Abstract
VioE, an unusual enzyme with no characterized homologues, plays a key role in the biosynthesis of violacein, a purple pigment with antibacterial and cytotoxic properties. Without bound cofactors or metals, VioE, from the bacterium Chromobacterium violaceum, mediates a 1,2 shift of an indole ring and oxidative chemistry to generate prodeoxyviolacein, a precursor to violacein. Our 1.21 Å resolution structure of VioE shows that the enzyme shares a core fold previously described for lipoprotein transporter proteins LolA and LolB. For both LolB and VioE, a bound polyethylene glycol molecule suggests the location of the binding and/or active site of the protein. Mutations of residues near the bound polyethylene glycol molecule in VioE have identified the active site and five residues important for binding or catalysis. This structural and mutagenesis study suggests that VioE acts as a catalytic chaperone, using a fold previously associated with lipoprotein transporters to catalyze the production of its prodeoxyviolacein product.
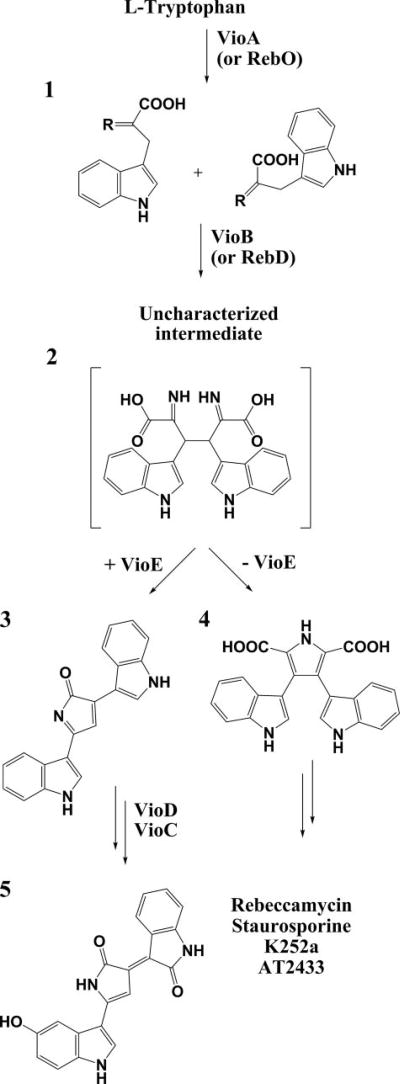
Violacein 5 (Fig. 1) is a purple pigment that gives tropical bacterium Chromobacterium violaceum its characteristic coloring (1–5). Violacein 5 has potential medical applications as an anti-bacterial (6–8), anti-tryptanocidal (8–10), anti-ulcerogenic (11), and anti-cancer drug (8, 12–17). The molecule, made of l-tryptophan and molecular oxygen (18–21), was originally thought to be synthesized by the action of just four enzymes VioABCD (22, 23), but an additional open reading frame (24), also in the operon, was shown to encode the enzyme VioE (25, 26). A22-kDa protein with few homologues based on sequence analysis, VioE is required for formation of prodeoxyviolacein 3, a precursor to violacein 5 (25–27). Without the presence of VioE, a shunt product, chromopyrrolic acid 4, is produced, rather than 3 (25–27). VioE is therefore required for the 1,2 shift of the indole ring and subsequent chemistry that gives violacein 5 its distinctive architecture. Surprisingly, despite carrying out this chemistry, VioE lacks any bound cofactors or metal ions (26).
FIGURE 1. Biosynthesis of violacein and indolocarbazoles.
Indole 3-pyruvic acid 1 (r = NH imine form, r = O ketone form) is generated biosynthetically from l-tryptophan by the action of the enzyme VioA or a homologue, such as RebO. In both the violacein and related indolocarbazole biosynthetic pathways, two molecules of IPA imine 1 are dimerized to generate an as yet uncharacterized intermediate, which has been proposed to be 2 (26). The presence of VioE routes this intermediate to the production of prodeoxyviolacein 3, whereas in the absence of VioE this intermediate converts to chromopyrrolic acid 4. Prodeoxyviolacein 3 is a precursor to violacein 5, whereas chromopyrrolic acid 4 derivatives are precursors to indolocarbazole compounds rebeccamycin, staurosporine, K252a, or AT2433 (30, 31, 57).
Using genetic and biochemical analysis, an enzymatic pathway for violacein 5 production has been determined (Fig. 1), and intermediates have been identified (26), with the major exception that the substrate of VioE (product of VioB) is still unknown. It has, however, been established that the product of VioB is the substrate for VioE; when VioA and VioB are incubated in a reaction mixture with the VioA substrate l-tryptophan for 10 min, passed through a 10-kDa filter (removing VioA and VioB), and then incubated with VioE, prodeoxyviolacein 3 is produced (26). By contrast, in the absence of VioE, no prodeoxyviolacein 3 is made (26). This result reveals three things: first, the VioB product is a small molecule that can pass through a 10-kDa filter; second, no interaction between VioB and VioE is necessary for product formation; and third, VioE is a catalyst. The small molecule passed through the filter (the product of VioB and substrate of VioE) has not yet been isolated, but it is known to convert spontaneously to chromopyrrolic acid 4 when VioE is not present; chromopyrrolic acid 4 itself is not a substrate for VioE (25, 26). Homologues of VioB are RebD (36% identity), StaD (38% identity), InkD (34% identity), and AtmD (38% identity) from the rebeccamycin, staurosporine, K252a, and AT2433 biosynthetic pathways, respectively (28–31). Each of these homologues produces chromopyrrolic acid 4 scaffolds as on-pathway products to generate indolocarbazole natural products, and each lacks a VioE homologue in its biosynthetic cluster (28–31). Yet, in vitro tests of RebD from the rebeccamycin biosynthetic pathway show that coincubation of a reaction mixture of RebD and IPA5 imine 1 with VioE results in the production of prodeoxyviolacein 3 rather than chromopyrrolic acid 4 (26). Thus, the presence of the enzyme VioE reroutes toward a violacein biosynthetic pathway and away from the related indolocarbazole biosynthetic pathways.
