ABSTRACT
Factors shaping the human intestinal microbiota range from environmental influences, like smoking and exercise, over dietary patterns and disease to the host's genetic variation. Recently, we could show in a microbiome genome-wide association study (mGWAS) targeting genetic variation influencing the β diversity of gut microbial communities, that approximately 10% of the overall gut microbiome variation can be explained by host genetics. Here, we report on the application of a new method for genotype-β-diversity association testing, the distance-based F (DBF) test. With this we identified 4 loci with genome-wide significant associations, harboring the genes CBEP4, SLC9A8, TNFSF4, and SP140, respectively. Our findings highlight the utility of the high-performance DBF test in β diversity GWAS and emphasize the important role of host genetics and immunity in shaping the human intestinal microbiota.
KEYWORDS: β diversity, GWAS, human gut microbiota, immunity, IBD
Introduction
The human gut microbiota as an important focus of medical research within the past few years, has been investigated in the context of numerous inflammatory and non-inflammatory disorders of the intestine, but also in other systemic diseases, rendering gut health and the underlying host-microbiota interactions as a key component of well-being. While changes in α- and β diversity, as well as changes in the presence or absence and the abundance of specific microbial taxa have been shown to be associated with numerous diseases, the processes and factors shaping a ‘healthy’ gut microbiota are still largely understudied. First studies could show connections between host genotypes and changes in the abundance of specific taxa. These studies were either rather underpowered, investigating only roughly one hundred individuals,1,2 or based on candidate genes to reduce multiple testing burden.3,4
An analysis approach, focusing on host-genetic influences on β diversity using the microbiomeGWAS framework,5 which uses linear models to correlate genotype distance data with pairwise β diversity data, correcting for skewness and kurtosis of the results, identified 2 loci on chromosome 9 and chromosome 4 to be associated with variation in weighted UniFrac distance and Bray-Curtis dissimilarity, respectively.4
Recently, we estimated in a host-microbiome genome-wide association study (mGWAS), linking β diversity to host genetic variation, that roughly 10% of the variation in the gut microbiota is explained by the host's genetic architecture (model with 42 loci) in a Northern German study population.6 This proportion of explained variation has about the same order of magnitude as the proportion explained by non-genetic factors (such as dietary and lifestyle factors) described elsewhere.7,8 Additionally, we could show correlations of serum bile- and fatty acids with the abundance of microbial traits. Especially variants in the gene encoding for the transcription factor Vitamin D Receptor (VDR), among whose ligands are also bile acids, were found to play an essential role in shaping of intestinal communities.6
Here, we present the application of an alternative analytical approach for the investigation of β diversity host-genomic associations with shaping the gut microbiota, which does not rely on extensive permutations, thus massively reducing the computational burden, while exhibiting high concordance with comparable permutation-based approaches.
Our findings highlight the role of the host's immune functions and signaling in the assembly and homeostasis of gut-associated microbial communities in humans. In addition, our identified loci are located near known inflammatory bowel disease (IBD) genetic susceptibility loci, previously identified through case-control GWAS, implicating the host-microbiome interplay in IBD disease etiology.
