Abstract
Breast cancer risk is intimately intertwined with exposure to estrogens. While more than 160 breast cancer risk loci have been identified in humans, genetic interactions with estrogen exposure remain to be established. Strains of rodents exhibit striking differences in their responses to endogenous ovarian estrogens (primarily 17β-estradiol). Similar genetic variation has been observed for synthetic estrogen agonists (ethinyl estradiol) and environmental chemicals that mimic the actions of estrogens (xenoestrogens). This review of literature highlights the extent of variation in responses to estrogens among strains of rodents and compiles the genetic loci underlying pathogenic effects of excessive estrogen signaling. Genetic linkage studies have identified a total of the 35 quantitative trait loci (QTL) affecting responses to 17β-estradiol or diethylstilbestrol in 5 different tissues. However, the QTL appear to act in a tissue-specific manner with 9 QTL affecting the incidence or latency of mammary tumors induced by 17β-estradiol or diethylstilbestrol. Mammary gland development during puberty is also exquisitely sensitive to the actions of endogenous estrogens. Analysis of mammary ductal growth and branching in 43 strains of inbred mice identified 20 QTL. Regions in the human genome orthologous to the mammary development QTL harbor loci associated with breast cancer risk or mammographic density. The data demonstrate extensive genetic variation in regulation of estrogen signaling in rodent mammary tissues that alters susceptibility to tumors. Genetic variants in these pathways may identify a subset of women who are especially sensitive to either endogenous estrogens or environmental xenoestrogens and render them at increased risk of breast cancer.
Keywords: Estrogen, Estrogen Receptors, Xenoestrogens, Quantitative Trait Loci, Breast Cancer
Estrogens and breast cancer risk
Estrogens have diverse actions in directing development and maintenance of tissues. However, prolonged exposure to estrogens has been implicated in initiation and progression of cancers primarily in breast, endometrium, and ovary (Burns and Korach 2012). Given the diverse roles, the health benefits and risks of estrogen supplementation was examined in postmenopausal women in the Women’s Health Initiative (WHI). This study demonstrated an increased incidence of breast cancer among women receiving postmenopausal hormone therapy and a lack of clear beneficial effects in other tissues (Prentice and Anderson 2008; Prentice et al. 2008). The risk associated with postmenopausal hormone therapy was also observed in the Million Women Study (Beral 2003). Both studies reported a modest increase in risk of breast cancer associated with estrogen alone therapies but was significantly higher when progestins were included. Exposure to estrogens is also significant for women prior to menopause. Higher levels of circulating estradiol were associated with increased breast cancer risk among premenopausal women (Eliassen et al. 2006). Breast cancer risk was also increased by early menarche and late menopause (Bernstein 2002; Tamimi et al. 2016). Together these associations suggest that lifetime exposure to estrogens play an important role in the pathogenesis of breast cancer (Pike 1987).
Although estrogens participate in the initiation and progression, the majority of women do not develop detectable breast cancers (American Cancer Society, 2017) despite exposures to both endogenous estrogens produced during each ovarian cycle. During pregnancy, estrogens reach concentrations that are approximately 10-fold higher than in postmenopausal women. Rather than increased risk, these levels of estrogen along with other hormones during pregnancy have an overall long-term protective effect reducing the risk of breast cancer by up to 50% (Albrektsen et al. 2005; Chie et al. 2000; MacMahon et al. 1970). The major estrogen produced by the ovaries is 17β-estradiol. Administration of 17β-estradiol alone or in combination with progesterone was to sufficient to mimic the protective effects of pregnancy in rodents (Dunphy et al. 2008; Guzman et al. 1999; Rajkumar et al. 2001; Rajkumar et al. 2007; Sivaraman et al. 1998). Similarly, for the subset of women in WHI without benign breast disease or family history of breast cancer, estrogen-only postmenopausal hormone therapies were associated with measurable decreases in breast cancer and an overall decrease in mortality (Anderson et al. 2012). While these observations are not sufficient to warrant use of estrogen treatments for risk reduction, they do highlight the paradoxical roles of estrogens in preventing as well as promoting breast cancer and suggest a complex balance of responses within breast tissue.
Genetic variation in estrogen signaling has the potential to determine the balance of responses to estrogens within breast tissues and modify the risk of breast cancer associated with exposure to estrogens. As estrogen receptor alpha (ERα) is expressed in a majority of breast cancers and is a target of therapies to block signaling, genetic variants within the ESR1 locus have been examined for associations with breast cancer risk in women (Dunning et al. 2016). While these genetic variants have a significant effect, the overall impact on breast cancer risk is modest with odds ratios of less than 1.2. In contrast, genetic differences rendering strains of rats sensitive or resistant to the tumorigenic effects of estrogens reveal the potency of the underlying regulatory pathways (Shull et al. 2001; Shull et al. 1997). Differences in sensitivity to estrogen signaling among strains of rodents have the potential to define critical pathways affecting breast cancer risk in humans as well as provide new therapeutic targets. In this review, we have summarized data examining differences in sensitivity to estrogens among strains of mice and rats and the genetic basis for the variation.
Variation in responses to estrogenic compounds among strains of rodents
PubMed was searched to identify publications where estrogenic compounds were evaluated in strains of rats or mice. The initial search retrieved 265 publications that included the terms “strain differences estrogen”. This set was refined by selecting those using rat or mouse models. The set was manually curated to identify studies in which at least 2 strains of rats or mice were compared. Studies that included treatments with 17β-estradiol are emphasized as this is the most active of the estrogens produced by ovaries and provides a reference for comparisons. Studies using well-characterized agonists of estrogen receptors (e.g ethinyl estradiol, diethylstilbestrol) were also included. Environmental chemicals were evaluated for estrogenic activity, referred to as xenoestrogens, were included when comparisons with either 17β-estradiol or estrogen agonists were available. While effects in mammary tissue were of primary interest, all tissues were included in the search to assess whether strain-effects were consistent across tissues. The data are also selected to emphasize inbred strains of mice (BALB/c, C3H, C57BL/6, DBA, 129) and rats (ACI, BN, COP, DA, F344, WKY). However, Sprague Dawley (SD) outbred rats are included because they were used across many studies. The Holzman strain (Holz) is outbred and was derived from SD. Long-Evans and SD outbred strains are related to the Wistar (Wis) strain from which they were derived. Similarly, outbred CD-1 mice were compared in some reports. Most studies examined only single doses of compounds. This limits the ability to detect non-monotonic differences in responses among strains and sensitivity would be overlooked if saturating doses were used. Therefore, the data may underestimate the extent of genetic variation. The selected studies are summarized in Table 1.
Table 1.
Variation in responses to estrogens among strains of rodents.
| Sp./Sex | Strainsa,b | Compoundc | Doses | Trt Duration | Phenotypes | Reference |
|---|---|---|---|---|---|---|
| Mammary | ||||||
| Mus, Female | C57BL/6J, C3H/HeJ | E2 | 40 μg/kg daily injections | Acute, 4 days | Proliferation: C57BL/6J > C3H/HeJ Apoptosis: C57BL/6J > C3H/HeJ |
(Wall et al., 2014a) |
| Mus, Female | C57BL/6J, CD-1 | E2 BPA |
0.25–50 μg/kg/d 250 ng/kg/d |
Acute, 10 days E2 | Ductal area, E2: C57BL/6J > CD-1 TEB/Ductal area; E2: C57BL/6J > CD-1 TEB/Ductal area: BPA: C57BL/6J > CD-1 |
(Wadia et al., 2007) |
| Mus, Female | BALB/cJ, 129/SvEv | DES | 15 ppb po | Chronic, 6 months | Ductal branching: 129/SvEv = BALB/cJ | (Bennett et al., 2000) |
| Ratus, Female | ACI, BN | E2 | 27 mg in silastic implant | Chronic, 1–12 weeks | Proliferation: ACI > BN Apoptosis: ACI = BN |
(Ding et al., 2013) |
| Ratus, Female | ACI, SD | DES | 5 mg implants | Chronic, 10–214 days | Mammary tumors: ACI > SD Chronic stimulation resulted in mammary tuors in ACI but not SD rats. |
(Stone et al., 1979) |
| Uterus and vagina | ||||||
| Ratus, Female | SD, F344 | EE DES-DP CE D4 |
1–30 μg/kg/d po 0.5–15 μg/kg/d po 10–150 mg/kg/d po |
Acute, 18–21 days old | Uterine weight, EE: SD = F344 Uterine weight, DES-DP: SD = F344 Uterine weight, CE: SD = F344 Uterine weight, D4: SD > F344 |
(McKim et al., 2001) |
| Ratus, Female | DA/Han, SD, Wis | EE BPA OCT Gen |
100 μg/kg/d po 5–200 mg/kg/d po 5–200 mg/kg/d po 25–100 mg/kg/d po |
Acute, 3 days | Uterine weight, EE: Wis = DA/HAN > SD Potency rank was similar for BPA, OCT, Gen. |
(Diel et al., 2004) |
