The imprinted gene Grb10 is expressed in the brain from the paternal copy only. Here, Dent et al. show that paternal Grb10 regulates impulsive choices, i.e. whether an animal chooses a smaller food reward...
Keywords: genomic imprinting, Nesp55, delayed reinforcement, intragenomic conflict, Grb10
Abstract
Imprinted genes are expressed from one parental allele only as a consequence of epigenetic events that take place in the mammalian germ line and are thought to have evolved through intragenomic conflict between parental alleles. We demonstrate, for the first time, oppositional effects of imprinted genes on brain and behavior. Specifically, we show that mice lacking paternal Grb10 make fewer impulsive choices, with no dissociable effects on a separate measure of impulsive action. Taken together with previous work showing that mice lacking maternal Nesp55 make more impulsive choices, this suggests that impulsive choice behavior is a substrate for the action of genomic imprinting. Moreover, the contrasting effect of these two genes suggests that impulsive choices are subject to intragenomic conflict and that maternal and paternal interests pull this behavior in opposite directions. Finally, these data may also indicate that an imbalance in expression of imprinted genes contributes to pathological conditions such as gambling and drug addiction, where impulsive behavior becomes maladaptive.
IMPRINTED genes are expressed from one parental allele only as a consequence of epigenetic events that take place in the mammalian germ line (Ferguson-Smith 2011). Functionally, imprinted genes converge on specific biological processes that have prominent importance in mammals, such as in utero growth, metabolism, and behavior (Peters 2014; Wilkins et al. 2016). Many of the effects that maternally and paternally expressed imprinted genes have on in utero growth and during the early postnatal period are oppositional in direction (Haig and Graham 1991; Madon-Simon et al. 2014), lending support to the theory that imprinting has evolved as a consequence of intragenomic conflict (Moore and Haig 1991; Wilkins and Haig 2003).
Grb10 is currently a unique example of an imprinted gene that is expressed from the maternal allele only in some tissues, and the paternal allele only in others (Arnaud et al. 2003; Hikichi et al. 2003; Monk et al. 2009). Consequently, different parental alleles of Grb10 mediate distinct physiological functions (Garfield et al. 2011). Maternal Grb10 is expressed in many peripheral body tissues, while being excluded from the central nervous system (Garfield et al. 2011), and plays a prominent role in controlling placental function (Charalambous et al. 2003, 2010) and metabolism (Smith et al. 2007). In contrast, paternal Grb10 is essentially restricted to the brain alone and impacts on behavioral phenotypes, including social dominance (Garfield et al. 2011). Grb10 expression in the brain is neuronal, showing a pattern of expression in discrete brain regions that is shared with the maternally expressed imprinted gene Nesp (Plagge et al. 2005). This overlap in brain expression of maternal Nesp (encoding Nesp55) and paternal Grb10 has led to speculation that these two genes may influence common adult behaviors, possibly in an opposite manner (Garfield et al. 2011; Dent and Isles 2014), which would be consistent with the prediction from the intragenomic conflict theory of genomic imprinting evolution (Haig 2000).
Here, we address this question directly. We have shown that mice lacking maternal Nesp55 (Nespm/+) make more impulsive choices (Dent et al. 2016). In contrast, here we show that Grb10+/p mice make less impulsive choices relative to their littermate controls. Furthermore, there were no dissociable effects on a separate measure of impulsive action, completely matching the specificity of effects seen previously in Nespm/+ mice (Dent et al. 2016). Our data provide new findings on Grb10 function in the brain and reveal the opposite effects of manipulating Grb10 and Nesp expression on discrete aspects of impulsive responding. Taken together, these data are consistent with the conflict theory of the evolution of imprinted genes and, as such, provide the first direct evidence for intragenomic conflict impacting adult behavior.
Materials and Methods
Animals
All procedures were conducted in accordance with the UK Animals (Scientific Procedures) Act 1986. Subjects were male mice, and were 4 months old at the start of testing, which was completed after 6 months. Standard laboratory chow was available ad libitum, but during the experiment water was restricted to 2 hr access per day. This regime maintained the subjects at ≈90% of free-feeding body weight. Due to potentially confounding phenotypes associated with Grb10m/+ mice, comparisons of Grb10+/p were made with wild-type (WT) littermate controls derived from eight separate litters. The Grb10-null line was maintained on an F1-hybrid (C57/Bl6 × CBA) background. The same cohort was used throughout testing but a number of animals were lost (due to death or inability to perform training stages) as testing progressed.
