Abstract
INTRODUCTION
Genetic associations for endophenotypes of Alzheimer’s disease (AD) in cognitive stages preceding Alzheimer disease have not been thoroughly evaluated.
METHODS
We conducted GWAS for AD-related endophenotypes including hippocampal volume (HPV), logical memory scores (LMT), and cerebrospinal fluid (CSF) amyloid β42 and total/phosphorylated tau (t-/p-Tau) in cognitively normal (CN), mild cognitive impairment (MCI), and AD dementia subjects from the Alzheimer’s Disease Neuroimaging Initiative study.
RESULTS
In CN subjects, study-wide significant (SWS; P<8.3×10−9) loci were identified for t-Tau near SRRM4 and C14orf79 and for HPV near MTUS1. In MCI subjects, SWS association was found with SNPs near ZNF804B for LMT-delayed recall. We found consistent expression patterns of C14orf40 and MTUS1 in carriers with risk alleles of eSNPs and in brains of AD patients, compared with in the non-carriers and in brains of controls.
DISCUSSION
Our findings for AD-related brain changes prior to AD provide insight about early AD-related biological processes.
Keywords: Alzheimer’s disease, Genome-wide association, endophenotypes, cerebrospinal fluid, MRI, logical memory, biomarker, ADNI, tau, co-expression network
1. Introduction
Alzheimer’s disease (AD) is the most common type of dementia and typically occurs after age 65 years. It is highly heritable, but the known genetic risk factors (currently numbering more than 25 loci including APOE) account for no more than 50% of the heritability of the disorder [1]. However, genetic association findings based on AD risk do not explain the whole genetic architecture of AD because the mechanistic complexity underlying AD is not captured entirely by disease status, especially in preclinical stages [2, 3]. To overcome this limitation and understand preclinical stages of AD, researchers have examined the genetic underpinnings of AD-related endophenotypes including cerebrospinal fluid (CSF) levels of beta amyloid peptide (Aβ42) and tau proteins, structural brain changes quantified by magnetic resonance imaging (MRI), and neuropsychological test measures of cognitive functioning, including memory loss [4,5]. Genome-wide association (GWA) studies for AD-related endophenotypes have identified novel loci in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study which enrolled appreciable numbers of subjects across three stages: AD dementia, mild cognitive impairment (MCI), and normal cognitive functioning (CN) [6]. Previous ADNI studies indicated the importance of delineating different stages of subjects [7, 8]. We hypothesized that some genes may contribute to AD-related processes specifically during stages prior to AD dementia onset. Genes and pathways that are strongly associated with AD-related endophenotypes in early disease stages may be promising targets for developing AD biomarkers and preventive medicines. To test this hypothesis, we conducted GWA analyses for AD-related endophenotypes in the ADNI sample stratified by stage. Here, we focused on the association tests in the CN and MCI subgroups because we were interested in identifying genes that may contribute to AD-related processes prior to AD dementia onset.
2. Subjects and Methods
2.1. Subjects
GWA and phenotype data for ADNI participants were downloaded from a public-access database (http://www.loni.usc.edu). A total of 1,189 subjects before QC were available with GWA data from two different chips (ADNI-1, n=757 and ADNI-GO/2, n=432). We stratified subjects by stage (CN, MCI, and AD dementia) based on diagnosis at the baseline assessment as defined by the standard ADNI protocol. Demographic information and mean endophenotype values stratified by stage as well as for the entire sample are presented in Supplementary Table 1. Age is similarly distributed in each subgroup. Sample sizes for analyses of CSF biomarkers were considerably smaller than for those of other traits.
2.2. Phenotypic Evaluation
Previously suggested AD-related endophenotypes including CSF biomarkers [9], MRI brain imaging measures [10] and episodic memory tests [11] were selected for GWA analyses in this study. CSF biomarkers of Aβ42, total tau (t-Tau), and phosphorylated tau (p-Tau), brain MRI measure for hippocampal volume (HPV), and scores for logical memory immediate (LMiT) and delayed (LMdT) recall tests which were all measured at baseline were analyzed in this study. Details about collection of CSF biomarkers, brain MRI scan data, and neuropsychological tests are reported elsewhere [12-15].
2.3. Genotyping, Quality Control, Imputation, and Population Substructure Analysis
Details of quality control, genotype imputation, and population substructure analyses are described in Supplementary Information. After QC, the ADNI-1 sample with genotype data consisted of 187 CN, 329 MCI, and 163 AD dementia subjects, and the ADNI-GO/2 sample contained 118 CN, 252 MCI, and 27 AD dementia subjects with genotype data.
2.4. Statistical Methods
2.4.1. Genome-wide association tests
Prior to the association tests, each of the six endophenotypes was adjusted for covariates using linear regression. Age and sex were used as covariates for the six endophenotypes. A term for education level was also included in the regression models for LMiT and LMdT, and the model for HPV was further adjusted for total intracranial volume. The residuals derived from the regression models were rank-transformed for normalization as previously described [16]. Analyses were conducted for all autosomal SNPs using the expected genotype dose, a quantitative measure between 0 and 2 of the number of effect alleles computed from the imputed genotype probabilities as the predictor. Association of the rank-normalized endophenotypes with each SNP was evaluated using a linear regression model including covariate terms for the first three principal components (PC) of population substructure using the R software package. The two ADNI datasets were analyzed independently for the CN and MCI subjects and the results from the two ADNI datasets were combined by meta-analysis using inverse variance weights as implemented in the METAL program [17]. AD cases from the two ADNI datasets were analyzed as one group because the ADNI-GO/2 sample included only 27 AD subjects, and regression models for this group included an extra covariate for ADNI dataset. The genome-wide significant (GWS) threshold was set at 5.0×10−8. We determined a conservative study-wide significant (SWS) level of 8.3×10−9 which was calculated as the GWS level divided by the effective number of two independent endophenotypes and three clinical subgroups. The effective number of independent endophenotypes was computed by counting the number of eigenvalues greater than one from the PC analysis of all six endophenotypes. A threshold of P<10−6 was considered as suggestive evidence of association in the functional/pathway analysis.
