Abstract
The biosynthesis of lignin monomers involves two methylation steps catalyzed by orthodiphenol-O-methyltransferases: caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferases (COMTs) and caffeoyl-coenzyme A (CoA)/5-hydroxyferuloyl-CoA 3/5-O-methyltransferases (CCoAOMTs). Two COMT classes (I and II) were already known to occur in tobacco (Nicotiana tabacum) and three distinct CCoAOMT classes have now been characterized. These three CCoAOMT classes displayed a maximum level of expression at different stages of stem development, in accordance with their involvement in the synthesis of lignin guaiacyl units. Expression profiles upon tobacco mosaic virus infection of tobacco leaves revealed a biphasic pattern of induction for COMT I, COMT II, and CCoAOMTs. The different isoforms were expressed in Escherichia coli and our results showed that CCoAOMTs and, more surprisingly, COMTs efficiently methylated hydroxycinnamoyl-CoA esters. COMT I was also active toward 5-hydroxyconiferyl alcohol, indicating that COMT I that catalyzes syringyl unit synthesis in planta may operate at the free acid, CoA ester, or alcohol levels. COMT II that is highly inducible by infection also accepted caffeoyl-CoA as a substrate, thus suggesting a role in ferulate derivative deposition in the walls of infected cells. Tobacco appears to possess an array of O-methyltransferase isoforms with variable efficiency toward the diverse plant o-diphenolic substrates.
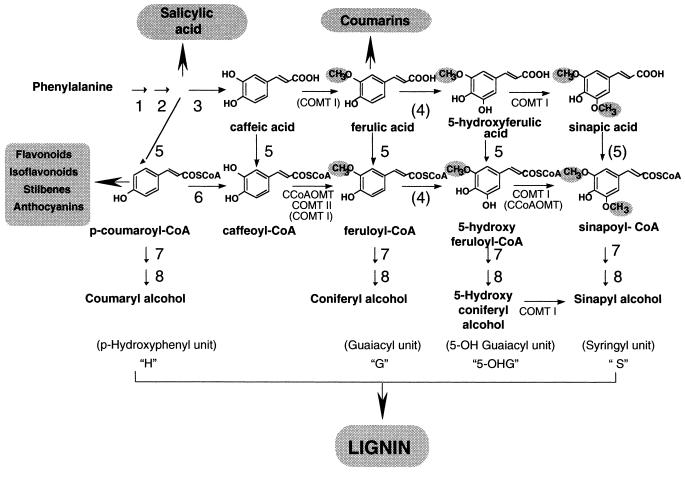
Lignin is a major cell wall polymer of vascular plants that provides mechanical strength and hydrophobicity to vascular vessels. It is a heteropolymer composed of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units derived from p-coumaryl, coniferyl, and sinapyl alcohols, respectively (Fig. 1). Lignin composition is known to change during plant development and under the influence of environmental factors (Boudet et al., 1995; Campbell and Sederoff, 1996; Baucher et al., 1998; Whetten et al., 1998). The phenylpropanoid pathway that provides the lignin-building units called monolignols is strongly activated upon infection by pathogens or treatment with elicitors (Legrand, 1983; Pakush and Matern, 1991; Pakush et al., 1991; Jaeck et al., 1992). In infected plants, the deposition of phenylpropanoid compounds participates in cell wall reinforcement that restricts pathogen invasion (Nicholson and Hammerschmitt, 1992; Kauss et al., 1993; Dixon and Paiva, 1995).
Figure 1.
OMT-catalyzed reactions in the general phenylpropanoid pathway. Nomenclature of lignin units is indicated in parentheses. 1, Phe ammonia-lyase; 2, cinnamate 4-hydroxylase; 3, p-coumarate 3-hydroxylase; 4, ferulate 5-hydroxylase; 5, coumarate CoA ligase; 6, coumaroyl-CoA 3-hydroxylase; 7, cinnamoyl-CoA reductase; 8, cinnamyl alcohol dehydrogenase. Methyl groups incorporated by OMTs are surrounded by gray areas. COMT and/or CCoAOMT are indicated according to substrate specificity assayed in vitro; brackets indicate that their role in vivo is questionable (Atanassova et al., 1995). Ferulate 5-hydroxylase substrate specificity is unknown (Chapple et al., 1992; Meyer et al., 1998). In some species, coumarate CoA ligase has very little activity with sinapic acid as the substrate (Lee and Douglas, 1996; Allina et al., 1998).
Most of the enzymes involved in monolignol biosynthesis have been characterized (Fig. 1). In particular, several caffeic acid 3-O-methyltransferases (COMTs) from dicots have been shown to catalyze the methoxylation of caffeic and 5-hydroxyferulic acids in vitro and were believed to be involved in guaiacyl and syringyl unit synthesis in vivo. However, caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), an enzyme that converts caffeoyl-CoA to feruloyl-CoA, has been characterized from parsley cell suspensions treated with an elicitor preparation (Kühnl et al., 1989; Pakush et al., 1989; Schmitt et al., 1991). The enzyme was thought to be involved in plant defense reactions by synthesizing wall-bound forms of ferulic acid. More recently, CCoAOMT has been proposed as the key element of an alternative pathway for lignin biosynthesis in zinnia cells during tracheary element formation (Ye et al., 1994; Ye and Varner, 1995).
Lignin analysis of transgenic plants or mutants down-regulated for COMT expression disclosed a decreased S to G ratio and the appearance of a new unit, the 5-hydroxyguaiacyl unit (Fig. 1) (Lapierre et al., 1988; Atanassova et al., 1995; Van Doorsselaere et al., 1995). These data demonstrated that in vivo, COMT activity controls the second methylation step leading to the syringyl unit, but left open the question of the redundancy between COMTs and CCoAOMTs with respect to their function in the first methylation step leading to the guaiacyl unit. Two classes of COMTs have been isolated and cloned from tobacco mosaic virus (TMV)-infected tobacco (Nicotiana tabacum) leaves (Legrand et al., 1978; Jaeck et al., 1992; Pellegrini et al., 1993): class I COMTs are highly expressed in lignified tissues, whereas class II COMTs are barely expressed in healthy tissues but are strongly stimulated upon infection. Recently, four CCoAOMT cDNA clones were isolated from tobacco leaves, and sequence comparison has shown that they belong to the same class of CCoAOMT (Martz et al., 1998).
