SUMMARY
Axonal regrowth is crucial for recovery from CNS injury but is severely restricted in adult mammals. We used a genome-wide loss-of-function screen for factors limiting axonal regeneration from cerebral cortical neurons in vitro. Knockdown of 16,007 individual genes identified 580 significant phenotypes. These molecules share no significant overlap with those suggested by previous expression profiles. There is enrichment for genes in pathways related to transport, receptor binding, and cytokine signaling, including Socs4 and Ship2. Among transport-regulating proteins, Rab GTPases are prominent. In vivo assessment with C. elegans validates a cell-autonomous restriction of regeneration by Rab27. Mice lacking Rab27b show enhanced retinal ganglion cell axon regeneration after optic nerve crush and greater motor function and raphespinal sprouting after spinal cord trauma. Thus, a comprehensive functional screen reveals multiple pathways restricting axonal regeneration and neurological recovery after injury.
In Brief
Sekine et al. conduct a genome-wide loss-of-function screen for factors limiting the success of CNS axonal regeneration in mice. They uncover a role for transport, receptor binding, and cytokine signaling pathways. In particular, in vivo loss of Rab27b expression increases axonal regeneration in worm and mouse optic nerve.
INTRODUCTION
Devastating and persistent functional deficits occur after spinal cord injury (SCI), despite survival of nearly all neurons. Because the primary cause of disability is disconnection of networks by axon transection, axon regrowth has the potential to provide recovery by restoring connectivity, without requiring “new” cells. It is clear that both cell-autonomous and environmental factors contribute to axon growth failure.
There have been genetic attempts to identify axon regeneration factors, but the field has not benefited from unbiased genome-wide functional approaches. Most efforts have started with expression surveys rather than functional studies. No functional screen has focused on endogenous genes in adult mammalian CNS at a level approaching the entire genome (Blackmore et al., 2010; Moore et al., 2009; Park et al., 2008), and existing efforts frequently use gain of function, initial outgrowth, and/or cell lines (Blackmore et al., 2010; Buchser et al., 2010; Loh et al., 2008; Moore et al., 2009; Sepp et al., 2008). Non-mammalian regeneration has been analyzed extensively in C. elegans by loss of function (LOF) with both small interfering RNA (siRNA) and mutant alleles, confirming regeneration mechanisms conserved between mammals and C. elegans (El Bejjani and Hammarlund, 2012). DLK-1, PTEN, and cAMP are important regulators of regeneration in mammals and have similar functions in worms (El Bejjani and Hammarlund, 2012; Hammarlund et al., 2009; Wang and Jin, 2011; Yan et al., 2009). Yet few C. elegans regeneration genes have been validated in mammals. Moreover, even in this model organism, using both mutant alleles and siRNA screening, less than 25% of the genome has been tested for axon regeneration. In summary, a loss of function screen for mammalian CNS regeneration has not been completed on a scale approaching the entire genome.
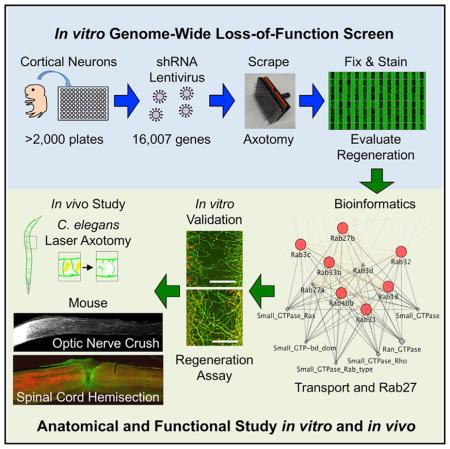
Here, we sought an unbiased genome-wide assessment of mammalian genes whose loss of function allows axonal sprouting and regeneration after CNS trauma. Critically, our approach was unbiased at the genome-wide level in mammalian species, focused on axonal regeneration and using cerebral cortical projection neurons. With single clones spanning a lentiviral short hairpin RNA (shRNA) library, we assessed the role of each gene to limit axonal regeneration. Our pilot screen restricted to 219 phosphatases had uncovered a role for Inpp5f in limiting axonal regrowth and neurological recovery from trauma (Zou et al., 2015). Here, a comprehensive genome-wide screen reveals about 500 genes with a regeneration phenotype, and the vast majority were not previously identified by expression surveys or previous limited functional studies. We validate these hits and show that protein transport function is the most highly enriched group limiting axon regeneration. The studies uncover multiple pathways with a role in limiting regeneration, and highlight Rab-dependent membrane trafficking as a key factor for enhancing neurological recovery.
RESULTS
Functional Genomic Screen of Mouse CNS Axon Regeneration
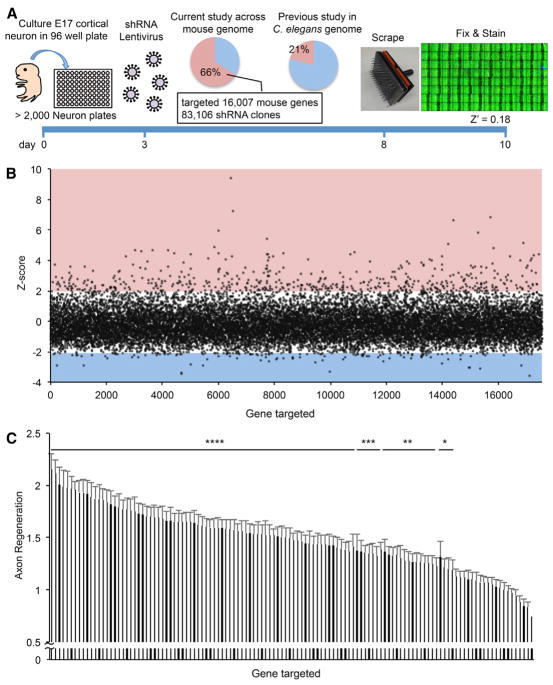
We conducted a loss of function genome-wide in vitro axon regeneration screen in primary mouse cortical neurons (Huebner et al., 2011; Zou et al., 2015). We reasoned that this in vitro model, although lacking features of the in vivo CNS such as environmental contributions from glia and matrix, would capture cell-intrinsic functions of CNS neurons that limit regeneration. In these cultures more than 80% of cells are NeuN-positive cells at day in vitro (DIV) 10 (Figures S1A and S1E). There are low percentages of astrocytic and oligodendrocytic lineage cells with about 10% of cells being detected with O4, anti-PDGFRα, and anti-GFAP antibodies, but there are essentially no detectable microglial cells detected with Iba1 antibodies (Figures S1B–S1E). At DIV 3, clones from a lentiviral mouse shRNA library were added to cortical neurons in 96-well microtiter plates at a titer of 104 to 105. Approximately 83,000 separate clones were tested, with about 20 no-virus controls per plate and each plate tested in two replicates. The resulting screen targeted more than 16,000 protein-coding genes with three to five shRNA species per gene, representing about 70% of the predicted protein-coding genes in the mouse genome. On DIV 8, by which time axon extension had ceased and neurons were quiescent, we initiated axon injury and potential regeneration in each well by using a 96-pin tool to generate a reproducible scrape lesion. After injury, neurons were allowed 2 days for axon regeneration. Then neurons were fixed and stained with anti-βIII tubulin antibody to visualize axons, rhodamine-conjugated phalloidin for growth cones, and DAPI for nuclei (Huebner et al., 2011). The regenerated zone contains axons but essentially no cell soma or dendrites. Stained plates were imaged using an ImageXpress fluorescent microscopy system with autofocus and motorized stage, and images were processed using scripts written in MATLAB (The MathWorks) to detect the injury zone and measure axonal regeneration with a Z′ of 0.18 (Figure 1A).
Figure 1. Mouse Cortical Axon Regeneration Analysis in a Genome-wide Loss-of-Function Screen.
(A) Schematic time line for this screen.
(B) The Z score for axonal regeneration is plotted from all measurements for each of 16,007 genes normalized to control shRNA. Red square shows Z score > 2.0, and blue square shows Z score < −2.0.