2.4.2. Expression SNP Analysis
We examined association of the SWS SNPs (allele counts) with transcript-level expression, i.e., expression SNP (eSNP), using a publically available database via the GTEx Portal (http://www.gtexportal.org; [18]).
2.4.3. Differential Gene Expression Analysis
Differential gene expression (DGE) was evaluated for genes containing or near significantly associated SNPs in two independent human brain expression datasets from the Eisai Bio Bank (EBB) and Mt. Sinai Hospital (MSH) (which was downloaded from Gene Expression Omnibus [GEO]: GSE44772). The EBB has gene expression measures obtained from RNA sequencing from the hippocampus (HIPP) of samples collected from autopsied brains from 35 AD cases and 16 normal subjects ascertained at the University of Miami and McLean Hospital (Belmont, MA). A measure of neurofibrillary tangles (Braak stage) in the same samples was assessed following an established protocol [19]. Details about sample collection and preparation, and demographic characteristics are provided in Supplementary Information. The association of log2-transformed transcript expression levels (outcome) with AD status (predictor) was evaluated using linear regression adjusting for site, age, sex, and RNA integrity number (RIN). A model testing the effect of Braak stage (0-6 stages) on transcript expression levels was also evaluated and included the same covariates. The MSH microarray expression data (GEO: GSE44772) were generated from autopsied brain tissue collected from dorsolateral prefontal cortex (DLPFC), visual cortex (VCX), and cerebellum (CER) regions in 129 AD patients and 101 controls. Samples were profiled on a custom-made Agilent 44K array containing 40,638 probes. Gene expression data were normalized using Rosetta Resolver gene-expression analysis software as previously described [20]. The association of log2-transformed gene expression level (outcome) and AD status (predictor) was tested using a linear regression model adjusting for RIN, postmortem interval (PMI), batch, preservation method, tissue pH, age, and sex. A significance threshold to correct for the number of eQTL analysis tests was applied which in this case was P=0.025 since only two genes (C14orf40 and MTUS1) were tested.
2.4.4. Co-expression Network Analysis of Human Brain RNA-Sequence Data
The top-ranked genes at or near (< 50kb) loci that achieved suggestive significance (P<10−6) in GWA tests of any trait in CN or MCI subjects or in the total sample were further evaluated for gene co-expression networks. We built gene co-expression networks in the EBB HIPP RNA-Seq data (41,249 transcripts) by weighted gene co-expression network analysis (WGCNA) [21], an approach which defines modules (or subsets) of genes that are highly co-expressed (or co-regulated). Details of this approach are described in Supplementary Information. In the HIPP co-expression network, we selected modules which carry both the top-ranked genes from this study and previously known AD genes [22-25]. The selected modules were functionally annotated by two enrichment analyses of ‘gene ontology (GO)’ and ‘disease-associated genes’ with a hypergeometric test. The ‘disease-associated genes’, genes involved in risk of diseases, were downloaded from GWASdb2 [26]. A P-value of significance for each enrichment test was calculated along with a false discovery rate (FDR), estimated using the Benjamini-Hochberg procedure [27]. We used an FDR threshold of 0.05 to define associated GOs or diseases. The selected modules were further examined for correlations with traits (AD status and Braak stages) using the EBB sample by calculating the Pearson’s correlations between the module eigengene and the traits.
2.4.5. Functional Analysis and Brain Cell Type-Specific Expression Profiling
We evaluated predicted functions of SNPs showing suggestive evidence for association (P<10−6) from the GWA tests using HaploReg (http://www.broadinstitute.org/mammals/haploreg/) [28]. ENCODE data [29] were used to evaluate potential regulatory function. We also investigated the expression profiles of the top-ranked gene from mouse and human brain from the cerebral cortex (http://web.stanford.edu/group/barres_lab/brainseqMariko/brainseq2.html) [30]. To identify shared functions among the top-ranked genes, we performed functional analysis using Ingenuity Pathway Analysis software (IPA, QIAGEN, Redwood, CA). IPA determines which molecular/cellular function terms (e.g., top-ranked genes in association tests) are statistically over represented, suggesting the GWA findings capture functional mechanisms underlying disease-related biological processes. We used a nominal P-value threshold of 0.05 to flag associated functions. We also examined neuronal cell type-specific expression for the top-ranked genes using single cell RNA sequencing data (which was downloaded from GEO: GSE67835). Further details of this analysis are described in Supplementary Information.
3. Results
3.1. Genome-wide Association Results
There was slight genomic inflation in GWA results for Aβ42 and LMdT in CN subjects (λ=1.02 for both traits; Supplementary Figs. 1-12). Associations of the APOE ε4 allele were SWS with the CSF biomarkers and suggestive with the other traits in MCI subjects, whereas ε4 was significantly associated only with Aβ42 level in CN subjects (Supplementary Table 2). Eleven of 25 other previously known AD loci [22-25] – CR1, INPP5D, MEF2C, HBEGF, HLA region, ZCWPW1, USP6NL, MS4A region, PICALM, SLC2A4A, and CASS4 – were nominally significant (P<0.05) with at least one trait in CN and/or MCI groups (Supplementary Table 3).
Novel SWS associations were observed for several endophenotypes in the CN and/or MCI groups (Table 1). Among CN subjects, t-Tau was associated with SNPs located 28 kb upstream of SRRM4 (best SNP: rs10775009, P=1.6×10−9; Fig. 1A) and 66.8 kb upstream of C14orf79 (rs2819438; P=6.9×10−9; Fig. 1B). In the same group, HPV was associated with rs4921790 in PDGFRL and near MTUS1 (P=4.6×10−9; Fig. 1C). This finding was supported by associations with many SNPs in high LD (r2>0.8 and D`>0.9) with rs4921790 which span MTUS1. In the MCI subgroup, LMdT was associated with three SNPs in ZNF804B (best SNP: rs73705514; P=2.9×10−9; Fig. 1D). Association was also observed between HPV and a SNP near LINC00271 and PDE7B (P=1.76×10−8)in AD subjects (Supplementary Fig. 13A). Several GWS associations are also noteworthy (Supplementary Table 4) including Aβ42 level with GRIN2B SNP rs74442473 (P=2.52×10−8; Supplementary Fig. 13B) and rs2378873 near BRIP1 and NACA2 (P=2.03×10−8) in CN subjects, LMdT with DAB1 SNP rs74834332 (P=4.20×10−8) and PRKG1 SNP rs12268753 (P=2.01×10−8) in CN subjects, LMdT with ARHGAP24 SNP rs111882035 (P=2.74×10−8) in MCI subjects, and LMiT with NRG1 SNP rs118130881 (P=1.72×10−8; Supplementary Fig. 13C) in MCI subjects, and Aβ42 level with rs55644114 near GFRA2 and LZTS1 in the total sample.
