Abstract
Purpose
Studies demonstrating bacterial DNA and cultivable bacteria in urine samples have challenged the clinical dogma that urine is sterile. Furthermore, studies now indicate that dysbiosis of the urinary microbiome is associated with pathological conditions. We propose that the urinary microbiome may influence chronic inflammation observed in the prostate, leading to prostate cancer development and progression. Therefore, we profiled the urinary microbiome in men with positive vs negative biopsies for prostate cancer.
Materials and Methods
Urine was collected from men prior to biopsy for prostate cancer. DNA was extracted from urine pellet samples and subjected to bacterial 16S rDNA Illumina® sequencing and 16S rDNA quantitative polymerase chain reaction. We determined the association between bacterial species and the presence or absence of cancer, cancer grade, and type and degree of prostate inflammation.
Results
Urine samples revealed diverse bacterial populations. There were no significant differences in α or β diversity and no clear hierarchical clustering of benign or cancer samples. We identified a cluster of pro-inflammatory bacteria previously implicated in urogenital infections in a subset of samples. Many species, including known uropathogens, were significantly and differentially abundant among cancer and benign samples, in low vs higher grade cancers and in relation to prostate inflammation type and degree.
Conclusions
To our knowledge we report the most comprehensive study to date of the male urinary microbiome and its relationship to prostate cancer. Our results suggest a prevalence of pro-inflammatory bacteria and uropathogens in the urinary tract of men with prostate cancer.
Keywords: prostatic neoplasms, urinary tract, microbiota, inflammation, bacteria
Advanced sequencing technologies for culture independent molecular microbiology have revealed the unexpected presence of microbial populations in areas of the human body once thought to be devoid of microbes. As a chief example, the long held clinical dogma that urine is sterile was disproved1 by the demonstration of populations of live bacteria that reside in the urinary tract.1-4 While there are shared species in the urinary tract and those found in microbial communities associated with skin, gastrointestinal tract and vagina, studies currently suggest that the urinary tract contains microbial populations that are distinct from those at other sites of the human body that harbor microbiota.1,5-7 Because the urinary microbiome was excluded from seminal studies such as HMP (Human Microbiome Project), which characterized the healthy human microbiota,8 studies profiling healthy urinary microbiota are less mature, as is our understanding of how urinary microbial populations may shift with disease.1
Chronic inflammation due to prostate infection is hypothesized to contribute to prostate cancer development and/or progression.9-11 Indeed, several recent studies highlight a mechanistic role for inflammation induced by infection or bacterial components (eg lipopolysaccharides) in the formation of hallmark prostate cancer ETS gene fusions12 and in the expansion of intermediate cells,13 the hypothesized cells where neoplastic transformation in prostate cancer may commence.14 Given this recent experimental evidence linking bacterial infections to prostate cancer development, there is a pressing need to understand how, when and what microorganisms may be introduced in the prostate.
To our knowledge no single microorganism to date has been recognized to contribute to prostate cancer etiology and yet several species of bacteria are known or suspected to induce prostatic inflammation, as in the case of symptomatic bacterial prostatitis related microbes, certain sexually transmitted infections and the pro-inflammatory bacterium Propionibacterium acnes.15-19 The recent discovery of a urinary microbiome sheds new light on the possibility of frequent exposure of the prostate to a diverse number of microorganisms due to anatomical proximity, eg the urethra runs through the prostate and the prostatic ducts feed into the prostatic urethra. Therefore, the urinary tract may serve as a route of exposure of the prostate to microorganisms contained in the urethra.
We hypothesized that microorganisms that may have an etiological role in prostate cancer originate from the urinary tract and men with prostate cancer may show dysbiotic urinary microbial signatures which are more likely to contain opportunistic pathogens. In the current study we characterized the urinary microbiota in a cohort of men with or without a biopsy proven diagnosis of prostate cancer. We identified species that were differentially abundant with features such as the presence of cancer, cancer grade and the degree of acute or chronic prostatic inflammation.
MATERIALS AND METHODS
Supplementary figure 1 (http://jurology.com/) shows an overview of the study schema.
Study Design and Patient Population
All specimens were obtained under a Johns Hopkins Medicine institutional review board approved protocol. Urine samples were collected from men prior to antibiotic prophylaxis and prostate biopsy at The Johns Hopkins Hospital for suspicion of prostate cancer. Urine samples were collected after digital rectal examination. The men were undergoing a first prostate biopsy or had not been biopsied for 1 year or more before urine sample collection. None of the men were on antibiotics at the time of sample collection. The table lists the clinical and pathological details of the men included in study. Notably the samples in the benign group were significantly larger according to average TRUS volume than in the cancer group, indicating that this group was enriched for patients with prostate enlargement.
table.
