Abstract
DNA sequence analysis of the bt2-7503 mutant allele of the maize brittle-2 gene revealed a point mutation in the 5′ terminal sequence of intron 3 changing GT to AT. This lesion completely abolishes use of this splice site, activates two cryptic splice sites, and alters the splicing pattern from extant splice sites. One activated donor site, located nine nt 5′ to the normal splice donor site, begins with the dinucleotide GC. While non-consensus, this sequence still permits both trans-esterification reactions of pre-mRNA splicing. A second cryptic site located 23 nt 5′ to the normal splice site and beginning with GA, undergoes the first trans-esterification reaction leading to lariat formation, but lacks the ability to participate in the second reaction. Accumulation of this splicing intermediate and use of an innovative reverse transcriptase-polymerase chain reaction technique (J. Vogel, R.H. Wolfgang, T. Borner [1997] Nucleic Acids Res 25: 2030–2031) led to the identification of 3′ intron sequences needed for lariat formation. In most splicing reactions, neither cryptic site is recognized. Most mature transcripts include intron 3, while the second most frequent class lacks exon 3. Traditionally, the former class of transcripts is taken as evidence for the intron definition of splicing, while the latter class has given credence to the exon definition of splicing.
The accurate removal of introns from the primary transcript is a fundamental process essential for the expression of eukaryotic genes. It is a two-step trans-esterification reaction. The first reaction involves cleavage of the 5′ terminal nucleotide of the intron with subsequent covalent linkage to an adenosine at the branch point within the 3′ portion of the intron. This results in formation of the so-called lariat structure. The second step involves cleavage at the 3′ intron splice site, release of the intron lariat, and ligation of the two adjacent exons. The lariat is then quickly de-branched and degraded (Moore and Sharp, 1993; Brown, 1996; Simpson and Filipowicz, 1996). This dynamic and complex process is carried out in association with a large ribonucleosome protein complex termed a spliceosome (for review, see Moore et al., 1993; Sharp, 1994).
Although introns are ubiquitous and share a high degree of structural/sequence similarity across species, the signals that specifically define splice sites are not completely understood. Some conserved but short terminal sequences within introns function in intron splicing. Virtually all introns begin with the dinucleotide GU and end with AG (Green, 1991; Moore et al., 1993). In yeast, introns possess a highly conserved branch point sequence UACUAAC 10 to 50 nt upstream of the 3′ splice site. This pairs with the U2 snRNP and plays an integral role in recognizing 3′ splice sites (Parker et al., 1987). In contrast, vertebrates possess a less-conserved branch point sequence, CURAC, located 18 to 40 nt upstream of the 3′ splice site (Green, 1991). This also pairs with U2 snRNP (Wu and Manley, 1989; Zhuang and Weiner, 1989). In addition, vertebrate introns possess a unique, 10- to 15-nt polypyrimidine tract located near the 3′ end that interacts with splicing factor U2AF during early spliceosome assembly. This aids the binding of U2 snRNP to the branch site (Ruskin et al., 1988).
The lack of an in vitro system capable of efficiently splicing plant introns has considerably hampered studies of plant pre-mRNA splicing (for review, see Simpson and Filipowicz, 1996; Brown and Simpson, 1998). Despite similarities in sequence and in the splicing process, animal introns are not efficiently spliced in plant cells and vice versa (Barta et al., 1986; van Santen and Spritz, 1987; Pautot et al., 1989). Splicing differences also distinguish monocots and dicots. A series of studies suggests that the monocot splicing machinery is more flexible or more complex, since dicot introns are efficiently spliced in monocots, whereas at least some monocot introns are not spliced in dicots (Keith and Chua, 1986; Goodall and Filipowicz, 1991). Splicing within dicots may in fact be species dependent, since Arabidopsis and tobacco apparently differ in the splicing of transcripts arising from a transgenic maize transposable element, Ac (Jarvis et al., 1997; Martin et al., 1997; for review, see Brown and Simpson, 1998).
Particular structural and sequence features distinguish plant introns from those of vertebrates and yeast. Plant introns lack the conserved branch point sequence of yeast and the 3′ polypyrimidine tract typical of vertebrate introns (Goodall and Filipowicz, 1991; Luehrsen and Walbot, 1994). The high A+U content of many plant introns likely plays a role in splicing, possibly through defining intron/exon junctions (Lou et al., 1993; McCullough et al., 1993; Carle-Urioste et al., 1994; Luehrsen and Walbot, 1994). However, some monocot introns are GC rich and, hence, the requirement for AU richness may be more stringent in dicots (Goodall and Filipowicz, 1989, 1991).
Identification of the branch point has also been problematic in plants. While branch point consensus motifs as defined in vertebrates can be found in the majority of plant introns (Brown, 1986; Simpson et al., 1996), an AU-rich synthetic plant intron devoid of a branch point sequence or a polypyrimidine tract is efficiently processed in tobacco protoplasts (Goodall and Filipowicz, 1989, 1991). However, Simpson et al. (1996) reported that mutation of the adenosine residue within the putative branch point dramatically decreased the splicing efficiency of several introns of both monocot and dicot origin when expressed in tobacco protoplasts. Only recently has the mapping of a branch point nucleotide and direct evidence for lariat formation in plants appeared (Liu and Filipowicz, 1996).
