Abstract
Porcine CL develop sensitivity to regression by PGF2α, termed luteolytic capacity, about 13 d after estrus. We postulated that PGF2α regulation of AP-1 transcriptional factor expression underlies acquisition of luteolytic capacity. Gilts on Day 9 (estrous cycle) or Day 17 (pseudopregnancy) had CL collected before or after PGF2α treatment with mRNA measured for FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, and JUND and AP-1 target genes CCL2 and SERPINE1. At 0.5h after PGF2α, both Day 9 and Day 17 CL had increased (P < 0.01) mRNA for FOS (2,225% and 1,817%), JUNB (237% and 358%), and FOSB (1,060% and 925%). Intriguingly, at 0.5 h after PGF2α there were increased (P < 0.01) mRNA encoding JUN (1099%) and JUND (300%) in Day 17 but not Day 9 CL. At 10 h after PGF2α there was elevated FOSB mRNA in Day 17 (771%) but not Day 9 CL and no PGF2α-induced change in FOS, JUN, JUND, and JUNB mRNA in Day 9 or Day 17 CL. Treatment with PGF2α increased mRNA for AP-1-responsive genes, CCL2, at 0.5 h (202%) and CCL2 and SERPINE1 at 10 h (719% and 1515%) only in Day 17 CL. Thus, many of the fos family of transcription factors are dramatically induced by PGF2α in CL with or without luteolytic capacity. However, PGF only induced JUN and JUND expression in CL with luteolytic capacity, a finding that may be key for understanding acquisition of luteolytic capacity given that JUN is the only AP-1 family member with strong N-terminal trans-activation activity.
Keywords: corpus luteum, luteolysis, ovary
1. Introduction
Luteolysis involves a decrease in progesterone (P4) production and induction of luteal cell death [1]. In most species, the early CL does not undergo luteolysis, even when challenged with the normal luteolysin, PGF2α [2]. Acquisition of luteal sensitivity to PGF2α has been termed luteolytic capacity [3,4]. The pig CL acquires luteolytic capacity later in the luteal phase (Day 12–13) and well after CL have reached mature size and maximum hormonal secretion [2].
The mechanisms involved in acquisition of luteolytic capacity are largely undefined. Active receptors for PGF2α (FP receptors- PTGFR) are present on luteal cells well before acquisition of luteolytic capacity [5,6]. For example, PGF2α induces a decrease in PTGFR (FP receptor) and HSD3B1 (3-beta hydroxysteroid dehydrogenase) mRNA in CL that have or do not have luteolytic capacity [3,4]. Nevertheless, some gene expression pathways are only induced by PGF2α in CL with luteolytic capacity. For example, PGF2α differentially regulates pathways involved in production of PGF2α [3,4], P4 [2,7], endothelin-1 (EDN1) [8], chemokine C-C motif ligand 2 (CCL2), also known as monocyte chemoattractant protein-1 [9], and estradiol biosynthesis and signaling [10]. Differential regulation of gene expression in CL with luteolytic capacity likely involves differences in activation of transcription factors and signaling pathways.
The activating protein-1 (AP-1) family of transcription factors contain characteristic basic leucine-zipper regions and includes FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, and JUND proteins. Genes for AP-1 are immediate early genes that regulate a wide range of physiological responses such as cell death, inflammation, and proliferation [11]. Responses to induction of AP-1 are dependent on specific gene promoter, cell type, and which AP-1 proteins are induced [12]. The Jun family members can homodimerize with other Jun proteins or heterodimerize with Fos proteins or with other basic leucine zipper-containing transcription factors such as activator transcription factor (ATF) family members. In contrast, Fos members do not homodimerize but can only heterodimerize with Jun family members to form active transcription complexes.
The AP-1 proteins have been localized within the pig CL [13]. Also, treatment with PGF2α was found to induce AP-1 proteins in pig CL [14] and in bovine luteal cells via a PKC-dependent MAP kinase pathway [15]. One indication that AP-1 proteins may be differentially regulated in CL without luteolytic capacity is that some AP-1-regulated genes are regulated differently in CL with or without luteolytic capacity. For example, PGF2α increases CYP19A1 [10] and CCL2 mRNA [9], and decreases STAR mRNA [7] only in CL with luteolytic capacity. All three of these genes (CYP19A1, STAR, and CCL2) are regulated by AP-1 transcriptional complexes [16–20]. Therefore, this study was undertaken to determine whether AP-1 transcription factors are differentially regulated by PGF2α in porcine CL before and after acquisition of luteolytic capacity. We hypothesized that PGF2α would induce AP-1 transcription factors only after acquisition of luteolytic capacity. Alternatively, specific AP-1 transcription factors may be differentially regulated, potentially providing insight into the underlying transcriptional mechanisms associated with acquisition of luteolytic capacity.
2. Materials and Methods
2.1. Chemicals and Reagents
Cloprostenol was purchased from Bayer Corporation (Shawnee Mission, KS), Ketamine was from Fort Dodge Animal Health (Fort Dodge, IO), and Xylazine was from Phoenix Pharmaceuticals (St. Joseph, MO). T7 RNA polymerase, Taq polymerase, Reverse Transcriptase, dNTPs, RNAsin and DNAase I were purchased from Promega (Madison, WI). Molecular weight markers were from Gibco/BRL (Gaithersburg, MD). Magnetight oligo(dt) beads were from Novagen (Madison, WI). Unless otherwise specified, other chemicals and reagents used in these studies were purchased from Sigma (St. Louis, MO).
2.2. Animals
Crossbred gilts (Cambrough × Line 19) 6–8 months of age were obtained from the university herd or purchased from Pig Improvement Company (PIC, Franklin, KY). Animals were kept in individual pens with free access to water and were fed a maintenance diet of corn and soybean meal. For all studies, animals were checked daily for standing estrus with a mature boar. First day of estrus was designated as Day 0. Pseudopregnancy was induced in some gilts with daily injections of estradiol benzoate (2 mg i.m.) on Days 11–15. On the day ovaries were collected, anesthesia was induced with i.m. injection of ketamine (15 mg/kg) and xylazine (0.3 mg/kg). Gilts were intubated and surgical plane of anesthesia maintained with halothane. Ovaries were collected via midventral laparotomy and CL were dissected away from ovarian stroma and either frozen in liquid nitrogen or transported to the laboratory in cold media (M199, 100 IU/mL penicillin, 10 mg/mL streptomycin, 0.1% BSA) for further processing. The Research Animal Resource Center Committee of the College of Agricultural and Life Sciences at University of Wisconsin-Madison approved all procedures performed on animals.
2.3. Experiment I
This experiment examined the acute (0.5 h) in vivo regulation of mRNA for FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, JUND, SERPINE1 and CCL2 by PGF2α. On Day 9 after estrus (n=4) or Day 17 of pseudopregnancy (n=4), gilts were anesthetized and one ovary collected (control CL). Following removal of the control ovary, 500 μg of PGF2α (cloprostenol i.m.) was given and the other ovary was collected 0.5 h later (treated CL). Corpora lutea were collected and immediately frozen in liquid nitrogen for later quantitation of specific mRNAs.