Patient clinical characteristics
| No. Pts | Mean Age (range) | p Value | Mean ng/ml PSA (range) | p Value | Mean cc TRUS Vol (range) | p Value | |
|---|---|---|---|---|---|---|---|
| Benign | 65 | 62.8 (43–78) | – | 6.9 (0.6–25.7) | – | 64.2 (24–227) | – |
| Ca | 65 | 62.8 (41–79) | >0.99 | 6.2 (1.7–30) | 0.380 | 38.1 (13.8–113.1) | <0.0001 |
| Biopsy Ca Gleason grade: | 65 | ||||||
| 6 | 43 | 63.5 (49–79) | 6.4 (1.7–30) | 40.1 (13.8–113.1) | |||
| 7+ | 22 | 61.4 (41–75) | 0.312 | 5.9 (3.8–16.7) | 0.604 | 31.3 (15–69) | 0.130 |
| Biopsy Ca clinical stage: | 65 | – | – | – | – | – | – |
| T1c | 57 | ||||||
| T2a | 4 | ||||||
| T2b | 2 | ||||||
| Tx | 2 | ||||||
| Subsequent biopsy Ca | 5 | 56.2 (49–64) | 0.045,* 0.069† | 3.7 (2.5–6.3) | 0.175,* 0.186† | 53.5 (44–66.7) | 0.598,* 0.130† |
Benign compared to cancer on subsequent biopsy.
Cancer compared to cancer on subsequent biopsy.
Sample Collection and DNA Isolation
The 30 ml urine samples were handled using sterile technique and were pelleted by centrifugation at 1,000 × gravity for 10 minutes within less than 4 hours of collection. Urine pellets were resuspended in a total volume of 500 μl 1 × phosphate buffered saline and DNA was extracted (supplementary Methods, http://jurology.com/). A total of 16 mock (500 μl 1 × phosphate buffered saline as starting material) DNA extractions were performed to control for contamination from DNA extraction through the full amplification and sequencing pipeline.
16S rDNA Gene Library Preparation and Sequencing
The V6 hypervariable region of the 16S rRNA gene was amplified using a 2-step PCR strategy (supplementary Methods, http://jurology.com/). PCR products were visualized on agarose gel, gel extracted and pooled before submission to the Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing Core at The Johns Hopkins Hospital for next generation sequencing on a HiSeq™ instrument.
Universal 16S rDNA Real-Time Polymerase Chain Reaction
A universal 16S rDNA qPCR assay was developed using the first round V6 primer set. The number of 16S rDNA copies were quantified in relation to a standard curve of known copies of Escherichia coli DNA. The supplementary Methods (http://jurology.com/) show additional details of sequence analysis, inflammation analysis, organism specific PCR and statistical analyses.
RESULTS
Urinary Microbiome Characterization
A total of 135 urine samples were analyzed, including 65 from men with benign biopsies, 65 from men diagnosed with prostate cancer and 5 from men with an initial benign biopsy who were later diagnosed with prostate cancer on subsequent biopsy (see table). Biopsy Gleason grade was used to differentiate low grade—Gleason 6 (ISUP Grade Group I) in 43 men from higher grade—Gleason 7+ (ISUP Grade Group II or greater) in 22 men. In the higher grade group 16 cancers were 3 + 4 = 7 (ISUP Grade Group II), 5 cancers were 4 + 3 = 7 (ISUP Grade Group III) and 1 cancer was 4 + 4 = 8 (ISUP Grade Group IV). More samples from men with high risk prostate cancer were not available in the current study. They will be the focus of followup studies.
After excluding samples with low sequencing reads we characterized the bacterial composition of urine samples from 129 men, including 63 diagnosed as benign, 61 with prostate cancer and 5 with cancer on SB. A total of 2,457 bacterial OTUs were identified in the urine samples after sequences were rarified to 5,000 reads per sample. An average of 64 OTUs were present in each sample with an average of 60 OTUs in benign samples, 67 OTUs in cancer samples and 79 OTUs in cancer on SB samples. No significant difference in α or β diversity was found between the bacterial profiles of cancer and benign urine samples (supplementary fig. 2, http://jurology.com/).
