Abstract
Carcinogenic effect of tobacco in oral cancer is through chewing and/or smoking. Significant differences exist in development of oral cancer between tobacco users and non-users. However, molecular alterations induced by different forms of tobacco are yet to be fully elucidated. We developed cellular models of chronic exposure to chewing tobacco and cigarette smoke using immortalized oral keratinocytes. Chronic exposure to tobacco resulted in increased cell scattering and invasiveness in immortalized oral keratinocytes. miRNA sequencing using Illumina HiSeq 2500 resulted in the identification of 10 significantly dysregulated miRNAs (4 fold; p ≤ 0.05) in chewing tobacco treated cells and 6 in cigarette smoke exposed cells. We integrated this data with global proteomic data and identified 36 protein targets that showed inverse expression pattern in chewing tobacco treated cells and 16 protein targets that showed inverse expression in smoke exposed cells. In addition, we identified 6 novel miRNAs in chewing tobacco treated cells and 18 novel miRNAs in smoke exposed cells. Integrative analysis of dysregulated miRNAs and their targets indicates that signaling mechanisms leading to oncogenic transformation are distinct between both forms of tobacco. Our study demonstrates alterations in miRNA expression in oral cells in response to two frequently used forms of tobacco.
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common cancers worldwide and remains the most common malignancy of the head and neck cancers. Tobacco use, alcohol consumption and human papilloma virus (HPV) 16/18 have been identified as the main risk factors for the initiation and progression of OSCC1. Tobacco is mainly consumed worldwide in the form of manufactured cigarettes. Tobacco is also consumed in the form of smokeless tobacco, especially chewing tobacco in South-East Asian countries2. Despite being one of the most common cancers in India, molecular alterations in oral cancer development in tobacco chewers and smokers is not well understood.
MicroRNAs have been established as key regulators of oncogenic potential in cells. Alterations at the genetic and epigenetic levels in the complex enzymatic machinery involved in miRNA biogenesis can result in aberrant miRNA expression3. Post-transcriptional regulation of gene expression by miRNAs has an influence on multiple pathways, including those involved in cellular transformation and proliferation4. miRNAs function as either oncogenes or tumor suppressors, playing crucial roles in tumorigenesis, tumor invasion and metastasis5. In recent years multiple studies have revealed the altered expression of miRNAs which play a role in the development and progression of diverse cancers including oral squamous cell carcinoma6–8. Increased expression of microRNAs including miR-155 and miR-23a, have been observed in oral cancer patients that are tobacco chewers compared to non-chewers9,10. Fanconi anemia complementation group G protein (FANCG) is a miR-23a target with a role in DNA double strand break repair pathway. Decreased expression of FANCG in normal oral fibroblasts contributes to the development of carcinogenesis on treatment with areca nut10. In contrast, microRNAs such as miR-145 are found to be significantly downregulated upon cigarette smoke condensate treatment in oral fibroblasts while its target protein MMP-2 is overexpressed which plays a key role in perturbation of stromal-epithelial communication and promotes pro-tumorogenic interactions11. Similarly, a decrease in miR-101-3p and a corresponding increase in expression levels of its target protein COX2 was observed in an esophageal non-tumorigenic cell line upon treatment with cigarette smoke condensate thereby facilitating cell transformation and cancer development12. Downregulation of miR-200c levels in human bronchial epithelial cells with increased expression of IL-6 and activation of nuclear factor-κB (NF-κB) pathway by cigarette smoke extract is also shown to regulate epithelial-mesenchymal transition and carcinogenesis13.
Taken together, these studies indicate that molecular mechanisms for cellular transformation may vary depending upon the form of tobacco used. Till date no study has systematically investigated the differences in molecular alterations induced in oral cells upon exposure to different forms of tobacco. To achieve this, we developed two cellular models where immortalized, oral keratinocytes (OKF6/TERT1) were chronically treated with either chewing tobacco or exposed to cigarette smoke for a period of six months. To understand specific molecular alterations brought about by each form of tobacco, we performed miRNA sequencing of oral keratinocytes chronically treated with chewing tobacco/cigarette smoke. miRNA dysregulation is in turn known to affect expression of their target proteins leading to diverse functional consequences. Hence, in addition to studying miRNA dysregulation, we have investigated proteomic alterations associated with exposure to these two forms of tobacco.
We observed that chronic treatment of, immortalized oral keratinocytes with either chewing tobacco extract or cigarette smoke condensate affected expression of distinct set of miRNAs and their corresponding protein targets.
Results
Chronic exposure to chewing tobacco and cigarette smoke results in phenotypic changes in oral cells
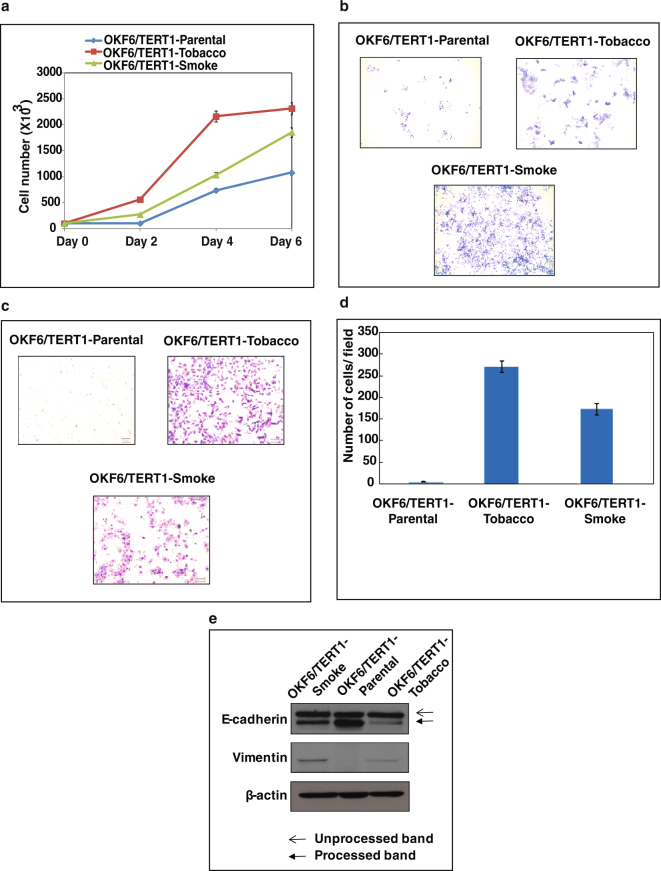
In this study, we chronically treated immortalized oral keratinocytes (OKF6/TERT1) with chewing tobacco extract and cigarette smoke condensate to model the effects of tobacco chewing and smoking in oral cancer. We observed an increase in proliferative capability of both OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells compared to OKF6/TERT1-Parental cells (Fig. 1a). Both tobacco treated and smoke exposed cells displayed increased cell scattering phenomena (Fig. 1b) which is one of the features of cancer cell phenotype. We also observed an increase in the invasive ability of oral keratinocytes upon chronic treatment with chewing tobacco/cigarette smoke (Fig. 1c,d). We also observed change in expression levels of markers associated with EMT in both chewing tobacco treated and cigarette smoke exposed cells compared to parental cells.There was a decrease in expression of E-Cadherin (processed band, lower) and a increase in vimentin expression in OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells compared to OKF6/TERT1-Parental cells (Fig. 1e). To further elucidate the molecular alterations imparted by each type of insult, we studied miRNA and proteomic expression pattern in these cells compared to untreated parental cells.
Figure 1.
Chronic treatment of oral keratinocytes with chewing tobacco and cigarette smoke. OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells were assessed for (a) cellular proliferation (b) colony formation (c,d) invasive ability and (e) epithelial–mesenchymal transition (EMT) using Western blotting. β-actin was used as a loading control.