2.4. Experiment II
This experiment examined the late (10 h) in vivo regulation of mRNA for FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, JUND, SERPINE1 and CCL2 by PGF2α. Gilts were checked for estrus daily with a mature boar. Animals were randomly assigned to one of four groups: Day 9 saline (n=5), Day 9 PGF2α (n=4), Day 17 saline (n=5) and Day 17 PGF2α (n=5). On Day 9 of the estrous cycle or 17 of pseudopregnancy, gilts received either saline or PGF2α (500 μg of cloprostenol i.m.) and ovaries were surgically removed 10 h later. Corpora lutea were collected and immediately frozen in liquid nitrogen.
2.5. Isolation of mRNA
Total RNA was isolated using the RNAgents total RNA isolation system (Promega, Madison, WI). Briefly, CL were ground in a mortar and pestle cooled with liquid nitrogen. Approximately 40 mg of tissue was transferred to a fresh tube containing 900 μL of denaturing solution (4M Guanidine thiocyanate, 0.01M Tris [pH 7.5], 0.97% β-Mercaptoethanol) and homogenized for 20 sec using a polytron tissue grinder. Ninety μL of 2 M sodium acetate and 900 μL of phenol/chloroform/IAA were added to the lysate and incubated on ice for 15 minutes. Samples were centrifuged for 20 min at 14,000 RPM in a refrigerated microcentrifuge. Supernate was transferred to a fresh tube and RNA was precipitated with an equal volume of isopropanol and incubated at −20°C for 1 h. Samples were then centrifuged at 14,000 RPM for 10 min to pellet RNA and washed with 1 mL 70% ethanol. RNA pellet was dried and resuspended in 30 μL DEPC-treated water and RNA purity and quantity was measured by absorbance at 260/280 nm in a spectrophotometer.
2.6. Quantification of mRNA using RT-PCR and real-time PCR
Evaluation of specific mRNAs was done using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control. Primers for GAPDH, FOS, FOSB, JUNB, FOSL1, CCL2 and JUN were synthesized from published genebank sequences (see Table 1) to produce the expected 285, 599, 129, 387, 377, 308, and 467 bp products. Reverse transcription was carried out with 19 μL of 1X master mix (1X RT buffer, 0.2 mM dNTPs, 100 pmol random primer and 40 U reverse transcriptase) and 1 μL of sample for 1.5 h at 37°C. For PCR, 4 μL of RT reaction were added to 1X PCR master mix (1X thermophilic buffer supplied with enzyme, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.4 μM each of forward and reverse primers and 0.5 U Taq DNA polymerase) in a 20 μL final volume and amplified with 25–30 cycles of PCR (95°C: 30 sec, 55°C: 30 sec and 72°C: 30 sec) followed by a final extension at 72°C for 5 min. Reactions were separated on 5% PAGE gel and stained with ethidium bromide. For each sample, the two products were quantified using the Collage imaging system (Fotodyne, Heartland, WI). Values were calculated as the ratio of the gene-specific band intensity/GAPDH band intensity.
Table 1.
Primer sequences used for PCR. All sequences are 5′ to 3′.
| Gene | Forward | Reverse | Reference |
|---|---|---|---|
| FOS | GGAAAGGAATAAGATGGCTG | AGTCTGCTGCATAGAAGG | AJ132510 |
| FOSB | CCGGGCATGAGTGGCTACAG | CGTCTCCTCTCGGGGTCTCCT | AF120155 |
| FOSL1 | GAGGAGCGCCGCCGAGTAAG | CAGGCTGGGGGTGAAAGGAG | X16707 |
| FOSL2 | CAGCATTGCTGGGGGCTTCTA | TGATTGGTCCCCGCTGCTACT | X16706 |
| JUN | TTCGCGGTCGCTGGTGAGGA | GGGTCGGCGTGGTGGTGATG | S83515 |
| JUNB | CTACACGACTACAAACTCCT | GGTGTCACGTGGTTCATCT | BC009466 |
| JUND | CGTTGGTTGTGTGTGTGTGTG | CAGGAATGTGGACTCGTAGCA | NM_005354 |
| CCL2 | TGAAGGTCTCTGCAGCCCTC | AGTCAGGCTTCAAGGCTTCG | X79416 |
| SERPINE1 | TTGCCCTTGTGTGCTTGTTAG | AAAGAGAGGAGCAATGGGGTT | NM_213910 |
| GAPDH | ATTGCCCTCAACGACCACTT | ACATGACGAGGCAGGTCTCC | X94251 |
| ACTB | CCCAGCACGATGAAGATCAAG | AGAAGCATTTGCGGTGGACGA | AY550069 |
Steady-state concentrations of investigated mRNAs for FOSL2, JUND, and SERPINE1 were quantified by real-time PCR using a GeneAmp® 5700 Sequence Detection System (PE Biosystems, Foster City, CA) with PCR products detected with SYBR Green I (Molecular Probes, Eugene, OR). Primers for amplification were designed using Primer Express (PE Biosystem, Foster, CA) and are listed in Table 1. Each PCR reaction mix (25 μL) contained 1× PCR Buffer (Promega, Madison, WI) with 1:20,000 dilution of SYBR Green I, 1.5 mM MgCl2, 200 μM dNTP, 250 nM forward primer, 250 nM reverse primer, 2 μL RT products, and 1.25 U GoToTaq polymerase (Promega, Madison, WI) [21]. Thermal cycling conditions were 94°C for 30 sec, followed by 40 cycles at 94° C for 30 sec, 57° C for 30 sec, and 72° C for 30 sec, and finally 72° C for 10 min. Melting curve analyses and agarose gel electrophoresis were performed after real-time PCR reactions to monitor PCR product purity.
The threshold cycle (CT) numbers were determined for the amplified cDNA for each investigated mRNA and for the housekeeping gene, ACTB (known as β-ACT), in each unknown sample during real-time PCR. The relative quantification of investigated gene expression was evaluated using a standard curve method [22]. For each sample, the amount of investigated mRNA and the housekeeping mRNA (ACTB) was determined from the standard curve. Then, the amount of investigated mRNA was divided by the amount of ACTB to obtain a normalized mRNA value for each investigated gene.
2.7. Statistical analyses
Results from experiment I (0.5 h) were analyzed by paired t-test. Results for experiment II (10 h) were analyzed by two way analysis of variance (ANOVA) using the general linear model (GLM) procedure of the Statistical Analysis System (SAS). If a positive F-test was detected, means were separated using Fisher’s least significant difference test (LSD). A P value < 0.05 was considered significant.
