 96 nano-crystallization trials can be done with just (sub)-milligram amounts of an organic salt to directly yield single crystal structures.
96 nano-crystallization trials can be done with just (sub)-milligram amounts of an organic salt to directly yield single crystal structures.
Abstract
The generation of solid salts of organic molecules is important to the chemical and pharmaceutical industry. Commonly used salt screening methods consume a lot of resources. We employed a combination of ion exchange screening and vapour diffusion for crystallization. This technique is semi-automatic and requires just nanoliters of the solution of the analyte to be crystallized. This high throughput screening yielded single crystals of sufficient size and quality for single-crystal X-ray structure determination using an in-house X-ray diffractometer. The broad scope of our method has been shown by challenging it with 7 very different organic cations, whose aqueous solubilities vary by a factor of almost 1000. At least one crystal structure for 6 out of 7 tested cations was determined; 4 out of the successful 6 ones had never been crystallized before. Our method is extremely attractive for high throughput salt screening, especially for active pharmaceutical ingredients (APIs), as about 40% of all APIs are cationic salts. Additionally, our screening is a new and very promising procedure for the crystallization of salts of organic cations.
Introduction
Organic salts are multi-component ionic compounds, in which at least one of the components is an organic ion. The focus of this paper is the organic, cationic part of salts. Different salts of the same organic cation can have significantly different physicochemical properties.1–6 The critical properties are solubility,7 crystal shape, hygroscopicity, melting point, and physical as well as chemical stability. The selection of a suitable anion can avoid problems during production, storage or shipping of an organic salt. For example, an appropriate anion can improve the purification and flow properties of the powder and/or reduce the hygroscopicity. Therefore, salt screening is a decisive step during the development and optimization of the production process of an organic salt.2 It can significantly reduce the production cost of the chemical or pharmaceutical, as about 40% of all active pharmaceutical ingredients (APIs) are cationic salts.8 However, there are also requirements for an ideal screening. It shall require the least possible amount of material, human work and time. At the same time, it should be as effective as possible in discovering new salts. Additionally, the ability to analyse the new crystalline forms directly from the basic screening would be a great advantage, ideally by single crystal X-ray diffraction (SCXRD) structure analysis, since fast access to SCXRD can save a lot of material and lab work, such as immediately identifying false positive screening hits.
Recently, several innovations have been described that, with the help of crystallography, allow the structure determination of compounds, which had previously been impossible by applying the methods of traditional crystal growth.9,10 The ‘crystalline sponge’ method was originally introduced in 2013 by Fujita and co-workers11 and has further been improved over the last few years.12,13 This method opened up new possibilities for determining the crystal structures of apolar compounds that are available only in minute amounts. However, this method is a 3 step procedure: (1) synthesis of the MOF host, (2) exchange of the previous solvent by an apolar one, and (3) soaking of the MOF crystal with the analyte. All steps are non-trivial to perform and especially the last step has to be optimized for every single compound. Alternative sponge methods have been described, avoiding the exchange of guest molecules; however, they require more analyte material and synchrotron radiation.14,15 Even the most recent optimisation of the crystal sponge method is still labour intensive, requires the analyte to be soluble in dichloromethane and can only in exceptional cases be used as an ab initio structure determination method without additional knowledge of the analyte.16 A different MOF lattice has recently been developed by the group of Yaghi for the co-crystallization of carboxylates or alcohol containing molecules.17 Summarizing the previous developments, a new method that could facilitate the crystallization of cationic, polar compounds would be highly desirable.
Results and discussion
Our new method addresses this need. Our screening technique is based on two simple ideas and combines basic screening with single crystal growth directly followed by structural analysis. The first hypothesis assumes that chloride salts of most cations have the highest solubility in water and analogously that sodium salts are the most common form of salts with a high solubility in water. The second hypothesis supposes that a slow increase of saturation might lead to the growth of single crystals. According to the first idea, if we mix solutions of organic chloride salts with various sodium salts, the least soluble combination of dissolved ions should crystallize, which should be a new organic salt. In addition, the mixture of the starting salts does not have to be equimolar, because all remaining ions will still be dissolved in water. Concerning the second idea, an increase of analyte concentration within a drop is generated by the method of vapour diffusion (see ESI Fig. 1†), which was previously developed for protein crystallization.18 This method basically consists of mixing 100–500 nl of the analyte in water with the same volume of a crystallization cocktail – in our case this is normally a sodium salt – to generate a small drop of 200–1000 nl volume. The drop is then equilibrated via the vapour phase against the big reservoir consisting of only the crystallization cocktail. Since the crystallization cocktail in the drop was diluted with the analyte, the water diffuses via the gas phase back to the reservoir. The mixing of the analyte and the sodium salt in the drop might already generate a supersaturated solution, while the diffusion of the water to the reservoir will increase this effect. In order to automate and miniaturize the whole procedure, the pipetting of the reservoir and the drop solutions, as well as the visual monitoring, is carried out by a robot. Such robots are commonly being used in most biochemical laboratories and pharmaceutical companies that are engaged in structural biology projects.