Chronic exposure to chewing tobacco/cigarette smoke results in altered miRNA expression in oral keratinocytes
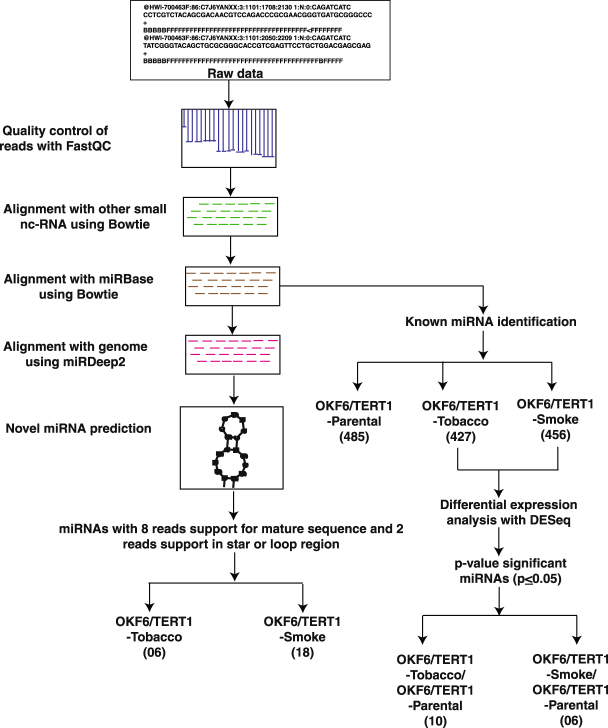
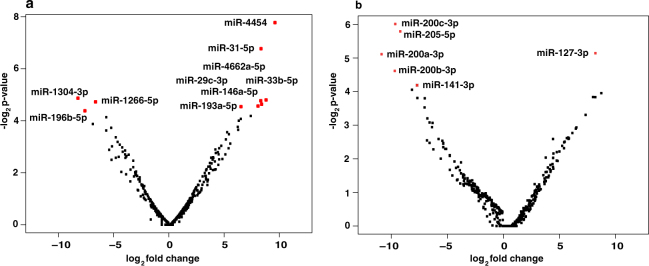
We carried out deep small RNA sequencing on Illumina HiSeq 2500 platform. We acquired 31 million reads for untreated control cells, 56 million reads for tobacco treated cells and 47 million reads for smoke exposed cells. A schematic representation of the workflow is provided in Supplementary Fig. S1. miRNA expression analysis resulted in identification of 427 and 456 miRNAs in chewing tobacco treated and cigarette smoke exposed oral keratinocytes, respectively (Fig. 2). Amongst these 10 miRNAs were significantly dysregulated (≥4 fold, p ≤ 0.05) in chewing tobacco treated cells and 6 miRNAs were significantly dysregulated in cigarette smoke exposed cells (Fig. 3a,b). A complete list of miRNAs along with corresponding fold-changes in OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells are provided in Supplementary Tables S1A,B.
Figure 2.
Bioinformatics workflow. Pipeline depicting the analysis of dysregulated miRNA in OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells and novel miRNA identification.
Figure 3.
A volcano plot showing distribution of miRNAs in (a) OKF6/TERT1- Tobacco cells (b) OKF6/TERT1-Smoke cells. The red dots represent significantly dysregulated miRNAs (p-value ≤ 0.05).
Altered protein expression pattern associated with chronic exposure to chewing tobacco/cigarette smoke in oral keratinocytes
We employed a TMT-based proteomic approach to investigate global proteomic alterations in response to chewing tobacco/cigarette smoke treatment. LC-MS - based quantitative proteomics led to quantification of 2,821 and 5,342 proteins in OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells, respectively. The proteomics workflow employed to carry out this investigation is outlined in Supplementary Fig. S1. Amongst the 311 significantly dysregulated proteins in OKF6/TERT1-Tobacco, 197 were overexpressed (≥1.5-fold, p-value ≤ 0.05) and 114 proteins were downregulated (≤0.6-fold, p-value ≤ 0.05). In case of OKF6/TERT1-Smoke cells, 229 proteins were overexpressed (≥1.5-fold, p-value ≤ 0.05) and 159 were downregulated (≤0.6-fold, p-value ≤ 0.05). The complete list of identified proteins and peptides is provided in Supplementary Tables S2A,B.
Correlation of miRNAs with protein expression
Since we observed altered expression of both miRNAs and proteins in response to two forms of tobacco, we compared expression pattern using experimentally proven targets of miRNAs from miRTarBase14. We fetched targets of 10 significantly dysregulated miRNAs identified in OKF6/TERT1-Tobacco cells from miRTarBase. There were 293 proteins that were identified in our proteomics data (Supplementary Table S3A). In case of OKF6/TERT1-Smoke cells, we identified 218 protein targets in our proteomic data for the 6 significantly dysregulated miRNAs (Supplementary Table S3B). To understand the correlation between significantly dysregulated miRNAs and their target proteins, miRNA-gene interaction network analysis was generated using miRNet tool15. The 10 miRNAs significantly dysregulated in OKF6/TERT1-Tobacco cells showed directional relationship with 36 target proteins identified in proteomics data (Fig. 4a). Reverse transcriptase PCR-based validation of some of the targets is provided in Supplementary Fig. S2A. Our proteomics data revealed downregulation of superoxidase dismutase, SOD2, in tobacco treated OKF6/TERT1 cells and in conjunction with this we observed a significant decrease in SOD2 expression in OKF6/TERT1-Tobacco cells compared to OKF6/TERT1-Parental cells (Supplementary Fig. S2A, see Western panel). The 6 miRNAs which were significantly dysregulated in OKF6/TERT1-Smoke cells showed directional relationship with 16 target proteins (Fig. 4b). Expression of some of these targets was validated using reverse transcriptase PCR and is provided in Supplementary Fig. S2B. We observed a 1.8 fold increase in TGFB2 expression in smoke exposed OKF6/TERT1 cells in our proteomics data and this increase in expression was confirmed by Western blot analysis of OKF6/TERT1-Smoke cells compared to OKF6/TERT1-Parental cells as depicted in Supplementary Fig. S2B (see Western panel). Significantly dysregulated miRNAs and their corresponding targets which showed a directional relationship are highlighted (miRNAs and protein targets which are upregulated are highlighted in red and miRNAs and protein targets which are downregulated are in blue color) in Supplementary Table S3A,B.
Figure 4.
