Abstract
Once urinary bladder cancer (UBC) develops into muscle-invasive bladder cancer, its mortality rate increases dramatically. However, the molecular mechanisms of UBC invasion and metastasis remain largely unknown. Herein, using 5637 UBC cells, we generated two sublines with low (5637 NMI) and high (5637 HMI) invasive capabilities. Mass spectrum analyses revealed that the Wnt family protein Wnt7a is more highly expressed in 5637 HMI cells than in 5637 NMI cells. We also found that increased Wnt7a expression is associated with UBC metastasis and predicted worse clinical outcome in UBC patients. Wnt7a depletion in 5637 HMI and T24 cells reduced UBC cell invasion and decreased levels of active β-catenin and its downstream target genes involved in the epithelial-to-mesenchymal transition (EMT) and extracellular matrix (ECM) degradation. Consistently, treating 5637 NMI and J82 cells with recombinant Wnt7a induced cell invasion, EMT, and expression of ECM degradation–associated genes. Moreover, TOP/FOPflash luciferase assays indicated that Wnt7a activated canonical β-catenin signaling in UBC cells, and increased Wnt7a expression was associated with nuclear β-catenin in UBC samples. Wnt7a ablation suppressed matrix metalloproteinase 10 (MMP10) expression, and Wnt7a overexpression increased MMP10 promoter activity through two TCF/LEF promoter sites, confirming that Wnt7a-mediated MMP10 activation is mediated by the canonical Wnt/β-catenin pathway. Of note, the microRNA miR-370-3p directly repressed Wnt7a expression and thereby suppressed UBC cell invasion, which was partially restored by Wnt7a overexpression. Our results have identified an miR-370-3p/Wnt7a axis that controls UBC invasion through canonical Wnt/β-catenin signaling, which may offer prognostic and therapeutic opportunities.
Keywords: Wnt signaling, cancer biology, invasion, mass spectrometry (MS), microRNA (miRNA), metalloprotease, bladder cancer, miR-370-3p, MMP10, Wnt7a
Introduction
As one of the most common malignancies in the urological system, urinary bladder cancer (UBC)5 ranks at the eighth position in cancer-related mortality in males in the United States, with an estimated 12,240 deaths in 2017 (1). Low-grade, nonmuscle-invasive tumors account for 75% of newly diagnosed UBC cases, among which 15–20% of these patients advance toward muscle-invasive bladder cancers (≥ stage T2). Unfortunately, patients with muscle-invasive bladder cancer will progress to metastasis, with a prognosis of a 5-year survival <50% (2). Hence, it is of great significance to characterize the molecular events that lead to UBC invasion and metastasis.
The Wnt family consists of 19 secreted glycoproteins that regulate many important cellular processes, including cell proliferation, differentiation, and cell-fate specification (3, 4). Wnts bind to the members of the seven transmembrane family of proteins called Frizzleds (Fzds) and their associated co-receptors to stimulate complex network events, which are either dependent on β-catenin (canonical pathway) or independent of β-catenin (noncanonical pathways) (4). Although frequent genetic alterations of the Wnt pathway have been detected in various cancers (5), somatic mutations of the Wnt pathway are seldom reported in UBCs, except the association of SNPs of Wnt-associated genes with UBC risk (6, 7). Nevertheless, combined constitutively activated Wnt and the AKT pathway in murine urothelial cells synergistically leads to UBC formation, reinforcing the importance of the Wnt pathway during bladder carcinogenesis (8). However, so far it is still unclear which components in the Wnt pathway may play key roles in UBC invasion and metastasis.
MicroRNAs are small noncoding RNAs ∼22 nucleotides in length. Through binding to 3′-UTR of the target gene, they can negatively regulate the expression of target genes, resulting in either the decreased mRNA stability or translational inhibition (9). A number of dysregulated miRNAs have been identified in UBC specimens (10). Although some miRNAs have been extensively characterized as tumor-suppressive or oncogenic roles in UBC cell proliferation, survival, and drug resistance (11, 12), the molecular functions of most miRNAs, and their functions on cancer cell invasion and metastasis are unclear.
In this study, we generated two subpopulations of 5637 UBC cells, one with high invasiveness (5637 HMI) and the other with low invasiveness (5637 NMI). Using MS, we found much higher expression of Wnt7a in 5637 HMI cells than that in 5637 NMI cells. Because Wnt7a plays contradictory roles as a tumor suppressor or an oncogenic driver in a cell context-related manner (13, 14) and its role in UBC has not been dissected yet, we carried out studies to prove that Wnt7a overexpression induced the activation of the canonical Wnt-signaling pathway and UBC cell invasion. We also confirmed that reduction of tumor-suppressive miR-370-3p in 5637 HMI cells induced Wnt7a overexpression.
Results
Establishment of UBC 5637 cell sublines with low- and high-invasive capabilities
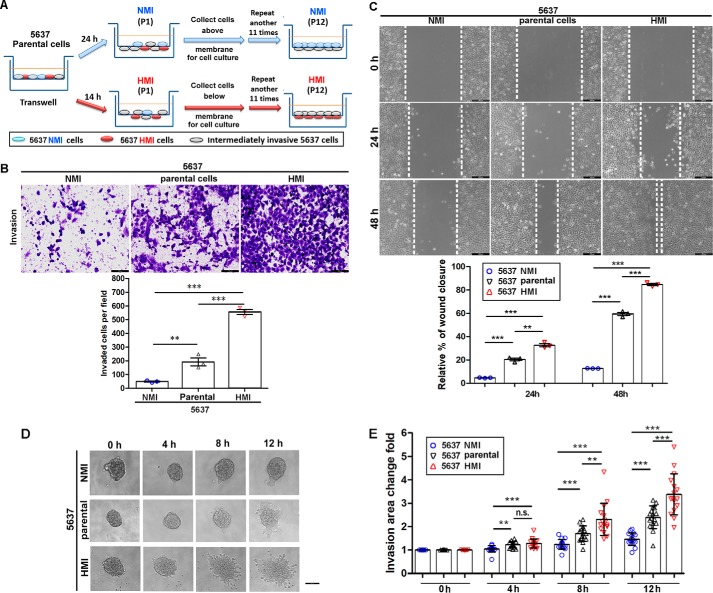
To investigate the mechanisms of UBC cell invasion, we isolated highly-invasive and noninvasive subpopulations from the UBC cell line 5637 by repeated transwell screening. As shown in Fig. 1A, 5637 cells that failed or succeeded to invade the Matrigel-coated membranes 24 h after NMI (P1) and 14 h after HMI (P1) seeding were harvested and cultured. Such separation was repeated for 12 rounds, and we obtained low-invasive (5637 NMI) and high-invasive (5637 HMI) sublines. To further validate their invasive properties, we performed Transwell invasion and wound-healing assays. As shown in Fig. 1B, 5637 HMI cells are more invasive than 5637 parental cells by 4.0-fold and 5637 NMI cells by 11.4-fold as early as 15 h after seeding. In addition, the wound-healing experiment also showed that 5637 HMI had a much higher capacity to migrate than parental cells by 5.2-fold and the 5637 NMI subline by 7.4-fold at 48 h (Fig. 1C). 3D Matrigel-invasive assay revealed that 5637 HMI cells invaded into Matrigel much earlier than its parental cells, whereas 5637 NMI cells invaded less (Fig. 1, D and E). However, cell proliferation rates of all of 5637 cells and their derivatives did not show significant changes, indicating that the difference of invasiveness among three cell lines is not due to cell proliferation rate (Fig. S1). Taken together, we successfully established two sublines derived from 5637 UBC cells with low- and high-invasive capability.
Figure 1.
Establishment and characterization of 5637 NMI and HMI cells. A, schematic illustration of the establishment of low-invasive (5637 NMI) and high-invasive (5637 HMI) sublines from primary 5637 cell line. B, transwell assay showed the invasive capacities of NMI 5637 and 5637 HMI cells. The upper photographs are representative fields of invaded cells 15 h after seeding. Scale bar, 100 μm. The lower graphs indicate the invaded cell number. C, wound healing assay show the migration capacities of 5637 NMI and 5637 HMI cells. The upper photographs are representative fields of wound closure at 0, 24, and 48 h, respectively. The lower graphs indicate the relative percent of wound closure at 24 and 48 h. Scale bar, 200 μm. The assays were performed in three independent biological replicates. D and E, invasive capacities of 5637 parental cells and derivatives in 3D Matrigel. 5637 parental cells, NMI, and HMI cells were formed spheroid in ultra-low 6-well plates and embedded in Matrigel for culture. The individual sphere in each group was monitored within 12 h and photographed (D), and the invasion area change fold was quantified (E) at the indicated time points. n = 16 for each group. Scale bar, 100 μm. **, p < 0.01; ***, p < 0.001; n.s., not significant.
