Abstract
Although single-cell RNA-seq is revolutionizing biology, data interpretation remains a challenge. We present SCENIC for the simultaneous reconstruction of gene regulatory networks and identification of cell states. We apply SCENIC to a compendium of single-cell data from tumors and brain, and demonstrate that the genomic regulatory code can be exploited to guide the identification of transcription factors and cell states. SCENIC provides critical biological insights into the mechanisms driving cellular heterogeneity.
The transcriptional state of a cell emerges from an underlying gene regulatory network (GRN) in which a limited number of transcription factors and co-factors regulate each other and their downstream target genes. Recent advances in single-cell transcriptome profiling have provided exciting opportunities for a high-resolution identification of transcriptional states, and to identify trajectories of transitions between states, for example during differentiation 1,2. Statistical techniques and bioinformatics methods have been optimized for single-cell RNA-seq, including methods for expression normalization, differential expression analysis, clustering, dimensionality reduction, rare cell type identification, and trajectory inference 3. Although these methods have led to significant new biological insights, it is still unclear whether specific and robust GRNs underlying stable cell states can be established. This may indeed be challenging given that at the single cell level, gene expression may be partially disconnected from the dynamics of transcription factor inputs due to stochastic variation of gene expression consecutive to, for example, transcriptional bursting 4. A few methods have been developed that infer co-expression networks from single-cell RNA-seq data 5–7, but they do not make use of regulatory sequence analysis to predict interactions between transcription factors and target genes.
We reasoned that linking the genomic regulatory code to single-cell gene expression variation could overcome drop-outs and technical variation, and could optimize the discovery and characterization of cellular states. To this end, we developed a new method, called SCENIC (Single-Cell rEgulatory Network Inference and Clustering), to map GRNs, and then identify stable cellular states by evaluating the activity of the GRNs in each cell. The SCENIC workflow consists of three steps (Fig. 1a, Supplementary Fig. 1 and Online Methods). In the first step, sets of genes that are co-expressed with transcription factors are identified using GENIE3 8 (Supplementary Fig. 1a). Being based only on co-expression, these modules may include many false positives and indirect targets. To identify putative direct-binding targets, each co-expression module is analyzed using cis-regulatory motif analyses using RcisTarget (Supplementary Fig. 1b and Online Methods). Only modules with significant motif enrichment of the correct upstream regulator are retained, and pruned to remove indirect target genes without motif support. Next, we score the activity of each of these regulons in each cell with AUCell (Supplementary Fig. 1c, Supplementary Fig. 2, and Online Methods). The relative scores of each regulon across the cells allow identifying which cells have a significantly high sub-network activity. The resulting binary activity matrix can be used as a biological dimensionality reduction for downstream analyses. For example, performing a clustering on this matrix allows identifying cell types and states based on the shared activity of a regulatory subnetwork. In addition, since the regulon is scored as a whole, instead of only the TF or individual genes, this approach is robust against drop-outs (Supplementary Fig. 3).
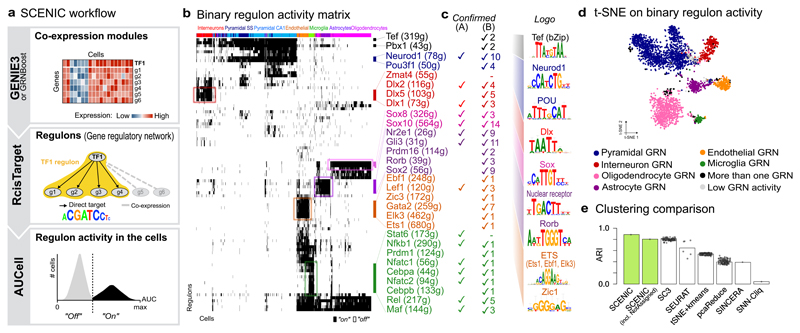
Figure 1. The SCENIC workflow and its application to the mouse brain.
(a) Co-expression modules between transcription factors and candidate target genes are inferred using GENIE3 or GRNBoost. RcisTarget identifies those modules where the regulator’s binding motif is significantly enriched across the target genes; and creates regulons with only direct targets. AUCell scores the activity of each regulon in each cell, yielding a binarized activity matrix. Cell states are based on the shared activity of regulatory subnetworks. (b) SCENIC results on the mouse brain 9; cluster labels correspond to 9; master regulators are color-matched with the cell types they control. (c) transcription factors confirmed by literature (A) or having brain phenotypes from MGI (B), and the enriched DNA motifs are shown. (d) t-SNE on the binary regulon activity matrix. Each cell is assigned the color of the most active GRN. (e) Accuracy of different clustering methods on this dataset.
