are processed by MUS81-EME1 during G2 and with the help of RAD52, POLD3 polymerase and SLX4-MUS81 perform mitotic DNA synthesis (MiDAS) to repair the collapsed forks (41–43). Furthermore, RECQL5 helicase is recruited to common fragile sites (CFSs) by MUS81 to remove RAD51 filaments from stalled replication forks to allow processing by MUS81-EME1 and enable MiDAS (44). Lack of MiDAS increases 53BP1 bodies, anaphase bridges and chromosomal rearrangements, promotes tumour growth and sensitizes cells to aphidicolin (41–43) (Fig. 2).
Targeting replication stress for cancer treatment
Importantly, RS can be exploited for cancer cell killing and thus has been described as an Achille’s heel of cancer. ATR or CHK1 inhibition induces RS and synthetic lethality in CYCLIN E (45), c-MYC (46) and H/K-RAS (47) overexpressing cells, as well as MYC-induced lymphomas (48), MLL-ENL or NRAS-driven acute myeloid leukaemias and HRAS-expressing fibrosarkomas (46). Furthermore, WEE1 inhibition confers synthetic lethality in H3K36me3-deficient cancer cells by instigating deoxyribonucleoside triphosphate (dNTP) starvation and RS (49). In an intriguing recent report, a combination of ATR and CHK1 inhibitors, result in fork slowing, replication catastrophe and synthetic lethality in cells overexpressing HRASV12 or c-MYC and in other cancer cell lines (50). In this case, CHK1 inhibition causes increased CDK-dependent origin firing that depletes dNTP pools, leading to fork slowing and ssDNA accumulation that is normally protected by an ATR-dependent deposition of RPA; the subsequent ATR inhibition deprotects the forks and kills the cell (50).
Reversing RS as a strategy to alleviate its tumourigenic effect has proven to be more complicated, as an extra CHK1 allele can reduce RS but surprisingly increases transformation (51). Nevertheless, replication fork stability is a significant contributing factor to chemotherapeutic drug resistance. This is exemplified by the fact that alleviation of MRE11-dependent GIN upon treatment with replication poisons in BRCA-deficient cells, by loss of PTIP, CHD4 or PARP1 confers synthetic viability and chemoresistance (52). All of the above, highlight the importance of RS as a hallmark and driver of cancer.
Oncogenes
Normal cells become cancerous through a complex process known as oncogenic transformation. Transformation is driven by altered expression of oncogenes, tumor suppressors or microRNAs that derail their normal physiological function (reviewed in ref. 53). A proto-oncogene is a gene that under unperturbed conditions generally encodes a protein implicated in cell growth, differentiation or apoptosis. Either through point mutation, chromosomal translocation or copy number amplification, expression of the proto-oncogene is misregulated resulting in an activated oncogene. Oncogenes are translated into oncoproteins, which are classified as growth factors, growth factor receptors, transcription factors, signal transducers, chromatin remodelers and apoptosis regulators. As such, oncogene activation may cause massive changes in the genome by deregulating cell cycle, metabolism, replication-timing or transcription, which ultimately drive GIN.
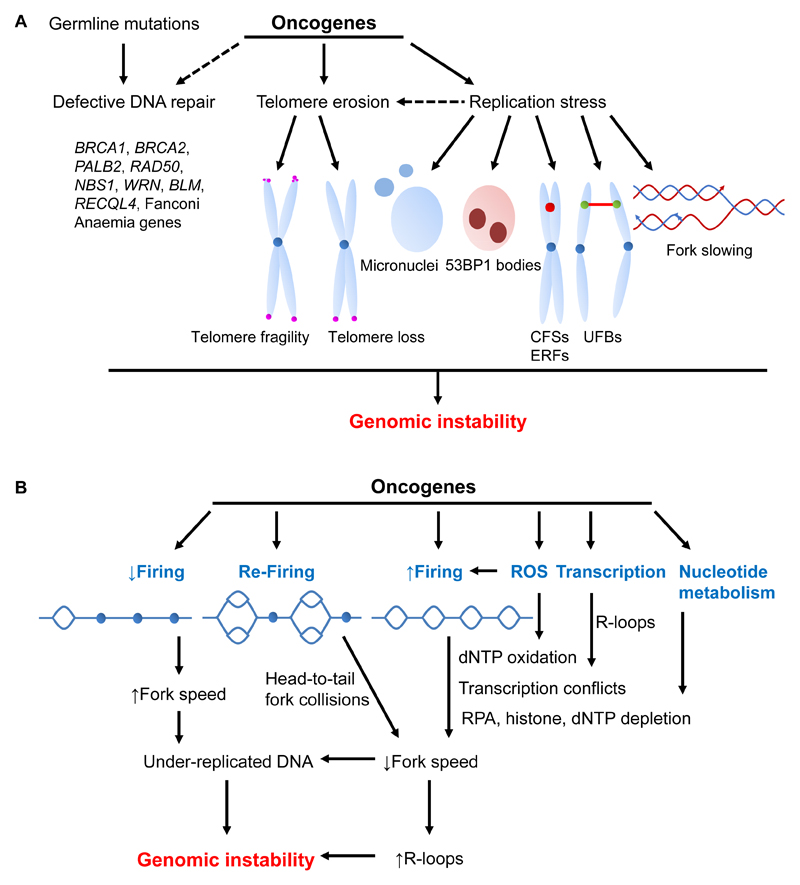
The principle mechanisms through which GIN is induced in cancer involve DNA repair defects, heightened replication stress and telomeric dysfunction (Fig. 3A). In hereditary cancers, GIN is commonly attributed to germline mutations in genes involved in DNA damage repair. These mutations give rise to unrepaired or inaccurately repaired DNA that lead to GIN (reviewed in ref. 54). Certain oncogenes can also cause a similar DNA repair defect. In particular, overexpression of oncogenic RAS and BRAF cause dissociation of BRCA1 from chromatin, which compromises DSB repair leading to DNA damage and senescence (55). Furthermore, wild type HRAS and NRAS modulate DDR to support the tumorigenic activity of oncogenic KRAS (56).
Figure 3.
Oncogene-induced genomic instability. A, germline mutations in DNA repair genes lead to GIN. Activated oncogenes elicit genomic instability by causing defects in DNA repair, telomere erosion or replication stress; see text for details. B, activated oncogenes induce a multifaceted set of intertwined activities that deregulate fork progression leading to under-replicated DNA and genomic instability. In particular, through deregulation of the Rb/E2F pathway licensing factors are increased which instigates origin re-firing that decreases fork speed in response to head-to-tail fork collisions. Deregulation of CDK activity can decrease or increase origin firing. In the first case, fork speed is initially increased, but the cell ends up with under-replicated DNA due to its inability to rescue endogenous RS by firing dormant origins. Increased origin firing raises the possibility of TRCs and simultaneously may cause depletion of dNTPs, histones or RPA. Oncogene-induced ROS either increase origin firing or oxidize nucleotides that potentially may affect fork progression. Oncogenes also increase transcription activity that either directly or through R-loops enhance TRCs.
Oncogenes can also induce DNA replication defects and under-replicated DNA, which leads to accumulation of mutations and GIN. RS manifests in various forms, the most obvious being perturbed fork extension rates and/or heighted fork stalling/collapse, which may prevent complete replication of the genome. Such sites are marked by 53BP1 (57) and often correspond to CFSs, which are more susceptible to breakage by MUS81-EME1 or XPF-ERCC1. Replication through CFSs is exacerbated by mild RS, partly due to the absence or aberrant activation of dormant origins within these regions (58). During chromosomal segregation in mitosis, under-replicated regions often present as ultra-fine anaphase DNA bridges (UFBs) (59) that are bound by PICH, BLM and RPA with FANCD2 associated with their extremities. Unresolved UFBs may lead to chromosome breakage and/or mis-segregation, resulting in micronuclei formation in daughter cells (59).
Finally, telomeric dysfunction can also lead to GIN. Telomeres are comprised of repeated sequences that maintain and protect chromosome ends from deleterious processing and/or unscheduled DNA repair. With each cell cycle, telomere repeats progressively shorten due to the ‘end-replication problem’, oxidative damage and RS associated with oncogenic activation. Telomeric dysfunction is linked to extended chromosomal shattering and rearrangements known as chromothripsis and kataegis, which drive GIN in cancer (60). Telomeric erosion is counteracted by the expression of telomerase that synthesizes de novo telomeric repeats at chromosome ends. Reports that HPV 16E6/E7-induced anaphase bridges (61) and oncogene-induced senescence (62) are ameliorated upon activation of telomerase activity, highlight the connection between telomeric integrity and GIN. Additionally, HRAS causes telomeric fork stalling, as well as telomeric fragility and loss of telomere repeats (63). Therefore, telomeric erosion can be linked to oncogene-induced telomeric replication stress, although it can also be attributed to oncogene-induced telomerase deregulation (61).