Another area of uncertainty concerns the initial recognition of exon/intron borders. Two models exist. One model is the exon definition for splice site selection, which argues that the interaction between factors bound to the 5′ and 3′ ends of one exon determines the splice site selection (Berget, 1995). A prediction of this model is that an alteration at either the 5′ or the 3′ splice site of an exon can disturb these interactions, resulting in blocked splicing at both sites. As a result, the splicing mechanism removes the two adjacent introns and the mutant exon in one splicing reaction (Antoniou, 1995). This gives rise to the absence of the exon in the mature transcript (“exon skipping”). The contrasting model is the intron definition of splicing. Here initial recognition involves sequences within introns. Once the initial recognition takes place, sliding occurs in both directions and sites competent for splicing are used in the processing event. A prediction of this model is that mutational removal of a wild-type splice site will sometimes activate normally silent or cryptic splice sites in close proximity to the mutant. This results in mature transcripts lacking exon termini or containing intron termini. If such cryptic splice sites are not activated, the entire intron is found in the mature transcript, giving rise to intron inclusion.
While most of the early studies of splicing mutants in vertebrates favored the exon definition of splicing (mutants exhibited exon skipping, as reviewed by Berget [1995]), initial reports from plants favored the intron definition of splicing (Goodall and Filipowicz, 1989; Lou et al., 1993; McCullough et al., 1993). One factor possibly important in explaining this difference concerns the relative size of exons and introns. Vertebrate introns can be quite large compared with adjacent exons. In contrast, plant introns are shorter relative to their counterparts in vertebrates and are comparable in size to their adjacent exons. If size is important, then mutants exhibiting exon skipping should be found in plants. In accordance, exon skipping has been demonstrated recently in a number of Arabidopsis mutants (Brown, 1996; Simpson et al., 1998; for review, see Brown and Simpson, 1998) and in one maize mutant (Lal et al., 1999).
The brittle-2 (Bt2) locus in maize encodes the small subunit of endosperm ADP-Glc pyrophosphorylase, a key regulatory heterotetrameric enzyme in the starch biosynthetic pathway. Null mutants dramatically reduce the starch content of the mature kernel, causing a collapsed, shrunken, or brittle phenotype (Hannah and Nelson, 1976). Northern analysis of several bt2 mutants showed that they produce multiple or non-wild-type-sized transcripts (Giroux and Hannah, 1994). Since Bt2 contains nine introns (J. Shaw and L.C. Hannah, unpublished data), it is likely that at least some of these mutants harbor RNA processing defects.
We have deciphered the bt2-7503 mutant and demonstrate that a single splice site mutation in bt2-7503 evokes both intron retention and exon skipping in the mutant transcript. We also note utilization of cryptic, non-canonical splice sites. One such site undergoes the first but not the second trans-esterification reaction of splicing. We took advantage of the resulting accumulation of the intron lariat in the mutant and used a novel reverse transcriptase (RT)-PCR technique to precisely map the first branch point nucleotide sequence in a monocot. This branch point bears little resemblance to branch points in other organisms including Arabidopsis.
MATERIALS AND METHODS
Plant Material
Plant material was grown and maintained in the greenhouse or in the field at the University of Florida (Gainesville). The mutant bt2-7503 (a kind gift from Dr. Oliver Nelson, University of Wisconsin, Madison) was described previously (Hannah et al., 1980; Giroux and Hannah, 1994). The wild-type Bt2 allele used in this study was originally isolated from McClintock's a1-m3 stock (Giroux et al., 1996).
RNA Isolation and Northern-Blot Analysis
Total RNA from 20-d post-pollinated kernels was isolated as described previously (McCarty, 1986). Total RNA (10 μg per lane) was resolved on 1.3% (w/v) agarose gels following standard protocols (Maniatis et al., 1982) and capillary transferred onto a Hybond N+ membrane (Amersham, Uppsala) as suggested by the manufacturer. Hybridization was performed in 0.5 m Na2HPO4 (pH 7.2), 7% (w/v) SDS, and 1% (w/v) BSA following the protocols of Church and Gilbert (1984). PCR products spanning exon 1 to 3 or exon 4 to 10 of Bt2 cDNA were labeled with [32P]dCTP using a DNA-labeling system (RTS RadPrime, Life Technologies, Cleveland). Blots were re-probed after two washes with 0.1× SSC and 1% (w/v) SDS for 15 min at 90°C, and an overnight exposure on film to ensure removal of the probe.
Branch Point Mapping
Identification of the intron 3 lariat branch point of bt2-7503 was fashioned after the protocol described for branch point determination of group II introns (Vogel et al., 1997). A RT kit (Superscript, Bethesda Research Laboratories, Gaithersburg, MD) and protocol were employed. Total RNA (5 μg) from bt2-7503 endosperm was subjected to first-strand synthesis using the primer Lo1 (5′-GGACCATTGTAGTG AGATTTTGG-3′) at 45°C for 50 min in a 20-μL reaction volume. This primer is complementary to 5′ sequence of intron 3. Following placement at 70°C for 15 min to terminate first-strand synthesis, half of the reaction (10 μL) was transferred to a fresh tube and subjected to RNase H (1.5 units, Bethesda Research Laboratories) digestion at 37°C for 20 min. RNase H (2 μL)-treated and non-treated first-strand reactions were separately subjected to PCR amplification in a 50-μL reaction containing sense Up1 primer (5′-GTCATCCTTTGT TCAAAGAG-3′) and nested antisense Lo2 primer (5′-GTCATGCAACTCTATTCCTACA-3′). A diagram of primer placement and the products synthesized is given in “Results.”
