Abstract
Systems biology is the study of complex living organisms, and as such, analysis on a systems-wide scale involves the collection of information-dense data sets that are representative of an entire phenotype. To uncover dynamic biological mechanisms, bioinformatics tools have become essential to facilitating data interpretation in large-scale analyses. Global metabolomics is one such method for performing systems biology, as metabolites represent the downstream functional products of ongoing biological processes. We have developed XCMS Online, a platform that enables online metabolomics data processing and interpretation. A systems biology workflow recently implemented within XCMS Online enables rapid metabolic pathway mapping using raw metabolomics data for investigating dysregulated metabolic processes. In addition, this platform supports integration of multi-omic (such as genomic and proteomic) data to garner further systems-wide mechanistic insight. Here, we provide an in-depth procedure showing how to effectively navigate and use the systems biology workflow within XCMS Online without a priori knowledge of the platform, including uploading liquid chromatography (LCLC)–mass spectrometry (MS) data from metabolite-extracted biological samples, defining the job parameters to identify features, correcting for retention time deviations, conducting statistical analysis of features between sample classes and performing predictive metabolic pathway analysis. Additional multi-omics data can be uploaded and overlaid with previously identified pathways to enhance systems-wide analysis of the observed dysregulations. We also describe unique visualization tools to assist in elucidation of statistically significant dysregulated metabolic pathways. Parameter input takes 5–10 min, depending on user experience; data processing typically takes 1–3 h, and data analysis takes ~30 min.
INTRODUCTION
The goal of systems biology is to decipher complex and interdependent biochemical processes to understand how a biological system operates on a mechanistic level and how it reacts to external factors1,2. Toward this goal, genomic, proteomic and metabolomic technologies have evolved to provide an impressive amount of comprehensive information on genes, proteins and metabolites, with the most recent of this trilogy, metabolomics, having joined as an interesting latecomer. This is in itself interesting, because this approach measures the furthest downstream products of the genes and proteins: metabolites. As metabolites are the most downstream biochemical products, metabolomic data can provide a readout of gene and protein function, thus representing a logical starting point for deciphering their activity.
Advances in high-resolution mass spectrometry (HR-MS) have enabled metabolomics to be used on a global scale, allowing for the detection of low-abundance metabolites in an unbiased manner3–5. However, global metabolomics is known to generate large data sets with thousands of metabolic features of great chemical diversity, making identification and analysis of biological relevance challenging and time-consuming6. Development of bioinformatic tools, such as XCMS (an abbreviation for various forms (X) of chromatography mass spectrometry)7, has helped alleviate processing times for metabolic feature detection, retention time alignment and statistical analysis, but further interpretation of the acquired data is necessary to garner biological insight on a systems level.
XCMS Online8, originally a data-processing platform, has recently been expanded to include multi-omic technology in which raw MS metabolomics data are superimposed directly onto pathway maps9. In addition, XCMS can be used to integrate these pathways with proteomics and transcriptomics results. Although mapping of metabolites can give an indication of gene and protein activity, integrating these metabolomics results with proteomics and transcriptomics data provides a more comprehensive and validated characterization of a system under study. XCMS Online can now be used to perform metabolomics-guided systems biology analysis as a cohesive and intuitive workflow that harnesses cloud-based multi-omic technology.
Development of the protocol
XCMS Online began as an automated cloud-based method to process raw metabolomic data, generating a list of statistically significant features that could then be used for biological interpretation8,10. For identifying potential metabolites, an algorithm was used to match the accurate masses of significant features (e.g., P value ≤ 0.01) at a minimum specified fold change (e.g., fold change ≥1.5) with metabolites listed in the METLIN database11 as an additional output. This results table can then be used to perform further biological analysis to identify changes in metabolism. As manual curation of these pathways is extremely time-consuming, pathway enrichment analysis began to appear in independent software applications12–17. There was a desire to make pathway enrichment easier for users who were not familiar with bioinformatics, thus necessitating the development of a facile approach to processing large quantities of data. Our answer to this was to automate pathway analysis by incorporating the mummichog algorithm16 into the workflow, producing a list of enriched (dysregulated) pathways directly from the raw metabolomic data.
This algorithm deconvolves large amounts of metabolic features, on the basis of their accurate m/z values and matching adducts, into two lists: a ‘significant list’ and a ‘reference list’. Using Fisher’s exact test (FET), the matched features are overlaid onto known metabolic pathways, curated from the BioCyc database (v20)18, and compared with a random sampling of features from the reference list. This process is repeated over many iterations, resulting in a significance P value for a given pathway (see Box 1 for more details). The current platform represents this in both a tabulated format and as a Pathway Cloud Plot (discussed below) to interpret the data. Each metabolite that was identified in a dysregulated pathway provides links to information on the biological importance of that molecule and its position within the metabolic pathway. Genes and proteins that interact with that metabolite are also present within the linked information. This brought about the idea to incorporate even more data into XCMS Online by adding gene and protein data integration. Interpreted metabolomics results can now be cross-referenced by uploading genomic, transcriptomic and/or proteomic data using a list of gene symbols or protein accession numbers (see Box 1 for more details). This subsequent analysis feature of XCMS Online allows researchers to take advantage of collaborative efforts, data sharing and even literature-curated information to make mechanistic interpretation of the identified dysregulated pathways. Differentially expressed genes and proteins that are found to overlap with pathways can be observed with the specific metabolites that have also undergone significant changes, thereby confirming or generating new hypotheses regarding mode of action.
Box 1. Systems biology tools.
This box describes the methods and algorithms used in the systems biology analysis.
Metabolite feature matching
Pathway analysis within XCMS Online predicts dysregulated metabolic pathways directly from the accurate mass (m/z) values of the features generated from the processed results. The m/z values in the results table are used to obtain matching compound identifications by searching against metabolites with predefined adducts in the BioCyc18 pathway database. This analysis can be specified for over 7,600 organisms, and the algorithm is described in more detail below.
Pathway prediction algorithm
A P value cutoff is applied to evaluate the statistical significance of each metabolite feature, dividing the results table into two lists, a significant metabolite list and a total metabolite list. To identify dysregulated pathways, the metabolite feature matches in both the significant and reference lists are correlated with the pathways to determine the degree of overlap. The probability of dysregulated features and their corresponding adducts being metabolites on a given pathway are evaluated using FET. Significance of the pathway fit is calculated with comparison to FET performed on numerous permutations of random features within the total feature list. Once performed for all identified pathways, a list of adjusted P values is tabulated.
Integrated omics
The systems biology platform implemented in XCMS Online contains an integrated omics method to superimpose gene and protein data on the predicted pathway results. Gene and protein lists are uploaded, and a secondary job is run to query the genes and/or proteins present within the biosource. This function allows users to quickly evaluate the overlap of other omic experimental data with metabolomic data via tabulated results or directly on the Pathway Cloud Plot visualization tool.
The systems biology platform was first used in a colon cancer study in human patients comparing normal versus tumor tissue samples19. The XCMS systems biology results implicated tumor progression with biofilm development via polyamine biosynthesis9. This platform has also been used on a phase I clinical trial drug for a Parkinson’s disease immunotherapy that was found to target the tryptophan pathway and later validated with targeted metabolomics20. More recently, cellular responses to chemical exposure of a xenoestrogen on breast cancer cells have been studied and found to alter tRNA charging and ribonucleoside salvage pathways21. In addition, our study of altering carbon sources in an Escherichia coli model system was crucial in the development of the XCMS Online systems biology platform and implicated glycolysis and amino acid biosynthesis pathways as significantly dysregulated9. Currently, the system has been optimized for >7,600 organisms and has been shown to be effective in cell culture and tissue-based studies9,21.
Comparison with other methods
Generation of raw metabolomics-integrated intensity matrices can be performed from a variety of different platforms22,23. These platforms perform a set of peak detection retention time alignments and statistical analysis. The results are typically produced in either information-dense tables or informative visualizations, the latter often including statistical plots such as principal component analysis (PCA) or box-and-whisker plots. Several freely available platforms that are commonly used to preprocess data include cloud-based XCMS Online8,10, or downloadable packages such as MZmine, which has its own user interface23, or mzMatch, which runs in R (ref. 22).
After detecting significantly dysregulated metabolic features, it is necessary to confirm metabolite identity and infer biological relevance by identifying the metabolic pathways in which they are involved. To expedite this process, pathway analysis can be achieved using algorithms that correlate dysregulated metabolites from an untargeted metabolomic analysis with known metabolic pathways in a biological model. The more identified metabolites in a pathway, the higher the confidence of that pathway being affected by the stressed condition under study. Some platforms that are capable of doing such analysis are MetaboAnalyst12,13, KEGG Mapper17 and MBRole24, but all require preprocessed data as input.
Further integration of metabolomic data with genomic, transcriptomic and proteomic data enables a systems-level understanding of the underlying biological mechanisms. Currently, there are concerted efforts in the field to build free-to-use multi-omic workflows, such as Galaxy25. This web-based platform was originally designed for genomic research, but now contains several bioinformatics tools for multi-omic integration and analysis. Galaxy metabolomics modules, such as Workflow4Metabolomics26 and Galaxy-M (ref. 27), can be used to analyze MS-based metabolomics data for systems biology interpretation, yet require XCMS to preprocess LC–MS data. At this stage, both of these are separate software installations that have not been integrated with other Galaxy modules to perform integration of multiple data types. There are also a series of stand-alone, web-based bioinformatics platforms that can perform multi-omics integration. IMPaLA maps dysregulated gene, protein and metabolite data onto pre-annotated pathways28; iPEAP integrates genomic, transcriptomic, proteomic and metabolomic data for pathway enrichment analysis29; iPATH2.0 is an interactive tool for visualization of cellular pathways14; MetExplore links metabolomics data within genome-scale metabolic networks30; Metscape is a Cytoscape plug-in for visualizing and interpreting metabolomic data within human metabolic networks31; and PIUMet takes untargeted metabolomics data (m/z values without IDs) and maps them onto biological networks to identify dysregulated pathways15. By contrast, the XCMS Systems Biology platform allows a fully integrated environment that requires no software installation and no previous programming experience, and has an intuitive workflow with no need to switch among multiple modules or platforms.