The urine samples were each often predominated by a single genus of bacteria, in particular Corynebacterium, Staphylococcus and Streptococcus. Also, select samples were predominated (greater than 80% of reads) by Anaerococcus, Lactobacillus, Actinobaculum and an unassigned genus in the family Enterobacreriaceae (fig. 1). The 16S rDNA qPCR analyses indicated that the bacterial load was low in most urine samples (fig. 2, A). The mock (negative control) samples were quantitatively lower than most urine pellet samples but they a had detectable 16S rDNA signal (fig. 2, A). Detectable signal in mock samples is not unexpected as the 16S rDNA PCR assay is sensitive and it is difficult or impossible to eliminate all bacterial DNA from reagents, tubes, etc.
Figure 1.
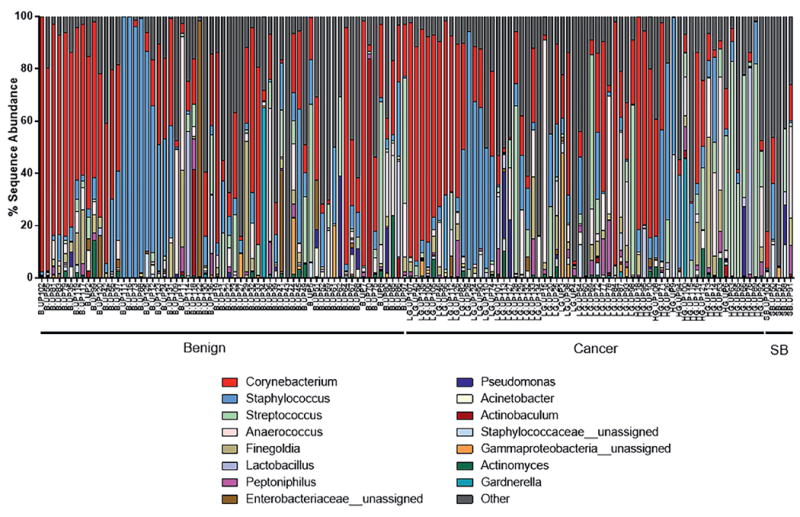
Genera identified in urine pellet samples from men with or without biopsy proven prostate cancer diagnosis. Stacked bar plots represent sequence abundances of 15 most abundant genus or family level taxa identified. Percent sequence abundance is shown as number of reads matching given genus per total reads for that sample. SB, prostate cancer diagnosed on subsequent biopsy.
Figure 2.
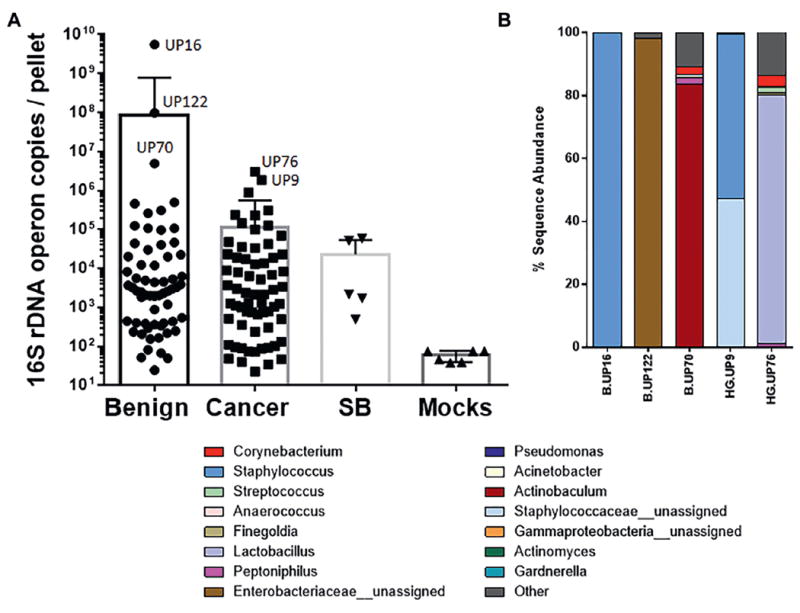
Bacterial load in urine pellet samples. A, load was determined by 16S rDNA qPCR and is shown as estimated number of operon copies of 16S rDNA present per urine pellet. Mocks, mock DNA extractions. B, percent sequence abundance of genus or family level taxa in samples with highest bacterial load (A).