| Ratus, Female | SD, Long-Evans | PEP | 25–100 μg sc | Acute, 1 day | Uterine weight: Long-Evans > SD | (Lawson et al., 1984) |
| Ratus, Female | SD, F344 | E2 BPA |
0.02–2.0 μg/kg ip 0.2–150 mg/kg ip |
Acute, 1 day | Proliferation, E2: SD = F344 Proliferation, BPA: F344 > SD |
(Long et al., 2000) |
| Ratus, Female | SD, Wis | EE Gen |
100 μg/kg/d 50–200 mg/kg/d |
Acute, 3 days | Uterine weight: EE: Wis > SD Uterine weight, Gen: Wis = SD |
(Geis et al., 2005) |
| Ratus, Female | Wis, WKY | E2 | 2 mg, ip | Chronic, 3 weeks | Uterine weight: Wis > WKY BrdU labeling: Wis > WKY |
(Mitsui et al., 2013) |
| Ratus, Female | F344, Holz | DES | 2.5 mg implants | Chronic, up to 8 weeks | Uterine growth: F344 = Holz | (Wiklund et al., 1981) |
| Mus, Female | 129, C57BL/6, C3H/He, C3H/JFe, DBA/1 | E2-DP | 1 μg/d ip | Acute, 3 days | Uterine weight: 129 > C3H/JFe = DBA/1 > C57BL/6 = C3H/He |
(Drasher, 1955) |
| Mus, Female | C57BL/6J, C3H/HeJ, B6C3 | E2 | 40 μg/kg/d sc | Acute, 2 days | BM8+ macrophages: C57BL/6J > C3H/HeJ | (Griffith et al., 1997) |
| Mus, Female | C57BL/6J, C3H/HeJ, B6C3-F1 | E2 | 40 μg/kg/d sc | Acute, 2 days | Uterine weight: C57BL/6J>C3H/HeJ=B6C3-F1 | (Roper et al., 1999) |
| Mus, Female | C57BL/6J, CD-1 | E2 BPA |
0.25–50 μg/kg/d 250 ng/kg/d |
Acute, 10 days E2 | Uterine weight, E2: CD-1 > C57BL/6J Uterine weight: BPA: CD-1 = C57BL/6J |
(Wadia et al., 2007) |
| Mus, Female | BALB/c* C57BL/6J, C57BL6.BALB/c-F1 | DES | 2.5–1000 ppb po | Acute, 6 days | Uterine weight: C57BL/6J = F1 > BALB/c | (Greenman et al., 1977) |
| Mus, Female | BALB/cJ, 129/SvEv | DES | 15 ppb | Chronic, 6 months | Neoplasia: 129/SvEv > BALB/cJ | (Bennett et al., 2000) |
| Pituitary | ||||||
| Ratus, Female | ACI, SD | DES | 5 mg implants | Chronic, 10–214 days | Pituitary weights: ACI > SD | (Stone et al., 1979) |
| Ratus, Male | F344, Holz | DES | 2.5 mg implants | Acute, 4–56 days | Proliferation: F344 > Holz | (Wiklund and Gorski, 1982) |
| Ratus, Female | SD, Long-Evans | PEP | 25–100 μg sc | Acute, 1 day | Prolactin secretion: Long-Evans > SD Pituitary weight: Long-Evans = SD |
(Lawson et al., 1984) |
| Ratus, Female | F344, Holz | DES | 2.5 mg implants | Chronic, up to 8 weeks | Pituitary weight: F344 > Holz | (Wiklund et al., 1981) |
| Ratus, Female | F344, Holz | E2 DES |
100 μg/kg/d 50–200 mg/kg/d |
Chronic, up to 8 weeks | Plasma prolactin, E2: F344 = Holz Plasma prolactin, DES: F344 > Holz |
(Moy and Lawson, 1992) |
| Ratus, Female | SD, Wis WAG/Rij, BN/BiRij | E2 | 20 mg implants | Chronic, 2–52 weeks | Plasma prolactin: SD > Wis WAG/Rij > BN/BiRij |
(Blankenstein et al., 1984) |
| Ratus, Female | Wis, WKY | E2 | 2 mg, ip | Chronic, 3 weeks | Pituitary weight: Wis > WKY BrdU labeling: Wis > WKY |
(Mitsui et al., 2013) |
| Ratus, Male | ACI, COP | DES | 5 mg implants | Chronic, 12 weeks | Pituitary weights: ACI > COP | (Spady et al., 1999) |
| Thymus | ||||||
| Mus | BALB/c* C57BL/6J, C57BL6.BALB/c-F1 | DES | 2.5–1000 ppb po | Acute, 6 days | Decrease thymic weight: BALB/c* ~ C57BL/6J = F1 Initial thymic weights significantly greater in BALB/c*. |
(Greenman et al., 1977) |
| Ratus, Female | BN, F344, SD | DES | 5 mg implants | Chronic, 1–28 days | Decrease thymic weight: SD > F344 = BN | (Gould et al., 2000) |
| Testis | ||||||
| Mus, Male | C17/Jls, C57BL/6J, CD1 | E2 | 0–40 μg implants | Chronic, 20 days | Inhibition of testes: C17/Jls = C57BL/6J > CD-1 | (Spearow et al., 2001) |
| Skin/Hair follicle | ||||||
| Mus, Female | C57BL/6, C3H, CD-1 | E2 | 10 nM topical treatment | Chronic, 20 weeks | Inhibition of hair growth: C57BL/6 = C3H = CD-1 | (Smart et al., 1999) |
Abbreviations:
Rat Strains: ACI=AxC-Irish; BN=Brown Norway; COP=Copenhagen; F344=Fischer 344; Holz=Holzman LE=Long-Evans; SD=Sprague-Dawley; Wis=Wistar; WKY=Wistar Kyoto
Mouse Strains: BALB/c*= BALB/c-StCrlfC3Hf/Nctr; B6C3-F1=C57BL/6JxC3H/HeJ
Compounds: BPA=Bisphenol A; CE=Coumestrol; D4=Octamethylcyclotetrasiloxane; DES=Diethylstilbestrol; E2=17β-estradiol; E2-DP=Estradiol Diproprionate; EE=Ethinyl Estradiol; Gen=Genestein; HMDS=Hexamethyldisiloxane; OCT=p-tert-octylphenol; PEP=Polyestradiol Phosphaste
Given the prominent role in breast cancer, effects of estrogen exposures in the mammary gland were of primary interest. However, only 5 publications compared responses across strains of mice or rats and differed greatly in doses and duration of treatments. Acute exposure to 17β-estradiol for 4 days was evaluated in C57BL/6J and C3H/H3J mice that were ovariectomized prior to puberty (Wall et al. 2014a). Branching of mammary ducts was greatest in C57BL/6J females, whereas C3H/HeJ exhibited a greater ductal length. The proliferative response was greater in C57BL/6J, but was balanced by higher levels of apoptosis. Acute treatment of C57BL/6J mice with 17β-estradiol also increased the ductal area to a greater extent than in CD-1 mice. However, the difference appears related to the near complete arrest of ductal elongation upon ovariectomy in C57BL/6J females whereas modest ductal growth continued in CD-1 even in the absence of ovaries (Wadia et al. 2007). Long-term exposure to diethylstilbestrol (DES) increased ductal branching to a similar extent in wild type BALB/cJ and 129/SvEv female mice but no mammary tumors were observed in either strain (Bennett et al. 2000). Effects of chronic treatment with 17β-estradiol were also compared in ACI and BN rats (Ding et al. 2013). ACI rats had dramatic proliferative responses after 1 week which were sustained throughout the 12-week treatment period. In contrast, BN rats exhibited a transient increase in proliferation during 1–3 weeks but returned to baseline levels by 12 weeks. Unlike C57BL/6J mice where increased proliferation was balanced by increased apoptosis, no difference in apoptosis was detected between ACI and BN rats using cleaved caspase as a marker. While 17β-estradiol stimulated proliferation and hyperplasia of the mammary epithelium in ACI rats, differentiation was the major effect in BN rats as indicated by the increased expression of milk proteins. ACI rats also developed mammary tumors with chronic exposure to 17β-estradiol or DES (Shull et al. 2001; Shull et al. 1997; Stone et al. 1979). While treatments and endpoints varied among these studies, they demonstrate clear differences in estrogen-induced proliferation, apoptosis and morphogenesis of the mammary ducts among the strains.
The uterus is also very sensitive to estrogens and responses have been compared in multiple strains of rats and mice. The outbred SD were used across 5 studies providing a common reference. To test acute responses, rats were ovariectomized then treated with 17β-estradiol, estrogen agonists or environmental compounds to test for xenoestrogen activity with uterine weight as a common endpoint. Increases in uterine weight in response to the estrogen agonist ethinyl estradiol (EE) were similar for the F344 and SD rats (McKim et al. 2001). EE increased uterine weights to a greater extent in Wis and DA/Han strains compared to SD (Diel et al. 2004; Geis et al. 2005). The outbred LE rats also had a greater increase in uterine weight in response to the estrogen analog PEP compared to SD rats (Lawson et al. 1984). The Wis rats were more sensitive to 17β-estradiol when compared to the WKY (Mitsui et al. 2013). As the SD strain is outbred and derived from the Wis strain, it appears that genetic variants determining uterine responses may be segregating within the populations used in the experiments. In mice treated with the estrogen analog E2-DP, the increase in uterine weight in the 129 strain was 2-fold that in the C57BL/6 and C3H/He strains with intermediate responses in C3H/JFe and DBA/1 (Drasher 1955). However, in subsequent studies using 17β-estradiol (Roper et al. 1999), responses were greater in C57BL/6J compared C3H/HeJ strains (3.5-fold vs 2.1-fold, respectively). The C57BL/6J strain also exhibited a stronger recruitment of macrophages compared to C3H/HeJ mice (Griffith et al. 1997).
The strain-differences in the uterus are more complex when comparing responses to environmental xenoestrogens. Although SD and F344 rats responded similarly to the estrogen analog EE, responses to octametthylcylotetrasioxane (D4) were greater in SD compared to F344 rats (McKim et al. 2001). However, bisphenol A (BPA) stimulated a 3-fold increase in proliferation of the vaginal epithelium in the F344 strain while responses in SD rats were negligible, yet responses to 17β-estradiol were similar for the two strains (Long et al. 2000). The strains did not differ in clearance of BPA, indicating genetic variation in intracellular signaling. In mice, low doses of DES increased uterine weights to a greater extent in C57BL/6J mice indicating that this strain is more sensitive than the BALB/c (Greenman et al. 1977). Prolonged treatment with DES induced neoplastic lesions in the uterus of 129/SvEv mice, but not BALB/cJ. But this response was limited to the uterus as there were no lesions detected in the mammary glands (Bennett et al. 2000). It is not clear from these studies whether the estrogenic responses are mediated by estrogen receptors as drugs to block these receptors were not included. Nonetheless, genetic variants determining sensitivity to xenoestrogens appear to be distinct from variants determining sensitivity to 17β-estradiol. Therefore, the relative potency of xenoestrogens across strains cannot be inferred from responses to 17β-estradiol.