Limitations
Support for systems biology analysis is currently available for pairwise and multigroup analyses (see Box 2 for current XCMS job types). For Systems Biology–supported jobs, the database for performing pathway analysis and multi-omic integration currently queries BioCyc18; pathways and networks from other sources (KEGG17, Reactome32 and Wikipathways33) will be included in the future to extend the pathway-mapping capabilities.
Box 2. XCMS data-processing job types.
This box describes XCMS Online data-processing job types.
Single
This job type is usually performed only for alignment, metabolite identification and MS/MS matching (if acquired) with the METLIN database. This is particularly useful if pooled samples were run as a method for metabolite validation. This job type does not currently support the Systems Biology platform.
Pairwise
The primary job type used in XCMS requires selection of two data sets, including control and ‘treatment’ sample classes. These data sets are contrasted on the basis of fold change and P value cutoffs that are user-defined. Statistical analysis can be parametric or nonparametric, as well as paired or unpaired. Pairwise jobs are capable of handling both MS and MS/MS data, and automatically perform predictive pathway analysis and metabolite feature matching with METLIN.
Meta XCMS
This is a secondary job type that compares two or more different perturbations to a single control. This is an excellent way to investigate overlap between placebo and drug effects, or similarities between disease states. Running this job type requires multiple pairwise jobs to be processed as primary jobs, all with the same control group. Each job is selected and results in a Venn diagram and detailed output of overlapping features between the perturbation groups. More details on this type of job can be found elsewhere69,70 and will not be discussed in detail here.
Multigroup
This job type can compare large data sets with multiple conditions and/or time points. Multiple data sets are uploaded, starting with a control data set. A QC data set can also be selected from this group, which is included in PCA analysis but not in the statistical or pathway analysis. Statistical analysis is done using ANOVA parametric statistical test or Kruskal–Wallis nonparametric statistical test. Multigroup jobs are capable of automatically generating pathway analysis results and processing both MS and MS/MS data. This job type does not currently support the Systems Biology platform.
Statistical analysis to identify dysregulated metabolites is limited to a select number of univariate parametric and nonparametric hypothesis tests. Once the statistical test is chosen in the parameter settings, each feature group is compared between sample classes. There is no provision for differentiating technical replicates from biological replicates at this time, and we recommend the use of biological replicates over technical replicates. Inclusion of technical replicates in data analysis should be done with caution. Technical replicates would be useful to investigators for inspecting the analytical reproducibility before committing data to the Systems Biology analysis. If their analytical system generating metabolomics data has poor replication, then they should make every attempt to improve it, otherwise, the biological interpretations from Systems Biology analysis will be compromised.
Pairwise analyses are performed in a simplified manner when a single control and perturbed condition exist. In reality, there are often biochemical feedback loops and cyclical processes within biological systems, and changes in metabolite concentration within these can alter gene and protein function. It is important to note that any metabolomics experiment is a snapshot of a system at a given time. In some instances, time-course sampling can be performed to assess how a system changes over time. However, if sampling is not done at sufficient frequency, changes can be missed. Time-series data can be analyzed using the multigroup analysis job type, but there is no specific function to include time series in the data analysis. Multigroup analysis does permit the addition of quality control (QC) samples34, typically a set of pooled samples measured throughout the analytical sequence at regular intervals, which is then removed from the statistical analysis, yet provides a means to assess the data quality in multivariate PCA and as a control group in the box-and-whisker plots. It should be noted that reported fold change values are the natural log of the median fold change, to account for variation in normally distributed data, and therefore, P values are uncorrected. Users are encouraged to look at the q value, which denotes the false-discovery rate35, before analyzing pathway analysis results and performing multi-omic integration.
The pathway analysis algorithm uses FET to evaluate statistical significance16. For metabolites that are significantly dysregulated in the identified pathways, it is recommended that further validation experiments be performed by MS/MS using the autonomous workflow36 or manually37. Users also need to pay attention to the interpretation of the predictive pathway analysis results and ensure that the raw data used in pathway analysis are accurate. Although FET is commonly used in pathway enrichment calculations38,39, we are also in the process of developing more sophisticated and advanced pathway prediction algorithms to replace FET. The systems biology platform uses the BioCyc database for predictive pathway analysis (https://biocyc.org). Metabolic pathway information archived in BioCyc is generated from literature-based curation (Tier 1 databases) or computational prediction (Tier 2 and Tier 3 databases). Neither approach is able to capture metabolic reactions that are not well defined, such as the complex biochemical interactions between organisms and diet, environmental exposures, xenobiotics and microbiota.
An important aspect of integrated omics in our systems biology platform is the preparation of transcriptomic40–42 and/or proteomic data43–45, which is not discussed in detail in this protocol. However, some considerations are given in Experimental design section. Currently, there is no direct statistical analysis performed during data integration and only overlap is shown. In addition, there is no value, such as log2 differential expression, that can be uploaded with the gene/protein lists. Future development will include statistical assessment of the metabolic pathways that overlap with significant genes and/or proteins, as well as incorporate the degree of differential expression.
As job sizes increase and, consequently, uploading times increase, the dead time between data collection and processing becomes more substantial. One way to alleviate delays in processing time is to use our data-streaming application XCMStream46, which directly uploads data from the instrument computer as it is generated and automatically initiates the job once complete. Analyzing data off-site can also be challenging, particularly if there is limited computer access. To improve accessibility to results, the XCMS Mobile app47 has recently been released to give XCMS users the ability to analyze data from the cloud; we are currently working to implement systems biology analysis and results view on the mobile platform.
There are many more unique functions available within XCMS Online and, as such, the protocols outlined in this paper will not be able to describe all possible permutations of the workflow beneficial for systems biology analysis. Video tutorials on the systems biology analysis and many additional features can be found on the XCMS Institute page within XCMS Online (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=institute).
Experimental design
Controls and replicates
An untargeted metabolomics workflow requires a robust experimental design with an appropriate metabolite extraction protocol, an optimized LC–MS or gas chromatography–MS method, and an effective data-analysis workflow to identify perturbations in metabolic pathways. When setting up the initial experimental conditions, metabolomic sample classes should be defined; these must include a control condition and at least one perturbation or treatment group. To determine the appropriate number of biological replicates, we typically recommend a rough statistical power estimation to ensure that there are enough samples48.
To ensure the quality of the MS data being generated, it is recommended to include a pooled sample (for QC purposes) containing an aliquot of all the samples, or, if the sample groups are large enough, a pooled sample can be prepared for each sample class. Pooled samples are run throughout the mass spectrometric sequence regularly (e.g., one in ten injections) as a QC check for signal intensity and retention time drift, but are also extremely valuable as a method for including preliminary metabolite validation using data-dependent or targeted MS/MS37. Further details on metabolomic sampling, extraction and chromatographic methods have been discussed elsewhere49–52.
Biological experimental design
When preparing samples for metabolomic analysis, it is recommended that samples use biological material that best represents the system under study. Tissue samples taken from the expressing phenotype tend to give more meaningful results versus plasma samples that will be more representative of the whole body. Urine samples, although the easiest to collect, consist of mostly metabolic breakdown products, may be far from the phenotype of interest and can vary greatly depending on dilution. Samples should also be collected in large enough quantities that multi-omics analyses can be performed on the same biological replicates used for the metabolomic analysis. This reduces the complexity of the data set and will typically produce more reproducible results. Biological samples prepared separately for each ‘omic’ analysis often suffer from minor variances during sample preparation that may result in data sets with detectable differences that are not relevant to the study. Preparation of separate samples may allow researchers to detect differences that are a result of natural biological variation within larger sample cohorts. However, in our experience, it is more important to keep the experimental conditions the same whenever possible, particularly during preliminary studies with limited sample size. To better account for natural biological variation and to confirm integrated multi-omic results, we recommend repeating the whole experiment in an independent manner.
An alternative to generating transcriptomic and proteomic data in-house is obtaining the data from publicly available data sets in which experimental conditions are either the same or very similar. This is useful for studies in which a large quantity of curated data is available, such as human cancer research53. This was demonstrated successfully with a colon cancer study in which 30 paired tumor and normal tissue samples were analyzed using untargeted metabolomics19 and then compared with existing transcriptomic and proteomic data obtained from The Cancer Genome Atlas and the Clinical Proteomic Tumor Analysis Consortium, respectively. The integrated omics module in XCMS Online resulted in excellent agreement with the automated pathway analysis of the metabolomics data9. The overlapping pathways included many pathways previously identified in colon cancer, including 1,25-dihydroxyvitamin D3 biosynthesis54, bile acid biosynthesis55, zymosterol biosynthesis56, ubiquinol-10 biosynthesis57 and the spermine–spermidine pathway identified in the original study19.
Modes of analysis
Some final considerations for running the systems biology platform relate to the type of analysis to be performed with respect to the prepared samples. The most commonly performed XCMS Online job is a pairwise analysis (‘Pairwise Job’), the purpose of which is to carefully contrast a control set of samples with a perturbed set in order to isolate the effects of a specific condition. This could include, for example, cell cultures exposed to a stressed condition, animal models perturbed by a specific drug or patients with a specific ailment compared with a healthy population. If more variables or time points are under study, a multigroup analysis (‘Multigroup Job’) can be performed. There are numerous parameters that can be defined when creating an XCMS Online job. In most cases, selecting the default parameters for the instrument platform used is adequate (e.g., time-of-flight detection after ultra-high-performance liquid chromatography separation); however, the user should have a good understanding of the function of the main parameters and when it is useful to change them; these will be highlighted in the protocol below.