Apoptosis and senescence act as protective mechanisms that eliminate or halt cells that present with RS and/or GIN. This cancer protection barrier is quite robust as affirmed by the fact that expression of oncogenes alone does not lead to oncogenic transformation, unless combined with other genetic events, most notably, additional expression of other oncogenes or mutation of tumour suppressor genes (64–66). Oncogene-induced senescence is ascribed to the actions of the tumour suppressor p53 and its positive regulator p14/p19 (ARF). ARF inhibits the ubiquitin ligase MDM2 that is normally responsible for p53 degradation, thereby stabilizing p53 levels. Amongst other oncogenes, p53 is activated in response to RAS, c-MYC, E1A and STAT5A overexpression either directly through ARF or RS-induced ATM activation. In addition, oncogenes RAS, MYC, E2F1, β-CATENIN and adenovirus E1A have been shown to upregulate ARF, while c-MYC causes ARF stabilization by inhibiting its ubiquitylation and subsequent degradation by ULF ubiquitin ligase (reviewed in ref. 67).
Of the 803 identified oncogenes in the ONGene database (http://ongene.bioinfo-minzhao.org/) only 27 have been assessed for their impact on RS, the majority of which are most commonly activated in human cancers. Additionally, for those oncogenes that have been studied there is variability in how RS is driven (Table 1). The concept that different oncogenes induce RS through different mechanisms is supported by many reports. In particular, RAS and CYCLIN E create unique landscapes of fragile sites that differ to the sites induced by aphidicolin treatment, which inhibits replication (68). Overexpression of CYCLIN E, but not A or D1, induces chromosomal instability in human cells (69). Overexpression of CYCLIN E and CDC25 also cause fork slowing and replication fork reversal at the same time point, but DSB formation and DDR signalling exhibit vastly different kinetics (70). It is worth noting that not all oncogenes lead to fork stalling, as exemplified by DEK that promotes fork progression under RS conditions (71) and E1A (72), SPI1/PU.1 (73) and LMO2 (74) that increase fork speed.
Table 1. List of oncogenes and their effect on replication stress-related response.
§: Indirectly, ↑: Positive effect, ↓: Negative effect, X: No effect, tr: translocation, pm: point mutation, oe: overexpression, kd: knockdown.
| oncogene | genetic alteration | effect on fork progression | ↑DSBs | ↑ROS | ↑transcription / R-loops | affects nucleotide pools | activates | induces senescence | ||
|---|---|---|---|---|---|---|---|---|---|---|
| γH2AX | ATR | ATM | ||||||||
| AKT-2 | pm, oe | §204 | §204 | |||||||
| AML1/ETO | oe | 205 | 205 | 205 | 205 | |||||
| AURORA A | oe | ↓206 | 207 | |||||||
| β-CATENIN | oe | 208 | 208 | |||||||
| BCL-2 | oe | ↓189 | 189 | 209 | ||||||
| BCR/ABL | oe, tr | 210, 205 | 210, 211 | X205 | X205 | 205 | ||||
| BRAF | oe, pm | 212 | 190 | 190 | 190, 213, 212, 214 | |||||
| B-MYB | kd | ↓215 | 215 | 215 | 216 | |||||
| CDC6 | oe | 217 | 3 | 3 | 3 | |||||
| CDC5A | oe | ↓70 | 70 | 2, 218, 70 | 2, 70 | 2, 70 | ||||
| CYCLIN D | oe, tr, kd | ↓108 | 108, 219 | X219 | 108 | 220 | ||||
| CYCLIN E | oe | ↓3, 86, 87, 70 | 3, 70 | §87 | 86 | 2, 3, 86, 87, 70, 219 | 2, 3, 70, 219 | 2, 70 | 3 | |
| DEK | kd | ↑71 | 221 | 221, X71 | X71 | X71 | 216, 221 | |||
| E1A | oe | ↑72 | 72 | X72 | 72 | |||||
| E2F1 | oe | 2, 218, 222 | 218, 222 | 223, 222, 224 | ||||||
| EGFR | oe | 225 | 225 | 225 | ||||||
| HPV E6/E7 | oe | ↓86 | 226, 202 | 202 | 86 | 86, 226, 224, 219 | ||||
| LMO1-4 | oe, kd | ↑74 | 176, 180 | |||||||
| MDM2 | oe | ↓227 | 227 | |||||||
| MOS | oe | 3 | 3 | 3 | 3 | |||||
| MYC | pm, oe | ↓200, 96 | 228, 199 | 200, 228, 199 | 177–179 | 191 | 229, 200, 96 | 229 | 228, 96 | 199 |
| MYH11/CBFB | oe | X205 | X205 | X205 | 205 | |||||
| NPM-ALK | oe | 230 | 231 | 232, 233 | 232 | 232 | 233 | |||
| RAS | pm, oe | ↓234, 97, 200 | 97, 205, 198 | 197, 225, 200, 203, 198 | 97 | 190 | 190, 94, 97, 225, 224, 200, 203, 205, 198 | 234, 94, 97, 224 | 224,94, 205 | 190, 94, 225, 224, 203, 66, 205 |
| SPI-1/PU.1 | oe | ↑73 | X73 | X73 | ||||||
| STAT5A | oe | 224 | 224 | 224 | 224 | |||||
| TAX | oe | ↓235 | 235, 236 | 236, 237 | 235, 236 | 236 | ||||
Mechanisms of Replication Stress Induction by Oncogenes
In the following section, we will discuss the various mechanisms through which oncogenes induce RS.
Origin firing dysregulation
As mentioned earlier, replication is a fine-tuned process that ensures faithful DNA duplication once and only once per cell cycle. Dysregulation of CDK activity or mutations in the retinoblastoma/E2F pathway lead to perturbation of licensing or initiation, which in turn cause the unscheduled firing of origins. This may involve increased or decreased firing or re-firing of the same origin, all of which compromise physiological fork progression and lead to cells entering mitosis bearing under- or over-replicated DNA that eventually leads to GIN.
Different origins are activated at different time points during S-phase and modulation of CDKs affect the number of replication clusters rather than the origins within them (75). Interestingly, the level of origin firing inversely correlates with the rate of replication fork progression, which is believed to reflect the availability of essential replication factors (76). Under physiological conditions, origin firing is regulated by ATM and ATR by controlling CDK2 and CDC7 through CDC25A (77). CHK1 is crucial in this regulation, as its inhibition increases origin firing and leads to RS (78).
Decreased firing
Inhibition of DNA licensing reduces origin firing and induces GIN and increased sensitivity to RS-inducing agents (79). Compromised MCM loading due to reduced CDT1, CDC6 and ORC or increased CDT1 inhibitor GEMININ may hinder licensing. In the absence of functional p53, inhibition of DNA licensing in cancer cells allows their entry into S-phase with reduced origin firing (80). Similarly, ORC1 deletion reduces origin firing and increases sensitivity to hydroxyurea in tumour or MYC-expressing cells (81), suggesting a possible therapeutic approach to target certain cancers. Moreover, CDK deregulation in G1 through inhibition of Cdh1 and Sic1 reduces origin firing and causes GIN (82). In addition, CDKs phosphorylate CDC6 and protect it from APC/C-dependent proteolysis during G1 (83), thus disruption of this mechanism would enable licensing and perhaps origin firing during S-phase.
Oncogenes have also been shown to affect replication origin licensing. In particular, MYC deregulates expression of both CDKs and E2Fs and modifies cell cycle progression (reviewed in ref. 84). On the other hand, there seems to be controversial data regarding CYCLIN E. In one report, CYCLIN E overexpression impairs MCM2, 4 and 7 loading onto chromatin during telophase and early G1, which reduces the number of active replication origins in early S-phase (85). In contrast to this, other groups have shown that CYCLIN E overexpression increases the number of origins firing in S-phase (86, 87). This discrepancy could be attributed to different properties of the cell lines used in these studies, as CYCLIN E can promote or inhibit pre-RC formation depending on cellular context (88).
Intriguingly, decreased origin firing increases fork speed progression (76), raising the question of how this causes GIN. In mouse, Chaos3 is a viable mutation that destabilizes MCM4. MCM4Chaos3/Chaos3 MEFs exhibit a 60% reduction of all MCM2-7 components, which does not affect origin firing but does reduce the number of dormant origins and increases fork speeds (89). Due to the inability of the cells to repair endogenous RS through firing of dormant origins, fork stalling occurs and RS markers such as γH2AX, pRAD17 and RPA are activated, suggesting that GIN can occur even in the absence of direct fork slowing (89). Following the same pattern, E1A expressing cells exhibit reduced origin firing and increased fork speeds (72).