Currently, the salt screening of active pharmaceutical ingredients (APIs) that can be protonated is mainly done by isolating and weighing the free base followed by addition of different acids in various solvents.19–21 This process is labour intensive and can be unreliable, as some free bases are hygroscopic, prone to oxidation, or may otherwise be chemically unstable. An additional disadvantage of this method is that the ratio of API to acid has a direct influence upon the pH in the chosen solvent and therefore also upon the propensity to crystallize. Furthermore, different solvents might change the degree of protonation of the API. Therefore, the amount of added acid must be chosen carefully in advance for each solvent.
In order to evaluate our new method of crystallization, we chose salts with seven quite different organic cations (Fig. 1). The chosen compounds include permanent cations (due to the presence of tetraalkylated ammonium groups), as well as compounds that can be protonated only at lower pH. Their solubilities in water range from 2.2 to 1800 mg ml–1 (Table 1). Furthermore, we included a racemic compound to evaluate the potential for enantiomeric resolution by chiral and sometimes even enantiomerically pure anions. Additionally, we tested the corresponding enantiomerically pure compound in order to compare the ease of crystallization of a racemate and of one of its enantiomers. Most of the chosen organic compounds have no reported crystal structure at all, while one, ephedrine, is known to form many different salt forms.22 5-Benzyloxytryptaminium chloride ([BaH]Cl) is an API, which acts as an antagonist of the TRPM8 ion-channel. It is used for the treatment of prostate cancer or benign prostate hyperplasia.23 The Cambridge Structural Database (CSD)24 does not contain any structure of 5-benzyloxytryptamine. Butyl scopolammonium bromide ([Bs]Br) is being used to treat crampy abdominal pain, renal colic, and esophageal and bladder spasms. The crystal structure its methanol solvate has been reported.25 (R,S)-Carnitinenitrile chloride ([(+/–)-Car]Cl) and (R)-(–)-carnitinenitrile chloride ([(–)-Car]Cl) are being used as intermediates for the preparation of carnitine or derivatives of carnitine, both of which are APIs with many different effects from neurology to diabetes mellitus.26–28 There is no structure of any carnitinenitrile in the CSD. 2-(3-(3,5-Bis(trifluoromethyl)phenyl)-thioureido)-N,N,N-trimethylethanaminium iodide ([Cat]I) is the achiral version of chiral thiourea catalysts,29 and the former can be used for the aminolysis of N-acyl homoserine lactones.30 No structure of any salt that contains [Cat]+ can be found in the CSD. (1S,2R)-(+)-Ephedrine hydrochloride ([(+)-EphH]Cl) is an active pharmaceutical ingredient used for the treatment of emphysema and bronchial asthma. Many crystal structures have been described for (+)-, (–)- or (+/–)-ephedrine salts.22,31 Ephedrine was also used as a test case for several salt screening studies.20,32 Trazodone hydrochloride ([TrH]Cl) is a selective inhibitor of serotonin and norepinephrine reuptake in a ratio of 25 : 1, and it is being used pharmaceutically as an antidepressant.33 Only the crystal structure of trazodone hydrochloride has been reported and can be found in the CSD.34
Fig. 1. Selected compounds (5-benzyloxytryptaminium chloride, butyl scopolammonium bromide, (R,S)-carnitinenitrile chloride, (R)-(–)-carnitinenitrile chloride, iodide salt of the catalyst, (1S,2R)-(+)-ephedrine hydrochloride and trazodone hydrochloride) for crystallization and salt screening.