(C) The top 122 genes from the first screen were retested for validation (see also Table S1). Data are mean with SEM for n = 16–20, four replicates of four or five shRNA species. Results for each gene were compared with non-targeting virus wells using ANOVA with the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In the primary screen, the Z score metric, (normalized regeneration − 1)/(SD for all genes), reveals a positive hit rate slightly less than 3%; 479 genes increased axonal regeneration by more than two SDs from control (Figure 1B; Table S1). Suppression of 100 genes showed decreased axon regeneration by more than two SDs, though either decreased survival or decreased axonal growth per se may explain this phenotype. We focused on genes whose suppression stimulates regeneration (Z score > 2.0), because future development of pharmacological reagents is feasible when antagonists might promote regeneration. The top 122 genes from the full screen were retested for the validity and reproducibility of the screen (Figure 1C). Even with correction for 122 pairwise comparisons, 63% of the retested hits showed strong statistical significance (p < 0.0001), and 82% achieved statistically significant increases in axon regeneration. Thus, the screen faithfully identifies genes for which loss of function enhances regeneration in our in vitro assay.
Axon Regeneration Genes Are Distinct from Expression Surveys or Invertebrate Screens
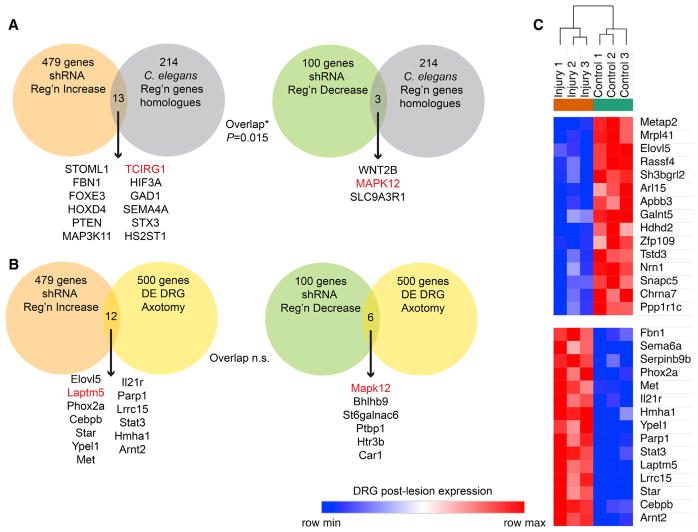
We asked whether the genes we found to functionally affect regeneration of cultured vertebrate CNS neurons after injury (Table S1) are similar to genes identified by other functional methods. Loss-of-function studies in vivo in peripheral nervous system (PNS) neurons (motor and sensory) completed in C. elegans have analyzed several thousand genes with 214 significant phenotypes (Chen et al., 2011; Nix et al., 2014). Murine orthologs for the 214 worm genes were identified bioinformatically and compared with the mouse regeneration gene lists (Figure 2A). There is a statistically significant overlap of these lists, with 16 of the 214 orthologs also affecting regeneration in our screen, suggesting that to some extent regeneration mechanisms are conserved between these two systems. Differences in neuron type, experimental method, or species may limit the degree of overlap that were detected.
Figure 2. Functional Regeneration Genes Are Distinct from Those Identified by Expression Profiling.
(A) The functional axonal regeneration gene list from Figure 1 was compared with the mouse orthologs of genes with axonal regeneration phenotypes in C. elegans (Chen et al., 2011; Nix et al., 2014). The genome-wide significance of the overlap between lists was compared using a chi-square test. Individual genes in both datasets are listed. Genes in red were also detected in other comparisons from Figure 2 or Figure S1.
(B) The functional axonal regeneration gene list from Figure 1 was compared with the list of genes differentially expressed in cultured DRG neurons preconditioned by sciatic nerve injury (Tedeschi et al., 2016). The genome-wide significance of the overlap between lists was compared using a chi-square test (p > 0.05). Individual genes in both datasets are listed. Genes in red were also detected in other comparisons from Figure 2 or Figure S1.
(C) For the functional axonal regeneration genes with Z scores > 2.0 from Figure 1, the expression level in cultured DRG neurons with or without preconditioning sciatic nerve injury was assessed from published values (Tedeschi et al., 2016). The top markers of injury-induced differential DRG expression within this set of 479 genes were identified by signal-to-noise ratio using the Morpheus website and plotted as a row-normalized expression map. Among genes limiting axonal regeneration, both up- and downregulated DRG genes are detected.
Next, we asked whether the genes we found to functionally affect regeneration of cultured vertebrate CNS neurons after injury are similar to genes found to change expression after injury. Such expression changes have been hypothesized to include genes that are functionally important for regeneration. The 500 most differentially expressed (DE) genes from a cultured dorsal root ganglion (DRG) neuron study of the effect of preconditioning axotomy (Tedeschi et al., 2016) were extracted from RNA sequencing (RNA-seq) data of the GEO repository. This list was compared with the functional shRNA cortical axon regeneration gene lists (Figure 2B). A total of 12 and 6 genes overlapped between the functional and expression studies, and this rate was not statistically significant on the basis of chi-square analysis of sampling across the mouse genome. A broad range of alternate expression studies have been performed (Chandran et al., 2016). Previously, we assessed lumbar DRG expression in vivo by Affymetrix array at 7 days post-sciatic nerve crush, identifying 279 genes with significantly altered expression (Tanabe et al., 2003). This study avoided any issues related to tissue culture prior to expression analysis, but the overlap with functional axon regeneration genes remained minimal with 5 genes in total, and was non-significant (Figure S2A). The functional regeneration genes were identified here in cortical neurons, so we also assessed overlap with our RNA-seq expression profile of sprouting corticospinal neurons after pyramidotomy (Fink et al., 2017). This expression survey also revealed minimal and statistically insignificant overlap with functional effects on axon regeneration, though the absolute numbers were higher than for the DRG studies (Figure S2B). Overall, we conclude that our functional screen identified genes largely distinct from analyses of gene expression after axon injury, suggesting that inhibition of regeneration is mediated largely by genes that are constitutively expressed rather than by injury-induced transcription.
To further assess any connection between our functional assay hits and differentially expressed genes, we examined expression levels from the RNA-seq GEO dataset analyzed in Figure 2B for each of the functional axon regeneration genes with Z scores > 2 and selected those with the most strongly altered expression (Figure 2C). Clearly some genes functionally limiting regeneration do show altered expression, even though they are not the most prominently altered in expression, and the directional effect on expression can be either increased or decreased.
Pathway Analysis of Functional Axonal Regeneration Genes
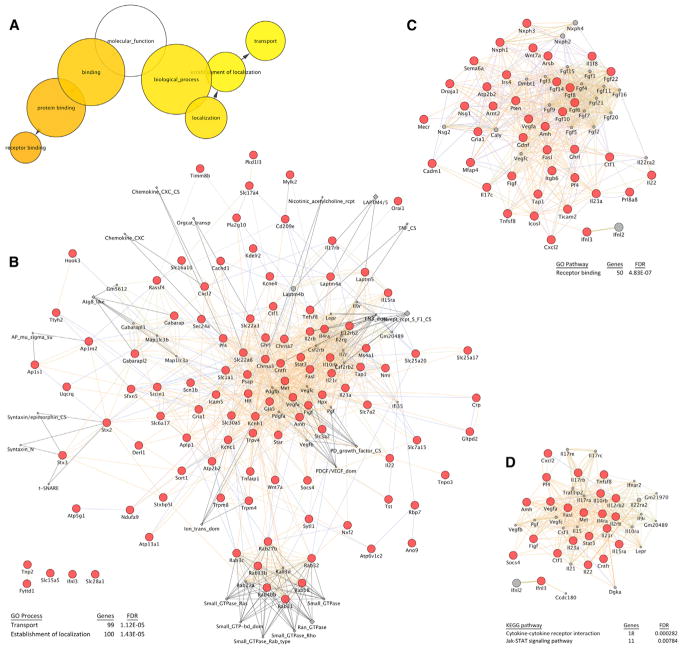
The 479 genes limiting axon regeneration with Z scores > 2.0 in the primary screen were analyzed bioinformatically to identify cellular pathways that limit regeneration. Three major pathways emerged from this analysis. Most strikingly, using Cytoscape and BINGO software (Maere et al., 2005; Shannon et al., 2003), the top Gene Ontology molecular function pathway enrichments include “transport” and “receptor binding” with a Bonferroni-corrected family-wise error rate (FWER) p value < 0.01 (Figure 3A). The protein-protein interactions, shared domain, and co-localization between the 99 genes linked to transport (false discovery rate [FDR] = 1.12E-05 by STRING; Szklarczyk et al., 2017) were assessed using GeneMania software (Montojo et al., 2010) (Figure 3B). Prominent among the transport group are the Rab GTPases, as detailed below. Also included are SNARES, ion channels, and transporters. The Rab GTPases are analyzed below. As a pathway, “transport” has not been associated with axonal regeneration mechanisms previously.
Figure 3. Functional Pathways Regulating Axonal Regeneration.