Table 1.
Genome-wide significant association (P<5.0×10−8) of novel genes in cognitively normal (CN), mild cognitive impairment (MCI), or AD dementia subjects with CSF protein levels (Aβ42 and total and phosphorylated taus), hippocampal volume (HPV), and logical memory tests of immediate (LMiT) and delayed (LMdT) recall.
| Group | Traits | CHR | BP | SNP | MA | MAF | BETA | SE | P | Closest Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | Aβ42 | 8 | 20647323 | rs55644114 | A | 0.16 | 0.41 | 0.07 | 2.54×10−8 | GFRA2, LZTS1 |
|
| ||||||||||
| CN | Aβ42 | 12 | 13870464 | rs74442473 | G | 0.07 | −1.02 | 0.18 | 2.53×10−8 | GRIN2B |
| 17 | 59687842 | rs2378873 | T | 0.44 | −0.53 | 0.10 | 2.03×10−8 | BRIP1, NACA2 | ||
|
| ||||||||||
| t-Tau | 12 | 119390525 | rs10775009 | T | 0.34 | 0.51 | 0.09 | 1.59×10−9 | SRRM4 | |
| 14 | 105385352 | rs2819438 | A | 0.13 | −0.80 | 0.14 | 6.94×10−9 | PLD4, C14orf79 | ||
|
| ||||||||||
| HPV | 8 | 17496561 | rs4921790 | C | 0.12 | 0.61 | 0.10 | 4.58×10−9 | PDGFRL, MTUS1 | |
|
| ||||||||||
| LMdT | 1 | 57739164 | rs74834332 | A | 0.03 | 0.79 | 0.14 | 4.30×10−8 | DAB1 | |
| 10 | 53818149 | rs12268753 | C | 0.21 | 0.30 | 0.05 | 2.01×10−8 | PRKG1 | ||
|
| ||||||||||
| MCI | LMdT | 4 | 86416554 | rs111882035 | G | 0.02 | −0.93 | 0.17 | 2.74×10−8 | ARHGAP24 |
| 7 | 88406552 | rs73705514 | C | 0.02 | −0.84 | 0.14 | 2.86×10−9 | ZNF804B | ||
|
| ||||||||||
| LMiT | 8 | 31228770 | rs118130881 | G | 0.04 | −0.61 | 0.11 | 1.72×10−8 | NRG1 | |
|
| ||||||||||
| AD | HPV | 6 | 136077929 | rs79846291 | T | 0.02 | 1.85 | 0.31 | 1.76×10−8 | LINC00271, PDE7B |
