Abstract
A lack of analytically robust and multiplexed assays has hampered studies of the large, branched phosphosignaling network responsive to DNA damage. To address this need, we developed and fully analytically characterized a 62-plex assay quantifying protein expression and post-translational modification (phosphorylation and ubiquitination) after induction of DNA damage. The linear range was over 3 orders of magnitude, the median inter-assay variability was 10% CV and the vast majority (~85%) of assays were stable after extended storage. The multiplexed assay was applied in proof-of-principle studies to quantify signaling after exposure to genotoxic stress (ionizing radiation and 4-nitroquinoline 1-oxide) in immortalized cell lines and primary human cells. The effects of genomic variants and pharmacologic kinase inhibition (ATM/ATR) were profiled using the assay. This study demonstrates the utility of a quantitative multiplexed assay for studying cellular signaling dynamics, and the potential application to studies on inter-individual variation in the radiation response.
INTRODUCTION
The DNA damage response (DDR) is mediated by cellular signaling through post-translational modification of proteins. A large phosphosignaling network (1), dependent on the ATM and ATR kinases, is critical for mediating the DDR, and phosphorylation is used to modulate protein activity, protein interactions and sub-cellular localization (2). Disruptions in the DDR signaling network affect human health, as they contribute to radiation sensitivity, cancer risk and cancer treatment responses, and they are associated with immunological and neurological disorders (3–6). Thus, functional assays for quantifying DDR proteins and phosphosignaling in response to DNA damage are valuable tools both for basic mechanistic studies as well as for the potential translation of basic science findings to clinical and population studies (7).
Functional studies of phosphosignaling networks have been hampered by a reliance on conventional immunoassay platforms (Western blot, IHC) that are semiquantitative at best and often suffer from poor specificity due to matrix interferences that can vary from one biospecimen to the next (8–11). Due to a lack of standardization, harmonization of results across laboratories is difficult using existing technologies, hindering reproducibility of results and transferability of methods (12). Furthermore, these approaches typically target one analyte at a time, which is less than ideal for studying the behavior of a complex and robust signaling network such as the DDR. Several new technologies have potential to address this issue. Technologies such as planar (13, 14) or bead-based (15) protein arrays and mass cytometry (16) have improved multiplexability and throughput of analysis. However, like all traditional antibody-based approaches, they rely on the most highly specific and validated antibodies for success, making generation of new assays costly, difficult and time consuming. Other promising approaches, like peptide arrays (e.g., kinome analysis) (17, 18), have demonstrated success in profiling kinase activity in a variety of systems and conditions. While this approach is powerful, the specificity of the interaction of a particular kinase and the peptide substrates can vary and the efficiency of phosphorylation of the spotted sequences is difficult to characterize, making this approach most useful for generating hypotheses for quantitative validation using other approaches.
To address this technological gap, the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC; assay portal: https://assays.cancer.gov/) has advanced a Nextgen platform for quantifying proteins (19–23), based on a targeted form of mass spectrometry (MS) called multiple reaction monitoring (MRM). While MRM is based on MS, it is completely different from MS, which is widely applied in discovery experiments to profile the proteome in an untargeted fashion. In contrast, MRM is a targeted mode of MS in which the full analytical capacity of the instrument is selectively focused on a set of specific analytes of biological interest, such as a panel of DDR proteins and phosphoproteins, enhancing the sensitivity of detection over untargeted modes of MS. Prior to MRM analysis, biospecimens are proteolyzed (typically with trypsin) to release peptide analytes that are then targeted for detection by MRM. Several peptides per protein may be targeted for MRM, enabling the distinction of different proteoforms. In quantitative measurements, stable isotope-labeled (“heavy”) synthetic versions of analyte peptides are spiked into each biospecimen to serve as internal standards, enabling standardization and transfer of assays among laboratories (24–26). Indeed, the NCI has developed an open-source CPTAC assay portal to distribute highly characterized, targeted MS assays to enable standardization of proteomic measurements across the research community (20).
While several thousand proteins can be quantified from neat mammalian cell lysates by MRM (26, 27), quantification of low abundance analytes and post-translational modifications, such as phosphorylation, usually requires an enrichment step. One robust method for enriching target peptide analytes is immunoaffinity precipitation using antipeptide antibodies (28–31). For these “immuno-MRM” assays, anti-peptide antibodies are used to capture the target analyte(s) as well as the spiked-in, isotopically-labeled internal standard(s) from the proteolyzed biospecimens. The antibody-analyte conjugates are recovered on magnetic beads, the peptides are eluted off the antibodies and the eluate is subjected to MRM analysis.
MRM assays have analytical advantages over conventional immunoassays. The presence of an internal standard, spiked-in at a known concentration for every peptide analyte, enables precise, relative quantification of the endogenous peptide in a highly standardized manner in which results can be harmonized across laboratories (26, 32). Furthermore, because the mass spectrometer is used as the detector, the specificity of MRM-based assays is exquisite, and is much higher than the specificity of conventional immunoassays that use surrogate signals (e.g., fluorescent, mass or enzymatic tags) to infer detection of the target analyte. The high specificity of the assays, coupled with the relatively large dynamic range of the mass spectrometer, readily enables multiplexing of MRM-based assays (26, 30, 33), which is very challenging on conventional platforms.
We recently reported the first demonstration of the feasibility of using multiplex MRM-based assays to quantify phosphosignaling, using immobilized metal affinity chromatography or polyclonal antibodies (34, 35). Herein, we describe the generation of a new panel of monoclonal antibodies to DDR proteins and configuration of a new 62-plex immuno-MRM assay for quantifying cellular phosphosignaling in response to DNA damage. The immuno-MRM assay is fully analytically characterized using fit-for-purpose method validation. To demonstrate the utility of the assay in profiling the DDR, proof-of-principle experiments are presented showing the applicability of the immuno-MRM panel in cell lines and primary human cells. We profile the effects of genomic mutations and/or pharmacologic inhibition on the ATM/ATR kinases using the assay. In addition, the potential for quantifying the person-to-person variation of DNA repair capacity is demonstrated using primary human cells exposed to ionizing radiation. The monoclonal antibodies used in the assay were further characterized for use in Western blotting, and all assays and 41 novel monoclonal antibodies have been made publically available as a resource to the community through the open-source CPTAC assay and antibody portal (https://proteomics.cancer.gov/antibody-portal).
MATERIALS AND METHODS
Materials and Reagents
Urea (cat. no. U0631), Trizma® base (cat. no. T2694), citric acid (cat. no. C0706), 4-nitroquinilone-1-oxide (4NQO, cat. no. N8141), dimethyl sulfoxide (DMSO, cat. no. D2438) and iodoacetamide (IAM, cat. no. A3221) were obtained from Sigma-Aldrich® (St. Louis, MO). Acetonitrile (cat. no. A955) and water (cat. no. W6, LCMS Optima® grade), tris(2-carboxyethyl)phosphine (TCEP, cat. no. 77720), phosphate buffered saline (PBS, cat. no. BP-399-20) and (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) (CHAPS, cat. no. 28300) detergent were obtained from Thermo Fisher Scientific™ (Waltham, MA). Formic acid (cat. no. 1.11670.1000) was obtained from EMD Millipore (Billerica, MA). Sequencing grade trypsin (cat. no. V5111) used for digestion of samples was obtained from Promega, Inc. (Madison WI). The ATM kinase inhibitor KU-55933 (cat. no. S1092) and the ATR kinase inhibitor AZD6738 (cat. no. S7693) were purchased from Selleckchem (Houston, TX) and dissolved at 10 mM in DMSO. Rabbit monoclonal antibodies were produced with Abcam® (Burlingame, CA). Light (unlabeled) synthetic peptides were obtained from Abcam and New England Peptide (NEP, Gardner, MA) as >95% purified by HPLC or passed through solid phase extraction. Stable isotope-labeled (heavy) peptides from NEP were purified >95% by HPLC, labeled with [13C and 15N] at the C-terminal Arg or Lys, and quantified by amino acid analysis. Aliquots were stored in 3% acetonitrile/0.1% formic acid at −80°C until use.
Cells and Culture Conditions
For assay characterization and proof-of-concept studies, the human lymphoblast cell lines (LCLs) GM07057 and GM01526 (Coriell Institute, Camden, NJ) and HeLa cell line (cat. no. CCL-2; ATCC®, Manassas, VA) were used. HeLa cell identity was authenticated by short tandem repeat (STR) profiles compared against the ATCC standard. For LCLs, we generated STR profiles for each of the cell lines. GM07057 allele profiles are Amelogenin: X, Y; CSF1PO: 11, 14; D13S317: 13, 14; D16S539: 10, 12; D18S51: 10, 13; D19S433: 13, 14.2; D21S11: 28, 30; D2S1338: 19, 24; D3S1358: 15, 17; D5S818: 10, 12; D7S820: 10, 12; D8S1179: 12, 13; FGA: 19, ; TH01: 6, 9.3; TPOX: 8, ; vWA: 16, 17. GM01526 allele profiles are Amelogenin: X; CSF1PO: 11, 12; D13S317: 12, ; D16S539: 12, ; D18S51: 13, 16; D19S433: 13, 15; D21S11: 28, 30; D2S1338: 17, 24; D3S1358: 12, 18; D5S818: 8, 11; D7S820: 11, 12; D8S1179: 11, 15; FGA: 22, 24; TH01: 7, 9.3; TPOX: 8, 10; vWA: 16, 18.