Behavioral testing
Operant testing apparatus:
All the sessions of the delayed reinforcement task and stop-signal reaction time (SSRT) task were performed in 9-hole operant chambers (Cambridge Cognition, Bottisham, UK) modified for use with mice, as described previously (Humby et al. 1999, 2013; Isles et al. 2003). For the delayed reinforcement task, holes 3, 5, and 7 were open, whereas only holes 4 and 6 were open for the SSRT task. The mice were presented with a visual stimulus (light) recessed into the holes and were trained to respond to this stimulus with a nose-poke, as recorded by infrared beams spanning the hole. Reward was presented in a recessed compartment on the wall opposite to the nose-poke/stimulus array. The control of the stimuli and recording of the responses were managed by an Acorn Archimedes computer with additional interfacing by ARACHNID (Cambridge Cognition). For all operant testing, animals were maintained on a restricted water access schedule with water provided for 2 hr immediately after testing.
Delayed reinforcement task:
Details of the shaping procedures and basic aspects of the delayed reinforcement task itself can be found elsewhere (Isles et al. 2003). Briefly, the task comprised of three sequential blocks of 12 trials, with each trial consisting of an initial nose-poke to the centrally located stimulus, followed by a second nose-poke to either the left or right apertures. Trials 1–4 in any block were “forced” information trials, where the initial nose-poke resulted in presentation of only one of the two choice options. This measure was designed to provide the subjects with prior notice of the extent of any delay associated with choosing the large reward. In the remaining eight trials of each block, designated as choice trials, the initial center nose-poke led to the option of a second nose-poke response to either the left or right apertures. One response resulted in the delivery of a large reward (50 μl 10% solution of condensed milk; Nestlé, Gatwick, UK), and the other in the delivery of small reward (25 μl 10% solution of condensed milk). The response contingencies were kept constant for each mouse, but were counterbalanced between subjects. In block 1, both responses led to the delivery of reward after a 1 sec delay. In blocks 2 and 3, increasing delays were introduced between the response and the delivery of the large reward (8 and 16 sec, respectively) whereas the delay between response and delivery of the small reward was fixed at 1 sec. As a probe to test the effect of the delays on behavior, sessions were conducted where the delay associated with the large reward was fixed at 1 sec, equivalent to that associated with the small reward, throughout all three blocks of the session.
The bias in choice of the larger reward at each block (whereby always choosing the large reward = 1; never choosing the large reward = 0) was the main measure used to determine impulsive responding. Additional measurements that related to general motoric competence and motivation within the task were also monitored, including the start and choice latencies, the time taken to initiate a trial, and the time taken to make a choice once a trial was initiated, respectively. Also measured were the number of “nonstarted” (no initial, central nose-poke) and “omitted” (no secondary, choice nose-poke, following central nose-poke initiating trial) trials.
SSRT task:
Details of the shaping procedures and basic aspects of the main SSRT task can also be found elsewhere (Humby et al. 2013). The SSRT task itself consisted of sessions of 100 trials, which involved both “go” and “stop” trials. Go trials consisted of rapid double nose-pokes (a go response) between two separate stimuli locations, which were rewarded with reinforcement (22 μl, 10% solution of condensed milk; Nestlé). Of the total number of trials, 20% were stop trials that were pseudorandomly distributed throughout each session, where a stop-signal (65 dB white noise for 0.3 sec) was presented between the first and second nose-poke responses. The aim of the stop-signal was to inhibit (stop) the mouse from making the second (go) nose-poke, and then wait for the reward. Failure to refrain from making this prepotent response was punished by the absence of reward and 5 sec time out (chamber light on). At baseline, the stop-signals were presented concurrently with the initial nose-poke response. To maintain high levels of performance of both go and stop responding, the go stimulus duration and wait period to reward delivery in a stop-signal trial were determined individually for each subject. To assess the ability to stop once an action had been initiated, sessions were implemented in which the onset of the stop-signal was presented at different positions within the individualized go response of each mouse. Thus, the stop-signal was pseudorandomly presented 10, 40, 50, 60, and 90% from the onset of the go response of each subject, with the assumption that stopping would be more difficult the closer the stop-signal presentation was to the termination of the go response.
The amount of correct stopping in stop-signal trials and the SSRT were the main measures of impulsive responding in this task. The SSRT was calculated by determining the 50% stopping ability for each subject from the range of sessions in which the stop-signal onset was varied from baseline [full details of this calculation can be found in the supplemental material and Davies et al. (2014)]. The proportion of correct go responses, and latency to respond, were also assessed. Additional measurements that related to general motoric competence and motivation within the task were also monitored, including the “initiation” and “magazine” latencies, the time taken to initiate a trial, and the time taken to collect the reward. Also measured was the number of trials completed for any given session.