Estrogen-stimulated proliferation of lactotrophs within the pituitary and secretion of prolactin have also been compared in strains of rats. Both 17β-estradiol and DES have been used and elicited consistent responses. The ACI and F344 strains are generally more responsive (Moy and Lawson 1992; Spady et al. 1999b; Stone et al. 1979; Wiklund and Gorski 1982) compared to the related outbred strains (SD, Holz, Wis). In contrast, the BN, COP and WKY strains had modest responses detectable at early time points, but were transient (Spady et al. 1999a). Tissue differences were especially striking in comparisons of Wis and WKY strains (Mitsui et al. 2013). While pituitary responses to 17β-estradiol were negligible in WKY rats, uterine weights were increased ~5-fold. Strain differences were also prominent following chronic stimulation with 17β-estradiol resulting in 5-fold higher levels of plasma prolactin in SD and Wis rats compared to BN (Blankenstein et al. 1984). This is similar to the relative sensitivity for estrogen-induced pituitary growth reviewed previously (Spady et al. 1999a) where F344 are most sensitive, SD being intermediate and BN being the least sensitive.
While estrogen agonists induce proliferative responses in mammary, uterine and pituitary, they cause regression in other tissues. DES induced thymic regression in both C57BL/6 and BALB/c strains (Greenman et al. 1977). Although the strains differed in initial thymus weights, both exhibited similar ~1.5 g decreases following DES treatment (Greenman et al. 1977). In rats, the F433 strain was most responsive in pituitary and uterine tissues, but DES-induced thymic regression was greatest in the SD strain compared to F344 and BN (Gould et al. 2000). Treatment with17β-estradiol elicited striking reductions in testicular weights in C57BL/6J males compared to outbred CD-1 mice (Spearow et al. 1999; Spearow et al. 2001). Similarly, hair regrowth is suppressed by 17β-estradiol in C57BL/6, C3H and CD-1 mice but did not differ among these strains (Smart et al. 1999). Therefore, it appears that genetic variants regulating estrogen-induced repression in these tissues differ from the variants mediating growth in other tissues.
The striking differences among strains in responses to estrogenic compounds demonstrates genetic diversity in the signaling pathways. It is especially notable that proliferation was transient in spite of continuous exposure in some strains. This was most notable in the pituitary and mammary tissues of BN and COP rats indicating an ability to attenuate signaling (Ding et al. 2013; Gould et al. 2000) but not in ACI and F344. In the case of mammary epithelium of C57BL/6J mice, proliferation in response to 17β-estradiol appeared to be balanced by an increase in apoptosis (Wall et al. 2014a). In contrast, ACI rats have a persistent proliferative response to DES in mammary epithelium without a compensatory increase in apoptosis and was associated with extensive hyperplasia (Ding et al. 2013). Thus, strains of rodents appear to have adopted different strategies to achieve tissue homeostasis. The ability to attenuate signaling also appears to differ among tissues. An example is the formation of neoplastic lesions in the uterus of 129/SvEv mice treated with DES, but no lesions developed in the mammary glands of these mice (Bennett et al. 2000). The diversity of responses among strains of rats and mice suggests that effects would be similarly variable among humans (Spearow and Barkley 2001). Thus, the genes and pathways regulating these responses can significantly affect whether exposure to endogenous levels of 17β-estradiol poses a significant risk. The genetic variation could also affect the risk associated with postmenopausal hormone therapies and exposures to environmental xenoestrogens.
Genetic variants determining responses to estrogen
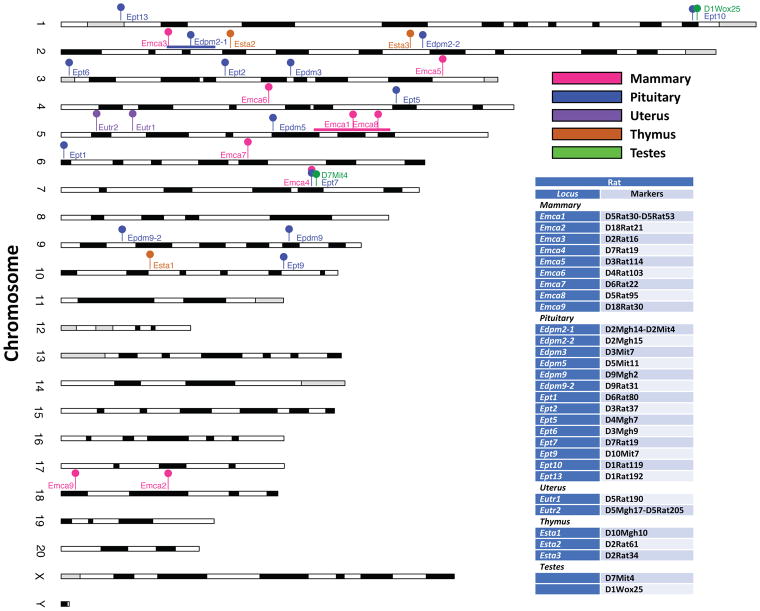
Linkage mapping has been used to identify the genetic basis for the variation in responses to estrogens among strains of rodents. Searches of rat and mouse genome databases and PubMed were conducted to compile relevant loci. These are summarized in Figure 1 with details provided in Supplementary Table 1.
Figure 1. Positions of QTL regulated responses to estrogens in 5 tissues in rats.
The phenotypes and strains used to define the quantitative trait loci (QTL) are indicated. The localization of QTL is based on the polymorphic markers used in mapping experiments. An interval is indicated using a horizontal line for Emca1 and Eutr2.
The pathogenic effects of prolonged estrogen stimulation on mammary tumorigenesis are best described in rats. ACI rats are highly susceptible to mammary tumors while COP are resistant and BN are highly resistant. In genetic crosses between these inbred rat strains, susceptibility to 17β-estradiol-induced mammary tumors behaves as an incompletely dominant trait (Shull et al. 2001). A total of 9 QTL (designated Emca) influencing susceptibility to mammary tumors (latency and/or number of mammary tumors) were identified (Figure 1). QTL mapped to rat chromosomes 5 and 18 in 17β-estradiol-treated F2 progeny generated in intercrosses between ACI and COP rats and chromosomes 2, 3, 4, 5, 6, 7, and 18 in progeny from intercrosses between ACI and BN rats (Gould et al. 2004; Schaffer et al. 2006; Shull 2007). The existence of each of these mammary tumor QTL has been confirmed through generation and characterization of congenic rat strains. The QTL exhibit orthology to a region of the human genome that has been linked to breast cancer risk and/or mammographic breast density in genome wide association studies, strongly suggesting these genetically defined rat models are relevant to understanding the genetic bases of breast cancer susceptibility in humans (Colletti et al. 2014; Schaffer et al. 2013).
Strain differences in the responsiveness of the anterior pituitary lactotroph to estrogens have also long been recognized (Spady et al. 1999a) allowing multiple QTL to be mapped (designated Edpm and Ept, Figure 1). F344 and ACI rats are highly sensitive to the stimulatory actions of estrogens on lactotroph proliferation, resulting in rapid development of lactotroph hyperplasia and adenoma. COP rats exhibit an intermediate sensitivity while BN rats are highly insensitive to estrogen-induced proliferation of lactotrophs. When pituitary weight was used as a phenotypic indicator of estrogen stimulated lactotroph proliferation, five QTL were identified upon characterization of DES treated female F2 progeny generated in an intercross between F344 and BN rats (Wendell and Gorski 1997). One additional QTL was mapped during characterization of backcross progeny generated using these same strains (Wendell et al. 2000). When ACI rats were utilized as the sensitive strain, six QTL were mapped upon characterization of DES treated male F2 progeny generated in reciprocal intercrosses between ACI and COP rats and two additional QTL were mapped upon characterization of 17β-estradiol-treated female F2 rats generated in a BNxACI intercross (Shull et al. 2007; Strecker et al. 2005). Each of the QTL mapped in these studies of estrogen action on the pituitary lactotroph resides in a distinct region of the rat genome, although the mapping resolution for Edpm3 and Ept2, both of which reside on rat chromosome 3, is insufficient to demonstrate conclusively that these two QTL are distinct entities. Several of these QTL that harbor genetic determinants of responsiveness of the rat pituitary gland to estrogens have been isolated as congenic rat strains, which will allow the genetic variants that impact responsiveness to estrogens to be mapped to higher resolution and their molecular actions to be further elucidated (Dennison et al. 2015; Kurz et al. 2014; Kurz et al. 2008; Pandey et al. 2004; Wendell et al. 2002). The presence of multiple loci regulating responses to estrogens and that these are distinct for mammary gland and uterus highlights the tissue-specific nature of estrogen signaling and its complexity.
In the rat uterus, estrogens exert strain specific actions on induction of pyometritis, which is thought to develop as a consequence of extensive over proliferation of the uterine epithelium. In this regard, BN rats are highly sensitive relative to ACI or F344 rats revealing 2 genetic loci (Figure 1). Eutr1, a QTL that influences pyometritis development, was mapped to the proximal region of rat chromosome 5 in a study in which F2 progeny generated in an intercross between BN and ACI rats were treated with 17β-estradiol (Gould et al. 2005). A second QTL, Eutr2 was mapped to the same region of chromosome 5 through characterization of DES treated congenic rats in which BN alleles across proximal chromosome 5 were introgressed onto the F344 genetic background (Pandey et al. 2005).
In contrast to the mammary epithelium, pituitary and uterus, estrogens inhibit proliferation of thymocytes in mice and rats (Gould et al. 2000; Greenman et al. 1977). QTL that modify the extent of the repression have been identified in rats (Figure 1). Treatment with DES identified QTL influencing repression on chromosome 10 (Esta1) and chromosome 2 (Esta2 and Esta3) in a study using male F2 progeny from a BN x ACI intercross (Gould et al. 2006). QTL associated with regression of testes induced by DES were identified on chromosomes 1 and 7 (Figure 1) in recombinant inbred male rats (Tachibana et al. 2006). Although no linkage was detected for pituitary adenoma development in male rats in the recombinant inbred strains, the markers associated with regression of testes (D1Wox25, D7Mit4) are in close proximity to a locus affecting pituitary adenomas on chromosome 1 (Ept10) and the locus involved in mammary tumors and pituitary adenomas (Emca4, Ept7) on chromosome 7 (Figure 1). The mammary tumor and pituitary phenotypes were identified in different crosses (ACI x COP and BN x ACI respectively) while the testicular phenotype was mapped in panel of 30 recombinant inbred strains derived from crosses of LE/Stm x F344/DuCrj rats (LEXF/FXLE). Therefore, while the locus may be the same, it is likely that the polymorphisms responsible for the tissue-specific effects differ among the strains.