To characterize VioE, we solved its crystal structure to 1.21 Å resolution. The structure, which confirms that VioE lacks bound cofactors or metals, reveals the similarity of VioE to the core structure of the lipoprotein carrier proteins LolA and LolB (32). Further, VioE resembles LolB in its binding of a PEG molecule in what appears to be the active site. To investigate whether the PEG molecule indeed identified the active site of VioE, we carried out site-directed mutagenesis on each of nine residues near the bound PEG molecule and further along the putative active site. Five of the altered proteins show significantly reduced production of prodeoxyviolacein 3 compared with wild-type VioE. This work suggests that the fold of a lipoprotein carrier protein has been co-opted to a catalytic function, enabling VioE to bind a chiefly hydrophobic molecule in a hydrophobic cavity and to catalyze a 1,2 shift reaction and oxidative chemistry, producing prodeoxyviolacein 3, rather than the unshifted shunt product chromopyrrolic acid 4.
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization
Eight liters of LB medium containing 30 mg/liter of kanamycin and 34 mg/liter of chloramphenicol were inoculated with a starting culture of Rosetta™ 2(DE3)pLysS (Novagen) transformed with pET24b-VioE (26). The cultures were grown at 21 °C to an A600 of 0.6 and then induced with 0.1 mm isopropyl β-d-thiogalactopyranoside and grown for an additional 17 h at 21 °C at 250 rpm. Cell pellets (40 g) were frozen at ×20 °C overnight, then sonicated with lysozyme in 300 mm NaCl, 25 mm Tris, 5 mm imidazole, pH 8.0, and then centrifuged at 50,000 × g to pellet insoluble material. The supernatant was incubated with nickel (II)-loaded Chelating Sepharase™ Fast Flow (GE Biosciences) for 40 min at 4 °C. Unbound material was removed, and then the column was washed with 25 column volumes of 300 mm NaCl, 25 mm Tris, 20 mm imidazole, pH 8.0. Bound protein was then eluted with 300 mm NaCl, 25 mm Tris, 200 mm imidazole, pH 8.0. This sample was then loaded onto a HiLoad Superdex™ 75 prep grade column (Amersham Biosciences) pre-equilibrated with 50 mm NaCl, 20 mm Tris, pH 8.0. Fractions containing VioE were pooled and concentrated to 17 mg/ml in 10% glycerol, 50 mm NaCl, 20 mm Tris, pH 8.0. Protein was then frozen in liquid nitrogen and stored at −80 °C. VioE was crystallized at room temperature from this sample using the hanging drop vapor diffusion method by incubating 1.5 µl of protein with 1.5 µl of a precipitant solution (composed of 25% (v/v) PEG 400, 20% (w/v) PEG 3350, 0.1 m MgCl2, and 0.1 m Tris, pH 8.5) over a 0.5-ml precipitant well solution. Crystals appeared overnight and grew larger over subsequent days. Native crystals were flash frozen in liquid nitrogen, without additional cryoprotection, for x-ray data collection.
Selenomethionine protein was prepared by inoculating minimal medium (6 g/liter Na2HPO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, 0.5 g/liter NaCl, 1 mm MgSO4, 0.4% (w/v) glucose, 30 mg/liter kanamycin, and 34 mg/liter chloramphenicol) with a starting culture of Rosetta™ 2(DE3)pLysS transformed with pET24b-VioE and growing cultures at 37 °C. At an A600 of 0.6, the cultures were supplemented with 100 mg/liter each l-lysine, l-threonine, and l-phenylalanine and 50 mg/liter each l-isoleucine, l-leucine, l-valine, and l-selenomethionine (33) and grown for an additional 20 min. Protein expression was then induced with 0.1 mm isopropyl β-d-thiogalactopyranoside, and the cultures were grown for an additional 20 h at 21 °C at 250 rpm. Selenomethionine protein was purified identically to native protein, except that all of the buffers, excluding the final storage buffer, were supplemented with 5 mm β-mercaptoethanol. A purified sample of the selenomethionine protein was assayed for incorporation of selenium using liquid chromatography-mass spectrometry, and strong peaks were found from a 23,028-atomic mass unit species, corresponding to the incorporation of four seleniums, with weak peaks found from a 23,104-atomic mass unit species, corresponding to the incorporation of four oxidized seleniums. A small number of crystals with defects were produced in the same condition as the native protein, and these were used in streak seeding experiments to obtain crystals appropriate for x-ray data collection. Single selenomethionine-VioE crystals were flash frozen directly into liquid nitrogen for x-ray data collection.
Data Collection and Processing
The data were collected at the Advanced Photon Source (Argonne, IL), beamline NE-CAT 24ID-C, with the Quantum 315 detector in a cryostream of 141.6 K. For the native crystal, the data were collected in two sweeps, one of 180° and one of 360°. A 180° sweep was first carried out in 1° oscillation steps to obtain data to 1.8 Å resolution. Next, a 360° sweep was carried out in 0.5° oscillation steps to full resolution. A small selenomethionine VioE crystal was used for a fluorescence scan to obtain the appropriate wavelengths for anomalous dispersion data collection. A larger selenomethionine VioE crystal was then used to carry out a single-wavelength anomalous dispersion data collection at the selenium peak wavelength of 0.9792 Å, with a 360° sweep in 0.5° oscillation steps. All of the data were indexed and integrated in DENZO and scaled in SCALEPACK (34). The data processing statistics are shown in Table 1. Data to 1.21 Å resolution for the native and to 1.86 Å resolution for the selenomethionine derivative were used, despite the low completeness in the highest resolution bins (54.3 and 58.7%, respectively). The values of other statistics in these highest resolution bins (including Rsym, I/σ, and redundancy) suggest that the data are of high quality to these resolutions.
TABLE 1.
Data collection and refinement statistics
| Native | Selenomethioninea | |
|---|---|---|
| Wavelength (Å) | 0.9795 | 0.9792 |
| Space group | P212121 | P212121 |
| Unit cell (Å) | a = 53.5, b = 82.5, c = 90.6 | a = 53.4, b = 82.3, c = 90.6 |
| Resolution (Å)b | 50-1.21 (1.25-1.21) | 50-1.86 (1.93-1.86) |
| Rsymb,c | 0.066 (0.258) | 0.060 (0.226) |
| Completeness (%)b | 90.6 (54.3) | 95.0 (58.7) |
| Unique reflections | 111,465 | 61,353 |
| Redundancyb | 13.9 (3.2) | 6.9 (3.8) |
| I/σb | 35.7 (3.4) | 27.8 (5.1) |
|
| ||
| Refinement statistics | ||
| Resolution range (Å) | 50–1.21 | |
| Rcryst(%)d | 16.8 | |
| Rfree(%)d | 19.5 | |
| No. nonhydrogen atoms | ||
| Protein | 3183 | |
| Water | 680 | |
| PEG | 23 | |
| Average B-factors (Å2) | ||
| Protein | 15.0 | |
| Water | 32.0 | |
| PEG | 44.2 | |
| Root mean squared deviation bond length (Å) | 0.006 | |
| Root mean squared deviation bond angle (°) | 1.209 | |
| Ramachandran plot (% residues) | ||
| Most favored | 91.6 | |
| Additionally allowed | 8.4 | |
| Generously allowed | 0 | |
| Disallowed | 0 | |
For this data set, Bijovet pairs were not merged during data processing.