We describe the cloning of two other CCoAOMTs, which, in view of the phylogenic data, are likely to belong to distinct gene classes defined as class 2 and class 3. To clarify the functions of the numerous OMT isoforms occurring in tobacco, we have compared their expression profiles during stem development and TMV infection and their affinities for various o-diphenolic substrates. The gene expression patterns of class 2 and 3 CCoAOMTs were found to be clearly different from those of class 1 CCoAOMT genes at various stages of stem development and during the hypersensitive response of tobacco leaves to TMV. Using specific antibodies, two CCoAOMT proteins were revealed in variable amounts in different plant tissues. Seven CCoAOMTs belonging to the three classes, one COMT I, and one COMT II were expressed in Escherichia coli. A preference for CoA esters was found for all enzymes tested, and was particularly pronounced with CCoAOMT isoforms. COMT I was demonstrated to efficiently methylate 5-hydroxyferulate, 5-hydroxyferuloyl-CoA, and 5-hydroxyconiferyl alcohol in vitro. These data indicate that the synthesis of lignin syringyl units that is catalyzed by COMT I in vivo (Atanassova et al., 1995) may proceed through various metabolic routes (Fig. 1). COMT II was also demonstrated to be very active toward caffeoyl-CoA as a substrate, suggesting that COMT II, which is known to be highly inducible upon infection or elicitor treatment, might catalyze the synthesis of ferulic derivatives whose deposition within the cell wall builds up a mechanical barrier, thus restricting pathogens (Nicholson and Hammerschmitt, 1992; Matern et al., 1995).
MATERIALS AND METHODS
Plant Material and Treatments
Tobacco plants (Nicotiana tabacum L. cv Samsun NN) were grown in the greenhouse under controlled conditions. One-month-old plants were infected by rubbing fully expanded leaves with a TMV suspension (0.5 μg/mL in the presence of an abrasive). At different times after infection, treated leaf tissues from three independent plants were harvested, quickly frozen in liquid nitrogen, and stored at −80°C.
Cloning of New Members of the Different Tobacco CCoAOMT Classes
Four CCoAOMT cDNA clones (CCoAOMT-1, CCoAOMT-2, CCoAOMT-3, and CCoAOMT-4) have been isolated from a library made from 48-h-infected tobacco leaves (accession nos. U38612, U62734, U62735, and U62736; Martz et al., 1998) and correspond to the CCoAOMT class 1. Primers were derived from sequences available in the database (accession nos. Z82982, corresponding to a premature transcript, and Z56282, from Busam et al., 1997a). The sense primer 5′-CTAGAACTAGTGGATCCCCC-3′ and the antisense primer 5′-CCAAAAGAGAAACAAAGAAAGAA-3′ derived from Z82982 sequence permitted the amplification of a new clone corresponding to the mature mRNA (CCoAOMT-5 class 2, accession no. AF022775). Using sense primer 5′-GAAACGAGAAAAGCTACAGA-3′ and the antisense primer 5′-TAGGGATAATCATGAGATACA-3′, we isolated a clone (CCoAOMT-6, class 3) basically similar to the Z56282 sequence. Finally, using primers 5′-CCCGAATTCTAGCAACCAATGGAGAAAATGG-3′ and 5′-TATAAAGCTTTTAACTAATGCGTCGGCAAAG-3′ (sense and antisense, respectively), we isolated a new clone of class 1, CCoAOMT-2tr (accession no. AF060180), which presents one nucleotide insertion (a thymidine in the position +247 after the start codon) compared with the CCoAOMT-2 sequence and encodes a truncated protein of 98 amino acids.
Sequence Comparison
The phylogenic tree of tobacco CCoAOMT protein sequences was drawn using the Treeview program (version 1.5, D.M. Rodery, Division of Environmental and Evolutionary Biology, IBLS University of Glasgow, UK, http://taxonomy.zoology.gla.ac. uk/rod/treeview.html).
Heterologous Expression in E. coli Cells, Purification of Recombinant Proteins, and Production of Polyclonal Antibodies
To study substrate specificity of all cloned CCoAOMTs and COMTs, their cDNAs were expressed in E. coli cells. Cloning in the pGEX-KG vector (Sigma), PCR screening for positive clones, and protein purification by chromatography on an agarose-glutathione matrix (Sigma) were performed as previously described (Martz et al., 1998). DNA sequencing was performed by the method of Sanger et al. (1977) using the rhodamin dye-terminator cycle ready kit with AmpliTaq DNA polymerase FS (no. P/N 402078, Perkin-Elmer) and a DNA sequencer (model 373A, Applied Biosystems).
The active purified CCoAOMT-1 (class 1) recombinant protein was used to raise polyclonal antibodies. About 50 to 100 μg of the purified protein was emulsified with 300 μL of either Freund's complete adjuvant for the first injection of antigen, or incomplete adjuvant for all the following boost injections, and was administrated in five intramuscular injections at 5-week intervals. Ten days after each boost immunization, serum was collected. After clot removal, the serum was clarified by centrifugation and stored in small aliquots at −20°C.
SDS-PAGE and Immunoblotting
The basic procedures for electrophoresis under denaturing conditions and for immunoblotting have been described previously (Legrand et al., 1987). The blots were scanned (Argus II AGFA scanner, Agfa-Gevaert, Mortsel, Belgium) and the protein amounts were quantified using imaging software (MacBAS version 2.2, Fuji Photo Film, Tokyo).
