Abstract
Inositol pyrophosphates (PP-InsPs) are “energetic” intracellular signals that are ubiquitous in animals, plants, and fungi; structural and biochemical characterization of PP-InsP metabolic enzymes provides insight into their evolution, reaction mechanisms, and regulation. Here, we describe the 2.35-Å-resolution structure of the catalytic core of Siw14, a 5-PP-InsP phosphatase from Saccharomyces cerevisiae and a member of the protein tyrosine-phosphatase (PTP) superfamily. Conclusions that we derive from structural data are supported by extensive site-directed mutagenesis and kinetic analyses, thereby attributing new functional significance to several key residues. We demonstrate the high activity and exquisite specificity of Siw14 for the 5-diphosphate group of PP-InsPs. The three structural elements that demarcate a 9.2-Å-deep substrate-binding pocket each have spatial equivalents in PTPs, but we identify how these are specialized for Siw14 to bind and hydrolyze the intensely negatively charged PP-InsPs. (a) The catalytic P-loop with the CX5R(S/T) PTP motif contains additional, positively charged residues. (b) A loop between the α5 and α6 helices, corresponding to the Q-loop in PTPs, contains a lysine and an arginine that extend into the catalytic pocket due to displacement of the α5 helix orientation through intramolecular crowding caused by three bulky, hydrophobic residues. (c) The general-acid loop in PTPs is replaced in Siw14 with a flexible loop that does not use an aspartate or glutamate as a general acid. We propose that an acidic residue is not required for phosphoanhydride hydrolysis.
Keywords: inositol phosphate, crystal structure, enzyme mechanism, phosphatase, dual-specificity phosphoprotein phosphatase
Introduction
Inositol pyrophosphates (PP-InsPs2; see Fig. 1) are diffusible, intracellular signaling molecules with a uniquely crowded arrangement of phosphate and “high-energy” diphosphate groups; up to seven or eight phosphates are tightly distributed around the six-carbon inositol ring, yielding InsP7 and InsP8, respectively (see Refs. 1–4). The highly electronegative and energetic properties of the PP-InsPs enable them to regulate protein functionality through both electrostatic interactions (1) and protein pyrophosphorylation (5). A number of biological effects have been attributed to the PP-InsPs, but most evidence favors a primary role in the regulation of metabolic circuitry (3, 6, 7). In particular, cellular rates of glycolysis and oxidative phosphorylation have recently been shown to be regulated by InsP8 (6). Thus, pharmacological manipulation of the enzymes that synthesize and metabolize PP-InsPs may offer new therapeutic approaches for cancer, diabetes, and obesity.
Figure 1.
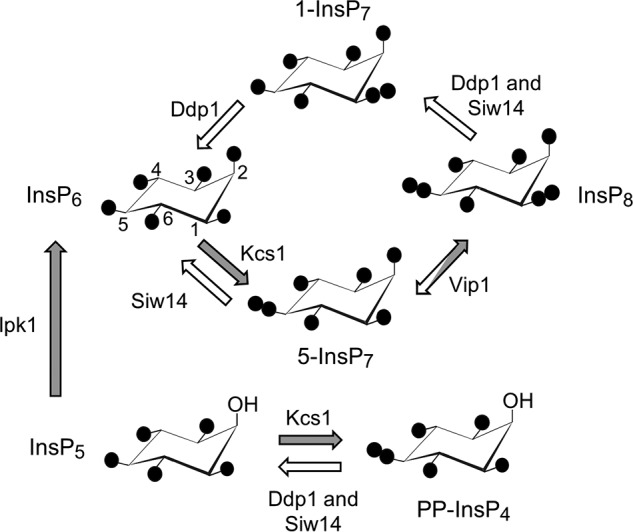
Inositol pyrophosphate metabolism in S. cerevisiae. A proposed cyclical pathway of PP-InsP turnover (see Ref. 1) is shown. Filled circles represent phosphate groups; their locants are given for InsP6, and these apply to the other molecules as each is oriented identically. The arrows depicting the various reactions are colored white to denote phosphatase activity and gray to describe kinase activity. Enzyme nomenclature is as follows: Ddp1, diphosphoinositol-polyphosphate phosphatase; Ipk1, inositol-pentakisphosphate kinase; Kcs1, inositol-hexakisphosphate kinase; Vip1, diphosphoinositol-pentakisphosphate kinase.
Two distinct families of enzymes synthesize PP-InsPs (see Fig. 1). The inositol-hexakisphosphate kinases (Kcs1 in Saccharomyces cerevisiae) utilize ATP as a phosphate donor to add a 5-β-phosphate to InsP6 (i.e. they exhibit a 5-kinase activity; see the numbering of the inositol ring in Fig. 1). A separate family of enzymes adds a β-phosphate to 5-InsP7 at C-1 (i.e. they exhibit 1-kinase activity); these are the diphosphoinositol-pentakisphosphate kinases (Vip1 in S. cerevisiae). There are data obtained in vitro that indicate each of these kinases shows some activity toward the other's substrate (8, 9). However, most current evidence indicates that sequential 5-kinase and 1-kinase activities comprise the major pathway of InsP8 synthesis (Fig. 1) in both S. cerevisiae and mammalian cells (1, 6, 10).
Control over PP-InsP function can also be elicited by PP-InsP phosphatases. One such group of enzymes is the diphosphoinositol polyphosphate phosphohydrolases (DIPPs; Ddp1 in S. cerevisiae) (11, 12). These are members of the “Nudix” superfamily, which all contain an ∼130-amino-acid β-grasp domain (13). The DIPPs can hydrolyze both the 1- and 5-diphosphate groups of PP-InsPs (14). However, no mechanisms to regulate DIPP activity have been described, which is peculiar considering the expectation that there should be highly regulated cellular turnover of cell-signaling molecules that are as important as InsP7 and InsP8. Another enigma enveloping the DIPPs is evidence of their catalytic promiscuity: alternative substrates include diadenosine polyphosphates (15), the 7-methylguanosine cap structure of mRNA (16), and inorganic polyphosphates (17). Such data raise questions as to how well-suited are the DIPPs for specifically regulating PP-InsP metabolism and function. Against this background, the discovery of a novel PP-InsP phosphatase in S. cerevisiae, Siw14 (10), promises to be a significant development. Siw14 is also of interest because it has a role in organization of the actin cytoskeleton (18). Additionally, Siw14-catalyzed PP-InsP phosphatase activity is implicated in suppressing prion propagation (19). Such an effect could be relevant to studies into basic mechanisms of amyloid formation and propagation that are applicable to human diseases (20).
Siw14 has a cysteine-based, class I CX5R(S/T) motif that defines the family of protein-tyrosine phosphatases (PTPs). Bioinformatic studies (21, 22) have led to Siw14 being classified as belonging within a specialist subgroup of PTPs, the “dual-specific” protein-tyrosine phosphatases (DUSPs). The DUSPs themselves include a distinct class of proteins that appears not to have substantial activity against phosphoproteins; hence these are usually described as “nonprotein-specific” or “atypical” phosphatases (23). The inclusion of Siw14 in this category is supported by biochemical analysis; the enzyme's catalytic activity against 5-InsP7 is several orders of magnitude greater than that against para-nitrophenyl phosphate (10), a generic protein phosphatase substrate. Other members of this atypical DUSP subgroup preferentially hydrolyze either phosphorylated carbohydrates, inositol lipids, or triphosphate groups in mRNA (23, 24). Thus, this DUSP subfamily exhibits catalytic site diversity that is not observed for classical PTPs (23).
There are considerable gaps in our structure-based understanding of the atypical DUSP catalytic mechanisms and substrate specificities. This topic is particularly complex when applied to the catalysis of PP-InsPs, which contain the most concentrated three-dimensional arrays of phosphate groups in nature. How has the basic PTP fold been repurposed to accommodate the steric bulk and intense electronegativity of PP-InsPs while also maintaining substrate specificity? In the current study, we pursue this question by providing detailed insight into new structural and catalytic properties of Siw14.