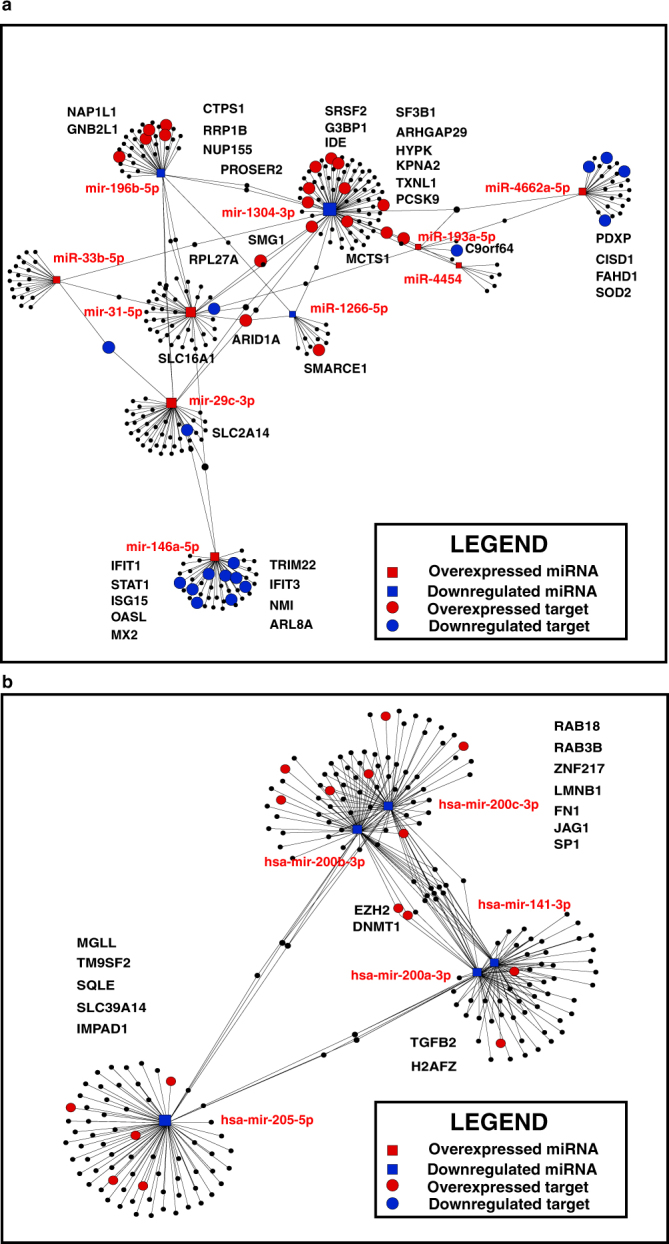
miRNA-mRNA interaction network of significantly dysregulated miRNAs and their targets in (a) OKF6/TERT1-Tobacco cells (b) OKF6/TERT1-Smoke cells. Square blocks represent dysregulated miRNAs and circles represent their target proteins showing inverse correlation. Overexpressed miRNAs and proteins are highlighted in red while downregulated miRNAs and proteins are highlighted in blue color.
Bioinformatics analysis
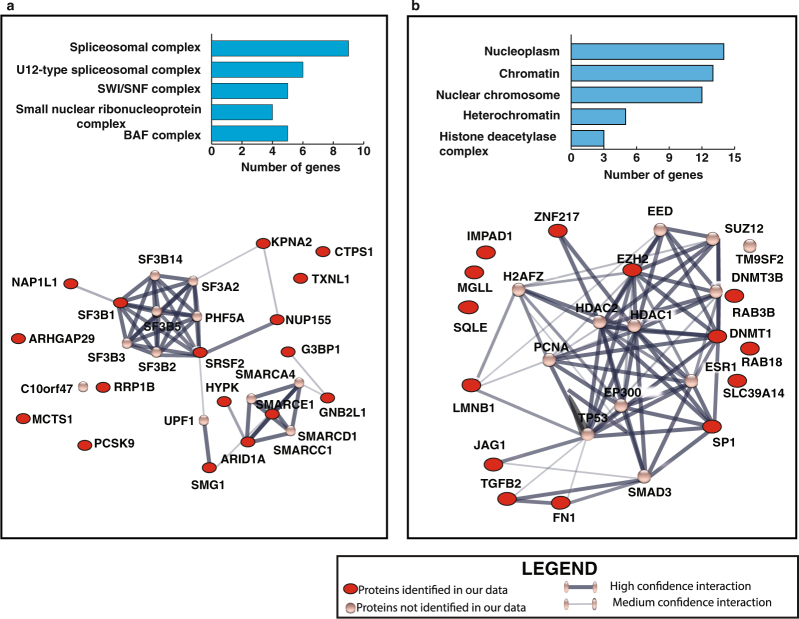
We used STITCH tool (version 5.0)16 to analyse the protein-protein interaction of targets which showed inverse relationship with their corresponding miRNAs in tobacco and smoke exposed cells. Functional enrichment analysis was performed for proteins which were overexpressed and targeted by significantly downregulated miRNAs in tobacco treated cells. This revealed enrichment of spliceosome-associated proteins that have a major role in processing of pre-mRNA/splicing into its mature forms (Fig. 5a). Similarly proteins which were overexpressed and are targeted by the significantly downregulated miRNAs in smoke exposed cells, showed enrichment of proteins which play an essential role in chromatin remodelling and transcriptional regulation (Fig. 5b).
Figure 5.
STITCH generated protein-protein interaction network of overexpressed protein targets of significantly downregulated miRNAs in (a) OKF6/TERT1-Tobacco cells and (b) OKF6/TERT1-Smoke cells. Red circles represent proteins identified in our proteomic data set.
Novel miRNA analysis
Small RNA-Seq datasets for OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells were analyzed for novel miRNAs using miRDeep217. Novel miRNA with miRDeep2 score ≥1, ≥8 reads supporting mature miRNA, and ≥2 reads supporting loop/star sequence were filtered to acquire high confidence list. This led us to identify 3 novel miRNAs in OKF6/TERT1-Parental, 6 novel miRNAs in OKF6/TERT1-Tobacco and 18 novel miRNAs in OKF6/TERT1-Smoke cells. The genomic locations of the novel miRNAs are provided in Supplementary Tables S4A–C. The secondary structures of these novel miRNAs are depicted in Supplementary Fig. S3A–C, Supplementary Fig. S4A–F and Supplementary Fig. S5A–R, respectively. Amongst the novel miRNAs, two miRNAs were common between OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells, but these were not identified in OKF6/TERT1-Parental cells (Fig. 6). To compare their expression pattern, we normalized read support of each novel miRNA with number of unique reads (length ≥18) from their respective sequence library. Novel miRNA NM-OKF6-ST-01-3p in tobacco treated cells showed a relatively higher expression with 1,726 reads compared to smoke exposed cells with 47 reads. Conversely, miRNA NM-OKF6-ST-02-5p showed higher expression in smoke exposed cells with 532 reads compared to tobacco treated cells with 53 reads.
Figure 6.
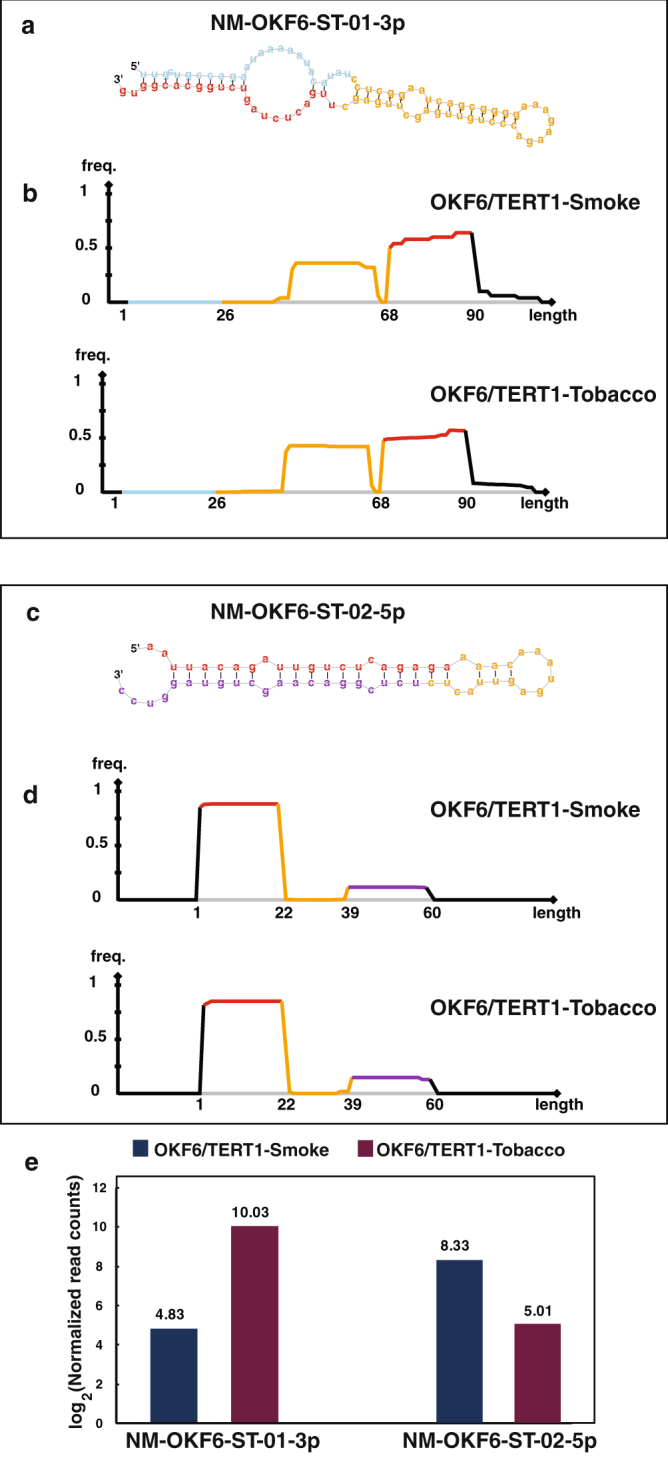
Expression pattern of novel miRNA common to OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells: (a) Secondary structure of novel miRNA NM-OKF6-ST-01-3p identified in both OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells (b) Read distribution frequency plot of NM-OKF6-ST-01-3p in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells (c) Secondary structure of novel miRNA NM-OKF6-ST-02-5p identified in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells (d) Read distribution frequency plot of NM-OKF6-ST-02-5p precursor in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells (e) Expression pattern of NM-OKF6-ST-01-3p and NM-OKF6-ST-02-5p in OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells using log transformed normalized read counts.