The values in parentheses indicate the highest resolution bin.
Rsym = (ΣiΣhkl|Ii(hkl) − <I(hkl)>|)/ Σhkl <I(hkl)>, where Ii(hkl) is the intensity of the ith measured reflection, and <I(hkl)> is the mean intensity for the reflection with the miller index (hkl).
Rcryst = (Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖)/Σhkl|Fobs(hkl)|; Rfree is calculated identically, using 5% of reflections omitted from refinement.
Structure Building and Refinement
Six selenium sites were found using SHARP (35); these include three selenomethionines from each molecule of the two in the asymmetric unit and do not include the N-terminal methionines, which are disordered. The overall figure of merit (acentric) of data to 1.87 Å was calculated by SHARP to be 0.438. The native and selenomethionine data were scaled in SHARP to generate a single file used for all initial building and refinement. 2.0-Å resolution experimental maps from the SHARP output, solvent flattened with SOLOMON (36), were used to build manually the entire main chain of the protein, with placement of most side chains. This model was then used in iterative rounds of CNS (37) refinement and manual adjustment of the model in COOT (38) to the full 1.21 Å resolution of the native data, to generate a nearly complete structure with placement of a first set of water molecules. Iterative rounds of refinement were then continued in Refmac (39) with manual adjustment of the model in COOT, using only native data (with identically flagged reflections). Alternate conformations of side chains, additional water molecules, and PEG molecules were incorporated at this stage. Refinement was then carried out with incorporation of anisotropic B-factors, and, in final rounds of refinement, with incorporation and refinement of hydrogen atoms. Crystallized VioE contains 199 residues, including the C-terminal His6 tail with two linking residues. In both chains A and B of VioE, residues 1–4 are disordered; in chain A, residues 190–199 are additionally disordered, and in chain B, residues 195–199 are absent. The refinement statistics are listed in Table 1.
Calculation of Buried Surface Area
The program AREAIMOL, from the CCP4i suite of programs (40), was used to calculate the accessible surface area for chain A alone, chain B alone, and the dimer, using a 1.4 Å probe radius. The buried surface area was taken as the sum of the accessible area for chain A and chain B, minus the accessible area for the dimer. The accessible area of chain A is 10,300 Å2, chain B is 10,700 Å2, and the dimer is 17,400 Å2.
Sedimentation Velocity Experiments
Sedimentation velocity experiments were carried out in a Beckman Coulter Optima XL-I analytical ultracentrifuge using a Beckman An60Ti rotor and a Beckman XL-A Monochrometer in the MIT Biophysical Instrumentation Facility. Absorbance data were collected during a velocity sedimentation run at 20.0 °C at 42,000 rpm. Protein samples were prepared as for crystallization, except that VioE eluting from a nickel (II)-loaded Chelating Sepharase™ Fast Flow (GE Biosciences) column was loaded onto a HiLoad Superdex™ 75 prep grade column (Amersham Biosciences) pre-equilibrated with 200 mm NaCl, 25 mm Tris, pH 8.0. VioE from the peak gel filtration fraction was then diluted in the identical buffer to concentrations of 8.6 and 2.3 µm, and the samples were loaded into one side of a sample cell, with buffer (200 mm NaCl, 25 mm Tris, pH 8.0) loaded into the other side of each sample cell. Sednterp (41) was used to derive values based on the experimental conditions and protein sequence for the density, partial specific volume, and mass extinction coefficient at 280 nm. These values were used to analyze the absorbance data from the sedimentation velocity experiments using SEDANAL (42).
Gel Filtration
The oligomeric state of purified VioE was additionally assayed via gel filtration using a HiLoad Superdex™ 75 prep grade column (Amersham Biosciences) pre-equilibrated with 200 mm NaCl, 25 mm Tris, pH 8.0. Gel filtration standards (Bio-Rad) were used to calibrate this column.
Mutagenesis and Mutant Protein Preparation
All VioE mutants were generated using splicing by overlap extension method (43) with the primers listed in supplemental Table S1. Two overlapping fragments (“a” and “b”) were generated from two initial PCRs, using 1 µl of miniprepped pET24b-VioE as a template, with 4 µm of each primer, 5% Me2SO, and 2× Phusion HF master mix (Invitrogen). After PCR purification using a GFX kit (GE Healthcare), the “a” and “b” fragments were mixed together and further amplified using the 5′-forward first primer from the “a” fragment reaction and the 3′-reverse second primer from the “b” fragment; all other components in the PCR were identical to those used in the first round of amplification. All of the PCRs were carried out using the following cycle parameters: 98 °C for 2 min, followed by 30 cycles of 98 °C for 15 s, 57 °C for 30 s, and 72 °C for 1.5 min, followed by cooling to 4 °C. The PCR products were once again purified using the GFX kit and then subjected to digestion with NdeI/XhoI for 3 h. The resulting DNA was purified and ligated into a similarly digested pET24b vector using Quick Ligase (New England Biolabs). The ligation reactions were then transformed into chemically competent TOP10 cells plated on LB-agar-kanamycin plates. The resulting colonies were miniprepped, and the plasmids were sequenced at the Molecular Biology Core Facilities of the Dana Farber Cancer Institute. The plasmids were then transformed into BL21(DE3) cells, and protein was prepared as described previously (26), using a dialysis rather than a gel filtration step to transfer protein into the final storage buffer.
Activity Assays
Activity assays were carried out, as described previously (26), by incubation of l-tryptophan (500 µm) with VioA (5 µm), VioB (5 µm), VioE (5 µm), and 50 units of catalase in 75 mm glycine, pH 9.25. The reactions were quenched after 1 h with 20% (v/v) Me2SO and 200% (v/v) methanol, and, after removal of precipitated protein, the samples were analyzed via high pressure liquid chromatography, as described previously (26). Prodeoxyviolacein 3 peaks at 590 nm were integrated with 32 Karat Software (version 5.0, Build 1021, Beckman Coulter, Inc.). All of the activity assays were run in triplicate and compared with authentic standards of prodeoxyviolacein 3.