3. Results
3.1. Experiment I
The mRNA for FOSL1 was not changed by PGF2α treatment in either Day 9 (Control 0.25±0.03; PGF2α 0.31±0.01 band intensity FOSL1 mRNA/GAPDH mRNA) or Day 17 (Control 0.26±0.02; PGF2α 0.25±0.02 band intensity FOSL1 mRNA/GAPDH mRNA) CL. Likewise, levels of FOSL2 mRNA were not changed in either Day 9 (Control = 2.33±1.00; PGF2α = 2.18±0.58; normalized to ACTB mRNA and X 10−3 for all values) or Day 17 (Control = 2.06±0.23; PGF2α = 2.02±0.59; normalized to ACTB mRNA and X 10−3 for all values) CL at 0.5 h after treatment. In contrast to FOSL1 and FOSL2 mRNA, the mRNA for FOS, FOSB, and JUNB were dramatically induced (P < 0.01) at 0.5 h after PGF2α treatment in both Day 9 and Day 17 CL. The most dramatic example was the increase (P < 0.01) in FOS mRNA (Figure 1, P < 0.01) that was observed in both Day 9 (22.3-fold increase) and Day 17 (18.2-fold) CL. The basal concentrations of FOS mRNA were low (0.04±0.02 and 0.06±0.03 for Day 9 and 17, respectively) prior to PGF2α. Likewise, the basal levels of FOSB were low prior to PGF2α treatment (0.05±0.01 and 0.08±0.05 for day 9 an d17, respectively). Treatment with PGF2α acutely induced (P < 0.01) FOSB mRNA in both Day 9 (10.6-fold) and Day 17 (9.3-fold increase) CL (Figure 2). In contrast, the basal levels of JUNB were moderately high under basal conditions (0.51 ± 0.10 and 0.48 ±0.15 for day 9 and 17, respectively). Nevertheless, the concentrations of JUNB mRNA were also increased (P < 0.01) in both Day 9 (2.37-fold) and Day 17 (3.58-fold) CL (Figure 3).
Figure. 1.
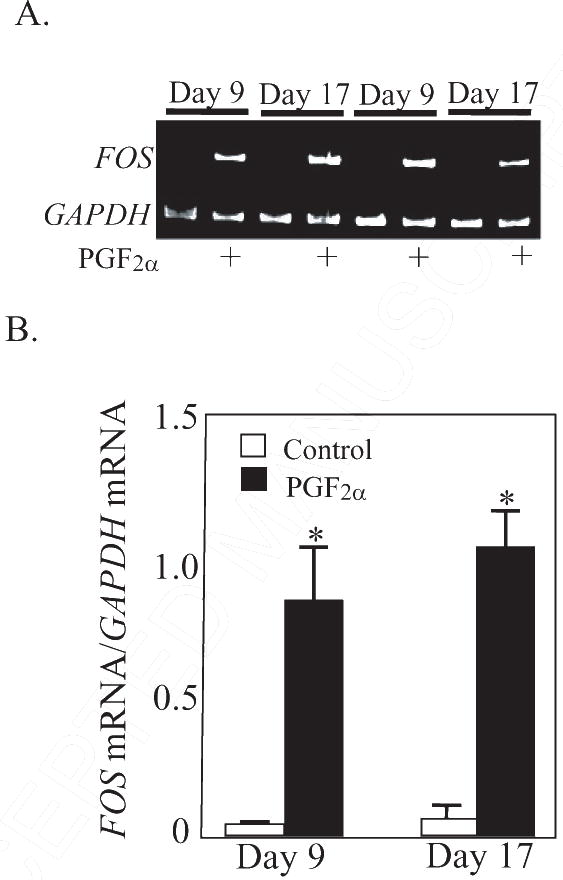
A. Representative gel photos of PCR products for FOS and GAPDH mRNA at 0.5 h after treatment with PGF2α. B. Steady state concentrations of FOS mRNA (ratio of FOS mRNA/GAPDH mRNA) at 0.5 h after PGF2α treatment in CL collected from gilts on Day 9 (n=4/5 per group) and Day 17 (n=5 per group) of pseudopregnancy. *Denotes differences from paired control, P < 0.01.
Figure. 2.
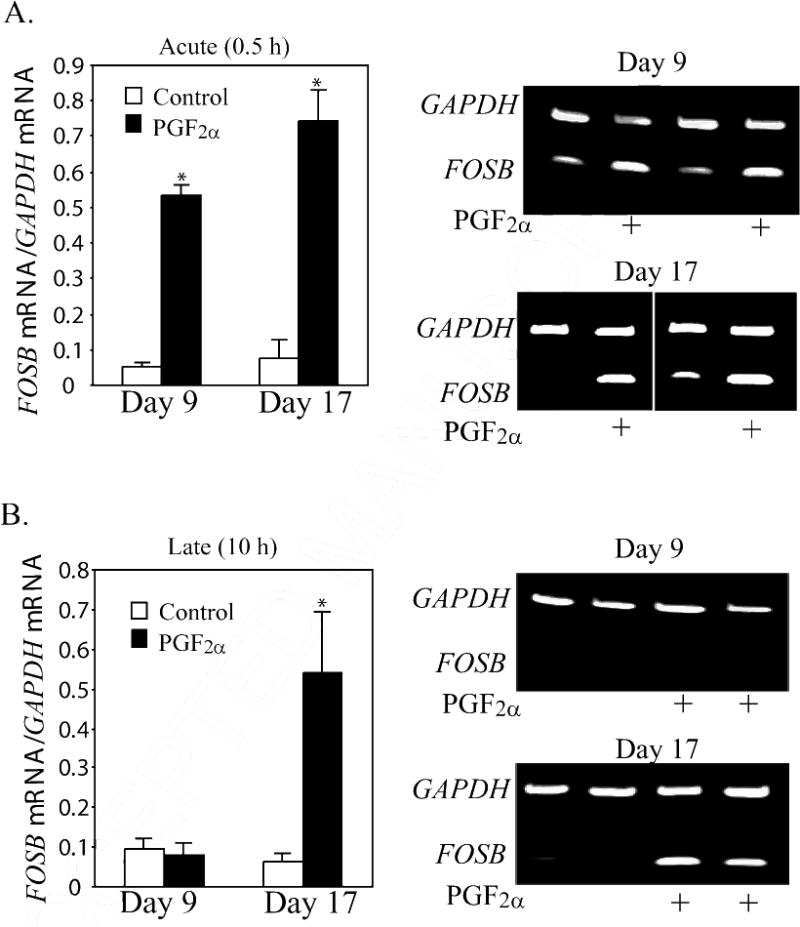
Steady state concentrations of FOSB mRNA (ratio of FOSB mRNA/GAPDH mRNA) at (A) 0.5 h and (B) 10 h after PGF2α treatment in CL from gilts on Day 9 (n=4/5 per group) or Day 17 (n=5 per group). Representative gel photos of PCR products for FOSB and GAPDH mRNA are shown on the right. *Denotes differences from control, P < 0.01.
Figure. 3.