Approximate inference of null distribution as an alternative to extensive permutative tests in β diversity GWAS
Permutative distance-based analysis of variance,9 as implemented in the adonis function of the vegan package10 for R,11 is an widely used approach to investigate differences in β diversity based on categorical variables. However, approaches relying on permutation are slow regarding computation time, and thus, not applicable to large data sets comprising several hundreds of samples and millions of genetic variants. The method of moment matching tries to overcome these problems by approximating an unknown null distribution based on known distributions. In this case a Pearson Type III distribution, and parameters estimated from the data itself,12 provide the opportunity to analyze large data sets in a GWAS setting comparably fast using this distance-based F test (DBF test). The Pearson Type III distribution was chosen as its properties as a 3-parameter Gamma distribution makes modeling of a multitude of other distributions possible, using its first 3 moments calculated from the data: mean, variance and skewness. While the DFB test has been shown to be applicable to different types of data sets and distance measures,12,13 it has not been used in large-scale studies investigating factors shaping microbial communities. We applied this method on β diversity data represented as Bray-Curtis dissimilarity on genus level abundance data, in analogy to the input data used in our previous publication.6 The genotype information used was the same as described in the previously published article.6 The data set consisted of 2 independent cohorts, PopGen and FoCus, from Northern Germany, comprising 830 and 937 individuals, respectively, and 1767 individuals in total. To account for influences of nutrition and anthropometrics, the Bray-Curtis dissimilarity was corrected for the covariates total energy intake, alcohol consumption, and water intake, as well as age, gender, and body mass index, respectively. Furthermore, β diversity data was corrected for variation in the first 3 genetic principal components. This was done fitting a distance-based Redundancy Analysis9 (capscale function of the vegan package10 for R11) using the aforementioned covariates as constraints. The residual variation of this model was subsequently used as distance matrix in the DBF-test. The DBF-test was performed in R11 using the snpStats package14 to import genotype data in plink format15 and applying the DBF.test function imported from the R source code file accompanying the original article describing the DBF test (https://wwwf.imperial.ac.uk/∼gmontana/software/dbf/dbf_test.R).12 To ensure the detection of robust signals and to account for the different sample sizes, a meta-analysis was performed only using genotype-information overlapping in both cohorts and using a weighted Z-score based test.16 Association results were classified as “significant,” if the meta-analysis P-value passed the genome-wide significance threshold of P < 5 × 10−8 in the meta-analysis, and both cohorts displayed a significant P-value (P < 0.05).
Genes involved in host-immunity are associated with shifts in β diversity
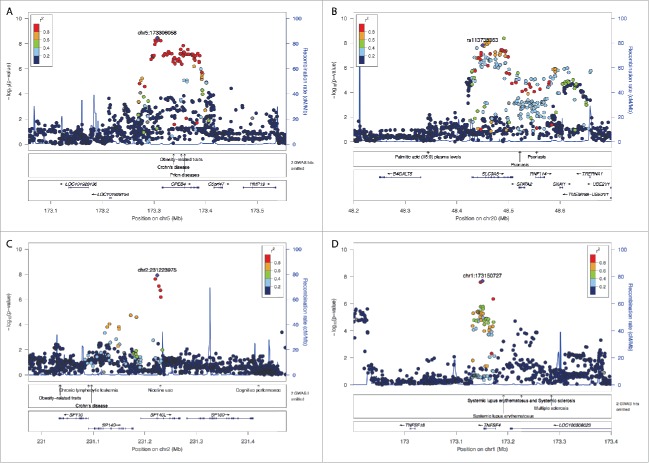
Using the afore-mentioned significance criteria, 4 loci were found as significantly associated with variation in β diversity in the meta-analysis. The locus with the strongest signal is located on chromosome 5 (rs67909753; chr5:173306058; Pmeta = 3.61 × 10−9; Fig. 1A in strong LD with the CPEB4 gene (Cytoplasmic Polyadenylation Element Binding Protein 4). CPEB4 is an effector by which RORγt, a key determinant in the cell differentiation of Th17 cells, inhibits proliferation of thymocytes.17 One variant at this locus (rs7705502; R2LeadSNP = 0.928) has previously been reported to be associated with Crohn's disease18,19 and obesity-related traits.20 The second signal is located on chromosome 20 (rs113738363; chr20:48449631; Pmeta = 1.54 × 10−8; Fig. 1B). A variant at this locus in strong linkage disequilibrium with the lead SNP (rs4809760; R2 = 0.765) has been identified in our previous mGWAS6 and is located in an intronic area of the SLC9A8 gene, encoding for NHE8 (cation proton antiporter 8). This protein is expressed in goblet cells in the intestine21 and is known to be essential for mucosal integrity, with loss of expression leading to increased bacterial adhesion and inflammation in mice following dextran sodium sulfate (DSS) treatment.22 Additionally, this locus was previously found to be associated with psoriasis,23-25 a chronic disorder of the skin with proposed links to the intestinal microbiota.26
Figure 1.
Regional association plots of the β diversity meta-analysis. (A) TNFSF4/OX40L, Chromosome 1: 173Mb-173.4Mb, Pmeta = 2.1 × 10−8; (B) SP140 and SP140L, Chromosome 2: 231Mb-231.4Mb, Pmeta = 1.19 × 10−8; (C) CBEP4, Chromosome 5: 173.1Mb-173.5Mb, Pmeta = 3.61 × 10−9; (D) SLC9A8/NHE8, Chromosome 20: 48.2Mb-48.7Mb, Pmeta = 1.54 × 10−8.