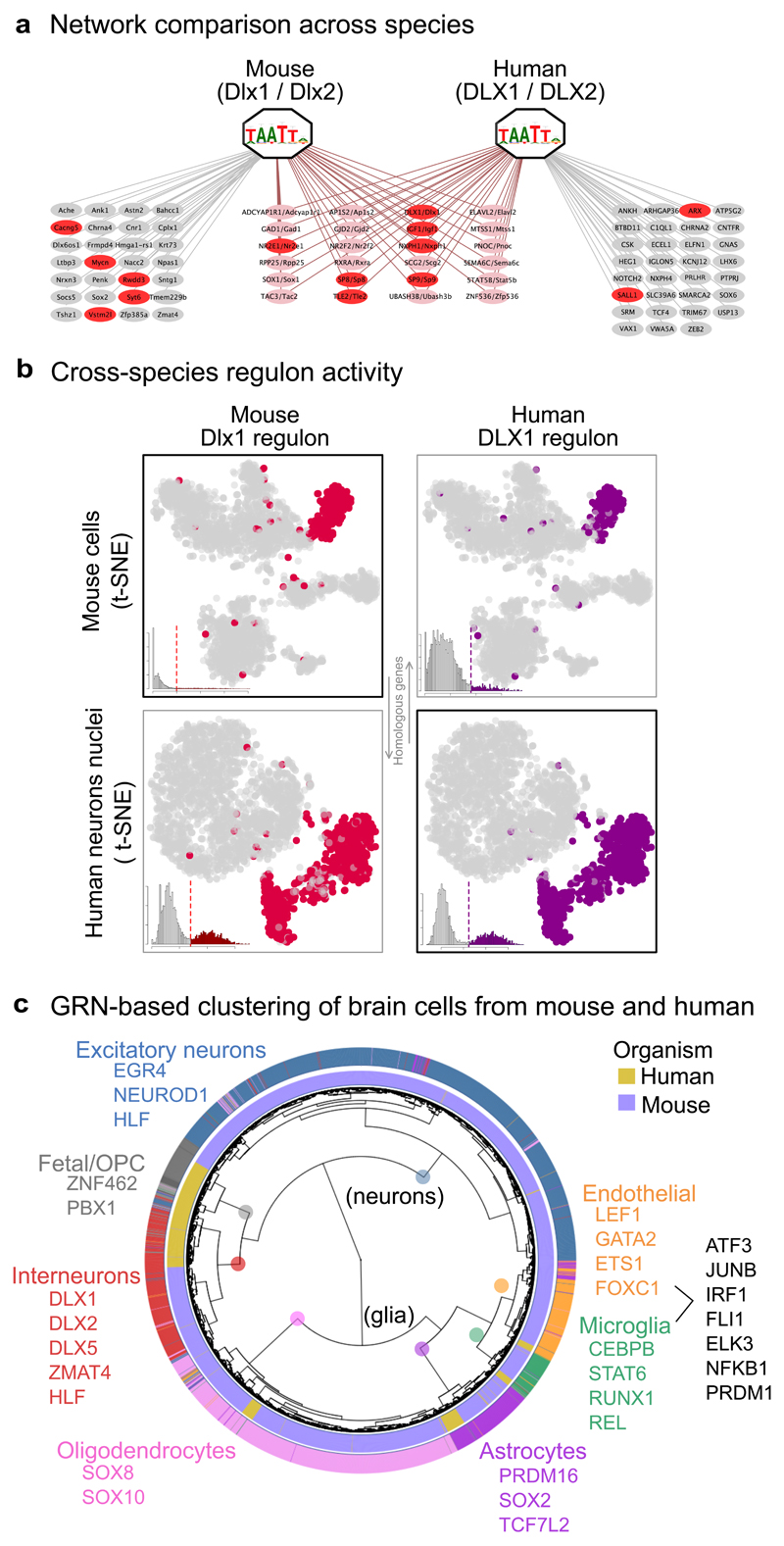
To evaluate the performance of SCENIC, we applied it to a scRNA-seq data set with well-known cell types from the adult mouse brain 9 (Fig. 1b-e). This analysis provided 151 regulons –out of 1046 initial co-expression modules– that presented significant enrichment of the motif of the corresponding transcription factor (7% of the initial TFs). Using the activity of these regulons to score each single cell revealed the expected cell types (Fig. 1d,e), alongside a list of potential master regulators per cell type (e.g., the microglia network in Supplementary Fig. 4). The clustering accuracy (cell-type overall sensitivity of 0.88, specificity of 0.99, and ARI > 0.80) is better than many dedicated single-cell clustering methods 10. To assess the robustness of SCENIC, we re-analyzed the mouse brain data, also including runs with only 100 randomly selected cells (to simulate small data sets), or 1/3 of the sequencing reads (to simulate low-coverage data sets). Interestingly, SCENIC identified cell types that are represented by only few cells (e.g. 2-6 cells from microglia, astrocytes or interneurons, Supplementary Fig. 5). In addition, the transcription factors predicted per cell type are consistent with previously established roles (Fig. 1c), and this accuracy outperforms standard analysis pipelines (Supplementary Fig. 3e). To validate the Dlx1/2 network identified for mouse interneurons, we analyzed a single-nuclei RNA-seq data set of the human brain 11 (Supplementary Fig. 6). On the human data, SCENIC also identifies a cluster of interneurons strongly driven by DLX1/2, having the same recognition motif as in mouse, and a set of conserved targets including DLX1 itself (Fig. 2a-b). Next, we expanded this cross-species analysis to other cell types. Interestingly, in contrast to standard clustering based on normalized expression, which yields a strong species-driven clustering (Supplementary Fig. 7), the SCENIC analysis effectively grouped cells by their cell type (Fig. 2c). This suggests that the scoring of network activity is robust and can be exploited to overcome batch or technical effects (Supplementary Fig. 3d).