Increased firing
Untimely origin firing is the result of disruption of initiation control. This has been shown in X. laevis where increasing CDK activity accelerates the origin firing pattern (75), as well as in other systems where dysregulation of ATR, CHK1 or WEE1 that control CDK or CDC7 activity, cause extensive origin firing (90, 78, 77). Dysregulation of origin activation increases the possibilities of conflicts with transcription (87) and may lead to enhanced depletion of replication building blocks such as dNTPs (90, 86, 91), histones (92) or RPA (93), which hinder fork progression (Fig. 3B).
Many oncogenes disrupt the physiological origin firing schedule. In particular, Di Micco et al. showed RAS to induce a hyper-proliferation phase accompanied by increased origin firing that contributes to RS (94). HPV E6/7 and CYCLIN E overexpression enhances origin firing leading to GIN (86), while c-MYC overexpression increases origin activity in a transcription-independent (95) but CDC45 and GINS-dependent manner (96). Interestingly, inhibition of origin firing failed to abrogate RAS-induced RS (97) but did rescue CYCLIN E–induced RS (98), which again hints at different mechanisms through which oncogene activation leads to RS.
Re-firing
Disruption of DNA licensing as well as CDK-dependent protective pathways enable re-replication events, which are associated with GIN and tumourigenesis (99). In particular, CDT1 (and to a lesser extent CDC6) overexpression induce re-firing that leads to GIN (100), while depletion of GEMININ induces re-firing and activation of the DDR (101). Moreover, compromising the CUL4-DDB1 ubiquitin ligase, which regulates CDT1 (102) or loss of EMI1 that indirectly regulates GEMININ, also leads to re-replication (103). The ATR-dependent S-phase checkpoint surveys the genome and prevents re-replication caused by overexpression of licensing factors (but not GEMININ loss), either through p53-dependent activation of p21 or through dephosphorylation of retinoblastoma (104, 101, 100). Circumventing the retinoblastoma/E2F pathway is another way to instigate re-replication and CYCLIN E and c-MYC facilitate this (105), while HPV E7 ubiquitinates retinoblastoma marking it for degradation (106).
The main consequences of origin re-firing entail the so-called head-to-tail collisions that occur between the leading strand of the secondary fork and unligated Okazaki fragments of an adjacent fork, that cause DSBs and DNA damage checkpoint activation (99). Deregulated origin firing produces ssDNA gaps that promote re-replication at these sites, leading to replication fork breakage (107). Re-replication also causes fork slowing, although it is not clear if this is a consequence of head-to-tail collisions or due to normal forks colliding with the re-replication-induced DSBs. Overexpression of oncogenic RAS, CYCLIN E or MOS upregulate CDC6 (2, 94, 83) and in the case of RAS this causes re-replication (94). Furthermore, overexpression of a constitutively nuclear mutant form of CYCLIN D1 induces CDT1 stabilization that promotes re-licensing and re-replication (108).
Transcription
Transcription-replication conflicts
DNA transcription is a physiological process that may turn into an intrinsic source of GIN upon interference with the replication fork machinery. Transcription-replication conflicts (TRCs) impede replication fork progression as a result of head-on or co-directional collisions between the two machines or between replication and R-loops (Fig. 4).
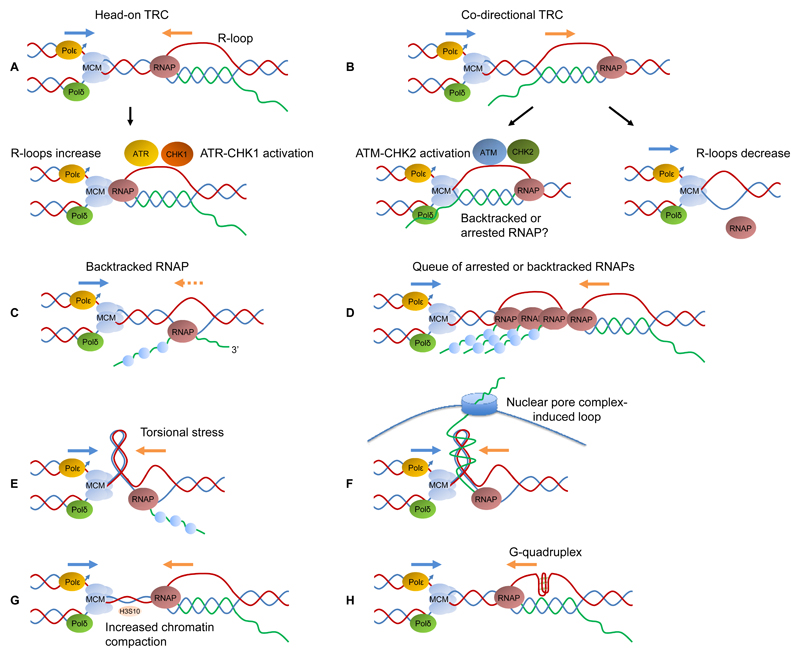
Figure 4.
Transcription-associated replication stress. A, upon head-on conflicts between replication and transcription, R-loop formation is increased and ATR/CHK1 are activated. B, upon co-directional conflicts between replication and a stalled or backtracked RNAP, DSBs are formed that activate ATM/CHK2 and R-loops are resolved. C, in response to any obstacle an RNAP may pause and backtrack and act as an impediment to fork progression; D, this can also occur as a result of an encounter with an R-loop. In both cases, this will cause accumulation of arrested or backtracked RNAPs that hinder fork progression. E, while the replication and transcription machineries move, positive supercoiling develops in front of them, that can impede progression of both. F, transcribed RNA may get trapped in the nuclear pore complex generating obstacles to fork progression. G, R-loops can cause chromatin condensation, marked by H3 phosphorylation at Ser-10, that impedes fork progression. H, G-quadruplexes can be formed co-transcriptionally or within the ssDNA part of an R-loop and act as obstacles to fork progression.
TRCs were first visualized by electron microscopy in E. coli, where an inducible origin was inserted upstream or downstream of an rRNA operon (109). Organisms have developed various strategies to avoid interactions between the two systems. In prokaryotes, where DNA is circular and there is a single origin of replication, co-orientation of the replisome and the transcription machinery is highly favoured (110). A similar orientation bias was also described in the human genome (111). In order to regulate replication through parts of the genome that are highly transcribed or difficult to replicate, both prokaryotes and eukaryotes have developed replication fork barriers comprising proteins bound tightly to DNA, which serve to limit TRCs. In eukaryotes where replication is mediated through multiple origins, transcription takes place throughout the cell cycle and is regulated spatially and temporally so that it does not clash with replication (112). Furthermore, in budding yeast, tRNA gene transcription is restrained during S-phase by Mec1/Mrc1/Rad53 in order to avoid collision with the replication machinery (113). RNApolII has a unique role in preventing and resolving TRCs, although it is not clear how this is achieved (114). Also, in E. coli the transcription factor DksA interacts with RNA polymerase (RNAP) and by altering the transcription elongation complex it prevents TRCs upon nutrition stress (115).
During transcription, RNAP may pause either as part of a regulatory checkpoint (promoter-proximal pausing) or in response to obstacles or misincorporated nucleotides that cause stalling and polymerase backtracking. Backtracking events involve RNAP sliding back and forth across DNA, which dislodges the 3’OH end of the RNA from the RNAP active site allowing it to exit through a secondary channel. Backtracked RNAPs can hinder replication, as well as transcription, and cause GIN (116) (Fig. 4C, D). To deal with arrested or backtracked RNAPs, cells employ various strategies, such as i) cleavage of the misplaced RNA transcript by GreA/B or TFIIS, ii) reduction of misincoroporated nucleotides by DksA, iii) RNAP removal by the combined actions of ppGpp, DksA, GreA and Mfd or UvrD and NusA (reviewed in ref. 117) or iv) transcription regulation by RECQL5 (118).