LCLs were grown in RPMI 1640 (cat. no. 11875-093; Gibco®, Grand Island, NY) plus 15% heat-inactivated FBS (cat. no. SH30071.03; HyClone™ Laboratories, Logan, UT) and 1% penicillin-streptomycin (cat. no. 15140-122; Gibco). HeLa were cultured in Eagle’s minimum essential medium (cat. no. 10370; Invitrogen™, Carlsbad, CA) supplemented with 10% heat-inactivated FBS (cat. no. SH30071.03HI; Hyclone), 1 mM sodium pyruvate (cat. no. 11360; Invitrogen), 2 mM l-glutamine (cat. no. 25030; Invitrogen), and 100 units/ml of penicillin-streptomycin (cat. no. 15140; Invitrogen,). LCLs were collected by centrifugation and diluted to one-million cells/ml in fresh growth media for 36 h prior to irradiation or 4-nitroquinilone-1-oxide (4NQO) treatment.
For peripheral blood mononuclear cells (PBMCs), whole blood was collected from healthy adult females with IRB approval (no. 8233; Fred Hutchinson Cancer Research Center and University of Washington) by venipuncture into K2EDTA tubes and maintained at ambient temperature during transport to the laboratory. PBMCs were isolated using Histopaque®-1077 (cat. no. 10771; Sigma-Aldrich) density gradient. Briefly, whole blood was diluted with an equal volume of PBS, layered over a one-third volume of Histopaque (density = 1.077 g/ml) at room temperature and centrifuged at 400g for 30 min with no brake. PBMCs were harvested from the Histopaque-plasma interface, washed twice in PBS and treated once with red blood cell lysis buffer (5 Prime). The T-cell population was activated and expanded using a 1:1 cell-to-bead ratio of CD3/CD28 Dynabeads™ (cat. no. 111-31D; Invitrogen). Culturing was done in Advanced RPMI 1640 (cat. no. 12633012; Gibco) supplemented with 10% heat-inactivated FBS (cat. no. SH30071.03HI; Hyclone), 100 units/ml of penicillin, 100 units/ml streptomycin (cat. no. 15070-063; Gibco), 2 mM l-glutamine (cat. no. 25030-081; Gibco), 100 units/ml IL2 (cat. no. PHC0027; Life Technologies, Grand Island, NY). Cells were cultured for 9 days at 37°C and 5% CO2 with fresh growth media and IL2 added every 2–3 days. On day 7, cells were collected by centrifugation and resuspended in fresh growth media at 1.25 × 106 cells/ml in T75 flasks and allowed to equilibrate at 37°C, 5% CO2 for 36–40 h before irradiation.
Induction of DNA Damage and Inhibition of DNA Damage Checkpoint Kinases
Irradiation was performed using a JL Shepherd Mark I irradiator with a 137Cs source delivering a dose rate of 4.7 Gy/min; mock-irradiated cells were handled in precisely the same manner as the irradiated cells, but the irradiator was not turned on. 4NQO was diluted in growth media just prior to addition to cell cultures at a final concentration of 2.5 µM. For inhibitor studies, LCLs were treated for 1 h prior to DNA damage by the addition of 1 µl of 10 mM KU-55933 per ml of culture media (final concentration 10 µM) or 1 µl of 10 mM of AZD6738 per ml of culture media (final concentration 10 µM). For controls in both kinase inhibitors, mock-treated cells received 1 µl DMSO (vehicle) per ml of culture media.
Sample Preparation
All samples were prepared and processed in a blinded fashion. Where indicated, biological replicates were performed by independent cultures processed on separate days. Cells were lysed at 5 × 107 cells/ml in freshly prepared ice-cold urea lysis buffer [6 M urea, 25 mM Tris (pH8.0), 1 mM EDTA, 1 mM EGTA containing protease and phosphatase inhibitors (cat. nos. P0044, P5726 and P8340; Sigma-Aldrich)]. Lysates were transferred to cryo-vials, stored in liquid nitrogen and thawed on ice. Protein concentrations of lysates were measured in triplicate using Micro BCA Protein Assay Kit (cat. no. 23235; Thermo Fisher Scientific). Lysates were reduced, alkylated with iodoacetamide and digested by the addition of trypsin at a 1:50 trypsin:protein ratio (by mass). After 2 h, a second trypsin aliquot was added at a 1:100 trypsin:protein ratio and incubated overnight at 37°C with shaking. After 16 h, the reaction was quenched with formic acid (final concentration 1% by volume). A mix of stable isotope-labeled peptide standards was added to the digest at 150 fmol/mg. Due to lower relative signal levels on the mass spectrometer, the concentration for the following seven peptides was adjusted to 600 fmol/mg (nomenclature gene product.peptide: CHEK1.YSSSQPEPR, CHE-K1. YSSpSQPEPR, MDC1.AQPFGFIDpSDTDAEEER, CLU.ASSIIDELFQDR, PCNA. AEDNADTDLALVFEAPNQEK, RAD50.LFDVCGSQDFESDLDR and RAD50.LFDVCGpSDFESDLDR). The mixture was desalted using Oasis HLB 96-well plates (cat. no. WAT058951; Waters® Corp., Milford, MA) and a positive pressure manifold (cat. no. 186005521; Waters Corp.). The eluates were aliquoted by volume, lyophilized and stored at −80°C.
Peptide Immunoaffinity Enrichment and Liquid Chromatography-Mass Spectrometry
Enrichment was performed as described elsewhere (36), with the following modifications. The final assay consisted of a mixture of 55 antibodies. Antibodies were crosslinked on Sepharose® protein G beads (cat. no. 28-9513-79; GE Healthcare Life Sciences, Logan, UT), and peptide enrichment was performed using 1 µg antibody-protein G magnetic beads for each target. For two targets (ATM and ATR peptides) 10 µg antibody-protein G beads was used. Unless indicated, 500 µg of trypsin-digested lysate resuspended in 200 µl PBS with 0.03% CHAPS (pH was adjusted to 8.0 with 5 µl of 1 M Tris) was inputted to each enrichment. Beads were mixed in the incubation plate, washed twice in PBS buffer with 0.03% CHAPS, washed once in 1/10× PBS, and peptides were eluted in 26 µl of 5% acetic acid/3% acetonitrile/50 mM citrate. The elution plate was covered with adhesive foil and frozen at −80°C until analysis.
LC-MS was performed with an Eksigent® Ultra nanoLC® system with a nano autosampler and chipFLEX system (Eksigent Technologies, Dublin, CA) coupled to a QTRAP® 6500 mass spectrometer (SCIEX, Foster City, CA). Peptides were loaded on a trap column (C18, 5 mm × 200 µm) at 5 µl/min for 3 min using mobile phase A (0.1% formic acid in water). The LC gradient was delivered at 300 nl/min and consisted of a linear gradient of mobile phase B (90% acetonitrile and 0.1% formic acid in water) developed from 3–14% B in 1 min, 14–34% B in 20 min, 34–90% B in 2 min and re-equilibration at 3% B on a 15 cm × 75 µm chip column (Reprosil AQ C18 particles, 3 µm; Dr. Maisch, Ammerbuch-Entringen, Germany). The nano electrospray interface was operated in the positive ion MRM mode. Parameters for declustering potential and collision energy were taken from a linear regression of previously optimized values found using Skyline software (37). Scheduled MRM transitions used a retention time window of 100 s and a desired cycle time of 0.5 s, enabling sufficient points across a peak for quantitation. A minimum of three transitions (six in total per peptide pair, including endogenous and spiked heavy peptides) were recorded for each light and heavy peptide. MRM data acquired on the QTRAP 6500 were analyzed using Skyline (37). Peak integrations were reviewed manually, and transitions from analyte peptides were confirmed by the same retention times of the light synthetic peptides and heavy stable isotope-labeled peptides, and with equivalent relative areas of recorded transitions. Transitions with detected interferences were not used in the data analysis.
Fit-for-Purpose Assay Validation
Four experiments (described below) were performed to characterize the analytical performance of the assays: 1. response curves, 2. repeatability, 3. stability and 4. endogenous detection.