After incubation at 94°C for 5 min, 40 cycles of amplification were followed by an extension at 72°C for 8 min. Products were resolved on a 1% (w/v) agarose gel, excised, and purified on columns (Ultrafree-MC, Millipore, Bedford, MA) according to the protocol provided by the manufacturer. Fragments were directly sequenced in both directions, as well as after cloning into pBluescript KS+. Sequencing was done at the University of Florida Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Laboratory using a dye-terminating system (Prism, Applied Biosystems, Foster City, CA).
PCR Amplification of bt2-7503 Genomic Sequence
Genomic DNA was isolated from bt2-7503 leaves using reagent (Plant DNAzol, Bethesda Research Laboratories) according to the manufacturer. DNA (1 μg) was used as a template in a 50-μL reaction containing primers 7503Up (5′-GGGCACATTAATCGCTATCAAGAC-3′) and primer 7503Lo (5′-AGGTGGCA GGTGTCGAGGTTG-3′). These are complementary to the intron 2 and exon 6 sequence of Bt2 gene, respectively (J. Shaw and L.C. Hannah, unpublished data). Optimization of the PCR conditions was done using an optimization kit (Opti-Prime PCR, Stratagene, La Jolla, CA). A single fragment of approximately 2.1 kb was amplified only when the reaction contained 1 unit of adjunct (Perfect Match, Stratagene). This fragment was cloned in pBluescript KS+ and sequenced in both directions.
RT-PCR Analysis of Mutant bt2-7503
Total RNA (1 μg) from the wild type and bt2-7503 were subjected to RT-PCR amplification using primers BT375 (5′-GCAAACCATGGCTTTGGCGTCTAAAGCCTCCCCTCCGCCATG GAATGCCACCGCCG-3) and BT378 (5′-CCCCGACGTCATCATATAACTGTTCCACTAGGG-3′). These primers are complementary to 5′ and 3′ non-translated sequences of Bt2 cDNA. Products were resolved on 1% agarose gel, blotted onto a membrane, and probed with full-length Bt2 cDNA probe. The fragments were excised from the gel and sequenced in both directions.
RESULTS
The Lesion of Mutant bt2-7503 Is a GT to AT Base Change at the 5′ Splice Site of Intron 3
The mutant bt2-7503 (obtained from Dr. O. Nelson) arose spontaneously and exhibits a severe bt2 phenotype. Previous northern analysis of this mutant demonstrated that it produces an abundant transcript slightly larger than wild type, and at least one smaller transcript of much lower abundance. It does not produce a detectable BT2 protein, as judged by western analysis (Bae et al., 1990; Giroux and Hannah, 1994). Genomic Southern analysis revealed no obvious insertion (data not presented). Since Bt2 contains nine introns, we investigated the possibility that the primary lesion of the mutant might involve RNA processing. RNA from the mutant and from the wild type was subjected to RT-PCR using oligo-dT to prime first-strand synthesis. Primers that amplify the entire coding region of Bt2 were then employed for PCR. Resulting PCR products were subjected to electrophoresis, blotted, and probed with a full-length Bt2 cDNA probe.
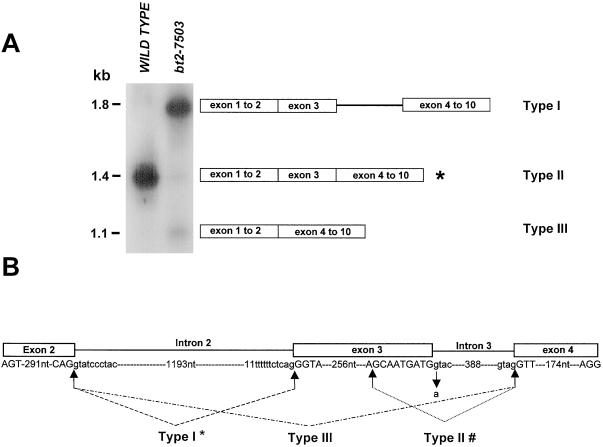
A major band of approximately 1.8 kb (type I) and two minor bands of approximately 1.4 kb (type II) and 1.1 kb (type III) were amplified from bt2-7503 RNA, whereas a single fragment of approximately 1.4 kb was amplified from the wild type (Fig. 1A). Sequencing of type I transcript showed that it retained intron 3. These sequence data also revealed a G → A alteration at the 5′ splice site of intron 3. Type II transcript lacked 9 nt normally present in the 3′ portion of exon 3. Here, splicing employed a cryptic, non-canonical donor splice site, GC, in exon 3, 9 nt upstream of the exon 3/intron3 junction and the normal acceptor site of exon 3. Type III lacked exon 3. The donor site following exon 2 and the acceptor site preceding exon 4 participated in the splicing event. A diagram of these splicing events is shown in Figure 1B.