Results and discussion
Characteristics of Siw14 catalytic activity
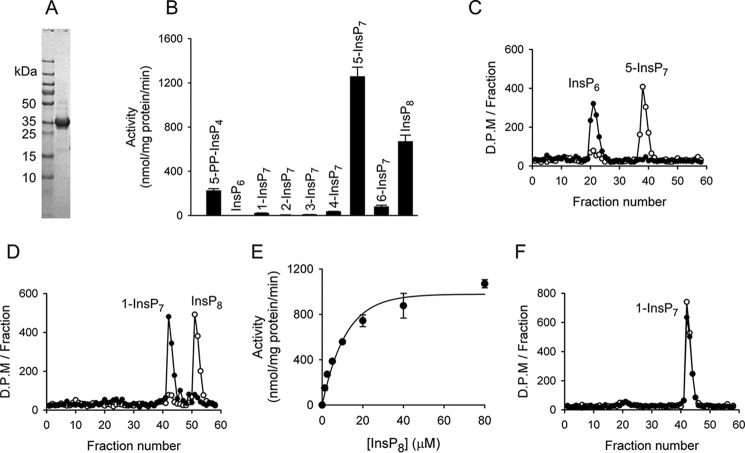
In a previous study with Siw14 (10), genetic manipulation of Siw14 expression in yeast cells was shown to have substantial effects upon cellular levels of 5-InsP7 (Fig. 1), a prominent member of the PP-InsP family. However, previous catalytic data obtained in vitro (10) indicated the activity of Siw14 toward 5-InsP7 is several orders of magnitude weaker than that catalyzed by members of the canonical PP-InsP phosphatase family, the DIPPs (Ddp1 in S. cerevisiae). It was suggested that those earlier preparations of Siw14 may have contained substantial quantities of misfolded protein (10); the possibility was also raised that Siw14 may require protein partners to express full activity (10). It is therefore significant that, in the current study, improvements to our purification procedures (Fig. 2A) have raised by 3 orders of magnitude our estimates of its catalytic activity against 5-InsP7 (measured as Kcat/Km) (Fig. 2, B and C, and Table 1). Removal of the 5-β-phosphate from 5-InsP7 leads to the accumulation of InsP6 product (Fig. 2C). Thus, we can be assured that our current experiments with Siw14 are a relevant model for furthering our understanding of PP-InsP metabolism in vivo.
Figure 2.
Catalytic activities of recombinant Siw14. A, SDS-PAGE of purified full-length Siw14. B, substrate specificity of Siw14. Each of the inositol phosphates was present at a concentration of 10 μm and incubated with 0.09–1.8 μg of Siw14 for 30 min. Vertical bars are means with S.E. (error bars) obtained from three to six experiments. C, representative HPLC analysis of assays containing 120 ng of Siw14 incubated with 5-[3H]InsP7 for 15 min (closed circles) plus a zero-time control (open circles). The elution peaks of the 5-[3H]InsP7 substrate and [3H]InsP6 product are observed in fraction numbers 38 and 21, respectively. D, representative HPLC analysis of assays containing 120 ng of Siw14 incubated with 1,5-[3H]InsP8 for 15 min (closed circles) plus a zero-time control (open circles). The peak elution time for the [3H]InsP7 product is observed in fraction number 42, and hence it is identified as 1-[3H]InsP7 (see F). E, substrate-saturation plot for Siw14-mediated dephosphorylation of 1,5-InsP8; data are means with S.E. (error bars) from five experiments. F, representative HPLC analysis of assays containing 120 ng of Siw14 incubated with 1-[3H]InsP7 for 15 min (closed circles) plus a zero-time control (open circles). The elution peak of 1-[3H]InsP7 is fraction number 42.
Table 1.
Kinetic parameters for Siw14-mediated dephosphorylation of naturally occurring PP-InsPs
Data for InsP8 dephosphorylation by full-length wildtype Siw14 are derived from Fig. 2F; similar experiments were performed with 5-PP-InsP4 and 5-InsP7 as substrates. Data are best fit values ±S.E. from three to six independent experiments using six to seven different substrate concentrations.
| Enzyme | Substrate | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm | s−1 | 106m−1 s−1 | ||
| Siw14 | 5-PP-InsP4 | 56.8 ± 5.1 | 0.77 ± 0.03 | 0.014 |
| Siw14 | InsP8 | 10.1 ± 1.5 | 0.63 ± 0.03 | 0.062 |
| Siw14 | 5-InsP7 | 10.4 ± 2.9 | 1.31 ± 0.14 | 0.125 |
| Siw14(116–281) | 5-InsP7 | 11.6 ± 2.7 | 1.32 ± 0.12 | 0.114 |
Two other PP-InsP isomers that are present in yeast and mammalian cells also contain a 5-β-phosphate: 5-PP-InsP4 and InsP8 (Fig. 1). Neither have previously been investigated as possible Siw14 substrates. A comparison of specificity constants (Kcat/Km) shows that 5-PP-InsP4 is a 9-fold weaker substrate than is 5-InsP7 (Table 1); this result indicates that the axial 2-phosphate contributes to substrate recognition. Another novel observation is that Siw14 actively dephosphorylates InsP8 (Fig. 2, B, D, and E, and Table 1). HPLC analysis of InsP8 dephosphorylation shows that the elution time for the InsP7 product (fraction number 42; Fig. 2D) coincides with that for 1-InsP7 (Fig. 2F) and not 5-InsP7 (fraction number 38; Fig. 2C). That is, the specificity of this enzyme for the 5-diphosphate group also extends to InsP8. The specificity constant for InsP8 is only 2-fold lower than the corresponding value for 5-InsP7 (Table 1); this comparison indicates the second diphosphate at the 1-position of InsP8 (Fig. 1) does not have a substantial effect on catalytic activity.
Yeast and mammalian cells also contain small quantities of 1-InsP7 (Fig. 1); we confirmed a previous demonstration (10) that Siw14 has negligible activity against 1-InsP7 (Figs. 1 and 2, B and F). Moreover, we have substantially extended our understanding of this enzyme's specificity profile by showing that 2-InsP7, 3-InsP7, 4-InsP7, and 6-InsP7 are also very poor substrates (Fig. 2B). None of these PP-InsPs are present in either yeast or mammalian cells. Nevertheless, an overarching conclusion that can be drawn from the data in Fig. 2B is that the 5-β-phosphate is an important determinant of Siw14 catalytic specificity.
Overall structure of Siw14
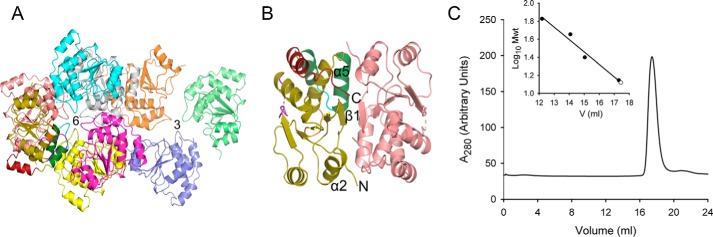
We were unable to produce crystals of the full-length protein, but we were successful with an N-terminally truncated construct (residues 116–281). The kinetic parameters for this construct are similar to those of the full-length protein (Table 1), indicating that our structure defines the catalytic core. We prepared crystals, which exhibit a space group of P6522, from a selenomethionine derivative of Siw14 and solved the structure by single-wavelength anomalous dispersion to a resolution of 2.35 Å (Table 2). The crystal structure reveals multiple molecules per asymmetric unit: nine in total, formed from a trimer, and a hexamer that is formed by three dimers (Fig. 3A). Macromolecular interface analysis (PDBePISA (25)) describes a loosely defined trimer interface with a solvent-accessible area of 530 Å2 (Fig. 3A). In contrast, the dimer interface involves 33 residues from N- and C-terminal loops, two α-helices, and a β-sheet. The interfacial, solvent-accessible area is 1230 Å2, indicating relatively tight binding. Nevertheless, analysis of recombinant Siw14(116–281) by gel filtration indicates a size of ∼14 kDa (Fig. 3C), indicating the protein is monomeric in solution. Therefore, we conclude that the oligomerization reflects crystallographic packing effects. Such artifacts have been observed for some other DUSPs, most of which also appear to be monomers in solution (26, 27).