MA is minor allele, and MAF is the minor allele frequency. Bold SNPs denote the study-wide significant association (P<8.33×10−9) with a trait.
Fig. 1.
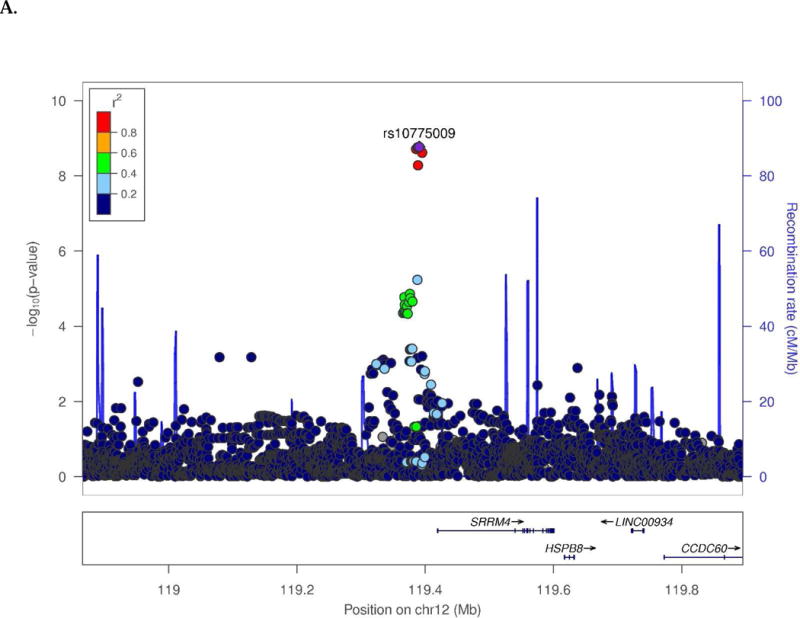
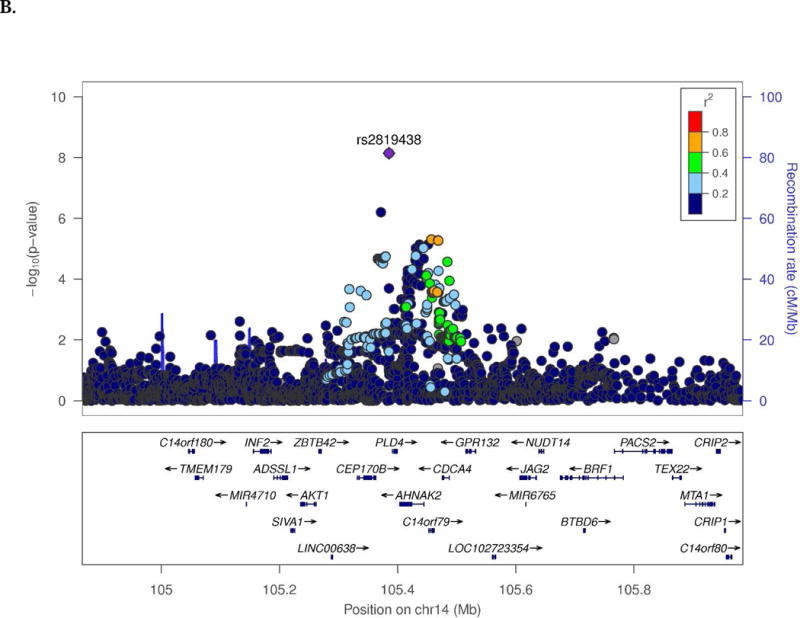
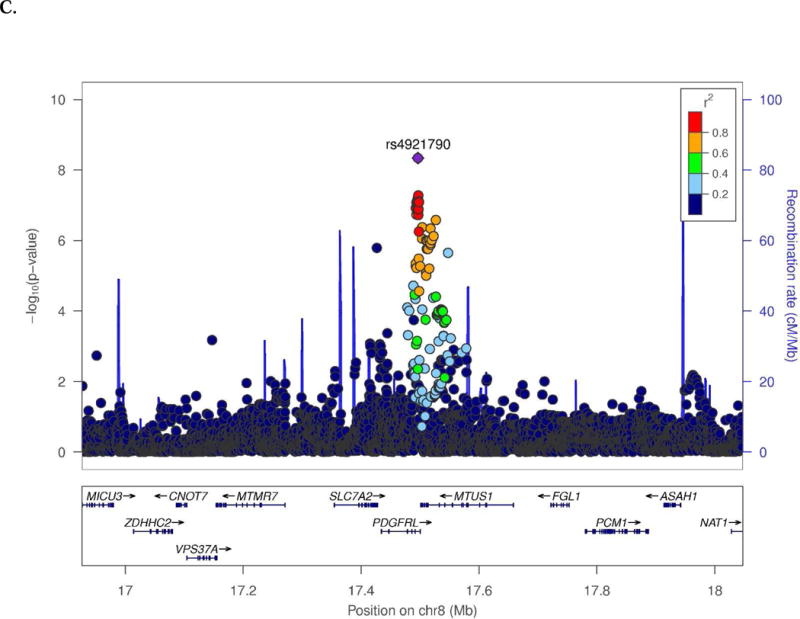
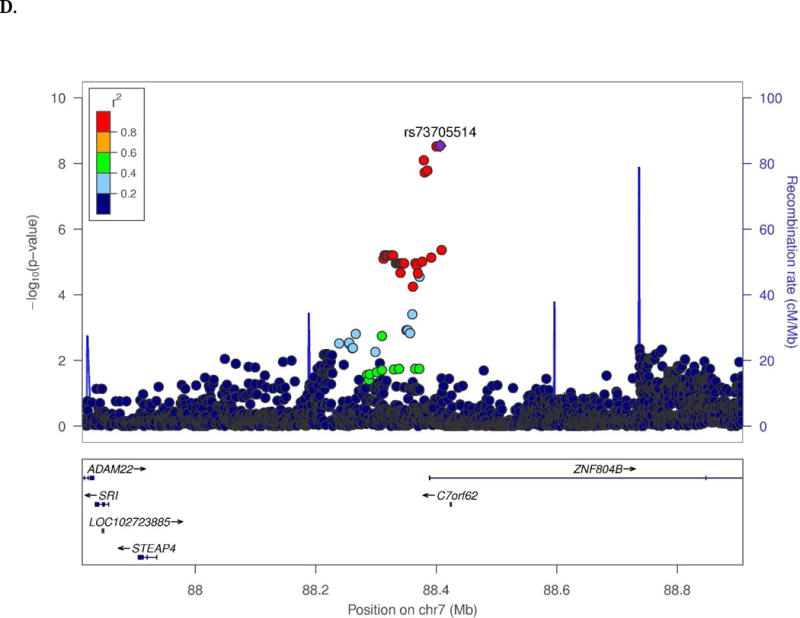
Regional association plots of (A) SRRM4 and (B) C14orf79 for CSF total tau in the CN subjects, (C) MTUS1 for hippocampal volume in the CN subjects, and (D) ZNF804B for logical memory delayed recall test in the MCI subjects.
3.2. Expression-SNP Association Results
According to the GTEx Portal database, several eSNPs under the SWS association peaks for the endophenotypes in the CN or MCI groups are significantly associated with expression of genes in those regions (Table 2). The major allele C of intergenic SNP rs2819438, which is associated with increasing CSF t-Tau, was significantly associated with lower expression of C14orf79 in brain (P-values in HIPP = 3.8×10−4 and CER=6.0×10−3; Supplementary Fig. 14A), but not with expression of PLD4 and AHNAK2 which are located between rs2819438 and C14orf79. Rs4921790 was not associated with expression of MTUS1 (P>0.05), however the major allele G of rs55653268 (a proxy SNP for rs4921790; r2 = 0.7; D’ = 1.0), which is associated with decreasing HPV, was significantly associated with increased expression of MTUS1 in the caudate (P=9.7×10−3; Supplementary Fig. 14B). PDGFRL expression was not tested, since its expression in brain is extremely low according to the GTEx database. SRRM4 SNP rs119390525 and ZNF804B SNP rs73705514 were not significantly associated with expression in any brain regions (P>0.05).
Table 2.