Careful measures were taken to filter potential contaminant reads from the data set (supplementary Methods, http://jurology.com/). We did not observe a significant association between bacterial load and TRUS volume, reference PSA or inflammation level (data not shown).
Bacterial Taxa Association with Pathological Features
Unsupervised clustering analyses did not indicate a clear clustering of benign or cancer samples at the genus or species level (figs. 3 and 4). Of interest is that we identified a clustered group of bacterial species that proportionally contained more cancer samples than the samples outside the cluster (17 of 24 or 70.8% vs 49 of 105 or 46.7%, Fisher exact test p = 0.041, fig. 5). This cluster contained the species Streptococcus anginosus, Anaerococcus lactolyticus, Anaerococcus obesiensis, Actinobaculum schaalii, Varibaculum cambriense and Propionimicrobium lymphophilum. Almost all of these species have been implicated as causative agents in urogenital infections, including prostatitis, bacterial vaginosis and urinary tract infections.20-24 Several species were found to be more abundant in cancer samples than in benign urine samples (fig. 6).
Figure 3.
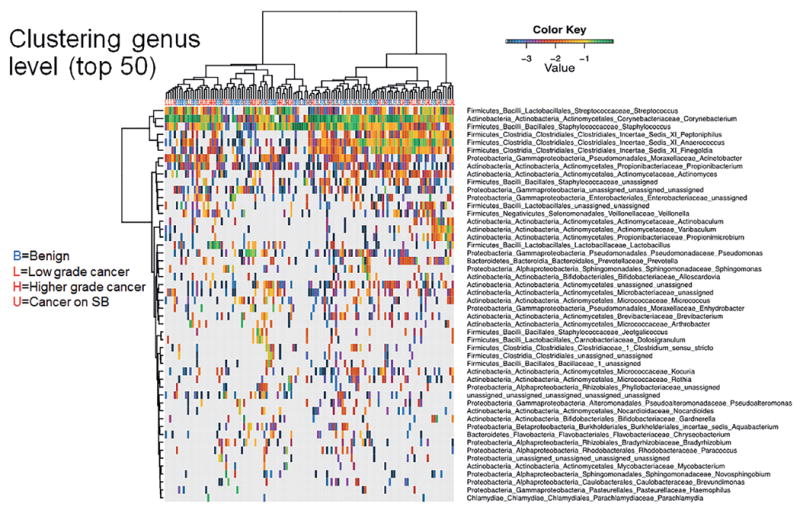
Dendrogram shows log transformed unsupervised clustering of 16S rDNA Illumina sequencing results from urine pellet samples by genus based on hierarchical clustering of Euclidean distance between samples in combined benign, cancer and SB groups.
Figure 4.

Dendrogram shows log transformed unsupervised clustering of 16S rDNA Illumina sequencing results from urine pellet samples by species based on hierarchical clustering of Euclidean distance between samples in combined benign, cancer and SB groups. Boxed area represents species level cluster (fig. 5).
Figure 5.
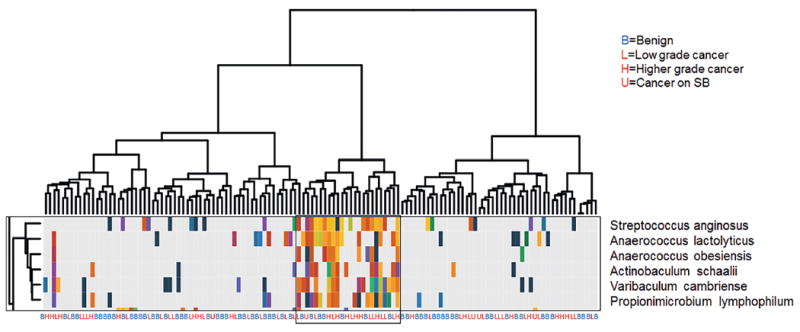
Cluster of pro-inflammatory bacterial species differentially present in cancer samples identified in hierarchical clustering of all samples (fig. 4).
Figure 6.