Data smoothing
XCMS Online allows the user to select and optimize from a set of base parameters; some of these settings are more sensitive to change than others. Algorithms for smoothing, correcting and aligning data are embedded within these parameters and can be tuned to best detect features for a given data set. For feature detection in global metabolomics data, HR-MS should be used, although both low- and high-resolution (i.e., resolution >10,000) (ref. 58) data can be used in XCMS Online (Step 6). Low-resolution data, in either centroid or profile, should be used with the matchedFilter algorithm7, whereas high-resolution centroid data should be used with the centWave algorithm59. Conversion of data from profile to centroid before upload may provide more robust feature detection and will have faster upload speeds.
Retention time correction
For retention time correction (Step 10), there are two algorithms to choose from. Obiwarp (option A) is the standard algorithm to select. Peaks are warped into groups when compared between samples; this method tends to be more global and smooth. The peakGroups algorithm (option B) can be used to optimize data processing for better control of alignment, but is more difficult to tune. For most analyses, Obiwarp will be sufficient. However, if there is trouble detecting features, peakGroups should be used. We recommend using the nonlinear retention time alignment ‘LOESS’ (locally weighted scatterplot smoothing) in peakGroups, which will also allow you to select the smoothing method ‘family’. This can be either ‘gaussian’ (if a normal distribution is expected and all the data are to be included), or ‘symmetrical’, which is based on a redescending M estimator used with a Tukey’s biweight function60 to allow for outlier removal. The ‘span’ parameter is based on degrees of freedom, is also very sensitive to change and should be selected with caution; the default is 0.6 and going larger (i.e., closer to 1) would obtain a more global smoothing result, but alignment may be diminished, whereas going smaller (i.e., closer to 0.05) may produce better alignment, but will have more-stringent peak selection within a group.
Statistical analysis
There are a select number of univariate statistics (Step 12) used for the generation of the significantly dysregulated m/z feature list that is used in the predictive pathway analysis. For pairwise data analysis, the choice of four standard statistical tests is available. Parametric test options are Welch’s t test61 and paired t test; nonparametric test options are Mann–Whitney U test62 and a paired Wilcoxon sum-ranked test61. For multigroup analysis, ANOVA63 and Kruskal–Wallis64 tests are given as choices, depending on whether the data are parametric or nonparametric.
Normalization
When setting the thresholds for significant features from the statistical test, the P value and natural log of the median fold change must be entered based on the quality of the data and a user-chosen level of stringency. Features considered ‘highly significant’ will be used for the metabolomic cloud plot and PCA, whereas a secondary threshold of ‘significant features’ is set as a cutoff for plotting extracted ion chromatograms (EICs) and box-and-whisker plots, as well as for performing database matching for peak annotation. Default values are provided as a starting point, but may be adjusted depending on confidence in the data. The fold change value is set to a default of 1.5. It is not recommended to use a value <1.5, as these data tend to be artifacts; however, to obtain features with greater dysregulation, a fold change ≥2 can be used.
Normalization, which can affect the outcome of the deciphered pathway, should be chosen carefully. Two normalization methods are also available to apply to the data set to compensate for analytical variances (Step 12). The ‘median fold change’ is well suited for normalizing dilution effects by adjusting the median log fold change of peak intensities in each sample in the whole data set to approximately zero34. The ‘LOESS’ or locally weighted scatterplot smoothing is a stronger polynomial regression normalization method in which the local median of the log fold change between peak intensities in each sample is adjusted to approximately zero across the entire peak intensity range65. LOESS is often applied to compensate for batch effects or systematic variation.
Multi-omics integration
The systems biology platform carries out predictive pathway analysis16 and multi-omic integration on metabolomic data sets using metabolic models from 7,627 unique organisms or biosources. To run predictive pathway analysis, a new parameter set must be defined and saved. The intensity filter in the predictive pathway analysis determines whether the signal intensity of a spectral peak is high enough to be considered as a confident and real metabolic feature for metabolite identification. This value can be determined by checking the signal-to-noise (S/N) ratio in the raw data, and we suggest setting it to at least 10×S/N ratio to avoid artifact metabolite identification from the background noise. When setting up the systems biology parameters, it is important to note that the mass tolerance and adduct forms set in Step 14 are applied only to identify potential metabolites within METLIN but not for predicting metabolic pathways. Metabolite identification in the predictive pathway analysis uses a different set of parameters, in which mass tolerance is defined in Step 14, and adduct forms are preset in the predictive pathway algorithm and cannot be changed. See Box 3 for the complete adduct list used in predictive pathway analysis. Currently, three different mass tolerance options are available: 5, 10 and 20 p.p.m., with 5 p.p.m. being selected for well-calibrated high-resolution MS data and generating the most precise results. A threshold value can also be set for minimal MS peak intensity for features to be considered in the pathway analysis. Users should also define a P value cutoff, which divides the entire metabolomic table into significant and nonsignificant metabolite lists. Typically, this value is the same or lower than the value for data processing; this field can also be left blank to use the default mode, which automatically assigns a P value cutoff based on the top quarter of statistically significant metabolic features.
Box 3. Adduct forms.
This box describes the default adduct forms used in the predictive pathway analysis.
Positive mode
[M+H]+, [M+2H]+, [M+Na]+, [M+H+Na]2+, [M−H2O+H]+, [M−NH3+H]+, [M+NH3+H]+, [M+K]+
Negative mode
[M−H]−, [M−2H]2−, [M+Na−2H]−, [M−H2O−H]−, [M−2H2O−H]−, [M+Cl]−, [M+HCOO]−, [M+CH3COO]−
Multi-omics integration takes place after the LC–MS data are processed, and the pathway analysis algorithm has run. Omics data are uploaded separately using a subjob parameter page found within the results summary page. This algorithm matches gene and protein data to the metabolic pathways identified as dysregulated. During the interpretation of multi-omics results, it may be necessary to sort by gene or protein to find overlap in less significant pathways. If differentially expressed gene or protein data do not directly match the observed up/downregulation of a metabolite, other processes are likely at work. This can include inhibition/activation by a small molecule or metabolite, or a rate-limiting enzymatic process (i.e., low enzymatic catalytic constant) that is up or downstream from a dysregulated metabolite. Interpretation outside the obvious connections should be considered and may require expertise and intuition beyond the immediate results. Given the well-known disconnect between gene expression and protein dynamics66, combining transcriptomic data with metabolomics data may prove useful in elucidating upstream mechanisms as relative metabolite concentrations provide information on protein function67. However, the more orthogonal data that can be included, the better the biological interpretation will be on a systems-wide scale.
MATERIALS
EQUIPMENT
Computer requirements
Browser requirements: XCMS Online supports many of the mainstream web browsers. For the best results, we recommend using the latest version of Google Chrome (v57+) or Mozilla Firefox (v51+).
Internet connection requirements: A fast upload connection is recommended, with a minimum of 5 Mbps, to upload files to data set storage. This can be done directly from the instrument computer or from a personal computer, provided there is adequate hard-drive space for data files. Physical Ethernet connections are normally preferred over wireless (wifi) connections.
Hardware requirements: To view and work with XCMS Online results, a minimum of a Pentium 4 processor with 8-GB of RAM and a screen resolution of 1,280 × 800 or higher is recommended.
Data files
XCMS Online currently supports upload of both raw data files and numerous converted MS data formats; see Box 4 for more details.
Gene and protein data format: Differentially expressed gene and protein data should be in the format of a comma-separated (.csv) or tab-separated (.tsv) file. Genes names should be in the format of gene symbols, and protein names should be in the format of gene symbols or Uniprot68 accession numbers. If multiple data sets are available, they must be uploaded individually.
The results for example data discussed in the ‘ANTICIPATED RESULTS’ section (see below) can be accessed after logging in to XCMS Online (https://xcmsonline.scripps.edu), clicking on the ‘XCMS Public’ menu (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=listPublicShares) and searching for the job number ‘1172567’ or name ‘Ecoli_glucose-vs-adenosine’. These data and multi-omics data files are also available online for download to users to test on their own using the two sample class data sets ‘Glucose.zip’ and ‘Adenosine.zip’ (MetaboLights, study identifier MTBLS572; https://www.ebi.ac.uk/metabolights/MTBLS572). For multi-omics integration, a demonstration transcriptomics data set is provided as ‘Ecoli_genes.csv’ (Supplementary Data 1) and a significant protein data set is provided as ‘Ecoli_proteins.csv’ (Supplementary Data 2). Information on the experimental design and XCMS Online parameter settings, including systems biology parameters and multi-omics integration settings, is provided in the Supplementary Methods and Supplementary Table 1.
Box 4. MS data conversion.
This box describes how to convert raw MS data to non-vendor-specific, open data types.
XCMS supports non-vendor-specific, open data types such as .mzXML, .mzML and .netCDF. Generally, users can use msConvert (http://proteowizard.sourceforge.net/tools.shtml), provided by ProteoWizard, to convert different MS raw data format into XCMS-supported data types. Direct support is coming soon for vendor-specific raw data upload without conversion or compression. In the meantime, raw data folders containing files with extensions such as .d and .raw can be compressed to a .zip file and uploaded to a data set. The following lists a short summary of how to convert MS raw data to appropriate data formats using some vendor software.
Agilent
Load Agilent MS raw data into Agilent MassHunter Qualitative Analysis. Click ‘File’ → ‘Export’ → ‘as mzData’ in the menu bar. In the pop-up dialog, highlight the MS raw data that must be converted. In the right-side ‘options’ window, select the ‘Entire data file’ as the export contents and assign an export location. Click ‘OK’ to export raw Agilent data to mzXML format.
Bruker
Load Bruker MS raw data into Bruker Compass DataAnalysis. Highlight the MS file in the Analysis List (one file at a time). Click ‘File’ → ‘Export’ → ‘Chromatogram Analysis’ in the menu bar. In the pop-up dialog, specify the exported file name. Select a compatible data format for file conversion and choose ‘Line spectra’ for the exported spectrum format. Click ‘Save’ to process the data conversion. Bulk file conversion can also be performed using Bruker’s free CompassXport command-line program. Once installed, open a command prompt window and type compassxport.exe –multi C:\your_datafile_folder\ to convert all files directly in that folder.