Figure 1.
RT-PCR analysis of mutant bt2-7503 transcript. A, Total endosperm RNA from the wild type and from bt2-7503 was subjected to RT-PCR using primers spanning exons 1 to 10 of the Bt2 transcript. Products were resolved on a 1% agarose gel, blotted, and probed with the full-length Bt2 cDNA probe. Fragments from the mutant were excised and sequenced. Schematic representations of the bt2-7503 RT-PCR products are shown. *, Region missing nine nucleotides from the 3′ portion of exon 3. B, Schematic representation of the genomic sequence bearing the splice site alteration of the mutant bt2-7503. Exon and intron sequences are given in capital and small letters, respectively. The mutant splice site nucleotide in intron 3 is marked by a downward arrow. Arrows joined by lines mark the donor and acceptor site utilized during splicing. #, Intron 2 is spliced normally in type II transcript; *, intron 3 is not spliced in type I transcript.
To confirm the lesion underlying the bt2-7503 mutation, genomic DNA corresponding to the region of alterations in the transcripts—intron 2 to exon 6—was isolated via PCR and sequenced. Comparison of the mutant sequence with the wild-type Bt2 allele revealed only the point mutation of G to A at the 5′ splice site seen in the type I transcripts.
Northern Analysis of Mutant bt2-7503 Transcripts Suggests Accumulation of Splicing Intermediates
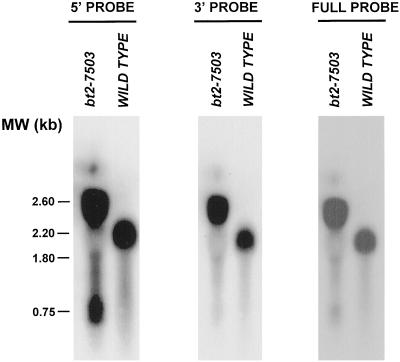
Because alterations in the invariant 5′ GT dinucleotide of introns sometimes lead to accumulation of splicing intermediates, we asked whether bt2-7503 exhibited this phenotype. Electrophoresed and blotted wild-type and bt2-7503 total RNAs were probed with portions of the Bt2 coding region. A full-length cDNA probe hybridized to three prevalent mutant transcripts of 0.75, 1.8, and 2.6 kb and a single 2.2-kb wild-type transcript (Fig. 2, right). A 5′ probe comprising exon 1 to 3 hybridized to the three mutant transcripts (Fig. 2, left), whereas the 3′ probe spanning exon 4 to 10 hybridized strongly only to the two larger mutant transcripts (Fig. 2, middle). The 2.6- and 1.8-kb bands correspond in size to type I and type III transcripts, respectively. The 0.75-kb transcript, however, contains only the 5′ end of the wild-type transcript, suggesting that it may represent transcripts somehow blocked in the splicing pathway.
Figure 2.
RNA blots of wild-type and bt2-7503 RNA. Total 20-d post-pollination endosperm RNA was subjected to electrophoresis, blotting, and hybridization with exons 1 to 3 of Bt2 cDNA (left). The blot was stripped and re-probed with exons 4 to 10 (middle) and then with the full-length Bt2 cDNA probe (right). Molecular mass markers are shown on the left.
Mapping of the Branch Point in Intron 3 of Mutant bt2-7503 Transcript
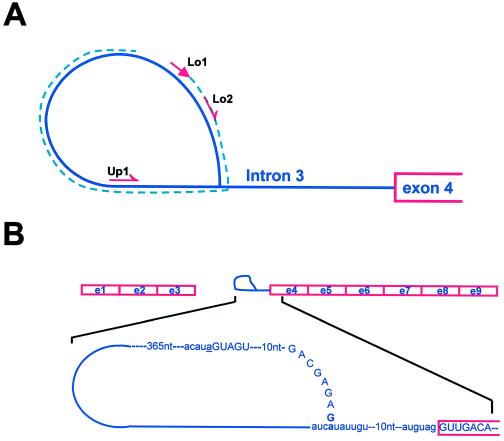
To investigate whether there is a population of bt2-7503 RNA blocked in splicing and possibly containing a lariat intermediate, we employed an RT-PCR procedure recently developed by Vogel et al. (1997) for use in their studies to map the branch point nucleotide of group II introns. This method exploits the property of RT to read through the 2′-5′ linkage in templates such as that involved in lariat formation of pre-mRNA splicing. The procedure is outlined in Figure 3A. An antisense primer (Lo1 in Figs. 3A and 4A) complementary to the 5′ end of the intron can synthesize DNA that continues through the branch point during first-strand synthesis. The resulting single-stranded DNA encompasses the branch point. This DNA can then be amplified by use of one primer lying in the same orientation as the first primer (Lo2 in Figs. 3A and 4A) and a second primer (Up1) lying in the sense orientation further 3′ relative to the RNA.
Figure 3.