Genotype specific effect of the expression level among study-wide significant (8.33×10−9) SNPs using the GTEx Portal database.
| eQTL | EA | RA | Gene | eSNP Association Summary
|
||
|---|---|---|---|---|---|---|
| Hippocampus
|
Other brain region
|
|||||
| P-value (β) | P-value (β) | Region | ||||
| rs10775009 | T | C | SRRM4 | 0.09 (0.09) | 0.16 (0.10) | Frontal Cortex |
|
| ||||||
| rs2819438 | A | C | PLD4 | 0.47 (0.12) | 0.10 (0.42) | Anterior cingulate cortex |
| AHNAK2 | 0.15 (0.23) | 0.06 (−0.28) | Cerebellar Hemisphere | |||
| C14orf79 | 3.8×10−4 (0.44) | 3.9×10−3 (0.31) | Cortex | |||
| 6.0×10−3 (0.44) | Cerebellum | |||||
|
| ||||||
| rs4921790 | C | A | MTUS1 | 1.0 (0.0) | 0.09 (−0.16) | Caudate |
| rs55653268 | T | G | 0.8 (0.04) | 9.7×10−3 (−0.33) | Caudate | |
| 0.02 (−0.41) | Nucleus accumbens | |||||
EA and RA indicate effect allele and reference allele, respectively. Positive effect (β) in eSNP association summary means that carriers with effect alleles of a SNP tend to have higher expression level of a gene. Rs55653268 is in high LD with rs4921790 (r2 = 0.7; D` = 1.0) and is significantly associated with the hippocampal volume (effective allele: T, β [SE]: 0.6 [0.1], P-value: 4.2×10−7)
3.3. Association of Gene Expression with AD Status and Braak Stage
Further examination of C14orf79 and MTUS1, whose expression levels were associated with SWS SNPs (or their proxy SNPs), in the EBB RNA-Seq and the MSH microarray datasets revealed multiple significant associations with AD status and Braak stage in several brain regions (Table 3). C14orf79 was not differentially expressed in HIPP (P=0.76), but its expression was significantly lower in AD cases than controls in CER (P=7.5×10−8), DLPFC (P=3.3×10−7, Fig 2A), and VCX (P=2.1×10−8). The association of C14orf79 expression with decreasing Braak stage was nearly nominally significant (β [SE]=−0.07 [0.03], P=0.06) (Fig. 2A). MTUS1 expression was nominally higher in AD cases than controls in HIPP (P=0.02), DLPFC (P=0.01, Fig. 2B) and VCX (P=0.01). MTUS1 expression was also positively correlated with Braak stage (β [SE]=0.16 [0.04], P=3.9×10−4; Fig. 2B). According to a publicly available murine brain expression profile data obtained from the cerebral cortex, Mtus1 is highly expressed in microglia and myeloid cells. Moreover, expression is highest in activated microglia (Supplementary Figure 15A). However, MTUS1 expression in mature human brain is ranked the second lowest in microglia/macrophage cells, whereas expression in adult mouse brain was ranked the second highest among glia, neurons, and vascular cells. This finding suggests that the role of MTUS1 may differ in mouse and human myeloid cells (Supplementary Figure 15B).
Table 3.
Summary statistics of association between expression levels of the genes (MTUS1 and C14orf79) and AD status as well as Braak Stage.
| Source | Brain region | Predictor |
MTUS1
|
C14orf79
|
||||
|---|---|---|---|---|---|---|---|---|
| β | SE | P | β | SE | P | |||
| Eisai Bio Bank (RNA-Seq) | HIPP | AD Status | 0.38 | 0.16 | 0.02 | 0.04 | 0.14 | 0.76 |
| HIPP | Braak Stages | 0.16 | 0.04 | 3.9×10−4 | −0.07 | 0.03 | 0.06 | |
|
| ||||||||
| Mt. Sinai Hospital (Microarray) | CER | AD Status | 0.00 | 0.02 | 0.78 | −0.13 | 0.02 | 7.5×10−8 |
| DLPFC | AD Status | 0.09 | 0.04 | 0.01 | −0.13 | 0.02 | 3.3×10−7 | |
| VCX | AD Status | 0.09 | 0.03 | 0.01 | −0.15 | 0.03 | 2.1×10−8 | |
Brain regions tested were hippocampus (HIPP), cerebellum (CER), dorsolateral prefontal cortex (DLPFC), visual cortex (VCX). Positive effect (β) for AD status means that a gene is up-regulated in AD cases vs. in controls. Positive effect for Braak stages means that the expression of a gene is positively correlated with the Braak stages.
Fig. 2.
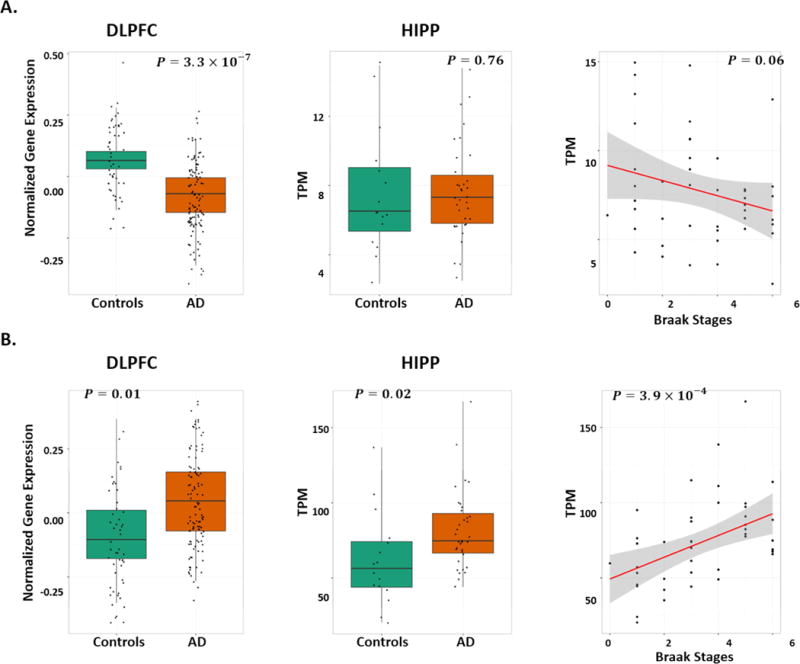
Expression studies of (A) C14orf79 and (B) MTUS1 including boxplots for differentially expressed genes in the brain dorsolateral prefontal cortex (DLPFC) in the microarray data (GEO: GSE44772; left column) and in the hippocampus (HIPP) in the RNA-Seq data (Eisai Bio Bank data; middle column), and regression plots of gene expression by Braak stage in the RNA-Seq data (Eisai Bio Bank data; right column). P-values in plots were computed from linear regression models after adjusting for covariates (details in the Methods).