R-loops
R-loops are DNA:RNA hybrids that are formed during transcription, when the newly synthesized RNA remains tangled around the template DNA, while the homologous ssDNA is displaced. Mapping of R-loops in mammals revealed that their formation is prevalent, conserved and dynamic across the genome and is favoured by increased transcription activity, polyA tracts and unmethylated CpG islands (reviewed in ref. 119). Information on R-loop forming sequences across the genome of various species can be found online at R-loopDB (http://rloop.bii.a-star.edu.sg). R-loops associate with specific epigenomic signatures at promoters and terminators and are involved in mitochondrial replication (120), transcription regulation (121), IgG class switch recombination (122), telomere maintenance in ALT cells (123) and homologous recombination-mediated DSB repair (124). Nevertheless, accumulation of R-loops can act as an obstacle to replication fork progression either through direct collisions (125) or indirectly through increased chromatin compaction (126), which leads to CFS formation (127) and GIN (128) (Fig. 4G). The ssDNA part of R-loops renders DNA more susceptible to DNA damaging agents, such as activation-induced cytidine deaminase (AID), which regulates class switch recombination and somatic hypermutation in mammalian B cells (reviewed in ref. 129). AID deaminates cytidine only in ssDNA with base or nucleotide excision repair of these alterations, giving rise to mutations, chromosomal translocations or breaks that in turn may facilitate oncogenic activation.
R-loops may inhibit transcription progression (130) and consequently either on their own or through stalled or backtracked RNAPs act as additional barriers to transcription or replication (Fig. 4D). Their length may vary from a few hundred to over 1 kb and recent evidence in S. cerevisiae show that it is not length but histone modifications that discriminate between ‘benign’ and ‘malignant’ (131). In particular, Garcia-Pichardo et al. proposed a two-step model, where at first R-loop formation is facilitated by an altered chromatin configuration, followed by chromatin modifications (including H3S10 phosphorylation (126)) that render them culpable for GIN (131). In addition, experiments in Drosophila showed that depletion of linker histone H1 facilitates transcription of heterochromatic transcripts enabling R-loop accumulation (132). Similarly, in C. elegans, H3K9 methylation suppresses transcription of repetitive elements that enhance R-loops (133). Despite the acquired knowledge of R-loop-induced GIN, the exact mechanisms behind it still remain elusive.
R-loops are regulated by a plethora of molecules. In particular, RNaseH1 (128), RNaseH2 (134), SETX (135), DDX19 (136), DDX21 (137), DDX23 (138), DinG (139) and Pif1/Rrm3 helicases (140) degrade or unwind R-loops. ASF/SF2 (141), TOP1 (142), RECQ5 (143), THO/TREX (130), Npl3 (144), AQR (145) and XRN2 (146) prevent R-loop formation. Also, BRCA1/2 (147) and Fanconi anaemia proteins remove R-loops (148) while BRCA1 forms a complex with SETX at transcription termination pause sites, which collaborate to suppress R-loop accumulation (149). In addition, the chromatin reorganizing complex FACT facilitates resolution of R-loop-dependent GIN, most likely through remodelling chromatin at the sites of conflict (150). Moreover, introns were recently identified as an unexpected source of R-loop regulation, as their presence within highly transcribed regions of yeast or human genomes protects against R-loop accumulation and DNA damage (151).
Since oncogenes enhance transcription, it is reasonable that they should also affect R-loop levels. Only recently, HRASV12 overexpression was shown to increase R-loop accumulation that was detrimental to fork progression (97). As part of the same study, HRASV12 caused stabilization of RNAseH1, which can be interpreted as an intrinsic response to increased R-loop levels. Likewise, CYCLIN E-induced RS was rescued upon RNaseH1 overexpression (87).
Orientation of transcription-replication conflicts
Orientation of TRCs has a profound effect on DNA integrity. Studies in yeast have shown that head-on collisions are more detrimental than co-directional ones with respect to fork slowing (152). Nevertheless, co-directional collisions also disrupt replication (153). Moreover, in bacteria co-directional collisions with backtracked transcription complexes cause DSBs, while head-on collisions do not (116). Further experiments in bacteria have shown that with both types of collision, the replisome resumes elongation after displacing RNAP from the DNA and in the case of co-directional collision it uses the nascent transcript as a primer (reviewed in ref. 154). Using a sophisticated episomal system to induce either head-on or co-directional TRCs, Hamperl et al. showed in human cells that replication itself modulates R-loop formation depending on the orientation of the TRC. In particular, co-directional TRCs resolve R-loops and activate the ATM-CHK2 pathway, while head-on conflicts between replication and an R-loop (but not an R-loop-less transcription machinery) promote R-loop accumulation and activation of the ATR-CHK1 pathway (125). They also showed that normal replication fork progression suppresses R-loop accumulation, so any type of fork slowing increases R-loop levels (125). According to their proposed model, head-on TRCs cause fork slowing, which leads to increased R-loops levels that in turn activate ATR, while co-directional TRCs resolve R-loops, but the replisome may collide with backtracked RNAPs left by the R-loops that causes DSBs and ATM activation. Similar results were obtained in B. subtilis, where head-on TRCs induce R-loop formation, which stall replication, increase mutagenesis and prevent fork restart (155). These findings were further corroborated in S. cerevisiae, where in unchallenged conditions, a mutation in the MCM2-7 complex modulates replication and causes accumulation of R-loops in S-phase (156). Taken together, these data disclose a conserved and complex mechanism that protects the genome from TRC-related GIN.
Topological stress
Topological constraints may also develop between the replication and transcription machines as they move along DNA, causing positive or negative supercoiling, that may act as an obstacle to fork progression (157) (Fig. 4E). Furthermore, it has been suggested that nascent RNA can become tangled around template DNA and form a loop during nuclear export (Fig. 4F). These loops can obstruct the release of torsional stress from incoming replication forks and thus impede fork progression (158). Torsional stress is released by the activity of topoisomerases I and II, which relax positive supercoiling by creating nicks in DNA (159). RECQ5 facilitates SUMOylation of TOP1 which enables TOP1 binding to RNAPolII and at the same time compromises its activity, indicative of a regulatory mechanism of TOP1-induced genotoxicity (143). Also, in yeast Mec1 and Rad53 phosphorylate nucleoporin Mlp1 and resolve the topological stress (158). Finally, according to a recent study, p53 loss causes stabilization of TOP2A on DNA, as well as fork retardation, suggesting a possible role in preventing torsional stress between the two machines (160).
G-quadruplexes
G-quadruplexes are four-stranded structures held by G-G base pairs that are formed co-transcriptionally and impede replication fork progression leading to breakage (Fig. 4H). G-quadruplexes can form within the displaced ssDNA part of an R-loop of an active transcription bubble (161), where they facilitate R-loop stabilization (162). G-quadruplexes are also found within start sites of highly transcribed genes such as MYC (163), KRAS (164) or PTEN and regulate transcription (165). Moreover, the ORC complex has been suggested to bind to G-quadruplexes at putative origin sites of replication initiation (166). In order to avoid potential collisions with the replication machinery, helicases such as Pif1, RTEL1 or DHX9 bind G-quadruplexes and unwind them to maintain genomic stability (reviewed in ref. 167).
TRCs-associated DNA damage response
Certain areas of the genome are more susceptible to TRCs. Barlow et al. identified highly unstable regions of the genome, named early replication fragile sites (ERFSs) that are more prone to breakage upon RS than CFSs (168). ERFSs are usually found within highly transcribed gene clusters and although they may break during normal replication, their fragility is increased in response to hydroxyurea-induced RS, ATR inhibition, c-MYC expression or increased transcription (168). Additionally, long genes are more prone to TRCs, with frequent R-loop accumulation and induction of CFSs instability (127).
In response to DNA damage or TRCs, the cell tends to shut off transcription to reduce further collisions and facilitate replication restart. This is evident in yeast, where genotoxic-induced RS causes tRNA gene transcription repression by Maf1 (113) and through the synergistic effect of checkpoint sensor Mec1-Ddc2, the chromatin remodelling complex INO80C and transcription complex PAF1C, RNAPolII is evicted from chromatin and degraded (169). Accordingly, Dcr1 releases RNApolII from highly transcribed genes that are sites of replication-transcription collisions to prohibit further instability (170). Moreover, in response to transcription-blocking DNA damage, R-loop-dependent ATM activation triggers spliceosome displacement from chromatin, causing widespread splicing changes that presumably facilitate DNA repair (171). Interestingly, the de Bruin lab has shown that the cell responds to RS by upregulating transcription of E2F target genes that maintain checkpoint activation and facilitate DNA damage tolerance (172, 173). These data suggest that there needs to be a fine tuning between decreasing deleterious transcription and retaining and/or upregulating levels of checkpoint proteins.
Various proteins, especially helicases, preserve genomic stability by preventing or resolving TRCs. Amongst them, RECQL5 has a dual role in resolving conflicts between replication and RNApolI/RNApolII-dependent transcription by either promoting RAD18-dependent PCNA ubiquitination or through its helicase activity resolves RAD51-dependent replication intermediates at these sites (174). In S. pombe, Pfh1, a member of the Pif1 family of helicases, facilitates replication across highly transcribed RNApolII- and RNApolIII-dependent genes (175). Likewise, in S. cerevisiae, another Pif1 family member, Rrm3 allows replication through TRCs (152). Moreover, in E. coli, helicases DinG, Rep and UvrD collaborate to promote replication through transcription units (139).