Circular Dichroism
Circular dichroism spectra of native and mutant proteins were collected on an Aviv model 202 CD Spectrometer (software by Aviv, version 3.00C) at the MIT Biophysical Instrumentation Facility from 190 to 250 nm with a 1-nm wavelength step, an averaging time of 3 s, and a settling time of 0.333 s. The samples were processed in a 0.1-cm-path length rectangular quartz cuvette. All of the proteins, which were stored in a buffer solution composed of 50 mm NaCl, 20 mm Tris, pH 8.4, and 10% glycerol, were diluted in degassed, double distilled water to 17 µm in 400-µl samples. A buffer solution, also diluted in degassed, double distilled water, with a final composition of 0.2% glycerol, 1 mm NaCl, and 0.4 mm Tris, pH 8.4, was analyzed between each sample, and the average values over 10 buffer samples were used for background subtraction before generation of a final graph.
RESULTS
VioE crystallizes as a dimer, with each concave side of the curved β-sheet facing away from the interface (Fig. 2A) and with an average of 1800 Å2 of buried surface area per monomer. The large dimeric interface of the two chains is chiefly hydrophobic (63% of interacting residues) with water largely excluded (only 28 molecules of water in the interface) and with 17 hydrogen bonds that pair polar atoms with one another across the interface. To investigate the oligomeric state in solution of VioE, a sedimentation velocity experiment was carried out at each of two concentrations (8.6 and 2.3 µm). The results of the sedimentation velocity experiment are consistent with a dimer in solution (supplemental Fig. S1). The sedimentation velocity experiments are in contrast to the result from preparative scale gel filtration, which suggested that VioE eluted as a monomer, based on the gel filtration standards (supplemental Fig. S2). However, given the results of sedimentation velocity analysis, the large size of the dimeric interface, the predominance of hydrophobic residues, the general exclusion of water, and the presence of specific interactions distributed across much of the interface, the dimer appears to be the preferred oligomeric state of purified VioE.
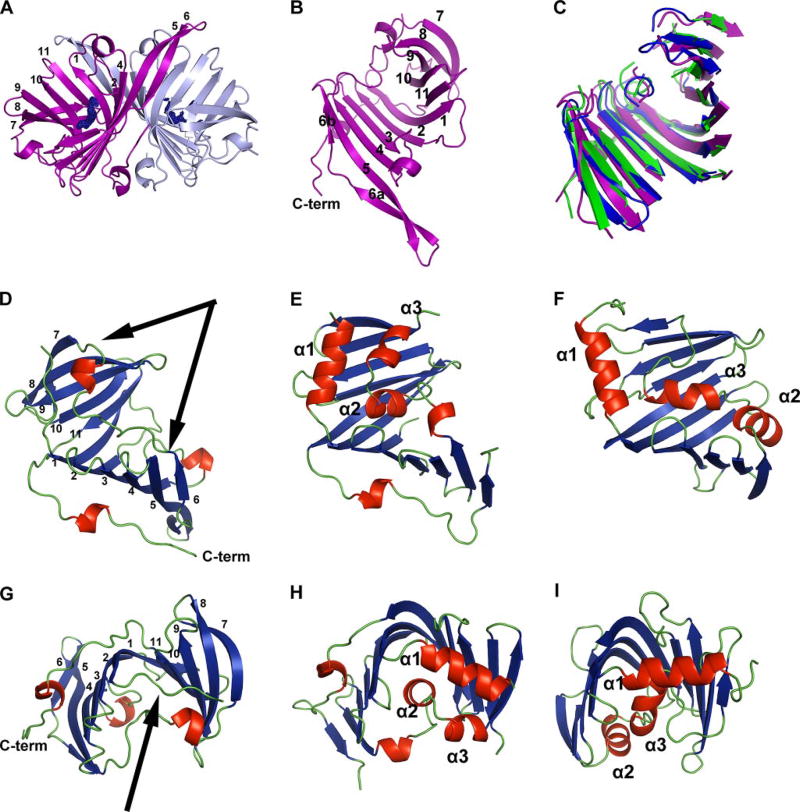
FIGURE 2. Fold of VioE and structural comparisons.
A, VioE crystallizes as a dimer, with the β-sheets positioned back-to-back. Chain A is shown in light blue, and chain B is shown in purple. A sigmaA-weighted 2Fo − Fc omit map that surrounds the PEG was generated by excluding PEG from the phase calculation, running 20 rounds of maximum likelihood refinement in Refmac and converting the structure factor file to a map file. Resulting density is shown in blue at 1.0 σ around the PEG modeled near each chain. β strands of chain B are numbered 1–11 based on their arrangement from the N to the C terminus of the protein; β3 is not visible from this view. B, one VioE monomer is shown, with β strands numbered 1–11 based on their arrangement from the N to the C terminus of the protein and with the C terminus labeled C-term. C, structurally aligned portions of VioE (purple), LolA (Protein Data Bank code 1UA8, green), and LolB (Protein Data Bank code 1IWM, blue) are shown, oriented identically to the VioE monomer shown in B. D–I,D and G show VioE (chain B), with arrows designating the loop region including amino acids 105–121 (in D) and the loop including amino acids 5–10 (in G), and with β strands numbered 1–11 based on their arrangement from the N to the C terminus of the protein and with the C terminus labeled C-term. E and H show LolA (Protein Data Bank code 1UA8), and F and I show LolB (Protein Data Bank code 1IWM). The models in D–F are identically positioned, and models in G–I are identically positioned. β-Strands are shown in blue, α-helices and short 310 helices are shown in red, and loop regions are shown in green. The three α-helices identified as forming a “lid” in LolA and LolB are labeled α1, α2, and α3. Figs. of protein models were prepared in PYMOL (http://pymol.sourceforge.net), with secondary structure assignment carried out using DSSP (58).
In each monomer, strand 1 begins in the center of the protein and initiates an anti-parallel β-sheet, building from β1 to β6 one half of the structure (Fig. 2B). Between β3 and β4 is an intervening loop, which contains a four-amino acid α-helical turn, and β6 is composed of two short β-strands, termed 6a and 6b, which are connected by a loop region. From β6, a loop extends to a four-amino acid α-helical turn, which then extends via another loop to β7 on the opposite side of the protein. β7 to β11 then build up, via anti-parallel β-strands, the second half of the sheet, with β11 and β1 hydrogen-bonding to each other, bringing the two halves of the structure together. A loop continues from the C terminus of β11 into a loop including 310 α-helical turn, which terminates the ordered portion of the molecule (Fig. 2, A and B). The long C-terminal structure is stabilized mainly with intrachain contacts, although there are also a small number of hydrophobic interactions between the two chains at their C termini, involving residues Ala180, Pro183, and Ala185.