Response Curves
Response curves were generated in a background matrix consisting of an equal mixture of protein lysate from four components: LCL GM07057 + 10 Gy irradiation (1 h), LCL GM07057 + mock irradiation (1 h), LCL GM01526 + 10 Gy irradiation (1 h), LCL GM01526 + mock irradiation (1 h). The pooled lysate was digested by trypsin, and the heavy stable isotope-labeled peptides were added to aliquots by serial dilution covering the concentrations 4,000, 400, 100, 25, 8.3, 2.8, 1.4, 0.7 fmol/mg (concentrations were 20,000, 2,000, 500, 125, 41.7, 13.9, 6.9, 3.5 for CHEK1.YSSSQPEPR, CHEK1. YSSpSQPEPR, MDC1.AQPFGFIDpSDTDAEEER, CLU.ASSIIDELFQDR, PCNA.AEDNADTDLALVFEAPNQEK, RAD50.LFDVCGSQDFESDLDR and RAD50.LFDVCGpSDFESDLDR). Light peptide was also spiked into the cell lysate pool at 200 fmol/mg. Blanks were prepared using background matrix with light peptide (no heavy spike). All points were analyzed by immunocapture and mass spectrometry in triplicate with nine replicates for the blank samples. Curves were analyzed using Skyline software (37). Linear regression was performed using a 1/x2 weighting on all points above the lower limit of quantification. The lower limit of quantifications (LLOQs) were obtained by empirically finding the lowest point on the curve that met two conditions: 1. CV <20% in the curve replicates; and 2. above the threshold for noise (determined by the average of the peak area ratio from nine blank measurements plus three times the standard deviation of the noise). All measurements were filtered by the LLOQ (i.e., all measurements were required to be above the LLOQ). The upper limit of quantification (ULOQ) was determined by the highest concentration point of the response curve that was maintained in the linear range of the response. For curves that maintained linearity at the highest concentration measured, the ULOQ is a minimum estimate.
Repeatability
Repeatability was determined using the same pooled lysate matrix used to generate the response curves. Heavy peptides were spiked in at three concentrations (0.75, 75, 750 fmol/mg) with the exception of the following seven peptides, which were added at 3.75, 375 and 3,750 fmol /mg: CHEK1.YSSSQPEPR, CHEK1.YSSpSQPEPR, MDC1.AQPFGFIDpSDTDAEEER, CLU.ASSIIDELFQDR, PCNA.AEDNADTDLALVFEAPNQEK, RAD50.LFDVCGSQDFESDLDR, and RAD50.LFDVCGpSDFESDLDR. All light peptides were added at 200 fmol/mg. Complete process triplicates (including digestion, capture and mass spectrometry) were prepared and analyzed on five independent days. Intra-assay variation was calculated as the mean CV obtained within each day. Inter-assay variation was the CV calculated from the mean values of the five days.
Peptide Stability
Stability of the enriched peptides was determined by analyzing aliquots of the medium spike level sample used in repeatability studies after storage at 4°C in the autosampler for approximately 6 and 24 h. Other aliquots were analyzed after one freeze-thaw, two freeze-thaws and storage at −80°C for approximately five months.
Repeatability of Endogenous Detection
Determination of variation in measuring endogenous analytes was conducted by analyzing the endogenous expression of target analytes in the LCL lysate pool by five independent replicates (including digestion) on five days.
Western Immunoassay
Western immunoassay was performed using the Wes™ system (ProteinSimple®, San Jose, CA) according to the manufacturer’s protocol and as described elsewhere (38), with the following modifications. Fluorescent master mix was prepared using 1 part 5× Fluorescent Mix and 5 parts protein preparation, primary antibodies were diluted 1× with PBS to a concentration of 1 µg/ml then diluted 1:5 1× with SignaLOCK™ Blocking Solution (cat. no. 50-58-00; Seracare (formerly KPL), Milford, MA), the antibody diluent time was changed from 5 to 30 min, and the detection profile was changed from 7 to 8 exposures (1, 5, 15 30, 60, 120, 240 and 480 s). Lysates for protein detection were generated from HeLa cells harvested 1 h after 10 Gy irradiation (see above for culture and treatment conditions).
Statistical Analysis
Integrated raw peak areas were exported from Skyline and total intensity was calculated using peak area and background. Peak area ratios were obtained by dividing peak areas of light peptides by those of the corresponding heavy peptides and ratios were log (base 2) transformed for statistical analysis. For PBMC results, linear mixed-effects models (lmer package for R) were used to characterize the between-blood-draws and between-individual variation of 40 analytes detected above the LLOQ. Statistical significance of estimated random effects of individuals was determined by a permutation test of 1,000 permutations. False discovery rates were then derived based on the Benjamini-Hochberg procedure.
Public Availability of Data
Raw data are available via Panorama Public (39), a database of targeted proteomics measurements (https://panoramaweb.org/project/home/begin.view?). Characterization data for assays can be found via the CPTAC assay portal (https://assays.cancer.gov/). Antibodies are available through the CPTAC antibody portal (https://proteomics.cancer.gov/antibody-portal).
RESULTS AND DISCUSSION
Development and Assembly of Assay Reagents
To develop an immuno-MRM assay panel capable of quantifying cellular signaling in response to DNA damage, we followed established workflows (40, 41) to generate rabbit anti-peptide monoclonal antibodies to phosphorylated and nonmodified peptides corresponding to proteins associated with the DDR signaling pathway. The final panel of reagents for assay development consisted of 54 monoclonal anti-peptide antibodies targeting 71 peptides (30 phosphorylated, 40 nonmodified and 1 ubiquitinated peptides). (Some monoclonal antibodies capture more than one proteoform, such as ±phosphorylation.)
MRM uses the combination of a specific precursor mass and characteristic fragment ion (i.e., transition), selected in a triple quadrupole mass spectrometer, to achieve high specificity and sensitivity (42, 43). To select the best transitions, light and heavy stable isotope-labeled synthetic peptides were obtained for each targeted sequence and analyzed by tandem mass spectrometry (MS/MS) and MRM. The most intense ~3–6 transitions free from noise were selected for use in the assay. Collision energy was optimized for each transition to produce the most complete fragmentation (i.e., the highest intensity) (44). The optimized transition ions and collision energies are reported in Supplementary Table S1 (http://dx.doi.org/10.1667/RR14963.1.S1).
The anti-peptide antibodies and synthetic peptide standards were used to develop the multiplexed immuno-MRM assay. An overview of the workflow for the assay is provided in Fig. 1. The 54 antibodies were individually coupled to protein G magnetic beads and multiplexed together by mixing. We started with a multiplexed assay targeting 71 peptides (30 phosphorylated, 40 nonmodified and one ubiquitinated).
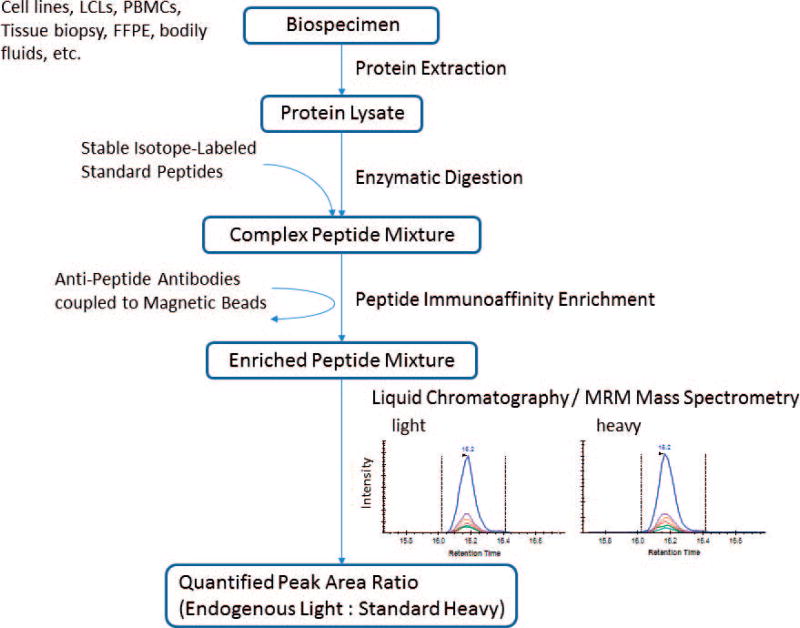
FIG. 1.
Overview of the immuno-MRM assay workflow. Proteins are extracted from the biological sample using procedures amenable to downstream mass spectrometry. The protein lysates are then enzymatically digested, typically using trypsin, and stable isotope-labeled standard (i.e., heavy) peptides corresponding to each analyte are added at known quantities to act as internal standards. The analyte peptides, along with their stable-isotope analogs, are enriched from the complex sample using anti-peptide antibodies coupled to magnetic beads. Measurement of the enriched analytes and standards is performed by liquid chromatography (LC) coupled to quantitative mass spectrometry (MS) using a targeted technique called multiple reaction monitoring (MRM). The peak area ratio (light endogenous analyte peptide to heavy peptide) is used for quantification. Specificity is confirmed by equivalent retention time and relative transition areas for light (endogenous) and heavy (standard) peptides.