3.4. Gene Co-expression Network Analysis in Hippocampus
Sixteen of 61 top-ranked genes (Supplementary Table 5) were clustered together with 17 known AD genes as co-expressed networks. Eight of 17 modules from WGCNA based on the human HIPP transcriptome (M1-M8) were enriched with the genes (P<1.0×10−6; Supplementary Table 5) identified in our GWAS analyses with AD-related endophenotypes (Supplementary Table 6). These eight modules were also significantly correlated with AD status or Braak stages (Fig. 3A). C14orf79 was co-expressed with MAPT-AS1 in M1. MTUS1 was co-expressed with ZCWPW1 in M4. SRRM4 and ZNF804B were co-expressed with previously known AD genes – APP, BZRAP1, MAPT, MEF2C, PLXNA4, PTK2B, and PSEN2 – in M7. The M7 module is enriched for genes involved in neuronal processes including ‘chemical synaptic transmission’ (FDR=1.8×10−84), ‘dendrite’ (FDR=1.7×10−33), and ‘axon guidance’ (FDR=1.6×10−24). Modules M2, 4, 5, 6, and 8 are enriched for genes involved in ‘protein binding’, ‘signal transduction’, and ‘microtubule binding’ (Supplementary Table 6). Among the eight modules enriched for previously known AD risk genes and genes identified in the current study as associated with AD endophenotypes, modules 1, 2, 5, 7, and 8 were also highly enriched with genes for other neurodegenerative and neuropsychiatric disorders including Parkinson disease and schizophrenia, as well as for cognitive performance (Supplementary Table 6). The correlation matrix of expression levels among the genes in Modules 4 and 7 (Fig. 3B) is shown in Supplementary Table 7. Further evaluation revealed that M2 and M7 contained the largest number of known AD genes among all modules co-regulated in the hippocampal region (Supplementary Table 8).
Fig. 3.
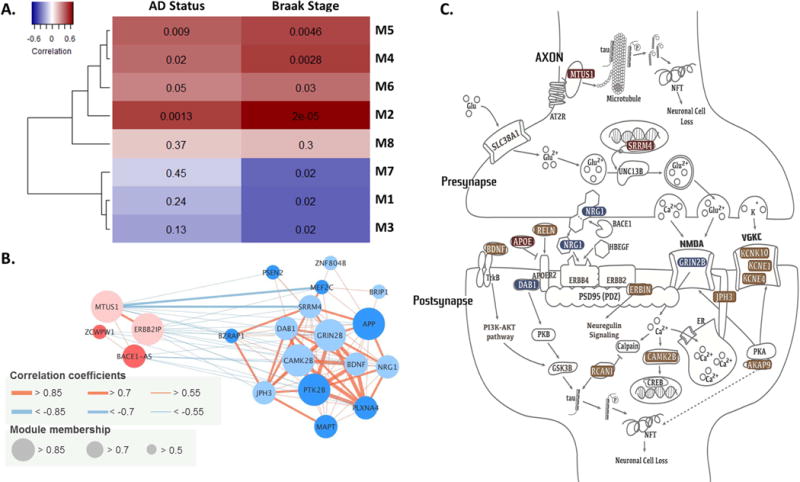
(A) A heat-map of correlations between AD traits and the first principal component values of genes in modules. Deeper colors indicate higher correlation with the traits (red: positive and blue: negative). Values in the heat-map are correlation P-values. (B) Connectivity plot of genes in modules M4 and M7, which were identified in this study or in previously reported in AD genome-wide association studies (GWAS). Light colored circles indicate the genes identified in this study, and deep colored circles indicate previously identified AD genes. (C) The role of genes identified in GWAS of AD-related endophenotypes among cognitively normal and mild cognitively impaired subjects in the top-ranked canonical pathways; colors indicate the level of association significance of the genes identified in this study (red = study-wide significance; blue = genome-wide significance; brown = P<1.0×10−6 (except for BDNF in Supplementary Fig. 13H).
3.5. Functional Analysis
The top-ranked genes from the GWA analyses in the CN and MCI groups (Supplementary Tables 5) are enriched in neuronal processes including synapse plasticity (P=3.4×10−5), axon quantity (P=2.6×10−4), microtubule dynamics (P=2.8×10−4), abnormal morphology of dentate gyrus (P=3.4×10−4), and neuronal development (P=3.5×10−4). These findings indicate that genes associated with AD-related endophenotypes among participants in clinical stages preceding AD dementia have particular roles in functioning of neuronal synapses (Fig. 3C). Single-cell transcriptome analysis of the human brain confirmed that the genes in Fig. 3C were highly expressed in neurons. Some genes were also expressed in other cell types: APOE, MTUS1, ERBIN, and AKAP9 in astrocytes; MTUS1, ERBIN, and AKAP9 in oligodendrocytes; and AKAP9 and RCAN1 in endothelial cells. Bioinformatic analysis using HaploReg and ENCODE suggests that GWS SNPs upstream of NRG1 may be located in a transcriptional regulatory site and rs111969453, a SNP in high linkage disequilibrium (r2=0.94 and D`=0.97) with the SNP (rs118130881) that is significantly associated with LMiT (P=5.9×10−8), is located on an enhancer histone mark in brain.
4. Discussion
A goal of this study was to identify genes that were both associated with AD-related endophenotypes in older, non-demented individuals and co-regulated with known AD genes. Using a GWA approach, we identified SWS associations in cognitively normal elders including CSF t-Tau level with SRRM4 and C14orf79, HPV with MTUS1, and CSF Aβ42 level with APOE. In MCI subjects we detected SWS associations for LMdT with ZNF804B, and for CSF levels of Aβ42, t-Tau, and p-Tau with APOE.