RNA Analysis
Relative amounts of CCoAOMT transcripts of different classes were estimated by RT-PCR after a limited number of cycles on a known amount of total RNA allowing quantitative amplification (Martz et al., 1998). The PCR products and known amounts of a quantified DNA Mr marker (MBI) were loaded on agarose gel (1%), stained with ethidium bromide (0.5 mg/mL), and photographed under UV light. Quantification was performed using imaging software. Class 1 CCoAOMTs were amplified using sense primer 5′-GGAAGACATCAAGAAGTTG-3′ and antisense primer 5′-TCATATGATCCATGGTATTT-3′ as previously described (Martz et al., 1998). For classes 2 and 3, the primers were chosen in non-conservative sequences to specifically amplify CCoAOMT transcripts of each class. For class 2, the sense primer was 5′-AATATCAAAGAAATGGCAGA-3′ and the antisense primer 5′-TACGTGCCATGATTCTTTTT-3′. For class 3 the sense and antisense primers were 5′-AACGGT GCAGCACAGGAAAA-3′ and 5′-GAAATCATATGTGCCATGATTA-3′, respectively. To confirm the specificity of each primer combination, we have performed RFLP analyses on the different PCR products. The 430-bp PCR products were purified from agarose gel using a kit (Qiaex-2, Qiagen) and re-suspended in water. An aliquot was digested with AvaI, BglI, FokI, or VspI in addition to EcoRV. The analysis of digestion products on 1% agarose gel stained with ethidium bromide allowed the identification of CCoAOMT sequences. These data confirmed the amplification of only one class of sequence in each case.
Chemical Synthesis of Substrates, Assays of Enzyme Activities, and TLC Analyses
5-Hydroxyferulic acid was synthesized according to the method of Legrand et al. (1978). CoA esters were prepared according to the method of Stöckigt and Zenk (1975), identified, and quantified spectrophotometrically as described by Lüderitz et al. (1982). 5-Hydroxyconiferyl alcohol β-d-glucoside was a generous gift of Professor Kazuhiko Fukushima (Matsui et al., 1996). OMT activities were determined according to the method of Ye et al. (1994). One unit of β-glucosidase (Sigma) was added in the reaction mixture for OMT assays with 5-hydroxyconiferyl alcohol β-d-glucoside. No activity was measured in the absence of β-glucosidase. Kinetic values (Vmax and Km) were determined with the Lineweaver-Burk method at a saturating concentration of S-adenosyl-l-Met. Vmax is expressed in nkat × 10−1 g−1 purified protein and Km in micromolar. The protein content was determined by the method of Bradford (1976) using the Bio-Rad reagent. TLC on cellulose (Polygram, Macherey-Nagel, Dueren, Germany) was performed as described in Martz et al. (1998), and reaction products were identified by comigration with reference compounds.
RESULTS
Different CCoAOMT Isoforms Are Expressed in Tobacco
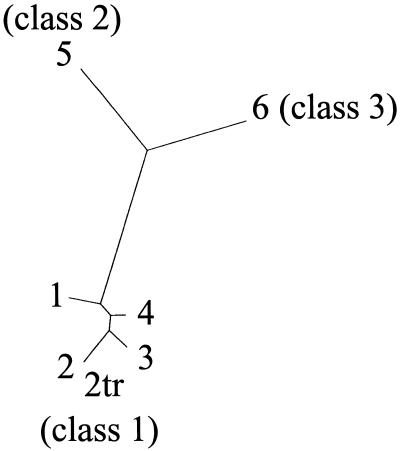
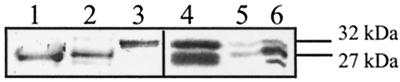
We have previously shown that several classes of CCoAOMT occur in tobacco, and Southern-blot experiments suggested the presence of six to eight genes (Martz et al., 1998). Four homologous CCoAOMT cDNAs (CCoAOMT-1, CCoAOMT-2, CCoAOMT-3, and CCoAOMT-4) that belong to class 1 have been isolated from TMV-infected tobacco leaves (Martz et al., 1998). From sequences available in the database (Busam et al., 1997a) two other complete CCoAOMT cDNAs were cloned by PCR. Phylogenic analysis of CCoAOMT sequences (Fig. 2) shows that these latter cDNAs belong to two other CCoAOMT classes. All of these CCoAOMTs have been expressed in E. coli. The CCoAOMT-1 recombinant protein has been purified and used to raise polyclonal antibodies. Figure 3 shows that these antibodies revealed recombinant proteins from the three CCoAOMT classes with similar efficiencies, as well as native CCoAOMTs extracted from plants.
Figure 2.
Phylogenic tree of CCoAOMT protein sequences of the tobacco cv Samsun NN. 1, 2, 2tr, 3, 4, 5, and 6 correspond to the different CCoAOMT proteins.
Figure 3.
Characterization of recombinant and native isoforms of CCoAOMTs. CCoAOMT recombinant proteins from E. coli cells were purified using a glutathione-agarose affinity matrix. Plant extracts were prepared from healthy tissues or from TMV-infected leaves. Proteins were separated on a SDS-polyacrylamide gel, and, after blotting, CCoAOMT isoforms were detected with polyclonal antibodies raised against tobacco recombinant CCoAOMT-1 (class 1). Lane 1, Recombinant CCoAOMT of class 1; lane 2, recombinant CCoAOMT of class 2; lane 3, recombinant CCoAOMT of class 3; lanes 4 to 6, native isoforms extracted from vascular tissues of stem (lane 4), healthy leaves (lane 5), and 45-h TMV-infected leaves (lane 6). The molecular mass of proteins is shown on the right.
The apparent molecular mass of class 1 and 2 recombinant proteins was 27 kD, close to the value deduced from the nucleotide sequences. Surprisingly, class 3 recombinant protein migrated as a 32-kD species, whereas its calculated mass was not significantly different from that of the two other enzymes. Two main bands of about 27 and 32 kD were also detected in plant extracts, indicating the same kinds of differences between native and recombinant proteins. These data suggest that the 27-kD protein originates from class 1 and 2 CCoAOMT genes, and that class 3 genes encode the 32-kD protein. However, sequence analysis of proteins purified from plant extracts would be needed to confirm this assumption. It is noteworthy that two protein bands were also revealed in tobacco stems with antibodies prepared against zinnia recombinant CCoAOMT (Zhong et al., 1998).