Table 1. Needed amount of material in milligrams for one 96-well crystallization screening (assuming 100 nl consumption per crystallization drop of a 90% saturated solution plus a 2 μl pipetting reserve for one 96-well plate).
| [BaH]Cl | [Bs]Br | [(+/–)-Car]Cl | [(–)-Car]Cl | [Cat]I | [(+)-EphH]Cl | [TrH]Cl | |
| Amount [mg] | 0.026 | 22 | 13 | 16 | 0.083 | 3.1 | 0.46 |
| Aqueous solubility [mg ml–1] | 2.2 ± 0.1 | 1800 ± 200 | 1120 ± 106 | 1300 ± 200 | 6.9 ± 0.1 | 257 ± 1 | 38.5 ± 0.2 |
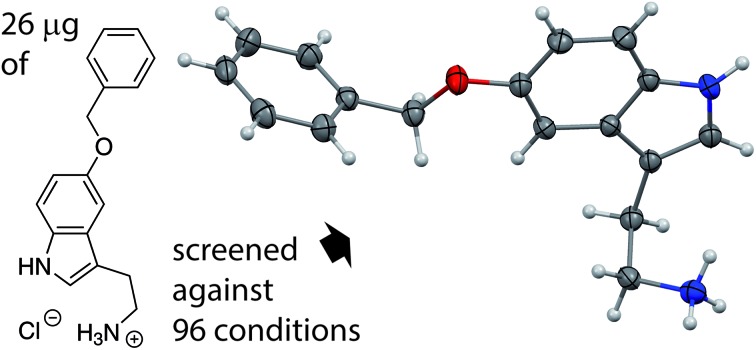
Our method employs a crystallization robot with a setup that is commonly used in protein crystallography, but only exceptionally in small molecule crystallography.35 Initially, we chose a crystallization setup that consisted of 500 nl of the aqueous nearly saturated solution of the analyte; however repetitions of the crystallizations with just 150 and 100 nl of the analyte solutions were similarly successful. The last measurement series with 100 nl analyte solution required really minute amounts of material and still yielded high quality crystals (ESI Fig. 3†) that directly led to publishable structures. As can be seen in Table 1, the required amount essentially depends upon the solubility of the compound in water. For our method, we work with solutions that are 90% saturated. We used two procedures to obtain these solutions: one was to determine the apparent solubility by dissolution of the solid material and observing the progress with a microscope. The other method was much simpler and used less material: we generated a saturated stock solution, separated it from the solid and diluted it then to 90% saturation.
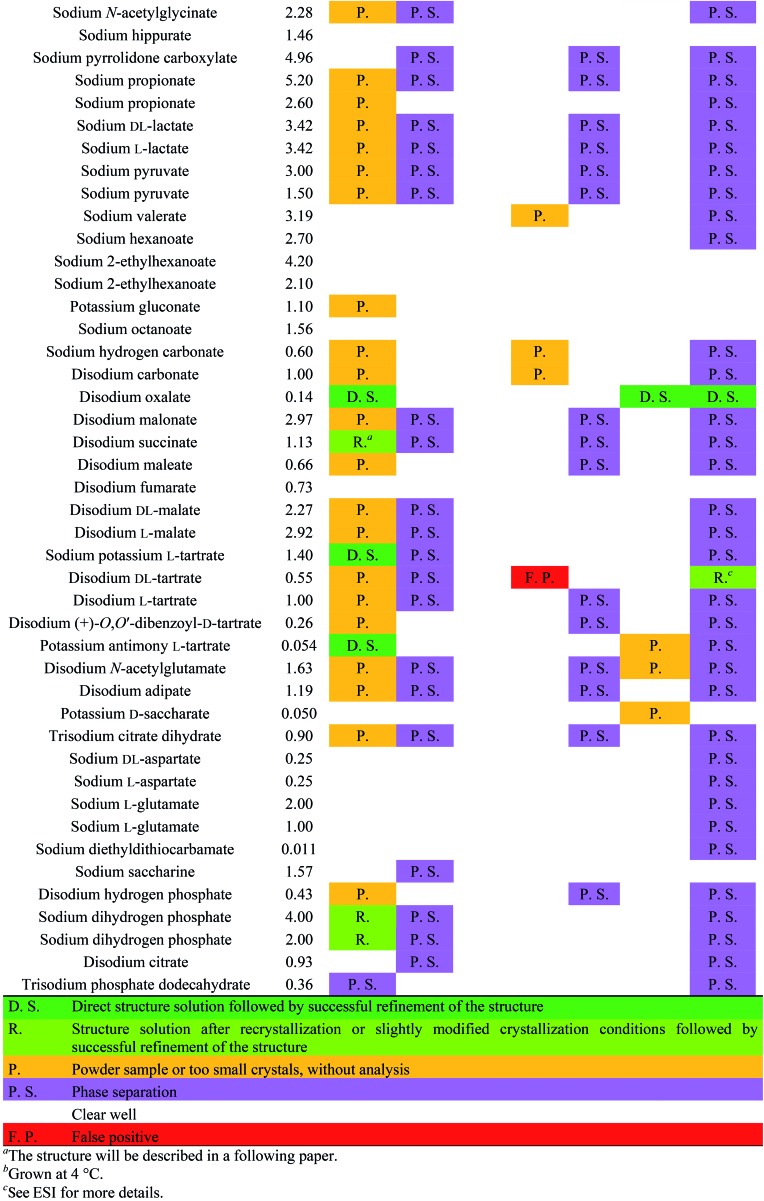
The screening was performed with the seven mentioned organic salts and mainly sodium salts of 77 different counterions (Table 2 and ESI†). The criteria for the selection of the counterions were a known propensity to crystallize and/or being a compound that is Generally Recognized As Safe (GRAS)36 for a potential later use in an API. Each