Figure 2. Cross-species comparison of neuronal networks and cell types.
(a) DLX1/2 regulons inferred from mouse and human brain scRNA-seq data. The genes highlighted in red have associations with Dlx1/2 in GeneMANIA. (b) Reciprocal activity of human and mouse Dlx1/2 regulons on mouse and human single-cell data. (c) Joint clustering of human and mouse brain scRNA-seq data based on GRN activity. Colored TF names correspond to regulons identified both in the human and mouse SCENIC runs.
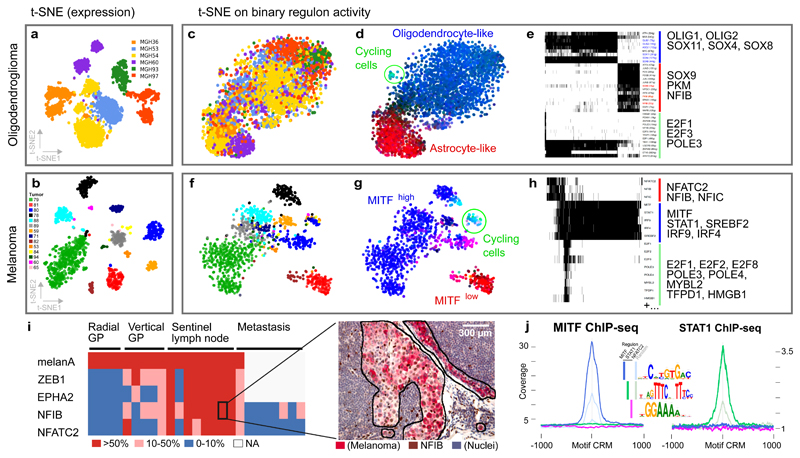
To evaluate SCENIC for the analysis of complex cancer cell states, we applied it to cancer cells from two scRNA-seq data sets, the first from oligodendroglioma 12 (4043 cells from six tumors), and the second from melanoma 13 (1252 cells from fourteen lesions). Due to tumor-specific mutations and complex genomic aberrations, the identification of cancer cell states is more challenging than characterizing normal cell types 14. Standard clustering of these data sets results in cells grouped by their tumor of origin (Fig. 3a,b). However, SCENIC reveals a different picture. On oligodendroglioma, three cancer cell states are identified across tumors (Fig. 3c-e), each state driven by expected/relevant TFs, including SOX10/4/8, OLIG1/2, and ASCL1 for the oligodendrocyte-like state; SOX9, NFIB, AP-1 for the astrocyte-like state; and E2F, FOXM1 for the cycling cells. Interestingly, when we used diffusion maps on the binary SCENIC matrix (Supplementary Fig. 8), the differentiation trajectory was reconstructed: from stem-like, undifferentiated, to the oligodendrocyte-like and astrocyte-like branches. Note that this path represents a different “trajectory” compared to normal oligodendrocyte differentiation (see Supplementary Fig. 9 for the SCENIC analysis of 5069 oligodendrocytes). We observed a similar tumor-effect correction on the melanoma data, where SCENIC identifies groups of cells across tumors (Supplementary Fig. 10), including a cluster of cycling cells, driven by similar transcription factors as in oligodendroglioma (e.g., E2F1/2/8 and MYBL2, Fig. 3f-h and Supplementary Fig. 10). In contrast to dedicated batch-effect removal methods such as Combat 15 and Limma 16, which require specifying the source of batch-effect a priori (Supplementary Fig. 11), SCENIC removes the tumor-effect automatically, using biologically-driven features.
Figure 3. SCENIC overcomes tumor effects and unravels relevant cell states and GRNs in cancer.
(a-b) t-SNEs on the expression matrices, colored by tumor of origin. (c-d, f-g) t-SNEs on the binary activity matrix (e,h) after applying SCENIC. In d and g, cells are colored by GRN activity. (i) Immunohistochemistry (IHC) on 25 human melanomas using NFATC2, NFIB, ZEB1, and EPHA2 antibodies. The heatmap shows the percentage of cells that are positive for each marker in the given sample. On the right, a representative example of IHC for NFIB on a sentinel lymph node is shown (for additional images, see Supplementary Fig. 13). (j) Aggregation plots for MITF and STAT1 ChIP-seq signal on the predicted target regions, and as control randomly selected genomic regions with MITF/STAT motif occurrences.