Identification of dysregulated proteins in 5637 HMI cells compared with 5637 NMI cells
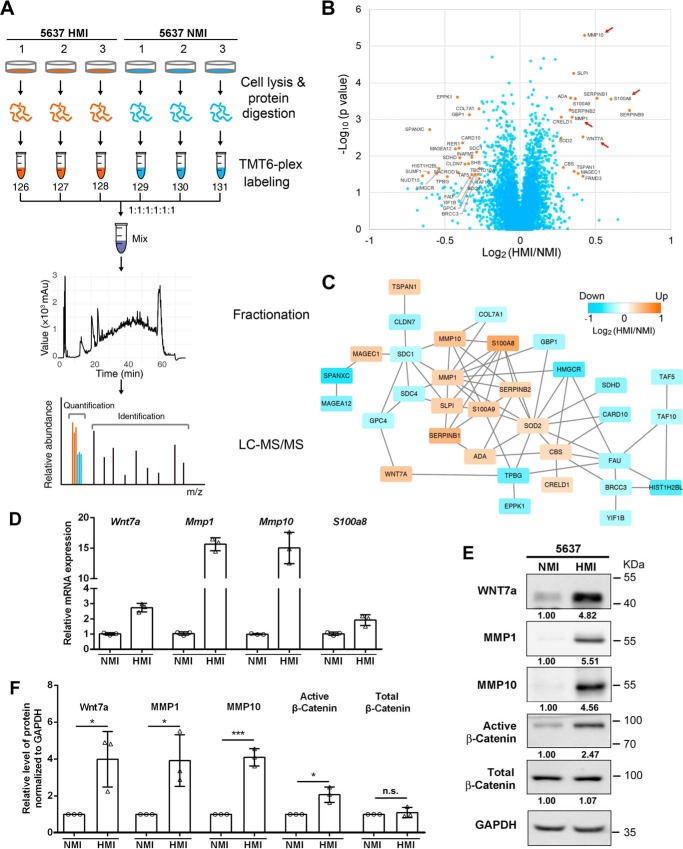
To unbiasedly identify dysregulated proteins involved in cancer cell invasion, we employed TMT6-plex labeling strategy to quantify fold changes of protein expression in 5637 HMI compared with 5637 NMI cells in biological triplicate (Fig. 2A). In total, 5,987 proteins were quantified across all six samples. Protein reporter intensities in each channel were adjusted by median normalization (Fig. S2A). Correlation coefficients among different channels were all nearly 1.00, suggesting good biological repeatability and minor differences between 5637 HMI and NMI samples (Fig. S2B). Hierarchical clustering analysis revealed that 5637 HMI and NMI samples can be clearly grouped, indicating the distinguishable proteome difference between 5637 HMI and NMI (Fig. S2C).
Figure 2.
Mass spectrum analysis of differentially expressed proteins in 5637 NMI and HMI cells. A, experimental workflow. 5637 HMI samples were labeled with 126, 127, and 128; 5637 NMI samples were labeled with 129, 130, and 131. The labeled samples were pooled and subjected to fractionation. Each fraction was analyzed on an Orbitrap Elite MS. B, volcano plot illustrating proteins with different abundances in 5637 HMI and NMI samples. It was displayed by −log10 (p value) versus log2 of the relative protein abundance of 5637 HMI to NMI cells. Orange points represent proteins with changes in abundance of greater than ±1.2-fold and p < 0.05. C, protein association network analysis of regulated proteins by STRING. Orange indicates up-regulation, and blue indicates down-regulation. Proteins were represented by log2 of the relative abundance of 5637 HMI to NMI cells. D and E, validation of mass spectrum data in 5637 NMI and HMI cells by qRT-PCR (D) and Western blotting. E, active β-catenin was also examined in 5637 NMI and HMI cells. GAPDH was used as loading control. F, quantification of Western blotting data in E. *, p < 0.05; ***, p < 0.001; n.s., not significant.
Proteins with both a significant p value < 0.05 and a fold change cutoff of >1.2 were determined to be differently expressed, resulting in 16 proteins up-regulated and 26 proteins down-regulated in 5637 HMI cells (Fig. 2B and Table S1). Wnt7a, a member of the WNT gene family, was of high abundance in 5637 HMI compared with 5637 NMI cells. The other three up-regulated proteins, MMP10, MMP1, and S100A8 as shown by protein association network analysis, indirectly interacted with Wnt7a and might be involved in the Wnt-signaling pathway (Fig. 2C).
We further validated those genes identified by mass spectrum that have also been reported to be involved in metastasis and invasion (15–18) by quantitative RT-PCR and Western blotting assays. mRNA levels of Wnt7a, MMP1, MMP10, and S100A8 significantly increased in 5637 HMI compared with 5637 NMI cells (Fig. 2D). Consistently, the remarkable up-regulations of Wnt7a, MMP1, and MMP10 were further confirmed at the protein level (Fig. 2, E and F). Because the Wnt-signaling pathway may contribute to metastasis and the controversial roles of Wnt7a in different cancer types (13, 14), we are eager to examine its clinical relevance and function on the invasive potential of UBC cells.
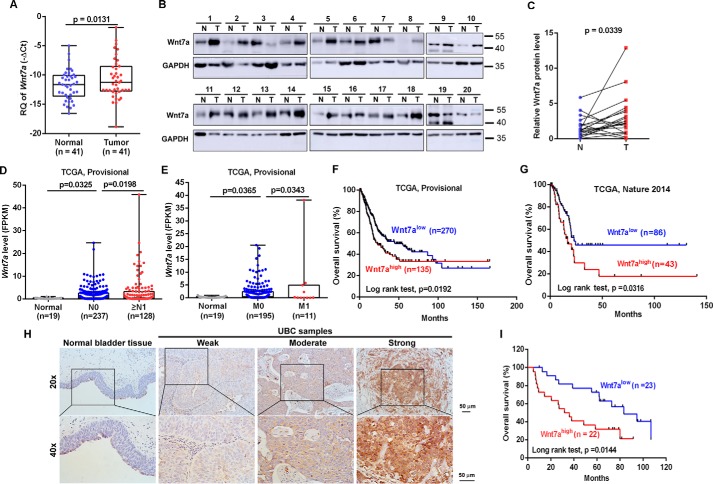
Overexpression of Wnt7a in UBC samples predicts poor clinical outcome
Because the Wnt7a gene localizes on human chromosome 3p25, which is frequently amplified in UBC samples and its role in UBC still remains unclear (18), at first we analyzed its expression level in UBC samples. As shown in Fig. 3A, Wnt7a was significantly increased in UBC samples (n = 41) from our cohort by 2.08-fold on average, compared with adjacent normal tissues. We further found that the Wnt7a protein level significantly increased in UBC tissues compared with their matched adjacent normal tissues (n = 20, p = 0.0339; Fig. 3, B and C). Using independent data from a public dataset (TCGA, Provisional), the Wnt7a mRNA level was 7.2-fold higher in UBC without lymph node metastatic lesion (N0) than normal bladder tissues, whereas it was even 1.62-fold higher in UBC with lymph node metastatic lesions (≥N1) than UBC with N0 (p < 0.05; Fig. 3D). Moreover, the Wnt7a mRNA level was 7.58-fold higher in UBC with M0 than normal bladder tissues, whereas it is even 2.44-fold higher in UBC with distant metastatic lesion (M1) than in UBC without distant metastatic lesion (M0) (p < 0.05; Fig. 3E). Notably, UBC patients with high Wnt7a mRNA levels have shorter overall survival time than those with low Wnt7a levels in two cohorts from the TCGA database (TCGA, Provisional and TCGA (6); Fig. 3, F and G). Immunohistochemical (IHC) staining revealed that Wnt7a was weakly detected in normal urothelial cells, whereas its overexpression could be easily detected in the UBC cells (Fig. 3H). Importantly, UBC patients with high Wnt7a protein levels had a shorter overall survival (Fig. 3I). In a word, Wnt7a may be a promising biomarker for UBC patients.
Figure 3.
Wnt7a overexpression is associated with canonical Wnt/β-catenin pathway and predicts clinical outcome. A, qRT-PCR showed Wnt7a mRNA expression level in matched clinical UBCs and corresponding normal tissues (n = 41). B, Western blotting results demonstrated the overexpression of Wnt7a in human UBC samples (n = 20). Wnt7a protein levels by quantitation of density of protein bands from Western blotting in UBCs (C) was relative to the matched normal tissues (n = 20) (B). N, nontumor; T, tumor. D, analysis of Wnt7a mRNA expression level fragments per kilobase million (FPKM) in normal bladder (n = 19), UBC without lymph node metastatic lesion (N0; n = 237), and UBC with lymph node metastatic lesions (≥N1; n = 128), which is derived from TCGA provisional database. E, analysis of Wnt7a mRNA expression level (FPKM) in normal bladder (n = 19), UBC without metastatic lesion (M0; n = 195), and UBC with distal metastatic lesions (M1; n = 11), which is derived from TCGA provisional database. F and G, correlation of Wnt7a mRNA expression with overall survival in TCGA provisional dataset (F) and one TCGA (6) dataset (G). H, representative IHC staining of Wnt7a in normal urothelial cells, and different staining intensities (weak, moderate, and strong) of Wnt7a in UBC samples. I, Kaplan-Meier plot of overall survival of 45 UBC patients, stratified by Wnt7a protein level.