Thermo and Sciex
XCMS Online allows direct upload of Thermo .raw and Sciex .wiff and .wiffscan data files in the HTML5 uploader. There is no need to convert the Thermo or Sciex MS raw data in advance.
PROCEDURE
Stage 1: data upload ● TIMING ~30 s–5 min per file, depending on the size of the data set
-
1|
Logging in. Go to the XCMS Online home page (https://xcmsonline.scripps.edu) and log in with your e-mail and password, or click ‘Sign Up’ to create a free user profile.
-
2|
Uploading data. It is recommended to upload data before starting a job. After logging in to XCMS Online, click ‘Stored Datasets’ from the top menu. Upload times may vary, depending on the type of Internet connection, file size, number of files, proximity to the XCMS server and how busy the servers are, but typically take ~30 s–5 min per file. Create one data set per sample class.
-
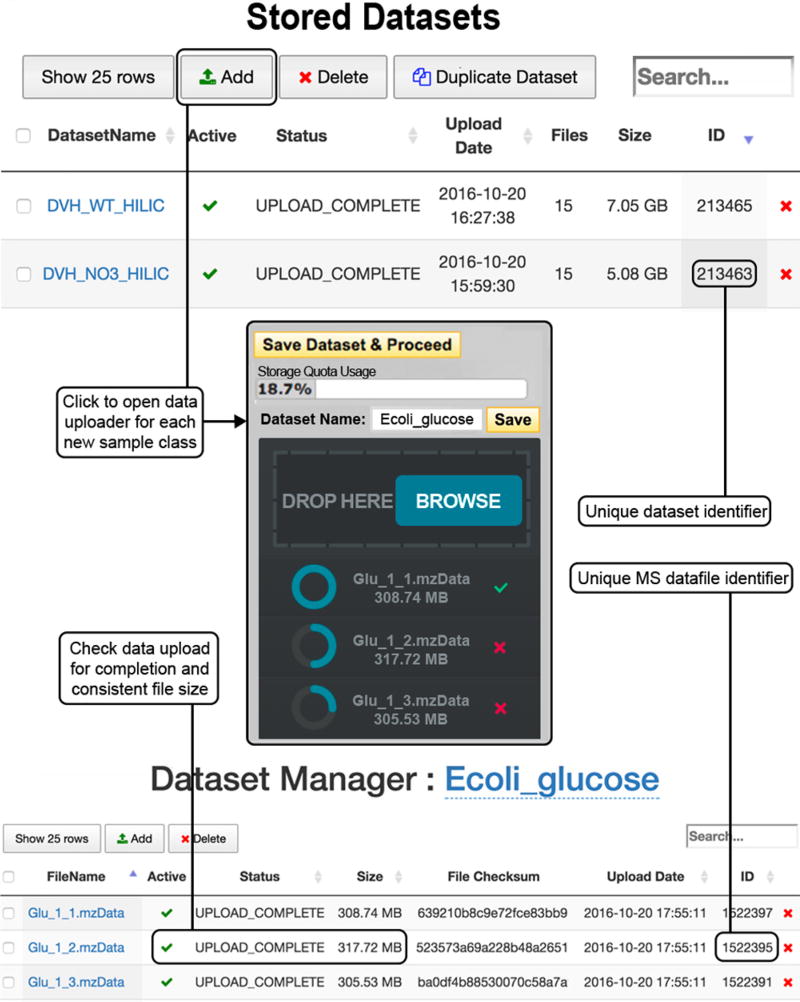
3|
To add data to a sample class, click ‘Add Dataset(s)’, as shown in Figure 1 (top). This opens the HTML5 uploader window (Fig. 1 (center)), in which files can be selected by clicking ‘BROWSE’ or can be dragged and dropped into the uploader window; follow Box 4 for acceptable file formats.
▲ CRITICAL STEP We also provide example data sets to test the Systems Biology platform (MetaboLights, study identifier MTBLS572; https://www.ebi.ac.uk/metabolights/MTBLS572), as well as transcriptomics (Supplementary Data 1) and protein (Supplementary Data 2) data for multi-omics integration.
? TROUBLESHOOTING
-
4|
Give the data set a meaningful name representative of the sample class and click ‘Save’. Once the files have finished uploading, click ‘Save Dataset & Proceed’.
-
5|
Click on the new data set name to open the ‘View/Edit Datasets’ window (Fig. 1 (bottom)), in which files should be checked for upload completion and that the file size is the same as that of the original file.
-
6|
Start an XCMS job. Click ‘Create Job’ from the top menu and select a pairwise or multigroup job. Data sets can be loaded directly via ‘Load New Dataset’ and following Steps 2–5 or can be selected from the previously prepared data sets via ‘Select Dataset’ (recommended). The control condition should be placed in Dataset 1 and the perturbed condition in Dataset 2. In multigroup analysis, the user can also define a QC data set.
Figure 1.
Upload of mass spectrometry data (Steps 2–5). To upload a data set for each sample class, go to the ‘Stored Datasets’ menu option (top). Previously stored data sets are found here, each with a unique data set identifier. Click ‘Add Dataset(s)’ to open the data uploader window (center). Files can be selected from the file directory or by dragging and dropping where indicated. Once the upload is complete, as indicated by a full blue circle and a green check mark, press ‘Save Dataset & Proceed’. Click on the name of the new data set to open the ‘View/Edit Dataset(s)’ window (bottom) and check that the upload is complete and the file sizes are equal to those of the original.
Stage 2: parameter settings ● TIMING 5–10 min
▲ CRITICAL Selection of parameters is imperative for accurate data processing and depends on the instrument used and the conditions in which the samples were run. If these parameters are not carefully set, this can result in low numbers of features.
-
7|
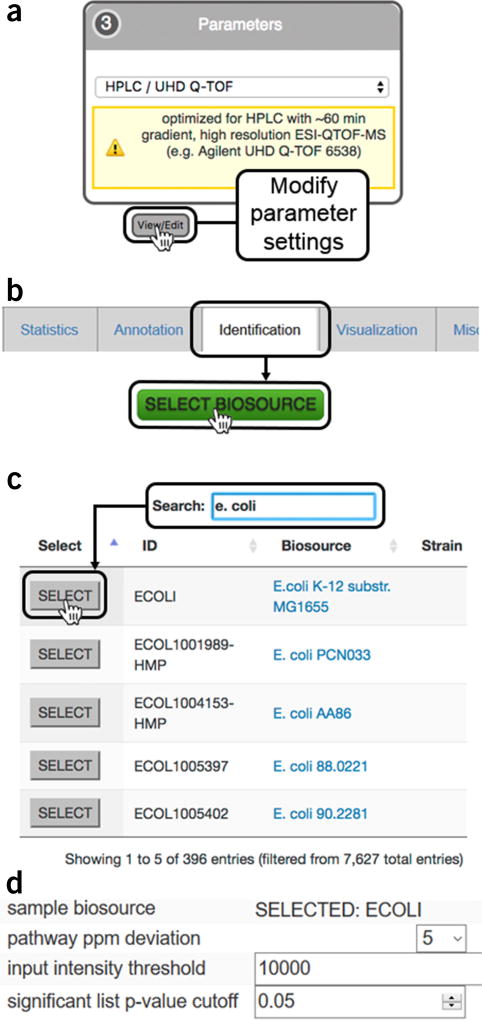
Select a base parameter set for data processing. Select a default parameter set that best represents your sample data from the parameter dropdown box. It is recommended to modify the parameters to make them specific to your acquisition parameters. Click ‘View/Edit’ (Fig. 2a) to open the parameter method details. Click ‘Create New’ to be able to modify the existing parameters. There are nine tabs with details pertaining to how data are to be processed (see Steps 8–16 for details on the parameters specified in the individual tabs).
-
8|
General. Give the parameter set a unique name. Once saved, this parameter set will be available only in the user’s parameter files. The ‘Retention time format’ can be changed to minutes or seconds, and polarity can be either positive or negative.
-
9|Feature detection. The method for feature detection is based on either the centWave algorithm59 (high-resolution centroid data) or the matchedFilter algorithm7 (low-resolution centroid or profile data). Define the following parameters:
Parameter Description ppm (Set mass accuracy) The deviation value should be slightly higher than that of the expected mass accuracy of the instrument. Guidelines are 5–15 p.p.m. for Orbitrap data, ~5 p.p.m. for lock mass quadrupole time of flight (QTOF) data and 10–20 p.p.m. for other QTOF instruments minimum/maximum peak width This depends mainly on the type of chromatographic separation performed. For standard reverse-phase separations, a general guideline is 20–60 s, whereas for hydrophilic interaction liquid chromatography (HILIC), in which run times tend to be longer with broader peaks, we recommend 25–90 s. When running with UPLC, these values should drop markedly because of shorter run times and higher resolution, with suggested starting values between 2 and 5 s to a maximum of 30 s. If in doubt of values, check the raw chromatographic run for peak widths of some common compounds from each end of the trace. These values are not hard cutoffs and may be detected slightly out of this range, depending on the quality of the peak data Click on ‘View Advanced Options’ and define the following advanced parameters:Parameter Description mzdiff This is the m/z tolerance allowed for spectral features; the ‘signal/noise’ threshold for peaks is set to a default of 6 and can be increased if data are noisy Integration method This can be chosen as a filtered method by selecting 1, which uses noise-reduced data, or raw data by selecting 2, which is more exact but prone to noise prefilter peaks This can be selected to apply a prefilter to mass traces; it specifies the minimum number of peaks a mass trace must contain in order to be retained prefilter intensity This defines the minimum scan intensity required for each peak Noise Filter This value can be entered for a minimum value that peaks must reach to be kept for analysis -
10|
Retention time correction. Select either the Obiwarp (option A, for data correction with well-behaved peak groups) or the peakGroups algorithm (option B, for more options to detect peaks that require more in-depth grouping) from the dropdown box (see also Experimental design).
Figure 2.