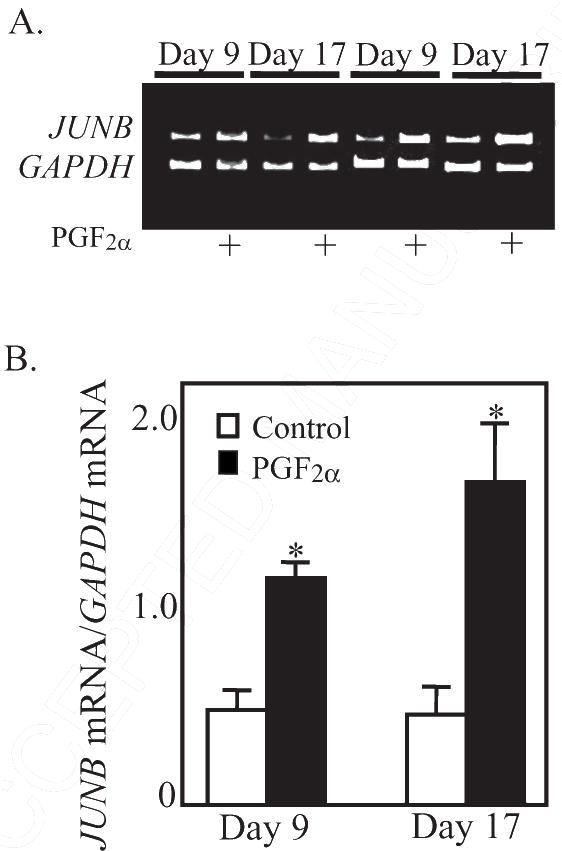
A. Representative gel photos of PCR products for JUNB and GAPDH mRNA. B. Steady state concentrations of JUNB mRNA (ratio of JUNB mRNA/GAPDH mRNA) at 0.5 h after PGF2α treatment in CL from gilts on Day 9 (n=4/5 per group) or Day 17 (n=5 per group).
*Denotes differences from paired control, P < 0.01.
In contrast to FOS, FOSB, and JUNB, there was an intriguing difference in the acute PGF2α-induced expression pattern for JUN and JUND mRNA in CL with or without luteolytic capacity (Figure 4A). Under basal conditions, JUN was at low concentrations (0.02 ± 0.003 – Day 9; 0.02 ± 0.004 – Day 17). The low concentration of JUN mRNA did not change after PGF2α in Day 9 CL (0.04 ± 0.01 – after PGF2α on Day 9). However, JUN mRNA was dramatically increased (P < 0.01) by PGF2α in Day 17 CL (11.0-fold increase). JUND mRNA concentrations were also increased (P < 0.01) by PGF2α in Day 17 CL (~2.5 fold) and not Day 9 CL (Figure 4B)
Figure. 4.
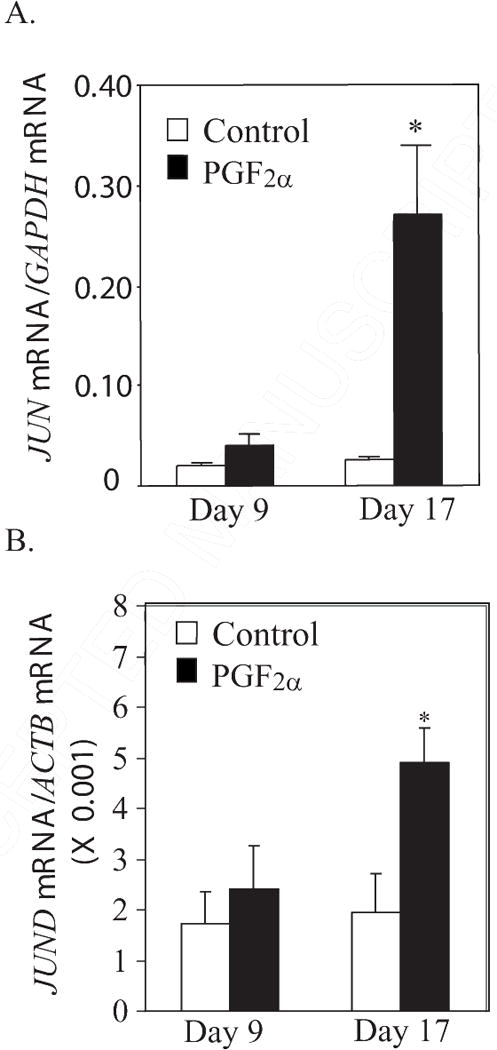
A. Steady state concentrations of JUN mRNA (ratio of JUN mRNA/GAPDH mRNA) at 0.5 h after PGF2α treatment in CL from gilts on Day 9 (n=4/5 per group) or Day 17 (n=5 per group). B. Steady state concentrations of JUND mRNA (normalized to ACTB mRNA) at 0.5 h after PGF2α treatment in CL from gilts on Day 9 (n=4/5 per group) or Day 17 (n=5 per group). *Denotes differences from paired control, P < 0.01.
The mRNAs for two AP-1-responsive genes were also evaluated in this study, CCL2 and SERPINE1. Treatment with PGF2α increased (P < 0.05) mRNA for CCL2 only in Day 17 (2.0-fold), but not Day 9 CL (Figure 5, P < 0.05). However, SERPINE1 mRNA concentrations were not affected by PGF2α in either Day 9 (Control 6.10± 4.09; PGF2α 9.40±4.26) or Day 17 (Control 6.14±4.25; PGF2α 7.52±2.25) CL at 0.5 h after treatment.
Figure. 5.