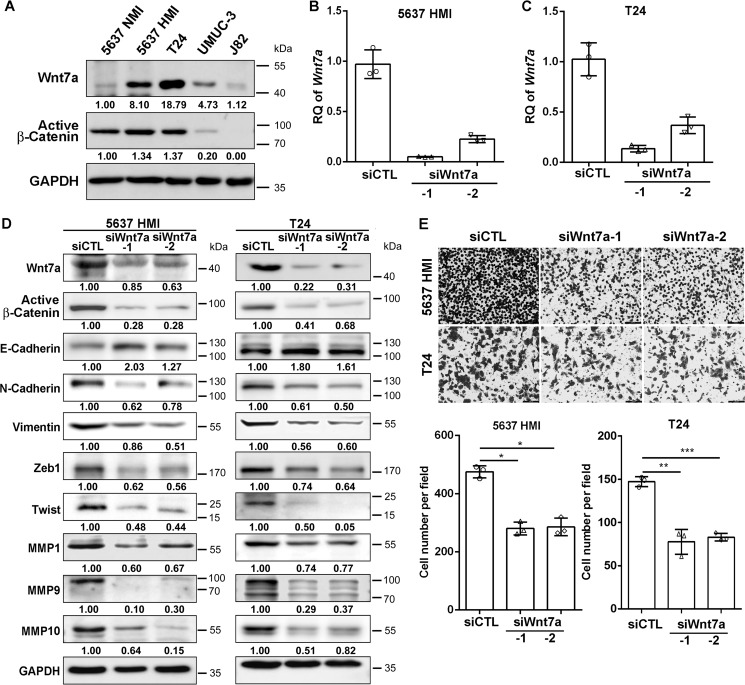
Depletion of Wnt7a inhibited UBC cell invasion
To validate the function of Wnt7a in UBC cells, we knocked down Wnt7a expression by the siRNA approach. First, we performed Western blotting assay and found T24 cells expressed Wnt7a at a high level, 5637 HMI and UMUC-3 cells expressed at an intermediate level, and 5637 NMI cells and J82 cells expressed at a low level. By using an antibody against active β-catenin, which recognizes nonphosphorylated β-catenin (Ser-33/37/Thr-41), we observed that the activation of the canonical Wnt/β-catenin pathway was roughly proportional to the Wnt7a level in these UBC cell lines (Fig. 4A), which is consistent with the finding in 5637 NMI and HMI cells (Fig. 2E). We then chose 5637 HMI cells and another UBC cell line T24 for the loss-of-function assay. Wnt7a was down-regulated in both cell lines by two different siRNAs (siWnt7a-1 and siWnt7a-2) compared with those transfected with control siRNA (siCTL; Fig. 4, B–D), as detected by qRT-PCR and Western blotting assays. In addition, transwell invasion assay demonstrated that Wnt7a depletion significantly decreased invasion capacities of 5637 HMI and T24 cells compared with control cells (Fig. 4E). As increasing evidence has shown that aberrant EMT is a characteristic feature of enhanced metastasis and two key EMT transcription factors ZEB1 and Twist are direct Wnt/β-catenin target genes (19, 20), we examined the EMT-associated proteins in our experimental setting. The ablation of Wnt7a significantly decreased ZEB1 and Twist, which facilitated EMT by activating mesenchymal genes and repressing epithelial genes (Fig. 4D). Consistently, we observed the reduced expression of vimentin and N-cadherin, with the increased level of E-cadherin. Matrix remodeling proteins, such as MMP1, MMP9, and MMP10, were also decreased by Wnt7a inhibition (Fig. 4D), suggesting that the expression of these proteins was regulated by Wnt7a signaling.
Figure 4.
Wnt7a deficiency decreased the invasiveness of 5637 HMI and T24 cells. A, expression level of Wnt7a and constitutively active β-catenin in UBC cell lines. B and C, Wnt7a mRNA expression levels in 5637 HMI (B) and T24 cells (C) transfected with control (siCTL) and two Wnt7a siRNAs (siWnt7a-1 and siWnt7a-2) by qRT-PCR. D, Western blotting showed EMT and invasiveness-associated protein expression in 5637 HMI and T24 cells transfected with siCTL, siWnt7a-1, and -2. E, transwell assay showed the invasive capacities of 5637 HMI and T24 cells followed by siCTL and Wnt7a siRNA treatment. The upper photograph are representative fields of staining invaded cells 16 h after seeding. Scale bar, 100 μm. The lower graphs indicated the counting number of invaded cells. The assays were performed in three independent biological replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Wnt7a promoted UBC cell invasion and metastasis
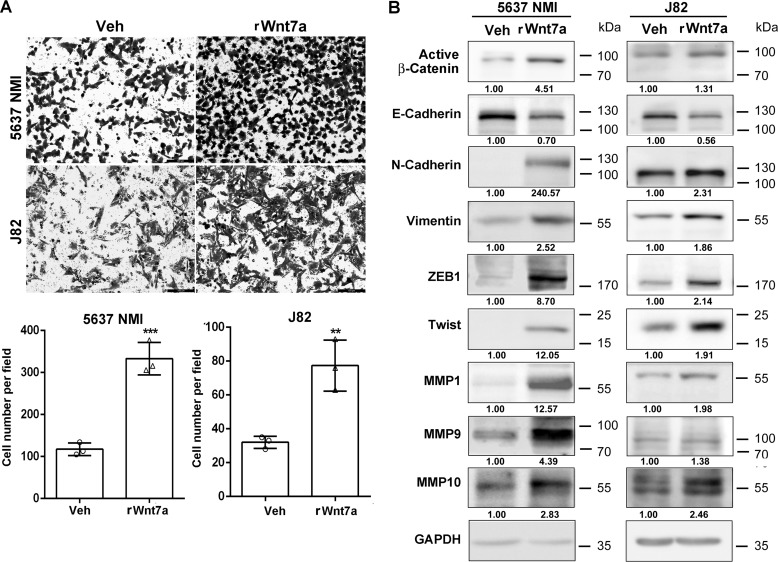
To determine the effects of Wnt7a overexpression, 5637 NMI and J82 cells that had low endogenous Wnt7a were treated with a recombinant human Wnt7a protein (rWnt7a, 100 ng/ml) for 24 h after serum starvation. Transwell assay demonstrated that rWnt7a protein significantly promoted the invasive capability of UBC cells, ∼3-fold in 5637 NMI and 2.5-fold in J82 cells compared with vehicle treatment (Fig. 5A). Consistently, the expression level of E-cadherin decreased, whereas the levels of N-cadherin and vimentin, as well as ZEB1, Twist, and matrix remodeling proteins (MMP1, MMP9, and MMP10), were elevated upon the treatment of the rWnt7a protein (Fig. 5B). Taken together, Wnt7a plays an important role in the regulation of UBC cell invasion.
Figure 5.
Wnt7a treatment induced invasive ability of 5637 NMI and J82. A, transwell assay showed the invasive capacities of 5637 NMI and J82 following treatment with recombinant Wnt7a protein (rWnt7a; 100 ng/ml) or vehicle (Veh) for 24 h. Photographs are representative fields of invaded cells 16 h after seeding. Scale bar, 100 μm. The graphs indicate the counting number of invaded cells in 5637 NMI and J82 cells. The assays were performed in three independent biological replicates. **, p < 0.01; ***, p < 0.001. B, Western blotting showed EMT and invasiveness associated protein expression in vehicle and rWnt7a (100 ng/ml) for 24 h in 5637 NMI and J82 cells.
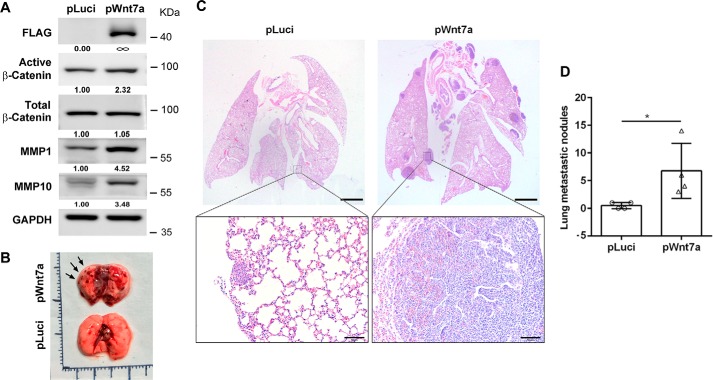
To test whether Wnt7a overexpression is sufficient for UBC metastasis, we established Wnt7a-overexpressing J82 cells by viral infection with the Wnt7a-overexpression construct (pWnt7a), whereas cells infected with luciferase expressing viral particle (pLuci) were used as control. Consistent with the data above, overexpression of Wnt7a activated the β-catenin pathway and induced MMP1 and MMP10 expression levels (Fig. 6A). By tail vein injection of pLuci and pWnt7a J82 cells into nude mice, we observed that Wnt7a overexpression cells showed more lung metastatic lesions than pLuci cells (Fig. 6, B–D).