Predictive pathway analysis parameter settings (Step 14). (a) The organism metabolic model, or biosource, for performing pathway analysis is selected during job creation while editing XCMS processing parameters under the ‘Identification’ tab during job creation; (b) clicking on the ‘SELECT BIOSOURCE’ button opens a new window with all the metabolic models available; (c) the search bar can be used to find the desired metabolic model. Clicking on the biosource link opens a link to BioCyc with a summary of the pathway information. Pressing the ‘SELECT’ button chooses the model and closes the window; (d) the correct metabolic model should now be visible in the ‘Identification’ tab under ‘sample biosource’ and ‘pathway ppm deviation’ can be selected from the dropdown menu; ‘input intensity threshold’ for peaks and P value for significant features can specified by the user.
(A) Retention time correction using Obiwarp
Set ‘profStep’, which defines the step size (in m/z) for profile generation from the raw data files. The default value is 0.5.
(B) Retention time correction using peakGroups
-
Define the following peak-grouping parameters:
Parameter Description non-linear/linear alignment Choose the alignment method from the dropdown box; this can be polynomial (nonparametric) using ‘LOESS’ (locally weighted scatterplot smoothing) or a ‘linear’ regression model; we recommend the LOESS method for most applications extra/missing These parameters are dependent on sample sizes and should be increased if the sample sizes are large (i.e., 1 if the data set contains five replicates or 5 if the data set has 25 replicates) Click on ‘View Advanced Settings’ to define further optional parameters for initial grouping performed with peakGroups:Parameter Description Ignore sample class Select TRUE/FALSE for ignoring sample class —selecting false will create bias to sample class bw The ‘bw’ or band width, is the peak width at half height, which describes the inclusiveness of the peak grouping in seconds; smaller values are less inclusive mzwid Enter the ‘mzwid’ or mass tolerance (m/z) between samples and across peak groups minfrac Enter the ‘minfrac’ or minimum fraction of samples required to accept a peak grouping, which is sample size–independent, whereas ‘minsamp’, the minimum number of samples required to accept a peak grouping, should be based on the sample size family The smoothing method ‘family’ should be selected when ‘LOESS’ alignment is performed and can be either ‘gaussian’, which will include outliers, or ‘symmetrical’, which will exclude them span Enter a ‘span’ value between 0 and 1. Again, this is only for ‘LOESS’ alignment, and values closer to 1 result in more global smoothing
-
11|Alignment. Once the peaks are grouped, align the peak features by defining the following parameters:
Parameter Description bw This is the band width, or peak width at half height, and the default is 5 s; this should be set to <10 s for HPLC and to 2–5 s for UPLC data minfrac This is the minimum fraction of samples required for a set of peaks to be called a group mzwid This is the difference in mass accuracy between samples Define the following additional parameters by clicking ‘View Advanced Options’:Parameter Description minsamp This is the minimum number of samples allowed for a set of peaks within the same m/z tolerance to be called a group max This is the maximum number of groups to be identified for a given m/z slice -
12|Statistics. Select the statistical test to be performed on metabolite features. Select from the following:
Analysis Description For unpaired pairwise analysis Choose between Welch’s t test (parametric) and Mann–Whitney test (nonparametric) For a paired pairwise analysis Select the paired parametric t test. If a nonparametric test is required, a Wilcoxon signed-rank test can be selected. This enables a new button to appear: ‘VIEW PAIRS2’, which opens a new window to select the pairs by dragging each sample in the correct order onto the list Multigroup job Choose either an ‘ANOVA’ parametric test or ‘Kruskal–Wallis’ nonparametric test Define the following P value and threshold parameters:Parameter Description P-value threshold (highly significant features) This generates plots such as the standard Cloud Plot and PCA Fold-change threshold This generates plots such as the standard Cloud Plot, EICs and box-and-whisker plots P-value threshold (significant features) This generates EICs and box-and-whisker plots and performs database matching for peak annotation Select additional options for how peaks are evaluated for statistical analysis:Parameter Description Value Select the type of intensity ‘value’ to be used for statistical tests. This can either be the feature peak maximum intensity value ‘maxo’ or peak area ‘into’ Normalization Select the normalization method, either ‘median fold change’ or ‘LOESS’ -
13|
Annotation. Define the parameters for matching isotopes and/or adducts to the features in the Results Table by selecting either ‘isotopes’ or ‘isotopes and adducts’ from the dropdown box, with the latter resulting in increased processing time but more identifications. Define the m/z tolerance in either absolute error or relative error values; the smaller deviation for each m/z feature will be used in the annotation process.
-
14|Identification. Define the parameters for matching significantly dysregulated features with known metabolites in the METLIN database11 and for performing pathway analysis16 as follows:
Parameter Description ppm Set the ‘ppm’ tolerance for labeling metabolite annotations; this matches the m/z values in the Results Table to the accurate mass in the METLIN database. Set to the same value as for feature detection or lower adducts Highlight the ionized forms, such as [M–H]−, [M–H2O–H]−, and [M+Cl]−, to be considered for database search Sample biosource Click ‘SELECT BIOSOURCE’ (Fig. 2b) to open a separate browser window as in Figure 2c. Biosources can be found by browsing or by using the search field; press ‘SELECT’ to confirm the biosource choice. Click ‘save’ to remember the biosource selection in the data-processing method (Fig. 2d) Pathway ppm deviation Define the mass tolerance ‘pathway deviation ppm’ for matching spectral peaks against metabolites in the BioCyc database Input intensity threshold Enter an ‘input intensity threshold’ or leave it blank to include all the metabolic features for pathway analysis Significant list P-value cutoff Enter a ‘significant list P-value cut-off’ for defining significantly dysregulated features for pathway analysis ▲ CRITICAL STEP Parameters for predictive pathway analysis can significantly alter the systems biology results. Each setting should be carefully considered on the basis of the type of instrument and the quality of data collected.
? TROUBLESHOOTING
-
15|
Visualization. Set the retention time window for visualization of the EIC of the statistically significant metabolic features. The default value of 200 s is recommended for HPLC data; this may be reduced to ~120 s for UPLC data.
-
16|
Miscellaneous. Leave the parameter settings in this tab unchecked for most data-processing cases. ‘Correct mass calibration gaps’ applies to MS data from Waters instruments to subtract lock mass scans from the data; ‘Bypass file sanity check’ disables the file completeness check to speed up data processing.
Stage 3: XCMS data processing and predictive pathway analysis ● TIMING 1–3 h, depending on data set size and server queue
-
17|
Job submission. Once all the parameter settings are defined, click ‘Submit Job’ to open the ‘Confirm Job Specifications’ window to view the job parameter summary. If all the information is correct, click ‘Submit Job’ in the window to launch the data processing, including pathway analysis.
-
18|
Confirm job. After the XCMS job is submitted, a notification email will be sent to the registered email address. Check the processing status by clicking the ‘View Results’ from the top menu to explore all the jobs submitted under the same user account. The status button indicates various data-processing situations, including ‘Not Submitted’, ‘Queued’, ‘Processing’ or ‘Error’. The progress bar indicates the percentage completion of the job. Refreshing the webpage updates the progress percentage. Once the job is completed, the status button will change to ‘View’ and the progress bar reaches 100%.
? TROUBLESHOOTING
Stage 4: interpretation of pathway analysis results ● TIMING 30–40 min
-
19|
Access results. Click ‘View Results’ from the top menu to open a list of in-progress or completed jobs. Refreshing the page will update the progress bar on in-progress jobs. Press ‘VIEW’ to view a finished job.
-
20|
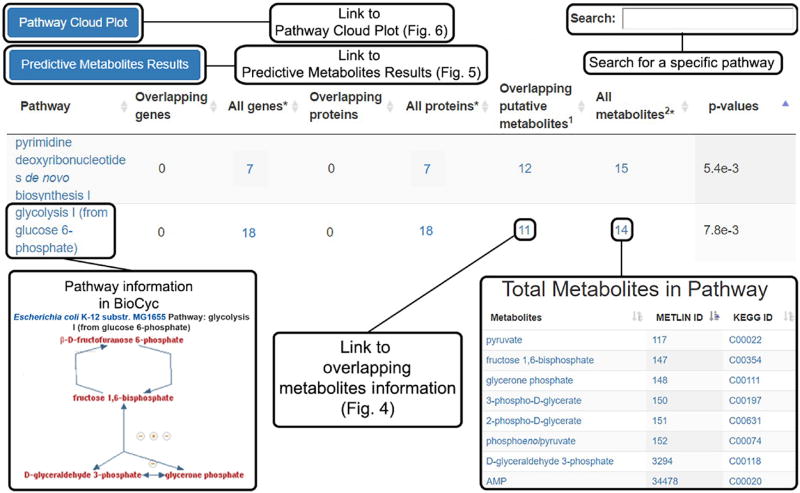
Predictive pathway analysis results. To view and interpret the pathway results table, click the ‘Systems Biology Results’ tab located on the left side of the Job Results Summary page. This table tabulates all the predicted pathways, together with the involved genes, proteins, metabolites and pathway P values (Fig. 3).
-
21|
Click the pathway names to view detailed pathway descriptions on the BioCyc website. Gene and protein information involved in the pathway are listed in the ‘Pathway Results Table’ but no overlapping information will be displayed until multi-omic integration is processed (see Stage 5).
-
22|
Click the number of ‘All genes’ links to the genes involved in the pathway with their names, enzyme activity and BioCyc reaction identities. Both the gene names and the reaction identities can be further linked to their detailed information in the BioCyc website.
-
23|
Similarly, click ‘All proteins’ to link to a list of proteins in the pathway with their names, the protein identifier as a UniProt accession number or gene symbol, and the number of pathways involved. The protein name links to the detailed information on the BioCyc website, whereas the protein identifier links to the protein information in the UniProt database. Clicking on the number of pathways involved opens a window to a new list detailing those pathways, each linking to detailed information on BioCyc.