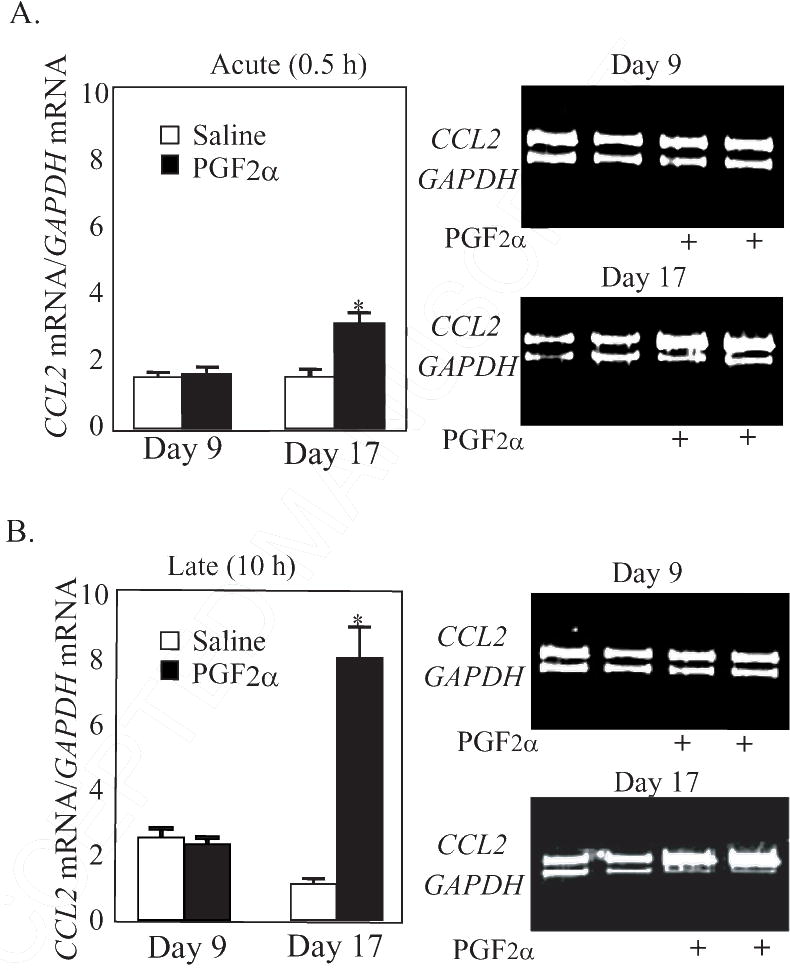
Steady state concentrations of CCL2 mRNA (ratio of CCL2 mRNA/GAPDH mRNA) at (A) 0.5 h and (B) 10 h after PGF2α treatment in CL from gilts on Day 9 (n=4/5 per group) and Day 17 (n=5 per group). Representative gel photos of PCR products for CCL2 and GAPDH mRNA are shown on the right. *Denotes differences from paired control, P < 0.05.
3.2. Experiment II
At 10 h after PGF2α treatment, the mRNA for these AP-1 factors (FOS, JUN, FOSL1, FOSL2, and JUNB) were not different from mRNA concentrations in Day 9 or Day 17 CL from untreated gilts (Table 2). In contrast to the mRNA for these AP-1 members, there was clear differential regulation of FOSB at 10 h after PGF2α treatment (Figure 2). In Day 9 CL, although there had been a dramatic induction of FOSB mRNA at 0.5 h after PGF2α treatment, there was no difference in FOSB mRNA at 10 h after treatment (0.09 ± 0.03 vs. 0.08 ± 0.03). However, in Day 17 CL (Figure 2B), there were much greater (P < 0.01) mRNA concentrations for FOSB in CL treated with PGF2α (10 h after treatment) than in control CL (7.7-fold increase). The mRNA concentrations for JUND were also higher (P < 0.05) in Day 17 than Day 9 CL at 10 h after PGF2α treatment (Table 2).
Table 2.
Effects of on mRNA concentrations for AP-1 factors in Day 9 and Day 17 porcine CL at 10 h after treatment.
| Day 9 | Day 17 | ||||
|---|---|---|---|---|---|
|
| |||||
| mRNA | Saline |
|
Saline | PGF2α | |
| FOS* | 0.09 ± 0.05 | 0.06 ± 0.01 | 0.07 ± 0.03 | 0.12 ± 0.08 | |
| FOSL1* | 0.25 ± 0.01 | 0.27 ± 0.02 | 0.25 ± 0.03 | 0.28 ± 0.02 | |
| FOSL2 (× 10−3)# | 2.14 ± 1.06 | 1.83 ± 0.55 | 1.80 ± 0.27 | 2.67 ± 0.33 | |
| JUN* | 0.02±0.004 | 0.02±0.003 | 0.02±0.004 | 0.05±0.012 | |
| JUNB* | 0.52±0.07 | 0.49±0.03 | 0.62±0.13 | 0.67±0.18 | |
| JUND (× 10−3)# | 1.82 ± 0.81ab | 1.38 ± 0.24a | 2.04 ± 0.55ab | 3.33 ± 0.49b | |
| SERPINE1 (× 10−3)# | 0.70 ± 0.19a | 1.63 ± 0.50a | 0.66 ± 0.30a | 10.0 ± 3.40a | |
Values are mean ± SEM of pixel intensity of specific mRNA/GAPDH mRNA.
Values are mean ± SEM of specific mRNA from real-time PCR/ACTB mRNA.
Groups within a row without common superscripts are different (P < 0.05).
The concentrations of mRNA for the two AP-1-responsive genes were increased (P < 0.01) at 10 h after PGF2α treatment. Expression of CCL2 mRNA was increased 7.2-fold (P < 0.01) at 10 h after PGF2α in Day 17 CL but there was no difference due to PGF2α treatment in Day 9 CL (Figure 5). Similarly, treatment with PGF2α increased (P < 0.01) concentrations of mRNA for SERPINE1 only in Day 17 (15.2-fold) but not Day 9 CL (Table 2).
4. Discussion
This research utilized the porcine CL model which is particularly interesting due to development of luteolytic capacity at a relative late state of luteal development (Day 12–13). However, development of luteolytic capacity occurs so close to the time of natural luteolysis that treatments to evaluate luteolytic capacity can be confounded with the natural luteolytic process [4]. To assure that CL had full luteolytic capacity but had not been exposed to uterine PGF2α, we utilized the pseudopregnant pig model in which treatment with estradiol is used to alter endometrial PGF2α production, similar to what occurs during pregnancy [10].
Acquisition of luteolytic capacity in pigs and other mammals is associated with changes in the PGF2α-induced transcriptional regulation of key genes involved in luteal function. It is logical that key transcriptional factors, such as the AP-1 factors, are not induced in CL without luteolytic capacity and that this would result in lack of changes in gene expression following PGF2α. However, many of the AP-1 transcriptional factors were dramatically induced by PGF2α in both the CL with (Day 17 pseudopregnant) as well as without (Day 9) luteolytic capacity. For example, FOS increased more than 20-fold after PGF2α treatment of the early porcine CL but yet this CL does not undergo regression in response to PGF2α. Similarly, FOSB and JUNB were also dramatically induced following PGF2α treatment. This is the first report of such a dramatic induction of genes following treatment with PGF2α of CL lacking luteolytic capacity and provides clear evidence that PGF2α can produce physiological responses in the early CL. Previous results indicated that receptors for PGF2α were abundantly present in the early CL [5,6] and that some genes were down-regulated by PGF2α in the early CL [3,4]. From our results it is clearly evident that PGF2α activates at least some of the intracellular signal transduction pathways associated with luteolysis in the early CL. Nevertheless, induction of these members of the AP-1 transcription complex appears to be insufficient to induce the luteolytic cascade. Thus, an intriguing paradox is that many of the critical genes associated with luteolysis are not activated by PGF2α in the early CL in spite of a clear activation of many intracellular regulators, such as FOS, FOSB, and JUNB.