Based on primary sequence, there are three putative homologues of C. violaceum VioE from related species (supplemental Fig. S3), all of which are uncharacterized proteins. The only partially characterized enzyme with any kind of sequence identity is the enzyme RifG (25, 44) (supplemental Fig. S4). RifG, a homologue of a dehydroquinoate synthase from Amycolatopsis mediterranei S699 (44), is proposed to function together with other enzymes from the Rif operon in the formation in 3-amino-5-hydroxybenzoic acid (44), specifically in the cyclization of 3,4-dideoxy-4-amino-d-arabino-heptulosonic acid 7-phosphate (45). However, the similarity of RifG to VioE is quite poor (E value = 0.055, with 31 identical residues, 8 clustered in the region between β6 and β7 of VioE), the sizes of the proteins differ dramatically (191 residues for VioE, and 351 residues for RifG), and the relevance of the similarity of RifG to VioE is unclear.
The story is different when one considers three-dimensional structure; VioE does have close structural homologues. VioE is structurally homologous to the unclosed β-barrel, core structure of both LolA (root mean square deviation of 1.73 Å over 124 residues) and LolB (root mean square deviation of 1.67 Å over 98 residues) (Fig. 2, C–I, and supplemental Fig. S5) (32). Unlike VioE, LolA and LolB are not enzymes. They are well characterized periplasmic lipoprotein transporters in E. coli. LolA binds lipoproteins released in an ATP-dependent manner by the ABC transporter LolCDE from the periplasmic side of the inner membrane and shuttles these lipoproteins through the periplasmic space, depositing them with LolB, a membrane-anchored protein, which incorporates them into the outer membrane (46).
LolA and LolB have “lid” regions, composed of α-helices (32, 47). Although the lid of LolB is thought to always be open (32), in LolA, these helices are thought to act as gates, controlling the access of hydrophobic cargo, the acyl chains of lipoproteins, to the unclosed β-barrel, where they are thought to be sequestered during lipoprotein transport (32, 47, 48). These LolA α-helices are thought to open and close, as lipoproteins enter and depart the hydrophobic core (32, 47, 48). In LolA, Arg43, positioned between β2 and β3, makes hydrogen bonding contacts with backbone carbonyls on the α1 (Leu10) and α2 (Ile93 and Ala94) helices (Fig. 3A) (32); Arg43 is thus thought to play a key role in stabilizing the closed form of LolA. Mutation of Arg43 to Leu leads to the accumulation of LolA-lipoprotein complexes and eliminates transfer of lipoproteins from LolA to LolB (49), suggesting that the closing of the lid via formation of hydrogen bonds to Arg43 may drive the energy-independent transfer of lipoproteins from LolA to LolB (32, 47, 48). LolB, which has a higher affinity for lipoproteins than LolA (50), lacks a residue playing this role; LolB is thought to be more stable in the “open” conformation, where its hydrophobic core is more accessible to lipoproteins (32).
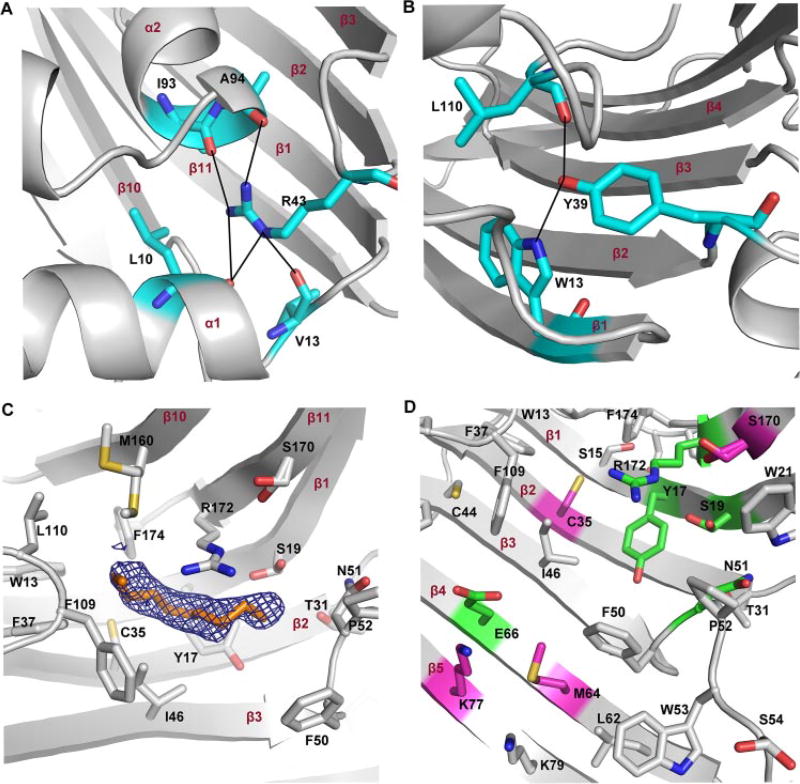
FIGURE 3. The putative lid and extended active site of VioE.
A, the proposed closed lid region of LolA is shown (Protein Data Bank code 1UA8), with Arg43 mediating a series of hydrogen-bonding interactions that close off the exposed side of the LolA structure. B, in VioE, the structurally superimposed residue to Arg43 is Tyr39, which is also positioned between β2 and β3 and also makes hydrogen bonding contacts to residues shown in blue. C, side chains within 6 Å of the PEG molecule of chain B are shown. Alternate conformations were refined for Ser19, Met160, and Ser170.D, mutated residues are shown within the context of surrounding residues, each of which are within 12 Å from the bound PEG molecule. Residues that were not targeted for mutagenesis are shown in gray. Those in pink, when mutated, had no significant effect on the production of prodeoxyviolacein 3, whereas those in green, when mutated, had significantly reduced production of prodeoxyviolacein 3. In all panels, secondary structural elements are labeled in red.