Interestingly, the melanoma cells fall largely into two groups, one corresponding to a MITFhigh state –the archetypical proliferative state– with MITF and STAT/IRF as key regulators, and an MITFlow state with up-regulated WNT5A, LOXL2 and ZEB1 expression, known markers of invasive states (Supplementary Fig. 10e-f). SCENIC identifies two new transcription factors in this MITFlow state, namely NFATC2 (114 predicted target genes) and NFIB (15 predicted target genes). NFATC2, a transcriptional repressor operating in the JNK/MAPK pathway, is involved in melanoma dedifferentiation to a stem cell fate, and in immune escape 17. NFIB on the other hand is linked to stem cell behavior of hair follicle and melanocyte stem cells 18, and plays an important role in metastatic progression of small cell lung cancer 19. To further explore the potential role of NFATC2 and NFIB this state, we performed immunohistochemistry on 25 melanoma specimens with varying tumor progression. Surprisingly, we found the highest NFIB and NFATC2 expression in the sentinel lymph nodes, co-localizing with ZEB1 expression, suggesting a relationship with the earliest metastatic events (Fig. 3i and Supplementary Fig. 12). When we knocked-down NFATC2 using siRNA in A375, a melanoma cell line that shows high NFATC2 and NFIB expression (Supplementary Fig. 13), we found that the predicted NFATC2 target genes are significantly upregulated (see Online Methods for details). This is consistent with previously established role as a repressor 20. In addition, genes involved in regulation of cell adhesion and extracellular matrix and several previously-published gene signatures representing the melanoma invasive state are also up-regulated (Supplementary Table 1), suggesting that NFATC2 may indeed play important role in the progression of the disease. As a second validation of the melanoma regulons we tested the predicted targets of MITF and STAT using ChIP-seq data (Fig. 3j).
As datasets with increasingly large numbers of single cells are becoming available, we suggest two complementary approaches to scale the network inference. The first approach is to infer the GRN from a sub-sampled data set, and to include all cells in the scoring step with AUCell. We illustrate this approach on a dataset with more than 40000 single-cells from the mouse retina (Supplementary Fig. 14). The second approach strives for using more efficient machine learning and big data handling solutions. Particularly, we implemented GRNboost, a new variant of GENIE3, in Scala on Apache Spark, replacing the Random Forest regression with gradient boosting. This new implementation drastically reduces the time needed to infer a GRN (Supplementary Fig. 15) and will pave the way to network inference on very large data sets, such as the soon available Human Cell Atlas 21.
In conclusion, SCENIC is a generally applicable method for the analysis of single-cell RNA-seq data that exploits transcription factors and cis-regulatory sequences to guide the discovery of cellular states. Our results show that gene regulatory networks constitute robust guides to identify cellular states, and that single-cell RNA-seq data are well-suited to trace gene regulatory programs in which specific combinations of transcription factors drive cell type-specific transcriptomes.
Online Methods
SCENIC workflow
SCENIC is a workflow based on three new R/bioconductor packages: (1) GENIE3, to identify potential TF targets based on co-expression, (2) RcisTarget, to perform the TF-motif enrichment analysis and identify the direct targets (regulons), and (3) AUCell, to score the activity of regulons (or other gene sets) on single cells. We also provide GRNboost, implemented on Spark 22, as scalable alternative to build the co-expression network on bigger datasets (step 1, replacing GENIE3).
The three R/bioconductor packages, and GRNboost, include detailed tutorials to facilitate their use within an automated SCENIC pipeline, as well as independent tools. Links to the tools, SCENIC code and tutorials are available at http://scenic.aertslab.org.
GENIE3
GENIE3 8 is a method for inferring gene regulatory networks from gene expression data. In brief, it trains random forest models predicting the expression of each gene in the dataset, using as input the expression of the transcription factors. The different models are then used to derive weights for the transcription factors, measuring their respective relevance for the prediction of the expression of each target gene. The highest weights can be translated into TF-target regulatory links 8. Since GENIE3 uses Random Forest regression, it has the added value of allowing complex (e.g., non-linear) co-expression relationships between a TF and its candidate targets. GENIE3 is available in Python, Matlab and R. To allow for inclusion in SCENIC workflow, we optimized the previous R implementation of GENIE3. The core of this new implementation is now written in C –which makes it orders of magnitude faster–, it requires lower memory, and supports execution in parallel. GENIE3 was the top-performing method for network inference in the DREAM4 and DREAM5 challenges 23. The new package provides similar results in the DREAM challenge to previously existing implementations, but with improved speed. The comparison is available at the following website: http://www.montefiore.ulg.ac.be/~huynh-thu/GENIE3.html.
The input to GENIE3 is an expression matrix. The preferred expression values are gene-summarized counts (which might or might not use unique molecular identifiers, UMI 24). Other measurements, such as counts or transcripts per million (TPM) and FPKM/RPKM are also accepted as input. However, note that the first network-inference step is based on co-expression, and some authors recommend avoiding within sample normalizations (i.e. TPM) for this task because they may induce artificial co-variation 25. To evaluate to what extent the normalization of the input matrix affects the output of SCENIC, we also ran SCENIC on Zeisel’s dataset after library-size normalization (using the standard pipeline from scran 26, which performs within-cluster size-factor normalization). The results are highly comparable, both in regards to resulting clusters/cell types (ARI between the cell types obtained from raw UMI counts or normalized counts: 0.90, ARI from normalized counts compared to the author’s cell types: 0.87), and the TFs identifying the groups (26 out of the 30 regulons highlighted in Fig. 1b). Furthermore, during the course of this project we have applied GENIE3 to multiple datasets using UMI counts (e.g. mouse brain and oligodendrocytes), and TPM (e.g. human brain and melanoma) and both units provided reliable results.