Comparisons of estrogen responses in uterus and mammary glands of C3H/HeJ and C57BL/6J female mice reveal a complex set of modifiers in each tissue as well. Treatment with 17β-estradiol increased uterine weights 3.3-fold in C57BL/6J compared to a 2.2-fold increase in C3H/HeJ (Roper et al. 1999; Wall et al. 2013). The greater sensitivity to estrogen in the C57BL/6J strain was dominant in F1 hybrids. Therefore, linkage analysis was performed using backcross of F1 progeny to C3H/HeJ which identified 5 QTL (designated Est, Supplementary Table 1). Differences in estrogen-stimulated uterine weight was linked to chromosomes 5 and 11. Infiltration of eosinophils in the uterus induced by 17β-estradiol administration was linked to loci on chromomosomes 4, 10 and 16 as well as interactions between D10Mit180 and the loci on chromosomes 4 and 5. The effects on uterine weight in C57BL/6J and C3H/HeJ were not due to proliferative responses, but rather a higher level of apoptosis in the C3H/HeJ mice (Wall et al. 2013). Treatment with 17β-estradiol also stimulates expansion of the of ductal branching in mammary glands. The extent of development was nearly 2-fold greater in C57BL/6J mice compared to C3H/HeJ (Wall et al. 2014a). Although estrogen stimulates increases in tissue mass in both the mammary epithelium and uterus, little overlap was observed in transcriptional profiles induced by 17β-estradiol in these tissues (Wall et al. 2014b). Therefore, it appears that the QTL regulating responses to estrogenic stimulation are distinct among mammary epithelium and uterus in these strains of mice.
These studies indicate that the genetic variants controlling responsiveness to estrogens do so in a tissue and/or cell-type specific manner. For example, the QTL that influence development of pituitary lactotroph hyperplasia/adenoma are physically distinct from those that influence susceptibility to mammary cancer; the only exception being Ept7 and Emca4, which both map to the same region of rat chromosome 7 (Kurz et al. 2014; Schaffer et al. 2006; Shull et al. 2007). The tissue-specific actions of these loci allowed development of a novel rat model of 17β-estradiol-induced mammary tumors that lacks the deleterious morbidities associated with pituitary lactotroph hyperplasia/adenoma (Dennison et al. 2015). As a result, studies of regulation of estrogen signaling must be studied in the appropriate tissue. Despite the limited genetic diversity in ACI, COP and BN strains, a total of 9 QTL were identified that modify sensitivity to mammary tumors induced by chronic exposure to 17β-estradiol. This suggests that a much larger number of loci and variants may participate in regulating the consequences of exposure to estrogenic compounds in the human population.
Effects of estrogens on mammary gland development and breast cancer risk
Estrogen is a primary driver of mammary gland development during puberty. Estrogen receptor alpha (ERα) plays a principal role as deletion of the gene encoding ERα (Esr1) causes a profound failure of mammary gland development (Feng et al. 2007). Therefore, differences among strains in mammary development during puberty offers an additional approach to evaluate estrogen sensitivity and actions. A survey of mammary gland architecture was performed using 43 recombinant inbred strains of mice that comprise the Mouse Diversity Panel (Hadsell et al. 2015). Tissues were evaluated during puberty (42 day old) and at maturity (12 weeks) for 15 different measures of area, branching and density of the mammary ducts. The phenotypes were largely consistent at both ages within strains, and therefore, not simply a consequence of differences in the onset of puberty. Hierarchical clustering defined 4 patterns of development represented by C3H/HeJ, 129S1/SvlmJ, C57BL/6J and BALB/cByJ (Clusters 1–4, respectively). This is consistent with the differences in ductal growth between C57BL/6J and C3H/HeJ strains (Table 1) following acute treatment with 17β-estradiol (Wall et al. 2014a). The analysis of mammary gland development identified 20 mammary ductal quantitative trait loci (Mdq, Table 3). Of these, 9 were found to be within, or close to, orthologous regions associated with risk for breast cancer in humans.
Differences in mammary structure among the strains in the Mouse Diversity Panel also appear to provide insights into pathways underlying breast density. Most notably, the lead SNP for Mdq15 maps to within 0.5 Mbp of a locus on 12q24 associated with mammographic density in women (Stevens et al. 2012). The most significant SNP (rs1265507) is in close proximity to the Tbx3 gene, which is required for the ERα expressing lineage of mammary epithelial cells (Davenport et al. 2003; Kunasegaran et al. 2014) and breast cancer (Douglas and Papaioannou 2013; Krstic et al. 2016; Stephens et al. 2012). In addition, the lead SNP in Mdq8, which was associated with ductal branch density, maps to within 0.11 Mbp of an orthologous region in the human containing the breast cancer associated SNP, rs11814448 (Michailidou et al. 2015; Michailidou et al. 2013). This particular SNP is located between the genes Dnajc1 and Bmi1. Bmi1 is regulated by ERα and has been implicated in a number of behaviors in breast cancer cells including epithelial-to-mesenchymal transition (Wang et al. 2014). Therefore, the genetic modifiers that regulate ductal development in mice appear to overlap with breast cancer risk loci in women. Although it is unclear whether the Mdq loci regulated by development or are dependent on estrogen exposure, A search of ChIP-seq databases for H3K4me2 binding was used to identify regulatory elements within the Mdq loci and colocalize binding of progesterone receptor within 15 (Mbp) of the loci suggesting hormonal regulation (Hadsell et al. 2015).
The role of the Mdq in mammary tumorigenesis has not been tested directly. As a means to address their relationship with susceptibility to mammary tumors, the authors combined previously published data on strain dependent variation in mammary tumorigenesis and the incidence of lung metastases in the polyoma middle T-antigen model with their own data on variation in normal mammary ductal development. Modifiers of mammary tumorigenesis and lung metastasis have been mapped using mice expressing the oncogenic polyoma middle T-antigen (MMTV-PyMT) in crosses with 28 strains (Le Voyer et al. 2000; Le Voyer et al. 2001; Lifsted et al. 1998). Of these previously studied 28 strains, measures of normal branch density were available for 21. By plotting strain means, mammary ductal branch density and incidence of lung metatsatsis were found to be highly correlated (R2>0.7) indicating that at least a portion of the loci involved in ductal structure were related to tumor development and/or metastasis (Hadsell et al. 2015).
Modifiers of estrogen signaling --- challenges and opportunities
The studies in rodents demonstrate the presence of genetic variants that can limit or amplify the pathogenic effects of estrogenic compounds in mammary glands. A total of 29 QTL have been identified that influence the effects of estrogens on development of the mammary ductal network and susceptibility to mammary tumors (Figure 1 and Table 2). These loci are orthologous to polymorphisms associated with breast cancer risk in humans suggesting that variation in estrogen signaling may be a common mechanism underlying breast cancer risk. Furthermore, the risk posed by chronic exposure to estrogens would be of greatest concern for a subset of individuals who are particularly sensitive. This highlights the potential impact of gene x environment interactions in determining risk associated with estrogen exposures.
Table 2.