Figure 6.
Wnt7a overexpression promoted UBC cell metastasis in vivo. A, ectopic expression of Wnt7a in J82 cells activated β-catenin pathway and induced MMP1 and MMP10, detected by Western blotting. B, photograph of lung tissues from vehicle control (pLuci) and experimental group (pWnt7a). Arrows indicated the metastatic lesions. C, histological analysis of lung metastatic lesions from pLuci and pWnt7a groups. Scale bar, 1 mm. Below are two enlarged fields from the sections above. Scale bar, 50 μm. D, quantification of lung metastatic nodules in pLuci (n = 4) and pWnt7a (n = 4) groups. *, p < 0.05.
Wnt7a activated canonical β-catenin pathway and induced MMP10 expression
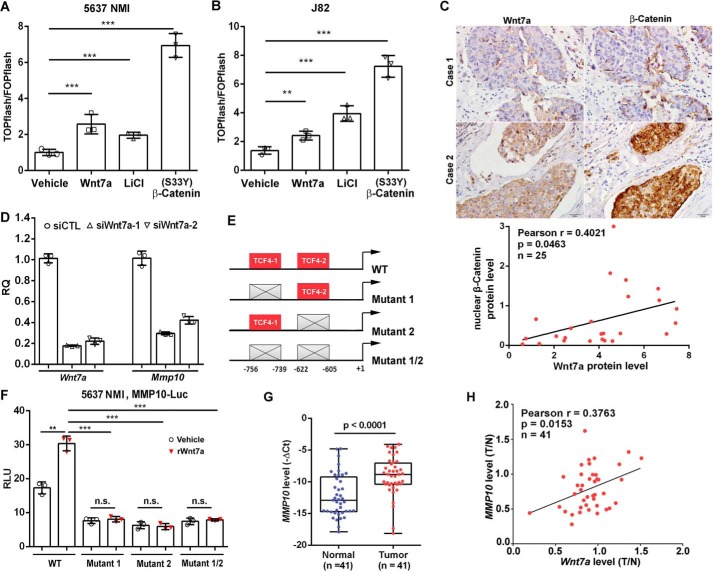
Next, we investigated whether Wnt7a activates the canonical β-catenin pathway in UBC cells. Using an antibody against the active form of β-catenin, we observed that depletion of Wnt7a inactivated β-catenin in both 5637 HMI and T24 cells, whereas rWnt7a treatment and Wnt7a overexpression activated β-catenin (Figs. 4D, 5B, and 6A). Furthermore, we utilized the TOP/FOPflash luciferase system to assess whether Wnt7a can transactivate the β-catenin–TCF complex. TOPflash reporter construct contains repeated TCF/LEF-binding elements, which can be induced by the β-catenin–TCF complex, whereas the FOPflash construct only contains repeated mutant TCF-binding elements, which can serve as a negative control. As shown in Fig. 7, A and B, Wnt7a treatment induced TOPflash activity by 2.5-fold in 5637 NMI and 2.4-fold in J82 cells. LiCl (a selective inhibitor of GSK-3β) and the constitutively activated mutant β-catenin (S33Y-β-catenin) plasmid were used as positive controls, respectively. Moreover, we stained adjacent UBC sections with antibody against total β-catenin. As a result, we found that Wnt7a overexpression was strongly associated with nuclear staining of β-catenin in a cohort of UBC patients (r = 0.4021, p = 0.0463, n = 25; Fig. 7C). In addition, we stained the same section with Wnt7a and β-catenin by immunofluorescence. We confirmed that there was less Wnt7a staining and strong membrane β-catenin staining in the urothelium at the earlier invasive stage (Case 1), whereas there were stronger cytosolic Wnt7a staining and more nuclear staining of β-catenin in the invasive UBC section (Case 2; Fig. S3). Overall, these data supported the notion that Wnt7a overexpression is associated with the activation of the Wnt/β-catenin pathway in UBC samples.
Figure 7.
Wnt7a/β-catenin regulated MMP10 expression in UBC cells. A and B, TOP/FOPflash reporter assay to measure WNT/β-catenin signaling in 5637 NMI (A) and J82 (B) cells with indicated treatment. The treatment with 10 mm LiCl and transfection of (S33Y) β-catenin plasmid served as positive controls, respectively. C, IHC staining showed Wnt7a and β-catenin expression in clinical UBC samples (n = 25). The representative photos of Wnt7a and β-catenin staining data in two cases of UBC samples are presented. Scale bar, 20 μm. The correlation between Wnt7a and β-catenin expression in UBC samples are shown below. D, qRT-PCR assay showed MMP10 and Wnt7a mRNA expression levels in Wnt7a knockdown 5637 HMI cells. E, schematic illustration of the promoter of human MMP10 gene. It showed wildtype (WT) contained two consensus TCF/LEF-binding sites, whereas mutants 1, 2, and 1/2 had indicated the mutations in TCF4-1 or/and TCF4-2 sites. F, luciferase assay showed relative luciferase activities of WT, mutant 1, mutant 2, and mutant 1/2 MMP10 promoters within the rWnt7a protein (100 ng/ml) treatment in 5637 NMI cells for 24 h. All the luciferase activity was calculated in triplicate and repeated three times. **, p < 0.01; ***, p < 0.001; n.s., not significant. G, relative expression level of MMP10 in a cohort of human UBCs and adjacent normal bladder tissues (n = 41). H, association of Wnt7a and MMP10 expression levels in a cohort of human UBC samples (n = 41).
To dissect potential downstream targets of Wnt7a/β-catenin pathway, we examined a matrix metalloproteinase MMP10, which is involved in degradation of extracellular matrix (13). At first, knockdown of MMP10 by two different siRNAs significantly reduced cell invasion of 5637 HMI cells (Fig. S4), which indicated that MMP10 is involved in UBC cell invasion. Next, depletion of Wnt7a in 5637 HMI cells remarkably reduced MMP10 at the mRNA level (Fig. 7D). To examine whether Wnt7a directly regulates MMP10 through β-catenin, bioinformatics analysis of its promoter was performed. The MatInspector program (genomatrix.de) revealed there were two putative TCF/LEF sites at −756 (site 1) and −622 (site 2), with matrix similarity scores of 0.885 and 0.981, respectively (Fig. 7E and Table S2). Wnt7a treatment transactivated the WT reporter of the MMP10 gene (WT MMP10-Luc). Mutated reporters of the MMP10 gene on two TCF/LCF sites (TCF4–1 and TCF4–2) were also generated, with mutations on TCF4-1 alone (mutant 1), TCF4-2 alone (mutant 2), and both (mutant 1/2), respectively. Three mutant reporters could not respond to Wnt7a stimulation in 5637 NMI cells (Fig. 7F). In addition, the MMP10 mRNA level was significantly increased in UBC tissues compared with normal tissues (n = 41, p < 0.0001; Fig. 7G), and the MMP10 mRNA level was also positively correlated with that of Wnt7a (r = 0.3763, p = 0.0153, n = 41; Fig. 7H). These data provided the first molecular evidence that Wnt7a induces MMP10 expression through canonical Wnt/β-catenin signaling in UBC cells.
miR-370-3p negatively regulated UBC cell invasion through repressing Wnt7a
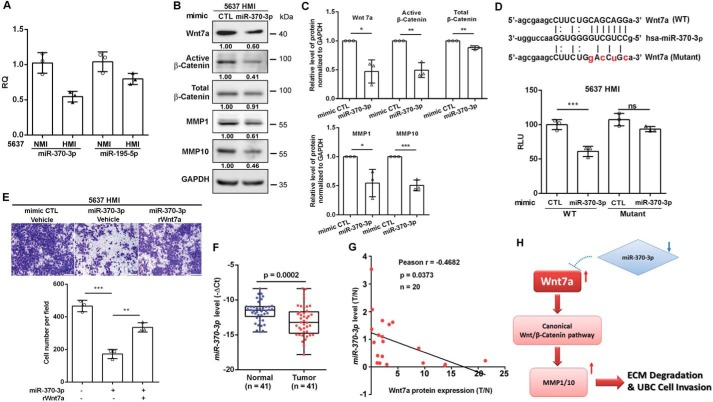
miRNAs have been implicated as key regulators for cancer cell invasion and metastasis, and they have been widely used for diagnostic and prognostic biomarkers (21). We thus utilized the miRanda algorithm and identified 12 putative miRNAs targeting Wnt7a (Fig. S5A). The expression levels of those miRNAs in UBC patients from TCGA provisional databases were analyzed. Two miRNAs (miR-370-3p and miR-195-5p) were identified to be down-regulated in UBC samples with lymph nodes invasion (N1; n = 128) compared with normal bladder samples (normal, n = 19, p < 0.05). Notably, statistically significant down-regulation of miR-195-5p, but not miR-370-3p, was detected in UBC samples without lymph node metastasis (N0; n = 236) compared with normal bladder samples (normal, n = 19, p < 0.05) (Fig. S5, B and C). The expression levels of miR-370-3p and miR-195-5p were also evaluated in 5637 HMI and 5637 NMI cells. Expression level of miR-370-3p and miR-195-5p was 50% and about 20% lower in 5637 HMI cells than that in 5637 NMI cells, respectively (Fig. 8A). These data suggest that miR-370 might play an important role in UBC invasion and metastasis, but not in tumorigenesis. Therefore, we focused on miR-370-3p for further study.