Immunohistochemistry
Dual-labeling immunofluorescence analysis of Nesp55 colocalization with Grb10 was carried out on brain sections from Grb10+/p mice to stain for the Lac-Z reporter gene, which is expressed in place of Grb10 and has been used as a faithful proxy for Grb10 expression previously (Garfield et al. 2011). Grb10+/p mice were transcardially perfused using 10% formalin (Sigma [Sigma Chemical], St. Louis, MO) and whole brains dissected, postfixed, and equilibrated with 30% sucrose in PBS. Brains were sectioned into 40-μm coronal slices using a freezing microtome. Sections were washed three times for 10 min each in 0.1% PBS before being incubated for 15 min in 0.3 M glycine in 0.1% PBS at room temperature, to neutralize endogenous aldehyde groups. Sections were washed, as before, in 0.1% PBS and then incubated at room temperature for 1 hr in 10% blocking solution; 0.5% BSA (BB International, Cardiff, UK) and 0.5% Triton X-100 (v/v; Sigma) in 0.1% PBS. Sections were then transferred to a 1% blocking solution containing a β-galactosidase (β-gal)-specific antibody (Abcam, Cambridge, UK), used at a 1:1000 dilution, and a Nesp55 primary antibody (1:1000). The Nesp-55 primary antibody was generated in house, and is a rabbit anti-Nesp55 polyclonal antibody recognizing the free terminal end (GAIPIRRH) of Nesp55. It has been successfully characterized and used previously (Ischia et al. 1997). Sections were incubated overnight at 4° while gently shaking. Sections were then washed three times for 10 min in 0.1% PBS and then incubated with the appropriate fluorescent secondary antibodies (Alexa Fluor; Life Technologies) (1:1000) in 1% blocking solution in the dark at room temperature for 2 hr, while gently shaking. Sections were then washed in 0.1% PBS as before (in the dark) and transferred to polysine-coated slides and allowed to dry overnight in a dark, dust-free environment. The mounted slides were then dehydrated through a process of incubation in a rising concentration of alcohol, followed by xylene, then cover-slipped and sealed using DPX mountant (Raymond Lamb DPX), and allowed to dry overnight. To control for nonspecific binding of the secondary antibodies, secondary-only negative controls were carried out alongside all experiments. Immunofluorescence slides were viewed and images captured using an upright fluorescence microscope (Leica DM5000 B). Dual-labeled immunofluorescence images were acquired through separate channels for different wavelengths (488 and 568 nm) then subsequently merged using ImageJ (Image > color > merge channels).
Data analysis and statistics
All behavioral data were analyzed using SPSS 20 (SPSS). Data were assessed for normality and then analyzed by Student’s t-test or mixed ANOVA, with between-subject factors of GENOTYPE (Grb10+/p vs. WT), and within-subject factors DELAY (1, 8, and 16 sec, or 1, 1, and 1 sec), CHOICE (choice of large or small reward during forced trials of the delayed reinforcement task), and STOP-SIGNAL POSITION (position of stop-signal relative to individualized go response). For repeated-measures analyses, Mauchly’s test of sphericity of the covariance matrix was applied; significant violations from the assumption of sphericity were subject to the Huynh–Feldt correction to allow more conservative comparisons through adjusted d.f. Variables that were expressed as a percentage (delayed reinforcement, choice bias; SSRT, % correct) were subjected to an arcsine transformation to limit the effect of an artificially imposed ceiling. All significance tests were performed at an α level of 0.05. Effect sizes (partial η2) were reported for the main measures in all tasks.
Data availability
The supplemental materials contain additional control measures in the delay discounting task (Figure 1 and Supplemental Material, Figure S1 and Table S1) and SSRT task (Figure 2); alongside further methodological detail relating to SSRT measurement. Individual animal data for the delayed reinforcement task can be found in Appendix S1 (Excel spread sheet). Individual animal data for the SSRT task can be found in Appendix S2 (Excel spread sheet). All other data sets in the current study are available from the corresponding author on reasonable request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5972224.
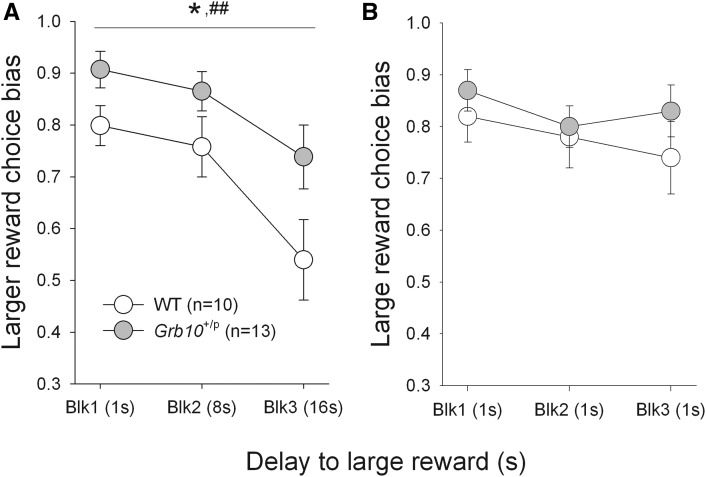
Figure 1.