Fit-for-Purpose Analytical Characterization of the Multiplexed Immuno-MRM Assay
The multiplexed immuno-MRM assay was characterized using fit-for-purpose method validation to determine the assay performance and analytical figures of merit (20, 22, 45). Response curves were used to characterize the linear range and limits of quantification. Heavy stable isotope-labeled peptide standards were serially diluted into 500 µg aliquots of a pooled background matrix of protein lysates from lymphoblast cell lines (equal amounts of four samples: ATM+/+ and ATM−/− LCLs, with/without ionizing radiation) spiked with a constant concentration of light synthetic peptide (200 fmol/mg light standards). This curve format allows measurement of the response in a true biological matrix. Specificity was determined by equivalent retention time of light and heavy peptides and consistent relative areas of multiple transitions for each peptide. The measured peak area ratios (sum of heavy peak area:sum of light peak area) were plotted as a function of spiked peptide concentration (curves available in Supplementary Fig. S1; http://dx.doi.org/10.1667/RR14963.1.S2). Assays to two of the 71 peptide targets (phospho FANCJ pS930 and phospho RIF1 pS1542) failed to validate in the response curves due to poor linearity (correlation coefficients R2 < 0.6) in the curves, likely emanating from poor overall response performance of the peptides in electrospray-MS. These peptides were omitted from the multiplexed panel. For the remaining 69 working assays, the median linear range was >3 orders of magnitude, with the median LLOQ 3.8 fmol/mg (ranging from 0.8–42 fmol/mg). The resulting figures of merit for working assays are shown in Supplementary Table S2.
Repeatability experiments were used to determine the intra-assay (within day) and inter-assay (between day) repeatability by assaying peptides over three concentrations and multiple days. The heavy stable isotope-labeled peptides were spiked into 500 µg aliquots of a pooled background matrix of protein lysates from LCLs (equal amounts of four samples: ATM+/+ and ATM−/− LCLs, with/without ionizing radiation) at three concentration levels (low, medium, high). An equal amount of light peptides (200 fmol/mg) was added to each sample. Three process replicates (including enzymatic digestion) were measured over five separate days. Specificity was determined by equivalent retention time of light and heavy peptides and consistent relative areas of transitions. For characterization, a peptide was required to have signal greater than the LLOQ on three or more days. Five out of the 69 target peptides did not meet this requirement (unmodified JUN.EEPV, unmodified CLUS.ASSI, phospho JUN pS243, phospho TOPBP1 pS1138, phospho RAD50 pS635) (nomenclature gene product.peptide first 4 amino acids). It is likely that these peptides failed to validate due to a number of factors, including insufficient affinity of the antibodies, high hydrophobicity of the peptides (contributing to instability over time), or poor electrospray performance of the peptides (decreasing sensitivity). The intra-assay (within day) and inter-assay (between day) variation for 64 qualifying assays is shown in Supplementary Table S3 (http://dx.doi.org/10.1667/RR14963.1.S1). The median intra-assay variability was 9%, 7% and 8% CV for the low, medium and high concentration samples, respectively. The median inter-assay variability was 19%, 10% and 7% CV for the low, medium and high concentration samples, respectively.
Stability of the enriched peptides was evaluated to determine appropriate handling conditions of the prepared samples. Heavy peptides were spiked into a pool of protein lysate digest derived from LCLs (equal amounts of four samples: ATM+/+ and ATM−/− LCLs, with/without ionizing radiation) and analyzed by immunoaffinity enrichment and LC-MRM under several conditions in duplicate. The control samples were analyzed immediately, and test samples were analyzed after ~6 h and ~24 h at 4°C. Additional aliquots of the enriched samples were analyzed after one and two freeze-thaw cycles and after five months of storage at −80°C. The peak area ratios of the 64 multiplexed analytes measured in the stored samples were compared to the peak area ratio for the control sample. Stability was determined by evaluating the variation (range of the duplicates divided by the mean) and the percentage difference relative to the fresh sample (Supplementary Table S4; http://dx.doi.org/10.1667/RR14963.1.S1). Overall, variation did not increase after storage for up to 24 h, as indicated by the median variation (4–10% for the conditions tested). However, the stability test provided important characterization for a subset of peptide analytes. Two analytes (unmodified PCNA.AEDN, unmodified RRM2.DIQH) failed the stability test due to excessive differences (>100%) in the stored samples or no signal above the detection threshold. Three analytes (phospho BRCA2 pS1680, phospho PALB2 pS376, and phospho TOPBP1 pT1062) showed large differences (>25%) upon extended storage at 4°C or after freeze-thaw, indicating that only a freshly prepared sample is adequate for analysis of these targets. An additional six analytes (phospho CHK1 pS317, phospho NBN pS432, phospho RAD18 pS99, phospho FANCI pS730, unmodified FEN1.SIEE, unmodified TOPBP1.QTVP) showed large (>25% or not detected) differences after storage at −80°C for five months, indicating that prepared samples should not be stored for long periods of time for analysis of these targets. After stability testing, the final validated multiplexed assay consisted of 50 antibodies targeting 62 peptides (25 phosphorylated, 26 nonmodified, 1 ubiquitinated) corresponding to 31 proteins (see Table 1 for a list of assay targets).
TABLE 1.
Sixty-Two Qualified Assay Targets
| Gene symbol | Description | Peptide analyte sequence | Modification site |
CPTAC assay portal ID |
|---|---|---|---|---|
| UBE2C | Ubiquitin-conjugating enzyme E2C | GISAFPESDNLFK | CPTAC-3259 | |
| UBE2C | Ubiquitin-conjugating enzyme E2C | LSLEFPSGYPYNAPTVK | CPTAC-3260 | |
| CHK1 | CHK1 checkpoint homolog (S. pombe) | YSSS[PO4]QPEPR | pS317 | CPTAC-3282 |
| CHK1 | CHK1 checkpoint homolog (S. pombe) | YSSSQPEPR | CPTAC-3223 | |
| FANCA | Fanconi anemia, complementation group A | QQAAPDADLSQEPHLF | CPTAC-3226 | |
| RAD51C | RAD51 homolog C (S. cerevisiae) | DLVSFPLS[PO4]PAVR | pS20 | CPTAC-3287 |
| RAD51C | RAD51 homolog C (S. cerevisiae) | DLVSFPLSPAVR | CPTAC-3250 | |
| REV3L | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | SGTLS[PO4]PEIFEK | pS1724 | CPTAC-3253 |
| REV3L | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | SGTLSPEIFEK | CPTAC-3252 | |
| NBN | Nibrin | IPNYQLS[PO4]PTK | pS432 | CPTAC-3239 |
| NBN | Nibrin | IPNYQLSPTK | CPTAC-3238 | |
| NBN | Nibrin | TTTPGPSLS[PO4]QGVSVDEK | pS343 | CPTAC-3237 |