The amounts of the two CCoAOMT isoforms varied in the different tobacco tissues analyzed (Fig. 3, lanes 4, 5, and 6), accumulating in vascular tissues (lane 4) but being only slightly detectable in healthy tobacco leaves (lane 5). Upon infection, the 27-kD species was strongly induced (lane 6). These data indicate that CCoAOMTs have differential patterns of expression in tobacco. An additional band of slightly smaller mass appeared in infected material but was not studied further.
OMT Expression during Stem Development
We have previously shown that COMT of class I is strongly expressed in lignified tissues of the stem, in good correlation with its implication in the synthesis of the syringyl units (Atanassova et al., 1995; Jaeck et al., 1996). Here developmental expression of both COMT I and CCoAOMTs was analyzed at the protein (Fig. 4A) and enzymatic activity levels (Fig. 4B) in different internodes of the stem. Both CCoAOMT and COMT I proteins accumulate throughout the stem but to a lower extent at both extremities (i.e. the oldest and youngest parts). Consistently, enzymatic activities were maximum in the middle of the stem (Fig. 4B). These results suggest a tight coordination between the two OMT activities during the development of the stem, which is in good agreement with their involvement in lignin biosynthesis.
Figure 4.
CCoAOMT and COMT I expression during stem development. Extracts were prepared from stem tissues harvested at different internodes numbered from the bottom to the top of the stem and were immunoblotted with antibodies recognizing CCoAOMTs and COMT I (A) or tested for enzymatic activities in the presence of caffeoyl-CoA or caffeic acid (B).
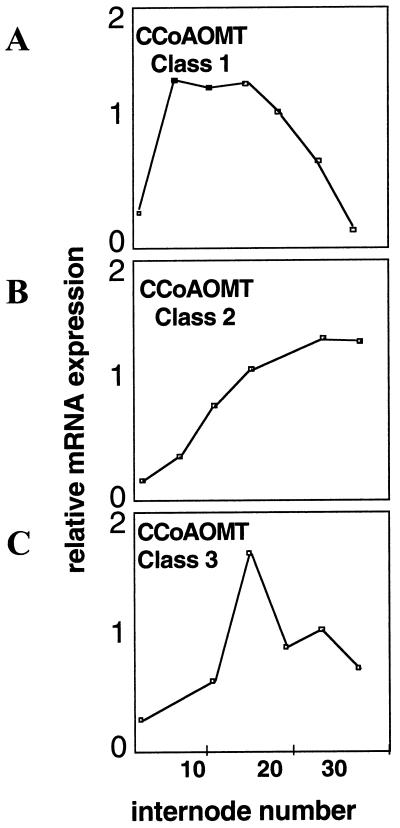
The expression profiles of the three CCoAOMT classes during stem development were studied by RT-PCR using primers specific for each type of sequences and shown to be strikingly different (Fig. 5). The expression level of class 2 (Fig. 5B) steadily increased from the bottom to the top of the plants, whereas class 1 (Fig. 5A) transcripts mostly accumulated in the bottom and middle parts of the stems. Class 3 (Fig. 5C) displayed an intermediate profile. These data indicate that each CCoAOMT class may be involved at specific stages of lignification. A similar observation has been made for two lignin-peroxidase genes in Populus kitakamiensis (Osakabe et al., 1995). These differential regulations may correlate with variations in lignin composition that are known to occur during development (Lewis and Yamamoto, 1990; Baucher et al., 1998).
Figure 5.
Differential expression of the CCoAOMT classes during stem development. CCoAOMT expression was analyzed by reverse transcriptase-PCR using total RNA extracted from different stem internodes numbered from the bottom to the top.
OMT Expression in TMV-Infected Leaves
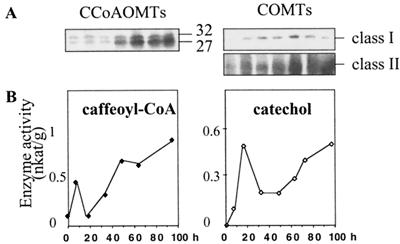
The expression of COMTs of class I and class II is strongly activated in tobacco leaves hypersensitively reacting to TMV (Jaeck et al., 1992; Pellegrini et al., 1993). Time-course curves of CCoAOMT induction by TMV infection were compared with those of COMTs I and II at the protein and activity levels (Fig. 6). Class I and II COMTs differ by their molecular mass and are distinguishable by their positions on immunoblots (Hermann et al., 1987). The intensity of CCoAOMT and COMT immunoreactive bands was evaluated in leaf extracts obtained at various times after inoculation (Fig. 6). Kinetics of accumulation of CCoAOMT isoforms (27- and 32-kD bands) and COMTs of class I and II were similar. A strong induction of enzymes was observed following the appearance of necrotic lesions about 48 h after inoculation. On the other hand, different levels of accumulation were observed for the two CCoAOMT isoforms and for COMT I and II isoforms (Fig. 6A). Assays of methylating activities against caffeoyl-CoA or catechol that are good substrates for CCoAOMT and COMT I and for COMT II, respectively (Legrand et al., 1978, and below), showed the occurrence of two peaks of induction. However, the first response to virus inoculation (about 8 h after inoculation) is likely to represent a wounding response, since it was also detected in mock-inoculated leaves (data not shown).
Figure 6.
Kinetics of CCoAOMT and COMT amounts and enzymatic activities in TMV-infected tobacco leaves. Extracts were prepared from leaf tissue harvested at different times after infection (0, 16, 32, 48, 64, 72, and 96 h). A, Immunoblots with antibodies recognizing the different isoforms of CCoAOMTs and COMTs. B, Enzymatic activities were measured in the presence of caffeoyl-CoA or catechol.
The expression profiles of each CCoAOMT class are presented in Figure 7. Time-course studies of individual gene expression revealed a biphasic pattern of induction similar for the three classes but with a much higher amplitude in class 1 genes (Fig. 7A). These patterns are consistent with those observed at the protein and activity levels (Fig. 6) and suggest that CCoAOMT expression is controlled at the RNA level. These data contrast with those reported in alfalfa cell suspensions, where the accumulation of OMT transcripts after treatment by a fungal elicitor did not lead to increased extractable enzyme activities (Ni et al., 1996).