Overlap between mouse Mdq loci and breast cancer risk loci in humans1
| QTL | Lead SNP | P | Effect Sizea | Human locus | Human Syntenic Locationb | Breast Cancer Risk Loci | Nearest BrCa SNP | GWAS P | Distance to Mouseb | iCOGS Pc | Combined GWAS iCOG Pc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mdq1 | rs29972830 | 9.E-12 | 0.26 | 7q32.3 | 132.73 | YESd | rs4593472 | 3.E-05 | 2.06 | 4.E-06 | 2.E-09 |
|
| |||||||||||
| Mdq2 | rs32398376 | 2.E-09 | 0.21 | 1q25.3 | 183.13 | NO | rs11249433 | 7.E-10 | 61.85 | ||
|
| |||||||||||
| Mdq3 | rs13483228 | 3.E-09 | 0.21 | 18q12.1 | 28.16 | YES | rs1436904 | 1.E-03 | 3.59 | 5.E-06 | 3.E-08 |
| Mdq4 | rs33051600 | 3.E-08 | 0.2 | 2q24.3 | 169.07 | YES | rs2016394 | 1.E-02 | 3.90 | 2.E-06 | 8.E-06 |
| Mdq5 | rs30095733 | 6.E-08 | 0.17 | 5q11.2 | 51.23 | YES | rs889312 | 7.E-20 | 4.81 | 3.E-27 | 1.E-39 |
|
| |||||||||||
| Mdq6 | rs46013670 | 8.E-08 | 0.18 | 3q21.1 | 122.88 | NO | rs1053338 | 2.E-02 | 58.91 | 2.E-07 | 9.E-09 |
| Mdq7 | rs50444438 | 1.E-07 | 0.17 | 7p15.3 | 21.90 | NO | rs6977610 | 2.E-04 | 6.64 | ||
|
| |||||||||||
| Mdq8 | rs27176282 | 1.E-07 | 0.17 | 10p12.3 | 22.21 | YES | rs11814448 | 4.E-05 | 0.11 | 3.E-13 | 6.E-17 |
| Mdq9 | rs36588132 | 1.E-07 | 0.16 | 3p22.3 | 33.06 | YES | rs12493607 | 4.E-02 | 2.38 | 7.E-08 | 1.E-08 |
|
| |||||||||||
| Mdq10 | rs29830755 | 2.E-07 | 0.16 | 7p15.1 | 28.64 | NO | rs6977610 | 2.E-04 | 13.38 | ||
| Mdq11 | rs3696903 | 4.E-07 | 0.17 | 11p15.2 | 16.42 | NO | rs909116 | 7.E-07 | 14.48 | ||
|
| |||||||||||
| Mdq12 | rs33753649 | 5.E-07 | 0.17 | 22q12.1 | 25.95 | YES | rs132390 | 1.E-03 | 3.67 | 4.E-07 | 3.E-09 |
|
| |||||||||||
| Mdq13 | rs30305626 | 7.E-07 | 0.15 | 7q31.2 | 117.30 | NO | rs2048672 | 6.E-06 | 13.35 | ||
| Mdq14 | rs27275156 | 8.E-07 | 0.15 | 20p13 | 4.75 | NO | rs6109595 | 1.E-04 | 8.11 | ||
|
| |||||||||||
| Mdq15 | rs29778211 | 8.E-07 | 0.14 | 12q24.21 | 114.97 | YES | rs1265507 | 1.E-08 | 0.54 | ||
| Mdq16 | rs31558147 | 8.E-07 | 0.15 | 4q12 | 56.08 | YES | rs11732323 | 6.E-05 | 0.43 | ||
|
| |||||||||||
| Mdq17 | rs46415121 | 1.E-06 | 0.15 | 12q13.13 | 52.83 | NO | rs10506095 | 1.E-05 | 20.03 | ||
| Mdq18 | rs27336088 | 1.E-06 | 0.16 | 20q12 | 40.01 | NO | rs6109595 | 1.E-04 | 27.15 | ||
| Mdq19 | rs29782631 | 2.E-06 | 0.14 | 11q22.1 | 100.83 | NO | rs948725 | 2.E-04 | 23.58 | ||
| Mdq20 | rs36274122 | 2.E-06 | 0.14 | 1q24.3 | 171.86 | NO | rs1538472 | 5.E-05 | 73.77 | ||
Mouse loci from Hadsell et al 2015 (Hadsell et al. 2015); Human loci from Phengeni (http://www.ncbi.nlm.nih.gov/gap/phegeni), and
Michailidou et al. (Michailidou et al. 2015; Michailidou et al. 2013)
r2,
Mbp
Less than 5Mbp apart; iCOGS data provide larger sample sizes
Several hurdles hamper the ability to detect genetic variants that interact with levels of estrogen exposure to influence breast cancer risk in humans. The difficulty of accurately estimating levels of estrogen exposure in large populations of women is a major challenge. The magnitude of risk associated with genetic variants is often modest and, without very large studies, the statistical power to detect interactions can be limiting (Rudolph et al. 2016). However, relationships between mammographic breast density and estrogen exposure suggest that genetic interactions may be important. Mammographic density is among the strongest predictors of breast cancer risk and is determined, in part, by estrogen exposure. Estrogen plus progestin hormone therapies increase breast density and can explain the increased risk associated with the use of exogenous hormones (Byrne et al. 2017; Chlebowski et al. 2013). Conversely, inhibition of signaling through estrogen receptors by Tamoxifen reduced mammographic density and was associated with decreased risk of subsequent breast cancer (Cuzick et al. 2011). Identification of genes involved in estrogen signaling among the 11 loci associated with differences in mammographic density in GWAS (Lindstrom et al. 2014) reinforces the potential interactions between estrogen exposure and breast cancer risk. Candidate genes linked to polymorphisms include the gene encoding ERα (ESR1) and estrogen-responsive target genes (Amphiregulin, Insulin-like growth factor 1) as well as lymphocyte-specific factor 1 (LSP1). A targeted screen of genes linked to breast cancer risk (Odefrey et al. 2010) also found significant associations between mammographic breast density and polymorphisms in LSP1. Similar to the effects in ACI rats (Ding et al. 2013), genetic variants in humans that impair the ability to attenuate estrogen signaling may render individuals susceptible to breast cancer associated with postmenopausal hormone therapies. The reduction in breast density in response to Tamoxifen has been shown to vary substantially among women (Cuzick et al. 2011) and genetic variants that are permissive to estrogen signaling may play a role in limiting the efficacy of Tamoxifen in these individuals. Thus, the challenge is to identify the rate-limiting steps in estrogen signaling and pathogenic responses in breast tissue.
Genetic variation in responses to estrogenic stimulation among strains of rodents provides an incisive approach to define critical pathways in mammary tissue. The dose, duration and types of estrogenic compounds can be controlled in experimental settings. Study designs allow effects of modest magnitude to be detected. The availability of genetically diverse strains of mice represented in the Collaborative Cross and Diversity Outbred panels (Harrill and McAllister 2017) offer new opportunities to expand the number of loci influencing sensitivity to elevated levels of endogenous estrogens or estrogen therapies. These studies would provide a foundation for selection of strains to evaluate the risks posed by environmental xenoestrogens. The loci in humans that are orthologous to the rodent loci regulating estrogen sensitivity can be used in targeted screens for interactions with estrogenic exposures (endogenous or environmental sources) providing greater statistical power to detect associations. The pathways identified in rodents would provide insights into risk alleles in humans rendering individuals sensitive to the pathogenic effects of estrogenic exposures as well as provide biomarkers for assessing risk in breast tissues.
Supplementary Material
Acknowledgments
Research reported in this publication was supported, in part, by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number U01ES026140 (DJJ, SSS), U01ES026127 (AT-D), U01ES026132 (CB), and R01-CA77876 and R01-CA204320 (JDS). Funding was also provided by the Department of Defense under contract # W81XWH-15-1-0217 (DJJ) and the Rays of Hope Center for Breast Cancer Research (DJJ, GM-J) and USDA/ARS Cooperative Agreement No. 6250-51000-052 (DLH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.” More info at https://grants.nih.gov/grants/acknow.htm.
Contributor Information
D. Joseph Jerry, Department of Veterinary & Animal Sciences, 661 North Pleasant Street, Integrated Life Sciences Building, Amherst, MA 01003; Pioneer Valley Life Sciences Institute, 3601 Main Street, Springfield, MA 01199.
James D. Shull, McArdle Laboratory for Cancer Research, Department of Oncology, and UW Carbone Cancer Center, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, Wisconsin 53705
Darryl L. Hadsell, USDA/ARS Children's Nutrition Research Center, Department of Pediatrics, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston Texas. 77030
Monique Rijnkels, Department of Veterinary Integrative Biosciences, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX 77843.
Karen A. Dunphy, Department of Veterinary & Animal Sciences, 661 North Pleasant Street, Integrated Life Sciences Building, Amherst, MA 01003
Sallie S. Schneider, Pioneer Valley Life Sciences Institute, Baystate Medical Center, 3601 Main Street, Springfield, MA 01199
Laura N. Vandenberg, Department of Environmental Health Sciences, School of Public Health and Health Sciences, University of Massachusetts, Amherst 01003
Prabin Dhangada Majhi, Department of Veterinary & Animal Sciences, 661 North Pleasant Street, Integrated Life Sciences Building, Amherst, MA 01003.
Celia Byrne, Department of Preventive Medicine, F. Edward Hébert School of Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814.
Amy Trentham-Dietz, Department of Population Health Sciences and the Carbone Cancer Center, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI.
References
- Albrektsen G, Heuch I, Hansen S, Kvale G. Breast cancer risk by age at birth, time since birth and time intervals between births: exploring interaction effects. Br J Cancer. 2005;92:167–175. doi: 10.1038/sj.bjc.6602302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GL, Chlebowski RT, Aragaki AK, Kuller LH, Manson JE, Gass M, Bluhm E, Connelly S, Hubbell FA, Lane D, Martin L, Ockene J, Rohan T, Schenken R, Wactawski-Wende J. Conjugated equine oestrogen and breast cancer incidence and mortality in postmenopausal women with hysterectomy: extended follow-up of the Women's Health Initiative randomised placebo-controlled trial. The lancet oncology. 2012;13:476–486. doi: 10.1016/S1470-2045(12)70075-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LM, McAllister KA, Malphurs J, Ward T, Collins NK, Seely JC, Gowen LC, Koller BH, Davis BJ, Wiseman RW. Mice heterozygous for a Brca1 or Brca2 mutation display distinct mammary gland and ovarian phenotypes in response to diethylstilbestrol. Cancer Res. 2000;60:3461–3469. [PubMed] [Google Scholar]
- Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362:419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- Bernstein L. Epidemiology of endocrine-related risk factors for breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:3–15. doi: 10.1023/a:1015714305420. [DOI] [PubMed] [Google Scholar]