Our third hit is located on chromosome 2 (rs11678791; chr2:231223975; Pmeta = 1.19 × 10−8; Fig. 1C) harboring the SP140 Nuclear Body Protein and the SP140L genes. This locus was previously associated with Crohn's disease19 and SP140L is a key regulator of the macrophage transcriptional program, whose depletion leads to a severely impaired microbe-induced activation.27 The fourth and last association finding is located on chromosome 1 (rs11811788; chr1:173150727; Pmeta = 2.1 × 10−8; Fig. 1D). This locus harbors the TNFSF4 (OX40L; CD252) gene that is located 2.1 kbp downstream of rs11811788. The OX40-OX40L signaling pathway has been shown to regulate cytokines in T-cells, antigen-presenting cells (APCs), NK cells and NKT cells, thus plays a central role in inflammation.28
Permutation-based analysis
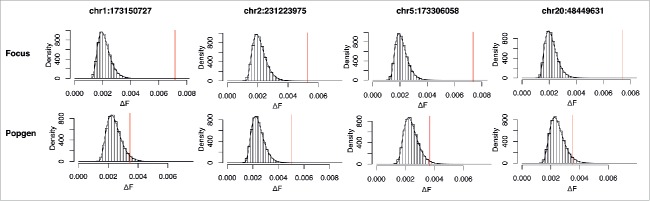
To confirm the validity of the signals, permutation based testing was performed for the 4 variants identified as genome-wide significant in the analysis based on approximate inference. Using the adonis function from the vegan10 package for R11 and 106 random permutations of the genotypes, the ΔF distribution was determined empirically. Comparing P-Values from DBF test and permutation based test, we see a large congruency of the results (Table 1). We could not find any systematic deviations exhibited by the permutation-free method, as all P-values are in the same order of magnitude as those obtained from a classical and widely used permutational approach (Table 1). This is also made evident by the good concordance of the empirical distribution with the approximated probability density function obtained from the DBF test for each of the respective variants under investigation (Fig. 2). While 106 permutations only allow to calculate P-values larger than 10−6, all variants with P-values below this threshold in the DBF test showed no permutations with stronger signals than the actual genotype.
Table 1.
Comparison of DBF test based [P(DBF)] and permutation based analysis [P(Perm)] of the 4 variants showing significant associations to changes in β diversity in 2 independent Northern-German cohorts. In the case that none of the permutations resulted in a larger ΔF than the actual genotype, P(Perm) is set to <10−6. Positions are given as chromosome and position (chr:pos) and are based on the hg19 version of the human genome annotation.
| Focus |
Popgen |
Meta | ||||||
|---|---|---|---|---|---|---|---|---|
| rsID | chr:pos | ΔF | P(DBF) | P(Perm) | ΔF | P(DBF) | P(Perm) | P(meta) |
| rs11811788 | chr1:173150727 | 0.0071569 | 1.08 × 10−8 | < 10−6 | 0.0034576 | 0.035664 | 0.033779 | 2.10 × 10−8 |
| rs11678791 | chr2:231223975 | 0.0052987 | 1.50 × 10−5 | 2.5 × 10−5 | 0.0050288 | 0.00019994 | 0.000234 | 1.19 × 10−8 |
| rs67909753 | chr5:173306058 | 0.0073541 | 4.10 × 10−9 | < 10−6 | 0.0036817 | 0.01813936 | 0.017608 | 1.45 × 10−8 |
| rs113738363 | chr20:48449631 | 0.0073984 | 5.82 × 10−9 | < 10−6 | 0.0035011 | 0.03922766 | 0.037279 | 1.54 × 10−8 |
Figure 2.
Comparison of the empirical distribution of ΔF from 106 permutations of each of the 4 variants in both cohorts with probability density function approximated by using moment matching to Pearson Type III distribution. Red lines indicate the ΔF of the actual genotype distribution in the cohorts.