(A) The list of regeneration genes from Figure 1 and Table S1 with Z scores > 2.0 was assessed for Gene Ontology (GO) pathway enrichment using BINGO software (Maere et al., 2005; Shannon et al., 2003). Those pathways significantly enriched (Bonferroni-corrected family-wise error rate [FWER] p < 0.01) are shown with colored circles of greater intensity for greater significance. The size of each circle reflects the number of genes in that category.
(B) The list of regeneration genes from Figure 1 and Table S1 with Z scores > 2.0 that are in the “transport” GO function group was analyzed using GeneMania software (Montojo et al., 2010) for interactions. Each red circle is regeneration gene, each gray diamond is a protein domain, and each gray circle is predicted regeneration gene on the basis of sequence homology. Protein-protein interactions, co-localization, and co-expression are shown by connecting lines. The “transport” pathways includes multiple Rab proteins. Statistical significance for enrichment of this process by genome-wide false discovery rate (FDR) was calculated using the STRING database (Szklarczyk et al., 2017).
(C) Similar analysis as in (B) but for the “receptor binding” GO process.
(D) Similar analysis as in (B) but for the “cytokine” plus “Jak-Stat signaling” KEGG pathways.
Second, we found numerous protein-protein, domain, and co-localization associations between the 50 receptor binding genes with axon regeneration phenotypes (FDR = 4.83E-07 by STRING; Figure 3C). Prominent among the receptor binding group are several growth factors, including Fgf family members. It may be that suppression of growth factors promotes differentiation and axon growth in this cortical neuron culture system.
Finally, analysis of KEGG pathway (Kanehisa et al., 2017) enrichment highlighted a role of cytokine and Jak-Stat signaling (FDRs = 2.82E-04 and 7.84E-03 by STRING; Figure 3D). These findings are consistent with previous work identifying Stat3 and Socs as critical regulators of regeneration in vivo (Qiu et al., 2005; Smith et al., 2009). Thus, our screen successfully identified known regeneration mechanisms, in addition to identifying a large number of genes and functions not previously associated with regeneration.
The presence of transcription factor binding sites within 2 kb of the translation start site or microRNA binding sites within the 3′UTR as collected by MSigDB was also analyzed using GeneMania for the axonal regeneration genes (Figure S3A). Binding sites for SP1, ATF3, MEF2, and FAC1 were each significantly enriched among the genomic sequence near the transcriptional start sites of the axon regeneration genes. The genes with binding site for these factors are illustrated in Figure S3A. Because ATF3 overexpression has been associated with greater axonal regeneration (Seijffers et al., 2006; Tanabe et al., 2003), the presence of these sites in genes limiting regeneration implies that ATF3 suppresses their expression, that increased transcription by ATF3 for these genes tends to counteract ATF3 action through other sites, or that ATF3 binding sites are non-functional in these genes.
The presence of binding sites for one microRNA binding site in the 3′UTR was enriched among axonal regeneration genes, namely miR-202, which may regulate both Stat3 and Pten expression (Figure S3B). This has the potential to provide a strong synergistic action in promoting axonal regeneration.
Pharmacological Targets in Axonal Regeneration
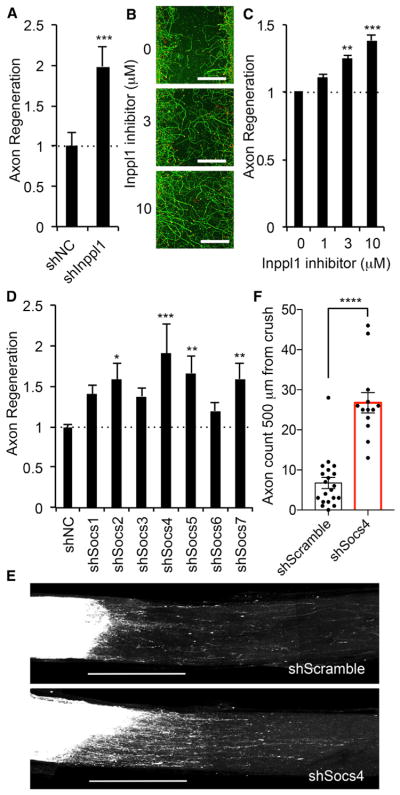
Among the axon regeneration gene list, a subset includes the targets of existing pharmacological agents. One such gene encodes inositol polyphosphate phosphatase-like 1, Inppl1, which was revalidated by a second production of Inppl1-shRNA-expressing lentivirus (Figure 4A). Innpl1 encodes Ship2 protein, which is known to decrease phosphatidylinositol-3,4,5, trisphosphate levels and whose function may overlap with PTEN (Vinciguerra and Foti, 2006). Innpl1 is also required for signaling by other regeneration genes identified here, such as the HGF receptor Met (Koch et al., 2005). The Ship2 inhibitor AS1949490 (Suwa et al., 2009) dose-dependently increased cortical axon regeneration (Figures 4B and 4C). Therefore, Ship2 is a potential drug target for axonal regeneration therapy. Although the in vivo effect of Ship2 inhibition is unknown, these data suggest that combining genetic screening and drug testing in our in vitro regeneration assay can be used to identify targets and compounds that increase regeneration.
Figure 4. Functional Analysis of Identified Protein Families for Axonal Regeneration.
(A) Quantification of axonal regeneration in shNC and shInppl1 transduced neuron is shown with SEM, n = 128 for shNC and n = 32 for shInppl1 from eight replicates of four shRNA species. ***p < 0.005, Student’s t test.
(B) Representative pictures of regenerated axons 3 days after axotomy with indicated amount of Inppl1 inhibitor. Neurons were stained with βIII tubulin (green) and phalloidin of F-actin (red) to illustrate growth cones. Scale bars represent 200 μm.
(C) The graph shows quantification of axonal regeneration. Neurons were treated with indicated amount of Inppl1 inhibitor right after axotomy for 3 days. Error bars represent SEM, n = 3 biological replicates. **p < 0.01 and ***p < 0.005, one-way ANOVA followed by Tukey’s test.
(D) The graph shows quantification of axonal regeneration after axotomy in shNC and shSocs transduced neurons. Error bars represent SEM, n = 30–116 from 10 replicates of four or five shRNA species. *p < 0.05, **p < 0.01, and ***p < 0.005, one-way ANOVA followed by Dunnett’s test.
(E) Representative confocal images of optic nerve at 17 days after crush injury from shScramble and shSocs4 AAV-injected mice. AAV was injected intraocularly 2 weeks before injury. The CTB-labeled RGC axons are white. The eye is to the left and the brain is to the right. Scale bars represent 500 μm.
(F) Quantification of regenerating axons at 500 μm distances distal to the lesion sites at 17 days after injury. Data are presented as mean with SEM for n = 20 shScramble and n = 13 shSocs4. ****p < 0.001, Student’s t test.
Socs Specificity in Axonal Regeneration
Next, we asked whether regeneration genes uncovered in our screen would have similar effects on axon regeneration in the CNS in vivo. We first analyzed the function of cytokine signaling, as its regeneration function in titrating axonal growth is documented in previous work. In the present dataset, Socs4 had the most prominent effect across the Socs family (Figure 4D), even though published studies have focused on Socs3 (Smith et al., 2009). The results are obtained from three to five different shRNA species for each gene, so knockdown efficiency could account for minor differences in regeneration results among Socs genes. Suppression of Socs2, Socs5, and Socs7 levels also yielded significantly increased cortical axon regeneration. These findings confirm the significance of the gene family and highlight the importance of a member not previously studied with respect to regeneration. We validated the regeneration role of Socs4 in vivo by creating an AAV2/2 vector expressing the Socs4 shRNA species with the most prominent effect and testing by optic nerve regeneration. Virus expressing Socs4 shRNA or non-targeting control was injected intravitreally 2 weeks prior to retro-orbital optic nerve crush. On day 14 after crush, the axonal tracer cholera toxin β (CTB) conjugated to a fluorescent dye was injected intravitreally, and optic nerve anatomy was assessed 3 days later. Few retinal ganglion cell axons regenerate to 500 μm past the crush in control mice, but 4 times greater axon regeneration is detected in the socs4-suppressed optic nerves (Figures 4E and 4F). We conclude that Socs4 contributes to limited axonal regeneration in the adult optic nerve and that our in vitro screen identified genes that modulate CNS regeneration in vivo.