Table 2.
Data collection and structure refinement statistics
r.m.s., root mean square.
| Protein Data Bank code 6BYF | |
|---|---|
| Data collection | |
| Space group | P6522 |
| Cell dimensions a, c (Å) | 92.97, 814.7 |
| Resolution (Å)a | 50–2.35 (2.38) |
| Rmeasa | 0.116 (0.544) |
| CC1/2 in the highest shell | 0.824 |
| I/σIa | 15.1 (1.5) |
| Completeness (%)a | 97.9 (87.1) |
| Redundancya | 5.2 (3.9) |
| Refinement | |
| Resolution (Å)a | 2.35 (2.43) |
| No. reflections | 88,286 (8,575) |
| Rworka | 0.206 (0.270) |
| Rfreea | 0.229 (0.312) |
| No. atoms | |
| Protein | 12,692 |
| Ligand | 131 |
| Solvent | 412 |
| B-factors (Å2) | |
| Protein | 46.8 |
| Ligand | 56.1 |
| Solvent | 44.1 |
| r.m.s. deviations | |
| Bond length (Å) | 0.005 |
| Bond Angle (°) | 0.80 |
a The numbers in parentheses are for the highest-resolution shell.
Figure 3.
Crystal packing of Siw14. A, shown are the interfaces of the trimer (denoted as 3) and hexamer (denoted as 6). Ribbon structures are shown with various colors for different chains. B, the dimer interface and structural elements involved are denoted. C, analysis of recombinant Siw14(116–281) purified by gel filtration. Mwt, molecular weight.
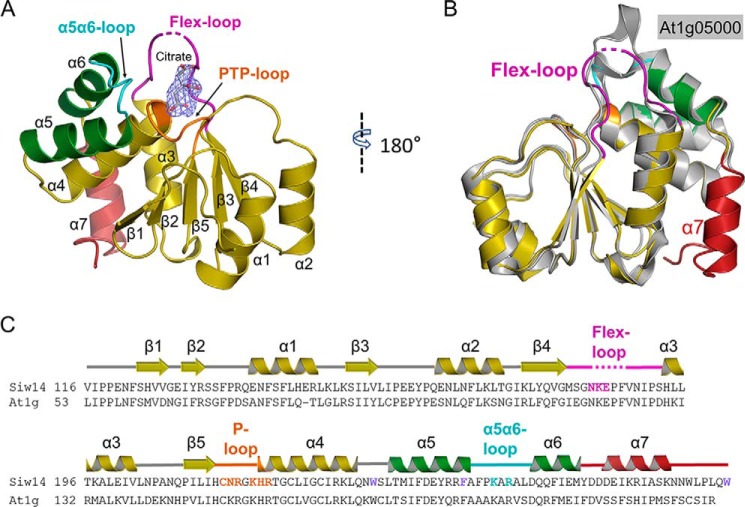
All of the residues in Siw14(116–281) can be traced without ambiguity except a flexible loop in which four amino acids are disordered (residues 184–187; Fig. 4, A, B, and C). The overall topology of the core Siw14 structure (Fig. 4, A, B, and C) is similar to that of other DUSPs (26) in that there is a canonical PTP fold comprising a central cluster of β-strands (β1 runs antiparallel to β2–β5; Fig. 4, A and B), which are asymmetrically flanked by α-helices (two on one side and five on the other; Fig. 4, A and B). Thus, we can also readily identify the various structural elements of Siw14 that establish the PTP-based active site (see below). Additionally, there are a number of structures of DUSPs in which an anion from the crystallization buffer is trapped in the active site, which has a stabilizing influence that aids crystallographic analysis (26). The equivalent region of our Siw14 structure is a 9.2-Å-deep pocket that contains a citrate ion (Fig. 5, A, B, and C).
Figure 4.
Crystal structure of Siw14. A, overall structure of Siw14(116–281). A citrate molecule was observed in the structure. The 2Fo − Fc electron density map was contoured at 1.0 σ. B, superimposition of Siw14 (in color) and At1g05000 (in gray; Protein Data Bank code 1XRI) catalytic domains; core root mean square deviation is 0.654 Å. C, alignment of residues 116–281 of Siw14 (KZV08605) with corresponding residues from At1g05000 (At1g; AAO63274). Those residues that were targeted for site-directed mutagenesis are highlighted in color. The secondary structural elements of Siw14 are also shown above the alignment and are color-coded to match A and B.
Figure 5.
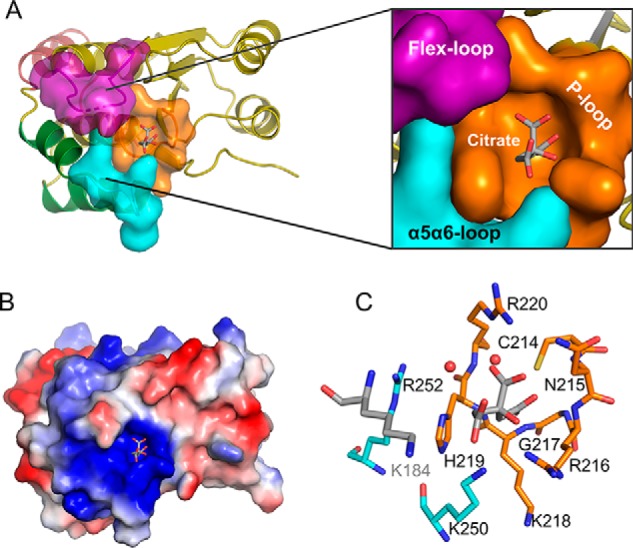
The architecture of the Siw14 active site. A, surface representation of the substrate-binding pocket with a zoom-out to show its position in the overall protein structure. The P-loop is colored orange, the α5-α6 loop is colored cyan, and the flex-loop is colored purple (omitting residues 184–187, which have low electron density in the structure). Other structure elements are shown in ribbon. B, electrostatic surface plot with blue and red coloration to denote positive and negative electrostatic potentials, respectively, at physiological pH; the captured citrate molecule is also shown. C, residues that surround the citrate are shown as sticks. Nitrogen is blue, sulfur is yellow, oxygen is red, and carbons are color-coded to indicate their parent structural feature (A). The two red spheres represent water molecules.
The closest structural homologue of Siw14(116–281) is the core structure of an Arabidopsis DUSP, At1g05000 (Fig. 4B), which was previously solved at a resolution of 3.3 Å (28). Our Siw14 structure offers improved clarity because of its higher resolution of 2.35 Å. Moreover, there are 13 C-terminal residues in At1g05000 that could not be observed in its crystal structure (28). In contrast, the corresponding region of Siw14 is included in our structure and contains both an extra helix (α7; Fig. 4, A, B, and C) and a functionally significant C-terminal residue (Trp-281; see below). At1g05000 has been reported to express relatively low activity against phosphotyrosine and some lipid phosphoinositides (21, 28), but there are no reports of PP-InsPs being tested as substrates.