Branch point mapping strategy and the structure of the intron lariat intermediate. A, The RT-PCR strategy employed for mapping branch point sequence is outlined. The intron 3 lariat is represented by a blue line. Red arrows represent the positions of the primers used during RT-PCR. First-strand DNA synthesized with primer Lo1 is represented by a broken blue line. Primers Lo2 and Up1 were employed for subsequent amplification. B, Schematic representation of the lariat intermediate during processing of mutant bt2-7503 transcript. The regions corresponding to exons 1 to 3 and to intron 3 in the form of a lariat attached to the downstream exons are shown. Informative sequences of the intron lariat and of exon 4 are presented. Exon and intron sequence are in capital and small letters, respectively. The G residue at the −23 position of exon 3, which forms a covalent linkage with branch point residue A at −23 position of intron 3, is shown in bold type. Residue a at the 5′ end of intron 3 is underlined.
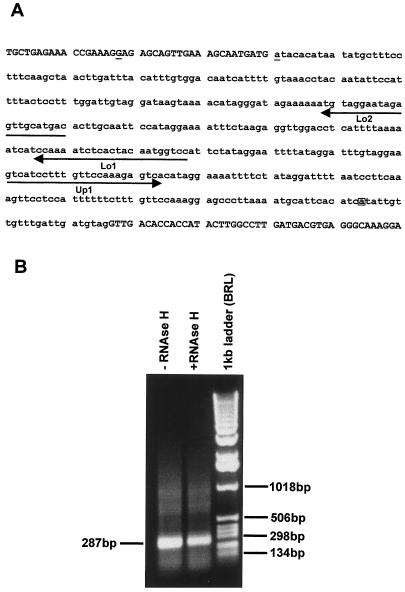
Figure 4.
Mapping of the branch point nucleotide during the processing of the mutant bt2 transcript. A, The genomic sequence spanning exon 3 to exon 4 of the mutant bt2-7503 is given with exons in uppercase letters and intron sequences in lowercase letters. Primers used during RT-PCR are underlined with arrows. The mutated 5′ end nucleotide of intron 3 is underlined and the branch point nucleotide is boxed. B, Ethidium-bromide-stained gel of the RT-PCR product resolved on 1% agarose gel resulting from subjecting total bt2-7503 endosperm RNA to RT-PCR as outlined in A. The size of the product is shown on the left. Marker mass markers are given on the right.
Total RNA from bt2-7503 was subjected to first-strand synthesis using the Lo1 primer and then amplified using the nested primers Lo2 and Up1 (Fig. 3A). This resulted in the synthesis of a 287-bp fragment (Fig. 4B). Omission of the RNase H digestion step resulted in a slight increase in the abundance of this fragment (Fig. 4B). Wild-type endosperm RNA used as a control in these experiments did not amplify any PCR product. The resulting 287-bp fragment from independent PCR reactions was directly subjected to several runs of sequencing in both directions. The resulting sequence chromatograms show that this fragment is a homogeneous population. This was further confirmed by sequencing several independent clones of this fragment. The branch point nucleotide A resides 23 nt 5′ to the splice site of intron 3. Furthermore, the donor site for this splicing event was not the mutant AT at the 5′ splice site of intron 3, but rather the dinucleotide GA located 23 nt 5′ to the 3′ splice site of exon 3. Here the G residue at position −23 of exon 3 was covalently linked to the branch point nucleotide. The sequences involved are shown in Figure 4A. We also noted the misincorporation of adenine instead of thymine at the branch point nucleotide, which Vogel et al. (1997) also found in their studies of group II introns. This 3′ splicing intermediate, while detectable, is apparently quite unstable. On northern blots, the 3′ portion with a size of 1.2 kb is barely detectable.
The Reason for the Lack of a BT2 Protein in This Mutant Is Not Obvious
While the type I transcript contains a premature termination codon within intron 3, and hence would not be expected to encode a protein bearing enzymatic activity or appreciable antigenic activity, type II and III transcripts lack 9 and 270 nt, respectively, and therefore maintain translational continuity. We note that while transcript type II differs from wild type only by the deletion of three amino acids, these amino acids lie in a region involved in binding of the substrate, Glc-1-P (Fu et al., 1998), and hence are likely to be critically important for enzymatic activity. Sequence analysis, however, does not provide a ready explanation for the absence of BT2 protein in this mutant. Perhaps none of the mutant proteins can form tetramers with the SH2 protein and so the BT2 proteins are more susceptible to protease cleavage. Evidence favoring rapid turnover of AGP monomeric subunits has been presented previously (Giroux and Hannah, 1994; Greene and Hannah, 1998).
DISCUSSION
Plant pre-mRNA processing is not well understood, primarily due to lack of an in vitro system efficient in splicing plant introns. Therefore, most of the research has progressed using transient assay systems. While transient assays have given significant insight into the process of plant pre-mRNA splicing, this approach has certain limitations (for review, see Brown and Simpson, 1998). In our efforts, we focus on naturally occurring splicing mutants in highly expressed genes. Our studies of the mutant bt2-7503 have uncovered a number of important features concerning splicing. The defining mutation in bt2-7503 is a GT to AT transition at the 5′ splice site of intron 3. This single mutation completely abolishes the use of this splice site, alters splicing from extant splice sites, and activates two non-canonical splice sites. Of these, only one can complete both steps in splicing. Splicing from extant sites gave rise to either intron inclusion or exon skipping.