Grb10+/p mice make less impulsive choices on a delayed reinforcement task. Behavior of both Grb10+/p and wild-type (WT) mice changed across session blocks (blk) with increasing delay, such that choice bias moved away from the response leading to the large reward toward the small reward, with increasing delay (A). However, there were systematic differences between the groups in their behavior, such that Grb10+/p animals switched their choice to the small, less delayed reward less quickly than WT mice. When the delay associated with the large and small rewards was equal (1 sec) throughout the session (B), choice bias was consistently high (large reward chosen ∼80% of the time). Under these task conditions there were no differences in choice bias between Grb10+/p and WT mice. Data shows mean ± SEM of three consecutive stable sessions; * represents P < 0.05 main effect of GENOTYPE; ## represents P < 0.01 main effect of DELAY.
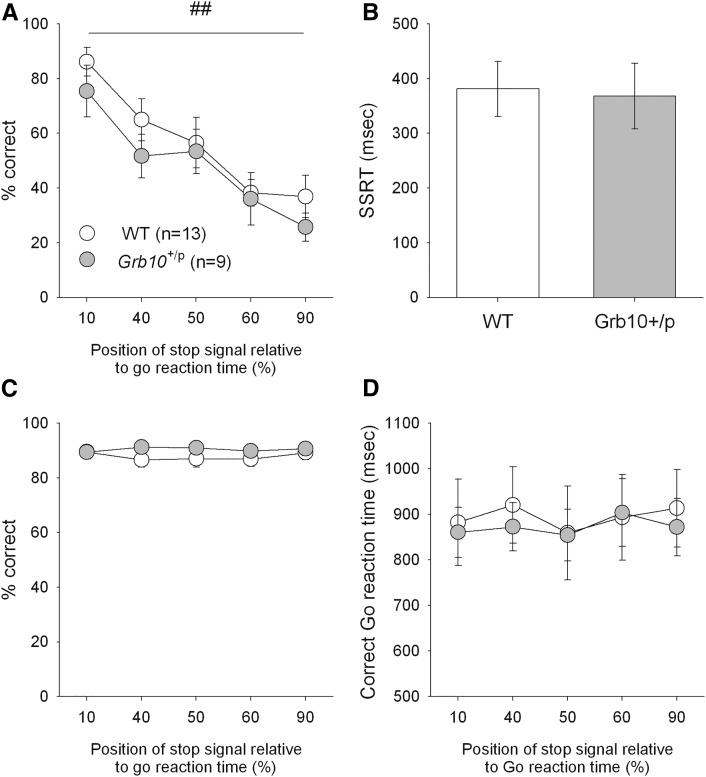
Figure 2.
No difference between Grb10+/p and wild-type (WT) mice in performance on a stop-signal reaction time (SSRT) task. Both WT and Grb10+/p mice showed an equivalent ability to perform the SSRT task, showing the expected change in percentage correct responding during a stop trial (A) as the position of the stop-signal was altered, but there were no differences between Grb10+/p and WT mice. Grb10+/p and WT mice also showed equivalent SSRTs at 50% correct stopping (B). There were no genotype differences for the go response for both groups of mice, in terms of percentage correct responding (C) or response speed (D). Data shows mean ± SEM; ## represents P < 0.01 main effect of stop-signal position.
Results and Discussion
Grb10+/p mice make less impulsive choices
We used the delayed reinforcement task to examine impulsive choice behavior (Isles et al. 2003), where subjects made a choice between receiving a small food reward after a short (1 sec) delay or a larger reward after a longer delay (1, 8, or 16 sec). The extent to which subjects make impulsive choices is indexed by a preference for choosing the immediate but smaller reward, vs. a larger delayed reward (delayed gratification). Increasing the delay to the larger reward within a session decreased the likelihood of choosing that reward across all subjects (Figure 1A; main effect of DELAY, F2,42 = 9.20, P = 0.001, partial η2 = 0.37). However, Grb10+/p mice were significantly more tolerant of an increased delay to the larger reward than WT littermate mice (Figure 1A; main effect of GENOTYPE, F1,21 = 6.45, P = 0.019, partial η2 = 0.25). However, while the data indicate that the experimental group diverges from WT increasingly as the delay increases, this effect failed to result in a significant interaction between GENOTYPE and DELAY (Figure 1A; F2,42 = 0.56, P = 0.946, partial η2 = 0.003), due largely to a preexisting group difference in the first 1-sec delay block.
A standard control measure to assess the extent to which behavior is sensitive to delay is a task manipulation where any differential delay associated with the larger and smaller reward was equalized (1 sec) across all three blocks. Here, all subjects now demonstrated the expected preference for the larger reward throughout the session (Figure 1B; no main effect of DELAY, F1.7,35.1 = 1.88, P = 0.17, partial η2 = 0.08); that is under these task conditions there were no differences in choice-bias between Grb10+/p and WT mice (no main effect of GENOTYPE, F1,21 = 0.63, P = 0.44, partial η2 = 0.03). Furthermore, there was no difference in the sessions taken to achieve a stable baseline in these conditions (WT 9 ± 0.8; Grb10+/p 8 ± 0.5; t21 = 1.51, P = 0.15).