Transport Pathway and GTPase Family Members Limit Axonal Regeneration
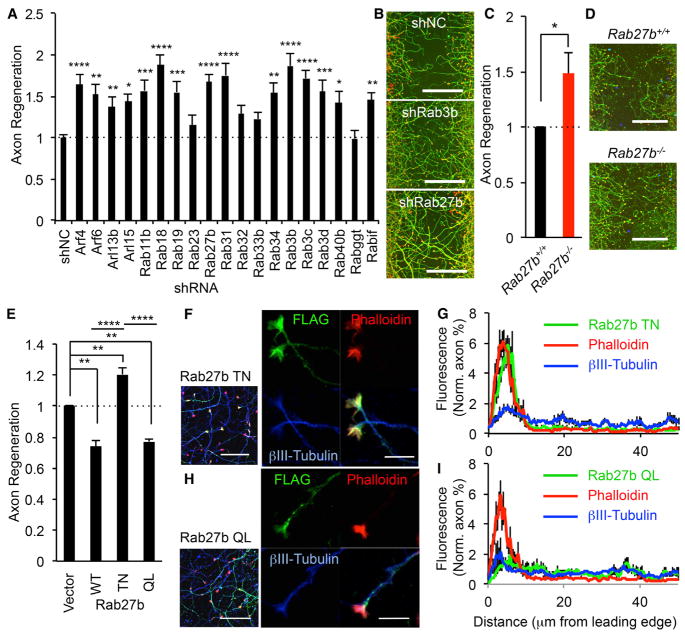
As noted, our network analysis identified intracellular transport as a key process that inhibits regeneration, and within this network were multiple Rab and Rab-related proteins (Figure 3B). Thus, we focused on Rabs and closely related monomeric GTPases involved in organelle traffic. Axon regeneration after suppression of each Rab or members of related Arf (ADP-ribosylation factor) and Arl (Arf-like) families from the primary full screen is shown in Figure S4A. The genes required for Rab prenylation (Rabggta, Rabggtb, Chm, and Chml) are also included. The 19 Rab and related genes for which axonal regeneration was >1.3 times control in the full screen were retested in the axonal regeneration assay (Figures 5A and 5B). The data from primary screening merged with a re-produced shRNA lentivirus study show that 15 Rab and related genes out of 19 genes exhibit statistically significant increased axon regeneration compared with non-targeting shRNA control. The GTPase enzymology allows the creation of point mutants that are constitutively active (CA) or dominant negative (DN), on the basis of oncogenic mutations for related Ras proteins. We generated a DN and CA form for each of six Rab and related proteins and assessed their effect on axonal regeneration. Compared with the DN form, neurons nucleofected with an expression vector for the CA form exhibit significantly suppressed axonal regeneration in Rab3b (p < 0.01), Rab3c (p < 0.005), and Rab27b (p < 0.005) but not Rab18 (p > 0.57), Rab31 (p > 0.9), and Arf4 (p > 0.15) (Figure S4B). Thus, activation of several Rab family members limits cortical axon regeneration.
Figure 5. Transport Pathway and Rab Proteins Limit Axonal Regeneration.
(A) Nineteen Rab-related proteins, with axon regeneration > 1.3 from genome-wide screen, were retested in the axonal regeneration scrape assay. Results for each gene were compared with control using ANOVA with Dunnett’s multiple comparisons test. n = 31–116; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent SEM.
(B) Photomicrographs of regenerating axons in shNC, shRab3b, and shRab27b transduced neurons stained with βIII tubulin (green) and phalloidin of F-actin (red) to illustrate growth cones. Scale bars represent 200 μm.
(C) Quantification of axonal regeneration in Rab27b+/+ and Rab27b−/− neurons is shown with SEM, n = 5 biological replicates. *p < 0.05, Student’s t test.
(D) Microphotographs of axonal regeneration assay in Rab27b+/+ and Rab27b−/− neuron. Scale bars represent 200 μm.
(E) Cortical neurons were nucleofected with vector, Rab27b WT, T23N, or Q78L. Neurons were scraped at DIV 8 and regenerated for 3 days. The graph shows quantification of axonal regeneration. Error bars represent SEM, n = 3 biological replicates. **p < 0.005 and ****p < 0.0001, one-way ANOVA followed by Tukey’s test.
(F–I) Localization of Rab27b TN and QL in regenerating neurons. (F and H) Cortical neurons were nucleofected with FLAG-Rab27b T23N or Q78L. Neurons were scraped at DIV 8 and regenerated for 3 days. Confocal microscope images of FLAG-Rab27b (FLAG; green), axon (βIII-tubulin; blue), and growth cones (rhodamine-phalloidin; red) are taken. Left pictures are 63× objective lens images, and scale bars represent 50 μm. Right pictures are 63× objective lens plus 3× digital zoom images, and scale bars represent 10 μm. (G and I) The graphs show quantification of distribution of Rab27b, βIII-tubulin, and F-actin in regenerating axon 3 days after axotomy. Data are presented as mean ± SE, n = 9.
Rab27b Suppresses Axonal Regeneration In Vitro
In selecting a gene to advance to in vivo studies, we were concerned that manipulating expression of a single Rab3 gene might not show a strong phenotype because of compensation by paralogs because Rab3 has four isoforms, Rab3a, Rab3b, Rab3c, and Rab3d, and it is thought their functions are overlapping and redundant (Schlüter et al., 2004). Rab27 has two isoforms, Rab27a and Rab27b, but the predominant form is Rab27b in cortical neurons (Figure S4C). On the basis of these considerations, we focused subsequent analysis on Rab27b.
In the primary screen, the regenerating Z score was combined three to five shRNA species per gene, and the knockdown efficiency was not verified across the genome. For further validation, we generated two different shRNA constructs in an AAV transfer vector targeting Rab27b and evaluated the reduction of endogenous protein expression levels in shRNA nucleofected neurons. Each of the shRNA constructs shows a drastic reduction of endogenous protein levels compared with control (Figure S4D). We also used these constructs in axonal regeneration assays for further confirmation. Rab27b-knockdown neurons showed significantly enhanced axonal regeneration compared with non-targeting control (Figures S4E and S4F).
Rab27b deletion mice are viable (Tolmachova et al., 2007), so we cultured Rab27b−/− cortical neurons. Consistent with the shRNA data, axonal regeneration from Rab27b−/− mouse neurons is enhanced significantly relative to wild-type (WT) (p < 0.05, Student’s t test) (Figures 5C, 5D, and S4G). Critically, this enhancement is rescued to WT levels by exogenous expression of FLAG-Rab27b WT in Rab27b−/− cortical neurons (Figures S4H and S4I). These data confirm that Rab27b is a suppressor of axonal regeneration after axotomy in vitro.
Localization of Rab27b in Regenerating Neuron
As mentioned above, neurons nucleofected with Rab27b T32N (DN) mutant show enhanced axonal regeneration mimicking the shRNA result (Figures 5E and S4J). In contrast, either Rab27b WT and Q78L (CA) mutant suppress regeneration (Figures 5E and S4J). Subcellular localization of Rab GTPase proteins to specific compartments is crucial to their function. To examine the axonal Rab27b localization, we examined the localization of FLAG-tagged Rab27b T32N and Q78L. Rab27b expressing neurons were axotomized mechanically on DIV 8 and incubated a further 3 days to allow regeneration. The inactive FLAG-Rab27b DN protein is highly enriched in regenerating growth cones and strongly co-localizes with F-actin visualized by rhodamine-conjugated phalloidin but not microtubules in the axon shaft detected by anti-βIII tubulin (Figures 5F and 5G). In marked contrast, the activated Rab27b CA mutant is most prominent in regenerating axon shafts and largely excluded from F-actin-positive growth cone structures (Figures 5H and 5I). Rab27b WT is present at similar intensity in both growth cones and axon shafts of regenerating axons (data not shown). Thus, the regenerating axon contains Rab27b and redistributes the protein on the basis of guanine nucleotide binding and activation state.