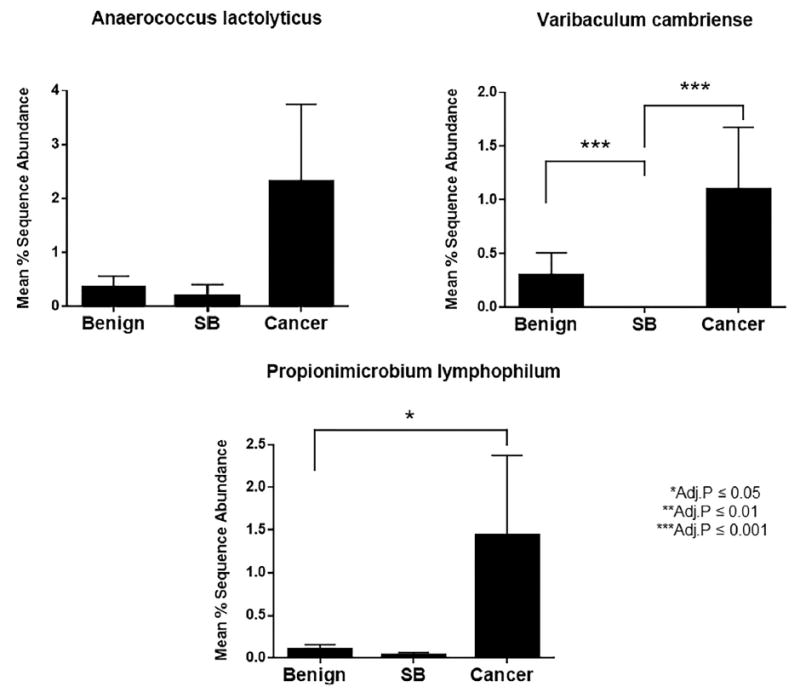
Mean percent differential abundance of select species of bacteria in benign, cancer and cancer SB samples. Adj., adjusted.
No difference in β diversity was found among the level of acute or chronic inflammation, Gleason score, reference PSA or TRUS volume (supplementary figs. 3 to 7 Almost all of these species have been implicated as causative agents in urogenital infections, including prostatitis, bacterial vaginosis and urinary tract infection, http://jurology.com/). Many additional species were significantly and differentially abundant among cancer and benign samples, low vs higher grade cancers, the presence or absence of acute inflammation and the degree of chronic inflammation. The supplementary table (http://jurology.com/) highlights the results.
Pathogen Detection
In addition to the described cluster of species implicated in urogenital infections, we identified other known uropathogens in the urine pellet samples. One notable example was 2 OTUs identified as Ureaplasma parvum or Ureaplasma urealyticum, which were differentially represented in cancer samples vs benign samples (supplementary table, http://jurology.com/). Another example was the identification of Gardnerella vaginalis in male urine samples. Although this species was not differentially abundant in cancer vs benign samples, it was more prevalent in samples from men in whom medium or high chronic inflammation was observed in the prostate biopsy (supplementary table, http://jurology.com/).
A species of particular interest to prostate cancer is P. acnes as we and others have reported the presence of this species in prostatectomy tissues.16,17,25 Furthermore, we reported that the strains of P. acnes isolated from prostate tissues were most similar to strains that are implicated in opportunistic infections or reside in the urinary tract as opposed to typical skin strains.18 Unfortunately P. acnes was highly prevalent in the mock negative control samples and many P. acnes reads were filtered out of the sequencing data as contaminants. Therefore, P. acnes was difficult to assess reliably in the Illumina sequencing data.
As an alternative strategy we performed qPCR for P. acnes using species specific primers. This assay confirmed that the level of P. acnes 16S rDNA reads was generally higher in urine pellet samples than in negative controls. However, no significant difference was observed between benign and cancer samples (supplementary fig. 8, http://jurology.com/).
Another organism of interest that would not be detected with the 16S rDNA sequencing approach is the protozoan Trichomonas vaginalis. We developed a qPCR assay for this organism and of the 135 urine pellet samples analyzed 1 was positive for T. vaginalis (data not shown).
Finally, in contrast to the dominant organisms observed in most samples (fig. 1), the samples with the highest bacterial load were often dominated by atypical species (fig. 2). Benign samples UP122 and UP70 were predominated by an unassigned Enterobacteriaceae (possibly E. coli) and Actinobaculum urinale (another potential uropathogen), respectively. Benign sample UP16 was also atypical in that 99.5% of the reads matched to an unassigned Staphylococcus species, indicating an unusually high presence of this species. Likewise, cancer samples UP9 and UP76 had a high abundance of an unassigned Staphylococcaceae and a Lactobacillus species, respectively.