Non-Consensus Splicing Donor Sites
We found that one cryptic splice does not start with the consensus dinucleotide GT, but rather begins with the dinucleotide GC. While not consensus, the use of GC in the donor site has been reported previously. Approximately 0.5% to 1% of both monocot and dicot donor sites start with the dinucleotide GC (Korning et al., 1996). In fact, this donor site is conserved across plant genera in myrosinase (Xue and Rask, 1995).
In contrast, activation of a cryptic splice site beginning with GA capable of the first but not the second step in splicing, and the total inability of the AU mutant site to participate in any splicing step, is without precedence. While the lack of an in vitro system capable of efficiently processing plant pre-mRNA has made studies of RNA branching extremely difficult, Liu and Filipowicz (1996) recently took advantage of a property of RT and of an Arabidopsis mutant (Orozco et al., 1993) bearing a GU to AU mutation at the 5′ donor site within the Rubisco activase gene to study lariat formation. Previous studies of accumulated lariats in other organisms had shown that RT will pause at the branch point during primer extension reactions. The pause is less apparent if the RNA is pretreated with HeLa cell-derived RNA debranching enzyme. Primer extension studies showed that the mutant AU donor site is used for lariat synthesis in this mutant. Previous studies in yeast and mammalian systems (Newman et al., 1985; Aebi et al., 1986) of the identical mutation gave similar results.
In contrast, we failed to obtain any evidence for the involvement of AU in splicing. Instead, a cryptic GA in exon 3 (23 nucleotides upstream of the 5′ splice site of the mutant intron 3) was covalently linked to the A residue in the branch point. This GA splice site apparently only functions when the wild-type site is mutant, since we could not isolate any lariat structures from wild-type RNA. Unlike the other cryptic 5′ splice site GC, the GA site is unable to proceed to the second step of splicing. In this regard, our results agree with those from yeast and mammalian introns, in which a GT to GA alteration at the 5′ splice site resulted in lariat accumulation (Aebi et al., 1986; Ruis et al., 1994). On the other hand, a synthetic intron with a GA donor site is efficiently spliced in tobacco protoplasts (Liu and Filipowicz, 1996). Furthermore, the use of an AT mutant at the 5′ splice site in yeast is increased by mutating the 3′ splice AG to AC, AA, or AU. This suggests that the terminal bases of introns may interact during the second step of splicing (Parker and Siliciano, 1993).
Plant Branch Point Sequences
Direct identification of sequences within the branch point of plant introns has been severely hampered by the fast turnover of wild-type lariat structures and by the lack of an in vitro splicing system. In the aforementioned studies of Liu and Filipowicz (1996), the underlined A of the sequence UUGAU located 32 nt from the 3′ end of the mutant intron formed the 5′ to 2′ linkage. These investigators also identified the branch point sequence, CUAAC, at position −31 in a synthetic intron spliced in tobacco. This branch, however, was resistant to RNA debranching activity. The comparable sequence identified here in intron 3 of Bt2 is AUCAU. Aside from the A residue that participates in the branch, these three sequences contain only one nucleotide in common: U in the second position.
The most frequent branch point nucleotide A in vertebrate introns usually exists 18 to 40 nt upstream from the 3′ splice site. We note that the branch point A residue of Bt2 intron 3 and the previously reported plant branch point A residues (Liu and Filipowicz, 1996) fall in this range. Previously, Goodall and Filipowicz (1989, 1991) reported that a synthetic intron devoid of potential branch point A residues is efficiently spliced in protoplasts. In vertebrates, nucleotides other than A can sometimes substitute for the branch point nucleotide (Hartmuth and Barta, 1988). Whether this is also true in plants will require more extensive analyses of plant branch point sequences.
A mammalian consensus branch point sequence of CU (A/G) A (C/T) has been proposed (Green, 1991). Recently, Simpson et al.(1996) suggested the identical branch point consensus sequence for plants using search keys derived from the vertebrate and fungi branch point consensus. This sequence lies in a region −15 to −50 from the splice acceptor site. Brown et al. (1996) analyzed 998 Arabidopsis introns and proposed consensus branch point sequence (C/T)UNAN located in a region between −18 to −60 from the 3′ splice site. Recently, a new branch point consensus CT (A/G) A(A/T) has been proposed for Arabidopsis introns using a model that does not rely on sequences derived from mammalian branch point consensus (Tolstrup et al., 1997). The branch point of Bt2 intron 3 does not fit any of these consensus sequences. In fact, the C residue at position −1 of the branch point of Bt2 intron 3 occurs in only approximately 15% of eukaryotic introns (Harris and Senapathy, 1989; Brown et al., 1996). Whether this reflects a monocot/dicot difference awaits further investigation.
Intron Definition versus Exon Definition of the Initial Recognition in Splicing
By far the vast majority of the bt2-7503 transcripts undergo splicing consistent with the intron definition of splicing. Transcripts exhibiting intron inclusion comprise the great bulk of the transcripts, while the use of upstream cryptic splice sites as seen in type II transcripts and in those undergoing incomplete splicing are also in accord with the intron definition of splicing. While rare, exon skipping as predicted by the exon definition of splicing was also noted in this mutant. Thus, intron inclusion and exon skipping are not mutually exclusive events within the context of a particular sequence, a pattern also noted by McCullough et al. (1997).