Increased Axonal Regeneration in C. elegans Lacking Rab27
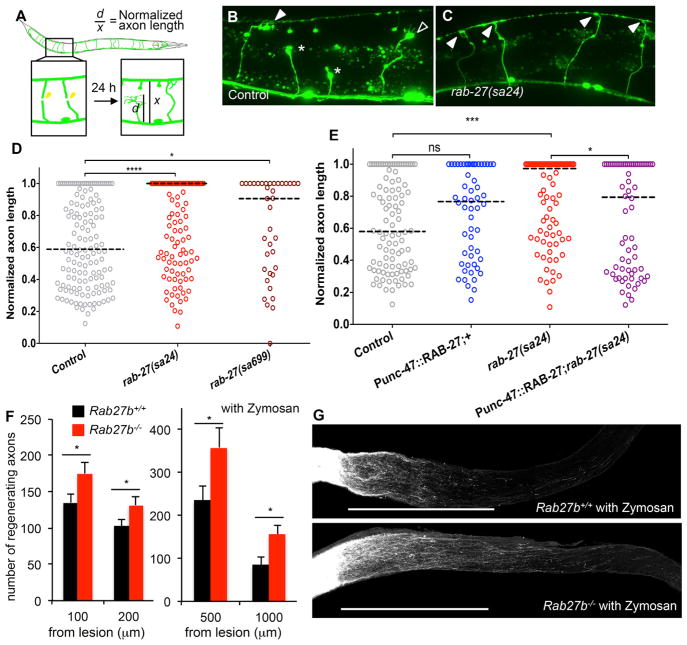
On the basis of the in vitro primary neuron findings, we sought to determine whether Rab27b regulates neural repair in vivo. C. elegans provide a robust system to score single axon regeneration, and we focused on GABAergic axons filled with a GFP reporter protein (Figure 6A). Two different rab-27 hypomorphic alleles were crossed onto the commissural neuron GABAergic reporter line and worms subjected to laser axotomy at the dorsal-ventral midline in young adult animals. No developmental aberration in axonal guidance was detected (not shown). Twenty-four hours after axotomy, the extent of regeneration was measured as the fraction of axon length from the dorsal nerve cord to the ventral nerve cord (Figures 6B–6D). Both rab-27 alleles, sa24 and sa699, significantly increase regeneration, with a majority of axons regenerating fully to the ventral surface. In contrast, the median axon length from control worms after cutting at the length of 0.5 reaches only a length of 0.6. Thus, endogenous rab-27 expression limits axon regeneration.
Figure 6. Rab27 Inhibits Axonal Regeneration In Vivo.
(A) Commissural axons of the GABAergic DD/VD neurons are severed using a pulsed laser, and regeneration is assessed after 24 hr in young adult (L4 stage + 24 hr at 20°C) animals.
(B) Normalized axon length in control and rab-27 mutant animals. Number of axons cut per genotype, left to right: 142, 148, and 37.
(C and D) Regenerating GABA axons 24 hr after axotomy in control (C) and rab-27-null (D) animals. Filled arrows indicate fully regenerated axons reaching the dorsal nerve cord, empty arrows indicate partial regeneration, and stars indicate nonregenerating axon stumps. All animals express Punc-47::GFP, which drives GFP expressing specifically in the GABA motor neurons.
(E) Normalized axon length in control, rab-27 mutants, and animals specifically expressing rab-27 cDNA in GABA neurons, in control and rab-27 mutant animals. Number of axons cut per genotype, left to right: 98, 56, 84, and 68. Kolmogorov-Smirnov test was used. ns, not significant; *p < 0.05, ***p < 0.0005, ****p < 0.0001.
(F) Age-matched (9–10 weeks old without zymosan, 14 weeks old with zymosan) animals underwent optic nerve crush (ONC). Quantification of regenerating RGC axons at indicated distances distal to the lesion sites at 17 days after injury from WT control mouse and Rab27b −/− mice. Data are presented as mean with SEM. Without zymosan, n = 29 Rab27b+/+ and n = 28 Rab27b−/−, and with zymosan, n = 10 Rab27b+/+ and n = 8 Rab27b−/− mice. *p < 0.05, Student’s t test.
(G) Representative confocal images of optic nerve at 17 days after crush injury with zymosan injection from WT control mouse and Rab27b−/− mouse. The CTB-labeled RGC axons are white. The eye is to the left and the brain is to the right. Scale bars represent 500 μm.
The rab-27 regeneration phenotype might be due to autonomous action within the injured GABA neuron or might be secondary to action in other cells. RAB-27 was overexpressed selectively in GABA neurons under the Punc-47 promotor to assess cell autonomy. High levels RAB-27 in GABA neurons generate no significant change in regeneration (Figure 6E). However, this expression significantly rescues the rab27 (sa24) increased regeneration phenotype. We conclude that RAB-27 acts cell-autonomously to restrict axon regeneration in worm GABA neurons.
Optic Nerve Axon Regeneration in Rab27b−/−
In order to evaluate the in vivo function of Rab27b in mammals, we used the optic nerve crush model of axon regeneration. Rab27b−/− mice are viable and fertile without any reported abnormalities (Tolmachova et al., 2007). Strong Rab27b expression is observed in WT retinal tissue but not in Rab27b−/− retina (Figure S5A). We subjected WT and Rab27b−/− mice to optic nerve crush injury and injected the anterograde tracer CTB into the retina 14 days after crush. At 3 days after crush, Rab27b protein levels in retina were similar for uninjured and injured WT mice (Figures S5B and S5C). Animals were sacrificed 3 days after CTB injection and dissected to collect the optic nerves. The total number of CTB-positive axons regenerating beyond the injury site in Rab27b−/− optic nerve is significantly increased compared with WT (Figure 6F). We examined synergy of this phenotype with zymosan-induced inflammation (Figures 6F and 6G). Substantial numbers of regenerating fibers are observed in zymosan-injected optic nerve up to 2,000 μm distal to the injury site. The number of CTB-labeled regenerating axon at 500 or 10,000 μm distal to the injury site in Rab27b−/− optic nerve after zymosan injection is significantly increased compared with WT with zymosan. Thus, Rab27b limits vertebrate axonal regeneration not only in vitro but also in vivo.
Enhanced Behavioral Recovery in Rab27b−/− Mice after T7 Dorsal Hemisection
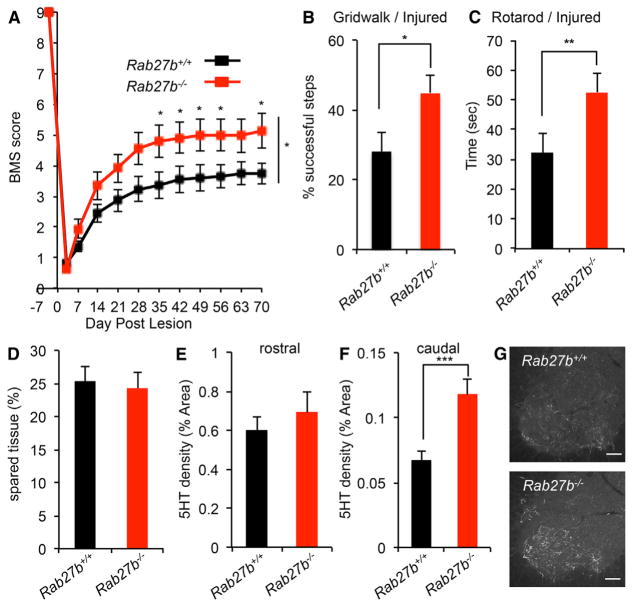
Because suppression of Rab27b expression enhanced neural repair in vitro and in vivo, we sought to determine whether functional recovery from traumatic spinal cord injury might be enhanced by Rab27b deletion. We verified that Rab27b protein is expressed in adult motor cortex (Figure S6A). Furthermore, motor cortex Rab27b protein levels were equal in uninjured and injured WT animals 7 days after spinal cord injury (Figures S6B and S6C). WT and Rab27b−/− mice received dorsal hemisection of the midthoracic spinal cord (n = 19 per genotype). Unfortunately, 2 of 19 Rab27b−/− animals died 1 day after surgery, presumably because of hemorrhagic complications of systemic platelet function (Tolmachova et al., 2007). The Basso Mouse Scale (BMS) score is the most reliable test to monitor locomotion in the open field after dorsal hemisection surgery (Basso et al., 2006). Recovery of hindlimb function is significantly improved in the BMS test of Rab27b−/− animals between 5 and 10 weeks after axotomy (at indicated days, p < 0.05, Student’s t test; between groups, p < 0.05, repeated-measures ANOVA) (Figure 7A). The same cohorts were subjected to additional functional outcomes. In the gridwalk test, uninjured mice made similar numbers of missed steps as did WT mice (p > 0.13), consistent with normal CNS development (Figure S6D). After injury and consistent with the BMS scores, Rab27b−/− mice group show a reduced rate of missed steps on the grid at 55 days post-lesion (dpl) compared with the WT group (p < 0.05, Student’s t test) (Figure 7B). The Rab27b−/− mice at 48 dpl are able to stay on the rotating rotarod drum longer than WT animals (p < 0.05, Student’s t test) (Figure 7C), although performance is equal before injury (p > 0.29) (Figure S6E). The behavioral improvement in Rab27b−/− mice are not due to differences in the degree of injury or in tissue sparing, because intact tissue was identical in the two groups by histological assessment with anti-GFAP staining at the end of the experiment (p > 0.74, Student’s t test) (Figure 7D).