The output of GENIE3 is a table with the genes, the potential regulators, and their “importance measure” (IM), which represents the weight that the transcription factor (input gene) has in the prediction of the target. We explored several ways to determine the threshold (e.g. looking at the rankings, distributions and outputs after pruning with RcisTarget), and finally opted for building multiple gene-sets of potential targets for each transcription factor: (a) setting several IM thresholds (IM > 0.001 and IM>0.005), (b) taking the 50 targets with highest IM for each TF, and (c) keeping only the top 5, 10 and 50 TFs for each target gene (then, split by TF). In all these cases, only the links with IM>0.001 were taken into account. Furthermore, each gene-set was then split into positive- and negative- correlated targets (i.e. Spearman correlation between the TF and the potential target) to separate likely activated and repressed targets. Finally, only the gene-sets (TF co-expression modules) with at least 20 genes were kept for the following step.
GRNboost
GRNboost is based on the same concept as GENIE3: inferring regulators for each target gene purely from the gene expression matrix. However, it does so using the gradient boosting machines (GBM) 27 implementation from the XGBoost library 28. A GBM is an ensemble learning algorithm that uses boosting 29 as a strategy to combine multiple weak learners, like shallow trees, into a strong one. This contrasts with random forest, the method used by GENIE3, which uses bagging (bootstrap aggregation) for model averaging to improve regression accuracy. GRNBoost uses gradient boosted stumps (regression trees of depth 1) 30 as the base learner. GRNBoost’s main contribution is casting this multiple regression approach into a Map/Reduce 31 framework based on Apache Spark 22. In GRNBoost, the core data entry is a tuple of a gene and a vector of TF expression values. Using a Spark RDD, GRNBoost first partitions the gene expression vectors over the nodes available in the compute cluster. Subsequently, it constructs a predictor matrix that contains the expression values for all candidate regulator genes. Using a Spark broadcast variable, the predictor matrix is broadcasted to the different compute partitions. In the map phase of the framework, GRNBoost iterates over the gene tuples (expression vector) and uses the predictor matrix to train the XGBoost regression models with the expression vectors as respective training labels. From the trained models, the strengths of the regulator-target relationships are extracted and emitted as a set of network edges. In the reduce phase, all sets of edges are combined into the final regulatory network.
The performance of GRNBoost and GENIE3 was compared on a workstation with 2 Intel Xeon E2696 V4 CPUs with in total 44 physical cores or 88 threads and 128 GB of 2133Ghz ECC memory. Large datasets and hence large predictor matrices cause the network inference to become memory-bound rather than CPU-bound. In order to comfortably fit the amount of memory required into the available 128 GB of memory, we decreased the number of partitions to 11, therefore having only a maximum of 11 predictor matrices in flight simultaneously. However, we increased the number of threads available to each individual XGBoost regression to 8, effectively using all available (88) threads in the workstation. GRNBoost is written in the Scala programming language and can be used as a software library or be submitted as a Spark job from the command line.
RcisTarget
RcisTarget is a new R/Bioconductor implementation of the motif enrichment framework of i-cisTarget and iRegulon. RcisTarget identifies enriched transcription factor binding motifs and candidate transcription factors for a gene list. In brief, RcisTarget is based on two steps. First, it selects DNA motifs that are significantly over-represented in the surroundings of the transcription start site (TSS) of the genes in the gene-set. This is achieved by applying a recovery-based method on a database that contains genome-wide cross-species rankings for each motif. The motifs that are annotated to the corresponding TF and obtain a Normalized Enrichment Score (NES) > 3.0 are retained. Next, for each motif and gene-set, RcisTarget predicts candidate target genes (i.e. genes in the gene-set that are ranked above the leading edge). This method is based on the approach described by Aerts et al. 32 which is also implemented in i-cisTarget (web interface) 33 and iRegulon (Cytoscape plug-in) 34. Therefore, when using the same parameters and databases, RcisTarget provides the same results as i-cisTarget or iRegulon, benchmarked against other TFBS-enrichment tools in Janky et al. 34. More details about the method and its implementation in R are given in the package documentation.
To build the final regulons, we merge the predicted target genes of each TF-module that show enrichment of any motif of the given TF. To detect repression, it is theoretically possible to follow the same approach with the negative-correlated TF modules. However, in the datasets we analyzed, these modules were less numerous and showed very low motif enrichment, suggesting that these are lower quality modules. For this reason, we finally decided to exclude the detection of direct repression from the workflow, and continue only with the positive-correlated targets. The databases used for the analyses presented in this paper are the "18k motif collection" from iRegulon (gene-based motif rankings) for human and mouse. For each species, we used two gene-motif rankings (10kb around the TSS or 500bp upstream the TSS), which determine the search space around the transcription start site.