Angiotensin II interacting (AT2) protein (ATIP) isoforms encoded by MTUS1 are highly expressed in most brain regions [31]. ATIP1 binds to AT2 proteins, mediating neuronal differentiation survival and regeneration in brain [31-33]. ATIP3 colocalizes with microtubules and regulates their polymerization, thereby regulating neuronal differentiation and neurite outgrowth [33]. Gene expression analysis revealed that the risk allele of eSNP near MTUS1 was associated with increased expression of MTUS1 in caudate. DGE analyses demonstrated that expression of MTUS1 was higher in AD cases than controls in HIPP, DLPFC, and VCX. Also, expression of MTUS1 was significantly greater in brains showing severe neurofibrillary tangle involvement. The ADNI GWAS findings together with these expression findings suggest that MTUS1 expression may be related to changes in hippocampal volume prior to onset of cognitive impairment.
SRRM4 encodes the neural-specific Ser/Arg repeat-related protein of 100kDa (nSR100) that promotes neurite outgrowth and alternative splicing and controls most neural microexons [34-36]. Down-regulated SRRM4 alters splicing of microexons in autism brains [34]. Association of SRRM4 SNP rs1997111 with t-Tau level in a subset of ADNI controls (i.e., from “ADNI 1” only) was previously identified at a significance level one order of magnitude less (P=1.1×10−8) than the result we obtained among CN subjects in our study (P=1.76×10−9) [37]. The functional relevance of C14orf79 and ZNF804B, the two other SWS but poorly characterized genes, to AD is unclear.
GRIN2B encodes the GluN2B subunit of the N-methyl-D-aspartate (NMDA) receptor (GluN2B-NMDA) which is involved in synaptic plasticity and memory function. GluN2B-NMDA is the target of memantine, a drug that provides symptomatic relief in patients with AD by antagonizing the GluN2B-NMDA receptor [38]. A suggestive association of GRIN2B with temporal lobe volume (P=1.3×10−7) was reported in a previous GWA study in the ADNI-1 sample [39]. NRG1 functions as an epidermal growth factor in the nervous system [40] and is involved in the neuregulin signal transduction pathway for synapse maturation and dendritic morphology [41, 42].
Among the known AD genes, we found suggestive evidence for association with several SNPs located 381 kb upstream of AKAP9 with p-Tau level in MCI subjects (Supplementary Table 2, Supplementary Fig. 13D). We also found a suggestive association between rs149454736, located between exons 45 and 46 of AKAP9, and HPV in AD dementia subjects (P=2.2×10−7). Previously, we identified significant association of AD with two rare coding AKAP9 missense mutations in exons 31 (rs144662445) and 46 (rs149979685) in African Americans [43], and our study is the first to report association with this gene for AD risk in European ancestry individuals. Rs149454736 is located 1.5 kb away from rs149979685. AKAP9 has functional similarity with the tau protein in terms of microtubule stability and assembly [44].
With the exception of APOE, there is little overlap in GWS findings between our study and an investigation of genetic determinants of CSF Aβ42, t-Tau, and p-Tau levels in a much larger sample which included 787 ADNI subjects [45]. Some of the differences may be explained by our study design which conducted analyses within separate groups of CN, MCI, and AD subjects. The prior study combined cognitive groups and included multiple cohorts with highly variable ascertainment and distributions of subjects across cognitive groups. In addition, we extended our findings by incorporating them in analyses of co-regulated transcriptional networks enriched with previously established AD genes. Further comparison of results across the two studies revealed that four of the six GWS SNPs (excluding the APOE region) in the Deming et al. report [45] were nominally significant in specific cognitive groups in our study (Supplementary Table 9). For example, the association of rs185031519 near GLIS1 CSF Aβ42 (P=2.1×10−8) in the prior study was nominally significant in the CN group (P=8.3×10−3) in our study. Several of our most significant findings (including SRRM4, MTUS1, and GRIN2B) are consistent with results of other studies of AD-related endophenotypes (Supplementary Table 10).
Our findings may provide important insights about the sequence of processes leading to AD. The SWS associations in CN subjects (SRRM4 with t-Tau and MTUS1 with HPV) implicate neuronal signaling, development and loss, but with the exception of APOE, they do not involve Aβ processing in the asymptomatic stage of AD. It is noteworthy that the APOE Ɛ4 allele was not associated with the CSF Tau biomarkers and other endophenotypes in cognitively normal subjects. The variants associated with CSF biomarkers and HPV could also be interpreted to be markers of cognitive reserve/resilience because they predict the extent of AD pathology in cognitively normal persons, but not in MCI or AD subjects. There is extensive evidence supporting the cognitive reserve hypothesis in AD [46]. Also, the variants identified for memory performance and HPV in cognitively normal individuals could be markers of inherent memory function and hippocampal volume, completely independent of AD.
Our study has several limitations. First, the sample sizes for analyses of all traits in each clinical group, especially AD cases, were relatively small. Thus, we had low power to identify variants having small effects. In addition, it is possible that our top findings are false positives. However, the significant SNPs were supported by evidence in both constituent datasets (ADNI-1 and -GO/2) and from expression data analysis. Second, in the transcriptomic analyses we did not consider differences in cellular composition between AD and control brain tissue. Therefore, our results may have resulted from the excessive neuronal death in brains from AD subjects compared to controls. However, this concern would not impact our finding of increased MTSU1 expression in AD brains. Third, the two expression datasets generated using different platforms (RNA-seq versus microarray) from different brain regions (HIPP versus CER/DLPFC/VCX) limit direct comparisons between these datasets. Finally, it is necessary to repeat analyses in independent samples to confirm our findings and increase power to elevate the significance of true associations which did not attain study-wide significance.
In summary, we identified novel genes associated with AD-related endophenotypes in cognitively normal and MCI subjects. These genes had not been previously identified with AD risk and most of them are involved in neuronal development and signaling. Our findings suggest that genes influencing AD-related processes in individuals with normal cognition or with mild cognitive impairment may differ from those influencing these processes in individuals with AD dementia, but regulated together in the transcription level.
Supplementary Material
Research highlights.