Figure 7. Improvement of Functional Recovery after Spinal Cord Injury in Rab27b−/− Mouse.
(A) Open-field locomotion performance measured by BMS of Rab27b+/+ and Rab27b−/− mice. Animals were scored on day post-lesion (DPL) −3, 3, 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70 by two experienced observers blinded to group. Data are mean ± SEM for n = 19 Rab27b+/+ and n = 17 Rab27b−/−. *p < 0.05, significant difference between genotypes through DPL 35 to 70, one-way repeated-measure ANOVA across time series followed by Student’s t test between genotypes at indicated times.
(B and C) Gridwalk test at DPL 55 (B) and RotaRod performance at DPL 48 (C) of Rab27b+/+ and Rab27b−/− mice. Data are mean with SEM for n = 19 Rab27b+/+ and n = 17 Rab27b−/−. *p < 0.05 and **p < 0.01, Student’s t test.
(D) Sagittal sections of thoracic cord were stained with anti-GFAP antibody, and the extent of spared tissue at the injury site was quantified. Data are presented as mean with SEM for n = 19 Rab27b+/+ and n = 17 Rab27b−/−. No significant differences between groups with Student’s t test.
(E and F) Serotonergic (5HT+) fiber density at coronal sections of rostral to the lesion (E) and caudal to the lesion (F) from Rab27b+/+ and Rab27b−/− mice 70 days after hemisection were quantified. Data are presented as mean with SEM for n = 19 Rab27b+/+ and n = 17 Rab27b−/−. No significant differences between groups with Student’s t test (E). *p < 0.05, Student’s t test (F).
(G) Representative image of raphespinal fibers stained with anti-5HT antibody in the spinal ventral horn.
The raphespinal serotonergic (5HT) axonal tract possesses a known ability for injury-induced axonal growth and contributes substantially to locomotion and is significantly lesioned by the dorsal hemisection trauma (Kim et al., 2004). Because of the in vitro regenerative efficacy and the improved behavioral performance after deletion of Rab27b, we assessed 5HT staining for axonal growth after injury. The density of proximal 5HT-positive fibers in the ventral horn rostral to the lesion site is similar between groups on day 70 after dorsal hemisection injury (Figure 7E). Caudal to the lesion site, the density of ventral horn distal 5HT-positive fibers is twice as great in the Rab27b−/− group compared with WT (p < 0.005, Student’s t test) (Figures 7F and 7G). This phenotype is not secondary to development changes, because the 5HT fibers in the ventral horn of either cervical or lumbar cord in uninjured WT and Rab27b−/− mouse is indistinguishable (Figures S6F and S6G).
We also examined the projection of corticospinal axons in Rab27b−/− after spinal cord injury. Biotin-dextran amine (BDA) anterograde tracing from injections in the motor cortex was conducted 8 weeks after spinal cord injury. BDA-labeled axons were visualized in fixed tissue collected 2 weeks after tracer injection using streptavidin Alexa Fluor 568. Equivalent numbers of BDA-labeled CST axons were detected rostral to the injury site in both groups, but no regenerating axons reached the caudal spinal cord in either genotype (Figures S6H and S6I). Immediately rostral to the injury epicenter, significantly greater numbers of CST axons were observed Rab27b−/− mice compared with WT mice (Figures S6H–S6J). The increased CST axon density in this region for spinal cord injury mice lacking Rab27b may be due either to reduced dieback from the axotomy site or to short range regeneration after dieback from spinal cord injury. Taken together, the improved functional recovery and greater descending axonal length of serotonin and CST fibers demonstrate that deletion of Rab27b is beneficial for neural repair after spinal cord injury.
DISCUSSION
In the present study, we screened the mouse genome for factors with a role in restricting axonal regeneration by suppression of expression. Importantly, our unbiased screen was based on functional analyses of regeneration after gene knockdown: we assess the ability of cultured cortical neurons to regrow after injury. We found approximately 500 genes that show a regeneration phenotype, and validation studies on more than 120 genes confirm reproducible effect axon regeneration in our in vitro system. Among these genes, most have not previously been linked to axonal regeneration or neural repair. Transport, receptor binding and cytokine signaling are enriched pathways. Most highly enriched was the membrane trafficking Rab GTPase family, and Rab27b was studied in detail. The inactive Rab27b protein is localized to regenerating growth cones and inhibition of regeneration requires the active GTP conformation. Adult worms and mice lacking this protein exhibit greater axonal regeneration. Moreover, mice null for Rab27b recover greater motor function after spinal cord trauma. The many other genes and cellular pathways identified in our screen await in vivo study, but for one of them (Inppl1), we found that a small-molecule inhibitor was able to replicate the in vitro regeneration effect.
Despite the limited data regarding a role for Rabs themselves in axonal regeneration prior to this work, there is pre-existing evidence that membrane traffic plays a key role in axonal extension. Rab 11 has been implicated in regulating the traffic of inhibitory proteins from axons to dendrites (Koseki et al., 2017). There is a link between endoplasmic reticulum (ER) and endosome contact in mediating axonal extension (Raiborg et al., 2015). Semaphorins, as extracellular cues inhibiting extension and collapsing growth cones, stimulate local and massive macropinocytosis at the growth cone (Fournier et al., 2000). Inpp5f regulates both axon regeneration and membrane traffic (Nakatsu et al., 2015; Zou et al., 2015). In C. elegans, loss of function in any of three endocytosis genes (unc-26/synaptojanin, unc-57/endophilin, and unc-41/stonin) results in decreased regeneration (Chen et al., 2011). Multiple studies have demonstrated that new membrane is added to the distal axon tip during growth, and the growth cone is known to be highly enriched in endomembranous stacks (Cheng and Reese, 1987; Diefenbach et al., 1999; Hazuka et al., 1999; Kolpak et al., 2009; Lockerbie et al., 1991; Tojima et al., 2007). Dendritic branching in Drosophila is intimately connected with Golgi outposts (Ye et al., 2007). Thus, Rab regulation of distal membrane traffic may be crucial for effective regeneration via regulation of membrane addition, a hypothesis that we favor. An alternative hypothesis stems from the role of retrograde transport to the cell soma for signaling from distal extracellular cues (Cosker and Segal, 2014). For both synaptic vesicle and non-synaptic vesicle Rabs, gene suppression is hypothesized to allow a net diversion of membrane delivery to axonal extension. This hypothesis explains the observation that suppression of multiple different intracellular trafficking events supports greater axonal extension. In this light, it is important to note that the surface area of the axon membrane of mature mammalian projection neuron of the corticospinal tract may be 300 times that of the cell soma, so that regeneration requires very substantial plasma membrane delivery.
Multiple Rab-family proteins have regeneration phenotypes when expression is suppressed. This includes Rab3b and Rab3c proteins, which are known to share synaptic vesicle regulation with Rab27b. As an effector, Rabphilin3 has been linked to Rab3s as well as Rab27b, and suppression of its expression phenocopies Rab27b loss of function both in vitro and in vivo. In addition, Arf4, Rab18, and Rabif were validated by repeat testing among 25 Rab-related genes identified as hits in the original screen. We expect that Rab27b and other regenerating-controlling Rabs are likely to play a role in modifying membrane delivery and retrieval to the cell surface in the distal axons. Although Rab27b and the Rab3s have been implicated in synaptic vesicle exocytosis in the distal axon (Fukuda, 2008; Pavlos et al., 2010), a loss of these Rabs may shift membrane traffic from synaptic function to permit greater plasma membrane addition for axon extension. Rab27b has also been associated with melanosome traffic, platelet degranulation and exosome release (Chen et al., 1997; Fukuda, 2008; Mizuno et al., 2007; Tolmachova et al., 2007). For both synaptic vesicle and non-synaptic vesicle Rabs, gene suppression is hypothesized to allow a net diversion of membrane delivery to axonal extension. This hypothesis explains the observation that suppression of multiple different intracellular trafficking events supports greater axonal extension. These findings highlight the critical role of membrane traffic for successful axonal extension. In this light, it is important to note that the surface area of the axon membrane of mature mammalian projection neuron of the corticospinal tract may be 300 times that of the cell soma, so that regeneration requires very substantial plasma membrane delivery.