AUCell
AUCell is a new method that allows identifying cells with active gene regulatory networks in single-cell RNA-seq data. The input to AUCell is a gene set, and the output the gene set “activity” (AUC) in each cell. In SCENIC, these gene sets are the regulons, consisting of the TF and their putative targets. AUCell calculates the enrichment of the regulon as an area under the recovery curve (AUC) across the ranking of all genes in a particular cell, whereby genes are ranked by their expression value. This method is therefore independent of the gene expression units and the normalization procedure. In addition, since the cells are evaluated individually, it can easily be applied to bigger datasets (e.g. subsetting the expression matrix if needed). In brief, the scoring method is based on a recovery analysis, where the x-axis (Supplementary Fig. 1c) is the ranking of all genes based on expression level (genes with the same expression value, e.g. '0', are randomly sorted); and the y-axis is the number of genes recovered from the input set. AUCell then uses the "Area Under the Curve" (AUC) to calculate whether a critical subset of the input gene set is enriched at the top of the ranking for each cell. In this way, the AUC represents the proportion of expressed genes in the signature and their relative expression value compared to the other genes within the cell. The output of this step is a matrix with the AUC score for each gene-set in each cell. We use either the AUC scores (across regulons) directly as continuous values to cluster single-cells, or we generate a binary matrix using a cutoff of the AUC score for each regulon. These cutoffs are determined either automatically, or manually adjusted by inspecting the distribution of the AUC scores. Some examples of AUC distributions are provided in Supplementary Fig. 2a. Supplementary Fig. 2b-c shows the validation of AUCell using previously published neuronal and glial gene signatures. The tutorial included in the package, also includes practical explanations and implications of each of the steps of the method.
Cell clustering based on GRNs
The cell-regulon activity is summarized in a matrix in which the columns represent the cells and the rows the regulons. In the binary regulon activity matrix, the coordinates of the matrix that correspond to active regulons in a given cell will contain a “1”, and “0” otherwise. The equivalent matrix, containing the continuous AUC values for each cell-regulon, is normally referred to as the AUC activity matrix. Clustering of either of the regulon activity matrices reveals groups of regulons (jointly a network) that are recurrently active across a subset of cells. The binary activity matrix tends to highlight higher-order similarities across cells (and therefore, highly reduces batch effects and technical biases), on the other hand, the AUC matrix allows to observe more subtle changes. For visualization, we have mostly used t-SNEs (Rtsne package35, always tested consistency across several perplexity values and distance metrics/number of PCs), and heatmaps with hierarchical clustering (although the heatmap figures feature selected regulons, the t-SNEs are always run on the whole matrices). In the tutorials, we have also included several options to explore the results. For example, to detect most likely stable states (higher-density areas in the t-SNE), and to help identify key regulators, known cell properties (based on the dataset annotation) and GO terms (GO enrichment analysis of the genes in the cluster of regulons) that might be associated to the detected states.
SCENIC runs on the different datasets
SCENIC was run on all the datasets using the expression matrices provided by the authors (downloaded from GEO or the authors website), including only the cells that passed their quality control, and the default gene filtering for GENIE3 (which resulted in 12-15k genes). The standard SCENIC workflow was run on all datasets (the version at the time of publication is available as supplementary file, updated versions can be found at http://scenic.aertslab.org). A more detailed description of the datasets and the any peculiarities for each analysis are available in Supplementary Note 1. Here we provide a brief description of the datasets:
Mouse cortex and hippocampus (Zeisel et al.9, GSE60361): single-cell RNA-seq of 3005 brain cells of juvenile mice (21-31 days old). It contains the main cell types in hippocampus and somatosensory cortex, namely neurons (pyramidal excitatory neurons, and interneurons), glia (astrocytes, oligodendrocytes, microglia), and endothelial cells. Expression matrix units: UMI counts.
Human neurons (Lake et al. 11): single-nuclei RNA-seq of 3083 neuronal cells from a normal human brain (retrieved postmortem from a 51-year old female, from six different Brodmann areas). Expression matrix units: TPM.
Human brain (Darmanis et al.36, GSE67835): scRNA-seq from 466 cells from adult and fetal human brains. The fetal samples were taken from four different individuals at 16 to 18 weeks post-gestation. The adult brain samples were taken from healthy temporal lobe tissue from 8 different patients (21 - 63 years old) during temporal lobectomy surgery for refractory epilepsy and hippocampal sclerosis. Expression matrix units: logged CPM.
Mouse oligodendrocytes (Marques et al. 37, GSE75330): scRNA-seq data of 5069 cells from the oligodendrocyte lineage. Cells were obtained from several different mouse strains and isolated from ten different regions of the anterior-posterior and dorsal-ventral axis of the mouse juvenile and adult CNS; including white and grey matter. Expression matrix units: UMI counts.
Oligodendroglioma (Tirosh et al. 38, GSE70630): scRNA-seq expression profiles for 4347 cells from 6 untreated grade II oligodendroglioma tumors with either IDH1 or IDH2 mutation, and 1p/19q co-deletion. Only the tumoral cells were used for the analysis (selected by the authors based on CNV profile). Expression matrix units: log2(TPM+1).
Melanoma (Tirosh et al. 13, GSE72056): scRNA-seq of 1252 melanoma cells from 14 different tumors. These include only the cells that are labeled as malignant by the authors, based on their CNV profiles. Expression matrix units: log2(TPM/10+1).
Mouse retina (Macosko et al. 39, GSE63472): scRNA-seq data of 44808 cells obtained through Drop-seq from mouse retina (14 days post-natal). Expression matrix units: log((UMI counts per gene in a cell/Total UMI counts in cell)*10000)+1)].