Active site architecture
An electrostatic surface plot shows that the catalytic pocket resides within a very positively charged region of the protein (Fig. 5B), providing an excellent complement for accommodating the extremely negatively charged PP-InsP substrates. Despite many attempts, we were unable to crystallize a substrate–enzyme complex, and we were also unable to soak any substrate into the active site.
The loops surrounding the substrate-binding pocket (Fig. 5A) are spatial equivalents for three structural motifs that are universal in the PTP family: the P-loop, Q-loop, and general-acid loop (29). The functional definition of the P-loop is a structural element that hosts the CX5R(S/T) catalytic motif of PTPs (23, 30). The corresponding P-loop region of Siw14 includes the residues that connect the β5 strand to the α4 helix plus two additional residues (His-219 and Arg-220) at the N terminus of the α4 helix. This structure includes an invariant nucleophile, Cys-214, which is located near the low point of the substrate pocket. Ionic interactions between this cysteine and an adjacent histidine stabilize the thiolate anion (31). Arg-220 corresponds to the invariant arginine that normally participates in substrate binding and stabilization of the cysteinyl-phosphate intermediate during the reaction cycle (30). The critical importance of both Cys-214 and Arg-220 was verified by site-directed mutagenesis: the catalytic activities of the C124S mutant and the R220A mutant were reduced >98% for all substrates (Table 3). In this respect, Siw14 shares critical catalytic residues with classical PTPs.
Table 3.
Mutagenic analysis of the specificity of PP-InsP hydrolysis by Siw14
All reaction rate data (i.e. specific activity) were obtained by phosphate release assays as described under “Experimental procedures” using a 10 μm concentration of each substrate. The reaction rate against 5-InsP7 for WT full-length Siw14 is 1858 ± 117 nmol/mg of protein/min (n = 9). Relative rate data are means from three to nine experiments; all S.E. values were <15% of the mean values. The dashes (—) indicate assays that were not performed.
| Mutation | Structural element | Relative rate; mutant/WT |
||||
|---|---|---|---|---|---|---|
| 5-InsP7 | 5-PP-InsP4 | 4-InsP7 | 6-InsP7 | 1,5-InsP8 | ||
| % | ||||||
| N183A | Flex-loop | 46 | 35 | 30 | 17 | 49 |
| K184A | Flex-loop | 9 | 3 | 3 | 3 | 5 |
| E185A | Flex-loop | 33 | 85 | 96 | 59 | 70 |
| C214S | P-loop | 1 | 0.4 | 2 | 2 | 1 |
| N215A | P-loop | 40 | 44 | 22 | 28 | 57 |
| R216A | P-loop | 88 | 46 | 17 | 16 | 103 |
| K218A | P-loop | 22 | 13 | 6 | 8 | 17 |
| H219A | P-loop/α4 helixa | 19 | 26 | 9 | 10 | 49 |
| R220A | P-loop/α4 helixa | 1 | 0.1 | 0.2 | 1 | 2 |
| K250A | α4-α5 loop | 7 | 15 | 20 | 12 | 6 |
| R252A | α4-α5 loop | 7 | 7 | 6 | 5 | 5 |
| W234A | Bulky and hydrophobic | 30 | — | — | — | — |
| F246A | Bulky and hydrophobic | 7 | — | — | — | — |
| W281A | Bulky and hydrophobic | 7 | — | — | — | — |
a Note that His-219 and Lys-220 lie in the N terminus of the α4 helix that adjoins the P-loop; we have included these two residues in the definition of the P-loop.
Small side-chain amino acids are normally found in the P-loop of PTPs, but these are replaced with larger residues with positive charges in certain DUSPs such as the RNA triphosphatase PIR1; it is thought that this specialization facilitates the hydrolysis of polyphosphates (32, 33). In agreement with that idea, the P-loop in Siw14 not only contains the catalytic Arg-220 (see above) but also three additional charged residues that are all candidates to interact with the polyphosphate PP-InsP substrate: Arg-216, Lys-218, and His-219. The significance of the latter three residues was studied by mutagenesis (Table 3). The K218A mutation substantially decreases catalytic activity against all substrates (Table 3), presumably because the native lysine binds substrates through its positive charge. The H219A mutant also exhibits reduced overall catalytic activity, although InsP8 dephosphorylation was less severely impacted (Table 3); the 1-phosphate of InsP8 may reduce the strength of its interaction with His-219. The fact that the H219A and R216A mutations have quantitatively different effects on substrate hydrolysis can be rationalized by the data in Fig. 5, A and C, which show that these two residues lie on opposite sides of the substrate-binding pocket.
Interestingly, the R216A mutant has little impact upon the metabolism of 5-InsP7 and InsP8, but the rate of 5-PP-InsP4 hydrolysis is approximately halved (Table 3). These data suggest that 5-PP-InsP4, which lacks a 2-phosphate group (Fig. 1), takes a slightly different orientation within the active site than do 5-InsP7 and InsP8. The weak activity of Siw14 toward both 4-InsP7 and 6-InsP7 is further compromised by the R216A mutation (Table 3). These particular data have no obvious biological significance in mammalian and yeast cells, which do not synthesize either 4-InsP7 or 6-InsP7 (34), although 6-InsP7 has been observed in the dictyostelids (35).
Another architectural element of the substrate pocket of PTPs is the “Q-loop,” which is defined by a glutamine residue that coordinates a catalytically important water molecule. Siw14 has a spatially equivalent loop between α5 and α6, although it does not contain a glutamine. Instead, the α5-α6 loop contains two positively charged residues, Lys-250 and Arg-252. The side chains of these two residues form finger-like projections into the catalytic pocket (Fig. 5A). Mutation of either Lys-250 or Arg-252 to alanine reduced catalytic activity by >90% for all substrates (Table 3), confirming the catalytic significance of these two residues, presumably because they bind substrates through their positive charge. These two residues both lie opposite the P-loop (Fig. 5C) and so may anchor phosphate groups in 5-InsP7 that are distal to the cleaved 5-β-phosphate.
Our structural data reveal that the α5 helix of Siw14 is displaced relative to the positions of the corresponding helices in other PTPs. Specifically, there is a 30° rotation of the α5 helix such that its C terminus is repositioned 8 Å more distal from the β1 strand (Fig. 6A). This displacement is enforced by intramolecular crowding due to bulky, aromatic residues Trp-234, Phe-246, and Trp-281 (Fig. 6B). This structural specialization can be viewed as squeezing together the ends of the α5 and α6 helices so that the intervening loop “pops out” relative to the positions of the corresponding loop in other PTPs (Fig. 6A). This rearrangement is associated with the aforementioned projection of Lys-250 and Arg-252 into the catalytic pocket (Figs. 5A and 6A). We tested this structural interpretation by attempting to relieve the intramolecular crowding with either of three separate mutations, W234A, F246A, and W281A. They each strongly inhibited catalytic activity against 5-InsP7 (Table 3). Both Trp-234 and Phe-246 in Siw14 are conserved in At1g05000, but Trp-281 is unique to the yeast protein (Fig. 4C). The functional significance of the C terminus of the Siw14 protein is underscored not just by Trp-281 but also by the adjacent α7 helix (Figs. 4C and 6B). It is not known if the latter structural element is present in At1g05000, because the positions of the most C-terminal 13 residues of the plant protein are not evident in its crystal structure (28).
Figure 6.