- Blankenstein MA, Broerse JJ, van Zwieten MJ, van der Molen HJ. Prolactin concentration in plasma and susceptibility to mammary tumors in female rats from different strains treated chronically with estradiol-17 beta. Breast Cancer Res Treat. 1984;4:137–141. doi: 10.1007/BF01806396. [DOI] [PubMed] [Google Scholar]
- Burns KA, Korach KS. Estrogen receptors and human disease: an update. Archives of toxicology. 2012;86:1491–1504. doi: 10.1007/s00204-012-0868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Ursin G, Martin CF, Peck JD, Cole EB, Zeng D, Kim E, Yaffe MD, Boyd NF, Heiss G, McTiernan A, Chlebowski RT, Lane DS, Manson JE, Wactawski-Wende J, Pisano ED. Mammographic Density Change With Estrogen and Progestin Therapy and Breast Cancer Risk. Journal of the National Cancer Institute. 2017;109 doi: 10.1093/jnci/djx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chie WC, Hsieh C, Newcomb PA, Longnecker MP, Mittendorf R, Greenberg ER, Clapp RW, Burke KP, Titus-Ernstoff L, Trentham-Dietz A, MacMahon B. Age at any full-term pregnancy and breast cancer risk. Am J Epidemiol. 2000;151:715–722. doi: 10.1093/oxfordjournals.aje.a010266. [DOI] [PubMed] [Google Scholar]
- Chlebowski RT, Manson JE, Anderson GL, Cauley JA, Aragaki AK, Stefanick ML, Lane DS, Johnson KC, Wactawski-Wende J, Chen C, Qi L, Yasmeen S, Newcomb PA, Prentice RL. Estrogen plus progestin and breast cancer incidence and mortality in the Women's Health Initiative Observational Study. Journal of the National Cancer Institute. 2013;105:526–535. doi: 10.1093/jnci/djt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti JA, 2nd, Leland-Wavrin KM, Kurz SG, Hickman MP, Seiler NL, Samanas NB, Eckert QA, Dennison KL, Ding L, Schaffer BS, Shull JD. Validation of six genetic determinants of susceptibility to estrogen-induced mammary cancer in the rat and assessment of their relevance to breast cancer risk in humans. G3 (Bethesda, Md) 2014;4:1385–1394. doi: 10.1534/g3.114.011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Warwick J, Pinney E, Duffy SW, Cawthorn S, Howell A, Forbes JF, Warren RM. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: a nested case-control study. Journal of the National Cancer Institute. 2011;103:744–752. doi: 10.1093/jnci/djr079. [DOI] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Dennison KL, Samanas NB, Harenda QE, Hickman MP, Seiler NL, Ding L, Shull JD. Development and characterization of a novel rat model of estrogen-induced mammary cancer. Endocrine-related cancer. 2015;22:239–248. doi: 10.1530/ERC-14-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diel P, Schmidt S, Vollmer G, Janning P, Upmeier A, Michna H, Bolt HM, Degen GH. Comparative responses of three rat strains (DA/Han, Sprague-Dawley and Wistar) to treatment with environmental estrogens. Archives of toxicology. 2004;78:183–193. doi: 10.1007/s00204-003-0535-y. [DOI] [PubMed] [Google Scholar]
- Ding L, Zhao Y, Warren CL, Sullivan R, Eliceiri KW, Shull JD. Association of cellular and molecular responses in the rat mammary gland to 17beta-estradiol with susceptibility to mammary cancer. BMC cancer. 2013;13:573. doi: 10.1186/1471-2407-13-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas NC, Papaioannou VE. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. Journal of mammary gland biology and neoplasia. 2013;18:143–147. doi: 10.1007/s10911-013-9282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drasher M. Strain differences in the response of the mouse uterus to estrogens. J Heredity. 1955;46:190–192. [Google Scholar]
- Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, Healey CS, Kar S, Pooley KA, Lopez-Knowles E, Dicks E, Barrowdale D, Sinnott-Armstrong NA, Sallari RC, Hillman KM, Kaufmann S, Sivakumaran H, Moradi Marjaneh M, Lee JS, Hills M, Jarosz M, Drury S, Canisius S, Bolla MK, Dennis J, Wang Q, Hopper JL, Southey MC, Broeks A, Schmidt MK, Lophatananon A, Muir K, Beckmann MW, Fasching PA, Dos-Santos-Silva I, Peto J, Sawyer EJ, Tomlinson I, Burwinkel B, Marme F, Guenel P, Truong T, Bojesen SE, Flyger H, Gonzalez-Neira A, Perez JI, Anton-Culver H, Eunjung L, Arndt V, Brenner H, Meindl A, Schmutzler RK, Brauch H, Hamann U, Aittomaki K, Blomqvist C, Ito H, Matsuo K, Bogdanova N, Dork T, Lindblom A, Margolin S, Kosma VM, Mannermaa A, Tseng CC, Wu AH, Lambrechts D, Wildiers H, Chang-Claude J, Rudolph A, Peterlongo P, Radice P, Olson JE, Giles GG, Milne RL, Haiman CA, Henderson BE, Goldberg MS, Teo SH, Yip CH, Nord S, Borresen-Dale AL, Kristensen V, Long J, Zheng W, Pylkas K, Winqvist R, Andrulis IL, Knight JA, Devilee P, Seynaeve C, Figueroa J, Sherman ME, Czene K, Darabi H, Hollestelle A, van den Ouweland AM, Humphreys K, Gao YT, Shu XO, Cox A, Cross SS, Blot W, Cai Q, Ghoussaini M, Perkins BJ, Shah M, Choi JY, Kang D, Lee SC, Hartman M, Kabisch M, Torres D, Jakubowska A, Lubinski J, Brennan P, Sangrajrang S, Ambrosone CB, Toland AE, Shen CY, Wu PE, Orr N, Swerdlow A, McGuffog L, Healey S, Lee A, Kapuscinski M, John EM, Terry MB, Daly MB, Goldgar DE, Buys SS, Janavicius R, Tihomirova L, Tung N, Dorfling CM, van Rensburg EJ, Neuhausen SL, Ejlertsen B, Hansen TV, Osorio A, Benitez J, Rando R, Weitzel JN, Bonanni B, Peissel B, Manoukian S, Papi L, Ottini L, Konstantopoulou I, Apostolou P, Garber J, Rashid MU, Frost D, Izatt L, Ellis S, Godwin AK, Arnold N, Niederacher D, Rhiem K, Bogdanova-Markov N, Sagne C, Stoppa-Lyonnet D, Damiola F, Sinilnikova OM, Mazoyer S, Isaacs C, Claes KB, De Leeneer K, de la Hoya M, Caldes T, Nevanlinna H, Khan S, Mensenkamp AR, Hooning MJ, Rookus MA, Kwong A, Olah E, Diez O, Brunet J, Pujana MA, Gronwald J, Huzarski T, Barkardottir RB, Laframboise R, Soucy P, Montagna M, Agata S, Teixeira MR, Park SK, Lindor N, Couch FJ, Tischkowitz M, Foretova L, Vijai J, Offit K, Singer CF, Rappaport C, Phelan CM, Greene MH, Mai PL, Rennert G, Imyanitov EN, Hulick PJ, Phillips KA, Piedmonte M, Mulligan AM, Glendon G, Bojesen A, Thomassen M, Caligo MA, Yoon SY, Friedman E, Laitman Y, Borg A, von Wachenfeldt A, Ehrencrona H, Rantala J, Olopade OI, Ganz PA, Nussbaum RL, Gayther SA, Nathanson KL, Domchek SM, Arun BK, Mitchell G, Karlan BY, Lester J, Maskarinec G, Woolcott C, Scott C, Stone J, Apicella C, Tamimi R, Luben R, Khaw KT, Helland A, Haakensen V, Dowsett M, Pharoah PD, Simard J, Hall P, Garcia-Closas M, Vachon C, Chenevix-Trench G, Antoniou AC, Easton DF, Edwards SL. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy KA, Blackburn AC, Yan H, O'Connell LR, Jerry DJ. Estrogen and progesterone induce persistent increases in p53-dependent apoptosis and suppress mammary tumors in BALB/c-Trp53+/- mice. Breast Cancer Res. 2008;10:R43. doi: 10.1186/bcr2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliassen AH, Missmer SA, Tworoger SS, Spiegelman D, Barbieri RL, Dowsett M, Hankinson SE. Endogenous steroid hormone concentrations and risk of breast cancer among premenopausal women. J Natl Cancer Inst. 2006;98:1406–1415. doi: 10.1093/jnci/djj376. [DOI] [PubMed] [Google Scholar]
- Feng Y, Manka D, Wagner KU, Khan SA. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci U S A. 2007;104:14718–14723. doi: 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis RB, Diel P, Degen GH, Vollmer G. Effects of genistein on the expression of hepatic genes in two rat strains (Sprague-Dawley and Wistar) Toxicology letters. 2005;157:21–29. doi: 10.1016/j.toxlet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Gould KA, Pandey J, Lachel CM, Murrin CR, Flood LA, Pennington KL, Schaffer BS, Tochacek M, McComb RD, Meza JL, Wendell DL, Shull JD. Genetic mapping of Eutr1, a locus controlling E2-induced pyometritis in the Brown Norway rat, to RNO5. Mammalian genome : official journal of the International Mammalian Genome Society. 2005;16:854–864. doi: 10.1007/s00335-005-0070-7. [DOI] [PubMed] [Google Scholar]
- Gould KA, Shull JD, Gorski J. DES action in the thymus: inhibition of cell proliferation and genetic variation. Molecular and cellular endocrinology. 2000;170:31–39. doi: 10.1016/s0303-7207(00)00336-1. [DOI] [PubMed] [Google Scholar]
- Gould KA, Strecker TE, Hansen KK, Bynote KK, Peterson KA, Shull JD. Genetic mapping of loci controlling diethylstilbestrol-induced thymic atrophy in the Brown Norway rat. Mammalian genome : official journal of the International Mammalian Genome Society. 2006;17:451–464. doi: 10.1007/s00335-005-0183-z. [DOI] [PubMed] [Google Scholar]
- Gould KA, Tochacek M, Schaffer BS, Reindl TM, Murrin CR, Lachel CM, VanderWoude EA, Pennington KL, Flood LA, Bynote KK, Meza JL, Newton MA, Shull JD. Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics. 2004;168:2113–2125. doi: 10.1534/genetics.104.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman DL, Dooley K, Breeden CR. Strain differences in the response of the mouse to diethylstilbestrol. Journal of toxicology and environmental health. 1977;3:589–597. doi: 10.1080/15287397709529591. [DOI] [PubMed] [Google Scholar]
- Griffith JS, Jensen SM, Lunceford JK, Kahn MW, Zheng Y, Falase EA, Lyttle CR, Teuscher C. Evidence for the genetic control of estradiol-regulated responses. Implications for variation in normal and pathological hormone-dependent phenotypes. The American journal of pathology. 1997;150:2223–2230. [PMC free article] [PubMed] [Google Scholar]