Discussion
Chewing tobacco is one of the known risk factors associated with oral cancer18. This is a common practice in India compared to the the western world2. Therefore, limited molecular level investigations have been carried out on the effect of chewing tobacco compared to tobacco smoking. In this study, using cellular models, we have systematically studied the alterations in miRNA expression and their targets in response to both chewing tobacco and cigarette smoke.
During the last few decades, miRNAs have emerged as one of the key regulators of cellular processes ranging from cell cycle regulation, differentiation, cell migration and apoptosis19. Their discovery has initiated a revolutionary change in cancer research, making them one of the important parameters to be elucidated in cancer. We identified several miRNAs previously reported to be dysregulated in oral cancer substantiating our data in both chewing tobacco treated and cigarette smoke exposed oral cells. We observed significant upregulation of miR-31-5p in OKF6/TERT1-Tobacco cells. Association of miR-31-5p dysregulation has been shown to result in initiation and progression of oral leukoplakia to full blown OSCC20,21. We also observed overexpression of miR-29c-3p and miR-146a-5p in chewing tobacco treated cells. High expression of miR-29c-3p has been reported in aggressive oral tongue tumours and in OSCC22,23. Also, miR-29c-3p was found to be upregulated with decreased expression level of its target protein PTX3 in meningiomas24. The role of miR-146a-5p remains ambiguous in oral cancer related studies. Loss or decreased expression of miR-146a-5p was found to be associated with aggressive behaviour in oral squamous cell carcinoma25. Contrary to this, Hung et al., reported that miR-146a expression contributes to oral carcinogenesis by targeting the IRAK1, TRAF6 and NUMB genes. The plasma levels of miR-146a in OSCC patients were also significantly higher as compared to control groups26. miR-1266-5p has been reported as an oncomiR and is reported to be over expressed in HNSCC patients who are users of alcohol27. Similarly, miR-196b-5p is reported to be overexpressed in oesophageal adenocarcinoma, gastric and oral cancer in smokers28–30. In contrary to this data we observed downregulation of miR-1266-5p and miR-196b-5p in oral keratinocytes chronically treated with tobacco, indicating that expression profile of miRNAs possibly depends on the type of insult which induces carcinoma. Interestingly, downregulation of miR-196b has been associated with increase in EMT31. This is in concordance with our cellular assays where we observed an increase in protein markers associated with EMT in tobacco treated cells. Similarly we observed increased expression of miR-193a-5p in the tobacco treated cells, where as previous studies have shown miR-193a-5p is silenced by DNA hypermethylation in HNSCC and OSCC in smokers, indicating that the expression profile of miRNAs in oral cells may depend on the form of tobacco used32.We identified several miRNAs in chewing tobacco treated oral keratinocytes which are reported to be dysregulated in other cancer types but not in OSCC. miR-4454 and miR-33b-5p were seen to be significantly overexpressed in chewing tobacco treated cells and have been previously reported to be highly expressed in bladder cancer patients33,34. Decreased expression of miR-1304-3p is reported in non-small cell lung carcinoma35.
Regulation of gene expression at the post-transcriptional level by miRNAs influences a wide variety of pathways, including oncogenic pathways. Since miRNAs are known to be involved in tumorigenesis by inhibiting target proteins, we also studied the expression profile of proteins targeted by significantly dysregulated miRNAs. miR-1304-3p identified as downregulated and its target protein ras GTPase-activating protein-binding protein 1 (G3BP1) was found to be upregulated (1.52 fold; p-value ≤ 0.05) in tobacco treated cells. Overexpression of G3BP1 has been reported in multiple cancers including oral squamous cell carcinoma36–38. Knockdown of G3BP1 induced programmed cell death and decreased cell viability in OSCC cell lines39. Chromatin remodelling complex protein, SMARCE1 (a target of miR-1266-5p) which was overexpressed (1.64 fold; p-value ≤ 0.05) in tobacco treated cells has been associated with both breast cancer invasion and metastasis in ovarian cell carcinoma40–42. We observed downregulation of signal transducer and activator of transcription 1, (STAT1) (0.35 fold; p-value ≤ 0.05) in OKF6/TERT1-Tobacco cells. STAT1 is a target protein of miR-146a-5p (significantly upregulated in tobacco treated cells) and is reported to be downregulated due to promoter methylation in HNSCC. Its exogenous expression in HNSCC cell lines led to growth inhibition and increased chemotherapy-induced apoptosis43. GNB2L1 (RACK1 – target of miR-196b-5p) has been reported to promote progression of oral squamous cell carcinoma via the AKT/mTOR pathway and induces OSCC cell migration; invasion and metastasis44. We observed a 1.55 fold (p-value ≤ 0.05) overexpression of GNB2L1 and downregulation of miR-196b-5p in the cells chronically treated with chewing tobacco.