Recently, we (Lal et al., 1999) noted that alteration of the 3′ splice site from AG to AA in an intron of the Shrunken2 (Sh2) locus of maize led to exon skipping, while the identical mutation in a different intron activated a downstream splice site. In the former case, each of the flanking introns was much larger than the intervening exon, whereas the opposite situation was found in the second mutant. We speculated that splice-altering mutants in small exons flanked by large introns, as found in the first mutant and generally in animal genes, gave rise to exon skipping, but in the case of relatively small introns flanked by large exons, splicing in line with intron definition was seen. However, this argument does not explain the splicing seen in bt2-7503. Exon 3 (270 nt) abuts introns of larger sizes, 1,213 and 396 nt. Furthermore, studies of Arabidopsis splicing mutants by Simpson et al. (1998) showed that exon skipping in this dicot is usually associated with small exons with an AU content very similar to their flanking exons. The AU contents of Bt2 exons 2 and 4 are 53% and 56%, respectively, which is very similar to the 56% found in exon 3. These factors predict that exon skipping in bt2-7503 should occur; however, this phenomenon is rare in this mutant.
One explanation for the low frequency of exon skipping in bt2-7503 may lie within exon 3. We note that the 5′ splice site is not a particularly good fit to the monocot consensus donor site and the exon is not GC rich. In the absence of the GT dinucleotide within the donor site, perhaps the splicing apparatus does not recognize this sequence as an exon. Splicing then removes intron 2, exon 3, and intron 3 in one reaction. We must also consider the possibility that neither the intron definition nor the exon definition adequately accounts for the early steps in pre-mRNA splicing, at least in maize endosperm.
ACKNOWLEDGMENTS
We thank Dr. Oliver Nelson for bt2-7503 and Drs. Robert Ferl and Karen Koch for critically reviewing an earlier version of this manuscript.
Footnotes
We gratefully acknowledge support from the National Science Foundation (grant nos. IBN–9316887 and MCB–9420422) and U.S. Department of Agriculture Competitive Grants Program (grant nos. 94–37300–453, 97–36306–4461, 95–37301–2080, and 98–01006). This is Florida Agricultural Experiment Station Journal Series no. R–06706.
LITERATURE CITED
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eucaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Antoniou M. Clinical defects in pre-mRNA processing. In: Lamond AI, editor. Pre-mRNA Processing. Austin, TX: RG Landes; 1995. pp. 187–201. [Google Scholar]
- Bae JM, Giroux M, Hannah LC. Cloning and characterization of the Brittle-2 gene of maize. Maydica. 1990;35:317–322. [Google Scholar]
- Barta A, Sommergruber K, Thompson D, Harmuth K, Matzke ME, Matzke AJM. The expression of Nopaline synthase-human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol Biol. 1986;6:347–357. doi: 10.1007/BF00034942. [DOI] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Brown JWS. A catalogue of splice junction and putative branchpoint sequences from plant introns. Nucleic Acids Res. 1986;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Simpson CG. Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:77–95. doi: 10.1146/annurev.arplant.49.1.77. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Smith P, Simpson CG. Arabidopsis consensus intron sequence. Plant Mol Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- Carle-Urioste JC, Ko CH, Bonito M, Walbot V. In vivo analysis of intron processing using splicing-dependent reporter gene assays. Plant Mol Biol. 1994;26:1785–1795. doi: 10.1007/BF00019492. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ballicora MA, Preiss J. Mutagenesis of the glucose-1-phosphate-binding site of potato tuber ADP-glucose pyrophosphorylase. Plant Physiol. 1998;117:989–996. doi: 10.1104/pp.117.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux MJ, Hannah LC. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Mol Gen Genet. 1994;243:400–408. doi: 10.1007/BF00280470. [DOI] [PubMed] [Google Scholar]
- Giroux MJ, Shaw GB, Cobb BG, Greene T, Okita T, Hannah LC. A single gene mutation increases maize seed weight. Proc Natl Acad Sci USA. 1996;93:5824–5829. doi: 10.1073/pnas.93.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991;10:2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Greene TW, Hannah LC. Maize endosperm ADP-glucose pyrophosphorylase SHRUNKEN 2 and BRITTLE 2 subunit interactions. Plant Cell. 1998;10:1295–1306. doi: 10.1105/tpc.10.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, Nelson OE. Characterization of ADP-glucose pyrophosphorylase from shrunken-2 and brittle-2 mutants of maize. Biochem Genet. 1976;14:547–560. doi: 10.1007/BF00485834. [DOI] [PubMed] [Google Scholar]
- Hannah LC, Tuschall DM, Man RJ. Multiple forms of maize endosperm ADP-glucose pyrophosphorylase and their control by Shrunken-2 and Brittle-2. Genetics. 1980;95:961–970. doi: 10.1093/genetics/95.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris NL, Senapathy P. Distribution and consensus of branch point signals in eucaryotic genes: a computerized statistical analysis. Nucleic Acids Res. 1989;18:3015–3019. doi: 10.1093/nar/18.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K, Barta A. Unusual branchpoint selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988;8:2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Belzile F, Dean C. Inefficient and incorrect processing of the Ac transposase transcript in iae1 and wild type Arabidopsis thaliana. Plant J. 1997;11:921–931. doi: 10.1046/j.1365-313x.1997.11050921.x. [DOI] [PubMed] [Google Scholar]