-
24|
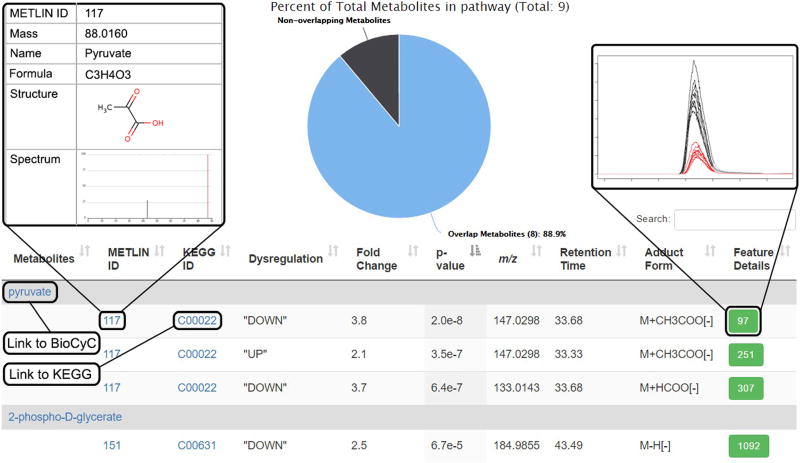
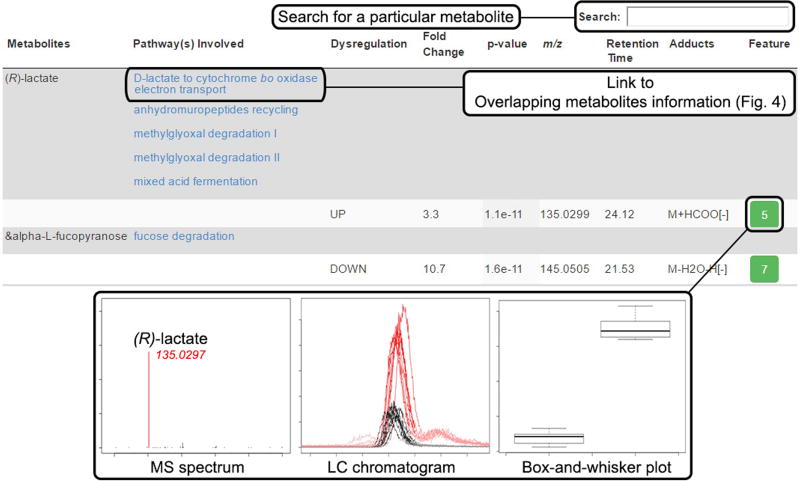
Click the number of ‘Overlapping putative metabolites’ to show the list of dysregulated metabolites involved in the pathway and a pie chart showing the percentage coverage of dysregulated metabolites in the pathway. This view is illustrated in Figure 4. Detailed metabolic information is provided, including METLIN ID, KEGG ID, up/downregulation, feature fold change, feature P value, m/z, retention time, matched adduct form and feature details for each unique metabolic feature ID.
-
25|
Click on a metabolite to open a new tab linking to detailed information on BioCyc.
-
26|
Click on a METLIN ID to open a new tab linking to the METLIN database metabolite entry.
-
27|
Click on a KEGG ID to open a new tab linking to the KEGG database metabolite entry.
-
28|
Click a feature ID number under ‘Feature Details’ to open a separate window displaying the EICs, the MS spectrum of the average feature m/z value and the box-and-whisker plot of that metabolic feature.
-
29|
Press the ‘Back’ button on the browser to return to the ‘Metabolic Pathway Results’.
-
30|
Click the number of ‘All metabolites’ to display a list of all metabolites found involved in that pathway.
-
31|
Click the name of the metabolite to open a new tab linking to BioCyc information.
-
32|
Click the numbers under ‘METLIN ID’ to open a new tab linking to the METLIN database metabolite entry.
-
33|
Click on a KEGG ID to open a new tab linking to the KEGG database metabolite entry.
-
34|
Press the ‘Back’ button on the browser to return to the ‘Metabolic Pathway Results’.
? TROUBLESHOOTING
-
35|
Assess pathway significance by P value; typically, ≤0.05 indicates significant dysregulation and implies that this pathway is worth further investigation.
-
36|
Predictive metabolite results. In the ‘Metabolic Pathway Results’ table, click the ‘Predictive Metabolites Results’ button (Fig. 3) to access information on all dysregulated putative metabolic identifications. This view is illustrated in Figure 5.
-
37|
Click the name of the ‘Pathway(s) Involved’ to display a list of all the dysregulated metabolites that are involved in the pathway.
-
38|
Click the column name ‘Fold Change’, ‘P-value’, ‘m/z’ or ‘Retention Time’ to sort the entire table by that column in an ascending order. Click the column name again to sort it in a descending order. Click the ‘Reset’ button above the table to return to the original view.
-
39|
Click the ‘Feature’ number to open a separate window to view the LC chromatogram, MS spectrum and box-and-whisker plot of that metabolic feature.
-
40|
Type the metabolite name in the ‘Search’ bar at the top right of the page to search for a specific metabolite in the metabolomics data set.
-
41|
Predictive pathway results download. Download the results of the data processing and pathway analysis via the ‘Download Result’ button on the top right of the ‘Job Results Summary’ page. In the downloaded folder, the pathway analysis results can be found in the zipped ‘results’ folder. All the predicted metabolic pathways and associated metabolic information are stored in the ‘mcg_pathwayanalysis_mummichog.tsv’ file. Metabolites that contribute to the statistically significant pathways (default P value ≤ 0.05) are stored in the ‘mcg_metabolite_worksheet_mummichog.tsv’ file. Putative metabolic identifications for all the m/z values in the results table are in the ‘tentative featurematch_mummichog.tsv’ file.
-
42|
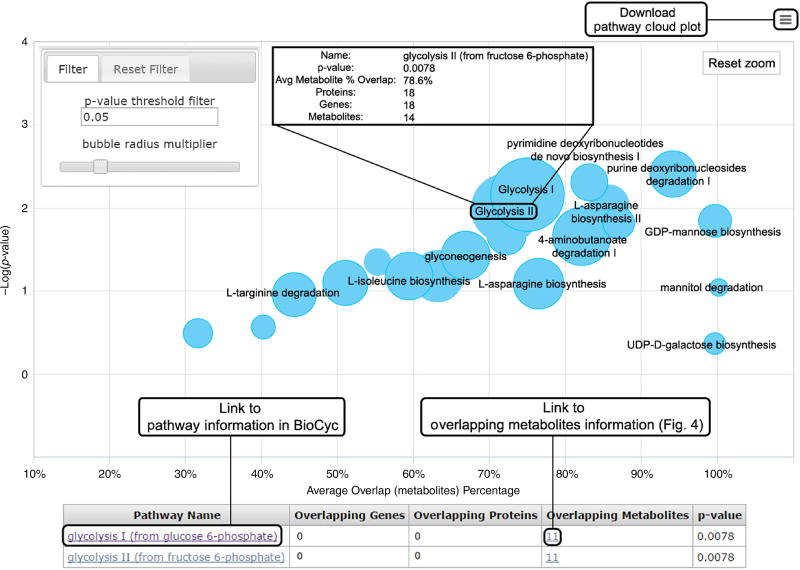
Pathway cloud plot. Access the pathway cloud (Fig. 6) from either the job summary page or inside the pathway results table. The x axis of the plot represents the percentage of overlapped metabolites, and the y axis represents the negative log of the P values. Each metabolic pathway is represented as a circle in the plot. The radius of each circle is proportional to the total number of metabolites identified in that pathway. The interactive pathway cloud plot allows users to zoom in on any part of the plot by drawing a rectangle with the cursor for a detailed view. Users can reset to the original plot by clicking on the ‘Reset zoom’ box in the top right of the graph.
-
43|
Adjust the P value threshold on the left side of the pathway cloud plot to refine and display pathways with P values smaller than that threshold.
-
44|
Adjust the bubble radius multiplier to optimize the plot view by tuning the bar on the top left side of the plot.
-
45|
Hover the cursor over the circle to show pathway name, P value, metabolite overlap percentage and total numbers of genes, proteins and metabolites involved in the pathway.
-
46|
Click on a pathway circle to display specific pathway result details underneath the pathway cloud plot with overlapping gene, protein and metabolite information. If multiple pathways are on a single point, they will all be tabulated.
Figure 3.
Predictive metabolic pathway results (Steps 20–24). Pairwise and multigroup jobs will automatically generate a table of predicted metabolic pathways; the total genes, proteins and metabolites known to be associated with the pathways; the putatively identified dysregulated metabolites; and the calculated P value for pathway significance. Clicking the name of the pathway opens a BioCyc pathway map, whereas the numbers in the table link to more detailed information about the respective total genes, proteins and metabolites. Overlapping metabolites lead to detailed information on the dysregulated metabolic features (Fig. 4). Overlapping genes and proteins will not be tabulated unless multi-omics integration is performed. At the top left of the table are links to the Predictive Metabolites Results (Fig. 5) and the Pathway Cloud Plot (Fig. 6). At the top right of the page is a search bar retrieving specific pathway information.
Figure 4.
Overlapping metabolite information (Steps 24–29). Dysregulated metabolic features involved in a pathway are opened when this link is clicked from the Systems Biology Results page. The pie chart at the top shows the percentage of dysregulated metabolites putatively identified in the pathway. The table underneath shows information for each metabolite identified by the predictive pathway algorithm. Each metabolite is followed by individual entries for detected adducts matching the accurate mass within the defined deviation threshold (p.p.m.). Information for each feature from the XCMS-processed results is provided: the ‘METLIN ID’, the ‘KEGG ID’, direction of dysregulation, fold change, P value, average accurate mass m/z of the peak, retention time, adduct form and ‘Feature Details’ (green boxes), which give the XCMS ‘feature ID’ number. Clicking on a feature ID number opens a pop-up window with an extracted ion chromatogram (shown), a mass spectrum and a box-and-whisker plot (not shown). Entries in blue font link to a new page when clicked, linking to more detailed information.
Figure 5.