Embryonic mouse brain (10X Genomics): Chronium Megacell demonstration dataset containing 1,306,127 cells from cortex, hippocampus and subventricular zone of two E18 mice (strain: C57BL/6).
Gene filtering
For gene filtering to run GENIE3, we applied a soft filter based on the total number of counts of the gene, and the number of cells in which it is detected. The first filter, the total number of reads per gene, is meant to remove genes that are most likely unreliable and provide only noise. The specific value depends on the dataset, for the ones used in this paper we set the thresholds at, for example, 3 UMI counts (slightly over the median of the non-zero values) multiplied by 1% of the number of cells in the dataset (e.g. in mouse brain: 3 UMI counts x 30 (1% of cells) = minimum 90 counts per gene). The second filter, the number of cells in which the gene is detected (e.g. >0 UMI, or >1 log2(TPM)), is to avoid that genes that are only expressed in one, or very few cells, gain a lot of weight if they happen to coincide in a given cell. In the workflow, we recommend to set a percentage lower than the smallest population of cells to be detected. For example, since microglia cells represent approximately 3% of the total cells in the dataset, we used a detection threshold of at least 1% of the cells.
Cross-species network comparisons
SCENIC was run independently for each of the three datasets used for the GRN comparison: Zeisel et al. (mouse brain cells), and Lake et al. (human neurons nuclei) and Darmanis et al. (human brain cells). To compare the networks across species, the genes in the human regulons were converted into the homologous mouse genes using Biomart (through biomaRt R package 40), and vice versa (the mouse regulons into human genes). In Fig. 2a, the genes highlighted in red also have associations with Dlx1/2 in GeneMANIA 41 (protein-protein interactions, genetic interactions, co-expression, or literature co-mentioning).
For the cross-species cell clustering (Fig. 2c), the genes in the mouse expression matrix were converted into the homologous human genes, and merged with Darmanis’ expression matrix by row (only genes available in both matrices are kept). The 259 human regulons from Darmanis’ dataset, and the human homologs of the mouse regulons were evaluated on this merged matrix to obtain the binary regulon activity containing 410 regulons. The cells were clustered based on the binary activity matrix using Ward’s hierarchical clustering with Spearman’s distance. Similar results were obtained for the reverse approach (converting the expression matrix into mouse genes, to evaluate the mouse regulons). In order to provide an alternative approach based only on expression (Supplementary Fig. 7), we also generated a merged expression matrix. Since the merged data sets use different measurement units (CPM in human and UMI in mouse), each matrix was Z-score normalized by gene before merging.
Method comparisons
We performed different evaluations and benchmark comparisons, each assessing a different aspect of SCENIC (e.g. cell type identification, TF identification, co-founding effect correction…). The detailed description of how these comparisons were performed is available in Supplementary Note 1. Here we provide a brief summary:
Cell clustering: To determine whether the clustering based on gene regulatory network activity matches real cell types, we compared the clustering based on the regulon activity matrices to the cell labels provided in the corresponding publications. To compare SCENIC performance to other methods, we re-used the benchmark presented in the SC3 publication 10, which provides the adjusted Rand index (ARI) for 6 clustering methods on the mouse brain dataset.
TF-motif discovery: The validation of the TFs identified by SCENIC was mainly done by confirming their role in the given cell type in literature (e.g. Fig. 1e). However, we also compared SCENIC to an alternative approach to identify TFs potentially regulating cell states: Applying transcription factor motif enrichment analysis on genes differentially expressed between clusters (i.e. gene signature, or markers for a cell type).
Batch effect correction: The results of SCENIC on the oligodendroglioma dataset (clustering of the full binary regulon activity matrix) were compared to Combat 15,42 and Limma 16,43 correcting for “patient of origin” as source of batch effect.
Cycling cells: The cycling cells were predicted based on consistent up-regulation of 46 sets related to the mitotic cell cycle from amiGO and cycleBase 1.0 and 2.0. We then compared the ability of different clustering methods to identify these cells (sensitivity and specificity). Since most of the methods provide multiple clusters as output, to compare their results, for each method we selected the cluster with the largest amount of CC cells.
Immunohistochemistry of melanoma biopsies
Immunohistochemistry with antibodies for melanA, EPHA2, ZEB1, NFATC2 and NFIB was performed on formalin-fixed, paraffin-embedded melanoma samples. The samples include biopsies of 9 primary melanomas (4 in radial growth phase and 5 in vertical growth phase), 8 melanoma-containing sentinel lymph nodes, and 8 melanoma metastases. A detailed description of how the immunohistochemistry was performed is available in Supplementary Note 1.
Knock-down of NFATC2 in melanoma cell culture
The A375 cell line was selected as representative of the MITFlow state based on expression of NFATC2, NFIB (Supplementary Fig. 13) and SOX10, after comparing 59 melanoma cell lines from the COSMIC Cancer Cell lines Project 44. Knockdown of NFATC2 was performed in A375 using NFATC2 siRNA, and total RNA was extracted 72 hours after the knockdown. The final libraries were pooled and sequenced on a combination of NextSeq 500 and HiSeq 4000 (Illumina). RNA-seq reads were mapped to the genome (hg19) for upstream analysis. A detailed description of the methods, including cell line source, knock-down of NFATC2, RNA-seq protocol and bioinformatics analysis, are available in Supplementary Note 1.