of the chosen 7 analytes was screened under 96 conditions, as some of the counterions were present in the screening at multiple, different concentrations. The screening was stopped after 16 days; however longer equilibration times could be applied. Quite often the crystals formed after a few days, see e.g. the case of 5-benzyloxytryptaminium antimony-l-tartrate, in which a change of crystal morphology could be observed (ESI Fig. 2†). The same crystals could sometimes be observed in several drops containing the same counterion at different concentrations. It was possible to determine 15 high quality single crystal structures of chemically different salts directly from the screening without the need for any further optimisation, and 14 of them were novel structures. Additionally, 14 lead hits consisting of too small crystals were manually recrystallized, which yielded in 7 cases big enough crystals that could be measured and refined by SCXRD to give 7 novel structures (Fig. 2 and Table 2). In one further case, covering the drop with oil induced the crystal growth of the free base trazodone. The crystal nucleated at the oil/water interface and then grew by penetrating into the oil. The three compounds – [Bs]Br, [(+/–)-Car]Cl and [(–)-Car]Cl – all having an unusually high solubility of more than one gram per milliliter were most challenging; obviously if the chloride is already present at such high concentrations, it is difficult to substitute it during crystallization from a mixed chloride/anion solution.
Table 2. Crystallization results.

|
|
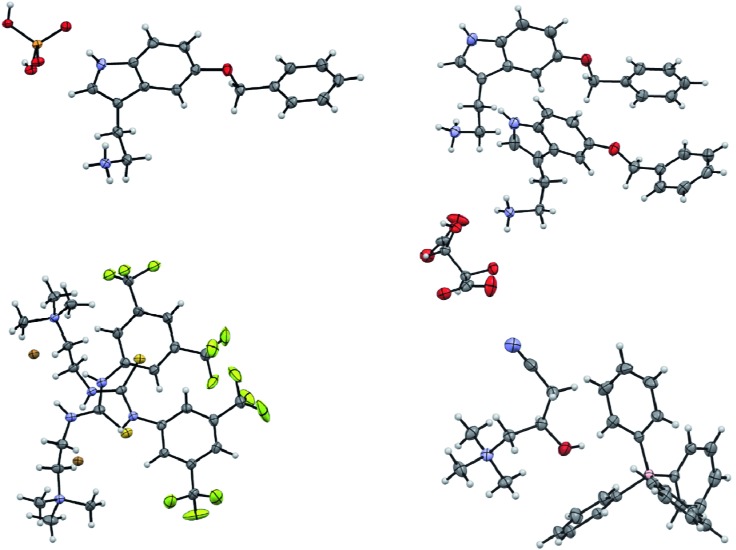
Fig. 2. Displacement ellipsoid representations of the crystal structures of 5-benzyloxytryptaminium dihydrogenphosphate (top left), 5-benzyloxytryptaminium tartrate (minor disordered parts and water molecules omitted for clarity, top right), 2-(3-(3,5-bis(trifluoromethyl)phenyl)-thioureido)-N,N,N-trimethylethanaminium bromide (lower left) and (R)-carnitinenitrile tetraphenylborate (lower right). Ellipsoids are drawn at 50% probability.