| NBN | Nibrin | TTTPGPSLSQGVSVDEK | CPTAC-3236 | |
| JUN | Jun oncogene | NSDLLTS[PO4]PDVGLLK | pS63 | CPTAC-3233 |
| JUN | Jun oncogene | NSDLLTSPDVGLLK | CPTAC-3232 | |
| RB1 | Retinoblastoma 1 | IPGGNIYIS[PO4]PLK | pS807 | CPTAC-3288 |
| RB1 | Retinoblastoma 1 | IPGGNIYISPLK | CPTAC-3251 | |
| GSTP1 | Glutathione S-transferase pi | YISLIYTNYEAGK | CPTAC-3231 | |
| PARP1 | poly (ADP-ribose) polymerase 1 | EELGFRPEYS[PO4]ASQLK | pS177 | CPTAC-3241 |
| PARP1 | poly (ADP-ribose) polymerase 1 | EELGFRPEYSASQLK | CPTAC-3240 | |
| PCNA | Proliferating cell nuclear antigen | DLSHIGDAVVISC[Cam]AK | CPTAC-3244 | |
| PCNA | Proliferating cell nuclear antigen | DLSHIGDAVVISC[Cam]AK[-GG]DGVK | uK164 | CPTAC-3245 |
| PCNA | Proliferating cell nuclear antigen | LVQGSILK | CPTAC-3242 | |
| RRM2 | Ribonucleotide reductase M2 polypeptide | IEQEFLTEALPVK | CPTAC-3255 | |
| BRCA1 | Breast cancer 1, early onset | NYPS[PO4]QEELIK | pS1524 | CPTAC-3219 |
| BRCA1 | Breast cancer 1, early onset | NYPSQEELIK | CPTAC-3218 | |
| FEN1 | Flap structure-specific endonuclease 1 | SIEEIVR | CPTAC-3230 | |
| BRCA2 | Breast cancer 2, early onset | TS[PO4]VSQTSLLEAK | pS1680 | CPTAC-3280 |
| BRCA2 | Breast cancer 2, early onset | TSVSQTSLLEAK | CPTAC-3279 | |
| RAD23B | RAD23 homolog B (S. cerevisiae) | IDIDPEETVK | CPTAC-3248 | |
| RAD23B | RAD23 homolog B (S. cerevisiae) | ILNDDTALK | CPTAC-3249 | |
| UBE2I | Ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | DHPFGFVAVPTK | CPTAC-3261 | |
| FAAP100 | Fanconi anemia core complex-associated protein 100 | APS[PO4]PLGPTR | pS667 | CPTAC-3225 |
| FAAP100 | Fanconi anemia core complex-associated protein 100 | APSPLGPTR | CPTAC-3224 | |
| ATM | Ataxia telangiectasia mutated | NLS[PO4]DIDQSFNK | pS2996 | CPTAC-3213 |
| ATM | Ataxia telangiectasia mutated | NLSDIDQSFNK | CPTAC-3212 | |
| ATM | Ataxia telangiectasia mutated | SLEIS[PO4]QSYTTTQR | pS367 | CPTAC-3211 |
| ATM | Ataxia telangiectasia mutated | SLEISQSYTTTQR | CPTAC-3210 | |
| ATR | Serine/threonine-protein kinase ATR | GVELC[Cam]FPENET[PO4]PPEGK | pT1989 | CPTAC-3215 |
| ATR | Serine/threonine-protein kinase ATR | GVELC[Cam]FPENETPPEGK | CPTAC-3214 | |
| MDC1 | Mediator of DNA-damage checkpoint 1 | AQPFGFIDS[PO4]DTDAEEER | pS329 | CPTAC-3235 |
| MDC1 | Mediator of DNA-damage checkpoint 1 | AQPFGFIDSDTDAEEER | CPTAC-3234 | |
| TERF2 | Telomeric repeat binding factor 2 | DLVLPTQALPAS[PO4]PALK | pS323 | CPTAC-3257 |
| TERF2 | Telomeric repeat binding factor 2 | DLVLPTQALPASPALK | CPTAC-3256 | |
| RIF1 | RAP1 interacting factor homolog (yeast) | ASQGLLSSIENSESDSSEAK | CPTAC-3254 | |
| PALB2 | Partner and localizer of BRCA2 | NENLQESEILS[PO4]QPK | pS376 | CPTAC-3284 |
| ATRIP | ATR interacting protein | LAAPSVSHVS[PO4]PR | pS224 | CPTAC-3216 |
| ATRIP | ATR interacting protein | LSDGDMTSALR | CPTAC-3217 | |
| TOPBP1 | Topoisomerase (DNA) II binding protein 1 | GVLT[PO4]QTLEMR | pT1062 | CPTAC-3290 |
| TOPBP1 | Topoisomerase (DNA) II binding protein 1 | GVLTQTLEMR | CPTAC-3258 | |
| TOPBP1 | Topoisomerase (DNA) II binding protein 1 | QTVPDVNTEPSQNEQIIWDDPTAR | CPTAC-3291 | |
| RAD50 | RAD50 homolog (S. cerevisiae) | LFDVC[Cam]GSQDFESDLDR | CPTAC-3286 | |
| FANCJ | BRCA1 interacting protein C-terminal helicase 1 | ATPELGSSENSASS[PO4]PPR | pS1032 | CPTAC-3222 |
| FANCJ | BRCA1 interacting protein C-terminal helicase 1 | ATPELGSSENSASSPPR | CPTAC-3221 | |
| FANCJ | BRCA1 interacting protein C-terminal helicase 1 | YST[PO4]PPYLLEAASHLSPENFVEDEAK | pT918 | CPTAC-3281 |
| FANCJ | BRCA1 interacting protein C-terminal helicase 1 | YSTPPYLLEAASHLSPENFVEDEAK | CPTAC-3220 | |
| UBE2T | Ubiquitin-conjugating enzyme E2T (putative) | AS[PO4]QLVGIEK | pS184 | CPTAC-3263 |
| UBE2T | Ubiquitin-conjugating enzyme E2T (putative) | ASQLVGIEK | CPTAC-3262 | |
| RAD18 | RAD18 homolog (S. cerevisiae) | NDLQDTEIS[PO4]PR | pS471 | CPTAC-3247 |
| RAD18 | RAD18 homolog (S. cerevisiae) | NHLLQFALES[PO4]PAK | pS99 | CPTAC-3285 |
| RAD18 | RAD18 homolog (S. cerevisiae) | NHLLQFALESPAK | CPTAC-3246 | |
| FANCI | Fanconi anemia, complementation group I | SADFS[PO4]QSTSIGIK | pS730 | CPTAC-3283 |
Notes. Modifications in the peptide analyte sequence are phosphorylation (PO4), ubiquitination (-GG) and carbamidomethylated cysteine (Cam). CPTAC assay portal identifiers are provided for reference.
To complete the characterization of the assay, the reproducibility in quantification of endogenous analytes was characterized by measuring aliquots of a common sample prepared and analyzed over multiple days. Aliquots of 500 µg of protein lysate from pooled LCLs (harvested 1 h after 5 Gy or mock irradiation) were analyzed by up to five complete process replicates (including protein digestion, immunoaffinity enrichment, and immediate LC-MRM analysis) over five days. Heavy (stable isotope labeled) peptides were spiked into the digests at a known concentration. Peptide detection was required to have signal greater than the LLOQ. Specificity was determined by equivalent retention time of light and heavy peptides and consistent relative transition areas. In total, 54 analyte peptides were detected at endogenous levels at least once in the pooled lysate. The median intra-assay variability was 12% CV (range 3–26% CV) and the median inter-assay variability was 13% CV (range 3–60% CV) for detection of the endogenous analytes (see Supplementary Table S5; http://dx.doi.org/10.1667/RR14963.1.S1).
Quantifying Cell Signaling in Immortalized Human Cells
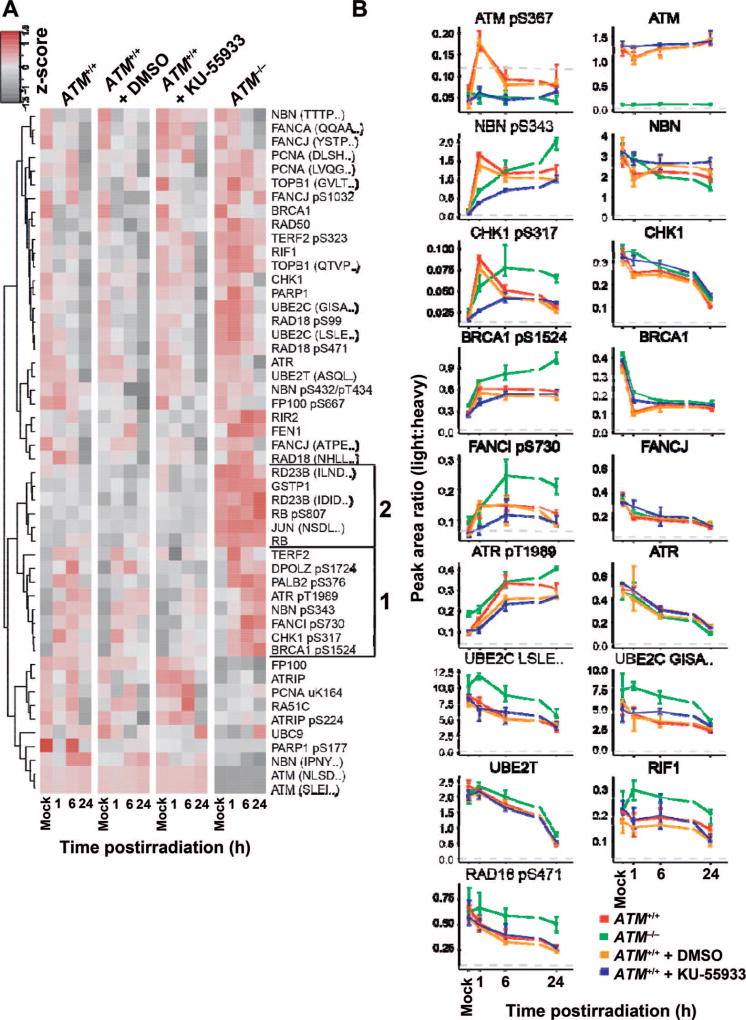
We conducted proof-of-principle experiments to demonstrate the utility of the quantitative multiplexed assay in profiling the response to DNA damage. LCLs derived from an ataxia telangiectasia (AT) patient (ATM−/−) and from a healthy control (ATM+/+) were used as a model system for profiling the effects of genomic mutation and pharmacological inhibition on kinase activity after DNA damage by ionizing radiation. The ATM gene encodes a serine/threonine protein kinase that activates checkpoint signaling in response to DNA double-strand breaks (46) through dissociation of the ATM homodimer to active monomers via autophosphorylation at several sites, including Ser1981 (47) and Ser367 (48). Cells from AT patients are sensitive to ionizing radiation and defective in phosphosignaling in response to DNA damage (49). Using the 62-plex assay, we profiled the time course (1, 6, and 24 h postirradiation) of cell signaling (i.e., pharmacodynamic profile) in 5 Gy irradiated cells. An aliquot of ATM+/+ cells were also treated with the ATM kinase inhibitor KU-55933 prior to 5 Gy irradiation. All experiments were performed in biological triplicate (measured peak area ratios are available in Supplementary Table S6; http://dx.doi.org/10.1667/RR14963.1.S1). A heatmap showing the response is plotted in Fig. 2A. The time courses of individual analytes with greater than twofold change (P < 0.05) are plotted in Fig. 2B.