VioE lacks the three longer α-helices of LolA and LolB, having only two short α-helices and one 310 α-helical turn, but it also may have a similar lid region. Amino acids 5–10 traverse a similar space in the VioE structure as the α1-helix of LolA. Further, amino acids 105–121 of VioE, a loop region extending from β6 to β7 and containing the second α-helix of VioE, traverse across similar space to that of the α2 and α3-helices of LolA and the α3-helix of LolB. Finally, the structurally super-imposed residue to Arg43 of LolA is Tyr39 of VioE. Tyr39, which is also positioned between β2 and β3 of VioE (like Arg43 of LolA), is in position to hydrogen bond with the indole nitrogen of Trp13 (positioned on β1) and with the backbone carbonyl of Leu110 (Fig. 3B). Although there are fewer hydrogen bonds to Tyr39 in VioE than to Arg43 in LolA, VioE, like LolA, appears to at least partially seclude its hydrophobic core from exposure to solvent, suggesting that VioE, like LolA, may have a dynamic lid region, involving Tyr39.
The 90-some liporoteins that are targeted for transfer by LolA and LolB are all thought to be recognized by a common feature: their acyl chains. A structure of LolB reported with a bound PEG MME molecule was used to identify the likely site of acyl chain binding in LolB (32), and, by extension, in LolA (48). Strikingly, a tube-like molecule corresponding to PEG was refined into each monomer in the VioE structure (Figs. 2A and 3C). This PEG molecule is located in the same area as the PEG MME molecule identified in the LolB structure (Protein Data Bank code 1IWN) (32). In VioE, the PEG molecule is bound at the base of the bowl-like curve of the β-sheet and is partially exposed to solvent. Specifically, one orientation of a tri(ethylene) glycol molecule was fit to one monomer and one orientation of a tetra(ethylene) glycol molecule was fit to the other monomer. Unlike the rest of the VioE structure, however, where the B-factors of the protein atoms are quite low, averaging 15.0 Å2, the average B-factors of the atoms of the PEG molecules are significantly higher, averaging 44.2 Å2 (Table 1). Further, the maps show diffuse electron density, indicating flexibility in the binding of the PEG molecules (Fig. 3C), in contrast to the sharp quality of electron density throughout most of the structure (supplemental Fig. S6). Despite the fact that the PEG is not well ordered in the structure, its positioning is intriguing. As was the case for LolB, it is tempting to speculate that the site of PEG binding also represents a binding site for a substrate, an acyl chain, in the case of LolB, or a substrate molecule, in the case of VioE.
Surrounding the PEG molecule in the VioE structure are a series of hydrophobic residues (Fig. 3C): Trp13, Phe37, Ile46, Phe50, Pro52, Phe109, Leu110, Met160, and Phe174. Any putative substrates of VioE would be hydrophobic; both IPA 1 (the substrate of VioB) and prodeoxyviolacein 3 (the product of VioE) are poorly soluble in water, and it is unlikely that any molecules spanning the chemical space between IPA 1 or prodeoxyviolacein 3 would be well soluble in water. These hydrophobic residues surrounding the PEG molecule in the putative VioE active site should form an ideal binding pocket for the hydrophobic substrate molecule, any intermediates, and the prodeoxyviolacein 3 product during catalysis.
Within 6 Å of the bound PEG molecule are also a small number of polar and charged residues, Tyr17, Ser19, Thr31, Cys35, Asn51, Ser170, and Arg172, which might have relevance in binding the polar parts of the substrate molecule or in catalysis (Fig. 3C). Further from the bound PEG molecule, within 12 Å, are additional polar or charged residues, including Ser21, Cys44, Ser54, Glu66, Lys77, and Lys79 (Fig. 3D), among others. To investigate the importance of particular residues to catalysis and to investigate whether the PEG molecule had indeed identified the active site of VioE, a series of nine site-directed mutants were generated, and their activities were tested (Fig. 4). To verify that each mutant was properly folded, each mutant was examined with CD spectrometry and compared with wild-type VioE. We found that each mutant shows a similar CD spectrum to that of wild-type VioE (supplemental Fig. S7), suggesting that all mutants are properly folded and that mutations reveal the importance of particular residues to binding or catalysis and not a general effect on the proper folding of the protein. All of the targeted side chains were mutated to alanine.
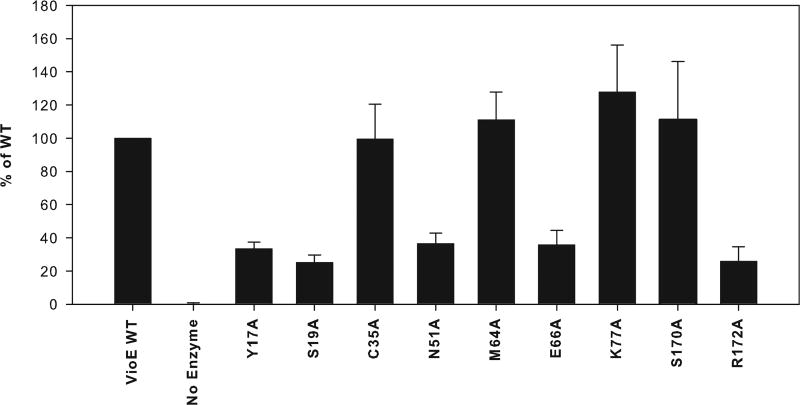
FIGURE 4. Activities of mutant VioE proteins.
The average percentage of production of prodeoxyviolacein 3 by each mutant relative to that of wild-type VioE is shown, with error bars representing the standard deviation over three trials. Y17A, S19A, N51A, E66A, and R172A show significantly reduced prodeoxyviolacein 3 production relative to wild type, whereas C35A, M64A, K77A, and S170A produce prodeoxyviolacein 3 at a level that is not significantly different from wild type.
Two groups of mutants were generated. The first group was in the immediate area of the PEG molecule (within 6 Å), and includes Arg172, closest to the PEG molecule (Fig. 3C). Mutation of Arg172 resulted in a significant decrease in prodeoxyviolacein 3 production (Fig. 4). Similarly, mutation of either Tyr17 or Ser19, each between 3 and 5 Å from the bound PEG molecule (Fig. 3C) resulted in a significant decrease in prodeoxyviolacein 3 production (Fig. 4). Additionally, mutation of Asn51, between 5 and 8 Å from the bound PEG molecule (depending on the monomer) (Fig. 3C), resulted in a significant decrease in prodeoxyviolacein 3 production (Fig. 4). By contrast, mutation of Cys35, ~4 Å from the bound PEG molecule (Fig. 3C), yet on a different side of the PEG from Tyr17 and Ser19, did not result in a significant change in prodeoxyviolacein 3 production (Fig. 4). Additionally, mutation of Ser170, which is out further from the PEG molecule (Fig. 3C) but positioned near Ser19, did not significantly alter prodeoxyviolacein 3 production (Fig. 4). These mutations thus delineate the active site adjacent to the bound PEG molecule (Fig. 3D).