Conclusion
Our screening method for the growth of single crystals containing organic cations was very effective: six out of seven tested cationic moieties yielded at least one crystal structure. Also, the primary screening used just a very small amount of material (12 μl of a 90% saturated aqueous solution of a salt with an organic cation) in order to test a total of 96 different conditions, and we were able to determine at least one unit cell directly in most of the positive crystalline hits using in-house X-ray diffractometers. Furthermore, the required manual work is quite low due to the usage of a pipetting robot and a crystal farm, both of which are available in many biochemical institutes and pharmaceutical companies. With just 26 μg of one organic salt employing 100 nl of analyte solution per crystallization experiment, we were able to directly determine the structure of 6 different salts and obtained the lead to another 5 salts. The new screening method requires compounds that can form stable cations in water and have a water solubility of at least 2 mg ml–1 of their salt form. We are currently working on a system, which can crystallize less water soluble cations. In contrast to the report of Berghausen,21 we were able to perform a salt screening with highly water soluble compounds in a purely aqueous environment. Performing the screening at temperatures other than room temperature is possible and yielded a second, unknown polymorph of enantiomerically pure ephedrinium iodide.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
We thank Prof. Dr Mario Waser for kindly providing compound [Cat]I, Lonza Ltd. Basel for providing the sodium dicyanamide, and Beat Blattmann as well as Céline Stutz for setting up our crystallization trials on the robot. We thank Mohammad Al-Qatanani for determining the aqueous solubility of various compounds by HPLC and Prof. Dr Anthony Linden for a critical reading of the manuscript. We thank the University of Zürich, the R'Equip programme of the Swiss National Science Foundation (project number 206021_164018) and the Czech Science Foundation (grant No. 16-10035S) for financial support.
Footnotes
†Electronic supplementary information (ESI) available: Details of experimental, crystallographic procedures and descriptions of the crystal structures. CCDC 1585747–1585765 and 1823199–1823201. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8sc00783g
References
- von Raumer M., Dannappel J., Hilfiker R. Chem. Today. 2006;24:41–44. [Google Scholar]
- Kumar L., Amin A., Bansal A. K. Drug Discovery Today. 2007;12:1046–1053. doi: 10.1016/j.drudis.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Tong W.-Q., in Developing Solid Oral Dosage Forms, ed. Y. Chen, G. G. Z. Zhang, L. Liu and W. R. Porter, Academic Press, San Diego, 2009, ch. 4, pp. 75–86. [Google Scholar]
- Wyttenbach N., Sutter B. and Hidber P., in Handbook of Pharmaceutical Salts, ed. P. H. Stahl and C. G. Wermuth, Verlag Helvetica Chimica Acta, 2nd edn., 2011, ch. 8, pp. 203–233. [Google Scholar]
- Pharmaceutical Salts and Co-crystals, ed. J. Wouters and L. Quéré, Royal Society of Chemistry, Cambridge, 2011.
- Byrn S. R., Zografi G., Chen X. J. Pharm. Sci. 2010;99:3665–3675. doi: 10.1002/jps.22215. [DOI] [PubMed] [Google Scholar]
- Elder D. P., Holm R., de Diego H. L. Int. J. Pharm. 2013;453:88–100. doi: 10.1016/j.ijpharm.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Paulekuhn G. S., Dressman J. B., Saal C. J. Med. Chem. 2007;50:6665–6672. doi: 10.1021/jm701032y. [DOI] [PubMed] [Google Scholar]
- Spingler B., Schnidrig S., Todorova T., Wild F. CrystEngComm. 2012;14:751–757. [Google Scholar]
- Nievergelt P. P., Spingler B. CrystEngComm. 2017;19:142–147. [Google Scholar]