Although Rab proteins and intracellular membrane traffic were highlighted bioinformatically as most enriched gene set among regeneration genes, many non-Rab-related genes were identified as limiting axonal regeneration. These do not constitute a single pathway but cover a range of pathways, some of which have been connected with axonal regeneration and many of which have not previously been identified as participating in axonal regeneration. Of the top hits revalidated by rescreening, xylt1, encoding xylosyl transferase, is central for chondroitin sulfate synthesis (Baker et al., 1972), so its role may fit with well-documented role of CSPG to inhibit regeneration. Hif3a encodes an inactive subunit that titrates Hif1 and Hif2 signaling in protective responses to hypoxic stress. The ability of Hif3a suppression to increase regeneration is consistent with HIF1 signaling in C. elegans axon regeneration (Alam et al., 2016). Parp1 was previously reported to have a role in C. elegans and mouse regeneration (Byrne et al., 2016), but in vivo evaluation of its role as a target for neural repair in mammals were disappointing (Wang et al., 2016).
We focused on those genes whose suppression increased regeneration. By our screening criteria, about 100 genes reduced axon regeneration when expression was suppressed. It is possible that these genes are required for endogenous regenerative potential. However, we have not excluded cell toxicity as a cause for the reduced number of βIII-tubulin regenerating axons in these cases. Thus, this group may contain both essential regeneration genes and genes required for cell survival non-specifically. Further studies will investigate these possibilities.
Importantly, our screen was based on functional analyses with loss of function. In contrast, the most common approach to identifying genes involved in regeneration has focused on expression surveys, most commonly at the mRNA level (Belin et al., 2015; Bonilla et al., 2002; Chandran et al., 2016; Fink et al., 2017; Tanabe et al., 2003; Tedeschi et al., 2016). Such previous work has the premise that genes involved in regulating regeneration are controlled transcriptionally by injury. Although this can be the case, there is no a priori basis for this assumption, and especially for those genes limiting regeneration, their physiological function and regulation may relate to alternate cellular functions, which must be suppressed for successful axon regrowth. As hypothesized above, this may the case with Rab proteins.
The approach described here examined one gene at a time for effects on functional axon regeneration. However, it is highly likely the combinations of different genes may have far greater effects in many cases, and examples of successful combinations have been reported (Bei et al., 2016; Benowitz et al., 2017; Sun et al., 2011; Wang et al., 2012; Zai et al., 2011). The screen identifies genes with unrelated cellular functions, thereby predicting that additive effects on regeneration may exist. For genes related to a single pathway, the consequence of dual suppression is not obvious. To the extent that the factors in the same pathway are redundant, dual inhibition is expected to be synergistic, while to the extent that they are epistatic, one will occlude that other’s effect.
Both the comprehensive screening results and the specific data for Rab trafficking events provide new directions for research and therapy based on axonal regeneration and neural plasticity after injury. Because a number of genes not previously associated with neural repair have been nominated by the loss of function screen, methods for rapid in vivo evaluation is essential. In this regard, species conservation allows implementation of secondary studies in tractable genetic organisms, and CNS regional conservation permits evaluation of genes relevant for spinal cord injury in more accessible injury models, such as optic nerve regeneration. The present study broadens the horizons for successful neural repair and neurological recovery after trauma.
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures. All animal studies were conducted with approval of the Yale Institutional Animal Care and Use Committee. All behavioral measurements and all imaging quantifications were conducted by experimenters unaware of experimental group. No data were excluded from the analysis. Both male and female mice were included, as mice were collected from sequential littermates of the appropriate genotypes in tissue culture experiments. Spinal cord injury studies were performed only with female mice to facilitate bladder management. Both male and female mice were used of optic nerve crush studies. The age of mice is specified in each figure legend, and for CNS injury was introduced at 10 weeks.
Primary Cortical Neuron Culture and Axon Regeneration Assay
Primary cortical neuron axon regeneration assay was performed as described previously (Huebner et al., 2011). For the shRNA-based regeneration screen, lentiviral particles targeting 16,007 mouse genes with 83,106 unique shRNA clones (Mission TM TRC Mouse Lentiviral shRNA Library 10180801; Sigma-Aldrich) were added to the neurons on DIV 3. On DIV 8, 96-well cultures were scraped and fixed on DIV 10.
C. elegans Laser Axotomy Studies
Laser axotomy was performed on late L4 C. elegans larvae as previously described (Byrne et al., 2011).
Mice and Surgery
Age-matched adult (10 weeks) C57BL/6 WT female mice or Rab27b−/− mice (Tolmachova et al., 2007) were subjected to dorsal hemisection as described previously (Zou et al., 2015). For optic nerve crush injury study, both male and female C57BL/6J mice or Rab27b−/− mice were used. AAV serotype 2/2 was produced and purified >1 × 1012 genome copies per milliliter and then injected intraorbitally to WT animals 2 weeks prior to crush surgery. The optic nerve was exposed intraorbitally with care taken to avoid damage to the ophthalmic artery. Alexa 555-CTB was injected intravitreally to trace axons 14 days after injury.
Quantification and Statistical Analyses
One-way ANOVA with post hoc Tukey pairwise comparisons, repeated-measures ANOVA, and Student’s t test as specified in the figure legends were performed using GraphPad Prism version 5.0d and SPSS Statistics version 22. Mean ± SEM and specific n values are reported in each figure legend. Data are considered to be statistically significant if p < 0.05. The assumption of Gaussian distribution was checked using the D’Agostino-Pearson omnibus test.
Supplementary Material
Highlights.
Comprehensive loss-of-function screen for mouse CNS axonal regeneration
Genes identified by functional screen are distinct from expression profiling
Transport, receptor binding, and cytokine signaling pathways limit regeneration
Rab27b loss increases axonal regeneration in worm and mouse optic nerve
Acknowledgments
We thank Stefano Sodi, Tomoko Sekine, Kristin DeLuca, and Yram Foli for expert technical assistance. This work was supported by grants from the Falk Medical Research Trust to S.M.S. and from the NIH (R35NS097283 and R01NS098817) to M.H. and S.M.S.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.058.
AUTHOR CONTRIBUTIONS
Y.S., A.L.-M., M.H., and S.M.S. designed most experiments. Y.S. and A.L.-M. collected most of the data. D.M.C., X.W., and Z.J. conducted optic nerve crush experiments. Y.S., A.L.-M., W.B.C., M.H., and S.M.S. analyzed results and wrote the manuscript.
DECLARATION OF INTERESTS
S.M.S. is a co-founder and consultant for ReNetX Bio, which seeks to develop NgR1-based therapies for neural repair.