Predictive metabolites results (Steps 36–40). This table is accessed from the Systems Biology Results page to display all the dysregulated metabolites used in the predicted pathway analysis and all the pathways within that organism that each metabolite is involved in. Metabolites are matched to one or more features on the basis of the detected accurate mass m/z adduct forms and the defined deviation threshold (p.p.m.) from the m/z value of the accurate mass. Information for each metabolic feature is also tabulated, including direction of dysregulation, fold change, P value, m/z, retention time, adduct form and the XCMS feature ID number (green box). Clicking the unique feature ID opens a pop-up window displaying the MS spectrum, LC chromatogram and box-and-whisker plot for that metabolic feature.
Figure 6.
Pathway cloud plot (Steps 42–46). This plot illustrates the results of the predictive pathway analysis. Each pathway is displayed as a circle, with the x axis representing the percentage of metabolite overlap within that pathway and the y axis representing increased pathway significance calculated from the pathway analysis. The radius of each circle is proportional to the total number of metabolites in the pathway. Drawing a rectangle with the cursor zooms into that part of the plot; clicking on the ‘Reset zoom’ button at the top right of the graph resets to the original plot. Adjusting the P value threshold in the filter box at the top left of the figure displays pathways with P values below that threshold. Sliding the ‘bubble radius multiplier’ adjusts the circle radius to better view and compare pathways. Hovering the cursor over the circle generates pop-up information on that pathway. Clicking on a circle displays pathway results below the plot, in which additional information can be accessed through the hyperlinks, such as overlapping metabolites information (Fig. 4). If multiple pathways are overlaid on the plot, they can be found in the table below.
Stage 5: multi-omic integration ● TIMING <1 min
-
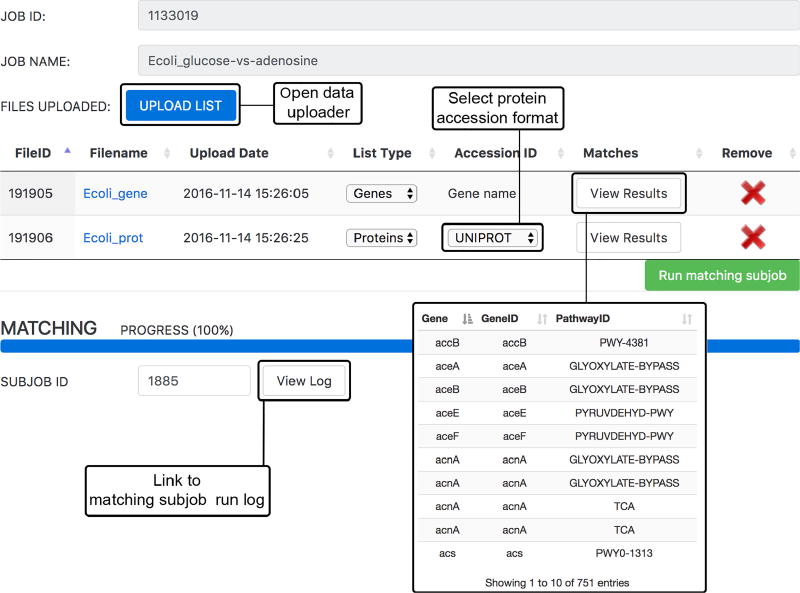
47|
Multi-omic data upload. Press the ‘+’ button beside ‘Multi-Omics Data’ to open the ‘Systems Biology Matching Parameters’ window (Fig. 7) to manage the subjob for omics integration. The XCMS job ID and name will be listed for the current job. Click ‘Upload’ to open the uploader window.
▲ CRITICAL STEP Make sure that only one omics file is uploaded at a time and the correct gene/protein format is selected. Data integration will not proceed if these are not set correctly.
-
48|
Uploader window. Ensure that gene data are listed as gene symbols, whereas protein data should be in either gene symbols or UniProt accession numbers. Both must be uploaded in either .csv or .tsv format, ensuring that there are no commas present in the names, if uploading, as this may result in improper matching. After selecting or dragging and dropping the file, wait for the upload to complete and click ‘Save and Proceed’ to close the window. If another file is to be uploaded, repeat this process.
-
49|
On the main window, indicate whether the file is gene or protein data under ‘List Type’. Check to make sure that all the files have the right designation (protein or gene). If the file type is a protein list, ensure that the correct format is selected. Click ‘Run matching subjob’.
-
50|
The progress bar will show 100% once the job is complete. Access the matched results by clicking ‘View Results’, and the run logs can also be checked for completion statistics or error messages by clicking ‘View Log’.
Figure 7.
Multi-omics integration (Steps 47–50). Uploading of multi-omics data occurs in the ‘Matching Parameter Sub-Job’ window. Gene, transcript and/or protein data can be uploaded in .csv or .tsv format using the ‘UPLOAD LIST’ button. ‘List Type’ must be selected from the dropdown box and, if uploading protein data, the ‘Accession ID’ format must also be selected before clicking ‘Run matching subjob’. Once the job is complete, the progress bar will be at 100%. The matched genes or proteins from the analysis can be viewed under ‘View Results’; the run log can be accessed by pressing ‘View Log’.
Stage 6: interpretation of multi-omic integration results ● TIMING ~20 min
-
51|
Systems biology results. Download the overlap list in .tsv or .pdf format at the top of the table. Differentially expressed genes and proteins that are overlapping with the predicted dysregulated pathways can be found in the ‘Systems Biology Results’ tab with the dysregulated pathways (Fig. 8).
-
52|
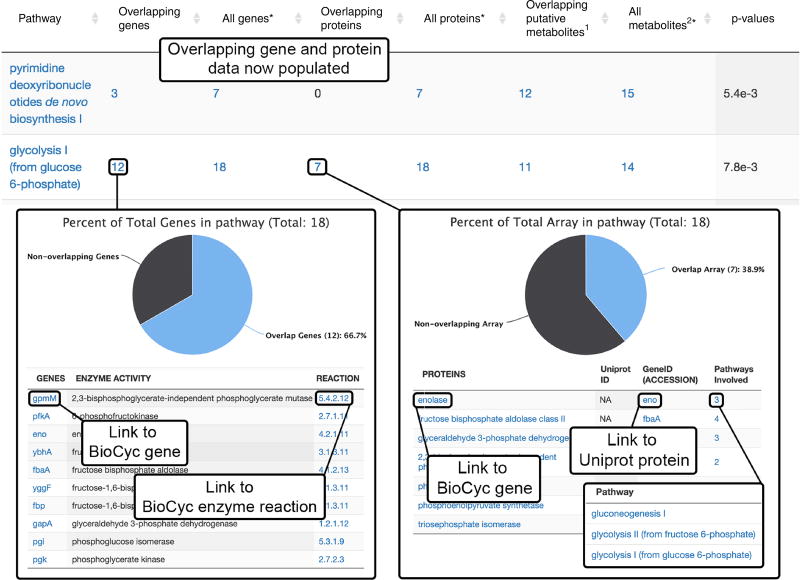
Gene results. Click on the overlapping gene number in a pathway to open a new window showing the percentage overlap and the list of overlapped genes.
-
53|
Click the gene name in order to link to the BioCyc page containing the gene information.
-
54|
Click on the reaction in order to open a page describing the enzymatic reaction for the encoded protein related to that gene.
-
55|
Protein results. In a similar manner to that used for gene overlap, click on the overlapping protein number to show the percentage overlap and a list of overlapped proteins
-
56|
Click on the proteins in the list in order to link to the BioCyc page that shows related protein information
-
57|
Click on the ‘gene ID (accession)’ in order to link to the UniProt protein information and the genes that encode for it.
-
58|
Click on the ‘pathways involved’ in order to open a list of pathways in which the protein is involved. Each pathway in that list opens a BioCyc page with metabolic pathway information.
-
59|
Initial biological interpretation. Correlate overlapping genes and proteins with up- and downregulation of the metabolites to determine if over- or underexpression is occurring as a result of the treatment applied in the experiment.
Figure 8.