Replication of 42 loci identified in mGWAS
The boundaries of the loci provided in Table 1 in Wang et al.6 were evaluated for their replicability using the DBF test. The major difference between both approaches is that the DBF test is based directly on the β diversity matrix, while the previously published approach is based on the ordination of this distance matrix. For 41 of the 42 loci we obtained a nominally significant P-value (P < 0.05) at the exact respective position of the lead SNPs. As mentioned earlier, the SLC9A8 locus on chromosome 20 shows a genome-wide significant association in both analysis strategies (see Table 2). Three more of the lead SNPs showing significant associations in the original article have P-values <10−5, and another 5 loci reached this threshold when considering SNPs in the neighborhood – using physical boundaries obtained from the DEPICT analysis – of the lead SNP of the original analysis (see Table 2). Among these loci is one that spans the BANK1 (B-Cell Scaffold Protein With Ankyrin Repeats 1; chr4:102901822) gene, which was previously reported to be associated with IBD19 and which is in line with the reported loci reaching genome-wide significance. One locus on chromosome 8 (rs138022915; chr8:19885934) covers the LPL (Lipoprotein Lipase) gene. Gene expression of LPL was shown to be influenced by the microbiota through altered expression of fasting-induced adipose factor (Fiaf) in mice. The only lead SNP not exhibiting a significant P-value < 0.05 is the variant rs225153 (chr11:8853177), however, within the only 0.94 kb spanning locus another variant reaches at least nominal statistical significance (chr11:8852400; Pmeta = 2.38 × 10−2).
Table 2.
Replication of the 42 genome-wide significant loci previously found to be associated with β diversity. We modified Table 1 from Wang et al.6 as follows: Lead SNP corresponds to position and P-value from the Meta-Analysis of the DBF-test applied to the Popgen and FoCus cohorts. Best in Locus: Position and lowest P-value of the DBF test meta-analysis in the locus defined by the columns ‘Locus Start’ and ‘Locus End’. Positions are based on the hg19 version of the human genome annotation. P-values in bold font indicate a value below of P < 0.05. Additional italic fonts indicate P-values < 10−5.
| Effect size | Lead SNP | Best in Locus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP_ID | Chr | A1 | A2 | Locus Start | Locus End | Nearest Gene | Genes in Locus | Wang et al. | Position | P(Meta DBF) | Position | P(Meta DBF) |
| rs804427 | 1 | A | C | 33538964 | 33623510 | AK2 | ADC; TRIM62; AK2 | 0.79% | chr1:33538964 | 4.96 × 10−3 | chr1:33595212 | 2.09 × 10−3 |
| rs1288616 | 1 | G | A | 53885577 | 53965248 | DMRTB1 | DMRTB1 | 0.76% | chr1:53952777 | 1.58 × 10−4 | chr1:53946485 | 9.29 × 10−5 |
| rs1102737 | 1 | G | A | 172700868 | 172779833 | FASLG | 0.66% | chr1:172777616 | 2.24 × 10−3 | chr1:172747021 | 1.97 × 10−3 | |