References
- Alam T, Maruyama H, Li C, Pastuhov SI, Nix P, Bastiani M, Hisamoto N, Matsumoto K. Axotomy-induced HIF-serotonin signalling axis promotes axon regeneration in C. elegans. Nat Commun. 2016;7:10388. doi: 10.1038/ncomms10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JR, Rodén L, Stoolmiller AC. Biosynthesis of chondroitin sulfate proteoglycan. Xylosyl transfer to Smith-degraded cartilage proteoglycan and other exogenous acceptors. J Biol Chem. 1972;247:3838–3847. [PubMed] [Google Scholar]
- Basso DM, Fisher LC, Anderson AJ, Jakeman LB, McTigue DM, Popovich PG. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- Bei F, Lee HHC, Liu X, Gunner G, Jin H, Ma L, Wang C, Hou L, Hensch TK, Frank E, et al. Restoration of visual function by enhancing conduction in regenerated axons. Cell. 2016;164:219–232. doi: 10.1016/j.cell.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin S, Nawabi H, Wang C, Tang S, Latremoliere A, Warren P, Schorle H, Uncu C, Woolf CJ, He Z, Steen JA. Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron. 2015;86:1000–1014. doi: 10.1016/j.neuron.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz LI, He Z, Goldberg JL. Reaching the brain: advances in optic nerve regeneration. Exp Neurol. 2017;287:365–373. doi: 10.1016/j.expneurol.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Blackmore MG, Moore DL, Smith RP, Goldberg JL, Bixby JL, Lemmon VP. High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44:43–54. doi: 10.1016/j.mcn.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchser WJ, Slepak TI, Gutierrez-Arenas O, Bixby JL, Lemmon VP. Kinase/phosphatase overexpression reveals pathways regulating hippocampal neuron morphology. Mol Syst Biol. 2010;6:391. doi: 10.1038/msb.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Edwards TJ, Hammarlund M. In vivo laser axotomy in C. elegans. J Vis Exp. 2011;(22):2707. doi: 10.3791/2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, McWhirter RD, Sekine Y, Strittmatter SM, Miller DM, Hammarlund M. Inhibiting poly(ADP-ribosylation) improves axon regeneration. eLife. 2016;5:5. doi: 10.7554/eLife.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, et al. A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron. 2016;89:956–970. doi: 10.1016/j.neuron.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Guo J, Miki T, Tachibana M, Gahl WA. Molecular cloning and characterization of rab27a and rab27b, novel human rab proteins shared by melanocytes and platelets. Biochem Mol Med. 1997;60:27–37. doi: 10.1006/bmme.1996.2559. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O’Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TP, Reese TS. Recycling of plasmalemma in chick tectal growth cones. J Neurosci. 1987;7:1752–1759. doi: 10.1523/JNEUROSCI.07-06-01752.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Segal RA. Neuronal signaling through endocytosis. Cold Spring Harb Perspect Biol. 2014;6:6. doi: 10.1101/cshperspect.a020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach TJ, Guthrie PB, Stier H, Billups B, Kater SB. Membrane recycling in the neuronal growth cone revealed by FM1-43 labeling. J Neurosci. 1999;19:9436–9444. doi: 10.1523/JNEUROSCI.19-21-09436.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bejjani R, Hammarlund M. Neural regeneration in Caenorhabditis elegans. Annu Rev Genet. 2012;46:499–513. doi: 10.1146/annurev-genet-110711-155550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink KL, López-Giráldez F, Kim IJ, Strittmatter SM, Cafferty WBJ. Identification of intrinsic axon growth modulators for intact CNS neurons after injury. Cell Rep. 2017;18:2687–2701. doi: 10.1016/j.celrep.2017.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, Nakamura F, Kawamoto S, Goshima Y, Kalb RG, Strittmatter SM. Semaphorin3A enhances endocytosis at sites of receptor-F-actin colocalization during growth cone collapse. J Cell Biol. 2000;149:411–422. doi: 10.1083/jcb.149.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol Life Sci. 2008;65:2801–2813. doi: 10.1007/s00018-008-8351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuka CD, Foletti DL, Hsu SC, Kee Y, Hopf FW, Scheller RH. The sec6/8 complex is located at neurite outgrowth and axonal synapse-assembly domains. J Neurosci. 1999;19:1324–1334. doi: 10.1523/JNEUROSCI.19-04-01324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner EA, Kim BG, Duffy PJ, Brown RH, Strittmatter SM. A multi-domain fragment of Nogo-A protein is a potent inhibitor of cortical axon regeneration via Nogo receptor 1. J Biol Chem. 2011;286:18026–18036. doi: 10.1074/jbc.M110.208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Liu BP, Park JH, Strittmatter SM. Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron. 2004;44:439–451. doi: 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Koch A, Mancini A, El Bounkari O, Tamura T. The SH2-domian-containing inositol 5-phosphatase (SHIP)-2 binds to c-Met directly via tyrosine residue 1356 and involves hepatocyte growth factor (HGF)-induced lamellipodium formation, cell scattering and cell spreading. Oncogene. 2005;24:3436–3447. doi: 10.1038/sj.onc.1208558. [DOI] [PubMed] [Google Scholar]
- Kolpak AL, Jiang J, Guo D, Standley C, Bellve K, Fogarty K, Bao ZZ. Negative guidance factor-induced macropinocytosis in the growth cone plays a critical role in repulsive axon turning. J Neurosci. 2009;29:10488–10498. doi: 10.1523/JNEUROSCI.2355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki H, Donegá M, Lam BY, Petrova V, van Erp S, Yeo GS, Kwok JC, Ffrench-Constant C, Eva R, Fawcett JW. Selective rab11 transport and the intrinsic regenerative ability of CNS axons. eLife. 2017;6:6. doi: 10.7554/eLife.26956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockerbie RO, Miller VE, Pfenninger KH. Regulated plasmalemmal expansion in nerve growth cones. J Cell Biol. 1991;112:1215–1227. doi: 10.1083/jcb.112.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh SH, Francescut L, Lingor P, Bähr M, Nicotera P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008;15:283–298. doi: 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Tolmachova T, Ushakov DS, Romao M, Abrink M, Ferenczi MA, Raposo G, Seabra MC. Rab27b regulates mast cell granule dynamics and secretion. Traffic. 2007;8:883–892. doi: 10.1111/j.1600-0854.2007.00571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Messa M, Nández R, Czapla H, Zou Y, Strittmatter SM, De Camilli P. Sac2/INPP5F is an inositol 4-phosphatase that functions in the endocytic pathway. J Cell Biol. 2015;209:85–95. doi: 10.1083/jcb.201409064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, Bastiani M. Axon regeneration genes identified by RNAi screening in C. elegans. J Neurosci. 2014;34:629–645. doi: 10.1523/JNEUROSCI.3859-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlos NJ, Grønborg M, Riedel D, Chua JJ, Boyken J, Kloepper TH, Urlaub H, Rizzoli SO, Jahn R. Quantitative analysis of synaptic vesicle Rabs uncovers distinct yet overlapping roles for Rab3a and Rab27b in Ca2+-triggered exocytosis. J Neurosci. 2010;30:13441–13453. doi: 10.1523/JNEUROSCI.0907-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cafferty WB, McMahon SB, Thompson SW. Conditioning injury-induced spinal axon regeneration requires signal transducer and activator of transcription 3 activation. J Neurosci. 2005;25:1645–1653. doi: 10.1523/JNEUROSCI.3269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Schmitz F, Jahn R, Rosenmund C, Südhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Allchorne AJ, Woolf CJ. The transcription factor ATF-3 promotes neurite outgrowth. Mol Cell Neurosci. 2006;32:143–154. doi: 10.1016/j.mcn.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, Perrimon N. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008;4:e1000111. doi: 10.1371/journal.pgen.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He Z. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa A, Yamamoto T, Sawada A, Minoura K, Hosogai N, Tahara A, Kurama T, Shimokawa T, Aramori I. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br J Pharmacol. 2009;158:879–887. doi: 10.1111/j.1476-5381.2009.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Dupraz S, Laskowski CJ, Xue J, Ulas T, Beyer M, Schultze JL, Bradke F. The calcium channel subunit Alpha2delta2 suppresses axon regeneration in the adult CNS. Neuron. 2016;92:419–434. doi: 10.1016/j.neuron.2016.09.026. [DOI] [PubMed] [Google Scholar]
- Tojima T, Akiyama H, Itofusa R, Li Y, Katayama H, Miyawaki A, Kamiguchi H. Attractive axon guidance involves asymmetric membrane transport and exocytosis in the growth cone. Nat Neurosci. 2007;10:58–66. doi: 10.1038/nn1814. [DOI] [PubMed] [Google Scholar]
- Tolmachova T, Abrink M, Futter CE, Authi KS, Seabra MC. Rab27b regulates number and secretion of platelet dense granules. Proc Natl Acad Sci U S A. 2007;104:5872–5877. doi: 10.1073/pnas.0609879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M, Foti M. PTEN and SHIP2 phosphoinositide phosphatases as negative regulators of insulin signalling. Arch Physiol Biochem. 2006;112:89–104. doi: 10.1080/13813450600711359. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jin Y. Genetic dissection of axon regeneration. Curr Opin Neurobiol. 2011;21:189–196. doi: 10.1016/j.conb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hasan O, Arzeno A, Benowitz LI, Cafferty WB, Strittmatter SM. Axonal regeneration induced by blockade of glial inhibitors coupled with activation of intrinsic neuronal growth pathways. Exp Neurol. 2012;237:55–69. doi: 10.1016/j.expneurol.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sekine Y, Byrne AB, Cafferty WB, Hammarlund M, Strittmatter SM. Inhibition of poly-ADP-ribosylation fails to increase axonal regeneration or improve functional recovery after adult mammalian CNS injury. eNeuro. 2016;3:3. doi: 10.1523/ENEURO.0270-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai L, Ferrari C, Dice C, Subbaiah S, Havton LA, Coppola G, Geschwind D, Irwin N, Huebner E, Strittmatter SM, Benowitz LI. Inosine augments the effects of a Nogo receptor blocker and of environmental enrichment to restore skilled forelimb use after stroke. J Neurosci. 2011;31:5977–5988. doi: 10.1523/JNEUROSCI.4498-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Stagi M, Wang X, Yigitkanli K, Siegel CS, Nakatsu F, Cafferty WB, Strittmatter SM. Gene-silencing screen for mammalian axon regeneration identifies Inpp5f (Sac2) as an endogenous suppressor of repair after spinal cord injury. J Neurosci. 2015;35:10429–10439. doi: 10.1523/JNEUROSCI.1718-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.