- Guzman RC, Yang J, Rajkumar L, Thordarson G, Chen X, Nandi S. Hormonal prevention of breast cancer: mimicking the protective effect of pregnancy. Proc Natl Acad Sci U S A. 1999;96:2520–2525. doi: 10.1073/pnas.96.5.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadsell DL, Hadsell LA, Olea W, Rijnkels M, Creighton CJ, Smyth I, Short KM, Cox LL, Cox TC. In-silico QTL mapping of postpubertal mammary ductal development in the mouse uncovers potential human breast cancer risk loci. Mammalian genome : official journal of the International Mammalian Genome Society. 2015;26:57–79. doi: 10.1007/s00335-014-9551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, McAllister KA. New Rodent Population Models May Inform Human Health Risk Assessment and Identification of Genetic Susceptibility to Environmental Exposures. Environmental health perspectives. 2017;125:086002. doi: 10.1289/EHP1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic M, Macmillan CD, Leong HS, Clifford AG, Souter LH, Dales DW, Postenka CO, Chambers AF, Tuck AB. The transcriptional regulator TBX3 promotes progression from non-invasive to invasive breast cancer. BMC cancer. 2016;16:671. doi: 10.1186/s12885-016-2697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunasegaran K, Ho V, Chang TH, De Silva D, Bakker ML, Christoffels VM, Pietersen AM. Transcriptional repressor Tbx3 is required for the hormone-sensing cell lineage in mammary epithelium. PloS one. 2014;9:e110191. doi: 10.1371/journal.pone.0110191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz SG, Dennison KL, Samanas NB, Hickman MP, Eckert QA, Walker TL, Cupp AS, Shull JD. Ept7 influences estrogen action in the pituitary gland and body weight of rats. Mammalian genome : official journal of the International Mammalian Genome Society. 2014;25:244–252. doi: 10.1007/s00335-014-9504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz SG, Hansen KK, McLaughlin MT, Shivaswamy V, Schaffer BS, Gould KA, McComb RD, Meza JL, Shull JD. Tissue-specific actions of the Ept1, Ept2, Ept6, and Ept9 genetic determinants of responsiveness to estrogens in the female rat. Endocrinology. 2008;149:3850–3859. doi: 10.1210/en.2008-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DM, Sensui N, Gala RR. Ovariectomized Sprague-Dawley and Long-Evans rats release prolactin differently in response to estrogen. Life sciences. 1984;34:455–459. doi: 10.1016/0024-3205(84)90500-9. [DOI] [PubMed] [Google Scholar]
- Le Voyer T, Lu Z, Babb J, Lifsted T, Williams M, Hunter K. An epistatic interaction controls the latency of a transgene-induced mammary tumor. Mamm Genome. 2000;11:883–889. doi: 10.1007/s003350010163. [DOI] [PubMed] [Google Scholar]
- Le Voyer T, Rouse J, Lu Z, Lifsted T, Williams M, Hunter KW. Three loci modify growth of a transgene-induced mammary tumor: suppression of proliferation associated with decreased microvessel density. Genomics. 2001;74:253–261. doi: 10.1006/geno.2001.6562. [DOI] [PubMed] [Google Scholar]
- Lifsted T, Le VT, Williams M, Muller W, Klein-Szanto A, Buetow KH, Hunter KW. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–644. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Lindstrom S, Thompson DJ, Paterson AD, Li J, Gierach GL, Scott C, Stone J, Douglas JA, dos-Santos-Silva I, Fernandez-Navarro P, Verghase J, Smith P, Brown J, Luben R, Wareham NJ, Loos RJ, Heit JA, Pankratz VS, Norman A, Goode EL, Cunningham JM, deAndrade M, Vierkant RA, Czene K, Fasching PA, Baglietto L, Southey MC, Giles GG, Shah KP, Chan HP, Helvie MA, Beck AH, Knoblauch NW, Hazra A, Hunter DJ, Kraft P, Pollan M, Figueroa JD, Couch FJ, Hopper JL, Hall P, Easton DF, Boyd NF, Vachon CM, Tamimi RM. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nature communications. 2014;5:5303. doi: 10.1038/ncomms6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Steinmetz R, Ben-Jonathan N, Caperell-Grant A, Young PC, Nephew KP, Bigsby RM. Strain differences in vaginal responses to the xenoestrogen bisphenol A. Environmental health perspectives. 2000;108:243–247. doi: 10.1289/ehp.00108243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMahon B, Cole P, Lin TM, Lowe CR, Mirra AP, Ravnihar B, Salber EJ, Valaoras VG, Yuasa S. Age at first birth and breast cancer risk. Bull World Health Organ. 1970;43:209–221. [PMC free article] [PubMed] [Google Scholar]
- McKim JM, Jr, Wilga PC, Breslin WJ, Plotzke KP, Gallavan RH, Meeks RG. Potential estrogenic and antiestrogenic activity of the cyclic siloxane octamethylcyclotetrasiloxane (D4) and the linear siloxane hexamethyldisiloxane (HMDS) in immature rats using the uterotrophic assay. Toxicological sciences : an official journal of the Society of Toxicology. 2001;63:37–46. doi: 10.1093/toxsci/63.1.37. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, Perkins BJ, Czene K, Eriksson M, Darabi H, Brand JS, Bojesen SE, Nordestgaard BG, Flyger H, Nielsen SF, Rahman N, Turnbull C, Fletcher O, Peto J, Gibson L, Dos-Santos-Silva I, Chang-Claude J, Flesch-Janys D, Rudolph A, Eilber U, Behrens S, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Khan S, Aaltonen K, Ahsan H, Kibriya MG, Whittemore AS, John EM, Malone KE, Gammon MD, Santella RM, Ursin G, Makalic E, Schmidt DF, Casey G, Hunter DJ, Gapstur SM, Gaudet MM, Diver WR, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Berg CD, Chanock SJ, Figueroa J, Hoover RN, Lambrechts D, Neven P, Wildiers H, van Limbergen E, Schmidt MK, Broeks A, Verhoef S, Cornelissen S, Couch FJ, Olson JE, Hallberg E, Vachon C, Waisfisz Q, Meijers-Heijboer H, Adank MA, van der Luijt RB, Li J, Liu J, Humphreys K, Kang D, Choi JY, Park SK, Yoo KY, Matsuo K, Ito H, Iwata H, Tajima K, Guenel P, Truong T, Mulot C, Sanchez M, Burwinkel B, Marme F, Surowy H, Sohn C, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Gonzalez-Neira A, Benitez J, Zamora MP, Perez JI, Shu XO, Lu W, Gao YT, Cai H, Cox A, Cross SS, Reed MW, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Lindblom A, Margolin S, Teo SH, Yip CH, Taib NA, Tan GH, Hooning MJ, Hollestelle A, Martens JW, Collee JM, Blot W, Signorello LB, Cai Q, Hopper JL, Southey MC, Tsimiklis H, Apicella C, Shen CY, Hsiung CN, Wu PE, Hou MF, Kristensen VN, Nord S, Alnaes GI, Giles GG, Milne RL, McLean C, Canzian F, Trichopoulos D, Peeters P, Lund E, Sund M, Khaw KT, Gunter MJ, Palli D, Mortensen LM, Dossus L, Huerta JM, Meindl A, Schmutzler RK, Sutter C, Yang R, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Hartman M, Miao H, Chia KS, Chan CW, Fasching PA, Hein A, Beckmann MW, Haeberle L, Brenner H, Dieffenbach AK, Arndt V, Stegmaier C, Ashworth A, Orr N, Schoemaker MJ, Swerdlow AJ, Brinton L, Garcia-Closas M, Zheng W, Halverson SL, Shrubsole M, Long J, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Radice P, Peterlongo P, Manoukian S, Bernard L, Bogdanova NV, Dork T, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Devilee P, Tollenaar RA, Seynaeve C, Van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Huzarski T, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Slager S, Toland AE, Ambrosone CB, Yannoukakos D, Kabisch M, Torres D, Neuhausen SL, Anton-Culver H, Luccarini C, Baynes C, Ahmed S, Healey CS, Tessier DC, Vincent D, Bacot F, Pita G, Alonso MR, Alvarez N, Herrero D, Simard J, Pharoah PP, Kraft P, Dunning AM, Chenevix-Trench G, Hall P, Easton DF. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015 doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Muller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guenel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Muller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dork T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. 361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui T, Ishida M, Izawa M, Arita J. Differences between rat strains in the development of PRL-secreting pituitary tumors with long-term estrogen treatment: In vitro insulin-like growth factor-1-induced lactotroph proliferation and gene expression are affected in Wistar-Kyoto rats with low estrogen-susceptibility. Endocrine journal. 2013;60:1251–1259. doi: 10.1507/endocrj.ej13-0245. [DOI] [PubMed] [Google Scholar]
- Moy J, Lawson D. Proceedings of the Society for Experimental Biology and Medicine. Vol. 200. Society for Experimental Biology and Medicine; New York, NY: 1992. Temporal effects of estradiol and diethylstilbestrol on pituitary and plasma prolactin levels in ovariectomized Fischer 344 and Holtzman rats: a comparison of radioimmunoassay and Nb2 lymphoma cell bioassay; pp. 507–513. [DOI] [PubMed] [Google Scholar]
- Odefrey F, Stone J, Gurrin LC, Byrnes GB, Apicella C, Dite GS, Cawson JN, Giles GG, Treloar SA, English DR, Hopper JL, Southey MC. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 2010;70:1449–1458. doi: 10.1158/0008-5472.CAN-09-3495. [DOI] [PubMed] [Google Scholar]
- Pandey J, Bannout A, Wendell DL. The Edpm5 locus prevents the 'angiogenic switch' in an estrogen-induced rat pituitary tumor. Carcinogenesis. 2004;25:1829–1838. doi: 10.1093/carcin/bgh192. [DOI] [PubMed] [Google Scholar]
- Pandey J, Gould KA, McComb RD, Shull JD, Wendell DL. Localization of Eutr2, a locus controlling susceptibility to DES-induced uterine inflammation and pyometritis, to RNO5 using a congenic rat strain. Mammalian genome : official journal of the International Mammalian Genome Society. 2005;16:865–872. doi: 10.1007/s00335-005-0071-6. [DOI] [PubMed] [Google Scholar]