Multi-omics results (Steps 51–59). After the multi-omics subjob has been completed, the overlapping genes and proteins will be populated in the Systems Biology Results table. Clicking on the overlapping gene number opens to a detailed list with specific genes that were found to overlap with the pathways. Links to BioCyc gene and enzyme reaction information are also provided. Clicking on the ‘Overlapping proteins’ number opens to a detailed list with specific proteins that were found to overlap with the pathway. Links to BioCyc encoding gene and Uniprot protein information are available. Clicking the number under ‘Pathways Involved’ opens a list of related pathways with links to BioCyc metabolic pathway information.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 3 | HTML 5 uploader cannot be opened for data upload | HTML 5 uploader is disabled | HTML 5 uploader can be activated under the ‘Account’ menu by selecting ‘Global Parameters’ from the right-side menu. Under the ‘Uploader Tech’ section there is a dropdown box in which ‘HTML5’ can be selected. Save the selection and return to performing data upload |
| 14 | The parameters for the predictive pathway analysis cannot be found in the parameter settings | The parameter set was created before the implementation of the Systems Biology platform on XCMS Online | Users must create a new parameter set based on one of the default parameter sets, which are displayed in bold in the parameter list dropdown box |
| 18 | A job is ‘QUEUED’ for >t;1 h | There may be an error with job submission | A job resubmission can be attempted by checking the box to the left of that job and selecting ‘Resubmit Job(s)’ from the top left menu under the ‘View Results’ page |
| A job appears to be stuck in ‘PROCESSING’ status for many hours | There are likely a substantial number of other jobs being processed | Please be patient and allow up to 24 h before taking further action | |
| A job fails, giving an ‘ERROR status’ within 7% of job completion | There was a problem with the file upload, such as a corrupted file | Always check that the uploaded files in your stored data set have a green check mark under the ‘Active’ column and that the status says ‘UPLOAD_COMPLETE’. Try removing corrupted files and uploading again. If this is still unsuccessful, create a new data set and upload all the data files again. Always check the raw chromatograms for data integrity before uploading. If the problem persists, select ‘Help’ from the main menu for further advice | |
| 34 | KEGG or METLIN IDs are not available for some metabolites and are reported as ‘NA’ in the table | For a small number of metabolites, there are some incomplete entries for METLIN and KEGG IDs within the BioCyc database | We are in the process of systematically identifying and updating any missing data entries |
● TIMING
Steps 1–6, Stage 1: data upload: 30 s–5 min per file, depending on the size of the data set
Steps 7–16, Stage 2: parameter settings: 5–10 min
Steps 17 and 18, Stage 3: XCMS data processing and predictive pathway analysis: 1–3 h, depending on data set size and server queue
Steps 19–46, Stage 4: interpretation of pathway analysis results: 30–40 min
Steps 47–50, Stage 5: multi-omic integration: <1 min
Steps 51–59, Stage 6: interpretation of multi-omic integration results: ~20 min
ANTICIPATED RESULTS
This protocol allows users to quickly generate pathway data directly from raw MS data and further interpret the dysregulated pathways on a systems level by implementing gene and/or protein information. An example was provided using E. coli K12 MG 1655 cultures grown on different carbon sources: glucose and adenosine9. Metabolomic data were generated in HILIC–MS in ESI negative mode, and transcriptomic data were generated using mRNA-seq technology on the same sample set (MetaboLights, study identifier MTBLS572; https://www.ebi.ac.uk/metabolights/MTBLS572; Supplementary Data 1). Dysregulated protein data were generated using literature search on proteome dysregulation under the same carbon source of the same E. coli strain (Supplementary Data 2). Predictive pathway analysis results were generated using XCMS Online, resulting in a list of dysregulated metabolic pathways (Fig. 3); among them, 16 metabolic pathways had predicted P values ≤0.05. Dysregulated metabolic features involved in these pathways, such as pyruvate, fructose 1,6-bisphosphate and 3-phospho-D-glycerate, were tabulated in a table linked to the pathway results, as shown in Figure 4. The most significantly disrupted pathways and metabolites are related to glucose and adenosine metabolism, reflecting the complex cellular response and modulation of major processes, particularly the TCA cycle, upon the change in media. All dysregulated metabolic pathways were plotted in a Pathway Cloud Plot as part of the workflow, illustrated in Figure 5, providing user-friendly visualization and interpretation. The pathway analysis results for the carbon source stress study can be accessed after logging in to XCMS Online (https://xcmsonline.scripps.edu), clicking on the ‘XCMS Public’ menu (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=listPublicShares) and searching for the job number ‘1172567’ or name ‘Ecoli_glucose-vs-adenosine’. All the displayed results can also be downloaded by clicking ‘Download Results’ in the ‘Results Summary page’ of the XCMS job. Upon the completion of pathway analysis, transcriptomic and proteomic data were uploaded for multi-omic integration (Fig. 6). Several dysregulated metabolic pathways were further confirmed by the integration with transcriptomic and proteomic data (Fig. 7). For example, in the glycolysis I pathway, 12 out of 18 genes (67%), 7 out of 18 proteins (39%) and 5 out of 10 metabolites (50%) were significantly dysregulated (Fig. 8). Our systems biology platform can capture metabolic regulation of how E. coli responds to a change in carbon source on a system-wide level. It has wide applicability in many different areas of study, including cell culture, toxicity screening, drug development and safety, epidemiological and exposome applications, and even personalized medicine. This platform provides a fast and efficient method to process MS-based metabolomics data, quickly assess pathway dysregulation and correlate with proteomic and genomic data for a comprehensive systems analysis.
Supplementary Material
Acknowledgments
The authors thank the following for funding assistance: Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research under contract number DE-AC02-05CH11231 (G.S.); and the National Institutes of Health (grants R01 GM114368 (G.S.) and PO1 A1043376-02S1 (G.S.)).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS E.M.F. and T.H. contributed equally to writing the manuscript. E.M.F., T.H., D.R., H.P.B., B.H. and G.S. contributed to platform development, and H.P.B., B.W. and G.S. contributed to manuscript writing.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Fondi M, Liò P. Multi-omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microbiol. Res. 2015;171:52–64. doi: 10.1016/j.micres.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zampieri M, Sekar K, Zamboni N, Sauer U. Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 2017;36:15–23. doi: 10.1016/j.cbpa.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016;88:524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 6.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith C, Want E, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 8.Gowda H, et al. Interactive XCMS online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014;86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huan T, et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods. 2017;14:461–462. doi: 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CA, et al. METLIN - a metabolite mass spectral database. Thera. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 12.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 14.Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–W415. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirhaji L, et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat. Methods. 2016;13:770–776. doi: 10.1038/nmeth.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li SZ, et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9:11. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendelman HE, et al. Evaluation of the safety and immunomodulatory effects of sargramostim in a randomized, double-blind phase 1 clinical Parkinson’s disease trial. Parkinson’s Dis. 2017;3:10. doi: 10.1038/s41531-017-0013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warth B, et al. Exposome-scale investigations guided by global metabolomics, pathway analysis, and cognitive computing. Anal. Chem. 2017;89:11505–11513. doi: 10.1021/acs.analchem.7b02759. [DOI] [PubMed] [Google Scholar]
- 22.Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 2011;83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 23.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27:730–731. doi: 10.1093/bioinformatics/btr001. [DOI] [PubMed] [Google Scholar]
- 25.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giacomoni F, et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31:1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson RL, Weber RJM, Liu HY, Sharma-Oates A, Viant MR. Galaxy-M: a Galaxy workflow for processing and analyzing direct infusion and liquid chromatography mass spectrometry-based metabolomics data. GigaScience. 2016;5:10. doi: 10.1186/s13742-016-0115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, et al. iPEAP: integrating multiple omics and genetic data for pathway enrichment analysis. Bioinformatics. 2014;30:737–739. doi: 10.1093/bioinformatics/btt576. [DOI] [PubMed] [Google Scholar]
- 30.Cottret L, et al. MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 2010;38:W132–W137. doi: 10.1093/nar/gkq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karnovsky A, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelder T, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gika H, Theodoridis G. Sample preparation prior to the LC-MS-based metabolomics/metabonomics of blood-derived samples. Bioanalysis. 2011;3:1647–1661. doi: 10.4155/bio.11.122. [DOI] [PubMed] [Google Scholar]
- 35.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benton HP, et al. Autonomous metabolomics for rapid metabolite identification in global profiling. Anal. Chem. 2015;87:884–891. doi: 10.1021/ac5025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z-J, et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith G, et al. Mutations in APC, Kirsten-ras, and p53 - alternative genetic pathways to colorectal cancer. Proc. Natl. Acad. Sci. USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan XQ, Desiderio DM. Signaling pathway networks mined from human pituitary adenoma proteomics data. BMC Med. Genom. 2010;3:26. doi: 10.1186/1755-8794-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 43.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 45.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 46.Montenegro-Burke JR, et al. Data streaming for metabolomics: accelerating data processing and analysis from days to minutes. Anal. Chem. 2017;89:1254–1259. doi: 10.1021/acs.analchem.6b03890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montenegro-Burke JR, et al. Smartphone analytics: mobilizing the lab into the cloud for omicscale analyses. Anal. Chem. 2016;88:9753–9758. doi: 10.1021/acs.analchem.6b02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trutschel D, Schmidt S, Grosse I, Neumann S. Experiment design beyond gut feeling: statistical tests and power to detect differential metabolites in mass spectrometry data. Metabolomics. 2015;11:851–860. [Google Scholar]
- 49.Causon TJ, Hann S. Review of sample preparation strategies for MS-based metabolomic studies in industrial biotechnology. Anal. Chim. Acta. 2016;938:18–32. doi: 10.1016/j.aca.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 50.Engskog MKR, Haglof J, Arvidsson T, Pettersson C. LC-MS based global metabolite profiling: the necessity of high data quality. Metabolomics. 2016;12:19. [Google Scholar]
- 51.Haggarty J, Burgess KEV. Recent advances in liquid and gas chromatography methodology for extending coverage of the metabolome. Curr. Opin. Biotechnol. 2017;43:77–85. doi: 10.1016/j.copbio.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Kohler I, Giera M. Recent advances in liquid-phase separations for clinical metabolomics. J. Sep. Sci. 2017;40:93–108. doi: 10.1002/jssc.201600981. [DOI] [PubMed] [Google Scholar]
- 53.Muzny DM, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 55.Payne CM, Bernstein C, Dvorak K, Bernstein H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin. Exp. Gastroenterol. 2008;1:19–47. doi: 10.2147/ceg.s4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Field AE, et al. Impact of overweight on the risk of developing common chronic diseases during a 10-year period. Arch. Intern. Med. 2001;161:1581–1586. doi: 10.1001/archinte.161.13.1581. [DOI] [PubMed] [Google Scholar]
- 57.Frei B, Kim MC, Ames BN. Ubiquinol-10 is an effective lipid-soluble antioxidant at physiological concentrations. Proc. Natl. Acad. Sci. USA. 1990;87:4879–4883. doi: 10.1073/pnas.87.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xian F, Hendrickson CL, Marshall AG. High resolution mass spectrometry. Anal. Chem. 2012;84:708–719. doi: 10.1021/ac203191t. [DOI] [PubMed] [Google Scholar]
- 59.Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shevlyakov G, Morgenthaler S, Shurygin A. Redescending M-estimators. J. Stat. Plan. Infer. 2008;138:2906–2917. [Google Scholar]
- 61.Welch BL. The generalisation of student’s problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 62.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Statist. 1947;18:50–60. [Google Scholar]
- 63.Fisher RA. On the probable error of a coefficient of correlation deduced from a small sample. Metron. 1921;1:3–32. [Google Scholar]
- 64.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952;47:583–621. [Google Scholar]
- 65.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 66.Maier T, et al. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011;7:511–511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirai MY, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- 68.Bateman A, et al. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tautenhahn R, et al. metaXCMS: second-order analysis of untargeted metabolomics data. Anal. Chem. 2011;83:696–700. doi: 10.1021/ac102980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.