Figure 7.
Differential expression of the CCoAOMT classes during the hypersensitive reaction of tobacco to TMV. CCoAOMT expression was analyzed by reverse transcriptase-PCR using total RNA extracted from TMV-infected leaves at various times after inoculation.
Comparison of Substrate Specificities of CCoAOMTs and COMTs
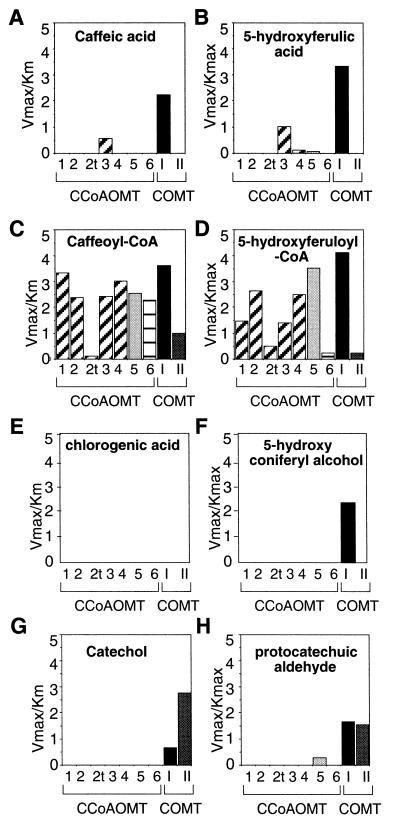
To understand the functional outcomes of the different patterns of expression observed for the distinct lignin O-methyltransferases of tobacco, we analyzed their efficiencies in vitro toward potential o-diphenolic-substrates (Fig. 8). As shown in Figure 1, five phenylpropanoid compounds, caffeic acid, 5-hydroxyferulic acid, their CoA esters, and 5-hydroxyconiferyl alcohol, are metabolic intermediates in the pathway and putative substrates of CCoAOMTs and COMTs. The Vmax/Km values that reflect the efficiency of an enzyme for a given substrate were calculated from Lineweaver-Burk plots. We also tested as substrates chlorogenic acid, since it represents the major pool of caffeate ester in leaves, and catechol and protocatechuic aldehyde, which have been shown previously to be substrates for COMTs of class I and II (Legrand et al., 1978).
Figure 8.
Substrate specificity of CCoAOMT and COMT recombinant proteins. The kinetic values of recombinant tobacco OMTs (seven CCoAOMTs and two COMTs) were determined by the Lineweaver-Burk method at a saturating concentration of S-adenosyl-l-Met, toward caffeic acid, 5-hydroxyferulic acid, the corresponding CoA esters, chlorogenic acid, 5-hydroxyconiferyl alcohol, protocatechuic aldehyde, and catechol. The calculated Vmax to Km ratios represent the mean value of four replicates. Vmax is expressed in nkat × 10 −1 g−1 of purified protein and Km in micromolar.
Among the seven CCoAOMTs tested, only CCoAOMT-3 (class 1) readily methylated free acids (Fig. 8, A and B). Although activity levels were low, TLC analysis confirmed the identity of the reaction products (data not shown). In all cases, CoA esters were better substrates of CCoAOMTs than the free forms. Caffeoyl-CoA was more efficiently methylated than 5-hydroxyferuloyl-CoA by all isoforms except CCoAOMT-2tr (class 1) and CCoAOMT-5 (class 2) (Fig. 8, C and D). CCoAOMT-6 (class 3) appeared to accept almost exclusively caffeoyl-CoA as a substrate. These data are in good agreement with the implication of the CCoAOMT in the synthesis of guaiacyl units in vivo (Fig. 1). The weak but significant activity of the 10-kD species encoded by CCoAOMT-2tr toward 5-hydroxyferuloyl-CoA, together with the occurrence of a 10-kD immunoreactive band in plant extracts (data not shown), supports the functionality of the CCoAOMT-2tr-encoded protein. With the exception of CCoAOMT-5 (class 2), which displayed some activity against protocatechuic aldehyde, CCoAOMTs do not accept chlorogenic acid or non-phenylpropanoid compounds as substrates (Fig. 8, E, G, and H).
With the exception of chlorogenic acid, COMT I was the sole enzyme that proved efficient against all substrates tested. In particular, this is the first report of the substrate specificity of a purified OMT toward 5-hydroxyconiferyl alcohol in vitro. Our results show that only class I COMT can efficiently methylate this substrate, as shown in Figure 8F. With all of the substrates tested, similar results were obtained with COMT I preparations purified from plant extracts (data not shown), suggesting that no posttranslational modifications are involved in substrate specificity. It is particularly striking that Vmax/Km values measured for COMT I were even higher with CoA esters than those calculated for free forms. It is also important to stress that the formation of feruloyl-CoA or sinapoyl-CoA after the incubation of COMTs in the presence of caffeoyl-CoA or 5-hydroxyferuloyl-CoA, respectively, was confirmed by TLC analysis (data not shown).
The preference for CoA esters versus free acid forms was even more pronounced for class II COMT, whose activity against free caffeic acid and 5-hydroxyferulic acid was undetectable under the conditions used (Fig. 8, A–D). These data confirm that catechol is the best substrate for COMT II, as was shown previously (Legrand et al., 1978). The activity of class II COMT against caffeoyl-CoA suggests that in infected leaves, where COMT II is strongly induced, this enzyme preferentially contributes to the synthesis of ferulic derivatives that are known to accumulate upon infection.