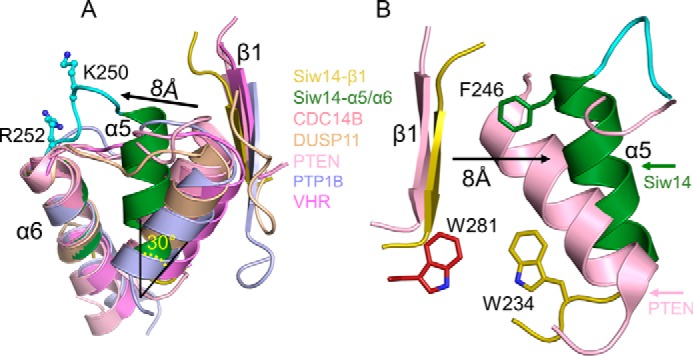
Orientation of the α5-α6 loop of Siw14 reflects intramolecular crowding. A, overlap of Siw14 β1 strand (shown in yellow) and α5 and α6 helices (shown in green) with class I PTPs, including CDC14B (Protein Data Bank code 1OHE; shown in dark pink), Dusp11 (Protein Data Bank code 4NYH; shown in wheat), PTEN (Protein Data Bank code 1D5R; shown in light pink), PTP1B (Protein Data Bank code 1PTY; shown in light blue), and VHR (Vaccinia H1-related phosphatase; Protein Data Bank code 1VHR; shown in light purple). Candidate substrate-binding residues Lys-250 and Arg-252 are shown in cyan with stick and ball modeling. We have highlighted the displacement of α5 relative to the corresponding α-helix in the other PTPs (by 30° and 8 Å). B, comparison of equivalent structural elements in both PTEN and Siw14. We have labeled the bulky, aromatic residues Phe-246, Trp-234, and Trp-281, which appear to contribute to the intramolecular crowding that underlies the displacement of the α5 helix. For Siw14, see Fig. 4 for the positions in the overall protein structure of the α5 and α6 helices and the β1 loop.
Between β4 and α3 in Siw14, there is a loop comprising13 residues, four of which exhibit weak electron density, which we attribute to the loop's flexibility; herein, we refer to this structural element as the “flex-loop” (Fig. 4). The relative positioning of the flex-loop is similar, with some displacement, to the relative positioning of the class I PTP general-acid loop (Fig. 7, A and B) which, interestingly, is typically also flexible; its movement into the active site during substrate binding is considered a classical example of an “induced fit” reaction mechanism (29). Two residues in the flex-loop of Siw14, Asn-183 and Lys-184, are both candidates for binding substrate; we separately mutated each of them to alanine. The N183A mutation has relatively minor effects on the hydrolysis of natural substrates (5-PP-InsP4, 5-InsP7, and InsP8; Table 3). In contrast, the catalysis against all substrates is impaired >90% by the Lys-184 mutation (Table 3); we propose that the positive charge of Lys-184 makes an important contribution to substrate binding.
Figure 7.
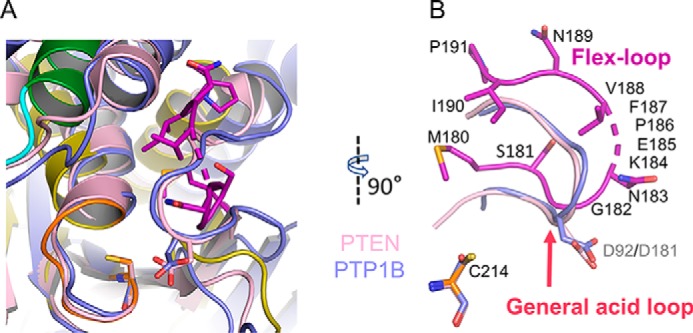
Absence of a general acid in Siw14. A, the catalytic center of Siw14 (green, α6 helix; orange stick, P-loop; purple stick, flex-loop) is superimposed upon PTEN (PDB code 1D5R; shown in light pink) and PTP1B (PDB code 1PTY; shown in light blue). B, highlight from A of the relative positions of the flex-loop of Siw14 compared with the general-acid loops of PTEN and PTP1B. Also shown, for the purposes of orientation, are each protein's catalytic cysteine residues, although in this perspective they cannot be individually distinguished; Cys-214 refers to Siw14. The numbers in gray font are the general-acid residues Asp-92 for PTEN and Asp-181 for PTP1B. The numbers in black font correspond to residues in Siw14; broken stick indicates the four residues that are disordered in our structure.
We also performed a kinetic analysis of 5-InsP7 hydrolysis and the effects of several mutations of residues in the catalytic pocket (Tables 1 and 4). It is notable that K184A, K218A, K250A, and K250A are all mutations that significantly elevate Km, indicating an impact upon substrate binding. In contrast, the N183A, N215A, and H219A mutations decrease catalytic efficiency primarily by reducing Kcat, which indicates that the native residues contribute to the reaction mechanism and/or the protein structure within the active site.
Table 4.
Mutagenic analysis of dephosphorylation of 5-InsP7 by Siw14
All data were obtained recording Pi release from 5-InsP7 by phosphate release assays as described under “Experimental procedures.” Relative values for Kcat/Km were calculated using the data for wildtype enzyme given in Table 1. Data are best fit values ±S.E. from three to five independent repeats. WT is full-length wildtype Siw14 for which the kinetic parameters for 5-InsP7 as substrate are described in Table 1.
| Enzyme | Km | Kcat | Kcat/Km | Kcat/Km; mutant/WT |
|---|---|---|---|---|
| μm | s−1 | 106m−1 s−1 | % | |
| N183A | 10.2 ± 1.9 | 0.57 ± 0.03 | 0.056 | 49 |
| K184A | 47.1 ± 12.0 | 0.46 ± 0.06 | 0.010 | 9 |
| E185A | 12.2 ± 2.1 | 0.80 ± 0.05 | 0.066 | 58 |
| N215A | 7.0 ± 1.2 | 0.29 ± 0.01 | 0.041 | 36 |
| R216A | 14.5 ± 2.3 | 1.26 ± 0.08 | 0.087 | 76 |
| K218A | 28.6 ± 4.9 | 0.48 ± 0.04 | 0.017 | 15 |
| H219A | 17.0 ± 2.7 | 0.38 ± 0.02 | 0.022 | 20 |
| K250A | 32.6 ± 7.3 | 0.21 ± 0.01 | 0.006 | 6 |
| R252A | 48.1 ± 8.8 | 0.35 ± 0.03 | 0.007 | 6 |
The catalytic pocket of Siw14 lacks the canonical “general acid,” which is catalytically essential for nearly all PTPs
In almost every PTP, the general-acid loop contains an aspartate residue (or, in rare cases, a glutamate residue (36)) that acts as an acid/base during the reaction (26, 29). The relative positions of this general-acid residue and the catalytic cysteine are virtually superimposable in PTPs such as PTEN and PTP1B (Fig. 7, A and B). There is not an equivalent residue in Siw14 (Fig. 7, A and B). The region of Siw14 that corresponds to the PTP general-acid loop is referred to as the flex-loop in our study (Fig. 7B). This loop does not contain any aspartate residues, although there is a glutamate at position 185 (Figs. 4C and 7B). This is the only residue to emerge from our structural analysis as a candidate for a PTP-like general acid in the catalytic pocket. We pursued its significance by mutagenesis.
We found that an E185A mutation reduced catalytic activity toward 5-InsP7 by 67% (Table 3). There was an even smaller effect of this mutation upon the dephosphorylation of 5-PP-InsP4 and InsP8 (15 and 30%, respectively; Table 3). The general requirement for a general acid in PTPs is sufficiently critical that its mutagenesis to alanine reduces catalytic activity >99%, resulting in a “trapped” enzyme–substrate complex (30, 36, 37). Thus, we propose that the degree of inhibition due to the mutation is too low to support the hypothesis that this residue can serve as a general acid.
General conclusions
We have performed a comprehensive biochemical, structural, and mutagenic analysis of Siw14 and gained atomic-level insight into the activities of this highly unusual member of the PTP family. Our work substantially improves our understanding of the active site of Siw14 (Figs. 4–7 and Tables 3 and 4), in particular by demonstrating the functional significance of the high degree of electrostatic complementarity between the substrate-binding pocket and the negatively charged PP-InsPs.