We identified significant downregulation of members of miR-200 family in OKF6/TERT1 cells chronically exposed to cigarette smoke. Multiple miR-200 members including miR-200a, miR-200b and miR-200c have been previously reported to be downregulated in head and neck cancers45–48. In addition, we also observed decrease in expression of miR-205 in smoke exposed cells. Loss of miR-205 expression has been associated with poor prognosis of head and neck cancer patients49,50. miR-200 family target protein, fibronectin 1, FN1 which was seen to be 1.6 fold overexpressed in OKF6/TERT1-Smoke cells is reported to be overexpressed in salivary gland carcinoma and in OSCC patients with lymph node metastasis51,52. Transforming growth factor-β 2, TGFB2 which is known to be overexpressed in multiple cancers including OSCC and induces malignancy was 1.8 fold overexpressed (p-value ≤ 0.05) in our data. It is targeted by miR-141-3p which was downregulated in our data. It is suggested that the ability of OSCC cells to secrete TGF-β2 could contribute to clinical progression by maintaining a microenvironment conducive for tumor growth and proliferation53,54. In addition to the proteins discussed above we have identified a number of proteins which were overexpressed in smoke exposed cells and inversely correlated to their respective miRNAs, but have not been reported in literature in the context of oral cancer. H2A histone family member Z, H2AFZ (1.7 fold overexpressed; p-value ≤ 0.05) which showed inverse relationship with miR-200a-3p and miR-141-3p has been reported to be highly expressed in breast, prostate, bladder and hepatocellular cancers and enhances tumorigenesis by accelerating cell cycle and epithelial to mesenchymal transition55–57. We also observed overexpression of a number of protein targets of miR-205-5p in smoke exposed cells. Reports indicate high expression of squalene monooxygenase (SQLE) in breast and hepatocellular carcinoma and plays a key role in induction of epithelial-mesenchymal transition in esophageal carcinoma58,59. Hypomethylation of inositol monophosphatase domain containing 1 (IMPAD1) has been associated with its overexpression in breast carcinoma60. Sharieha et al., reported upregulation of transmembrane 9 superfamily member 2 (TM9SF2) in breast carcinoma cells when compared to normal cells which in turn promoted growth and survival of carcinoma cells. Laminin B1 (LMNB1) (1.6 fold overexpressed; p-value ≤ 0.05) which is targeted by both miR-200b-3p and miR-200c-3p (downregulated in smoke exposed cells) is reported to be overexpressed in colorectal and hepatocellular carcinoma. It is also observed that its elevation in plasma could be used for early detection of hepatocellular carcinoma61,62. Loss of expression of miR-200 family plays a crucial role in the repression of E-Cadherin and promotes EMT13,63. We observed change in expression of markers associated with EMT progression in smoke exposed oral cells.
Our data indicates dysregulation of proteins which play a key role in mRNA processing and splicing in tobacco treated cells. We and others have shown in the past that aberrant activation of spliceosome complex-associated proteins has been shown to influence cellular transformation leading to development of cancer, including HNSCC64,65.Our proteomic data shows significant overexpression of splicing proteins serine and arginine rich splicing factor 2 (SRSF2) and splicing factor 3b (SF3B1) (1.9 and 1.5 fold overexpressed respectively; p-value ≤ 0.05) in tobacco treated cells. Increased expression of SRSF2 is associated with progression and poor prognosis in human hepatocellular carcinoma66. mRNA processing/splicing and chromosome remodelling were thought to traverse different paths but studies are revealing that there is a strong cross talk between the two in executing their vital processes67. Mutations in several components of chromatin remodelling complex, SWI/SNF have been linked to malignant transformation and progression of cancer. SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (SMARCE1) is known to play an essential role in breast cancer metastasis by protecting cells against anoikis through the HIF1A/PTK2 pathway41. We observed a significant increased expression of SMARCE1 in tobacco treated oral cells.
In contrast, we have observed increased expression of proteins responsible for transcriptional regulation and nucleosome assembly in smoke exposed oral keratinocytes. Acrolein which is a major component of cigarette smoke, forms adducts with histone proteins and specifically inhibits appropriate covalent modifications of cytosolic histone H3 and H4 by inhibiting acetylation and results in aberrant nucleosome assembly. Electrophiles, such as metabolically activated polycyclic aromatic hydrocarbons and alkylating agents are assumed to have the same effect on nucleosome assembly68. DNA methyltransferase 1 (DNMT1) which is reported to be overexpressed in response to cigarette smoke in non-small cell lung carcinoma and was found significantly overexpressed (1.9 fold overexpressed; p-value ≤ 0.05) in smoke exposed cells69. Overexpression of DNMT1 in lung cancer is associated with increased expression of histone deacetylases (HDACs) which increases the stability of DNMT1 and in turn stimulates cancer progression70. Sp1 transcription factor (Sp1) which plays a role in multiple cellular processes including chromatin remodelling is significantly upregulated in the smoke exposed cells. Nicotine promotes lung cancer proliferation via α7-nicotinic acetylcholine receptor (α7-nAChR). It has been unveiled that nicotine induces binding of Sp1 on α7-nAChR promoter, leading to transcriptional activation of α7-nAChR in lung cancer, which in turn facilitates tumor growth and progression71.
To the best of our knowledge, this is the first global study employing an integrative analysis approach on high-throughput proteomic and miRNA profiling data to investigate the molecular alterations brought about in immortal oral keratinocytes in response to chewing tobacco and cigarette smoke. Our results indicate that immortal oral keratinocytes treated with chewing tobacco or exposed to cigarette smoke show distinct alterations in both miRNA and their protein targets. In addition, we have also identified a set of high confidence novel miRNAs in both tobacco treated and smoke exposed oral keratinocytes. Taken together, these findings imply that altered expression of miRNAs and their target proteins may be prominently involved in the initiation and progression of oral squamous cell carcinoma. Functional and clinical validation of this data is essential before these findings can be taken forward as potential diagnostic biomarkers in OSCC patients segregated based on the history of tobacco usage.
Materials and Methods
Cell culture
Human oral keratinocytes, OKF6/TERT1 cells were a generous gift from Dr. James Rheinwald (Brigham and Women’s Hospital, Boston, MA). OKF6/TERT1 cells are normal oral mucosal epithelial cells immortalized by hTERT72. OKF6/TERT1 were cultured and maintained in keratinocytes serum free medium (KSFM) supplemented with 1% penicillin/streptomycin, CaCl2 (0.4 mM), bovine pituitary extract (25 µg/ml) and epidermal growth factor (EGF) (0.2 ng/ml). Cells were grown at 37 °C in a humidified 5% CO2 incubator.
Treatment of OKF6/TERT1 cells with chewing tobacco extract and cigarette smoke condensate
To study the chronic exposure effect of chewing tobacco, OKF6/TERT1 cells were treated with 1% chewing tobacco extract for a period of 6 months73. To study the chronic effect of cigarette smoke condensate (CSC Murty Pharmaceuticals, Inc, KY) OKF6/TERT1 cells were subjected to chronic exposure with 0.1% CSC in a smoke dedicated incubator for 6 months74. All cells were cultured for equal duration of time. Throughout the study, OKF6/TERT1 cells treated with chewing tobacco extract and cigarette smoke condensate have been referred to as “OKF6/TERT1-Tobacco” and “OKF6/TERT1-Smoke” cells, respectively. The parental control cells have been referred to as “OKF6/TERT1-Parental”.
Cell proliferation assays
OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells were seeded at a density of 25 × 103 cells. Cellular proliferation was monitored for 6 days where the cells were counted every 48 h using trypan blue exclusion method. All assays were performed in replicates.
Western blotting
Cells were grown to 80% confluence and the proteins were harvested in RIPA lysis buffer (10 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton-X-100, 0.1% SDS containing protease and phosphatase inhibitor cocktails) and sonicated. Western blot analysis was carried out as described previously74 VIM antibody was obtained from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX). Antibodies for E-cadherin were purchased from Cell Signaling (Cell Signaling Technology, Danvers, MA). SOD2 and TGFB2 antibodies were purchased from Cusabio (Baltimore Avenue, MD). β-actin antibody was procured from Sigma (Sigma Aldrich, USA) and was used as loading control.
Colony formation assays
Colony formation assays were carried out as described previously75.OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells were seeded at a cell density of 8.0 × 103 cells per well in 6-well dishes. The colonies formed were fixed with methanol and stained with 4% methylene blue. Colonies formed was counted for ten randomly selected viewing fields and representative images were photographed at 3x magnification. All experiments were carried out in replicates.