- Keith B, Chua N-H. Monocot and dicot pre-mRNAs are processed with different efficiencies in transgenic tobacco. EMBO J. 1986;5:2419–2425. doi: 10.1002/j.1460-2075.1986.tb04516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korning PG, Hebsgaard SM, Rouze P, Brunak S. Cleaning the GenBank Arabidopsis thaliana data set. Nucleic Acids Res. 1996;24:316–320. doi: 10.1093/nar/24.2.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal SK, Choi JH, Hannah LC. The AG dinucleotide terminating introns is important but not always required for pre-mRNA splicing in maize endosperm. Plant Physiol. 1999;120:65–72. doi: 10.1104/pp.120.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-X, Filipowicz W. Mapping of branchpoint nucleotide in mutant pre-mRNAs expressed in plant cells. Plant J. 1996;9:381–389. doi: 10.1046/j.1365-313x.1996.09030381.x. [DOI] [PubMed] [Google Scholar]
- Lou H, McCullough AJ, Schuler MA. 3′ Splice site selection in dicot plant nuclei is position dependent. Mol Cell Biol. 1993;13:4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Intron creation and polyadenylation in maize are directed by AU-rich RNA. Genes Dev. 1994;8:1117–1130. doi: 10.1101/gad.8.9.1117. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Martin DJ, Firek S, Moreau E, Draper J. Alternative processing of the maize Ac transcript in Arabidopsis. Plant J. 1997;11:933–943. doi: 10.1046/j.1365-313x.1997.11050933.x. [DOI] [PubMed] [Google Scholar]
- McCarty DR. A simple method for extraction of RNA from maize tissues. Maize Genet Coop Newslett. 1986;60:61. [Google Scholar]
- McCullough AJ, Baynton CE, Schuler MA. Interactions across exons can influence splice site recognition in plant nuclei. Plant Cell. 1997;8:2295–2337. doi: 10.1105/tpc.8.12.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ, Lou H, Schuler MA. Factors affecting authentic 5′ splice site selection in plant nuclei. Mol Cell Biol. 1993;13:1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to messenger RNAs by the spliceosome. In: Gestland R, Atkins J, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- Moore MJ, Sharp PA. Evidence of two active sites in the spliceosome provided by stereochemistry of pre-mRNA. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Ogden RC, Abelson J. Molecular consequence of specific mutants on yeast mRNA splicing in vivo and in vitro. Cell. 1985;42:335–342. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Orozco BM, Robertson MC, Werneke JM, Ogren WL. Molecular basis of the ribulose-1,5-biphosphate carboxylase/oxygenase activase mutation in Arabidopsis thaliana is a guanine-to-adenine transition at the 5′ splice junction of intron 3. Plant Physiol. 1993;102:227–232. doi: 10.1104/pp.102.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG. Evidence for an essential non-Watson-Crick interaction between the first and the last nucleotide of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base-pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pautot V, Brzezinski R, Tepfer M. Expression of mouse metallothionein gene in transgenic plant tissue. Gene. 1989;77:133–140. doi: 10.1016/0378-1119(89)90367-3. [DOI] [PubMed] [Google Scholar]
- Ruis BL, Kivens WL, Siliciano PG. The interaction between the first and the last intron nucleotide in the second step of pre-mRNA splicing is dependent on other conserved intron nucleotide. Nucleic Acids Res. 1994;22:5190–5195. doi: 10.1093/nar/22.24.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Greene JM, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Sharp PJ. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Simpson CG, Clark G, Davidson D, Smith F, Brown JWS. Mutation of putative branchpoint consensus sequence in plant introns reduces splicing efficiency. Plant J. 1996;9:369–380. doi: 10.1046/j.1365-313x.1996.09030369.x. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Simpson CG, McQuade C, Lyon J, Brown JWS. Characterization of exon skipping mutants of the COP1 gene from Arabidopsis. Plant J. 1998;15:125–131. doi: 10.1046/j.1365-313x.1998.00184.x. [DOI] [PubMed] [Google Scholar]
- Tolstrup N, Rouze P, Brunak S. A branch point consensus from Arabidopsis found by non-circular analysis allows for better prediction of acceptor sites. Nucleic Acids Res. 1997;25:3159–3163. doi: 10.1093/nar/25.15.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen VL, Spritz RA. Splicing of plant pre-mRNAs in animal system and vice versa. Gene. 1987;56:253–265. doi: 10.1016/0378-1119(87)90142-9. [DOI] [PubMed] [Google Scholar]
- Vogel J, Wolfgang RH, Borner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Manley J. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Xue J, Rask L. The unusual 5′ splicing donor GC is used in myrosinase genes of the Brassicaceae. Plant Mol Biol. 1995;29:167–171. doi: 10.1007/BF00019128. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in U2 snRNP suppresses a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]