The contrasting patterns of choice behavior between the Grb10+/p and WT mice were also not due to any differences between the groups in terms of basic motivation to carry out the task as Grb10+/p animals acquired the task at the same rate as WT littermates (sessions to last day of baseline; WT 50.4 ± 1.1 SEM, Grb10+/p 47.7 ± 1.2 SEM, t21 = 1.73, P = 0.10). Additionally, variability between Grb10+/p and WT mice was not related to differences in experiencing the information trial contingencies. In the forced trials (where no choice was available), both Grb10+/p and their WT littermates made equal responses to the large and small reward-related stimuli at all delays (no interaction GENOTYPE × DELAY × CHOICE, F2,42 = 0.44, P = 0.649; data not shown). Finally, there was no difference between Grb10+/p and WT mice on general measures of task performance (see supplemental material).
In large part, choice bias behavior in delayed reinforcement tasks is governed by three potentially interacting psychological processes; the perceived value of the rewards, and the perceived length and aversive nature of the delay (Ho et al. 1999). The difference in choice between Grb10+/p and WT mice in the first block of the task (where delays for the larger and smaller rewards are both 1 sec) may suggest a contribution to behavior in the Grb10+/p from a relatively less pronounced generalized conditioned place aversion to the delayed larger reward response [see Isles et al. (2004)]. However, importantly, such an effect did not lead to an inflexible bias in responding as demonstrated by the continued sensitivity to delay, both within a baseline session and in response to the equal-delay probe manipulation. Therefore, while the between-group behavioral differences were clearly influenced to a large degree by a differential tolerance to delay, the present data do not rule out influences due to aversion and reward perception
Grb10+/p mice show no difference in impulsive action
In contrast to impulsive choice, there was no difference between Grb10+/p and WT mice on a measure of impulsive action. The SSRT task measures the ability to stop an action once initiated by presenting a stop-signal (in this case an auditory tone) during a rapid response between two stimuli locations (the go response). Correctly inhibiting the go response will earn reward in a stop trial, whereas reward was also presented on completing a go response in trials where no stop-signal was presented. Throughout the training stages of the SSRT task, all subjects showed equivalent behavior in learning the task, and Grb10+/p and WT mice acquired the task at the same rate (sessions taken to complete the task, Grb10+/p: 42.4 ± 9.8, WT: 40.8 ± 4.5, t20 = 0.54, P = 0.59).
As expected (Humby et al. 2013; Davies et al. 2014; Dent et al. 2016), presenting the stop-signal progressively closer to the execution of the response (10, 40, 50, 60, and 90% into the individualized go response) led to systematic reductions in the ability to stop for all mice (Figure 2A; ANOVA, main effect of STOP, F4,80 = 24.37, P < 0.001, partial η2 = 0.55). However, there were no differences in stopping efficiency between Grb10+/p and WT mice (Figure 2A, main effect of GENOTYPE, F1,20 = 0.94, P = 0.34, partial η2 = 0.05). The lack of genotype differences in stopping ability was further demonstrated by another measure of response inhibition, the speed of stopping or SSRT derived in sessions where the subjects exhibited 50% correct stopping (Bari et al. 2009; Humby et al. 2013; Davies et al. 2014; Dent et al. 2016). Equivalent SSRTs were observed in both Grb10+/p mice and their WT littermates (Figure 2B; t = 17, P = 0.87).
There were also no differences in the go response between Grb10+/p and WT mice, in terms of the amount (Figure 2C, main effect of GENOTYPE, F1,20 = 1.14, P = 0.30) or speed (Figure 2D, main effect of GENOTYPE, F1,20 = 0.03, P = 0.86) of correct responding in go trials. These parameters were not affected in sessions when the stop-signal position was moved from baseline (main effect of STOP-SIGNAL POSITION, F4,80 = 0.28, P = 0.89 and F4,80 = 0.85, P = 0.50, for the amount and speed of correct responding in go trials, respectively). Additionally, there was no difference between Grb10+/p and WT mice on general measures of task performance during individualized SSRT sessions (see supplemental material), indicating a high degree of stimulus control for both groups in the task.