DISCUSSION
We have shown previously that tobacco possesses two classes of COMTs (class I and II) that have been purified and cloned (Hermann et al., 1987; Jaeck et al., 1992; Pellegrini et al., 1993). Substrate specificity studies of enzyme preparations purified from plant extracts (Legrand et al., 1978; Colendaveloo et al., 1981) and differential patterns of expression of the two COMT classes in tobacco tissues (Pellegrini et al., 1993; Jaeck et al., 1996) have suggested a specific role for class I enzyme in lignin biosynthesis and the involvement of class II COMT in the production of defense-related compounds. Lignin analysis of plants silenced in COMT expression (Atanassova et al., 1995) demonstrated that in planta COMT I is involved in the second methylating step leading to the syringyl unit, and that CCoAOMT activity is likely responsible for the synthesis of the lignin guaiacyl unit via the feruloyl-CoA ester (Fig. 1). Southern-blot experiments have indicated that several gene classes encode CCoAOMTs of tobacco, and four members of class 1 have been characterized (Martz et al., 1998).
In the present study, our major objective was to better define the role of the different tobacco OMTs. We show that tobacco possesses three CCoAOMT classes and have compared their expression patterns during development and defense responses. Furthermore, heterologous expression of seven distinct CCoAOMT cDNAs, together with cDNAs of COMTs I and II, enabled us to compare substrate specificities of the nine purified recombinant proteins. This led to the very surprising finding that caffeoyl-CoA and 5-hydroxyferuloyl-CoA are the best substrates for all enzymes, even for COMT I (recombinant and native enzymes), which is also very active against free acids (Fig. 8). COMT I activity against 5-hydroxyconiferyl alcohol (Fig. 8) was demonstrated for the first time and raises the possibility that the second methylation step of the pathway catalyzed by COMT I (Atanassova et al., 1995) can occur at the level of 5-hydroxyferulic acid, the corresponding CoA ester, or 5-hydroxyconiferyl alcohol (Fig. 1). The fact that in some species p-coumarate CoA ligases have been reported to have low activity toward sinapic acid (Lee and Douglas, 1996; Allina et al., 1998) argues in favor of the methylation at the level of the CoA ester or the alcohol.
Recent experiments using labeled precursors have demonstrated that coniferyl alcohol may be the precursor of lignin syringyl units in vivo (Matsui et al., 1994; Chen et al., 1999), in agreement with the efficient methylation of 5-hydroxy-coniferyl alcohol by COMT I (see Fig. 1). Recombinant class I COMTs from other plant species have been shown to accept both free acids and CoA esters, but free acids were preferentially accepted compared with cognate CoA esters (Meng and Campbell, 1996, 1998; Inoue et al., 1998). Recently, a COMT from a gymnosperm species, loblolly pine, was reported to share about 60% homology with COMTs of angiosperms and to accept hydroxycinnamoyl-CoA esters and free acids as substrates (Li et al., 1997). In fact, such multifunctionality appears to be a common feature of plant COMTs, with the notable exception of the zinnia enzyme, which did not methylate CoA esters (Ye and Varner, 1995).
A preference for CoA esters versus free acids was also observed for COMT II. Therefore, COMT II, which was known to accept a wide variety of phenolic substrates and was thought to participate in the synthesis of lignin-like compounds (Legrand et al., 1978), may also be involved in the increased deposition of hydroxymethoxycinnamate esters in the walls of plant cells responding to infection or elicitor treatment (Nicholson and Hammerschmitt, 1992; Kauss et al., 1993; Dixon and Paiva, 1995).
It has been reported that CCoAOMTs are encoded by one to two members in parsley (Grimmig and Matern, 1997), alfalfa (Inoue et al., 1998), aspen (Meng and Campbell, 1998), grapevine (Busam et al., 1997b), and five to 10 members in zinnia (Ye et al., 1994). Similar genomic complexity associated with distinct regulation of the different members of a gene family (Liang et al., 1989; Shufflebottom et al., 1993; Osakabe et al., 1995; Allina et al., 1998) and distinct substrate specificity of the different isoforms (Goffner et al., 1998; Hu et al., 1998) have also been described for different phenylpropanoid enzymes. These observations could explain, at least in part, the complex distribution of the phenylpropanoid compounds throughout the plant.
Our results suggest a role for each class of CCoAOMT genes at a specific stage(s) of lignification. The isolation of typical member(s) of each CCoAOMT class would be the first step toward the characterization of regulatory elements and transcriptional factors involved in spatiotemporal control of CCoAOMT gene expression. Recent studies have demonstrated that MYB-like factors control phenylpropanoid gene expression (Rushton and Somssich, 1998; Tamagnone et al., 1998) and are induced in TMV-infected tobacco leaves (Yang and Klessig, 1996). In situ hybridization experiments have demonstrated the accumulation of COMT I transcripts mainly in young xylem cells of tobacco stems and in the epidermis of TMV-infected leaves (Jaeck et al., 1996). In contrast, COMT II expression was undetectable in healthy tobacco (Pellegrini et al., 1993), but after infection by TMV, COMT II transcripts accumulated in all cell types of leaf tissues (L. Pellegrini, unpublished data) as observed for Phe ammonia-lyase transcripts (Pellegrini et al., 1994).
Infection of tobacco leaves by TMV induced COMT I, COMT II, and CCoAOMT expression at the protein and activity levels. Detailed kinetics studies uncovered two peaks of induction: an early one that was also detectable in wounded leaves, and a second one emerging at the time of necrotic lesion appearance. Moreover, the analysis of individual expression of CCoAOMT classes demonstrated that the three CCoAOMT classes were induced with similar kinetics in infected leaves, and that CCoAOMT transcripts of class 1 accumulated predominantly (Fig. 7A). Thus, the coordinated expression of CCoAOMTs and COMTs may provide the hypersensitively reacting plant cells with a variety of metabolites needed for wall reinforcement (Lagrimini, 1991; Campbell and Ellis, 1992a, 1992b; Nicholson and Hammerschmitt, 1992; Kauss et al., 1993). The fact that all tobacco OMTs accept at least one hydroxycinnamoyl-CoA ester as a substrate fosters the view that feruloyl-CoA and sinapoyl-CoA play a central role in the esterification process of cell wall polymers (Grisebach, 1981; Legrand, 1983; Schmitt et al., 1991; Ishii, 1997).