The second group of amino acids mutated is further away from the PEG molecule (>8 Å) but still in the general area of the concave β-sheet where substrate might bind. Glu66, positioned a distance of 9 Å from Arg172 (Fig. 3D), was a primary target, because its distance from Arg172 is within range of the size of putative substrates and products; for instance, the distance in prodeoxyviolacein 3 from the ketone oxygen to the furthest indole nitrogen is ~7 Å. Mutation of Glu66 resulted in a significant decrease in prodeoxyviolacein 3 production (Fig. 4). Two nearby residues to Glu66, Met64 and Lys77, were then targeted for mutagenesis. Although the average production of prodeoxyviolacein 3 was increased for both mutants relative to wild-type, based on the standard deviation, the increase was nonsignificant for both mutants (Fig. 4). This group therefore delineates a second part of the putative active site of VioE that includes charged residue Glu66 (Fig. 3D).
DISCUSSION
The unexpected result that VioE adapts a fold normally associated with lipoprotein carrier proteins to carry out an enzymatic function suggests that a crucial part of its function is, like a lipoprotein carrier protein, to bind a hydrophobic molecule. In the case of VioE, the binding of a hydrophobic substrate in a particular conformation may effectively chaperone the production of a desired product. The conversion that VioE facilitates involves oxidative chemistry, but the most unusual aspect of the reaction, a 1,2 shift of an indole ring, is a reaction that is not known to be catalyzed by other characterized enzymes. Other shift reactions have been characterized (51–54), and each of the responsible enzymes utilize iron as a cofactor (as a heme or non-heme iron), whereas VioE requires no cofactors or metals. The lack of cofactors of VioE might mean that this indole shift reaction is mechanistically more facile than other observed, enzyme-mediated shift reactions. Perhaps the main role of VioE is to stabilize its substrate in a favorable conformation to enable the 1,2 shift reaction to occur through classic acid-base or oxidative chemistry, and the fold adapted by VioE works well for this. Other enzymes thought to use stabilization of an alternate, higher energy orientation of a substrate to promote catalysis include the peptidyl-prolyl isomerases (55, 56), although a precise delineation of the mechanisms of these enzymes is still under investigation. One proposed substrate for VioE is the Cβ-Cβ benzylically coupled iminophenylpyruvate dimer 2 (Fig. 1 and Ref. 26), a molecule that could collapse spontaneously to chromopyrrolic acid 4 (26). Stabilization of this molecule (or a related intermediate) by VioE in an orientation suitable for promotion of a 1,2 indole shift might reroute it toward prodeoxyviolacein 3.
Our site-directed mutagenesis studies have identified a number of residues likely to be involved in binding polar parts of the substrate molecule or in catalysis: Tyr17, Ser19, Asn51, Glu66, and Arg172. Although mutation of any of these residues does not completely abolish VioE activity, the significant decrease in activity by each of these mutants implies that they play a role in enzymatic acceleration of prodeoxyviolacein 3 production by VioE. Our findings do suggest that the PEG molecule cocrystallized with VioE has indeed identified the active site of the enzyme. However, it appears that the active site extends beyond the site of PEG binding to include the side chain of Glu66. An extended active site is not surprising given the larger size of the known product molecule.
In conclusion, our crystal structure of VioE has revealed that an unusual enzyme, a cofactorless, metal-free protein known to catalyze an unusual 1,2 shift reaction, also adapts an unusual fold, that of a lipoprotein carrier protein. This finding indicates that the main role of VioE is to sequester a hydrophobic molecule from the milieu of the cell. Once bound, VioE likely uses acid-base chemistry, probably coupling transformations to spontaneous oxidative processes, to carry out its reaction, without the need for metals or cofactors. Further studies of this intriguing enzyme, including elucidation of its as yet unknown substrate, will help unravel how each residue identified in this study enables VioE to reroute an unknown intermediate away from spontaneous formation of chromopyrrolic acid 4 and toward production of the violacein 5 scaffold.
Supplementary Material
Acknowledgments
The MIT Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems and the Northeastern Collaborative Access Team facility at the Advanced Photon Source at Argonne National Laboratory are gratefully acknowledged. We thank Deborah Pheasant and Nelson Olivier for assistance with sedimentation velocity experiments and Kanagalaghatta Rajashankar and Thomas Schwartz for helpful discussions during x-ray data collection.
Footnotes
This work was supported in part by National Institutes of Health Grants GM65337 (to C. L. D.) and GM 20011 (to C. T. W.) and the MIT Center for Environmental Health Sciences NIEHS P30 ES002109. This work was also supported in part by National Science Foundation Grant 0070319, National Institutes of Health Grant GM68762, and other funds from the National Institutes of Health.
The atomic coordinates and structure factors (code 3BMZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and supplemental Figs. S1–S7.
The abbreviations used are: IPA, indole 3-pyruvic acid; PEG, polyethylene glycol.