Genetic factors for Alzheimer disease related endophenotypes influencing prediagnosis stages is evaluated
Genetic factors affecting early changes for Alzheimer disease via expression are proposed.
Co-regulated gene expression network containing genetic signatures associated with early changes in Alzheimer disease related endophenotypes that are interconnected with known AD risk genes.
Genes and pathways in early biological processes in Alzheimer disease are enriched in neuronal or synaptic function.
Acknowledgments
This study was supported in part by National Institutes of Health (NIH) grants R01-AG048927, U01-AG032984, UF1-AG046198 and P30-AG13846. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (NIH Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. The authors thank Eisai Bio Bank, University of Miami Brain Endowment Bank, and the Harvard Brain Tissue Resource Center, funded through NIH-NeuroBiobank HHSN-271-2013-00030C (The National Institute of Mental Health (NIMH), National Institute of Neurological Diseases and Stroke (NINDS) and Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and brain donors and their families for the tissue samples used in these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ertekin-Taner N, De Jager PL, Yu L, Bennett DA. Alternative Approaches in Gene Discovery and Characterization in Alzheimer’s Disease. Current Genetic Medicine Reports. 2013;1:39–51. doi: 10.1007/s40142-013-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glahn DC, Paus T, Thompson PM. Imaging genomics: Mapping the influence of genetics on brain structure and function. Human Brain Mapping. 2007;28:461–3. doi: 10.1002/hbm.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottesman II, Gould TD. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. American Journal of Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 6.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging and Behavior. 2014;8:183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–7. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roostaei T, Nazeri A, Felsky D, De Jager PL, Schneider JA, Pollock BG, et al. Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer/’s disease. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, Shaw LM, et al. Genome-wide association study of CSF biomarkers Abeta1-42, t-tau, and p-tau181p in the ADNI cohort. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010;53:1051–63. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedraza O, Allen M, Jennette K, Carrasquillo M, Crook J, Serie D, et al. Evaluation of memory endophenotypes for association with CLU, CR1, and PICALM variants in black and white subjects. Alzheimer’s & Dementia. 2014;10:205–13. doi: 10.1016/j.jalz.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal Fluid Biomarker Signature in Alzheimer’s Disease Neuroimaging Initiative Subjects. Annals of neurology. 2009;65:403–13. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16:1130–8. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Jr, Bernstein Ma, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner MW, Veitch Dp, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception [Google Scholar]
- 16.Jun G, Guo H, Klein BEK, Klein R, Wang JJ, Mitchell P, et al. <italic>EPHA2</italic> Is Associated with Age-Related Cortical Cataract in Mice and Humans. PLoS Genet. 2009;5:e1000584. doi: 10.1371/journal.pgen.1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov Alexei A, et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun G, Asai H, Zeldich E, Drapeau E, Chen C, Chung J, et al. PLXNA4 is associated with Alzheimer disease and modulates tau phosphorylation. Ann Neurol. 2014;76:379–92. doi: 10.1002/ana.24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2017 doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MJ, Liu Z, Wang P, Wong MP, Nelson MR, Kocher JP, et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic acids research. 2016;44:D869–76. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 28.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–D4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guimond M-O, Gallo-Payet N. How does angiotensin AT2 receptor activation help neuronal differentiation and improve neuronal pathological situations? Frontiers in Endocrinology. 2012;3 doi: 10.3389/fendo.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J-M, Mogi M, Tsukuda K, Tomochika H, Iwanami J, Min L-J, et al. Angiotensin II-Induced Neural Differentiation via Angiotensin II Type 2 (AT2) Receptor-MMS2 Cascade Involving Interaction between AT2 Receptor-Interacting Protein and Src Homology 2 Domain-Containing Protein-Tyrosine Phosphatase 1. Molecular Endocrinology. 2007;21:499–511. doi: 10.1210/me.2006-0005. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues-Ferreira S, le Rouzic E, Pawlowski T, Srivastava A, Margottin-Goguet F, Nahmias C. AT2 Receptor-Interacting Proteins ATIPs in the Brain. International Journal of Hypertension. 2013;2013:6. doi: 10.1155/2013/513047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–23. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nature reviews Neuroscience. 2016;17:265–81. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohnishi T, Shirane M, Nakayama KI. SRRM4-dependent neuron-specific alternative splicing of protrudin transcripts regulates neurite outgrowth. Scientific reports. 2017;7:41130. doi: 10.1038/srep41130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han MR, Schellenberg GD, Wang LS, Alzheimer’s Disease Neuroimaging I Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC neurology. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia P, Chen H-sV, Zhang D, Lipton SA. Memantine Preferentially Blocks Extrasynaptic over Synaptic NMDA Receptor Currents in Hippocampal Autapses. The Journal of Neuroscience. 2010;30:11246–50. doi: 10.1523/JNEUROSCI.2488-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS) NeuroImage. 2010;53:1160–74. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo R-S, Lee J-H, Yu H-N, Song D-Y, Baik T-K. Expression of ErbB4 in the apoptotic neurons of Alzheimer’s disease brain. Anat Cell Biol. 2010;43:332–9. doi: 10.5115/acb.2010.43.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao Y, Dai P, Liu Y, Marchetto S, Xiong W-C, Borg J-P, et al. Erbin regulates NRG1 signaling and myelination. Proceedings of the National Academy of Sciences. 2009;106:9477–82. doi: 10.1073/pnas.0901844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadugu B, Kühn B. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. American Journal of Physiology - Heart and Circulatory Physiology. 2012;302:H2139–H47. doi: 10.1152/ajpheart.00063.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logue MW, Schu M, Vardarajan BN, Farrell J, Bennett DA, Buxbaum JD, et al. Two rare AKAP9 variants are associated with Alzheimer’s disease in African Americans. Alzheimer’s & Dementia. 2014;10:609–18.e11. doi: 10.1016/j.jalz.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. The EMBO Journal. 2009;28:1016–28. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K, et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017;133:839–56. doi: 10.1007/s00401-017-1685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. The Lancet Neurology. 2012;11:1006–12. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