DISCUSSION
Our studies profiling the urinary microbiome in men with and without a biopsy proven diagnosis of prostate cancer revealed that the urinary microbiota of most men is predominated by a single genus and notably by species of Corynebacterium, Staphylococcus and Streptococcus. While similar trends have been reported in urine samples from women, female urine samples differ in that the predominant organisms are Lactobacillus and Gardnerella.2 Interestingly we identified a subset of men with predominant urine representation from Lactobacillus or Gardnerella species (fig. 1). The presence G. vaginalis was associated with chronic inflammation in corresponding prostate biopsies (supplementary table, http://jurology.com/). This raises the intriguing possibility that some men may harbor urinary microbiota associated with inflammatory conditions in women (eg bacterial vaginosis).
Several additional species of pro-inflammatory bacteria and/or known uropathogens were differentially represented in men with prostate cancer. Notable examples included A. schaalii, an emerging uropathogen of potentially underestimated clinical significance due to difficulty with phenotypic identification.22 A. schaalii was found in men with and without prostate cancer but this species was included in the cluster of pro-inflammatory bacteria more prevalent in men with cancer (fig. 5). As mentioned, species of Ureaplasma were also differentially abundant in the urinary microbiota of men with prostate cancer. Although only 5 samples were available in the current study, the cancer on SB samples had the highest average number of OTUs and none showed a predominance of Corynebacterium, Staphylococcus or Streptococcus (fig. 1).
While additional followup studies are needed to validate the finding of differentially represented bacterial species in the urine of men with cancer vs those with a negative biopsy, to our knowledge this is the first study to identify such differential bacterial species in men with prostate cancer.
There were several limitations to the current study, including the fact that all men were being seen for some indication for prostate biopsy. Although men in the benign group were biopsy negative for prostate cancer, they nevertheless represented a population with elevated PSA and were likely to have BPH and/or prostatic inflammation (see table). Indeed, the men in the benign group had a larger average prostate TRUS volume than the men with cancer or cancer on SB (see table), indicating prostate enlargement in this group. As BPH is also associated with chronic inflammation,26-28 future studies warrant an association of urinary microbiota with the presence of BPH. In addition, it is likely that a fraction of the men with a negative biopsy actually had prostate cancer because the false-negative rate of TRUS guided prostate biopsy is commonly reported to be up to 30%.
Another major limitation is that we did not have I-PSS (International Prostate Symptom Score) data on all of the men in the study. Therefore, we could not correlate urinary bacterial species to prostate symptoms. We relied on a large specimen repository of retrospectively collected urine sample in the current series so that we could begin to define the urinary flora in men with prostate cancer. Future followup studies will necessitate a true control population of men without an indication for prostate cancer to determine whether the urinary microbiome profile is unique in those without prostate disease or rather consistent with the control group in the current study.
The key hypothesis that emerged from the current study is that pro-inflammatory species that reside in the urinary tract may serve as a potential source of inciting chronic inflammation in the prostate. Ultimately establishing the link between the urinary microbiome and chronic inflammation in the prostate may be keenly important in terms of developing strategies for prostate cancer prevention.
CONCLUSIONS
To our knowledge we report the most comprehensive study to date of the urinary microbiome in men with and without a biopsy proven diagnosis of prostate cancer. Our results suggest a prevalence of pro-inflammatory bacteria and uropathogens in the urinary tract of men with prostate cancer. Future work will ideally include longitudinal studies to determine whether microbiome signatures associated with disease preexist the development of cancer.
Supplementary Material
Acknowledgments
Supported by NCI (National Cancer Institute) Grant P30CA006973 (Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing Core), the V Foundation for Cancer Research, the Patrick C. Walsh Prostate Cancer Research Fund, the Prostate Cancer Foundation and NCI SPORE P50CA058236.
Abbreviations and Acronyms
- BPH
benign prostatic hyperplasia
- ISUP
International Society of Urological Pathology
- OUT
operational taxonomic unit
- PCR
polymerase chain reaction
- PSA
prostate specific antigen
- QPCR
quantitative real-time PCR
- SB
subsequent biopsy
- TRUS
transrectal ultrasound
Footnotes
No direct or indirect commercial incentive associated with publishing this article.
The corresponding author certifies that, when applicable, a statement(s) has been included in the manuscript documenting institutional review board, ethics committee or ethical review board study approval; principles of Helsinki Declaration were followed in lieu of formal ethics committee approval; institutional animal care and use committee approval; all human subjects provided written informed consent with guarantees of confidentiality; IRB approved protocol number; animal approved project number.