In response to oncogenic signalling, transcription activity is enhanced (176–178, 38, 179, 180) and collisions between replication and transcription are more likely to occur. This is usually attributed to point mutations in the promoter region of proto-oncogenes that increase transcription activity. c-MYC is itself a transcription factor that under normal conditions drives a transcriptional programme that controls cell growth and division. Mitogenic growth signalling may deregulate c-MYC which can directly enhance RNApolI- (178), RNApolII- (179) and RNApolIII-dependent transcription (177). c-MYC increases transcription indirectly, by up- and/or downregulating selective target genes that in turn modify the cellular state and enable global RNA production (181). This transcription amplification has been documented in patient-derived tumours with elevated c-MYC thus establishing a link to tumorigenesis (179). CYCLIN E overexpression enhances both transcription activity and origin firing, causing increased TRCs and subsequent RS that can be rescued by either transcription or CDK inhibition (87). This is not the case with RAS overexpression, which causes RS, which can be rescued by transcription but not origin firing inhibition (97). RAS-induced enhanced transcription is driven through upregulation of transcription factors such as TBP, which is required for transcription of all promoters (97). Interestingly, overexpression of TBP alone causes RS, DNA damage, senescence and contributes to oncogenesis (182, 97).
A very recent report shed light on the mechanisms underlying oncogene-induced TRCs and GIN. Using sensitive seq-based assays Macheret and Halazonetis mapped replication initiation sites in human cells (183). They showed that under physiological conditions, transcription suppresses intragenic origin firing, but upon overexpression of CYCLIN E or MYC, G1-phase shortening drives earlier S-phase entry before completion of transcription. This allows origin firing within transcribing genes which leads to TRCs, fork collapse and DSB formation specifically at these sites. Moreover, this study revealed that fork collapse at oncogene-induced intragenic fired origins causes chromosomal translocations in a transcription-dependent manner both in their experimental setup as well as in a large cohort of human cancers (183). This highlights the importance of TRCs amongst the other mechanisms in inducing GIN.
Nucleotide metabolism
Nucleotides are the building blocks of the replication machine, therefore any dysregulation of their structure or levels has an immediate effect on fork progression. In mammals, dNTPs are either synthesized through the de novo pathway in the cytoplasm or by recycling nucleosides and nucleobases from nucleotide degradation through the salvage pathway, which acts both in the cytoplasm and in mitochondria (Fig. 5). The crucial step in dNTP synthesis is the reduction of ribonucleoside diphosphates to deoxyribonucleoside diphosphates, which is catalysed by the ribonucleotide reductase (RNR). RNR is a tetrameric protein composed of two catalytic (RRM1) and two regulatory (RRM2, RRM2B) subunits. RRM2 levels are rate-limiting for RNR activity and consequently dNTP levels, so in order to retain balanced dNTP pools, RRM2 expression increases in S-phase and is low during G1, G2 and M-phases (184). There is increasing evidence that part of RNR associates with the replication fork and that in response to DNA damage RNR components re-localize towards repair foci to increase dNTP availability (reviewed in ref. 185). In addition, ATR regulates dNTP pools by increasing RRM2 levels in response to DNA damage (11, 49). Similarly, MEFs from mice carrying an extra allele of RRM2 display increased RNR activity, which reduces chromosomal fragility following ATR inhibition (186).
Figure 5.
Nucleotide and ROS metabolism as a cause of replication stress. dNTPs are formed either through the de novo pathway from glutamine, glycine, folic acid, aspartate, 5-phosphoribosyl-1-pyrophosphate or by nucleoside degradation through the salvage pathway. The reduction of ribonucleosides to dNTPs is catalysed by the ribonucleotide reductase (RNR), which is compromised by two catalytic (RRM1) and two regulatory (RRM2, RRM2B) subunits. Oncogenes may target RNR activity or induce hyper-proliferation that will both reduce dNTP pool affecting fork progression. Oncogenes also induce production of ROS, including O2ˉ, H2O2 and OH•. O2ˉ is produced either by oxidation of NADPH by NOX enzymes or through aerobic respiration in mitochondria. H2O2 is generated by O2ˉ which is converted by superoxide dismutase (SOD), while OH• are produced from H2O2 in the presence of Fe+2. It is not clear if oxidized dNTPs affect replication fork speed.
dNTP levels are inversely related to origin firing and are key determinants of replication fork speed (90, 86, 91). Increased RNR activity accelerates fork progression, while reduced nucleotide pools instigate a transition to a slow replication mode with reduced origin usage, minutes after entry into S-phase (86, 91). RRM2/RRM2B-induced increase in replication fork speed causes misincorporated uracil into DNA and breaks at fragile sites and it has been suggested that dUTPase can sanitize this to avoid GIN (187). Nevertheless, dNTPs need to be regulated within certain levels, as increased dNTP pools will cause aberrant replication and lead to mutations (188).
With regards to the effect of oncogenes on nucleotide metabolism, BCL2 binds to RRM2 and disrupts the RRM1/RRM2 complex inhibiting RNR activity, which reduces nucleotide pool levels and leads to fork slowing (189). Moreover, overexpression of RAS downregulates RRM2 through binding of transcription repressor E2F7 to the RRM2 promoter, causing dNTP pool depletion, RS and senescence (190). In addition, CYCLIN E or HPV-16 E6/E7 overexpression activate cell proliferation but do not affect nucleotide biosynthesis, resulting in dNTP pool depletion and impaired fork progression (86). In contrast, c-MYC upregulates genes involved in nucleotide biosynthesis, increasing dNTP pools and enabling cell proliferation (191).
Upon reduced dNTP levels, there is increased incorporation of ribonucleotides (rNTPs) into DNA during replication. Since rNTPs are more prone to hydrolysis than dNTPs, their integration into DNA affects fork progression and may lead to GIN. Misincorporated rNTPs can be detrimental to genomic stability, as depletion of RNaseH2 that removes rNTPs causes RS (192). Despite the fact that there is no record of oncogenes promoting misincorporation of rNTPs on DNA, it seems quite likely that this may occur during the hyper-proliferation stage following oncogenic activation.
The integrity of dNTPs is another critical factor that may cause GIN. Reactive oxygen species (ROS) can oxidize nucleotides that when incorporated into DNA trigger breaks, mutations and senescence (193). Oxidized dNTPs trigger fork slowing in X. laevis, but this is attributed to a protein kinase C-mediated reduction in DNA replication and not to an actual fork retardation (194). So, although it seems reasonable for oxidized dNTPs to cause fork slowing it still has not been shown definitively (Fig. 5). A hint towards this view lies with MTH1, which is a non-essential enzyme that sanitizes oxidized dNTPs, hindering them from being incorporated into DNA, thus preserving genomic integrity.
Reactive oxygen species metabolism
ROS are an assorted class of free radical species produced under normal conditions as byproducts of aerobic metabolism (reviewed in ref. 195). ROS include superoxide anion (O2ˉ), hydrogen peroxide (H2O2) and hydroxyl radicals (OH•) and their biological effects are proportional to their levels. Basal ROS levels uphold physiological cellular functions as part of redox biology, while increased levels of ROS cause oxidative stress by damaging lipids, proteins and DNA or upregulating protein kinase Cδ and inducing senescence or apoptosis. O2ˉ is produced either by oxidation of nicotinamide adenine dinucleotide phosphate (NADPH) by NADPH oxidase enzymes (NOXs) or through aerobic respiration in mitochondria. H2O2 is generated from O2ˉ by superoxide dismutases and exerts its effect on redox biology or oxidative stress or may be converted to H2O by antioxidant proteins. Finally, OH• are produced from H2O2 in the presence of Fe+2 (196) (Fig. 5).
Cancer cells exhibit abrupt alterations in their metabolism to support their rapid growth. These include ATP generation to maintain energy levels, increased synthesis of macromolecules and maintenance of cellular redox status. Oncogenes are known to induce ROS and cause DNA damage. Expression of activated RAS increases intracellular and mitochondrial ROS and leads to senescence (197), which is partially achieved by upregulating NOX4-p22phox (198). c-MYC overexpression also induces ROS and causes DNA damage (199). A comparative study between RAS and MYC showed that both oncogenes evoke changes in the cellular metabolism patterns and induce different degrees of oxidative stress and RS, highlighting again that different oncogenes follow different mechanisms to cause RS (200). Moreover, in response to KRASG12D, B-RAFV619E and MYCERT2 primary murine cells respond by upregulating transcription factor Nrf2 that regulates an antioxidant program used to detoxificate the organism (201). In addition, expression of E6/E7 promotes NOX-dependent generation of ROS and subsequent DNA damage in head and neck cancer cells (202).