FIG. 2.
Use of the multiplexed assay to quantify radiation-induced changes in cellular signaling due to genomic mutations and/or pharmacologic inhibition of kinases. Human lymphoblast cell lines derived from wild-type (ATM+/+, GM07057) and ATM-deficient (ATM−/−, GM01526) patients were harvested in a time course (1, 6, 24 h) after perturbation with 5 Gy irradiation. Cultures of ATM+/+ cells were additionally treated with ATM kinase inhibitor KU-55933 (or control vehicle, DMSO). Panel A: Heatmap for the mean peak area ratio from three biological replicates normalized across samples for each peptide. Analytes detected at all timepoints above the LLOQ are plotted. Group “1”: analytes with decreased response in the presence of the inhibitor compared to the genomic variant. Group “2”: analytes with increased expression levels in the genetic variant (ATM−/−) compared to wild-type cells treated in the presence of kinase inhibitor. Panel B: The peak area ratio (light:heavy) for selected nonphosphorylated and phosphorylated peptides in the DDR panel with changes greater than twofold (P < 0.05). Error bars are the standard deviation of three biological replicates. The LLOQ is indicated by the gray dotted line.
As expected, ATM expression was detected in the ATM+/+ cells but not in the ATM−/− cells, due to the production of truncated and unstable protein in the AT patient cells (50, 51). In contrast, exposure of ATM+/+ cells to the inhibitor KU-55933 did not affect basal expression of the ATM kinase. For ATM+/+ cells, activation of the DDR can be seen by autophosphorylation of ATM pS367 proceeding rapidly after irradiation, with detection of the activated kinase at the earliest time point sampled after DNA damage (Fig. 2B). Both chemical inhibition of the ATM kinase and mutation of the ATM gene result in a failure of cells to autophosphorylate ATM in response to ionizing radiation. Multiple abnormalities can also be seen in the phosphosignaling of downstream pathway components. In ATM wild-type cells, an increase in phospho NBN pS343 (52) and phospho CHK1 pS317 (53) was detected within 1 h postirradiation, whereas phosphorylation of these targets was delayed in cells with ATM mutation and in wild-type cells treated with the inhibitor (Fig. 2B). The residual phosphorylation of these sites in cells exposed to the inhibitor or harboring ATM mutation is presumably achieved through the action of other DDR kinases and/or incomplete inhibition. The response of several components is muted in the inhibited samples compared to the genetic variant. For example, ATR pT1989 (54) is measured at higher levels in the ATM-deficient cells. This difference is also highlighted in the heatmap clustergram shown in Fig. 2A, indicating a distinct group of targets with decreased response in the presence of the inhibitor compared to the genomic variant (Fig. 2A, group 1). The heatmap also shows a cluster of analytes with increased basal levels of expression in the genetic variant (ATM−/−) compared to the wild-type cells (Fig. 2A, group 2). These findings demonstrate the utility of the multiplexed assay for profiling quantitative changes in the pharmacodynamic response of cells after perturbation.
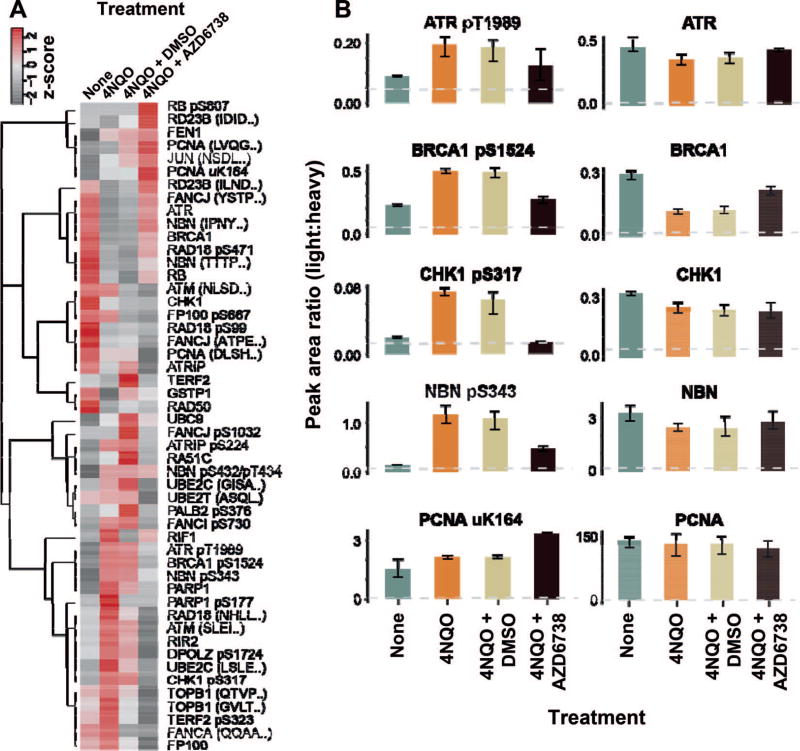
Next, we profiled the DNA damage response of LCLs exposed to 4-nitroquinoline 1-oxide (4NQO). Human LCLs were harvested after 2 h incubation with 2.5 µM 4NQO in biological triplicate (i.e., different cultures on different days), and control samples were mock-treated. The cells were lysed, digested and analyzed by the DDR 62-plex assay (measured peak area ratios are available in Supplementary Table S7; http://dx.doi.org/10.1667/RR14963.1.S1). A heatmap of the responses is shown in Fig. 3A and the responses of individual analytes with greater than twofold change (P < 0.05) are plotted in Fig. 3B. The ATR kinase is autophosphorylated after DNA damage at multiple sites, including Thr1989 (55). The activity of the DDR can be seen by an increase in the levels of several phosphosites [e.g., phospho BRCA1 pS1524 (56), phospho CHK1 pS317 (57), phospho NBN pS343 (58)]. The pharmacologic inhibition of the ATR kinase was profiled using the ATR kinase inhibitor AZD-6738 in conjunction with the 4NQO treatment. A decrease in phosphorylation of phospho ATR pT1989 is evident in the samples treated with inhibitor (Fig. 3B). Less than full suppression of the phospho ATR pT1989 may be due to partial inactivation or activity of another kinase. Activity of the DDR signaling is notably altered with the ATR kinase inhibitor, since phosphorylation of several sites have a marked decrease in phosphorylation in the presence of the inhibitor compound (e.g., phospho NBN pS343, phospho CHK1 pS317, phospho BRCA1 pS1524). There is an increase in ubiquitinated PCNA uK164 after 4NQO treatment in the presence of the inhibitor compound, consistent with increased replication stress in the absence of ATR activity (59–61).
FIG. 3.
Use of the multiplexed assay to quantify the response to 4-nitroquinoline 1-oxide (4NQO). Human lymphoblast cell lines were treated with 4NQO, and the DDR was profiled with the 62-plex immuno-MRM assay in biological triplicate. Control samples received no treatment, 4NQO-treated samples were harvested after 2 h. Samples labeled “AZD6738” refer to treatment in the presence of the ATR kinase inhibitor, and samples labeled “DMSO” contain vehicle only. Panel A: Heatmap showing the response to DNA damage induced by 4-nitroquinoline 1-oxide and the effects of AZD6738. Analytes detected above the LLOQ in all conditions are plotted in the heatmap. Panel B: The peak area ratio (light:heavy) for control (green), 4NQO (orange), 4NQO with vehicle (tan) and 4NQO with inhibitor (black) treatments. Error bars are the standard deviation of three biological replicates. The LLOQ is indicated by the gray dotted line.