Recently, transgenic tobacco plants with reduced CCoAOMT expression have been shown to display a marked decrease in lignin content with no abnormal visible phenotype (Zhong et al., 1998). Our unpublished results (S. Maury, P. Geoffroy, and M. Legrand), however, demonstrate that CCoAOMT inhibition may affect plant growth and resistance to pathogens. Analysis of phenolic content of such plants should bring new insights into the functions of phenylpropanoids in plant physiology and stress response.
ACKNOWLEDGMENTS
We thank Dr. B. Fritig for helpful discussions and continuous interest. We are grateful to Prof. Kazuhiko Fukushima for providing samples of 5-hydroxyconiferyl alcohol β-d-glucoside. We thank P. Keltz and R. Wagner for taking good care of tobacco plants, M. Meyer and B. Jessel for technical assistance in antibody production, and P. Hammann for DNA sequencing.
Footnotes
This work was supported by the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (grant no. ACC–SV14) and by the Commission of European Communities (FAIR-TIMBER grant no. CT 95–0424).
LITERATURE CITED
- Allina SM, Pri-Hadash A, Theilmann DA, Ellis BE, Douglas CJ. 4-Coumarate: coenzyme A ligase in hybrid poplar. Plant Physiol. 1998;116:743–754. doi: 10.1104/pp.116.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier MT, Monties B, Fritig B, Legrand M. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995;8:465–477. [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W. Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci. 1998;17:125–197. [Google Scholar]
- Boudet AM, Lapierre C, Grima-Pettenati J. Biochemistry and molecular biology of lignification. New Phytol. 1995;129:203–236. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Busam G, Grimmig B, Kneusel RE, Matern U. Isolation of tobacco cDNAs encoding caffeoyl-CoA 3-O-methyltransferase. Plant Physiol. 1997a;113:1003. [Google Scholar]
- Busam G, Junghanns KT, Kneusel RE, Kassemeyer HH, Matern U. Characterization and expression of caffeoyl-coenzyme A 3-O-methyltransferase proposed for the induced resistance response of Vitis vinifera L. Plant Physiol. 1997b;115:1039–1048. doi: 10.1104/pp.115.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MM, Ellis BE. Fungal elicitor-mediated responses in pine cell cultures. I. Induction of phenylpropanoid metabolism. Planta. 1992a;186:409–417. doi: 10.1007/BF00195322. [DOI] [PubMed] [Google Scholar]
- Campbell MM, Ellis BE. Fungal elicitor-mediated responses in pine cell cultures: cell wall-bound phenolics. Phytochemistry. 1992b;31:737–742. [Google Scholar]
- Campbell MM, Sederoff RR. Variation in lignin content and composition: mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CCS, Vogt T, Ellis BE, Somerville CR. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Yasuda S, Fukushima K. Evidence for a novel biosynthetic pathway that regulates the ratio of syringyl to guaiacyl residues in lignin in the differentiating xylem of Magniolia kobus DC. Planta. 1999;207:597–603. [Google Scholar]
- Collendavelloo J, Legrand M, Geoffroy P, Barthelemy J, Fritig B. Purification and properties of the three o-diphenol O-methyltransferases of tobacco leaves. Phytochemistry. 1981;20:611–616. [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffner D, Van Doorsselaere J, Yahiaoui N, Samaj J, Grima-Pettenati J, Boudet AM. A novel aromatic alcohol dehydrogenase in higher plants: molecular cloning and expression. Plant Mol Biol. 1998;36:755–765. doi: 10.1023/a:1005991932652. [DOI] [PubMed] [Google Scholar]
- Grimmig B, Matern U. Structure of the parsley caffeoyl-CoA O-methyltransferase gene, harbouring a novel elicitor responsive cis-acting element. Plant Mol Biol. 1997;33:323–341. doi: 10.1023/a:1005780529457. [DOI] [PubMed] [Google Scholar]
- Grisebach H (1981) Lignins. In EE Conn, ed, The Biochemistry of Plants. Academic Press, New York, pp 457–478
- Hermann C, Legrand M, Geoffroy P, Fritig B. Enzymatic synthesis of lignin: purification to homogeneity of the three O-methyltransferases of tobacco and production of specific antibodies. Arch Biochem Biophys. 1987;253:367–376. doi: 10.1016/0003-9861(87)90190-1. [DOI] [PubMed] [Google Scholar]
- Hu WJ, Kawaoka A, Tsai CJ, Lung J, Osakabe K, Ebinuma H, Chiang VL. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides) Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Murray Ballance G, Ni W, Stürzer C, Dixon RA. Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 1998;117:761–770. doi: 10.1104/pp.117.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127:111–127. [Google Scholar]
- Jaeck E, Dumas B, Geoffroy P, Favet N, Inzé D, Van Montagu M, Fritig B, Legrand M. Regulation of enzymes involved in lignin biosynthesis: induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol Plant-Microbe Interact. 1992;5:294–300. doi: 10.1094/mpmi-5-294. [DOI] [PubMed] [Google Scholar]
- Jaeck E, Martz F, Stiefel V, Fritig B, Legrand M. Expression of tobacco class I O-methyltransferase in healthy and TMV-infected tobacco. Mol Plant-Microbe Interact. 1996;9:681–688. doi: 10.1094/mpmi-9-0681. [DOI] [PubMed] [Google Scholar]
- Kauss H, Franke R, Krause K, Conrath U, Jeblick W, Grimmig B, Matern U. Conditioning of parsley (Petroselinum crispum L.) suspension cells increases elicitor-induced incorporation of cell wall phenolics. Plant Physiol. 1993;102:459–466. doi: 10.1104/pp.102.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E. Elicitor-induced S-adenosyl-l-methionine:caffeoyl-CoA 3-O-methyltransferase from carrot cell suspension cultures. Plant Sci. 1989;60:21–25. [Google Scholar]
- Lagrimini LM. Wound-induced deposition of polyphenols in transgenic plants overexpressing peroxidase. Plant Physiol. 1991;96:577–583. doi: 10.1104/pp.96.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Tollier M-T, Monties B. Mise en évidence d'un nouveau type d'unité constitutive dans les lignines d'un mutant de maïs bm3. C R Acad Sci Paris. 1988;307:723–728. [Google Scholar]