Figure 8.
miR-370-3p reduced Wnt7a to regulate UBC cell invasion. A, qRT-PCR showed the expression levels of two miRNAs: miR-370-3p and miR-195-5p in 5637 NMI and HMI cells. B, Western blotting showed protein expression of Wnt7a, active β-catenin, total β-catenin, MMP1, and MMP10 in 5637 HMI cells transfected with mimic miR-370-3p and control mimic. C, quantification of Western blotting is shown. D, schematic representation of the putative miR-370-3p-binding site in WT Wnt7a 3′UTR (WT). Wnt7a (WT) and Wnt7a (mutant) indicated the psiCHECK2 plasmid containing the Wnt7a 3′UTR WT sequence and mutated sequence. HMI 5637 cells were co-transfected with reporter vector (WT or mutant Wnt7a 3′UTR) and mimic miR-370-3p or control (CTL) as indicated. The relative luciferase activity of WT and mutant Wnt7a 3′UTR was measured 24 h after transfection. E, transwell assay showed invasive capacities of 5637 HMI cells with indicated treatment. The representative fields of invaded cells 16 h after seeding are shown. Scale bar, 100 μm. The graphs indicated the counting number of invaded cells in 5637 HMI cells with the indicated treatment. All assays were performed in three independent biological replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. F, relative expression level of miR-370-3p in a cohort of human UBC samples (n = 41). G, negative association of Wnt7a protein level and miR-370-3p level in the same human UBC samples (n = 20). H, hypothetical working model of the miR-370-3p/Wnt7a axis activating canonical Wnt/β-catenin pathway and inducing matrix metalloproteinases (MMPs) to promote UBC cell invasion.
Transient transfection of the miR-370-3p mimic in 5637 HMI cells remarkably reduced Wnt7a, subsequently inactivated β-catenin, and reduced its downstream targets MMP1 (40) and MMP10. We also observed a small but significant reduction of total β-catenin in miR-370-3p-treated 5637 HMI cells (Fig. 8, B and C). To determine whether Wnt7a is a direct downstream target of miR-370-3p, the 3′UTR of Wnt7a transcript containing the putative miR-370-3p-binding site was inserted downstream of the firefly luciferase reporter gene.
Mutant Wnt7a reporter with the point mutations on the putative binding site of miR-370-3p was also generated (Fig. 8D). After the miR-370-3p mimic was introduced into 5637 HMI cells, we observed about 50% reduction of luciferase activity compared with cells transfected with the mimic control; however, miR-370-3p mimic did not show a suppressive effect on the mutant Wnt7a reporter (Fig. 8D). Forced expression of miR-370-3p reduced the invasive capability of 5637 HMI cells, whereas addition of Wnt7a abrogated miR-370-3p-mediated suppression of the invasion phenotype (Fig. 8E).
In addition, we found that miR-370-3p was significantly reduced in 41 UBC samples from our cohort (Fig. 8F). As shown in Fig. 8G, we detected the negative correlation of the miR-370-3p level and the Wnt7a protein level in UBC samples (n = 20; r = −0.4682, p = 0.0373). Overall, these data indicate that miR-370-3p suppresses UBC cell invasion at least partially by down-regulating the Wnt7a protein level.
Discussion
Because the gain of invasive capacity increases aggressiveness and cancer mortality of UBC, the identification of invasive biomarkers and the understanding of its molecular mechanism will be of great importance. Elevated nuclear β-catenin in clinical samples and transgenic mouse models revealed the significance of Wnt/β-catenin in UBC development (8, 23). However, few somatic mutations of components in the Wnt pathway has been detected in UBCs, though APC and Axin1 in Wnt pathway are frequently mutated in gastrointestinal cancers (24, 25). Epigenetic silencing of Wnt inhibitors, such as WIF1 and SFRPs, is associated with aberrant Wnt/β-catenin pathway in UBCs (26, 27); so far, we have not known which ligand may activate Wnt pathway to promote UBC cell invasion and metastasis. In this study, we successfully established the UBC cell lines with high invasion (5637 HMI) and low invasion (5637 NMI) from parental 5637 cells. The proteomic analysis by mass spectrum unbiasedly revealed that Wnt7a was one of the proteins up-regulated in 5637 HMI cells, compared with 5637 NMI cells. Knockdown of Wnt7a, rWnt7a protein treatment, and ectopic expression of Wnt7a further demonstrated that Wnt7a was necessary and sufficient for UBC cell invasion and metastasis. We confirmed that Wnt7a overexpression predicted poor clinical outcome in UBC patients, suggesting that Wnt7a may function as a promising prognostic biomarker.
Wnt7a can activate the canonical Wnt pathway through β-catenin, as well as the noncanonical Wnt pathway, which is cell context-dependent (13, 14). To dissect the potential mechanism, our study revealed that Wnt7a activated the canonical Wnt/β-catenin pathway, which was based on several pieces of evidence below. First, Wnt7a activated β-catenin and resulted in the increased level of its downstream target genes involved in EMT and ECM degradation, whereas Wnt7a deficiency inactivated the Wnt/β-catenin pathway. Second, we confirmed that Wnt7a transactivated TOPflash reporter in UBC cells. Third, we first proved that Wnt7a regulated MMP10 expression through two TCF/LEF elements in the MMP10 promoter. Fourth, we observed the positive correlation between Wnt7a and nuclear β-catenin in UBC samples. Given that the activation of the Wnt/β-catenin pathway has been shown to play a role in the maintenance of bladder epithelium stem cell and tumor-initiating cells (28, 29), we speculate that Wnt7a overexpression may also be essential for UBC aggressiveness through the Wnt/β-catenin pathway. In fact, Wnt7a overexpression was reported to induce the canonical Wnt pathway for drug resistance of ovarian and pancreatic cancer cells (30, 31). Overall, our data indicate that Wnt7a activates the Wnt/β-catenin signaling pathway and promotes UBC cell invasion.
The role of Wnt7a in cancer development is cancer type-dependent. The overexpression of Wnt7a was identified in ovarian cancer and endometrial cancer (32), where it could induce cancer cell proliferation and promote cancer progression by activating the canonical Wnt pathway or remodeling of the tumor microenvironment (16, 33–35). On the contrary, in lung cancer, the clear cell renal cell carcinoma, and sporadic malignant pancreatic endocrine tumors, Wnt7a was frequently down-regulated through promoter methylation (36–40). The interaction of Wnt7a with Fzd-9 in lung cancer cells induced Sprouty-4 expression and the JNK pathway but not β-catenin/TCF activity (40). In addition, Wnt7a and Fzd-9 coordinately induced peroxisome proliferator-activated receptor γ activation through ERK5 (17). Furthermore, Wnt7a deficiency significantly increased carcinogen-induced lung cancer incidence in vivo through bypassing cellular senescence via the activation of S-phase kinase-associated protein 2 (41). Consistently, such a tumor-suppressive effect of Wnt7a was not dependent on the canonical Wnt-signaling pathway. Taken together, Wnt7a works through the canonical Wnt pathway to promote cancer aggressiveness in ovarian cancer and UBC, although it suppresses lung cancer progression by inducing the noncanonical Wnt pathway.
Our proteomic data from mass spectrum analysis identified the up-regulation of Wnt7a and another two MMP family members, MMP1 and MMP10, in 5637 UBC cells with highly invasive capability. Herein, we first proved that Wnt7a activated the canonical Wnt pathway through direct regulation of the MMP10 gene. Because MMP1 is also another direct target of the canonical Wnt pathway (22), Wnt7a overexpression up-regulated MMP1/10 to degrade the extracellular matrix and to facilitate UBC cell invasion. Besides, MMP1/10 overexpression may also remodel the microenvironment by promoting angiogenesis and infiltration/activation of macrophages (42–44). This evidence reinforces the notion that Wnt7a overexpression may confer invasive capability on UBC cells.