There may be wider significance to our conclusion that the catalytic site of Siw14 does not contain an aspartate (or glutamate) that acts as a general acid because this is also the case for one other group of DUSPs, the RNA triphosphatases (32, 33, 38). These enzymes also share with Siw14 an enzymatic characteristic that is unusual for a PTP: they hydrolyze a phosphoanhydride bond. We propose that it is this specific form of PTP-mediated catalytic activity that can proceed in the absence of either an aspartate or a glutamate acting as a general acid.
In this report, we have provided a comprehensive kinetic analysis of Siw14 activity against all PP-InsPs that contain the 5-diphosphate group: 5-PP-InsP4, 5-InsP7, and InsP8 (Tables 1, 3, and 4). These data unequivocally demonstrate that Siw14 has considerable activity as a 5-PP-InsP phosphatase. The Arabidopsis protein At1g05000 is proposed to be an ortholog of Siw14 (22). However, the possibility that any PP-InsP might be a substrate for At1g05000 has yet to be tested. Additionally, the mutants that we have analyzed provide insight into the determinants of Siw14 catalytic specificity and place enzyme catalysis in a biological context. In contrast, prior mutagenic analysis of At1g05000 targeted just three residues (Glu-117, Cys-150, and Asp-191) and was restricted to studying the impact on relatively weak activity against para-nitrophenyl phosphate and inositol lipids (22), none of which contain a diphosphate group.
Our new Siw14 structure depicts its catalytic core at a considerably higher resolution (2.35 Å) than the previous structure of the corresponding region of At1g05000, which was solved at a resolution of 3.3 Å (28). Furthermore, the previous At1g05000 structure is missing 13 residues from its C terminus (28). The corresponding region of Siw14 is not only present in our crystal structure but is also functionally significant; it contains Trp-281, a residue that makes an indirect but significant contribution to the active site (Fig. 6). Thus, our structural data provide more detailed insight into this class of protein phosphatase (Figs. 4–6 and Tables 3 and 4) than was previously possible. This information includes an enhanced topological description of the catalytic pocket (Fig. 4) by defining the functional equivalent of the PTP P-loop and by demonstrating the significance of two other structural elements, the flex-loop and the α5-α6 loop (Fig. 5 and Tables 3 and 4). Interestingly, we have also deciphered the participation of some bulky hydrophobic residues in driving functionally significant intramolecular interactions in Siw14, a novelty compared with other PTPs (Fig. 6 and Table 3). Armed with all this new information, we now possess an improved picture of the structural and functional relationships of Siw14 as an exemplar of the DUSP subfamily of PTPs, and we are now far better equipped to search for possible mammalian orthologs of this intriguing class of PP-InsP phosphatases.
Experimental procedures
Protein expression and purification
For expression of Siw14 in Escherichia coli, we purchased a codon-optimized cDNA (Genscript Inc.) with the following sequence: ATGGGCCTGTACCAAGCGAAAAATGACGAGGGCAGCGACCCGAAAAGCAGCAGCAAGATTGATGACCTGATTGAAAACGAAGCGGAGATTATCCGTCTGATTAAAGAGGACGGCAAACTGCTGATCGACAACGGTGATGGCCGTGACATTCACAACATCATTCAGGAAGACAAGCTGCTGAGCGTGGAGTTCAACGAAGTTCTGAAACGTTTTCACGGTGAAGAAAAGAGCGATATCCCGCGTAAAGAGTTCGACGAGGATGAAGACGATGGCTACGACAGCAACGAGCACCACCAGAAGACCATTGAAGTGATGAACACCCTGAACCACGTGATCAACAAAGAGGTTATTCCGCCGGAAAACTTCAGCCACGTGGTTGGCGAGATCTATCGTAGCAGCTTTCCGCGTCAAGAGAACTTCAGCTTTCTGCACGAACGTCTGAAGCTGAAAAGCATCCTGGTGCTGATTCCGGAGGAATACCCGCAGGAAAACCTGAACTTCCTGAAGCTGACCGGCATCAAACTGTATCAAGTGGGTATGAGCGGCAACAAGGAGCCGTTTGTTAACATCCCGAGCCACCTGCTGACCAAAGCGCTGGAAATTGTTCTGAACCCGGCGAACCAGCCGATCCTGATTCACTGCAACCGTGGCAAGCACCGTACCGGTTGCCTGATCGGCTGCATTCGTAAACTGCAAAACTGGAGCCTGACCATGATCTTCGATGAGTACCGTCGTTTCGCGTTTCCGAAGGCGCGTGCGCTGGACCAGCAATTTATTGAAATGTATGACGACGACGAAATCAAACGCATCGCGTCTAAGAACAACTGGCTGCCGCTGCAATGGTAA.
The Gateway expression system (Invitrogen) was used to subclone Siw14 into the pDest-566 vector. This vector encodes a His6 tag, maltose-binding protein tag, and tobacco etch virus protease cleavage site at the N terminus. Mutants were prepared using a site-directed mutagenesis kit (Stratagene) or a Q5 site-directed mutagenesis kit (Biolabs); all mutants were verified by sequencing. The sequences of the primers are as follows (mutagenic codons are in lowercase): N183A: forward, CAAGTGGGTATGAGCGGCgccAAGGAGCCGTTTGTTAACATCCC; reverse, GTTAACAAACGGCTCCTTGgcgCCGCTCATACCCACTTGATAC; K184A: forward, GGGTATGAGCGGCAACgcgGAGCCGTTTGTTAACATCCCGAG; reverse,GGATGTTAACAAACGGCTCCgcgTTGCCGCTCATACCCACTTG; E185A: forward, GTATGAGCGGCAACAAGgcgCCGTTTGTTAACATCCCGAGCC; reverse, CGGGATGTTAACAAACGGcgcCTTGTTGCCGCTCATACCCAC; C214S: forward, CCAGCCGATCCTGATTCACtccAACCGTGGCAAGCACCGTAC; reverse, GGTGCTTGCCACGGTTccaGTGAATCAGGATCGGCTGGTTCG; N215A: forward, GATTCACTGCgccCGTGGCAAGCACC; reverse, ATCAGGATCGGCTGGTTC; R216A: forward, TCACTGCAACgctGGCAAGCACCG; reverse, ATCAGGATCGGCTGGTTC; K218A: forward, CAACCGTGGCgcgCACCGTACCG; reverse, CAGTGAATCAGGATCGGC; H219A: forward, CCGTGGCAAGgccCGTACCGGT; reverse, TTGCAGTGAATCAGGATCG; R220A: forward, TGGCAAGCACgccACCGGTTGCC; reverse,CGGTTGCAGTGAATCAGG; K250A: forward, GTCGTTTCGCGTTTCCGgcgGCGCGTGCGCTGGAC; reverse, CAGCGCACGCGCCgccGGAAACGCGAAACGACGG; R252A: forward, GCGTTTCCGAAGGCGgctGCGCTGGACCAGCAATTTATTG; reverse, GCTGGTCCAGCGCagcCGCCTTCGGAAACGCGAAAC; W234A: forward, ACTGCAAAACgcgAGCCTGACCATG; reverse, TTACGAATGCAGCCGATC; F246A: forward, GTACCGTCGTgccGCGTTTCCG; reverse, TCATCGAAGATCATGGTCAGG; W281A: forward, GCCGCTGCAAgcgTAACCGACCC; reverse, AGCCAGTTGTTCTTAGACG.