Software packages, code and data availability
Updated links to the packages and tutorials related to SCENIC are available at http://scenic.aertslab.org; the package versions at the time of publication are provided as Supplementary Software.
The NFATC2 knock-down RNA-seq data have been deposited in NCBI's Gene Expression Omnibus 45 and are accessible through GEO Series accession number GSE99466.
Supplementary Material
Acknowledgements
This work is funded by The Research Foundation - Flanders (FWO, www.fwo.be; grants G.0640.13 and G.0791.14 to SAe; G092916N to JCM), Special Research Fund (BOF) KU Leuven (http://www.kuleuven.be/research/funding/bof; grant PF/10/016 and OT/13/103 to SAe), Foundation Against Cancer (http://www.cancer.be; 2012-F2, 2016-070, and 2015-143 to SAe) and ERC Consolidator Grant (724226_cis-CONTROL to SAe). SAi is funded by a PDM Postdoctoral Fellowship from the KU Leuven. ZKA and JW are funded by postdoctoral research fellowships from Kom op Tegen Kanker, and VAH by the F.R.S.-FNRS Belgium. HI has a PhD Fellowships from the agency for Innovation by Science and Technology (IWT, www.iwt.be). TM and JA funding is provided by Symbiosys and IMEC HI^2 Data Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. TM would like to thank Jaak Simm for helpful comments and suggestions regarding gradient boosting.
Footnotes
Author contributions
SAe and SAi conceived the study; SAi implemented SCENIC and related packages with help of VA and PG for GENIE3, and GH for RcisTarget; SAi and CBG analyzed the data, with help of ZKA and HI; TM and JA implemented GRNBoost; JW performed the IHC and knock-down experiments; FR, JCM, and JvdO contributed reagents and helped with the interpretation of the melanoma analyses; SAi, JW, and SAe and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Linnarsson S, Teichmann SA. Single-cell genomics: coming of age. Genome Biol. 2016;17:97. doi: 10.1186/s13059-016-0960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 4.Raj A, van Oudenaarden A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pina C, et al. Single-Cell Network Analysis Identifies DDIT3 as a Nodal Lineage Regulator in Hematopoiesis. Cell Rep. 2015;11:1503–1510. doi: 10.1016/j.celrep.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo M, Wang H, Potter SS, Whitsett JA, Xu Y. SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis. PLOS Comput Biol. 2015;11:e1004575. doi: 10.1371/journal.pcbi.1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PloS One. 2010;5 doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 10.Kiselev VY, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirosh I, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perotti V, et al. NFATc2 is an intrinsic regulator of melanoma dedifferentiation. Oncogene. 2016;35:2862–2872. doi: 10.1038/onc.2015.355. [DOI] [PubMed] [Google Scholar]
- 18.Chang C-Y, et al. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. doi: 10.1038/nature11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denny SK, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 21.Regev A, et al. The Human Cell Atlas. bioRxiv. 2017 doi: 10.1101/121202. [DOI] [Google Scholar]
- 22.Zaharia M, et al. Resilient Distributed Datasets: A Fault-tolerant Abstraction for In-memory Cluster Computing; Proceedings of the 9th USENIX Conference on Networked Systems Design and Implementation 2–2; USENIX Association; 2012. [Google Scholar]
- 23.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 25.Crow M, Paul A, Ballouz S, Huang ZJ, Gillis J. Exploiting single-cell expression to characterize co-expression replicability. Genome Biol. 2016;17:101. doi: 10.1186/s13059-016-0964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JH. Greedy Function Approximation: A Gradient Boosting Machine. Ann Stat. 2001;29:1189–1232. [Google Scholar]
- 28.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System; Proceedings of the 22Nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794; ACM; 2016. [DOI] [Google Scholar]
- 29.Freund Y, Schapire R. A short introduction to boosting. J-Jpn Soc Artif Intell. 1999;14:1612. [Google Scholar]
- 30.Sławek J, Arodź T. ENNET: inferring large gene regulatory networks from expression data using gradient boosting. BMC Syst Biol. 2013;7:106. doi: 10.1186/1752-0509-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean J, Ghemawat S. MapReduce: Simplified Data Processing on Large Clusters. Commun ACM. 2008;51:107–113. [Google Scholar]
- 32.Aerts S, et al. Robust Target Gene Discovery through Transcriptome Perturbations and Genome-Wide Enhancer Predictions in Drosophila Uncovers a Regulatory Basis for Sensory Specification. PLOS Biol. 2010;8:e1000435. doi: 10.1371/journal.pbio.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann C, Sande BV, de Potier D, Aerts S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012;40:e114–e114. doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janky R, et al. iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLOS Comput Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krijthe J. Rtsne: T-Distributed Stochastic Neighbor Embedding using Barnes-Hut Implementation. 2015 [Google Scholar]
- 36.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirosh I, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 41.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leek J. sva: Surrogate Variable Analysis [Google Scholar]
- 43.Smyth G. limma: Linear models for microarray data [Google Scholar]
- 44.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.