Dysregulation of miRNAs has been reported in various cancer types. Here, we identified that miR-370-3p was the most markedly down-regulated miRNAs in 5637 HMI cells, compared with 5637 NMI cells, as long as down-regulated in UBC samples. miR-370-3p is localized in chromosome 14q32, which is frequently lost in UBC patients (45). Down-regulation of miR-370-5p, which is transcribed under the same promoter with miR-370-3p, has been reported in UBCs (46). Nevertheless, the role of miR-370-3p in UBC patients has not been dissected previously. The reduction of miR-370-3p was more than other Wnt7a-targeting miRNAs in 5637 HMI cells. Re-introduction of miR-370-3p inhibited Wnt7a expression and suppressed 5637 HMI cell invasion, which could be rescued by the addition of rWnt7a. Because miRNA usually has multiple targets, we also investigated whether miR-370-3p can directly inhibit Wnt/β-catenin target genes in our study. From the protein list of mass spectrum data, we did not find evidence that miR-370-3p can target MMP1 and MMP10 by using the miRanda algorithm. It convinced us that it was Wnt7a that was down-regulated by miR-370-3p, leading to the up-regulation of β-catenin target genes, but not the direct regulation of miR-370-3p on these two target genes. In contrast, a recent study revealed miR-370-3p also directly targeted β-catenin in glioma cells (47). However, in our UBC cell model, we did not find significant changes of β-catenin in 5637 NMI and HMI cells and only half-reduction of miR-370-3p in 5637 HMI compared with NMI cells. Although transient transfection of miR-370-3p cells mimic slightly reduced total β-catenin levels in 5637 HMI cells, it induced more striking and significant reduction of Wnt7a, arguing that in UBC cells Wnt7a is the major target of miR-370-3p. Overall, these data suggested that miR-370-3p down-regulation may be essential for UBC cell invasion through the Wnt/β-catenin pathway.
In summary, we first identified that Wnt7a overexpression is significantly associated with UBC cell invasiveness and predicts poor clinical outcome in UBC patients. Wnt7a activates canonical Wnt pathway to induce EMT and the expression of MMP1 and MMP10. We also proved that Wnt7a directly regulates MMP10 through two TCF/LEF-binding elements in its promoter. In addition, down-regulation of miR-370-3p in invasive UBC cells accounts for Wnt7a overexpression and β-catenin/Wnt activation (Fig. 8H). Overall, our study indicated that Wnt7a and its associated Wnt/β-catenin pathway are promising drug targets for UBC cell invasion and metastasis.
Experimental procedures
Cell lines, stable cell line establishment, and clinical tissue samples
All human UBC cell lines (5637, T24, UMUC-3, and J82) were obtained from Cell Bank of Type Culture Collection, Chinese Academy of Sciences (Shanghai, China). All cancer cell lines were maintained in RPMI 1640 medium (Life Technologies, Inc.) containing 10% fetal bovine serum (FBS; FBS-12A, Capricorn Scientific, Ebsdorfergrund, Germany) at 37 °C in the humidified incubator containing 5% CO2. To establish low-invasive (5637 NMI) and high-invasive (5637 HMI) sublines, parental 5637 cells were sorted using six-well polycarbonate Transwell filters with 8-μm pore (catalog no. 3428, Corning, Corning, NY). After 24 h starvation in RPMI 1640 medium without FBS, 1 ml of cell suspension without FBS was seeded into the Matrigel (BD Biosciences)-coated top chamber, and the lower chamber was prepared with 1 ml of RPMI 1640 medium with 10% FBS as a chemoattractant. The noninvasive and invasive cells on the top and underside of the membrane were harvested following 24 and 14 h of incubation, respectively. After 12 rounds of repeated Transwell selection, the noninvaded and invaded cells were obtained with designation of 5637 NMI and 5637 HMI cells. For knockdown assay, 20 nm siRNAs, including two different siRNAs targeting Wnt7a and control siRNA (siCTL), were transfected into cells with Lipofectamine 2000 (Life Technologies, Inc.), according to the manufacturer's instruction. The sequences of siRNAs are listed in Table S3. For treatment with recombinant human Wnt7a, cells were cultured without FBS for 24 h, and then 100 ng/ml Wnt7a (catalog no. 120-31, PeproTech, Rocky Hill, NJ) was added to RPMI 1640 medium without FBS for another 24 h. Plasmid pCDH-3xFLAG-human Wnt7a-T2A-puromycin and empty vector were co-transfected with pVSVG and Gag-Pol into 293FT cells, respectively. The supernatants containing viral particles were harvested and infected J82 cells for 3 days, followed by 1 μg/ml puromycin selection for 7 days.
All fresh surgically removed paired cancer specimens and adjacent nontumorous bladder tissues from UBC patients were obtained from Drum Tower Hospital, affiliated with Nanjing University, and Shanghai General Hospital, affiliated with Shanghai Jiaotong University. The studies were approved by the review board/Ethics Committee of Nanjing Drum Tower Hospital and the review board of Shanghai General Hospital, and informed consent for research use was obtained from each patient. The studies abide by the Declaration of Helsinki principles. All specimens were collected within 24 h after resection, snap-frozen in liquid nitrogen, and confirmed by histological analysis. The clinicopathological parameters of UBC samples used for qRT-PCR and IHC staining were listed in Tables S4 and S5.
Mass spectrum analysis
Cell lysis preparation, mass spectrum assay, and bioinformatics analyses were described in details in the supporting information. The MS proteomics data have been deposited under the title “Proteomic analysis of the invasiveness of human bladder cancer cells” to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier (Project accession no. PXD007709).
RNA isolation and real-time qRT-PCR
Total RNA from the frozen tissues and cells were isolated with TRIzol reagent (catalog no. 15596018, Life Technologies, Inc.). cDNAs were synthesized by the Goldenstar RT6 cDNA synthesis kit (TSK301, TsingKe Biological Technology, Beijing, China). qRT-PCR was performed by SYBR Green (high ROX) MIX kit (catalog no. DRR041A, TaKaRa, Dalian, China), according to the manufacturer's instruction. Each reaction was performed in triplicate. Data analysis was performed using the ΔΔCt method. Fold change was determined in relative quantification units using ACTB for normalization. Primers are listed in Table S3.
Western blotting
Total protein in cells or tissues was extracted by RIPA buffer containing phosphatase inhibitor tablet (PhosStop, Roche Applied Science, and protease inhibitor tablet (cOmplete Roche Applied Science)). 20 μg of protein was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 5% nonfat milk or BSA. Then the membranes were incubated overnight at 4 °C with the primary antibodies (Table S6). The membranes were incubated by the corresponding secondary antibodies for 2 h at room temperature, visualized by Tanon High-sig ECL Western blotting substrate (catalog no. 180-501, Tanon, Shanghai, China) and detected by Amersham Biosciences Imager 600 (GE Healthcare). The intensities of bands were qualified by Image J software and calculated by normalization with the internal control (GAPDH).
Transwell invasion assays
Invasive capacity of T24, 5637, and J82 cells was evaluated using 8-μm Transwell filters (Corning, catalog no. 3422). 1 × 105 cells were resuspended in 100 μl of serum-free medium and then seeded in the upper chamber. The lower chamber was filled with 500 μl of medium containing 10% (v/v) FBS (Capricorn, FBS-12A). After incubation for 12 h (T24 cells), 16 h (5637 cells), or 20 h (J82 cells), the cells that had invaded onto the lower surface of the filter were fixed with 4% (v/v) formaldehyde solution for 8 min and then stained with crystal violet staining solution (Sangon Biotech, Shanghai, China). Images of stained cells were captured with a Leica DM IL LED inverted microscope (Leica Microsystems, Wetzlar, Germany).
Wound healing assay
Briefly, 5 × 106 cells were seeded into 6-well plates per well, and wounds were made with a 100-μl pipette tip on the cell monolayer 1 day later. Images of the scratch were taken at the indicated time points to evaluate the wound closure.
3D spheroid Matrigel invasion assay
5637 cells (1 × 105) were resuspended in an ultra-low 6-well plate (Corning) to allow the formation of spheroids. Afterward, spheroids were harvested and embedded in Matrigel, which was 1:1 diluted with medium. Medium was added to the top of polymerized Matrigel, and photographs were taken at the indicated time points. ImageJ software was used to measure the area occupied by cells.
Experimental metastatic mouse model
J82 cells with Wnt7a overexpression (pWnt7a) and control (pLuci) cells (5 × 104) were injected into the tail vein of 6-week-old male nude mice using sterile 28-gauge needles, respectively. All mice were sacrificed 24 days later to collect lung tissues. Total metastatic foci were counted in hematoxylin and eosin-stained lung sections under a microscope. All the experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the review board of the MARC at Nanjing University.
IHC staining
Formalin-fixed, paraffin-embedded specimens from UBC patients were from Drum Tower Hospital, affiliated with Nanjing University. 5-μm-thick paraffin sections were deparaffinized for antigen retrieval using Tris/EDTA (pH 9.0), followed by the incubation with primary antibody against Wnt7a (Abcam, ab100792) and β-catenin (BD Biosciences, 610153), respectively. The slides were stained with the DAB kit (Maixin Bio, DAB-0031) and counterstained by hematoxylin. Slides were scored by two pathologists to estimate percentage and intensity of stained signaling in carcinoma cells, as described previously (48). The proportion score and intensity score were multiplied to get the final score.