DE3 competent E. coli cells (Stratagene) that were pretransformed with chaperone plasmid pGro7 (Takara Clontech) were transformed with the resultant plasmid. An overnight culture of the transformed E. coli cells was inoculated into nutrient-rich 2× YT medium (16 g/liter Tryptone, 10 g/liter yeast extract, 5.0 g/liter NaCl) supplemented with 0.07% (w/v) l-arabinose at pH 7.5 and grown at 37 °C to an A595 of 0.7. Isopropyl β-d-thiogalactopyranoside (0.1 mm) was then added, and cultures were continued at 15 °C for 20 h. For preparation of selenomethionine-labeled Siw14, the cells were inoculated into a synthetic M9 minimal medium that was supplemented with the following amino acids: 100 mg/ml Lys, 100 mg/ml Thr, 100 mg/ml Phe, 50 mg/ml Leu, 50 mg/ml Ile, 50 mg/ml Val, and 50 mg/ml selenomethionine.
The cells were disrupted using a constant cell disruption system (Constant Systems Ltd.) under 20 k.p.s.i. Recombinant wildtype (WT) and mutant proteins were purified by several chromatographic procedures performed at 4 °C. First, the protein was applied to a nickel-nitrilotriacetic acid-agarose column (Qiagen); washed with buffer containing 300 mm NaCl, 20 mm Tris-HCl (pH 7.5), 20 mm imidazole; and then eluted by increasing the imidazole concentration to 400 mm. Second, the eluate was applied to a HiTrapTM Heparin HP column (GE Healthcare) and eluted with 10 column volumes of a 50–2000 mm NaCl gradient in 20 mm Tris-HCl (pH 7.5). Next, after cleavage using the tobacco etch virus protease, the protein was further purified using another HiTrap Heparin HP column and finally a SuperdexTM 200 gel filtration column (GE Healthcare) that was eluted with 150 mm NaCl, 20 mm Tris-HCl (pH 7.5). Purified proteins were concentrated to either 0.5–2 (for assaying catalytic activities) or 15 mg/ml (for crystallization); storage was at −80 °C.
The gel filtration was performed with a Superdex 200 Increase 10/300 GL column (GE Healthcare) with 50 mm NaCl, 20 mm Tris-HCl (pH 7.5) as the running buffer. The column was calibrated with RNase A (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), and BSA (67 kDa).
Crystallization
The core catalytic domain of Siw14 (residues 116–281; Siw14(116–281)) was initially screened for optimum crystallization conditions using the mosquito lipidic cubic phase (TTP Labtech). Multiple conditions with different salts were identified. Final conditions were optimized by hanging drop vapor diffusion against a well buffer of 0.7 m sodium citrate, 50 mm β-mercaptoethanol at 25 °C (2 μl of 15 mg/ml protein plus 2 μl of well buffer in the crystallization drop).
Data collection, structure determination, and refinement
Diffraction data were collected using Advanced Photon Source beam line 22-ID. All data were processed with the program HKL2000 (39). Initial phases for the structure were determined by Autosol in PHENIX followed by an autobuild procedure (40). This initial structure was manually rebuilt with Coot (41) and refined with PHENIX. The molecular graphics representations were prepared with the program PyMOL (Schrödinger, LLC). Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6BYF.
Enzyme assays and HPLC analysis
Enzyme activity was performed at 30 °C. Phosphatase activity was measured after 30-min assays in 100-μl reaction mixtures containing 20 mm HEPES (pH 7.2), 100 mm KCl, 0.6 mm MgCl2, 20 μm EDTA. Pi release from individual chemically synthesized PP-InsPs (42, 43) was determined with a colorimetric assay (44). Kinetic parameters were determined using six to seven different concentrations of the indicated substrate within the range of 1.3–80 μm. GraphPad Prism was used to analyze the data.
To assay phosphatase activity of Siw14 by HPLC, 120 ng of recombinant protein was incubated with 1 μm chemically synthesized 5-InsP7, 1-InsP7, or 1,5-InsP8 in 100 μl of assay buffer (20 mm HEPES, 100 mm KCl, 0.6 mm MgCl2, 20 μm EDTA) for 15 min at 30 °C. Each incubation also contained the corresponding 3H-labeled substrate (∼2000 dpm). Reactions were quenched with 100 μl of 1 m perchloric acid to which was added 500 μl of the supernatant from a centrifuged, perchloric acid–quenched extract of 5 × 106 HCT116 cells (to optimize recovery of inositol phosphates). Radiolabeled inositol phosphates were then purified with TiO2 beads (45). Inositol phosphates were separated and quantified using a CarboPacTM PA200 HPLC column as described (46).
Author contributions
H. W., C. G., R. J. R., H. J. J., and S. B. S. conceptualization; H. W. and C. G. data curation; H. W., C. G., and R. J. R. formal analysis; H. W., C. G., and R. J. R. investigation; H. W. and C. G. methodology; H. W., C. G., and S. B. S. writing-original draft; H. W. and S. B. S. project administration; H. W., C. G., R. J. R., H. J. J., and S. B. S. writing-review and editing; H. J. J. and S. B. S. resources.
Acknowledgments
We thank the NIEHS Collaborative crystallography group and the Advanced Photon Source Southeast Regional Collaborative Access Team 22-ID and 22-BM beam lines for assistance with crystallographic data collection.
This work was supported by the Intramural Research Program of the NIH, National Institutes of Environmental Health Sciences, and a Georgetown University Pilot Grant (to R. J. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 6BYF) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PP-InsP
- inositol pyrophosphate
- DIPP
- diphosphoinositol polyphosphate phosphohydrolase
- DUSP
- dual-specific protein-tyrosine phosphatase
- InsP6
- inositol hexakisphosphate
- 1-InsP7
- 1-diphosphoinositol 2,3,4,5,6-pentakisphosphate
- 5-InsP7
- 5-diphosphoinositol 1,2,3,4,6-pentakisphosphate
- 1,5-InsP8
- 1,5-bisdiphosphoinositol 2,3,4,6-tetrakisphosphate
- 5-PP-InsP4
- 5-diphosphoinositol 1,3,4,6-tetrakisphosphate
- PTP
- protein-tyrosine phosphatase.