| rs72853661 | 2 | T | C | 25323083 | 25453968 | POMC | POMC; EFR3B | 0.79% | chr2:25439262 | 6.57 × 10−5 | chr2:25439758 | 1.42 × 10−6 |
| rs7567349 | 2 | A | G | 61384324 | 61853037 | XPO1 | AHSA2; USP34;XPO1; KIAA1841 | 0.76% | chr2:61839853 | 1.49 × 10−6 | chr2:61486628 | 2.22 × 10−7 |
| rs2010917 | 2 | T | C | 135172338 | 135197891 | MGAT5 | MGAT5 | 0.74% | chr2:135194856 | 5.04 × 10−5 | chr2:135183686 | 6.13 × 10−6 |
| rs71415332 | 2 | G | A | 102309520 | 102616128 | — | IL1R2; MAP4K4 | 0.68% | chr2:102499952 | 2.56 × 10−5 | chr2:102529630 | 2.85 × 10−6 |
| rs4670302 | 2 | T | G | 33808725 | 34068392 | FAM98A | FAM98A | 0.92% | chr2:34068392 | 6.53 × 10−3 | chr2:34033733 | 7.43 × 10−4 |
| rs6711771 | 2 | C | G | 34339420 | 34491584 | — | — | 0.71% | chr2:34339420 | 1.77 × 10−2 | chr2:34421584 | 8.92 × 10−4 |
| rs13099587 | 3 | G | A | 146250561 | 146275555 | PLSCR1 | PLSCR1 | 0.70% | chr3:146268616 | 3.60 × 10−3 | chr3:146275555 | 1.09 × 10−3 |
| rs9647379 | 3 | G | C | 171759410 | 171833266 | FNDC3B | FNDC3B | 0.75% | chr3:171785168 | 8.98 × 10−5 | chr3:171785168 | 8.98 × 10−5 |
| rs143050036 | 3 | C | T | 49898318 | 50208819 | SEMA3F | RBM5; MST1R; CAMKV; MON1A; RBM6; SEMA3F | 0.75% | chr3:50071965 | 1.15 × 10−2 | chr3:49987475 | 1.33 × 10−5 |
| rs60500975 | 4 | A | T | 102769693 | 102929034 | — | BANK1 | 0.82% | chr4:102901822 | 2.03 × 10−6 | chr4:102885147 | 1.67 × 10−6 |
| rs62367773 | 5 | A | G | 74171398 | 74220999 | FAM169A | 0.67% | chr5:74179975 | 1.55 × 10−4 | chr5:74193565 | 6.08 × 10−5 | |
| rs1292672 | 6 | C | T | 87217958 | 87509434 | HTR1E | 0.70% | chr6:87432577 | 9.91 × 10−5 | chr6:87242812 | 4.85 × 10−5 | |
| rs35148810 | 7 | C | T | 151515842 | 151530983 | — | PRKAG2 | 0.83% | chr7:151520485 | 8.69 × 10−4 | chr7:151520550 | 3.77 × 10−4 |
| rs12705241 | 7 | A | C | 104219681 | 104381102 | — | LHFPL3 | 0.76% | chr7:104258313 | 2.01 × 10−3 | chr7:104258313 | 2.01 × 10−3 |
| rs13260600 | 8 | C | T | 3705807 | 3713004 | CSMD1 | CSMD1 | 0.77% | chr8:3705807 | 8.45 × 10−4 | chr8:3705807 | 8.45 × 10−4 |
| rs138022915 | 8 | T | C | 19815256 | 19939049 | LPL | LPL | 0.73% | chr8:19885934 | 2.19 × 10−4 | chr8:19876234 | 4.45 × 10−6 |
| rs11986935 | 8 | T | A | 10576753 | 10732050 | SOX7 | SOX7; PINX1 | 0.97% | chr8:10691549 | 1.05 × 10−5 | chr8:10695125 | 6.63 × 10−6 |
| rs7818750 | 8 | G | A | 135273640 | 135299611 | ZFAT | 0.74% | chr8:135274269 | 1.83 × 10−3 | chr8:135273640 | 4.42 × 10−4 | |
| rs1325919 | 9 | C | T | 37626956 | 37650386 | FRMPD1 | 0.67% | chr9:37642802 | 4.34 × 10−3 | chr9:37638047 | 1.93 × 10−3 | |
| rs7082134 | 10 | A | G | 87865009 | 87884110 | GRID1 | GRID1 | 0.84% | chr10:87865009 | 4.05 × 10−4 | chr10:87884110 | 3.71 × 10−4 |
| rs2251536 | 11 | G | C | 8852239 | 8853177 | — | ST5 | 0.76% | chr11:8853177 | 1.57 × 10−1 | chr11:8852400 | 2.38 × 10−2 |
| rs4472950 | 11 | C | T | 120798714 | 120853675 | — | GRIK4 | 0.69% | chr11:120807892 | 4.56 × 10−4 | chr11:120798714 | 3.16 × 10−4 |