Quantifying Individual Variations in DDR Capacity in Primary Human Cells
To demonstrate the potential for applying the assay to primary human cells, we measured the DDR in peripheral blood mononuclear cells exposed to ionizing radiation. PMBCs from four healthy adults were isolated from three separate blood draws per donor (the first and final blood draws for an individual were separated by 8–13 weeks). Upon isolation, PBMCs were expanded in culture and harvested 1 h after 10 Gy irradiation. Protein lysates (500 µg aliquots) were analyzed by the 62-plex immuno-MRM assay in triplicate process replicates (including digestion). A total of 40 peptides (12 phosphorylated, 27 nonmodified, 1 ubiquitinated) were detected at endogenous levels above LLOQ in the irradiated PBMCs (peak area ratios for detected peptides are available in Supplementary Table S8; http://dx.doi.org/10.1667/RR14963.1.S1), showing that the assay is capable of measuring endogenous protein expression and profiling phosphorylation events in a primary human sample. The median assay variation, measured from process triplicate analysis of each sample, was 8% CV (with a range of 3–36% CV), showing good analytical performance of the assay.
We next investigated whether there were differences in the level of expression for the DDR targets between individuals or within individuals (i.e., between different blood draws). The variation for each analyte is shown in Supplementary Table S9 (http://dx.doi.org/10.1667/RR14963.1.S1). There are a number of peptides that showed significant inter-person variation [e.g., unmodified ATM (2 peptides), phospho ATM pS367, unmodified RAD50 and unmodified RAD23B]. Interestingly, there are also several targets that showed a large degree of variation within a person over time [e.g., unmodified UBE2C (2 peptides) and phospho RAD18 pS471]. This dataset contains a limited number of individuals and is intended for a proof-of-principle demonstration; however, the results indicate that: 1. the assay is capable of quantitative measurements of DDR-related components in primary human specimens; and 2. there are significant inter-individual differences in the expression of DDR components.
A potential consideration of using the approach in clinical applications is the amount of material required for quantification of low-abundance phosphopeptides. We chose to use an input material requirement of 500 µg protein, which is easily obtained from expanding cell cultures in biological studies, but can be difficult to achieve in fine needle aspirates or core biopsies. To address this, further work is underway to improve sensitivity and enable scaling of the technique to smaller inputs. These approaches include obtaining the highest-affinity antibodies, optimizing for total recovery in protein extraction and trypsin digestion, and further optimizing the chromatography system for specific classes of peptides. Approaches for improving the sensitivity of the assay for individual analytes (e.g., increasing the number of individual antibodies to improve recovery or optimizing the chromatography system to improve detection) are currently possible, but at the expense of greater multiplexing.
Characterization of Anti-Peptide Antibodies for Western Blot
We previously demonstrated that some immuno-MRM affinity reagents also work for Western blotting (41), increasing the utility of the reagents. Therefore, we assessed the utility of the monoclonal anti-peptide antibodies generated for this assay in Western blotting. The antibodies were tested in two stages. First, antibodies were tested for Western blot detection of purified recombinant proteins spiked into a cell lysate. Antibodies that tested positive against recombinant proteins were subsequently tested in lysates from HeLa cells harvested 1 h after 10 Gy irradiation. Overall, 22 out of 54 (41%) antibodies were positive against the recombinant protein. Of these 22 antibodies, 11 were also successful in detecting endogenous protein in the cell lysates that were tested. Supplementary Fig. S2 (http://dx.doi.org/10.1667/RR14963.1.S2) shows the blot images and Supplementary Table S10 summarizes the Western blot results. Monoclonal antibodies, Western blot images, standard operating protocols and results are also publicly available as a resource to the community at the NCI’s CPTAC antibody portal
CONCLUSION
In this work, we report on the de novo development of 54 monoclonal antibodies targeting proteins involved in the DNA damage response, including post-translational modifications (phosphorylation, ubiquitination) induced by DNA damaging agents. The monoclonals were configured into a novel 62-plex immuno-MRM assay for quantification of phosphosignaling pharmacodynamics in response to ionizing radiation and other genotoxic stressors. All assays underwent fit-for-purpose characterization, and all assay protocols, as well as 41 monoclonal antibodies, are made available to the community via the NCI’s assay and antibody portals.
The multiplex immuno-MRM assay described in this report enables precise and highly specific quantification of 62 analytes in the DDR, essentially replacing 62 Western blots with a <1 h MRM-MS run, and enabling the DDR to be rigorously quantified. Based on the proof-of-concept data presented herein, this assay has many potential applications in basic and translational research. For example, signal transduction networks, such as the DDR, demonstrate biological robustness (62). Thus, determining pathway signaling based on a single analyte, or a small number of analytes, in a semi-quantitative fashion (e.g., using Western blotting) provides a very restricted readout of the network state. Multiplex, analytically robust assays, such as the one described herein, provide a potential platform for a less restrictive look at network activity, enabling studies of pathway cross-talk. Due to the highly quantitative nature of immuno-MRM (coupled with the use of stable isotope-labeled internal standards), DDR activity can be quantified as a continuous variable, which is very difficult and often imprecise when less quantitative techniques are used. This enables robust pharmacodynamic studies that can greatly facilitate characterization of lead compounds, such as radiosensitizing agents and potential chemotherapeutic compounds targeting DDR proteins (e.g., PARP and ATM inhibitors). Additionally, the 62-plex assay provides a functional readout of kinase activity after DNA damage, raising the interesting possibility that it could be useful for determining whether variants of undetermined significance in critical signaling kinases such as ATM or ATR have functional consequences on the DDR network, and to what extent. Similarly, the assay could have application in testing and molecular classification of DNA repair disorders. The proof-of-principle data from human PBMCs demonstrate the feasibility and potential utility of the assay to study inter- and intra-individual variation in the radiation response, which could have implications for biodosimetry as well as for personalized radiation therapy. Thus, there are many fertile areas of future studies in which this assay could advance the field of radiation research.
Supplementary Material
Table S1. MRM parameters for targeted peptides.
Table S2. Response curve performance figures of merit.
Table S3. Repeatability characterization figures of merit.
Table S4. Stability characterization figures of merit for qualified assays.
Table S5. Characterization of day-to-day variation in endogenous measurement.
Table S6. Peak area ratios for analytes measured above LLOQ in human LCLs. Rows 2–6 indicate genomic variants, inhibitors or vehicle, ionizing radiation, time point and biological replicates for each datapoint.
Table S7. Peak area ratios for analytes measured above LLOQ in human LCLs. Rows 2–6 indicate cells, DNA damaging agent, inhibitor or vehicle, time point, and biological replicates for each datapoint.
Table S8. Peak area ratios for analytes measured above LLOQ in human PBMCs.
Table S9. Variation of analytes measured in human PBMCs.
Table S10. Characterization of monoclonal antibodies in Western blot.
Fig. S1. Response curves for immuno-MRM assays.
Fig. S2. Images of Western blots from screening monoclonal antibodies.
Acknowledgments
The research reported on here was supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) Clinical Proteomics Tumor Analysis Consortium Initiative (nos. U24CA160034 and U01CA214114), the NCI Research Specialist Program (award no. R50CA211499) and a grant from the Athena Partners Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Editor’s note. The online version of this article (DOI: 10.1667/RR14963.1) contains supplementary information that is available to all authorized users.