It is unclear how ROS affect fork progression. According to the D’adda di Fagagna lab, RAS induces ROS and this triggers the hyper-proliferation that leads to RS and senescence (94, 197, 203). On the other hand, CYCLIN E-induced hyper-proliferation is independent of ROS (2). Another case involves ROS oxidizing dNTPs, as was covered in the previous chapter.
Concluding Remarks
Oncogene-induced RS has long been recognised as an early driver of cancer and investigating the mechanics of this process has been established as a field in its own right. Understanding RS has accelerated cancer diagnosis and assisted the development of more sophisticated anticancer treatments. Nevertheless, the mechanics of how oncogene activation lead to RS present a complicated kaleidoscope of intertwined pathways and to make matters worse, various oncogenes may differentially affect these pathways based on the cell type or the time point their effect is assessed. So, the consensus that different oncogenes exert a different effect at different time points needs to be more widely adopted.
Despite the importance of each of the different mechanisms of oncogene-induced RS presented here, oncogene-induced deregulation of transcription appears to play the most significant role in cancer development, in part, due to its multifaceted nature in inducing GIN. The role of TRCs is especially intriguing, as currently little is known regarding the proteins and pathways involved in their detection and repair, as well as the implication of oncogenes in them. Although there is growing interest in R-loops and their involvement in GIN, there are a lot to be explored on their regulation and role in genome wide RS and telomere erosion and potential connections between them. Finally, there are still unanswered questions on the role of G-quadruplexes, rNTP missincorporation and oxidized nucleotides in oncogene-induced RS that need to be addressed.
Significance.
Replication stress is a fundamental step and an early driver of tumourigenesis and has been associated with many activated oncogenes. Deciphering the mechanisms that contribute to the replication stress response may provide new avenues for targeted cancer treatment. In this review, we discuss the latest findings on the DNA replication stress response and examine the various mechanisms through which activated oncogenes induce replication stress.
Acknowledgments
The authors would like to apologize to all researchers whose important work could not be cited due to space restrictions. The authors would also like to thank R. Bellelli, G. Hewitt, M. Maric, A. Milona, S. Panier and J. Stingele for comments and discussions.
Grant Support
The Boulton lab is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC0010048), the UK Medical Research Council (FC0010048), and the Wellcome Trust (FC0010048). S.J.B. is also funded by a European Research Council (ERC) Advanced Investigator Grant (TelMetab) and a Wellcome Trust Senior Investigator Grant.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016;17:185. doi: 10.1186/s13059-016-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16:360–74. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 8.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18:622–36. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass TE, Luzwick JW, Kavanaugh G, Carroll C, Dungrawala H, Glick GG, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol. 2016;18:1185–95. doi: 10.1038/ncb3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haahr P, Hoffmann S, Tollenaere MA, Ho T, Toledo LI, Mann M, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol. 2016;18:1196–207. doi: 10.1038/ncb3422. [DOI] [PubMed] [Google Scholar]
- 11.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–24. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–6. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo C, Kumagai A, Schlacher K, Shevchenko A, Dunphy WG. Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol Cell. 2015;57:492–505. doi: 10.1016/j.molcel.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge XQ, Blow JJ. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol. 2010;191:1285–97. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, et al. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 19.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–79. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–23. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 21.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–46. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 22.Betous R, Glick GG, Zhao R, Cortez D. Identification and characterization of SMARCAL1 protein complexes. PLoS One. 2013;8:e63149. doi: 10.1371/journal.pone.0063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105:16107–12. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fugger K, Mistrik M, Neelsen KJ, Yao Q, Zellweger R, Kousholt AN, et al. FBH1 Catalyzes Regression of Stalled Replication Forks. Cell Rep. 2015;10:1749–57. doi: 10.1016/j.celrep.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Kile AC, Chavez DA, Bacal J, Eldirany S, Korzhnev DM, Bezsonova I, et al. HLTF's Ancient HIRAN Domain Binds 3' DNA Ends to Drive Replication Fork Reversal. Mol Cell. 2015;58:1090–100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell. 2017;67:882–90. doi: 10.1016/j.molcel.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Liu Z, Wang F, Temviriyanukul P, Ma X, Tu Y, et al. FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res. 2015;43:8325–39. doi: 10.1093/nar/gkv737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su F, Mukherjee S, Yang Y, Mori E, Bhattacharya S, Kobayashi J, et al. Nonenzymatic role for WRN in preserving nascent DNA strands after replication stress. Cell Rep. 2014;9:1387–401. doi: 10.1016/j.celrep.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell. 2015;59:462–77. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Kim TM, Son MY, Dodds S, Hu L, Luo G, Hasty P. RECQL5 and BLM exhibit divergent functions in cells defective for the Fanconi anemia pathway. Nucleic Acids Res. 2015;43:893–903. doi: 10.1093/nar/gku1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leuzzi G, Marabitti V, Pichierri P, Franchitto A. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 2016;35:1437–51. doi: 10.15252/embj.201593265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–62. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–9. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 36.Olmezer G, Levikova M, Klein D, Falquet B, Fontana GA, Cejka P, et al. Replication intermediates that escape Dna2 activity are processed by Holliday junction resolvase Yen1. Nat Commun. 2016;7:13157. doi: 10.1038/ncomms13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–23. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotsantis P, Jones RM, Higgs MR, Petermann E. Cancer therapy and replication stress: forks on the road to perdition. Adv Clin Chem. 2015;69:91–138. doi: 10.1016/bs.acc.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Ait Saada A, Teixeira-Silva A, Iraqui I, Costes A, Hardy J, Paoletti G, et al. Unprotected Replication Forks Are Converted into Mitotic Sister Chromatid Bridges. Mol Cell. 2017;66:398–410. doi: 10.1016/j.molcel.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Dehe PM, Gaillard PH. Control of structure-specific endonucleases to maintain genome stability. Nat Rev Mol Cell Biol. 2017;18:315–30. doi: 10.1038/nrm.2016.177. [DOI] [PubMed] [Google Scholar]
- 41.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64:1117–26. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 42.Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, et al. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–90. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 43.Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell. 2016;64:1127–34. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Marco S, Hasanova Z, Kanagaraj R, Chappidi N, Altmannova V, Menon S, et al. RECQ5 Helicase Cooperates with MUS81 Endonuclease in Processing Stalled Replication Forks at Common Fragile Sites during Mitosis. Mol Cell. 2017;66:658–71. doi: 10.1016/j.molcel.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122:241–52. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell. 2015;28:557–68. doi: 10.1016/j.ccell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjiv K, Hagenkort A, Calderon-Montano JM, Koolmeister T, Reaper PM, Mortusewicz O, et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016;17:3407–16. doi: 10.1016/j.celrep.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–61. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–7. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–48. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 54.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 55.Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, et al. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21:1077–91. doi: 10.1016/j.devcel.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, et al. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25:243–56. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–53. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 58.Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–31. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 59.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–60. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 60.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–54. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plug-DeMaggio AW, Sundsvold T, Wurscher MA, Koop JI, Klingelhutz AJ, McDougall JK. Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E6. Oncogene. 2004;23:3561–71. doi: 10.1038/sj.onc.1207388. [DOI] [PubMed] [Google Scholar]
- 62.Patel PL, Suram A, Mirani N, Bischof O, Herbig U. Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence. Proc Natl Acad Sci U S A. 2016;113:E5024–33. doi: 10.1073/pnas.1602379113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31:2839–51. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–6. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 65.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 66.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 67.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 68.Miron K, Golan-Lev T, Dvir R, Ben-David E, Kerem B. Oncogenes create a unique landscape of fragile sites. Nat Commun. 2015;6 doi: 10.1038/ncomms8094. [DOI] [PubMed] [Google Scholar]
- 69.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 70.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deutzmann A, Ganz M, Schonenberger F, Vervoorts J, Kappes F, Ferrando-May E. The human oncoprotein and chromatin architectural factor DEK counteracts DNA replication stress. Oncogene. 2015;34:4270–7. doi: 10.1038/onc.2014.346. [DOI] [PubMed] [Google Scholar]
- 72.Singhal G, Leo E, Setty SK, Pommier Y, Thimmapaya B. Adenovirus E1A oncogene induces rereplication of cellular DNA and alters DNA replication dynamics. J Virol. 2013;87:8767–78. doi: 10.1128/JVI.00879-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rimmele P, Komatsu J, Hupe P, Roulin C, Barillot E, Dutreix M, et al. Spi-1/PU.1 oncogene accelerates DNA replication fork elongation and promotes genetic instability in the absence of DNA breakage. Cancer Res. 2010;70:6757–66. doi: 10.1158/0008-5472.CAN-09-4691. [DOI] [PubMed] [Google Scholar]