References
- 1.Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract—a role beyond infection. Nat Rev Urol. 2015;12:81. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 2.Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5:e01283. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis DA, Brown R, Williams J, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. 2013;3:41. doi: 10.3389/fcimb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DE, Van Der Pol B, Dong Q, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson DE, Dong Q, Van Der Pol B, et al. Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS One. 2012;7:e36298. doi: 10.1371/journal.pone.0036298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abernethy MG, Rosenfeld A, White JR, et al. Urinary microbiome and cytokine levels in women with interstitial cystitis. Obstet Gynecol. 2017;129:500. doi: 10.1097/AOG.0000000000001892. [DOI] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium: Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sfanos KS, Isaacs WB, De Marzo AM. Infections and inflammation in prostate cancer. Am J Clin Exp Urol. 2013;1:3. [PMC free article] [PubMed] [Google Scholar]
- 10.De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sfanos KS, De Marzo AM. Prostate cancer and inflammation: the evidence. Histopathology. 2012;60:199. doi: 10.1111/j.1365-2559.2011.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani RS, Amin MA, Li X, et al. Inflammation-induced oxidative stress mediates gene fusion formation in prostate cancer. Cell Rep. 17:2620. doi: 10.1016/j.celrep.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon OJ, Zhang L, Ittmann MM, et al. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci U S A. 2014;111:E592. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Leenders GJ, Gage WR, Hicks JL, et al. Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am J Pathol. 2003;162:1529. doi: 10.1016/S0002-9440(10)64286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutcliffe S, Zenilman JM, Ghanem KG, et al. Sexually transmitted infections and prostatic inflammation/cell damage as measured by serum prostate specific antigen concentration. J Urol . 2006;175:1937. doi: 10.1016/S0022-5347(05)00892-X. [DOI] [PubMed] [Google Scholar]
- 16.Sfanos KS, Sauvageot J, Fedor HL, et al. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate. 2008;68:306. doi: 10.1002/pros.20680. [DOI] [PubMed] [Google Scholar]
- 17.Cohen RJ, Shannon BA, McNeal JE, et al. Pro-pionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J Urol. 2005;173:1969. doi: 10.1097/01.ju.0000158161.15277.78. [DOI] [PubMed] [Google Scholar]
- 18.Mak TN, Yu SH, De Marzo AM, et al. Multilocus sequence typing (MLST) analysis of Propionibacterium acnes isolates from radical prostatectomy specimens. Prostate. 2013;73:770. doi: 10.1002/pros.22621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinohara DB, Vaghasia AM, Yu SH, et al. A mouse model of chronic prostatic inflammation using a human prostate cancer-derived isolate of Propionibacterium acnes. Prostate. 2013;73:1007. doi: 10.1002/pros.22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiley RA, Beighton D, Winstanley TG, et al. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243. doi: 10.1128/jcm.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kline KA, Lewis AL. Gram-positive uropath-ogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.UTI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattoir V. Actinobaculum schaalii: review of an emerging uropathogen. J Infect. 2012;64:260. doi: 10.1016/j.jinf.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Chu YW, Wong CH, Chu MY, et al. Varibaculum cambriense infections in Hong Kong, China, 2006. Emerg Infect Dis. 2009;15:1137. doi: 10.3201/eid1507.081291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams GD. Two cases of urinary tract infection caused by Propionimicrobium lymphophilum. J Clin Microbiol. 2015;53:3077. doi: 10.1128/JCM.00438-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sfanos KS, Isaacs WB. An evaluation of PCR primer sets used for detection of Propionibacterium acnes in prostate tissue samples. Prostate. 2008;68:1492. doi: 10.1002/pros.20820. [DOI] [PubMed] [Google Scholar]
- 26.Nickel JC. Inflammation and benign prostatic hyperplasia. Urol Clin North Am. 2008;35:109. doi: 10.1016/j.ucl.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fibbi B, Penna G, Morelli A, et al. Chronic inflammation in the pathogenesis of benign prostatic hyperplasia. Int J Androl. 2010;33:475. doi: 10.1111/j.1365-2605.2009.00972.x. [DOI] [PubMed] [Google Scholar]
- 28.Gandaglia G, Briganti A, Gontero P, et al. The role of chronic prostatic inflammation in the pathogenesis and progression of benign prostatic hyperplasia (BPH) BJU Int. 2013;112:432. doi: 10.1111/bju.12118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