Cell invasion assays
Invasion assays were performed in a transwell system (BD Biosciences) with Matrigel-coated filters, and cellular invasion was evaluated after 48 h as described previously75. Briefly, invasiveness of the cells was assayed in the membrane invasion culture system using polyethylene terephthalate (PET) membrane (8-μm pore size) in the upper compartment of a transwell coated with Matrigel (BD BioCoat Matrigel Invasion Chamber; BD Biosciences). The cells were seeded at 2.0 × 104 cells per 500 μl of serum free media on the Matrigel-coated PET membrane in the upper compartment. The lower compartment was filled with complete growth media and the plates were maintained at 37 °C for 48 h. At the end of the incubation time, the upper surface of the membrane was wiped with a cotton-tip applicator to remove non migratory cells. Cells that migrated to bottom side of membrane were fixed and stained using 4% methylene blue. The number of cells that penetrated was counted for ten randomly selected viewing fields at 10x magnification. All experiments were performed in triplicates.
RNA isolation and miRNA enrichment
Total RNA, including small RNA, was isolated using the Qiagen RNAeasy isolation kit from OKF6-TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells. The quantity and quality of RNA was analyzed on denaturing agarose gel as well as on Bioanalyzer RNA 6000 Pico chip. RNA isolated from each condition was used to construct sequencing libraries with the Illumina TruSeq Small RNA Sample Prep Kit (Illumina, USA) as per manufacturer’s instructions. Briefly, 3′ and 5′ adapters were sequentially ligated to small RNA molecules and ligation products were subjected to reverse transcription to create single stranded cDNA. To selectively enrich fragments with adapter molecules on both ends, the cDNA was amplified with 50 PCR cycles using a common primer and a primer containing an index tag to allow sample multiplexing. The amplified cDNA constructs were gel purified, and validated by checking the size, purity, and concentration of the amplicons on the Agilent Bioanalyzer High Sensitivity DNA chip (#5067-4626, Genomics Agilent, Santa Clara, CA). The libraries were pooled in equimolar amounts, and sequenced on an Illumina HiSeq 2500 instrument to generate 50-base pair reads.
miRNA sequencing and data analysis
The quality of the raw reads was checked using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) for OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke samples. Cutadapt (http://cutadapt.readthedocs.io/en/stable/index.html) was used to remove low quality bases with Phred score ≤30 and adaptor sequences from 5′ and 3′ end of the read. The adaptor trimmed reads were aligned against nc-RNA sequence databases (GtRNA, piRNA, Rfam& snoRNA)76–79 using Bowtie (version 1.1.2)80. Remaining reads were used for known miRNA identification. These reads were first aligned to mature and precursor sequence from miRBase 21 miRNA database (Human)81 using Bowtie. Unaligned reads were further aligned against human genome hg19 using miRDeep2 (version 2.0.0.6). Known miRNA that are quantified with at least 10 reads in both samples of a pairs i.e. OKF6/TERT1-Tobacco vs. OKF6/TERT1-Parental and OKF6/TERT1-Smoke vs. OKF6/TERT1-Parental were further investigated for differential expression using DESeq82.
Novel miRNAs were assessed for the genomic location using UCSC BLAT83. They were also queried against database YM500V384 using ‘genomic location search’ option. YM500V3 is a repository of novel miRNA built using >8,000 smRNA data-sets pertaining to various cancers along with their CLIP-Seq results from different cell lines.
Sample preparation, in-solution digestion and TMT labelling
OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells were cultured and grown till 80% confluence. The cells were serum starved for 12 hours in ice-cold 1X PBS, lysed in 0.5% of SDS buffer followed by sonication (Branson Sonifier, Danbury, CT) and centrifugation. Supernatant was collected and protein estimation was carried out by using bicinchoninic acid assay (BCA)85. In-solution trypsin digestion of all three conditions was carried out as previously described86. Equal amounts of protein was taken from each condition, reduced using 5 mM dithiothreitol (DTT) and kept for incubation for 20 min at 60 °C. Alkylation was carried out using iodoacetamide (IAA) (20 mM) and incubated for 10 min in the dark at room temperature. The reduced and alkylated proteins were then precipitated using chilled acetone (1:5 v/v of sample) and incubated at −80 °C for 4–6 h to ensure complete removal of SDS from sample. Trypsin digestion was carried out at 37 °C for 12–16 h with an enzyme to substrate ratio of 1:50 (catalogue no. V5111; Promega). After trypsin digestion, the peptides were labelled with TMT reagents as per the manufacturer’s instructions. Briefly, peptide samples were dissolved in 50 mM TEABC (pH 8.0) and added to TMT reagents dissolved in anhydrous acetonitrile. Peptides from OKF6/TERT1-Parental cells and OKF6/TERT1-Tobacco were labelled with 126 and 129 C, respectively. In a separate experiment, OKF6/TERT1-Parental cells and OKF6/TERT1-Smoke cells were labelled with 128 N and 129 C, respectively. After incubation at room temperature for 1 h, the reaction was quenched with 5% hydroxylamine. The labelled samples from all conditions were pooled and subjected to fractionation.
Basic pH reverse-phase liquid chromatography
TMT labelled peptides were fractionated using a basic pH reverse HPLC system (Agilent,1290 series) by injecting 900 µL of the sample reconstituted in solvent A (10 mM TEABC in water; pH 8.5) on Xbridge (4.6 × 250 mm, 5 µm; Waters) column. The peptides were resolved by using a gradient of organic solvent; 2% solvent A (10 mM TEABC in water; pH 8.5) to 45% solvent B (10 mM TEABC in 90% acetonitrile, pH 8.5) over 100 min and further taken to 100% solvent B before coming back to 2% solvent A for re-equilibration87. A total of 96 fractions were collected from 96-well plate, concatenated to 12 fractions, and dried under vacuum for LC-MS2/MS3 analysis.
LC-MS2/MS3 analysis
Briefly all the 12 fractions were reconstituted in 0.1% formic acid and analyzed in triplicate on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) that was interfaced with an Easy-nLC II nanoflow liquid chromatography system (Thermo Scientific). Peptides reconstituted in 0.1% formic acid were loaded onto a trap column that was packed (75 µm × 2 cm) with Magic C18 AQ (Michrom Bioresources, Inc.) at a flow rate of 300 nL/min. Peptides were separated on an analytical column (75 µm × 20 cm) at a flow rate of 300 nL/min by using a linear gradient of 7–25% solvent B (0.1% formic acid in 95% acetonitrile) over 75 min which was increased to 35% for next 25 minutes. The total run time was set to 120 min and acquisition was carried out.
Data analysis
Proteome Discoverer (version 2.1.0.81) software suite (Thermo Fisher Scientific) was used for MS/MS searches using SEQUEST and Mascot (version 2.4.1; Matrix 617 Science) algorithms againt NCBI RefSeq human protein database (version 75 containing 36,709 protein sequences and known contaminants). The search parameters included trypsin as the protease with a maximum of 2 missed cleavages allowed; oxidation of methionine was set as a dynamic modification, carbamidomethylation of cysteine and TMT modification at peptide N-terminus and lysine were set as static modifications. Precursor ion mass tolerance and fragment ion mass tolerance were allowed with 10 ppm and 0.05 Da, respectively and all the PSMs were identified with 1% FDR88. Proteins were identified at MS2 level and quantification was done at MS3 level.
Data availability
Raw sequencing data is available in SRA (Sequencing Read Archive) database with accession number PRJNA398169.
Electronic supplementary material
Acknowledgements
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics (IOB), Bangalore. This work was supported by Department of Science and Technology (DST) grants (SERC/LS-439/2011 and SR/SO/HS/0208/2012) and FAMRI-funded 072017_YCSA. IOB is supported by DBT Program Support on Neuroproteomics and infrastructure for proteomic data analysis (BT/01/COE/08/05). MYB is a recipient of Junior Research Fellowship from DBT. JA and PR are recipients of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India.