| rs7974353 | 12 | T | C | 48256280 | 48270596 | — | VDR | 0.75% | chr12:48269798 | 4.69 × 10−3 | chr12:48263162 | 1.22 × 10−3 |
| rs4760399 | 12 | T | C | 93011759 | 93081307 | C12orf74 | 0.67% | chr12:93047282 | 1.80 × 10−2 | chr12:93021626 | 2.30 × 10−3 | |
| rs6573564 | 14 | T | A | 65119676 | 65157187 | PLEKHG3 | 0.73% | chr14:65142395 | 1.72 × 10−5 | chr14:65141759 | 1.72 × 10−5 | |
| rs12910631 | 15 | G | T | 26603288 | 26622999 | — | 0.79% | chr15:26606605 | 1.42 × 10−4 | chr15:26606605 | 1.42 × 10−4 | |
| rs8040493 | 15 | T | G | 101414167 | 101418682 | — | 0.65% | chr15:101414659 | 6.27 × 10−4 | chr15:101418335 | 5.03 × 10−5 | |
| rs293377 | 15 | G | C | 89623490 | 89635268 | ABHD2 | ABHD2 | 0.70% | chr15:89634414 | 3.83 × 10−3 | chr15:89623490 | 1.89 × 10−3 |
| rs8055365 | 16 | T | C | 84566729 | 84581275 | KIAA1609 | KIAA1609 | 0.70% | chr16:84580531 | 8.98 × 10−5 | chr16:84580531 | 8.98 × 10−5 |
| rs59986499 | 16 | G | A | 3065924 | 3097940 | CLDN6 | MMP25; TNFRSF12A; CLDN6; CCDC64B; HCFC1R1; THOC6 | 0.68% | chr16:3069752 | 8.73 × 10−3 | chr16:3082157 | 6.93 × 10−3 |
| rs12931878 | 16 | A | G | 11031741 | 11207817 | CLEC16A | DEXI; CLEC16A | 0.65% | chr16:11042194 | 2.02 × 10−3 | chr16:11082874 | 9.66 × 10−5 |
| rs62085746 | 17 | T | C | 66166300 | 66213540 | AMZ2 | 0.69% | chr17:66196145 | 2.04 × 10−3 | chr17:66196145 | 2.04 × 10−3 | |
| rs16969051 | 17 | C | T | 32248813 | 32258877 | ACCN1 | ACCN1 | 0.65% | chr17:32258877 | 5.12 × 10−4 | chr17:32258877 | 5.12 × 10−4 |
| rs12601692 | 17 | A | G | 782416 | 794333 | — | NXN | 0.68% | chr17:782416 | 1.57 × 10−2 | chr17:782416 | 1.57 × 10−2 |
| rs2267922 | 19 | C | G | 18217350 | 18289634 | IFI30 | MAST3; IFI30;PIK3R2 | 0.77% | chr19:18278766 | 3.32 × 10−7 | chr19:18278766 | 3.32 × 10−7 |
| rs273647 | 19 | C | G | 51739767 | 51766748 | C19orf75 | CD33; C19orf75 | 0.84% | chr19:51751858 | 1.38 × 10−3 | chr19:51766748 | 1.97 × 10−5 |
| rs4809760 | 20 | A | G | 48428863 | 48591125 | SLC9A8 | RNF114; SLC9A8; SPATA2 | 0.85% | chr20:48454671 | 9.28 × 10−9 | chr20:48490801 | 4.15 × 10−9 |
| rs2835692 | 21 | A | G | 38657572 | 38704886 | DSCR3 | 0.68% | chr21:38670335 | 2.11 × 10−4 | chr21:38657572 | 1.13 × 10−4 | |
| rs9917541 | 22 | C | A | 31520338 | 31531133 | PLA2G3 | PLA2G3; INPP5J | 0.71% | chr22:31529043 | 1.30 × 10−2 | chr22:31529043 | 1.30 × 10−2 |
Discussion
The effect of host-genetic variation on the complex phenotype of β diversity of the intestinal microbiota is still largely unknown. We could show, that our adapted method is applicable to microbiome data and yields results in line with classical permutation approaches, without the need of doing millions of permutations per variant, as at least 2 × 107 permutations would be needed to approach the threshold of genome-wide significance. For a typical data set of several millions of imputed genetic variants, this number would easily exceed 1014 necessary permutations.