The present study provides evidence that an early step in this differential regulation of gene expression may involve induction of the key AP-1 transcription factors, JUN and JUND. There was a dramatic difference in PGF2α-induction of JUN and JUND mRNA concentrations with this induction only occurring in CL with luteolytic capacity. In addition, there was an unexpected difference in FOSB expression at 10 h after PGF2α treatment with PGF2α induction of FOSB only in CL with luteolytic capacity. Differences in mRNA expression of AP-1 factors may translate into differences in expression of a variety of luteolysis-related gene products such as the AP-1-responsive genes, CCL2 and SERPINE1, which were induced by PGF2α only in Day 17 CL with luteolytic capacity. Nevertheless, a key limitation was that only mRNA was evaluated in this study. Obviously, this experimental approach can introduce the concept that regulation of AP-1 proteins, and particularly members of the jun family, could be key to molecular regulation of luteal resistance, however analysis of proteins and functional assays of AP-1 regulated transcription will be required to validate or invalidate the importance of these pathways. Transcriptional regulation and DNA binding studies were beyond the scope of the present study.
The fundamental role of an increase in transcription of fos and jun family members in activation of the AP-1 transcriptional complex has been extensively demonstrated in previous studies [12]. It is also clear that both fos and jun family members must be present to allow transcriptionally competent heterodimers to form [11,12,23]. Of particular interest to our research findings, upregulation of JUN activity has a critical role in induction of AP-1 transcriptional activity [24]. All of the Jun proteins have a DNA binding domain and a basic leucine-zipper region that allows heterodimerization with fos family members [12,24]. However, JUN is unique in having an N-terminal trans-activation domain whereas, JUNB and JUND exhibit only weak trans-activation activity [24]. The unexpected finding in our research of a lack of induction of JUN mRNA in CL without luteolytic capacity, although other elements of the AP-1 transcriptional complex have been induced, provides strong associative data for a key role of this protein in luteolytic capacity. Thus, given the potential importance of AP-1 activation in inducing the cellular pathways involved in luteolysis, the lack of JUN induction in the early CL may be crucial to preventing specific AP-1 mediated transcriptional events in the early CL.
Induction or lack of induction of JUN has been found to be crucial for other types of cellular regression [11,23,25]. For example, overexpression of JUN induces cell death in vitro [26]. Conversely, removal of JUN by either expression of a dominant negative form of JUN [27] or specific inactivation of JUN in the central nervous system using a cre/lox system [28] delayed or completely abolished injury-induced neuronal cell death. These results highlight the central role of JUN in normal cell death mechanisms and are consistent with the idea that JUN expression may be fundamental to initiation of the luteolysis process.
A number of other AP-1 proteins, in addition to JUN, also seem to be critical in cell death [11,23]. For example, evidence is accumulating that an AP-1 response element is critical for induction of FasL-mediated cell death in T-cells [29] and hepatocellular carcinoma cells [30]. Luteolysis is an apoptotic process [31] with Fas/FasL implicated in luteolysis-related cell death in rat [32] and bovine [33] CL. Thus, it seems likely that AP-1 also has a critical role in the FasL-mediated cell death associated with luteolysis. In particular, FOSB was found to be critical for the induction of cell death in T-cells [29]. Our results are consistent with these findings. Expression of FOSB remained elevated at 10 h after PGF2α treatment of Day 17 CL when the initial stages of luteal cell death would be observed (between 6–12 h after cloprostenol; [31]). Thus, specific and acute induction of JUN as well as sustained induction of FOSB is clearly associated with cellular capacity to respond to PGF2α with complete regression of the CL, or luteolytic capacity.
Unfortunately, the results of this in vivo study do not allow determination of the mechanisms that underlie the differential expression of AP-1 transcriptional factors in CL with or without luteolytic capacity. Given the vital role of P4 in luteal function [34–36], it is possible that high intraluteal concentrations of P4 could regulate induction of AP-1 proteins. Supplementation of pregnant rats with P4 blocks parturition and blocks the normal induction of FOS, FOSB, FOSL1, FOSL2, and JUNB mRNA occurring in the uterus at parturition [37]. However, a number of AP-1 proteins (FOS, FOSB, JUNB) were induced by PGF2α in the presence of high P4 in either Day 9 or Day 17 CL. In addition, JUN and JUND were induced in the Day 17 CL (Figure 4) in spite of high intraluteal P4 concentrations [4]. It seems possible that the sustained induction of FOSB could be related to lowered P4 because intraluteal P4 had dramatically decreased by 10 h after PGF2α treatment in Day 17 but not Day 9 CL [4]. However, the induction of JUN and JUND at only 0.5 h after PGF2α was not associated with changes in intraluteal P4 and is much more likely to be related to differences in factors present in the DNA promoter/enhancer region of JUN and JUND in Day 9 compared to Day 17 CL. The lack of sustained elevation in mRNA for FOS, JUN and JUNB at 10 h after PGF2α is probably due to autoregulatory mechanisms designed to rapidly terminate AP-1 signaling [38,39]. In the case of JUN, we speculate that once the luteolytic cascade is initiated continued induction of JUN is unnecessary. In vitro evaluations are likely to be required in order to elucidate the molecular mechanisms that underlie the switch during acquisition of luteolytic capacity that produces JUN and JUND responsiveness to PGF2α treatment.
We also chose to evaluate the effect of luteolytic capacity on 2 AP-1 regulated genes, one that has previously been evaluated in the CL, CCL2, and the other that has not been previously examined, SERPINE1. Luteolysis has clearly been associated with induction of CCL2 in many species [9,40,41]. In the bovine CL, CCL2 is induced only after acquisition of luteolytic capacity [9]. Likewise, we observed a similar response in the porcine CL with a dramatic and rapid increase in CCL2 mRNA only in CL with luteolytic capacity. Expression of CCL2 is regulated by AP-1 proteins in many cell types [42] including luteal cells [16]. A simplified scenario could be that lack of AP-1 activation due to lack of JUN induction leads to lack of CCL2 induction in CL without luteolytic capacity. A lack of CCL2 induction in the early CL could prevent premature recruitment and activation of immune cells in the CL and subsequent luteolysis. Similarly, the serine protease inhibitor E1 (SERPINE1) is regulated by AP-1 promoter activity [43]. This gene was induced only in CL with (Day 17) but not without (Day 9) luteolytic capacity. SERPINE1 may be a critical regulator of the tissue remodeling occurring during structural degradation of the CL by modulating the activity of tissue plasminogen activator [44,45].
In addition, two of the genes that were previously found to be differentially regulated in CL with versus without luteolytic capacity are also clearly regulated by AP-1 promoters. The AP-1 proteins are transcriptional repressors of STAR gene [19,20]. In addition, CYP19A1 is induced by binding of AP-1 proteins to the aromatase gene promoter [18]. These 2 genes appear to only be regulated by PGF2α in CL with luteolytic capacity. The mRNA and protein for STAR is decreased only in CL with luteolytic capacity [7]. In addition, PGF2α specifically induces expression of CYP19A1 mRNA only in CL with luteolytic capacity [10]. Thus, pathways that allow induction of AP-1 transcriptional pathways by PGF2α, most likely due to PGF2α-induced expression of JUN and/or JUND mRNA, may have a fundamental role in development of the cellular pathways associated with luteolysis.