Author Contributions
A.C. and H.G. conceptualized and designed the entire study. V.N. and P.R. performed all cell culture experiments. J.A. carried out miRNA sequencing data analysis and bioinformatics data analysis. K.P. carried out novel miRNA prediction. M.Y.B., J.A., P.R., K.P., H.S.S., A.H.P. and F.A.B., were involved in data analysis and interpretation. M.Y.B., J.A. and K.P. prepared the manuscript and manuscript figures. A.C., H.G., D.S., J.A.C., T.S.K.P., B.N. and P.P.M. critically reviewed and edited the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Mohd Younis Bhat and Jayshree Advani contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25498-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Harsha Gowda, Email: harsha@ibioinformatics.org.
Aditi Chatterjee, Email: aditi@ibioinformatics.org.
References
- 1.Saraiya M, et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 2015;107:djv086. doi: 10.1093/jnci/djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sreeramareddy CT, Pradhan PM, Mir IA, Sin S. Smoking and smokeless tobacco use in nine South and Southeast Asian countries: prevalence estimates and social determinants from Demographic and Health Surveys. Popul Health Metr. 2014;12:22. doi: 10.1186/s12963-014-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Pan X, Cobb GP, Anderson T. A. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Chen J, Huang Z. miR-372 promotes breast cancer cell proliferation by directly targeting LATS2. Exp Ther Med. 2018;15:2812–2817. doi: 10.3892/etm.2018.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Y, Rao Z, Chen C. miR-30a suppresses lung cancer progression by targeting SIRT1. Oncotarget. 2018;9:4924–4934. doi: 10.18632/oncotarget.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Ruan H, Hu X, Cao A, Song L. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res. 2017;7:913–922. [PMC free article] [PubMed] [Google Scholar]
- 9.Manikandan M, et al. Altered levels of miR-21, miR-125b-2*, miR-138, miR-155, miR-184, and miR-205 in oral squamous cell carcinoma and association with clinicopathological characteristics. J Oral Pathol Med. 2015;44:792–800. doi: 10.1111/jop.12300. [DOI] [PubMed] [Google Scholar]
- 10.Tsai YS, et al. Areca nut induces miR-23a and inhibits repair of DNA double-strand breaks by targeting FANCG. Toxicol Sci. 2011;123:480–490. doi: 10.1093/toxsci/kfr182. [DOI] [PubMed] [Google Scholar]
- 11.Pal A, et al. Cigarette smoke condensate promotes pro-tumourigenic stromal-epithelial interactions by suppressing miR-145. J Oral Pathol Med. 2013;42:309–314. doi: 10.1111/jop.12017. [DOI] [PubMed] [Google Scholar]
- 12.Gong J, Chu Y, Xu M, Huo J, Lv L. Esophageal squamous cell carcinoma cell proliferation induced by exposure to low concentration of cigarette smoke extract is mediated via targeting miR-101-3p/COX-2 pathway. Oncol Rep. 2016;35:463–471. doi: 10.3892/or.2015.4379. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, et al. NF-kappaB-mediated inflammation leading to EMT via miR-200c is involved in cell transformation induced by cigarette smoke extract. Toxicol Sci. 2013;135:265–276. doi: 10.1093/toxsci/kft150. [DOI] [PubMed] [Google Scholar]
- 14.Chou CH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niaz K, et al. Smokeless tobacco (paan and gutkha) consumption, prevalence, and contribution to oral cancer. Epidemiol Health. 2017;39:e2017009. doi: 10.4178/epih.e2017009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96:Suppl, R40–44. [PubMed] [Google Scholar]
- 20.Lu WC, et al. miR-31 targets ARID1A and enhances the oncogenicity and stemness of head and neck squamous cell carcinoma. Oncotarget. 2016;7:57254–57267. doi: 10.18632/oncotarget.11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maimaiti A, Abudoukeremu K, Tie L, Pan Y, Li X. MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene. 2015;558:271–277. doi: 10.1016/j.gene.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Hilly O, et al. Distinctive pattern of let-7 family microRNAs in aggressive carcinoma of the oral tongue in young patients. Oncol Lett. 2016;12:1729–1736. doi: 10.3892/ol.2016.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrano NA, et al. Integrative analysis in oral squamous cell carcinoma reveals DNA copy number-associated miRNAs dysregulating target genes. Otolaryngol Head Neck Surg. 2012;147:501–508. doi: 10.1177/0194599812442490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalan AB, et al. Simultaneous analysis of miRNA-mRNA in human meningiomas by integrating transcriptome: A relationship between PTX3 and miR-29c. BMC Cancer. 2017;17:207. doi: 10.1186/s12885-017-3198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z, Johnson JJ, Jiang R, Liu Y, Stack MS. Decrease of miR-146a is associated with the aggressiveness of human oral squamous cell carcinoma. Arch Oral Biol. 2015;60:1416–1427. doi: 10.1016/j.archoralbio.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung PS, et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS One. 2013;8:e79926. doi: 10.1371/journal.pone.0079926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saad MA, et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol Cancer. 2015;14:181. doi: 10.1186/s12943-015-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SW, et al. Dysregulation of MicroRNA-196b-5p and MicroRNA-375 in Gastric Cancer. J Gastric Cancer. 2016;16:221–229. doi: 10.5230/jgc.2016.16.4.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou YY, et al. Aberrant DNA hypomethylation of miR-196b contributes to migration and invasion of oral cancer. Oncol Lett. 2016;11:4013–4021. doi: 10.3892/ol.2016.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warnecke-Eberz U, Chon SH, Holscher AH, Drebber U, Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36:4643–4653. doi: 10.1007/s13277-015-3112-0. [DOI] [PubMed] [Google Scholar]
- 31.Bai X, et al. MicroRNA-196b Inhibits Cell Growth and Metastasis of Lung Cancer Cells by Targeting Runx2. Cell Physiol Biochem. 2017;43:757–767. doi: 10.1159/000481559. [DOI] [PubMed] [Google Scholar]
- 32.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modestou MA, Manzel LJ, El-Mahdy S, Look DC. Inhibition of IFN-gamma-dependent antiviral airway epithelial defense by cigarette smoke. Respir Res. 2010;11:64. doi: 10.1186/1465-9921-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li CG, et al. MicroRNA-1304 suppresses human non-small cell lung cancer cell growth in vitro by targeting heme oxygenase-1. Acta Pharmacol Sin. 2017;38:110–119. doi: 10.1038/aps.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou N, Chen J, Yu S, Gao Y, Li Y. G3BP1 contributes to tumor metastasis via upregulation of Slug expression in hepatocellular carcinoma. Am J Cancer Res. 2016;6:2641–2650. [PMC free article] [PubMed] [Google Scholar]
- 37.Min L, et al. Overexpression of Ras-GTPase-activating protein SH3 domain-binding protein 1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2015;67:677–688. doi: 10.1111/his.12695. [DOI] [PubMed] [Google Scholar]
- 38.Lo WY, Wang HJ, Chiu CW, Chen SF. miR-27b-regulated TCTP as a novel plasma biomarker for oral cancer: from quantitative proteomics to post-transcriptional study. J Proteomics. 2012;77:154–166. doi: 10.1016/j.jprot.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 39.Xu C, et al. Integrative genomics in combination with RNA interference identifies prognostic and functionally relevant gene targets for oral squamous cell carcinoma. PLoS Genet. 2013;9:e1003169. doi: 10.1371/journal.pgen.1003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol ES, et al. SMARCE1 is required for the invasive progression of in situ cancers. Proc Natl Acad Sci USA. 2017;114:4153–4158. doi: 10.1073/pnas.1703931114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethuraman A, et al. SMARCE1 regulates metastatic potential of breast cancer cells through the HIF1A/PTK2 pathway. Breast Cancer Res. 2016;18:81. doi: 10.1186/s13058-016-0738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giannakakis A, et al. Overexpression of SMARCE1 is associated with CD8+ T-cell infiltration in early stage ovarian cancer. Int J Biochem Cell Biol. 2014;53:389–398. doi: 10.1016/j.biocel.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 43.Xi S, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98:181–189. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, et al. Receptor for activated C kinase 1 (RACK1) promotes the progression of OSCC via the AKT/mTOR pathway. Int J Oncol. 2016;49:539–548. doi: 10.3892/ijo.2016.3562. [DOI] [PubMed] [Google Scholar]