By applying this new method, the DBF test, to β diversity data of 2 independent Northern German cohorts, consisting of a total of almost 1,800 individuals, we could show that variants in genes primarily involved in immune related functions and inflammatory processes showed an association with changes in the gut microbial community. While all for loci are sensible targets with respect to the interactions between host and associated microbes, especially the SLC9A8/NHE8 gene locus is an intriguing candidate for future studies. This is due to its high expression in goblet cells,17 its crucial role for mucosal integrity22 and its potential role in selective bacterial adherence.29
The association signal in the TNFSF4 locus and its role in regulation of cytokines is in line with recent findings underlining the links of the gut microbiota to cytokine production.30
Furthermore, 3 of the 4 loci found in our re-analysis are also known to be overlapping with loci associated to different kinds of chronic inflammatory disorders, namely Crohn's disease and psoriasis. Especially for Crohn's disease it was proposed, that host-microbe interactions were, and probably are, a driving factor in the manifestation of the disorder.18 Moreover, it was shown, that loci associated with Crohn's disease and psoriasis are overlapping to a certain extent31 and comorbidities of the 2 diseases are widely reported.32
Our findings emphasize the role of gut microbes as potential triggers of these diseases, and possibly additional chronic disorders.
The observed differences in significance of the results highlight the difficulties and challenges accompanying mbQTL (microbiome quantitative trait) association analyses of, for example, microbial diversity in connection to host-genetics. The ordination-based analysis described in Wang et al.6 reduces the dimensions of the high-dimensional data to principal coordinates, which has the benefit of removing stochastic noises and pathways with relatively smaller contributions, and reveals the most important pathways affecting the major variable patterns of microbial β diversity, in this case, vitamin-related pathways and bile-acid related genes centered by VDR. However, variation not necessarily displayed by the 2 major axes of the ordination might not be detected by this method. Thus, the DBF test serves as an addition to the previously published results on the connection between β diversity and host-genetics, strengthening especially the importance of those loci exhibiting strong to intermediate results in both analyses.
However, while these results are intriguing, they should mainly serve as a starting point and perspective for subsequent analyses in larger and hence better powered cohorts, investigating the genetic effects of host-microbiota interactions, leading to additional and potentially more robust signals for the complex trait of β diversity, overcoming the challenges of small effect sizes, sensitivity to technical differences and confounding environmental factors. In a recent review, Zhernakova and colleagues further discuss the phenomenon that there is little overlap in the findings between all the mbQTL studies with more than 1000 samples analyzed published so far, likely because there were many significant differences between the data sets and methods that were used.33 In summary, classical GWAS methodology cannot be used for mbQTL studies, given the complexity of the trait under study, and the development of best-practice workflows and stringent thresholds are in its infancy. As shown in this study, the DBF test deserves a careful consideration for future studies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the German Research Foundation (DFG) Collaborative Research Center (CRC) 1182, “Origin and Function of Metaorganisms” and the DFG Excellence Cluster 306, “Inflammation at Interfaces,” and the German Federal Ministry of Education and Research (BMBF) project CP3 in “SysINFLAME.”
References
- [1].Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, et al.. Host genetic variation impacts microbiome composition across human body sites. Genome Biology. 2015; 16(1):191. PMID:26374288. doi: 10.1186/s13059-015-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. Plos One. 2015; 10(11):e0140301. PMID:26528553. doi: 10.1371/journal.pone.0140301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al.. Human genetics shape the gut microbiome. Cell. 2014; 159(4):789-99. PMID:25417156. doi: 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host & Microbe. 2016; 19(5):731-43. doi: 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hua X, Song L, Yu G, Goedert JJ, Abnet CC, Landi MT, Shi J. MicorbiomeGWAS: a tool for identifying host genetic variants associated with microbiome composition. BioRxiv 2015; doi: 10.1101/031187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen F-A, Rühlemann MC, Szymczak S, et al.. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genetics. 2016; 48(11):1396-406. PMID:27723756. doi: 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al.. Population-level analysis of gut microbiome variation. Science. 2016; 352(6285):560-4. PMID:27126039. doi: 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- [8].Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila A V, Falony G, Vieira-Silva S, et al.. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016; 352(6285):565-9. PMID:27126040. doi: 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001; 82(1):290-7. doi: 10.1890/0012-9658(2001)082%5b0290:FMMTCD%5d2.0.CO;2 [DOI] [Google Scholar]