- 74.Sincennes MC, Humbert M, Grondin B, Lisi V, Veiga DF, Haman A, et al. The LMO2 oncogene regulates DNA replication in hematopoietic cells. Proc Natl Acad Sci U S A. 2016;113:1393–8. doi: 10.1073/pnas.1515071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomson AM, Gillespie PJ, Blow JJ. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol. 2010;188:209–21. doi: 10.1083/jcb.200911037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, et al. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol. 2013;201:373–83. doi: 10.1083/jcb.201208060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–55. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 78.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–5. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–8. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 80.Nevis KR, Cordeiro-Stone M, Cook JG. Origin licensing and p53 status regulate Cdk2 activity during G(1) Cell Cycle. 2009;8:1952–63. doi: 10.4161/cc.8.12.8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimmerman KM, Jones RM, Petermann E, Jeggo PA. Diminished origin licensing capacity specifically sensitises tumour cells to replication stress. Mol Cancer Res. 2013;11:370–80. doi: 10.1158/1541-7786.MCR-12-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ayuda-Duran P, Devesa F, Gomes F, Sequeira-Mendes J, Avila-Zarza C, Gomez M, et al. The CDK regulators Cdh1 and Sic1 promote efficient usage of DNA replication origins to prevent chromosomal instability at a chromosome arm. Nucleic Acids Res. 2014;42:7057–68. doi: 10.1093/nar/gku313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–26. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 84.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–16. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 85.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2012;32:3744–53. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 88.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:778–86. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 89.Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–53. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beck H, Nahse-Kumpf V, Larsen MS, O'Hanlon KA, Patzke S, Holmberg C, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–36. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, et al. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–94. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 94.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 95.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 96.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep. 2013;3:1629–39. doi: 10.1016/j.celrep.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kotsantis P, Silva LM, Irmscher S, Jones RM, Folkes L, Gromak N, et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun. 2016;7:13087. doi: 10.1038/ncomms13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–53. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 99.Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24:433–43. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 101.Melixetian M, Ballabeni A, Masiero L, Gasparini P, Zamponi R, Bartek J, et al. Loss of Geminin induces rereplication in the presence of functional p53. J Cell Biol. 2004;165:473–82. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–9. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 103.Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–94. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu E, Lee AY, Chiba T, Olson E, Sun P, Wu X. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J Cell Biol. 2007;179:643–57. doi: 10.1083/jcb.200704138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–33. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4. [PubMed] [Google Scholar]
- 107.Neelsen KJ, Zanini IM, Mijic S, Herrador R, Zellweger R, Ray Chaudhuri A, et al. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013;27:2537–42. doi: 10.1101/gad.226373.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–22. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–5. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 110.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, et al. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–85. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–6. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 113.Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol. 2010;17:976–81. doi: 10.1038/nsmb.1857. [DOI] [PubMed] [Google Scholar]
- 114.Felipe-Abrio I, Lafuente-Barquero J, Garcia-Rubio ML, Aguilera A. RNA polymerase II contributes to preventing transcription-mediated replication fork stalls. EMBO J. 2015;34:236–50. doi: 10.15252/embj.201488544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, et al. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–43. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–49. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chedin F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet. 2016;32:828–38. doi: 10.1016/j.tig.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 1996;15:3135–43. [PMC free article] [PubMed] [Google Scholar]
- 121.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–9. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 123.Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell. 2016;167:1001–13. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 125.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:774–86. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52:583–90. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 127.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–77. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 128.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–88. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Di Noia JM. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat Rev Immunol. 2016;16:164–76. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–21. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 131.Garcia-Pichardo D, Canas JC, Garcia-Rubio ML, Gomez-Gonzalez B, Rondon AG, Aguilera A. Histone Mutants Separate R Loop Formation from Genome Instability Induction. Mol Cell. 2017;66:597–609. doi: 10.1016/j.molcel.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 132.Bayona-Feliu A, Casas-Lamesa A, Reina O, Bernues J, Azorin F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat Commun. 2017;8:283. doi: 10.1038/s41467-017-00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet. 2016;48:1385–95. doi: 10.1038/ng.3672. [DOI] [PubMed] [Google Scholar]
- 134.Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–43. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hodroj D, Recolin B, Serhal K, Martinez S, Tsanov N, Abou Merhi R, et al. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017;36:1182–98. doi: 10.15252/embj.201695131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Song C, Hotz-Wagenblatt A, Voit R, Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017;31:1–12. doi: 10.1101/gad.300624.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sridhara SC, Carvalho S, Grosso AR, Gallego-Paez LM, Carmo-Fonseca M, de Almeida SF. Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation. Cell Rep. 2017;18:334–43. doi: 10.1016/j.celrep.2016.12.050. [DOI] [PubMed] [Google Scholar]
- 139.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–57. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tran PLT, Pohl TJ, Chen CF, Chan A, Pott S, Zakian VA. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun. 2017;8:15025. doi: 10.1038/ncomms15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–78. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 142.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li M, Pokharel S, Wang JT, Xu X, Liu Y. RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat Commun. 2015;6:6720. doi: 10.1038/ncomms7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27:2445–58. doi: 10.1101/gad.229880.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–85. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, et al. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016;12:e1006107. doi: 10.1371/journal.pgen.1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–5. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 148.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, et al. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol Cell. 2015;60:351–61. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–47. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014;28:735–48. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, et al. Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability. Mol Cell. 2017;67:608–21. doi: 10.1016/j.molcel.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 152.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–76. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–7. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–43. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, et al. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell. 2017;170:787–99. doi: 10.1016/j.cell.2017.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vijayraghavan S, Tsai FL, Schwacha A. A Checkpoint-Related Function of the MCM Replicative Helicase Is Required to Avert Accumulation of RNA:DNA Hybrids during S-phase and Ensuing DSBs during G2/M. PLoS Genet. 2016;12:e1006277. doi: 10.1371/journal.pgen.1006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–8. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 158.Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–46. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–36. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yeo CQ, Alexander I, Lin Z, Lim S, Aning OA, Kumar R, et al. p53 Maintains Genomic Stability by Preventing Interference between Transcription and Replication. Cell Rep. 2016;15:132–46. doi: 10.1016/j.celrep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 161.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–29. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Abraham KJ, Chan JN, Salvi JS, Ho B, Hall A, Vidya E, et al. Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA-DNA hybrids. Nucleic Acids Res. 2016;44:8870–84. doi: 10.1093/nar/gkw752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–8. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–49. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48:1267–72. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 166.Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, et al. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem. 2013;288:30161–71. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–32. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 2016;30:337–54. doi: 10.1101/gad.273813.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Castel SE, Ren J, Bhattacharjee S, Chang AY, Sanchez M, Valbuena A, et al. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159:572–83. doi: 10.1016/j.cell.