Luciferase activity assay
The luciferase activity was detected using a dual-luciferase reporter assay kit (E1960, Promega, Madison, WI), according to the manufacturer's instructions. TOP/FOPflash and four different MMP10 promoter reporter activities from firefly luciferase were normalized with transfection efficiency control reporter activity from Renilla luciferase. Among these, firefly luciferase-tagged TOPflash reporter, FOPflash reporter, and active β-catenin (S33Y–β-catenin) expression plasmid were kindly provided by Dr. Ying Cao at Nanjing University. The luciferase reporter vectors carrying the promoter of WT, mutant 1, mutant 2, and mutant 1/2 MMP10 were cloned into a pGL3-basic vector by ClonExpress II One-Step Cloning kit (C112 −01/02, Vazyme, Nanjing, China) to compare the relative luciferase activity from putative TCF/LEF-binding sites containing promoter regions from nucleotides −756, −739, −622, and −605 to +1, respectively. A 296-bp fragment of widetype and the mutant 3′UTR of Wnt7a (starting from 1,392 to 1,673 bp of NM_004625.3), which contains the predicted miR-370-3p-binding sites, were synthesized by General Biosystems (Chuzhou, Anhui, China). These two fragments then were cloned into psiCHECK2 vector (Promega) at XhoI and NotI sites. Cell lysates were harvested to measure firefly and Renilla luciferase activities, and the value was calculated by Renilla luciferase value normalization to firefly luciferase value.
Statistical analysis
Statistical analysis was performed by GraphPad Prisms 6.0 software. Data were presented as means ± S.D. with at least three biological independent experiments. Statistical significance was assessed by Student's t test to compare the means of two groups. Pearson correlation test was used to determine the correlation of different gene expressions. Survival was analyzed by the log-rank (Mantel-Cox) test. p value less than 0.05 was considered statistically significant.
Author contributions
X. H., H. Zhu, Z. G., J. L., Y. D., M. L., and J. Y. data curation; X. H., H. Zhu, and J. Z. formal analysis; X. H., Z. G., and J. Z. validation; X. H., H. Zhu, J. L., and H. Zhou methodology; X. H., H. Zhu, Z. G., J. L., H. G., R. H., and J. Y. writing-original draft; H. Zhu, Z. G., J. L., J. Z., B. S., H. Zhou, H. G., and R. H. resources; H. Zhu, Z. G., J. L., and H. Zhou software; H. Zhu, Y. D., B. S., M. L., H. Zhou, R. H., and J. Y. investigation; H. G., R. H., and J. Y. supervision; H. G., R. H., and J. Y. funding acquisition; R. H. and J. Y. conceptualization; R. H. and J. Y. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Ying Cao (Nanjing University), Dr. Wen Liu and Yichen Liu (Xiamen University) for reagents and bioinformatics supports. We also appreciate the assistance of staff from the Institutional Technology Service Center of Shanghai Institute of Materia Medica for their technical support.
This work was supported in part by National Natural Science Foundation of China Grants 81372168, 81572519, 81672873, and 81771890; Open Foundation of State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University Grant KF-GN-201602, and State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences Grant SIMM1705KF-02 and SIMM1705KF-06. The authors declare that they have no conflicts of interest with the contents of this article.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD007709.
This article contains Figs. S1–S5 and Tables S1–S6.
- UBC
- urinary bladder cancer
- EMT
- epithelial-to-mesenchymal transition
- EMC
- extracellular matrix
- FBS
- fetal bovine serum
- qRT-PCR
- quantitative RT-PCR
- IHC
- immunohistochemistry.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2017) Cancer statistics. CA Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2. Wu X. R. (2005) Urothelial tumorigenesis: a tale of divergent pathways. Nat. Rev. Cancer 5, 713–725 10.1038/nrc1697 [DOI] [PubMed] [Google Scholar]
- 3. Nusse R., and Clevers H. (2017) Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 4. Niehrs C. (2012) The complex world of WNT receptor signaling. Nat. Rev. Mol. Cell Biol. 13, 767–779 10.1038/nrm3470 [DOI] [PubMed] [Google Scholar]
- 5. Klaus A., and Birchmeier W. (2008) Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 10.1038/nrc2389 [DOI] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research Network. (2014) Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierzynski J. A., Hildebrandt M. A., Kamat A. M., Lin J., Ye Y., Dinney C. P., and Wu X. (2015) Genetic variants in the Wnt/β-Catenin signaling pathway as indicators of bladder cancer risk. J. Urol. 194, 1771–1776 10.1016/j.juro.2015.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmad I., Morton J. P., Singh L. B., Radulescu S. M., Ridgway R. A., Patel S., Woodgett J., Winton D. J., Taketo M. M., Wu X. R., Leung H. Y., and Sansom O. J. (2011) β-Catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene 30, 178–189 10.1038/onc.2010.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasquinelli A. E. (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- 10. Han Y., Chen J., Zhao X., Liang C., Wang Y., Sun L., Jiang Z., Zhang Z., Yang R., Chen J., Li Z., Tang A., Li X., Ye. J., Guan Z., Gui Y., and Cai Z. (2011) MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS ONE 6, e18286 10.1371/journal.pone.0018286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q., Zhao W., Ye C., Zhuang J., Chang C., Li Y., Huang X., Shen L., Li Y., Cui Y., Song J., Shen B., Eliaz I., Huang R., Ying H., Guo H., and Yan J. (2015) Honokiol inhibits bladder tumor growth by suppressing EZH2/miR-143 axis. Oncotarget 6, 37335–37348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhuang J., Shen L., Yang L., Huang X., Lu Q., Cui Y., Zheng X., Zhao X., Zhang D., Huang R., Guo H., and Yan J. (2017) TGFβ1 promotes gemcitabine resistance through regulating the lncRNA-LET/NF90/miR-145 signaling axis in bladder cancer. Theranostics 7, 3053–3067 10.7150/thno.19542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshioka S., King M. L., Ran S., Okuda H., MacLean J. A. 2nd., McAsey M. E., Sugino N., Brard L., Watabe K., and Hayashi K. (2012) WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/β-catenin pathway. Mol. Cancer Res. 10, 469–482 10.1158/1541-7786.MCR-11-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winn R. A., Van Scoyk M., Hammond M., Rodriguez K., Crossno J. T. Jr., Heasley L. E., and Nemenoff R. A. (2006) Antitumorigenic effect of Wnt 7a and Fzd 9 in non-small cell lung cancer cells is mediated through ERK-5-dependent activation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 281, 26943–26950 10.1074/jbc.M604145200 [DOI] [PubMed] [Google Scholar]
- 15. Shi X., Chen Z., Hu X., Luo M., Sun Z., Li J., Shi S., Feng X., Zhou C., Li Z., Yang W., Li Y., Wang P., Zhou F., Gao Y., and He J. (2016) AJUBA promotes the migration and invasion of esophageal squamous cell carcinoma cells through upregulation of MMP10 and MMP13 expression. Oncotarget 7, 36407–36418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim M. Y., Oskarsson T., Acharyya S., Nguyen D. X., Zhang X. H., Norton L., and Massagué J. (2009) Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 10.1016/j.cell.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim S. Y., Yuzhalin A. E., Gordon-Weeks A. N., and Muschel R. J. (2016) Tumor-infiltrating monocytes /macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene 35, 5735–5745 10.1038/onc.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon R., Richter J., Wagner U., Fijan A., Bruderer J., Schmid U., Ackermann D., Maurer R., Alund G., Knönagel H., Rist M., Wilber K., Anabitarte M., Hering F., Hardmeier T., et al. (2001) High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 61, 4514–4519 [PubMed] [Google Scholar]
- 19. Howe L. R., Watanabe O., Leonard J., and Brown A. M. (2003) Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 63, 1906–1913 [PubMed] [Google Scholar]
- 20. Sánchez-Tilló E., de Barrios O., Siles L., Cuatrecasas M., Castells A., and Postigo A. (2011) β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. U.S.A. 108, 19204–19209 10.1073/pnas.1108977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palma Flores C., García-Vázquez R., Gallardo Rincón D., Ruiz-García E., Astudillo de la Vega H., Marchat L. A., Salinas Vera Y. M., and López-Camarillo C. (2017) MicroRNAs driving invasion and metastasis in ovarian cancer: Opportunities for translational medicine (Review). Int. J. Oncol. 50, 1461–1476 10.3892/ijo.2017.3948 [DOI] [PubMed] [Google Scholar]