- [10].Oksanen J, Blanchet F, Kindt R, Legendre P, Minchin P, O'Hara R, Simpson G, Solymos P, Stevens M, Wagner H. vegan: community ecology package. R package version 2.0-10. R package version 2013; 1 [Google Scholar]
- [11].R Development Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna Austria; 2016; 0:{ISBN} 3-900051-07-0 [Google Scholar]
- [12].Minas C, Montana G. Distance-based analysis of variance: Approximate inference. Statistical Analysis and Data Mining: The ASA Data Sci J. 2014; 7(6):450-70. doi: 10.1002/sam.11227 [DOI] [Google Scholar]
- [13].Winkler AM, Ridgway GR, Douaud G, Nichols TE, Smith SM. Faster permutation inference in brain imaging. NeuroImage. 2016; 141:502-16. PMID:27288322. doi: 10.1016/j.neuroimage.2016.05.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clayton D. snpStats: SnpMatrix and XSnpMatrix classes and methods. R package version 1.26.0 2015 [Google Scholar]
- [15].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J, Sklar P, de Bakker P, Daly MJ, et al.. PLINK: a tool set for whole-genome and population-based linkage analyses. Am J Hum Genetics. 2007; 81:559-75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010; 26(17):2190-1. PMID:20616382. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006; 24(6):813-26. PMID:16782036. doi: 10.1016/j.immuni.2006.03.023 [DOI] [PubMed] [Google Scholar]
- [18].Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al.. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491(7422):119-24. PMID:23128233. doi: 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al.. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics. 2015; 47(9):979-86. PMID:26192919. doi: 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al.. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015; 518(7538):187-96. PMID:25673412. doi:20953189 10.1038/nature14132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu H, Li Q, Zhao Y, Li J, Ghishan FK. Intestinal NHE8 is highly expressed in goblet cells and its expression is subject to TNF-α regulation. Am J Physiol Gastrointestinal Liver Physiol. 2016; 310(2):G64-9. doi: 10.1152/ajpgi.00367.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang A, Li J, Zhao Y, Johansson MEV, Xu H, Ghishan FK. Loss of NHE8 expression impairs intestinal mucosal integrity. Am J Physiol Gastrointestinal Liver Physiol. 2015; 309(11):G855-64, ajpgi.00278.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Capon F, Bijlmakers M-J, Wolf N, Quaranta M, Huffmeier U, Allen M, Timms K, Abkevich V, Gutin A, Smith R, et al.. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum Mol Genetics. 2008; 17(13):1938-45. doi: 10.1093/hmg/ddn091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stuart PE, Nair RP, Ellinghaus E, Ding J, Tejasvi T, Gudjonsson JE, Li Y, Weidinger S, Eberlein B, Gieger C, et al.. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genetics. 2010; 42(11):1000-4. PMID:20953189. doi: 10.1038/ng.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Baurecht H, Hotze M, Brand S, Büning C, Cormican P, Corvin A, Ellinghaus D, Ellinghaus E, Esparza-Gordillo J, Fölster-Holst R, et al.. Genome-wide comparative analysis of atopic dermatitis and Psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genetics. 2015; 96(1):104-20. doi: 10.1016/j.ajhg.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, et al.. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015; 67(1):128-39. doi: 10.1002/art.38892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mehta S, Cronkite DA, Basavappa M, Saunders TL, Adiliaghdam F, Amatullah H, Morrison SA, Pagan JD, Anthony RM, Tonnerre P, et al.. Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140 Sci Immunol. 2017; 2(9). doi: 10.1126/sciimmunol.aag3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev. 2009; 229(1):173-91. PMID:19426222. doi: 10.1111/j.1600-065X.2009.00766.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu C, Xu H, Zhang B, Johansson ME V, Li J, Hansson GC, Ghishan FK. NHE8 plays an important role in mucosal protection via its effect on bacterial adhesion. AJP: Cell Physiol. 2013; 305(1):C121-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Horst R ter, Jansen T, Jacobs L, Bonder MJ, et al.. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016; 167(7):1897. PMID:27984736. doi: 10.1016/j.cell.2016.11.046 [DOI] [PubMed] [Google Scholar]
- [31].Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hübenthal M, et al.. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nature Genetics. 2016; 48(5):510-8. PMID:26974007. doi: 10.1038/ng.3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee FI, Bellary S V, Francis C. Increased occurrence of psoriasis in patients with Crohn's disease and their relatives. Am J Gastroenterol. 1990; 85(8):962-3. PMID:2375323 [PubMed] [Google Scholar]
- [33].Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 2017. [Epub ahead of print] PMID:28669638. doi: 10.1016/j.it.2017.06.003 [DOI] [PubMed] [Google Scholar]