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–8. doi: 10.1038/nature14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, de Bruin RA. Sustained E2F-Dependent Transcription Is a Key Mechanism to Prevent Replication-Stress-Induced DNA Damage. Cell Rep. 2016;15:1412–22. doi: 10.1016/j.celrep.2016.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bertoli C, Klier S, McGowan C, Wittenberg C, de Bruin RA. Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell-cycle transcription. Curr Biol. 2013;23:1629–37. doi: 10.1016/j.cub.2013.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Urban V, Dobrovolna J, Huhn D, Fryzelkova J, Bartek J, Janscak P. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J Cell Biol. 2016;214:401–15. doi: 10.1083/jcb.201507099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 2012;26:581–93. doi: 10.1101/gad.184697.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gerby B, Tremblay CS, Tremblay M, Rojas-Sutterlin S, Herblot S, Hebert J, et al. SCL, LMO1 and Notch1 reprogram thymocytes into self-renewing cells. PLoS Genet. 2014;10:e1004768. doi: 10.1371/journal.pgen.1004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 178.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–8. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 179.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–83. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 181.Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Johnson SA, Dubeau L, Kawalek M, Dervan A, Schonthal AH, Dang CV, et al. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol Cell Biol. 2003;23:3043–51. doi: 10.1128/MCB.23.9.3043-3051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018;555:112–16. doi: 10.1038/nature25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–34. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Techer H, Koundrioukoff S, Nicolas A, Debatisse M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet. 2017;18:535–50. doi: 10.1038/nrg.2017.46. [DOI] [PubMed] [Google Scholar]
- 186.Lopez-Contreras AJ, Specks J, Barlow JH, Ambrogio C, Desler C, Vikingsson S, et al. Increased Rrm2 gene dosage reduces fragile site breakage and prolongs survival of ATR mutant mice. Genes Dev. 2015;29:690–5. doi: 10.1101/gad.256958.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen CW, Tsao N, Huang LY, Yen Y, Liu X, Lehman C, et al. The Impact of dUTPase on Ribonucleotide Reductase-Induced Genome Instability in Cancer Cells. Cell Rep. 2016;16:1287–99. doi: 10.1016/j.celrep.2016.06.094. [DOI] [PubMed] [Google Scholar]
- 188.Davidson MB, Katou Y, Keszthelyi A, Sing TL, Xia T, Ou J, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31:895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xie M, Yen Y, Owonikoko TK, Ramalingam SS, Khuri FR, Curran WJ, et al. Bcl2 induces DNA replication stress by inhibiting ribonucleotide reductase. Cancer Res. 2014;74:212–23. doi: 10.1158/0008-5472.CAN-13-1536-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–65. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, et al. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet. 2015;24:649–58. doi: 10.1093/hmg/ddu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rai P, Onder TT, Young JJ, McFaline JL, Pang B, Dedon PC, et al. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc Natl Acad Sci U S A. 2009;106:169–74. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kai T, Matsunaga R, Eguchi M, Kamiya H, Kasai H, Suzuki M, et al. An oxidized nucleotide affects DNA replication through activation of protein kinases in Xenopus egg lysates. Nucleic Acids Res. 2002;30:569–73. doi: 10.1093/nar/30.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 196.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, et al. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274:7936–40. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 198.Weyemi U, Lagente-Chevallier O, Boufraqech M, Prenois F, Courtin F, Caillou B, et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2012;31:1117–29. doi: 10.1038/onc.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–44. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 200.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol. 2015;9:601–16. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Marullo R, Werner E, Zhang H, Chen GZ, Shin DM, Doetsch PW. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis. 2015;36:1397–406. doi: 10.1093/carcin/bgv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ogrunc M, Di Micco R, Liontos M, Bombardelli L, Mione M, Fumagalli M, et al. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ. 2014;21:998–1012. doi: 10.1038/cdd.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, et al. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14:458–70. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wajapeyee N, Wang SZ, Serra RW, Solomon PD, Nagarajan A, Zhu X, et al. Senescence induction in human fibroblasts and hematopoietic progenitors by leukemogenic fusion proteins. Blood. 2010;115:5057–60. doi: 10.1182/blood-2009-09-245928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tsunematsu T, Takihara Y, Ishimaru N, Pagano M, Takata T, Kudo Y. Aurora-A controls pre-replicative complex assembly and DNA replication by stabilizing geminin in mitosis. Nat Commun. 2013;4:1885. doi: 10.1038/ncomms2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang D, Shimizu T, Araki N, Hirota T, Yoshie M, Ogawa K, et al. Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice. Oncogene. 2008;27:4305–14. doi: 10.1038/onc.2008.76. [DOI] [PubMed] [Google Scholar]
- 208.Xu M, Yu Q, Subrahmanyam R, Difilippantonio MJ, Ried T, Sen JM. Beta-catenin expression results in p53-independent DNA damage and oncogene-induced senescence in prelymphomagenic thymocytes in vivo. Mol Cell Biol. 2008;28:1713–23. doi: 10.1128/MCB.01360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Crescenzi E, Palumbo G, Brady HJ. Bcl-2 activates a programme of premature senescence in human carcinoma cells. Biochem J. 2003;375:263–74. doi: 10.1042/BJ20030868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Koptyra M, Falinski R, Nowicki MO, Stoklosa T, Majsterek I, Nieborowska-Skorska M, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108:319–27. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sattler M, Verma S, Shrikhande G, Byrne CH, Pride YB, Winkler T, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem. 2000;275:24273–8. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 212.Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–12. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 213.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 214.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 215.Lorvellec M, Dumon S, Maya-Mendoza A, Jackson D, Frampton J, Garcia P. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells. 2010;28:1751–9. doi: 10.1002/stem.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–16. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sideridou M, Zakopoulou R, Evangelou K, Liontos M, Kotsinas A, Rampakakis E, et al. Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J Cell Biol. 2011;195:1123–40. doi: 10.1083/jcb.201108121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tort F, Bartkova J, Sehested M, Orntoft T, Lukas J, Bartek J. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–63. doi: 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- 220.Brown NE, Jeselsohn R, Bihani T, Hu MG, Foltopoulou P, Kuperwasser C, et al. Cyclin D1 activity regulates autophagy and senescence in the mammary epithelium. Cancer Res. 2012;72:6477–89. doi: 10.1158/0008-5472.CAN-11-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39:7465–76. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liontos M, Niforou K, Velimezi G, Vougas K, Evangelou K, Apostolopoulou K, et al. Modulation of the E2F1-driven cancer cell fate by the DNA damage response machinery and potential novel E2F1 targets in osteosarcomas. Am J Pathol. 2009;175:376–91. doi: 10.2353/ajpath.2009.081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20:273–85. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Leikam C, Hufnagel A, Schartl M, Meierjohann S. Oncogene activation in melanocytes links reactive oxygen to multinucleated phenotype and senescence. Oncogene. 2008;27:7070–82. doi: 10.1038/onc.2008.323. [DOI] [PubMed] [Google Scholar]
- 226.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62:7075–82. [PubMed] [Google Scholar]
- 227.Frum RA, Singh S, Vaughan C, Mukhopadhyay ND, Grossman SR, Windle B, et al. The human oncoprotein MDM2 induces replication stress eliciting early intra-S-phase checkpoint response and inhibition of DNA replication origin firing. Nucleic Acids Res. 2014;42:926–40. doi: 10.1093/nar/gkt944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Reimann M, Loddenkemper C, Rudolph C, Schildhauer I, Teichmann B, Stein H, et al. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood. 2007;110:2996–3004. doi: 10.1182/blood-2007-02-075614. [DOI] [PubMed] [Google Scholar]
- 229.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–59. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 230.Thornber K, Colomba A, Ceccato L, Delsol G, Payrastre B, Gaits-Iacovoni F. Reactive oxygen species and lipoxygenases regulate the oncogenicity of NPM-ALK-positive anaplastic large cell lymphomas. Oncogene. 2009;28:2690–6. doi: 10.1038/onc.2009.125. [DOI] [PubMed] [Google Scholar]
- 231.Leventaki V, Drakos E, Medeiros LJ, Lim MS, Elenitoba-Johnson KS, Claret FX, et al. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood. 2007;110:1621–30. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- 232.Ceccon M, Merlo ME, Mologni L, Poggio T, Varesio LM, Menotti M, et al. Excess of NPM-ALK oncogenic signaling promotes cellular apoptosis and drug dependency. Oncogene. 2016;35:3854–65. doi: 10.1038/onc.2015.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Martinelli P, Bonetti P, Sironi C, Pruneri G, Fumagalli C, Raviele PR, et al. The lymphoma-associated NPM-ALK oncogene elicits a p16INK4a/pRb-dependent tumor-suppressive pathway. Blood. 2011;117:6617–26. doi: 10.1182/blood-2010-08-301135. [DOI] [PubMed] [Google Scholar]
- 234.Carlos AR, Escandell JM, Kotsantis P, Suwaki N, Bouwman P, Badie S, et al. ARF triggers senescence in Brca2-deficient cells by altering the spectrum of p53 transcriptional targets. Nat Commun. 2013;4:2697. doi: 10.1038/ncomms3697. [DOI] [PubMed] [Google Scholar]
- 235.Chaib-Mezrag H, Lemacon D, Fontaine H, Bellon M, Bai XT, Drac M, et al. Tax impairs DNA replication forks and increases DNA breaks in specific oncogenic genome regions. Mol Cancer. 2014;13:205. doi: 10.1186/1476-4598-13-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kinjo T, Ham-Terhune J, Peloponese JM, Jr, Jeang KT. Induction of reactive oxygen species by human T-cell leukemia virus type 1 tax correlates with DNA damage and expression of cellular senescence marker. J Virol. 2010;84:5431–7. doi: 10.1128/JVI.02460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Takahashi M, Higuchi M, Makokha GN, Matsuki H, Yoshita M, Tanaka Y, et al. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood. 2013;122:715–25. doi: 10.1182/blood-2013-03-493718. [DOI] [PubMed] [Google Scholar]