Opposite effects to maternal Nesp
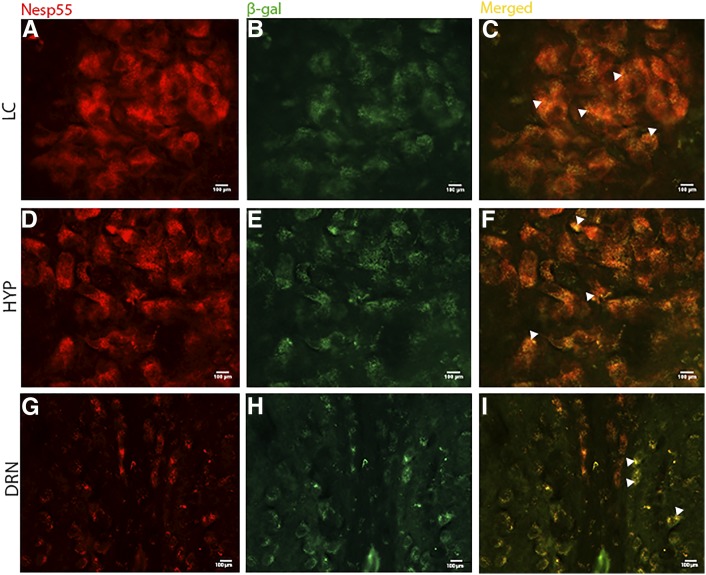
The current data obtained from knocking out the paternal copy of Grb10 are essentially opposite to our previously published effects of knocking out the maternal copy of Nesp55 (Dent et al. 2016). That is, under identical task conditions, mice lacking maternal Nesp55 make more impulsive choices, whereas mice lacking paternal Grb10 make fewer impulsive choices, relative to their littermate controls. These effects were highly specific in that they occurred in the absence of deficits in either model on responding in the SSRT task. The idea that these oppositely imprinted genes converge on discrete aspects of impulse control is further supported by their general overlapping expression patterns in the brain (Plagge et al. 2005; Garfield et al. 2011). In the present work, we extend these neuroanatomical data in showing that Nesp55 and Grb10 are colocalized in cells in the locus coeruleus (Figure 3, A–C), hypothalamus (Figure 3, D–F), and the dorsal raphe nucleus (Figure 3, G–I). Together, these studies indicate that impulsive choice behavior is a substrate for the action of genomic imprinting, and an extension of this idea is the speculation that an imbalance in expression of imprinted genes, such as GRB10 or NESP, may also contribute to pathological conditions, such as pathological gambling, where choices become highly maladaptive. In this regard, it should be noted that neither GRB10 nor NESP have associated SNPs identified as significant variants in recent genome-wide association studies (GWAS) studies of gambling behavior (Lang et al. 2016), or indeed delay discounting (Sanchez-Roige et al. 2018). However, it is worth noting also that genes with known roles in impulsive behavior (e.g., DAT, DRD1, or DRD2) were not associated either, suggesting that a lack of signal in GWAS does not necessarily diminish the suggestion of a role for GRB10 and/or NESP in these disorders of impulse control.
Figure 3.
Dual-labeling immunofluorescence histochemistry of Nesp55 and Grb10 in coronal sections of adult brain. Sections were dual-labeled with antibodies against Nesp55 and β-gal, where the reporter gene LacZ is expressed in place of Grb10 in tissue from Grb10+/p mice, and can be used to identify Grb10-positive cells. Images were viewed at different light intensities (568 and 488 nm for Nesp55 and β-gal, respectively), and were then merged to gauge cellular colocalization of the two target proteins, depicted by white arrows in the merged figures. The majority of cells showed evidence of colocalization: within the locus coeruleus (LC) (A–C), the hypothalamus (HYP) (D–F), and the dorsal raphe nuclei (DRN) (G–I). LC and HYP images at ×40 magnification, DRN at ×20.
We also suggest these data are consistent with the intragenomic conflict theory of the evolution of imprinted genes, providing the first direct evidence for oppositional effects of imprinted genes on an adult behavior. In this context, the examples of Nesp55- and Grb10-null mice are particularly important, as these models are free from confounding effects on in utero or preweaning growth (Plagge et al. 2005; Garfield et al. 2011). Hence, this strengthens the argument that imprinted gene effects on adult behavior have adaptive significance and are not simply epiphenomena, that is, enduring into adulthood but resulting from the more familiar imprinted gene substrate of resource allocation between mother and offspring (Constancia et al. 2004).
Imprinted genes are thought to have evolved as a consequence of conflicting phenotypic interests between maternal and paternal genes that cause an escalating arms race in relation to allelic expression, eventually leading to silencing of one or other parental allele (Moore and Haig 1991; Wilkins and Haig 2003). Evidence of this parental conflict is seen in the contrasting action of maternal and paternal imprinted genes during in utero growth and early postnatal life (Haig 2000). Although the present work is the first demonstration of opposing parental interests on behavior, the specificity of imprinting effects on impulsive choice behavior described here would fit with previous suggestions that maternal and paternal genomes may have a contrasting impact on decision-making in social animals (Haig 2000; Brandvain et al. 2011). Patterns of impulsive choice behavior with increasing delay, as measured by delay-discounting in the delayed reinforcement task here, are often thought of as “irrational,” in that the shape of behavior is not consistent with simple models of optimization (such as expected utility maximization) (Schonberg et al. 2011). It may be that the so-called “parliament of the mind” (Haig 2000), caused by opposing parental genomes pulling impulsive choice in different directions, is an additional factor that should be taken into consideration in the context of more complex models of apparently irrational behaviors (Fawcett et al. 2014).