- Pike MC. Age-related factors in cancers of the breast, ovary, and endometrium. Journal of chronic diseases. 1987;40(Suppl 2):59s–69s. doi: 10.1016/s0021-9681(87)80009-7. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Anderson GL. The women's health initiative: lessons learned. Annual review of public health. 2008;29:131–150. doi: 10.1146/annurev.publhealth.29.020907.090947. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Chlebowski RT, Stefanick ML, Manson JE, Langer RD, Pettinger M, Hendrix SL, Hubbell FA, Kooperberg C, Kuller LH, Lane DS, McTiernan A, O'Sullivan MJ, Rossouw JE, Anderson GL. Conjugated equine estrogens and breast cancer risk in the Women's Health Initiative clinical trial and observational study. Am J Epidemiol. 2008;167:1407–1415. doi: 10.1093/aje/kwn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar L, Guzman RC, Yang J, Thordarson G, Talamantes F, Nandi S. Short-term exposure to pregnancy levels of estrogen prevents mammary carcinogenesis. Proc Natl Acad Sci U S A. 2001;98:11755–11759. doi: 10.1073/pnas.201393798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar L, Kittrell FS, Guzman RC, Brown PH, Nandi S, Medina D. Hormone-induced protection of mammary tumorigenesis in genetically engineered mouse models. Breast Cancer Res. 2007;9:R12. doi: 10.1186/bcr1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, Griffith JS, Lyttle CR, Doerge RW, McNabb AW, Broadbent RE, Teuscher C. Interacting quantitative trait loci control phenotypic variation in murine estradiol-regulated responses. Endocrinology. 1999;140:556–561. doi: 10.1210/endo.140.2.6521. [DOI] [PubMed] [Google Scholar]
- Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer. Br J Cancer. 2016;114:125–133. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BS, Lachel CM, Pennington KL, Murrin CR, Strecker TE, Tochacek M, Gould KA, Meza JL, McComb RD, Shull JD. Genetic bases of estrogen-induced tumorigenesis in the rat: mapping of loci controlling susceptibility to mammary cancer in a Brown Norway x ACI intercross. Cancer Res. 2006;66:7793–7800. doi: 10.1158/0008-5472.CAN-06-0143. [DOI] [PubMed] [Google Scholar]
- Schaffer BS, Leland-Wavrin KM, Kurz SG, Colletti JA, Seiler NL, Warren CL, Shull JD. Mapping of three genetic determinants of susceptibility to estrogen-induced mammary cancer within the Emca8 locus on rat chromosome 5. Cancer prevention research. 2013;6:59–69. doi: 10.1158/1940-6207.CAPR-12-0346-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull JD. The rat oncogenome: comparative genetics and genomics of rat models of mammary carcinogenesis. Breast disease. 2007;28:69–86. doi: 10.3233/bd-2007-28108. [DOI] [PubMed] [Google Scholar]
- Shull JD, Lachel CM, Murrin CR, Pennington KL, Schaffer BS, Strecker TE, Gould KA. Genetic control of estrogen action in the rat: mapping of QTLs that impact pituitary lactotroph hyperplasia in a BN x ACI intercross. Mammalian genome : official journal of the International Mammalian Genome Society. 2007;18:657–669. doi: 10.1007/s00335-007-9052-2. [DOI] [PubMed] [Google Scholar]
- Shull JD, Pennington KL, Reindl TM, Snyder MC, Strecker TE, Spady TJ, Tochacek M, McComb RD. Susceptibility to estrogen-induced mammary cancer segregates as an incompletely dominant phenotype in reciprocal crosses between the ACI and Copenhagen rat strains. Endocrinology. 2001;142:5124–5130. doi: 10.1210/endo.142.12.8530. [DOI] [PubMed] [Google Scholar]
- Shull JD, Spady TJ, Snyder MC, Johansson SL, Pennington KL. Ovary-intact, but not ovariectomized female ACI rats treated with 17beta-estradiol rapidly develop mammary carcinoma. Carcinogenesis. 1997;18:1595–1601. doi: 10.1093/carcin/18.8.1595. [DOI] [PubMed] [Google Scholar]
- Sivaraman L, Stephens LC, Markaverich BM, Clark JA, Krnacik S, Conneely OM, O'Malley BW, Medina D. Hormone-induced refractoriness to mammary carcinogenesis in Wistar-Furth rats. Carcinogenesis. 1998;19:1573–1581. doi: 10.1093/carcin/19.9.1573. [DOI] [PubMed] [Google Scholar]
- Smart RC, Oh HS, Chanda S, Robinette CL. Effects of 17-beta-estradiol and ICI 182 780 on hair growth in various strains of mice. The journal of investigative dermatology Symposium proceedings. 1999;4:285–289. doi: 10.1038/sj.jidsp.5640231. [DOI] [PubMed] [Google Scholar]
- Society AC. Breast cancer facts & figures 2017–2018. American Cancer Society; Atlanta: 2017. [Google Scholar]
- Spady TJ, McComb RD, Shull JD. Estrogen action in the regulation of cell proliferation, cell survival, and tumorigenesis in the rat anterior pituitary gland. Endocrine. 1999a;11:217–233. doi: 10.1385/ENDO:11:3:217. [DOI] [PubMed] [Google Scholar]
- Spady TJ, Pennington KL, McComb RD, Shull JD. Genetic bases of estrogen-induced pituitary growth in an intercross between the ACI and Copenhagen rat strains: dominant mendelian inheritance of the ACI phenotype. Endocrinology. 1999b;140:2828–2835. doi: 10.1210/endo.140.6.6757. [DOI] [PubMed] [Google Scholar]
- Spearow JL, Barkley M. Reassessment of models used to test xenobiotics for oestrogenic potency is overdue. Human reproduction. 2001;16:1027–1029. doi: 10.1093/humrep/16.5.1027. [DOI] [PubMed] [Google Scholar]
- Spearow JL, Doemeny P, Sera R, Leffler R, Barkley M. Genetic variation in susceptibility to endocrine disruption by estrogen in mice. Science. 1999;285:1259–1261. doi: 10.1126/science.285.5431.1259. [DOI] [PubMed] [Google Scholar]
- Spearow JL, O'Henley P, Doemeny P, Sera R, Leffler R, Sofos T, Barkley M. Genetic variation in physiological sensitivity to estrogen in mice. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2001;109:356–364. doi: 10.1034/j.1600-0463.2001.090504.x. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van 't Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KN, Lindstrom S, Scott CG, Thompson D, Sellers TA, Wang X, Wang A, Atkinson E, Rider DN, Eckel-Passow JE, Varghese JS, Audley T, Brown J, Leyland J, Luben RN, Warren RM, Loos RJ, Wareham NJ, Li J, Hall P, Liu J, Eriksson L, Czene K, Olson JE, Pankratz VS, Fredericksen Z, Diasio RB, Lee AM, Heit JA, DeAndrade M, Goode EL, Vierkant RA, Cunningham JM, Armasu SM, Weinshilboum R, Fridley BL, Batzler A, Ingle JN, Boyd NF, Paterson AD, Rommens J, Martin LJ, Hopper JL, Southey MC, Stone J, Apicella C, Kraft P, Hankinson SE, Hazra A, Hunter DJ, Easton DF, Couch FJ, Tamimi RM, Vachon CM. Identification of a novel percent mammographic density locus at 12q24. Human molecular genetics. 2012;21:3299–3305. doi: 10.1093/hmg/dds158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JP, Holtzman S, Shellabarger CJ. Neoplastic responses and correlated plasma prolactin levels in diethylstilbestrol-treated ACI and Sprague-Dawley rats. Cancer Res. 1979;39:773–778. [PubMed] [Google Scholar]
- Strecker TE, Spady TJ, Schaffer BS, Gould KA, Kaufman AE, Shen F, McLaughlin MT, Pennington KL, Meza JL, Shull JD. Genetic bases of estrogen-induced pituitary tumorigenesis: identification of genetic loci determining estrogen-induced pituitary growth in reciprocal crosses between the ACI and Copenhagen rat strains. Genetics. 2005;169:2189–2197. doi: 10.1534/genetics.104.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Lu L, Hiai H, Tamura A, Matsushima Y, Shisa H. Quantitative trait loci determining weight reduction of testes and pituitary by diethylstilbesterol in LEXF and FXLE recombinant inbred strain rats. Experimental animals. 2006;55:91–95. doi: 10.1538/expanim.55.91. [DOI] [PubMed] [Google Scholar]
- Tamimi RM, Spiegelman D, Smith-Warner SA, Wang M, Pazaris M, Willett WC, Eliassen AH, Hunter DJ. Population Attributable Risk of Modifiable and Nonmodifiable Breast Cancer Risk Factors in Postmenopausal Breast Cancer. American journal of epidemiology. 2016;184:884–893. doi: 10.1093/aje/kww145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadia PR, Vandenberg LN, Schaeberle CM, Rubin BS, Sonnenschein C, Soto AM. Perinatal bisphenol A exposure increases estrogen sensitivity of the mammary gland in diverse mouse strains. Environ Health Perspect. 2007;115:592–598. doi: 10.1289/ehp.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall EH, Case LK, Hewitt SC, Nguyen-Vu T, Candelaria NR, Teuscher C, Lin CY. Genetic control of ductal morphology, estrogen-induced ductal growth, and gene expression in female mouse mammary gland. Endocrinology. 2014a;155:3025–3035. doi: 10.1210/en.2013-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall EH, Hewitt SC, Case LK, Lin CY, Korach KS, Teuscher C. The role of genetics in estrogen responses: a critical piece of an intricate puzzle. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014b;28:5042–5054. doi: 10.1096/fj.14-260307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall EH, Hewitt SC, Liu L, del Rio R, Case LK, Lin CY, Korach KS, Teuscher C. Genetic control of estrogen-regulated transcriptional and cellular responses in mouse uterus. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:1874–1886. doi: 10.1096/fj.12-213462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu H, Li X, Zhao J, Zhang H, Mao J, Zou Y, Zhang H, Zhang S, Hou W, Hou L, McNutt MA, Zhang B. Estrogen receptor alpha-coupled Bmi1 regulation pathway in breast cancer and its clinical implications. BMC cancer. 2014;14:122. doi: 10.1186/1471-2407-14-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell DL, Daun SB, Stratton MB, Gorski J. Different functions of QTL for estrogen-dependent tumor growth of the rat pituitary. Mammalian genome : official journal of the International Mammalian Genome Society. 2000;11:855–861. doi: 10.1007/s003350010168. [DOI] [PubMed] [Google Scholar]
- Wendell DL, Gorski J. Quantitative trait loci for estrogen-dependent pituitary tumor growth in the rat. Mammalian genome : official journal of the International Mammalian Genome Society. 1997;8:823–829. doi: 10.1007/s003359900586. [DOI] [PubMed] [Google Scholar]
- Wendell DL, Pandey J, Kelley P. A congenic strain of rat for investigation of control of estrogen-induced growth. Mammalian genome : official journal of the International Mammalian Genome Society. 2002;13:664–666. doi: 10.1007/s00335-002-2183-6. [DOI] [PubMed] [Google Scholar]
- Wiklund JA, Gorski J. Genetic differences in estrogen-induced deoxyribonucleic acid synthesis in the rat pituitary: correlations with pituitary tumor susceptibility. Endocrinology. 1982;111:1140–1149. doi: 10.1210/endo-111-4-1140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.