References
- 1.Reilly J, Pyne G. Biochem. J. 1927;21:1059–1064. doi: 10.1042/bj0211059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobie WC. J. Bacteriol. 1935;29:223–227. doi: 10.1128/jb.29.3.223-227.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong FM. Science. 1944;100:287. doi: 10.1126/science.100.2596.287. [DOI] [PubMed] [Google Scholar]
- 4.Ballantine JA, Beer RJ, Crutchley DJ, Dodd GM, Palmer DR. Proc. Chem. Soc. 1958;1:232–234. [Google Scholar]
- 5.Laatsch H, Thomson RH, Cox PJ. J. Chem. Soc. Perkin Trans. II. 1984;8:1331–1339. [Google Scholar]
- 6.Lichstein HC, Van De Sand VF. J. Bacteriol. 1946;52:145–146. [PMC free article] [PubMed] [Google Scholar]
- 7.Durán N, Erazo S, Campos V. An. Acad. Brasil. Cienc. 1983;55:231–234. [Google Scholar]
- 8.Durán N, Menck CF. Crit. Rev. Microbiol. 2001;27:201–222. doi: 10.1080/20014091096747. [DOI] [PubMed] [Google Scholar]
- 9.Durán N, Campos V, Riveros R, Joyas A, Pereira MF, Haun M. An. Acad. Bras. Cienc. 1989;61:31–36. [PubMed] [Google Scholar]
- 10.Leon LL, Miranda CC, De Souza AO, Duran N. J. Antimicrob. Chemother. 2001;48:449–450. doi: 10.1093/jac/48.3.449. [DOI] [PubMed] [Google Scholar]
- 11.Durán N, Justo GZ, Melo PS, De Azevedo MB, Brito AR, Almeida AB, Haun M. Can. J. Physiol. Pharmacol. 2003;81:387–396. doi: 10.1139/y03-033. [DOI] [PubMed] [Google Scholar]
- 12.Melo PS, Maria SS, Vidal BC, Haun M, Durán N. In Vitro Cell. Dev. Biol. Anim. 2000;36:539–543. doi: 10.1290/1071-2690(2000)036<0539:vcaioa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Melo PS, Justo GZ, de Azevedo MB, Durán N, Haun M. Toxicology. 2003;186:217–225. doi: 10.1016/s0300-483x(02)00751-5. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira CV, Bos CL, Versteeg HH, Justo GZ, Durán N, Peppelenbosch MP. Blood. 2004;104:1459–1464. doi: 10.1182/blood-2004-02-0594. [DOI] [PubMed] [Google Scholar]
- 15.Saraiva VS, Marshall J-C, Cools-Lartigue J, Burnier MN., Jr Melanoma Res. 2004;14:421–424. doi: 10.1097/00008390-200410000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Kodach LL, Bos CL, Durán N, Peppelenbosch MP, Ferreira CV, Hardwick JC. Carcinogenesis. 2006;27:508–516. doi: 10.1093/carcin/bgi307. [DOI] [PubMed] [Google Scholar]
- 17.de Carvalho DD, Costa FT, Durán N, Haun M. Toxicol. in Vitro. 2006;20:1514–1521. doi: 10.1016/j.tiv.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 18.DeMoss RD, Evans NR. J. Bacteriol. 1959;78:583–588. doi: 10.1128/jb.78.4.583-588.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeMoss RD, Evans NR. J. Bacteriol. 1960;79:729–733. doi: 10.1128/jb.79.5.729-733.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshino T, Takano T, Hori S, Ogasawara N. Agric. Biol. Chem. 1987;51:2733–2741. [Google Scholar]
- 21.Momen AZ, Hoshino T. Biosci. Biotechnol. Biochem. 2000;64:539–549. doi: 10.1271/bbb.64.539. [DOI] [PubMed] [Google Scholar]
- 22.August PR, Grossman TH, Minor C, Draper MP, MacNeil IA, Pemberton JM, Call KM, Holt D, Osburne MS. J. Mol. Microbiol. Biotechnol. 2000;2:513–519. [PubMed] [Google Scholar]
- 23.Brady SF, Chao CJ, Handelsman J, Clardy J. Org. Lett. 2001;3:1981–1984. doi: 10.1021/ol015949k. [DOI] [PubMed] [Google Scholar]
- 24.Brazilian National Genome Project Consortium. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez C, Braña AF, Méndez C, Salas JA. ChemBio-Chem. 2006;7:1231–1240. doi: 10.1002/cbic.200600029. [DOI] [PubMed] [Google Scholar]
- 26.Balibar CJ, Walsh CT. Biochemistry. 2006;45:15444–15457. doi: 10.1021/bi061998z. [DOI] [PubMed] [Google Scholar]
- 27.Asamizu S, Kato Y, Igarashi Y, Onaka H. Tetrahedron Lett. 2007;48:2923–2926. [Google Scholar]
- 28.Howard-Jones AR, Walsh CT. Biochemistry. 2005;44:15652–15663. doi: 10.1021/bi051706e. [DOI] [PubMed] [Google Scholar]
- 29.Asamizu S, Kato Y, Igarashi Y, Furumai T, Onaka H. Tetrahedron Lett. 2006;47:473–475. [Google Scholar]
- 30.Kim S-Y, Park J-S, Chae C-S, Hyun C-G, Choi BW, Shin J, Oh K-B. Appl. Microbiol. Biotechnol. 2007;75:1119–1126. doi: 10.1007/s00253-007-0924-x. [DOI] [PubMed] [Google Scholar]
- 31.Gao Q, Zhang C, Blanchard S, Thorson JS. Chem. Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Takeda K, Miyatake H, Yokota N, Matsuyama S, Tokuda H, Miki K. EMBO J. 2003;22:3199–3209. doi: 10.1093/emboj/cdg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 36.Abrahams JP, Leslie AG. Acta Crystallogr. D Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- 41.Laue TM, Shah BD, Ridgeway TM, Pelletier S. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton JC, editors. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 42.Stafford WF, Sherwood PJ. Biophys. Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene (Amst.) 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 44.Yu T-W, Müller R, Müller M, Zhang X, Draeger G, Kim C-G, Leistner E, Floss HG. J. Biol. Chem. 2001;276:12546–12555. doi: 10.1074/jbc.M009667200. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe K, Rude MA, Walsh CT, Khosla C. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9774–9778. doi: 10.1073/pnas.1632167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narita S, Matsuyama S, Tokuda H. Arch. Microbiol. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi N, Matsuyama S, Tokuda H. J. Biol. Chem. 2005;280:34481–34488. doi: 10.1074/jbc.M507388200. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe S, Matsuyama S, Tokuda H. J. Biol. Chem. 2006;281:3335–3342. doi: 10.1074/jbc.M509596200. [DOI] [PubMed] [Google Scholar]
- 49.Miyamoto A, Matsuyama S, Tokuda H. Biochem. Biophys. Res. Commun. 2001;287:1125–1128. doi: 10.1006/bbrc.2001.5705. [DOI] [PubMed] [Google Scholar]
- 50.Matsuyama S, Yokota N, Tokuda H. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li R, Reed DW, Liu E, Nowak J, Pelcher LE, Page JE, Covello PS. Chem. Biol. 2006;13:513–520. doi: 10.1016/j.chembiol.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Hashim MF, Hakamatsuka T, Ebizuka Y, Sankawa U. FEBS Lett. 1990;271:219–222. doi: 10.1016/0014-5793(90)80410-k. [DOI] [PubMed] [Google Scholar]
- 53.Bassan A, Blomberg MR, Siegbahn PE. Chemistry. 2003;9:4055–4067. doi: 10.1002/chem.200304768. [DOI] [PubMed] [Google Scholar]
- 54.Moran GR. Arch. Biochem. Biophys. 2005;433:117–128. doi: 10.1016/j.abb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Ranganathan R, Lu KP, Hunter T, Noel JP. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 56.Fischer G. Chem. Soc. Rev. 2000;29:119–127. [Google Scholar]
- 57.Howard-Jones AR, Walsh CT. J. Am. Chem. Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 58.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.