References
- 1. Shears S. B. (2018) Intimate connections: inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 233, 1897–1912 10.1002/jcp.26017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thota S. G., and Bhandari R. (2015) The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J. Biosci. 40, 593–605 10.1007/s12038-015-9549-x [DOI] [PubMed] [Google Scholar]
- 3. Azevedo C., and Saiardi A. (2017) Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 42, 219–231 10.1016/j.tibs.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 4. Chakraborty A. (2017) The inositol pyrophosphate pathway in health and diseases. Biol. Rev. Camb. Philos. Soc. 10.1111/brv.12392 10.1111/brv.12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saiardi A. (2016) Protein pyrophosphorylation: moving forward. Biochem. J. 473, 3765–3768 10.1042/BCJ20160710C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu C., Nguyen H. N., Ganini D., Chen Z., Jessen H. J., Gu Z., Wang H., and Shears S. B. (2017) KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl. Acad. Sci. U.S.A. 114, 11968–11973 10.1073/pnas.1702370114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szijgyarto Z., Garedew A., Azevedo C., and Saiardi A. (2011) Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 10.1126/science.1211908 [DOI] [PubMed] [Google Scholar]
- 8. Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., and York J. D. (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 10.1126/science.1139099 [DOI] [PubMed] [Google Scholar]
- 9. Weaver J. D., Wang H., and Shears S. B. (2013) The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci. Rep. 33, e00022 10.1042/BSR20120115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steidle E. A., Chong L. S., Wu M., Crooke E., Fiedler D., Resnick A. C., and Rolfes R. J. (2016) A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem. 291, 6772–6783 10.1074/jbc.M116.714907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Safrany S. T., Caffrey J. J., Yang X., Bembenek M. E., Moyer M. B., Burkhart W. A., and Shears S. B. (1998) A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 17, 6599–6607 10.1093/emboj/17.22.6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caffrey J. J., Safrany S. T., Yang X., and Shears S. B. (2000) Discovery of molecular and catalytic diversity among human diphosphoinositol polyphosphate phosphohydrolases: an expanding NUDT family. J. Biol. Chem. 275, 12730–12736 10.1074/jbc.275.17.12730 [DOI] [PubMed] [Google Scholar]
- 13. Srouji J. R., Xu A., Park A., Kirsch J. F., and Brenner S. E. (2017) The evolution of function within the Nudix homology clan. Proteins 85, 775–811 10.1002/prot.25223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kilari R. S., Weaver J. D., Shears S. B., and Safrany S. T. (2013) Understanding inositol pyrophosphate metabolism and function: kinetic characterization of the DIPPs. FEBS Lett. 587, 3464–3470 10.1016/j.febslet.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Safrany S. T., Ingram S. W., Cartwright J. L., Falck J. R., McLennan A. G., Barnes L. D., and Shears S. B. (1999) The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase: overlapping substrate specificities in a MutT motif. J. Biol. Chem. 274, 21735–21740 10.1074/jbc.274.31.21735 [DOI] [PubMed] [Google Scholar]
- 16. Grudzien-Nogalska E., Jiao X., Song M. G., Hart R. P., and Kiledjian M. (2016) Nudt3 is an mRNA decapping enzyme that modulates cell migration. RNA 22, 773–781 10.1261/rna.055699.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., and Saiardi A. (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 10.1074/jbc.M111.266320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., Mannazzu I., and Sudbery P. E. (2004) A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166, 707–719 10.1534/genetics.166.2.707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wickner R. B., Kelly A. C., Bezsonov E. E., and Edskes H. K. (2017) [PSI+] prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. U.S.A. 114, E8402–E8410 10.1073/pnas.1714361114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liebman S. W., and Chernoff Y. O. (2012) Prions in yeast. Genetics 191, 1041–1072 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romá-Mateo C., Sacristán-Reviriego A., Beresford N. J., Caparrós-Martín J. A., Culiáñez-Macià F. A., Martín H., Molina M., Tabernero L., and Pulido R. (2011) Phylogenetic and genetic linkage between novel atypical dual-specificity phosphatases from non-metazoan organisms. Mol. Genet. Genomics 285, 341–354 10.1007/s00438-011-0611-6 [DOI] [PubMed] [Google Scholar]
- 22. Romá-Mateo C., Ríos P., Tabernero L., Attwood T. K., and Pulido R. (2007) A novel phosphatase family, structurally related to dual-specificity phosphatases, that displays unique amino acid sequence and substrate specificity. J. Mol. Biol. 374, 899–909 10.1016/j.jmb.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 23. Hobiger K., and Friedrich T. (2015) Voltage sensitive phosphatases: emerging kinship to protein tyrosine phosphatases from structure-function research. Front. Pharmacol. 6, 20 10.3389/fphar.2015.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Worby C. A., Gentry M. S., and Dixon J. E. (2006) Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281, 30412–30418 10.1074/jbc.M606117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 26. Jeong D. G., Wei C. H., Ku B., Jeon T. J., Chien P. N., Kim J. K., Park S. Y., Hwang H. S., Ryu S. Y., Park H., Kim D. S., Kim S. J., and Ryu S. E. (2014) The family-wide structure and function of human dual-specificity protein phosphatases. Acta Crystallogr. D Biol. Crystallogr. 70, 421–435 10.1107/S1399004713029866 [DOI] [PubMed] [Google Scholar]
- 27. Ku B., Hong W., Keum C. W., Kim M., Ryu H., Jeon D., Shin H. C., Kim J. H., Kim S. J., and Ryu S. E. (2017) Structural and biochemical analysis of atypically low dephosphorylating activity of human dual-specificity phosphatase 28. PLoS One 12, e0187701 10.1371/journal.pone.0187701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aceti D. J., Bitto E., Yakunin A. F., Proudfoot M., Bingman C. A., Frederick R. O., Sreenath H. K., Vojtik F. C., Wrobel R. L., Fox B. G., Markley J. L., and Phillips G. N. Jr. (2008) Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000. Proteins 73, 241–253 10.1002/prot.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tonks N. K. (2013) Protein tyrosine phosphatases—from housekeeping enzymes to master regulators of signal transduction. FEBS J. 280, 346–378 10.1111/febs.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tautz L., Critton D. A., and Grotegut S. (2013) Protein tyrosine phosphatases: structure, function, and implication in human disease. Methods Mol. Biol. 1053, 179–221 10.1007/978-1-62703-562-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Z. Y., and Dixon J. E. (1993) Active site labeling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry 32, 9340–9345 10.1021/bi00087a012 [DOI] [PubMed] [Google Scholar]
- 32. Sankhala R. S., Lokareddy R. K., and Cingolani G. (2014) Structure of human PIR1, an atypical dual-specificity phosphatase. Biochemistry 53, 862–871 10.1021/bi401240x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lokareddy R. K., Bhardwaj A., and Cingolani G. (2013) Atomic structure of dual-specificity phosphatase 26, a novel p53 phosphatase. Biochemistry 52, 938–948 10.1021/bi301476m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin H., Fridy P. C., Ribeiro A. A., Choi J. H., Barma D. K., Vogel G., Falck J. R., Shears S. B., York J. D., and Mayr G. W. (2009) Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J. Biol. Chem. 284, 1863–1872 10.1074/jbc.M805686200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laussmann T., Reddy K. M., Reddy K. K., Falck J. R., and Vogel G. (1997) Diphospho-myo-inositol phosphates from Dictyostelium identified as D-6-diphospho-myo-inositol pentakisphosphate and D-5,6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem. J. 322, 31–33 10.1042/bj3220031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oganesian A., Poot M., Daum G., Coats S. A., Wright M. B., Seifert R. A., and Bowen-Pope D. F. (2003) Protein tyrosine phosphatase RQ is a phosphatidylinositol phosphatase that can regulate cell survival and proliferation. Proc. Natl. Acad. Sci. U.S.A. 100, 7563–7568 10.1073/pnas.1336511100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flint A. J., Tiganis T., Barford D., and Tonks N. K. (1997) Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 10.1073/pnas.94.5.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martins A., and Shuman S. (2000) Mechanism of phosphoanhydride cleavage by baculovirus phosphatase. J. Biol. Chem. 275, 35070–35076 10.1074/jbc.M005748200 [DOI] [PubMed] [Google Scholar]
- 39. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 40. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 42. Capolicchio S., Wang H., Thakor D. T., Shears S. B., and Jessen H. J. (2014) Synthesis of densely phosphorylated bis-1,5-diphospho-myo-inositol tetrakisphosphate and its enantiomer by bidirectional P-anhydride formation. Angew. Chem. Int. Ed. Engl. 53, 9508–9511 10.1002/anie.201404398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Capolicchio S., Thakor D. T., Linden A., and Jessen H. J. (2013) Synthesis of unsymmetric diphospho-inositol polyphosphates. Angew. Chem. Int. Ed. Engl. 52, 6912–6916 10.1002/anie.201301092 [DOI] [PubMed] [Google Scholar]
- 44. Hoenig M., Lee R. J., and Ferguson D. C. (1989) A microtiter plate assay for inorganic phosphate. J. Biochem. Biophys. Methods 19, 249–251 10.1016/0165-022X(89)90031-6 [DOI] [PubMed] [Google Scholar]
- 45. Wilson M. S., Bulley S. J., Pisani F., Irvine R. F., and Saiardi A. (2015) A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open. Biol. 5, 150014 10.1098/rsob.150014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu C., Wilson M. S., Jessen H. J., Saiardi A., and Shears S. B. (2016) Inositol pyrophosphate profiling of two HCT116 cell lines uncovers variation in InsP8 levels. PLoS One 11, e0165286 10.1371/journal.pone.0165286 [DOI] [PMC free article] [PubMed] [Google Scholar]