Acknowledgments
This work was funded by a Leverhulme Trust project grant (F/00 407/BF) to A.R.I. and J.F.W., which supported C.L.D. The work was also funded by the Medical Research Council Centre for Neuropsychiatric Genetics and Genomics (MR/L010305/1), of which A.R.I., T.H., and L.S.W. are members, and which also supported K.L. via a PhD studentship.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5972224.
Communicating editor: A. Palmer
Literature Cited
- Arnaud P., Monk D., Hitchins M., Gordon E., Dean W., et al. , 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12: 1005–1019. [DOI] [PubMed] [Google Scholar]
- Bari A., Eagle D. M., Mar A. C., Robinson E. S., Robbins T. W., 2009. Dissociable effects of noradrenaline, dopamine, and serotonin uptake blockade on stop task performance in rats. Psychopharmacology (Berl.) 205: 273–283. 10.1007/s00213-009-1537-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., Van Cleve J., Ubeda F., Wilkins J. F., 2011. Demography, kinship, and the evolving theory of genomic imprinting. Trends Genet. 27: 251–257. 10.1016/j.tig.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Charalambous M., Smith F. M., Bennett W. R., Crew T. E., Mackenzie F., et al. , 2003. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl. Acad. Sci. USA 100: 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous M., Cowley M., Geoghegan F., Smith F. M., Radford E. J., et al. , 2010. Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 337: 1–8. 10.1016/j.ydbio.2009.10.011 [DOI] [PubMed] [Google Scholar]
- Constancia M., Kelsey G., Reik W., 2004. Resourceful imprinting. Nature 432: 53–57. [DOI] [PubMed] [Google Scholar]
- Davies W., Humby T., Trent S., Eddy J. B., Ojarikre O. A., et al. , 2014. Genetic and pharmacological modulation of the steroid sulfatase axis improves response control; comparison with drugs used in ADHD. Neuropsychopharmacology 39: 2622–2632. 10.1038/npp.2014.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent C. L., Isles A. R., 2014. Brain-expressed imprinted genes and adult behaviour: the example of Nesp and Grb10. Mamm. Genome 25: 87–93. 10.1007/s00335-013-9472-0 [DOI] [PubMed] [Google Scholar]
- Dent C. L., Humby T., Lewis K., Plagge A., Fischer-Colbrie R., et al. , 2016. Impulsive choices in mice lacking imprinted Nesp55. Genes Brain Behav. 15: 693–701. 10.1111/gbb.12316 [DOI] [PubMed] [Google Scholar]
- Fawcett T. W., Fallenstein B., Higginson A. D., Houston A. I., Mallpress D. E., et al. , 2014. The evolution of decision rules in complex environments. Trends Cogn. Sci. 18: 153–161. 10.1016/j.tics.2013.12.012 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith A. C., 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12: 565–575. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]
- Garfield A. S., Cowley M., Smith F. M., Moorwood K., Stewart-Cox J. E., et al. , 2011. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 469: 534–538. 10.1038/nature09651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D., 2000. Genomic imprinting, sex-biased dispersal, and social behavior. Ann. N. Y. Acad. Sci. 907: 149–163. [DOI] [PubMed] [Google Scholar]
- Haig D., Graham C., 1991. Genomic imprinting and the strange case of the insulin-like growth-factor II receptor. Cell 64: 1045–1046. [DOI] [PubMed] [Google Scholar]
- Hikichi T., Kohda T., Kaneko-Ishino T., Ishino F., 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M. Y., Mobini S., Chiang T. J., Bradshaw C. M., Szabadi E., 1999. Theory and method in the quantitative analysis of “impulsive choice” behaviour: implications for psychopharmacology. Psychopharmacology (Berl.) 146: 362–372. [DOI] [PubMed] [Google Scholar]
- Humby T., Laird F. M., Davies W., Wilkinson L. S., 1999. Visuospatial attentional functioning in mice: interactions between cholinergic manipulations and genotype. Eur. J. Neurosci. 11: 2813–2823. [DOI] [PubMed] [Google Scholar]
- Humby T., Eddy J. B., Good M. A., Reichelt A. C., Wilkinson L. S., 2013. A novel translational assay of response inhibition and impulsivity: effects of prefrontal cortex lesions, drugs used in ADHD, and serotonin 2C receptor antagonism. Neuropsychopharmacology 38: 2150–2159. 10.1038/npp.2013.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischia R., Lovisetti-Scamihorn P., Hogue-Angeletti R., Wolkersdorfer M., Winkler H., et al. , 1997. Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5–HT1B receptor antagonist activity. J. Biol. Chem. 272: 11657–11662. [DOI] [PubMed] [Google Scholar]
- Isles A. R., Humby T., Wilkinson L. S., 2003. Measuring impulsivity in mice using a novel operant delayed reinforcement task: effects of behavioural manipulations and d-amphetamine. Psychopharmacology (Berl.) 170: 376–382. [DOI] [PubMed] [Google Scholar]