In summary, this study has used an in vivo model to clearly demonstrate that PGF2α acutely and dramatically induces the mRNA expression of specific luteolysis-related AP-1 transcription factors, such as FOS, FOSB, and JUNB, even in CL that will not undergo luteolysis. However, the lack of acute induction of mRNA for JUN and JUND, and possibly sustained FOSB mRNA expression, may prevent AP-1-mediated gene expression events and may represent a physiologically important functional lesion underlying luteolytic capacity.
Acknowledgments
This research was supported by NIH HD-50616, USDA-00-35203-9134, and an Advanced Opportunity Fellowship to F.J. Diaz. The authors thank Tom Crenshaw and Terry Jobsis for help with obtaining and caring for the gilts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niswender GD, Juengel JL, Mcguire WJ, Belfiore CJ, Wiltbank MC. Luteal Function -the Estrous-Cycle and Early-Pregnancy. Biol Reprod. 1994;50:239–247. doi: 10.1095/biolreprod50.2.239. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie HD, Polge C. Luteal function and oestrus in gilts treated with a synthetic analogue of prostaglandin F-2alpha (ICI 79,939) at various times during the oestrous cycle. J Reprod Fertil. 1976;48:423–425. doi: 10.1530/jrf.0.0480423. [DOI] [PubMed] [Google Scholar]
- 3.Tsai SJ, Wiltbank MC. Prostaglandin F2alpha regulates distinct physiological changes in early and mid-cycle bovine corpora lutea. Biol Reprod. 1998;58:346–352. doi: 10.1095/biolreprod58.2.346. [DOI] [PubMed] [Google Scholar]
- 4.Diaz FJ, Crenshaw TD, Wiltbank MC. Prostaglandin f(2alpha) induces distinct physiological responses in porcine corpora lutea after acquisition of luteolytic capacity. Biol Reprod. 2000;63:1504–1512. doi: 10.1095/biolreprod63.5.1504. [DOI] [PubMed] [Google Scholar]
- 5.Wiltbank MC, Shiao TF, Bergfelt DR, Ginther OJ. Prostaglandin F2 alpha receptors in the early bovine corpus luteum. Biol Reprod. 1995;52:74–78. doi: 10.1095/biolreprod52.1.74. [DOI] [PubMed] [Google Scholar]
- 6.Gadsby JE, Balapure AK, Britt JH, Fitz TA. Prostaglandin F2 alpha receptors on enzyme-dissociated pig luteal cells throughout the estrous cycle. Endocrinology. 1990;126:787–795. doi: 10.1210/endo-126-2-787. [DOI] [PubMed] [Google Scholar]
- 7.Diaz FJ, Wiltbank MC. Acquisition of luteolytic capacity involves differential regulation by prostaglandin F2alpha of genes involved in progesterone biosynthesis in the porcine corpus luteum. Domest Anim Endocrinol. 2005;28:172–189. doi: 10.1016/j.domaniend.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Levy N, Kobayashi S, Roth Z, Wolfenson D, Miyamoto A, Meidan R. Administration of prostaglandin f(2 alpha) during the early bovine luteal phase does not alter the expression of ET-1 and of its type A receptor: a possible cause for corpus luteum refractoriness. Biol Reprod. 2000;63:377–382. doi: 10.1095/biolreprod63.2.377. [DOI] [PubMed] [Google Scholar]
- 9.Tsai SJ, Juengel JL, Wiltbank MC. Hormonal regulation of monocyte chemoattractant protein-1 messenger ribonucleic acid expression in corpora lutea. Endocrinology. 1997;138:4517–4520. doi: 10.1210/endo.138.10.5577. [DOI] [PubMed] [Google Scholar]
- 10.Diaz FJ, Wiltbank MC. Acquisition of luteolytic capacity: changes in prostaglandin F2alpha regulation of steroid hormone receptors and estradiol biosynthesis in pig corpora lutea. Biol Reprod. 2004;70:1333–1339. doi: 10.1095/biolreprod.103.020461. [DOI] [PubMed] [Google Scholar]
- 11.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 12.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 13.Rusovici R, LaVoie HA. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol Reprod. 2003;69:64–74. doi: 10.1095/biolreprod.102.013995. [DOI] [PubMed] [Google Scholar]
- 14.Burne TH, Murfitt PJ, Gilbert CL. c-fos mRNA expression associated with PGF(2alpha)-induced nest-building behaviour in female pigs. Brain Res Mol Brain Res. 2002;104:31–37. doi: 10.1016/s0169-328x(02)00199-7. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Fong HW, Davis JS. Induction of c-fos and c-jun messenger ribonucleic acid expression by prostaglandin F2alpha is mediated by a protein kinase C-dependent extracellular signal-regulated kinase mitogen-activated protein kinase pathway in bovine luteal cells. Endocrinology. 2001;142:887–895. doi: 10.1210/endo.142.2.7938. [DOI] [PubMed] [Google Scholar]
- 16.Nagaosa K, Aikoshi I, Hasegawa Y, Nakanishi Y. Activator protein 1-mediated expression of monocyte chemoattractant protein 1 in cultured rat luteal cells. Mol Reprod Dev. 2008;75:1077–1084. doi: 10.1002/mrd.20849. [DOI] [PubMed] [Google Scholar]
- 17.Hu E, Mueller E, Oliviero S, Papaioannou VE, Johnson R, Spiegelman BM. Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J. 1994;13:3094–3103. doi: 10.1002/j.1460-2075.1994.tb06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Nichols JE, Valdez R, Mendelson CR, Simpson ER. Tumor necrosis factor-alpha stimulates aromatase gene expression in human adipose stromal cells through use of an activating protein-1 binding site upstream of promoter 1.4. Mol Endocrinol. 1996;10:1350–1357. doi: 10.1210/mend.10.11.8923461. [DOI] [PubMed] [Google Scholar]
- 19.Shea-Eaton W, Sandhoff TW, Lopez D, Hales DB, McLean MP. Transcriptional repression of the rat steroidogenic acute regulatory (StAR) protein gene by the AP-1 family member c-Fos. Mol Cell Endocrinol. 2002;188:161–170. doi: 10.1016/s0303-7207(01)00715-8. [DOI] [PubMed] [Google Scholar]
- 20.Manna PR, Eubank DW, Stocco DM. Assessment of the role of activator protein-1 on transcription of the mouse steroidogenic acute regulatory protein gene. Mol Endocrinol. 2004;18:558–573. doi: 10.1210/me.2003-0223. [DOI] [PubMed] [Google Scholar]
- 21.Luo W, Wiltbank MC. Distinct regulation by steroids of messenger RNAs for FSHR and CYP19A1 in bovine granulosa cells. Biol Reprod. 2006;75:217–225. doi: 10.1095/biolreprod.105.047407. [DOI] [PubMed] [Google Scholar]