- 22. Jean C., Blanc A., Prade-Houdellier N., Ysebaert L., Hernandez-Pigeon H., Al Saati T., Haure M. J., Coluccia A. M., Charveron M., Delabesse E., and Laurent G. (2009) Epidermal growth factor receptor/beta-catenin/T-cell factor 4/matrix metalloproteinase 1: a new pathway for regulating keratinocyte invasiveness after UVA irradiation. Cancer Res. 69, 3291–3299 10.1158/0008-5472.CAN-08-1909 [DOI] [PubMed] [Google Scholar]
- 23. Ahmad I., Patel R., Liu Y., Singh L. B., Taketo M. M., Wu X. R., Leung H. Y., and Sansom O. J. (2011) Ras mutation cooperates with β-catenin activation to drive bladder tumourigenesis. Cell Death Dis. 2, e124 10.1038/cddis.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoehr R., Krieg R. C., Knuechel R., Hofstaedter F., Pilarsky C., Zaak D., Schmitt R., and Hartmann A. (2002) No evidence for involvement of β-catenin and APC in urothelial carcinomas. Int. J. Oncol. 20, 905–911 [PubMed] [Google Scholar]
- 25. Mazzoni S. M., and Fearon E. R. (2014) AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 355, 1–8 10.1016/j.canlet.2014.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urakami S., Shiina H., Enokida H., Kawakami T., Tokizane T., Ogishima T., Tanaka Y., Li L. C., Ribeiro-Filho L. A., Terashima M., Kikuno N., Adachi H., Yoneda T., Kishi H., Shigeno K., et al. (2006) Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin. Cancer Res. 12, 383–391 10.1158/1078-0432.CCR-05-1344 [DOI] [PubMed] [Google Scholar]
- 27. Marsit C. J., Karagas M. R., Andrew A., Liu M., Danaee H., Schned A. R., Nelson H. H., and Kelsey K. T. (2005) Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 65, 7081–7085 10.1158/0008-5472.CAN-05-0267 [DOI] [PubMed] [Google Scholar]
- 28. Shin K., Lee J., Guo N., Kim J., Lim A., Qu L., Mysorekar I. U., and Beachy P. A. (2011) Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 472, 110–114 10.1038/nature09851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan K. S., Espinosa I., Chao M., Wong D., Ailles L., Diehn M., Gill H., Presti J. Jr, Chang H. Y., van de Rijn M., Shortliffe L., and Weissman I. L. (2009) Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14016–14021 10.1073/pnas.0906549106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. King M. L., Lindberg M. E., Stodden G. R., Okuda H., Ebers S. D., Johnson A., Montag A., Lengyel E., MacLean Ii J. A., and Hayashi K. (2015) WNT7A/β-catenin signaling induces FGF1 and influences sensitivity to niclosamide in ovarian cancer. Oncogene 34, 3452–3462 10.1038/onc.2014.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jung D. B., Yun M., Kim E. O., Kim J., Kim B., Jung J. H., Wang E., Mukhopadhyay D., Hammond E., Dredge K., Shridhar V., and Kim S. H. (2015) The heparan sulfate mimetic PG545 interferes with Wnt/β-catenin signaling and significantly suppresses pancreatic tumorigenesis alone and in combination with gemcitabine. Oncotarget 6, 4992–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y., Meng F., Xu Y., Yang S., Xiao M., Chen X., and Lou G. (2013) Overexpression of Wnt7a is associated with tumor progression and unfavorable prognosis in endometrial cancer. Int. J. Gynecol. Cancer 23, 304–311 10.1097/IGC.0b013e31827c7708 [DOI] [PubMed] [Google Scholar]
- 33. Zhao L., Ji G., Le X., Luo Z., Wang C., Feng M., Xu L., Zhang Y., Lau W. B., Lau B., Yang Y., Lei L., Yang H., Xuan Y., Chen Y., et al. (2017) An integrated analysis identifies STAT4 as a key regulator of ovarian cancer metastasis. Oncogene 36, 3384–3396 10.1038/onc.2016.487 [DOI] [PubMed] [Google Scholar]
- 34. Avgustinova A., Iravani M., Robertson D., Fearns A., Gao Q., Klingbeil P., Hanby A. M., Speirs V., Sahai E., Calvo F., and Isacke C. M. (2016) Tumour cell-derived Wnt7a recruits and activates fibroblasts to promote tumour aggressiveness. Nat. Commun. 7, 10305 10.1038/ncomms10305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sundqvist A., Morikawa M., Ren J., Vasilaki E., Kawasaki N., Kobayashi M., Koinuma D., Aburatani H., Miyazono K., Heldin C. H., van Dam H., and Ten Dijke P. (2018) JUNB governs a feed-forward network of TGFβ signaling that aggravates breast cancer invasion. Nucleic Acids Res. 46, 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Calvo R., West J., Franklin W., Erickson P., Bemis L., Li E., Helfrich B., Bunn P., Roche J., Brambilla E., Rosell R., Gemmill R. M., and Drabkin H. A. (2000) Altered HOX and WNT7A expression in human lung cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 12776–12781 10.1073/pnas.97.23.12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tennis M. A., Vanscoyk M. M., Wilson L. A., Kelley N., and Winn R. A. (2012) Methylation of Wnt7a is modulated by DNMT1 and cigarette smoke condensate in non-small cell lung cancer. PLoS ONE 7, e32921 10.1371/journal.pone.0032921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kondratov A. G., Kvasha S. M., Stoliar L. A., Romanenko A. M., Zgonnyk Y. M., Gordiyuk V. V., Kashuba E. V., Rynditch A. V., Zabarovsky E. R., and Kashuba V. I. (2012) Alterations of the WNT7A gene in clear cell renal cell carcinomas. PLoS ONE 7, e47012 10.1371/journal.pone.0047012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindberg D., Akerström G., and Westin G. (2007) Mutational analyses of WNT7A and HDAC11 as candidate tumour suppressor genes in sporadic malignant pancreatic endocrine tumours. Clin. Endocrinol. 66, 110–114 [DOI] [PubMed] [Google Scholar]
- 40. Winn R. A., Marek L., Han S. Y., Rodriguez K., Rodriguez N., Hammond M., Van Scoyk M., Acosta H., Mirus J., Barry N., Bren-Mattison Y., Van Raay T. J., Nemenoff R. A., and Heasley L. E. (2005) Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J. Biol. Chem. 280, 19625–19634 10.1074/jbc.M409392200 [DOI] [PubMed] [Google Scholar]
- 41. Bikkavilli R. K., Avasarala S., Van Scoyk M., Arcaroli J., Brzezinski C., Zhang W., Edwards M. G., Rathinam M. K., Zhou T., Tauler J., Borowicz S., Lussier Y. A., Parr B. A., Cool C. D., and Winn R. A. (2015) Wnt7a is a novel inducer of β-catenin-independent tumor-suppressive cellular senescence in lung cancer. Oncogene 34, 5317–5328 10.1038/onc.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saunders W., Bayless K. J., and Davis G. E. (2005) MMP-1 activation by serine proteases and MMP-10 induces human capillary tubular network collapse and regression in 3D collagen matrices. J. Cell Sci. 118, 2325–2340 10.1242/jcs.02360 [DOI] [PubMed] [Google Scholar]
- 43. Rohani M. G., McMahan R. S., Razumova M. V., Hertz A. L., Cieslewicz M., Pun S. H., Regnier M., Wang Y., Birkland T. P., and Parks W. C. (2015) MMP-10 regulates collagenolytic activity of alternatively activated resident macrophages. J. Invest. Dermatol. 135, 2377–2384 10.1038/jid.2015.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steenport M., Khan K. M., Du B., Barnhard S. E., Dannenberg A. J., and Falcone D. J. (2009) Matrix metalloproteinase (MMP)-1 and MMP-3 induce macrophage MMP-9: evidence for the role of TNF-alpha and cyclooxygenase-2. J. Immunol. 183, 8119–8127 10.4049/jimmunol.0901925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panani A. D., Ferti A. D., Raptis S. A., and Roussos C. (2004) Novel recurrent structural chromosomal aberrations in primary bladder cancer. Anticancer Res. 24, 2967–2974 [PubMed] [Google Scholar]
- 46. Wang C., Chen Z., Ge Q., Hu J., Li F., Hu J., Xu H., Ye Z., and Li L. C. (2014) Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett. 588, 4654–4664 10.1016/j.febslet.2014.10.037 [DOI] [PubMed] [Google Scholar]
- 47. Peng Z., Wu T., Li Y., Xu Z., Zhang S., Liu B., Chen Q., and Tian D. (2016) MicroRNA-370–3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting β-catenin. Brain Res. 1644, 53–61 10.1016/j.brainres.2016.04.066 [DOI] [PubMed] [Google Scholar]
- 48. Zhao W., Chang C., Cui Y., Zhao X., Yang J., Shen L., Zhou J., Hou Z., Zhang Z., Ye C., Hasenmayer D., Perkins R., Huang X., Yao X., Yu L., Huang R., Zhang D., Guo H., and Yan J. (2014) Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible factor 1α. J. Biol. Chem. 289, 11219–11229 10.1074/jbc.M113.535989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.