- 45.Brito BL, et al. Expression of stem cell-regulating miRNAs in oral cavity and oropharynx squamous cell carcinoma. J Oral Pathol Med. 2016;45:647–654. doi: 10.1111/jop.12424. [DOI] [PubMed] [Google Scholar]
- 46.Jamali Z, et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51:321–331. doi: 10.1016/j.oraloncology.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Wiklund ED, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One. 2011;6:e27840. doi: 10.1371/journal.pone.0027840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun L, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31:432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]
- 49.Kim JS, et al. MicroRNA-205 directly regulates the tumor suppressor, interleukin-24, in human KB oral cancer cells. Mol Cells. 2013;35:17–24. doi: 10.1007/s10059-013-2154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CJ, et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26:1003–1010. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 51.Morita Y, et al. Cellular fibronectin 1 promotes VEGF-C expression, lymphangiogenesis and lymph node metastasis associated with human oral squamous cell carcinoma. Clin Exp Metastasis. 2015;32:739–753. doi: 10.1007/s10585-015-9741-2. [DOI] [PubMed] [Google Scholar]
- 52.Leivo I, et al. Characterization of gene expression in major types of salivary gland carcinomas with epithelial differentiation. Cancer Genet Cytogenet. 2005;156:104–113. doi: 10.1016/j.cancergencyto.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 53.Cirillo N, et al. Cancer-associated fibroblasts regulate keratinocyte cell-cell adhesion via TGF-beta-dependent pathways in genotype-specific oral cancer. Carcinogenesis. 2017;38:76–85. doi: 10.1093/carcin/bgw113. [DOI] [PubMed] [Google Scholar]
- 54.Arantes DA, Costa NL, Mendonca EF, Silva TA, Batista AC. Overexpression of immunosuppressive cytokines is associated with poorer clinical stage of oral squamous cell carcinoma. Arch Oral Biol. 2016;61:28–35. doi: 10.1016/j.archoralbio.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Yang HD, et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7:11412–11423. doi: 10.18632/oncotarget.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baptista T, et al. Regulation of histone H2A.Z expression is mediated by sirtuin 1 in prostate cancer. Oncotarget. 2013;4:1673–1685. doi: 10.18632/oncotarget.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rangasamy D. Histone variant H2A.Z can serve as a new target for breast cancer therapy. Curr Med Chem. 2010;17:3155–3161. doi: 10.2174/092986710792231941. [DOI] [PubMed] [Google Scholar]
- 58.Qin Y, Zhang Y, Tang Q, Jin L, Chen Y. SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:138–148. doi: 10.1093/abbs/gmw127. [DOI] [PubMed] [Google Scholar]
- 59.Brown DN, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep. 2016;6:19435. doi: 10.1038/srep19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parris TZ, et al. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis. 2014;3:e95. doi: 10.1038/oncsis.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marshall KW, et al. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer. 2010;126:1177–1186. doi: 10.1002/ijc.24910. [DOI] [PubMed] [Google Scholar]
- 62.Sun S, Xu MZ, Poon RT, Day PJ, Luk JM. Circulating Lamin B1 (LMNB1) biomarker detects early stages of liver cancer in patients. J Proteome Res. 2010;9:70–78. doi: 10.1021/pr9002118. [DOI] [PubMed] [Google Scholar]
- 63.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radhakrishnan A, et al. Dysregulation of splicing proteins in head and neck squamous cell carcinoma. Cancer Biol Ther. 2016;17:219–229. doi: 10.1080/15384047.2016.1139234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–1093. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo C, et al. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res. 2017;77:1168–1178. doi: 10.1158/0008-5472.CAN-16-1919. [DOI] [PubMed] [Google Scholar]
- 67.Kornblihtt AR, Schor IE, Allo M, Blencowe BJ. When chromatin meets splicing. Nat Struct Mol Biol. 2009;16:902–903. doi: 10.1038/nsmb0909-902. [DOI] [PubMed] [Google Scholar]
- 68.Chen D, Fang L, Li H, Tang MS, Jin C. Cigarette smoke component acrolein modulates chromatin assembly by inhibiting histone acetylation. J Biol Chem. 2013;288:21678–21687. doi: 10.1074/jbc.M113.476630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon YM, et al. Different susceptibility of increased DNMT1 expression by exposure to tobacco smoke according to histology in primary non-small cell lung cancer. J Cancer Res Clin Oncol. 2007;133:219–226. doi: 10.1007/s00432-006-0160-2. [DOI] [PubMed] [Google Scholar]
- 70.Brodie SA, et al. Class I HDACs are mediators of smoke carcinogen-induced stabilization of DNMT1 and serve as promising targets for chemoprevention of lung cancer. Cancer Prev Res (Phila) 2014;7:351–361. doi: 10.1158/1940-6207.CAPR-13-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown KC, et al. Nicotine induces the up-regulation of the alpha7-nicotinic receptor (alpha7-nAChR) in human squamous cell lung cancer cells via the Sp1/GATA protein pathway. J Biol Chem. 2013;288:33049–33059. doi: 10.1074/jbc.M113.501601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass ap16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/MCB.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nanjappa V, et al. Chronic exposure to chewing tobacco selects for overexpression of stearoyl-CoA desaturase in normal oral keratinocytes. Cancer Biol Ther. 2015;16:1593–1603. doi: 10.1080/15384047.2015.1078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang X, et al. Adenylate kinase 3 sensitizes cells to cigarette smoke condensate vapor induced cisplatin resistance. PLoS One. 2011;6:e20806. doi: 10.1371/journal.pone.0020806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subbannayya Y, et al. Calcium calmodulin dependent kinase kinase 2 - a novel therapeutic target for gastric adenocarcinoma. Cancer Biol Ther. 2015;16:336–345. doi: 10.4161/15384047.2014.972264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daub J, Eberhardt RY, Tate JG, Burge SW. Rfam: annotating families of non-coding RNA sequences. Methods Mol Biol. 2015;1269:349–363. doi: 10.1007/978-1-4939-2291-8_22. [DOI] [PubMed] [Google Scholar]
- 77.Zhang P, et al. piRBase: a web resource assisting piRNA functional study. Database (Oxford) 2014;2014:bau110. doi: 10.1093/database/bau110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kent, W. J. BLAT–the BLAST-like alignment tool. Genome Res12, 656–664, 10.1101/gr.229202. Article published online before March2002 (2002). [DOI] [PMC free article] [PubMed]
- 84.Chung IF, et al. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45:D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker JM. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol Biol. 1994;32:5–8. doi: 10.1385/0-89603-268-X:5. [DOI] [PubMed] [Google Scholar]
- 86.Marimuthu A, et al. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim Biophys Acta. 2013;1834:2308–2316. doi: 10.1016/j.bbapap.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baker PR, Medzihradszky KF, Chalkley RJ. Improving software performance for peptide electron transfer dissociation data analysis by implementation of charge state- and sequence-dependent scoring. Mol Cell Proteomics. 2010;9:1795–1803. doi: 10.1074/mcp.M110.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data is available in SRA (Sequencing Read Archive) database with accession number PRJNA398169.