- Isles A. R., Humby T., Walters E., Wilkinson L. S., 2004. Common genetic effects on variation in impulsivity and activity in mice. J. Neurosci. 24: 6733–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M., Lemenager T., Streit F., Fauth-Buhler M., Frank J., et al. , 2016. Genome-wide association study of pathological gambling. Eur. Psychiatry 36: 38–46. 10.1016/j.eurpsy.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Madon-Simon M., Cowley M., Garfield A. S., Moorwood K., Bauer S. R., et al. , 2014. Antagonistic roles in fetal development and adult physiology for the oppositely imprinted Grb10 and Dlk1 genes. BMC Biol. 12: 771 10.1186/s12915-014-0099-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D., Arnaud P., Frost J., Hills F. A., Stanier P., et al. , 2009. Reciprocal imprinting of human GRB10 in placental trophoblast and brain: evolutionary conservation of reversed allelic expression. Hum. Mol. Genet. 18: 3066–3074. 10.1093/hmg/ddp248 [DOI] [PubMed] [Google Scholar]
- Moore T., Haig D., 1991. Genomic imprinting in mammalian development - a parental tug-of-war. Trends Genet. 7: 45–49. [DOI] [PubMed] [Google Scholar]
- Peters J., 2014. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 15: 517–530. 10.1038/nrg3766 [DOI] [PubMed] [Google Scholar]
- Plagge A., Isles A. R., Gordon E., Humby T., Dean W., et al. , 2005. Imprinted nesp55 influences behavioral reactivity to novel environments. Mol. Cell. Biol. 25: 3019–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S., Fontanillas P., Elson S. L., Pandit A., Schmidt E. M., et al. , 2018. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat. Neurosci. 21: 16–18. 10.1038/s41593-017-0032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg T., Fox C. R., Poldrack R. A., 2011. Mind the gap: bridging economic and naturalistic risk-taking with cognitive neuroscience. Trends Cogn. Sci. 15: 11–19. 10.1016/j.tics.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. M., Holt L. J., Garfield A. S., Charalambous M., Koumanov F., et al. , 2007. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol. Cell. Biol. 27: 5871–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins J. F., Haig D., 2003. What good is genomic imprinting: the function of parent-specific gene expression. Nat. Rev. Genet. 4: 359–368. [DOI] [PubMed] [Google Scholar]
- Wilkins J. F., Ubeda F., Van Cleve J., 2016. The evolving landscape of imprinted genes in humans and mice: conflict among alleles, genes, tissues, and kin. BioEssays 38: 482–489. 10.1002/bies.201500198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The supplemental materials contain additional control measures in the delay discounting task (Figure 1 and Supplemental Material, Figure S1 and Table S1) and SSRT task (Figure 2); alongside further methodological detail relating to SSRT measurement. Individual animal data for the delayed reinforcement task can be found in Appendix S1 (Excel spread sheet). Individual animal data for the SSRT task can be found in Appendix S2 (Excel spread sheet). All other data sets in the current study are available from the corresponding author on reasonable request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5972224.
Figure 1.
Grb10+/p mice make less impulsive choices on a delayed reinforcement task. Behavior of both Grb10+/p and wild-type (WT) mice changed across session blocks (blk) with increasing delay, such that choice bias moved away from the response leading to the large reward toward the small reward, with increasing delay (A). However, there were systematic differences between the groups in their behavior, such that Grb10+/p animals switched their choice to the small, less delayed reward less quickly than WT mice. When the delay associated with the large and small rewards was equal (1 sec) throughout the session (B), choice bias was consistently high (large reward chosen ∼80% of the time). Under these task conditions there were no differences in choice bias between Grb10+/p and WT mice. Data shows mean ± SEM of three consecutive stable sessions; * represents P < 0.05 main effect of GENOTYPE; ## represents P < 0.01 main effect of DELAY.
Figure 2.
No difference between Grb10+/p and wild-type (WT) mice in performance on a stop-signal reaction time (SSRT) task. Both WT and Grb10+/p mice showed an equivalent ability to perform the SSRT task, showing the expected change in percentage correct responding during a stop trial (A) as the position of the stop-signal was altered, but there were no differences between Grb10+/p and WT mice. Grb10+/p and WT mice also showed equivalent SSRTs at 50% correct stopping (B). There were no genotype differences for the go response for both groups of mice, in terms of percentage correct responding (C) or response speed (D). Data shows mean ± SEM; ## represents P < 0.01 main effect of stop-signal position.