- Lee D, Douglas CJ. Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. Plant Physiol. 1996;112:193–205. doi: 10.1104/pp.112.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M. Phenylpropanoid metabolism and its regulation in disease. In: Callow JA, editor. Biochemical Plant Pathology. Chichester, UK: John Wiley & Sons; 1983. pp. 367–384. [Google Scholar]
- Legrand M, Fritig B, Hirth L. o-Diphenol O-methyltransferases of healthy and tobacco-mosaic-virus-infected hypersensitive tobacco. Planta. 1978;144:101–108. doi: 10.1007/BF00385014. [DOI] [PubMed] [Google Scholar]
- Legrand M, Kauffmann S, Geoffroy P, Fritig B. Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci USA. 1987;84:6750–6754. doi: 10.1073/pnas.84.19.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Li L, Popko JL, Zhang XH, Osakabe K, Tsai CJ, Joshi CP, Chiang VL. A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc Natl Acad Sci USA. 1997;94:5461–5466. doi: 10.1073/pnas.94.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Dron M, Cramer CL, Dixon RA, Lamb CJ. Differential regulation of phenylalanine ammonia lyase genes during plant development and by environmental cues. J Biol Chem. 1989;264:14486–14492. [PubMed] [Google Scholar]
- Lüderitz T, Shatz G, Grisebach H. Enzymic synthesis of lignin precursors: purification and properties of 4-coumarate:CoA ligase from cambial sap of spruce (Picea abies L.) Eur J Biochem. 1982;123:583–586. [PubMed] [Google Scholar]
- Martz F, Maury S, Pinçon G, Legrand M. cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyltransferase, a lignin biosynthetic enzyme. Plant Mol Biol. 1998;36:427–437. doi: 10.1023/a:1005969825070. [DOI] [PubMed] [Google Scholar]
- Matern U, Grimmig B, Kneusel RE. Plant cell wall reinforcement in the disease resistance response: molecular composition and regulation. Can J Bot. 1995;73:551–571. [Google Scholar]
- Matsui N, Fukushima K, Terashima N, Yasuda S. Synthesis of radio-labeled monolignol glucosides that have 3- and 5-hydroxy group on aromatic ring. Mokuzai Gakkaishi. 1996;42:1020–1024. [Google Scholar]
- Matsui N, Fukushima K, Yasuda S, Terashima N. On the behavior of monolignol glucosides in lignin biosynthesis. Holzforschung. 1994;48:375–380. [Google Scholar]
- Meng H, Campbell WH. Characterization and site-directed mutagenesis of aspen lignin-specific O-methyltransferase expressed in Escherichia coli. Arch Biochem Biophys. 1996;330:329–341. doi: 10.1006/abbi.1996.0260. [DOI] [PubMed] [Google Scholar]
- Meng H, Campbell WH. Substrate profiles and expression of caffeoyl coenzyme A and caffeic acid O-methyltransferases in secondary xylem of aspen during seasonal development. Plant Mol Biol. 1998;38:513–520. doi: 10.1023/a:1006071708728. [DOI] [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Sewalt VJH, Korth KL, Blount JW, Ballance GM, Dixon RA. Stress responses in alfalfa. Plant Physiol. 1996;112:717–726. doi: 10.1104/pp.112.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R. Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol. 1992;30:369–389. [Google Scholar]
- Osakabe K, Koyama H, Kawai S, Katayama Y, Morohoshi N. Molecular cloning of two tandemly arranged peroxidase genes from Populus kitakamiensis and their differential regulation in the stem. Plant Mol Biol. 1995;28:677–689. doi: 10.1007/BF00021193. [DOI] [PubMed] [Google Scholar]
- Pakusch A-E, Kneusel RE, Mattern U. S-Adenosyl-l-methionine:trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys. 1989;271:488–494. doi: 10.1016/0003-9861(89)90299-3. [DOI] [PubMed] [Google Scholar]
- Pakusch A-E, Matern U. Kinetic characterization of caffeoyl-coenzyme A specific 3-O-methyltransferase from elicited parsley cell suspensions. Plant Physiol. 1991;96:327–330. doi: 10.1104/pp.96.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakusch A-E, Matern U, Schiltz E. Elicitor-inducible caffeoyl-coenzyme A 3-O-methyltransferase from Petroselinum crispum cell suspensions. Plant Physiol. 1991;95:137–143. doi: 10.1104/pp.95.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M. Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco leaves by infection or elicitor treatment. Plant Physiol. 1993;103:509–517. doi: 10.1104/pp.103.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Rohfritsch O, Fritig B, Legrand M. Phenylalanine ammonia-lyase in tobacco: molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor. Plant Physiol. 1994;106:877–886. doi: 10.1104/pp.106.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt D, Pakusch A-E, Matern U. Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem. 1991;266:17416–17423. [PubMed] [Google Scholar]
- Shufflebottom D, Edwards K, Schuch W, Bevan M. Transcription of two members of a gene family encoding phenylalanine ammonia-lyase leads to remarkably different cell specificities and induction patterns. Plant J. 1993;3:835–845. doi: 10.1111/j.1365-313x.1993.00835.x. [DOI] [PubMed] [Google Scholar]
- Stöckigt J, Zenk MH. Chemical synthesis and properties of hydroxycinnamoyl-coenzyme A derivatives. Z Naturforsch. 1975;30c:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorsselaere J, Baucher M, Chignot E, Chabbert B, Tollier MT, Petit-Conil M, Leplé JC, Pilate G, Cornu D, Monties B, Van Montagu M, Inzé D, Boerjan W, Jouanin L. A novel lignin in poplar trees with a reduced caffeic acid/5-hydroxyferulic acid O-methyltransferase activity. Plant J. 1995;8:855–864. [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff RR. Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]
- Yang Y, Klessig DF. Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc Natl Acad Sci USA. 1996;93:14972–14977. doi: 10.1073/pnas.93.25.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Kneusel RE, Matern U, Varner JE. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell. 1998;10:2033–2046. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]