- 22.Robert C, McGraw S, Massicotte L, Pravetoni M, Gandolfi F, Sirard MA. Quantification of housekeeping transcript levels during the development of bovine preimplantation embryos. Biol Reprod. 2002;67:1465–1472. doi: 10.1095/biolreprod.102.006320. [DOI] [PubMed] [Google Scholar]
- 23.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 24.Raivich G, Behrens A. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 2006;78:347–363. doi: 10.1016/j.pneurobio.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N, Verna L, Hardy S, Zhu Y, Ma KS, Birrer MJ, Stemerman MB. c-Jun triggers apoptosis in human vascular endothelial cells. Circ Res. 1999B;85:387–393. doi: 10.1161/01.res.85.5.387. [DOI] [PubMed] [Google Scholar]
- 27.Crocker SJ, Lamba WR, Smith PD, Callaghan SM, Slack RS, Anisman H, Park DS. c-Jun mediates axotomy-induced dopamine neuron death in vivo. Proc Natl Acad Sci U S A. 2001;98:13385–13390. doi: 10.1073/pnas.231177098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, Lipp HP, Aguzzi A, Wagner EF, Behrens A. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Baumann S, Hess J, Eichhorst ST, Krueger A, Angel P, Krammer PH, Kirchhoff S. An unexpected role for FosB in activation-induced cell death of T cells. Oncogene. 2003;22:1333–1339. doi: 10.1038/sj.onc.1206126. [DOI] [PubMed] [Google Scholar]
- 30.Eichhorst ST, Muller M, Li-Weber M, Schulze-Bergkamen H, Angel P, Krammer PH. A novel AP-1 element in the CD95 ligand promoter is required for induction of apoptosis in hepatocellular carcinoma cells upon treatment with anticancer drugs. Mol Cell Biol. 2000;20:7826–7837. doi: 10.1128/mcb.20.20.7826-7837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bacci ML, Barazzoni AM, Forni M, Costerbosa GL. In situ detection of apoptosis in regressing corpus luteum of pregnant sow: evidence of an early presence of DNA fragmentation. Domest Anim Endocrinol. 1996;13:361–372. doi: 10.1016/0739-7240(96)00049-5. [DOI] [PubMed] [Google Scholar]
- 32.Kuranaga E, Kanuka H, Furuhata Y, Yonezawa T, Suzuki M, Nishihara M, Takahashi M. Requirement of the Fas ligand-expressing luteal immune cells for regression of corpus luteum. FEBS Lett. 2000A;472:137–142. doi: 10.1016/s0014-5793(00)01426-5. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi H, Yokomizo Y, Okuda K. Fas-Fas ligand system mediates luteal cell death in bovine corpus luteum. Biol Reprod. 2002;66:754–759. doi: 10.1095/biolreprod66.3.754. [DOI] [PubMed] [Google Scholar]
- 34.Duffy DM, Hess DL, Stouffer RL. Acute administration of a 3 beta-hydroxysteroid dehydrogenase inhibitor to rhesus monkeys at the midluteal phase of the menstrual cycle: evidence for possible autocrine regulation of the primate corpus luteum by progesterone. J Clin Endocrinol Metab. 1994;79:1587–1594. doi: 10.1210/jcem.79.6.7989460. [DOI] [PubMed] [Google Scholar]
- 35.Rueda BR, Hendry IR, Hendry IW, Stormshak F, Slayden OD, Davis JS. Decreased progesterone levels and progesterone receptor antagonists promote apoptotic cell death in bovine luteal cells. Biol Reprod. 2000;62:269–276. doi: 10.1095/biolreprod62.2.269. [DOI] [PubMed] [Google Scholar]
- 36.Kuranaga E, Kanuka H, Hirabayashi K, Suzuki M, Nishihara M, Takahashi M. Progesterone is a cell death suppressor that downregulates Fas expression in rat corpus luteum. FEBS Lett. 2000B;466:279–282. doi: 10.1016/s0014-5793(00)01090-5. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell JA, Lye SJ. Differential expression of activator protein-1 transcription factors in pregnant rat myometrium. Biol Reprod. 2002;67:240–246. doi: 10.1095/biolreprod67.1.240. [DOI] [PubMed] [Google Scholar]
- 38.Schonthal A, Buscher M, Angel P, Rahmsdorf HJ, Ponta H, Hattori K, Chiu R, Karin M, Herrlich P. The Fos and Jun/AP-1 proteins are involved in the downregulation of Fos transcription. Oncogene. 1989;4:629–636. [PubMed] [Google Scholar]
- 39.Konig H, Ponta H, Rahmsdorf U, Buscher M, Schonthal A, Rahmsdorf HJ, Herrlich P. Autoregulation of fos: the dyad symmetry element as the major target of repression. Embo J. 1989;8:2559–2566. doi: 10.1002/j.1460-2075.1989.tb08394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petroff MG, Petroff BK, Pate JL. Mechanisms of cytokine-induced death of cultured bovine luteal cells. Reproduction. 2001;121:753–760. [PubMed] [Google Scholar]
- 41.Bowen JM, Keyes PL, Warren JS, Townson DH. Prolactin-induced regression of the rat corpus luteum: expression of monocyte chemoattractant protein-1 and invasion of macrophages. Biol Reprod. 1996;54:1120–1127. doi: 10.1095/biolreprod54.5.1120. [DOI] [PubMed] [Google Scholar]
- 42.Finzer P, Soto U, Delius H, Patzelt A, Coy JF, Poustka A, zur Hausen H, Rosl F. Differential transcriptional regulation of the monocyte-chemoattractant protein-1 (MCP-1) gene in tumorigenic and non-tumorigenic HPV 18 positive cells: the role of the chromatin structure and AP-1 composition. Oncogene. 2000;19:3235–3244. doi: 10.1038/sj.onc.1203643. [DOI] [PubMed] [Google Scholar]
- 43.Guo B, Inoki K, Isono M, Mori H, Kanasaki K, Sugimoto T, Akiba S, Sato T, Yang B, Kikkawa R, Kashiwagi A, Haneda M, Koya D. MAPK/AP-1-dependent regulation of PAI-1 gene expression by TGF-beta in rat mesangial cells. Kidney Int. 2005;68:972–984. doi: 10.1111/j.1523-1755.2005.00491.x. [DOI] [PubMed] [Google Scholar]
- 44.Smith MF, McIntush EW, Ricke WA, Kojima FN, Smith GW. Regulation of ovarian extracellular matrix remodelling by metalloproteinases and their tissue inhibitors: effects on follicular development, ovulation and luteal function. J Reprod Fertil Suppl. 1999;54:367–381. [PubMed] [Google Scholar]
- 45.Curry TE, Jr, Osteen KG. The Matrix Metalloproteinase System: Changes, Regulation, and Impact throughout the Ovarian and Uterine Reproductive Cycle. Endocr Rev. 2003;24:428–465. doi: 10.1210/er.2002-0005. [DOI] [PubMed] [Google Scholar]