- Inokuma Y., Yoshioka S., Ariyoshi J., Arai T., Hitora Y., Takada K., Matsunaga S., Rissanen K., Fujita M. Nature. 2013;495:461–466. doi: 10.1038/nature11990. [DOI] [PubMed] [Google Scholar]
- Inokuma Y., Yoshioka S., Ariyoshi J., Arai T., Fujita M. Nat. Protoc. 2014;9:246–252. doi: 10.1038/nprot.2014.007. [DOI] [PubMed] [Google Scholar]
- Hoshino M., Khutia A., Xing H., Inokuma Y., Fujita M. IUCrJ. 2016;3:139–151. doi: 10.1107/S2052252515024379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadhar T. R., Zheng S.-L., Chen Y.-S., Clardy J. Acta Crystallogr., Sect. A: Found. Adv. 2015;71:46–58. doi: 10.1107/S2053273314019573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadhar T. R., Zheng S.-L., Chen Y.-S., Clardy J. CrystEngComm. 2017;19:4528–4534. doi: 10.1039/C7CE00885F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai F., Khutia A., Kikuchi T., Fujita M. Chem.–Eur. J. 2017;23:15035–15040. doi: 10.1002/chem.201704176. [DOI] [PubMed] [Google Scholar]
- Lee S., Kapustin E. A., Yaghi O. M. Science. 2016;353:808–811. doi: 10.1126/science.aaf9135. [DOI] [PubMed] [Google Scholar]
- McPherson A., Gavira J. A. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2014;70:2–20. doi: 10.1107/S2053230X13033141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collman B. M., Miller J. M., Seadeek C., Stambek J. A., Blackburn A. C. Drug Dev. Ind. Pharm. 2013;39:29–38. doi: 10.3109/03639045.2012.656272. [DOI] [PubMed] [Google Scholar]
- Thorson M. R., Goyal S., Schudel B. R., Zukoski C. F., Zhang G. G. Z., Gong Y. C., Kenis P. J. A. Lab Chip. 2011;11:3829–3837. doi: 10.1039/c1lc20645a. [DOI] [PubMed] [Google Scholar]
- Tarsa P. B., Towler C. S., Woollam G., Berghausen J. Eur. J. Pharm. Sci. 2010;41:23–30. doi: 10.1016/j.ejps.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Collier E. A., Davey R. J., Black S. N., Roberts R. J. Acta Crystallogr., Sect. B: Struct. Sci. 2006;62:498–505. doi: 10.1107/S0108768106012018. [DOI] [PubMed] [Google Scholar]
- DeFalco J., Steiger D., Dourado M., Emerling D., Duncton M. A. J. Bioorg. Med. Chem. Lett. 2010;20:7076–7079. doi: 10.1016/j.bmcl.2010.09.099. [DOI] [PubMed] [Google Scholar]
- Groom C. R., Bruno I. J., Lightfoot M. P., Ward S. C. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger J. M., Gadret M., Carpy A. Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1978;34:3705–3709. [Google Scholar]
- Puetter H., Roske E. and Pander H. J., German Pat., DE3419723A1, 1985.
- Nakayama K., Honda H., Ogawa Y., Ohta T. and Ozawa T., US Pat., US5041375, 1991.
- Malaguarnera M. Curr. Opin. Gastroenterol. 2012;28:166–176. doi: 10.1097/MOG.0b013e3283505a3b. [DOI] [PubMed] [Google Scholar]
- Tiffner M., Novacek J., Busillo A., Gratzer K., Massa A., Waser M. RSC Adv. 2015;5:78941–78949. doi: 10.1039/C5RA14466C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci M. A., Lee S. J., Gagné M. R. Chem. Commun. 2013;49:2055–2057. doi: 10.1039/c3cc00268c. [DOI] [PubMed] [Google Scholar]
- Wu H., West A. R., Vickers M., Apperley D. C., Jones A. G. Chem. Eng. Sci. 2012;77:47–56. [Google Scholar]
- Black S. N., Collier E. A., Davey R. J., Roberts R. J. J. Pharm. Sci. 2007;96:1053–1068. doi: 10.1002/jps.20927. [DOI] [PubMed] [Google Scholar]
- Davidoff G., Guarracini M., Roth E., Sliwa J., Yarkony G. Pain. 1987;29:151–161. doi: 10.1016/0304-3959(87)91032-3. [DOI] [PubMed] [Google Scholar]
- Fillers J. P., Hawkinson S. W. Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1979;35:498–500. [Google Scholar]
- Mandal P. K., Kauffmann B., Destecroix H., Ferrand Y., Davis A. P., Huc I. Chem. Commun. 2016;52:9355–9358. doi: 10.1039/c6cc04466b. [DOI] [PubMed] [Google Scholar]
- SCOGS (Select Committee on GRAS Substances), https://www.accessdata.fda.gov/scripts/fdcc/?set=SCOGS, accessed 23rd October 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.