References
- 1.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKinnon PJ. Ataxia-telangiectasia: an inherited disorder of ionizing-radiation sensitivity in man. Progress in the elucidation of the underlying biochemical defect. Hum Genet. 1987;75:197–208. doi: 10.1007/BF00281059. [DOI] [PubMed] [Google Scholar]
- 5.Taalman RD, Jaspers NG, Scheres JM, de Wit J, Hustinx TW. Hypersensitivity to ionizing radiation, in vitro, in a new chromosomal breakage disorder, the Nijmegen Breakage Syndrome. Mutat Res. 1983;112:23–32. doi: 10.1016/0167-8817(83)90021-4. [DOI] [PubMed] [Google Scholar]
- 6.Aurias A, Antoine JL, Assathiany R, Odievre M, Dutrillaux B. Radiation sensitivity of Bloom’s syndrome lymphocytes during S and G2 phases. Cancer Genet Cytogenet. 1985;16:131–6. doi: 10.1016/0165-4608(85)90006-8. [DOI] [PubMed] [Google Scholar]
- 7.Reinlib L, Friedberg EC. Working Group on Integrated Translational Research in DNA Repair. Report of the Working Group on Integrated Translational Research in DNA Repair. DNA Repair. 2007;6:145–7. doi: 10.1016/j.dnarep.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Janes KA. An analysis of critical factors for quantitative immunoblotting. Sci Signal. 2015;8:rs2. doi: 10.1126/scisignal.2005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker RA. Quantification of immunohistochemistry–issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49:406–10. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- 10.Gown AM. Diagnostic immunohistochemistry: What can go wrong and how to prevent it. Arch Pathol Lab Med. 2016;140:893–8. doi: 10.5858/arpa.2016-0119-RA. [DOI] [PubMed] [Google Scholar]
- 11.Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics MCP. 2013;12:2381–2. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 13.Akbani R, Becker K-F, Carragher N, Goldstein T, de Koning L, Korf U, et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol Cell Proteomics MCP. 2014;13:1625–43. doi: 10.1074/mcp.O113.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF, III, Pierobon M. Reverse phase protein microarrays. Methods Mol Biol. 2017;1606:149–69. doi: 10.1007/978-1-4939-6990-6_11. [DOI] [PubMed] [Google Scholar]
- 15.Houser B. Bio-Rad’s Bio-Plex(R) suspension array system, xMAP technology overview. Arch Physiol Biochem. 2012;118:192–6. doi: 10.3109/13813455.2012.705301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Wu P, Yang L, Hou X, Lv Y. Inductively coupled plasma mass spectrometry-based immunoassay: A review. Mass Spectrom Rev. 2014;33:373–93. doi: 10.1002/mas.21391. [DOI] [PubMed] [Google Scholar]
- 17.Lemeer S, Jopling C, Naji F, Ruijtenbeek R, Slijper M, Heck AJ, et al. Protein-tyrosine kinase activity profiling in knock down zebrafish embryos. PloS One. 2007;2:e581. doi: 10.1371/journal.pone.0000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arsenault R, Griebel P, Napper S. Peptide arrays for kinome analysis: new opportunities and remaining challenges. Proteomics. 2011;11:4595–609. doi: 10.1002/pmic.201100296. [DOI] [PubMed] [Google Scholar]
- 19.Whiteaker JR, Halusa GN, Hoofnagle AN, Sharma V, MacLean B, Yan P, et al. Using the CPTAC Assay Portal to identify and implement highly characterized targeted proteomics assays. Methods Mol Biol. 2016;1410:223–36. doi: 10.1007/978-1-4939-3524-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteaker JR, Halusa GN, Hoofnagle AN, Sharma V, MacLean B, Yan P, et al. CPTAC Assay Portal: a repository of targeted proteomic assays. Nat Methods. 2014;11:703–4. doi: 10.1038/nmeth.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbatiello SE, Schilling B, Mani DR, Zimmerman LJ, Hall SC, MacLean B, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. 2015;14:2357–74. doi: 10.1074/mcp.M114.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–17. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoofnagle AN, Whiteaker JR, Carr SA, Kuhn E, Liu T, Massoni SA, et al. Recommendations for the generation, quantification, storage, and handling of peptides used for mass spectrometry-based assays. Clin Chem. 2016;62:48–69. doi: 10.1373/clinchem.2015.250563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, Zhao L, Pope ME, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013854. M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat Methods. 2014;11:149–55. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, Carapito C, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiteaker JR, Zhao L, Abbatiello SE, Burgess M, Kuhn E, Lin C, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol Cell Proteomics. 2011;10(4) doi: 10.1074/mcp.M110.005645. M110.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteaker JR, Paulovich AG. Peptide immunoaffinity enrichment coupled with mass spectrometry for peptide and protein quantification. Clin Lab Med. 2011;31:385–96. doi: 10.1016/j.cll.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiteaker JR, Zhao L, Lin C, Yan P, Wang P, Paulovich AG. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015347. M111.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 32.Netzel BC, Grant RP, Hoofnagle AN, Rockwood AL, Shuford CM, Grebe SK. First steps toward harmonization of LC-MS/MS thyroglobulin assays. Clin Chem. 2016;62:297–9. doi: 10.1373/clinchem.2015.245266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ippoliti PJ, Kuhn E, Mani DR, Fagbami L, Keshishian H, Burgess MW, et al. Automated microchromatography enables multiplexing of immunoaffinity enrichment of peptides to greater than 150 for targeted MS-based assays. Anal Chem. 2016;88:7548–55. doi: 10.1021/acs.analchem.6b00946. [DOI] [PubMed] [Google Scholar]
- 34.Whiteaker JR, Zhao L, Yan P, Ivey RG, Voytovich UJ, Moore HD, et al. Peptide immunoaffinity enrichment and targeted mass spectrometry enables multiplex, quantitative pharmacodynamic studies of phospho-signaling. Mol Cell Proteomics. 2015;14:2261–73. doi: 10.1074/mcp.O115.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy JJ, Yan P, Zhao L, Ivey RG, Voytovich UJ, Moore HD, et al. Immobilized metal affinity chromatography coupled to multiple reaction monitoring enables reproducible quantification of phospho-signaling. Mol Cell Proteomics MCP. 2016;15:726–39. doi: 10.1074/mcp.O115.054940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L, Whiteaker JR, Pope ME, Kuhn E, Jackson A, Anderson NL, et al. Quantification of proteins using peptide immunoaffinity enrichment coupled with mass spectrometry. J Vis Exp. 2011;53 doi: 10.3791/2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinforma Oxf Engl. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Valdez A, Chen Y. Evaluation of automated Wes system as an analytical and characterization tool to support monoclonal antibody drug product development. J Pharm Biomed Anal. 2017;139:263–8. doi: 10.1016/j.jpba.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 39.Sharma V, Eckels J, Taylor GK, Shulman NJ, Stergachis AB, Joyner SA, et al. Panorama: a targeted proteomics knowledge base. J Proteome Res. 2014;13:4205–10. doi: 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenherr RM, Zhao L, Whiteaker JR, Feng LC, Li L, Liu L, et al. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring-mass spectrometry. J Immunol Methods. 2010;353:49–61. doi: 10.1016/j.jim.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenherr RM, Saul RG, Whiteaker JR, Yan P, Whiteley GR, Paulovich AG. Anti-peptide monoclonal antibodies generated for immuno-multiple reaction monitoring-mass spectrometry assays have a high probability of supporting Western blot and ELISA. Mol Cell Proteomics. 2015;14:382–98. doi: 10.1074/mcp.O114.043133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 44.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 2010;82:10116–24. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant RP, Hoofnagle AN. From lost in translation to paradise found: enabling protein biomarker method transfer by mass spectrometry. Clin Chem. 2014;60:941–4. doi: 10.1373/clinchem.2014.224840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 47.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 48.Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006;25:3504–14. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu Rev Pathol. 2012;7:303–21. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 50.Lakin ND, Weber P, Stankovic T, Rottinghaus ST, Taylor AM, Jackson SP. Analysis of the ATM protein in wild-type and ataxia telangiectasia cells. Oncogene. 1996;13:2707–16. [PubMed] [Google Scholar]
- 51.Becker-Catania SG, Chen G, Hwang MJ, Wang Z, Sun X, Sanal O, et al. Ataxia-telangiectasia: phenotype/genotype studies of ATM protein expression, mutations, and radiosensitivity. Mol Genet Metab. 2000;70:122–33. doi: 10.1006/mgme.2000.2998. [DOI] [PubMed] [Google Scholar]
- 52.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–9. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 53.Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, et al. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem. 2003;278:14806–11. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 54.Nam EA, Zhao R, Glick GG, Bansbach CE, Friedman DB, Cortez D. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J Biol Chem. 2011;286:28707–14. doi: 10.1074/jbc.M111.248914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–82. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckta JM, Dever SM, Gnawali N, Khalil A, Sule A, Golding SE, et al. Mutation of the BRCA1 SQ-cluster results in aberrant mitosis, reduced homologous recombination, and a compensatory increase in non-homologous end joining. Oncotarget. 2015;6:27674–87. doi: 10.18632/oncotarget.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 58.Olson E, Nievera CJ, Lee AY-L, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–52. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 59.Lee K, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 60.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16125–30. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–52. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. MRM parameters for targeted peptides.
Table S2. Response curve performance figures of merit.
Table S3. Repeatability characterization figures of merit.
Table S4. Stability characterization figures of merit for qualified assays.
Table S5. Characterization of day-to-day variation in endogenous measurement.
Table S6. Peak area ratios for analytes measured above LLOQ in human LCLs. Rows 2–6 indicate genomic variants, inhibitors or vehicle, ionizing radiation, time point and biological replicates for each datapoint.
Table S7. Peak area ratios for analytes measured above LLOQ in human LCLs. Rows 2–6 indicate cells, DNA damaging agent, inhibitor or vehicle, time point, and biological replicates for each datapoint.
Table S8. Peak area ratios for analytes measured above LLOQ in human PBMCs.
Table S9. Variation of analytes measured in human PBMCs.
Table S10. Characterization of monoclonal antibodies in Western blot.
Fig. S1. Response curves for immuno-MRM assays.
Fig. S2. Images of Western blots from screening monoclonal antibodies.