Abstract
The concept of allosteric interaction was initially proposed to account for the inhibitory feedback mechanism mediated by bacterial regulatory enzymes. In contrast with the classical mechanism of competitive, steric, interaction between ligands for a common site, allosteric interactions take place between topographically distinct sites and are mediated by a discrete and reversible conformational change of the protein. The concept was soon extended to membrane receptors for neurotransmitters and shown to apply to the signal transduction process which, in the case of the acetylcholine nicotinic receptor (nAChR), links the ACh binding site to the ion channel. Pharmacological effectors, referred to as allosteric modulators, such as Ca2+ ions and ivermectin, were discovered that enhance the transduction process when they bind to sites distinct from the orthosteric ACh site and the ion channel. The recent X-ray and electron microscopy structures, at atomic resolution, of the resting and active conformations of several homologues of the nAChR, in combination with atomistic molecular dynamics simulations reveal a stepwise quaternary transition in the transduction process with tertiary changes modifying the boundaries between subunits. These interfaces host orthosteric and allosteric modulatory sites which structural organization changes in the course of the transition. The nAChR appears as a typical allosteric machine. The model emerging from these studies has led to the conception and development of several new pharmacological agents.
This article is part of a discussion meeting issue ‘Allostery and molecular machines’.
Keywords: allosteric protein, nicotinic acetylcholine receptor, molecular machine, allosteric modulation
1. Introduction and definitions
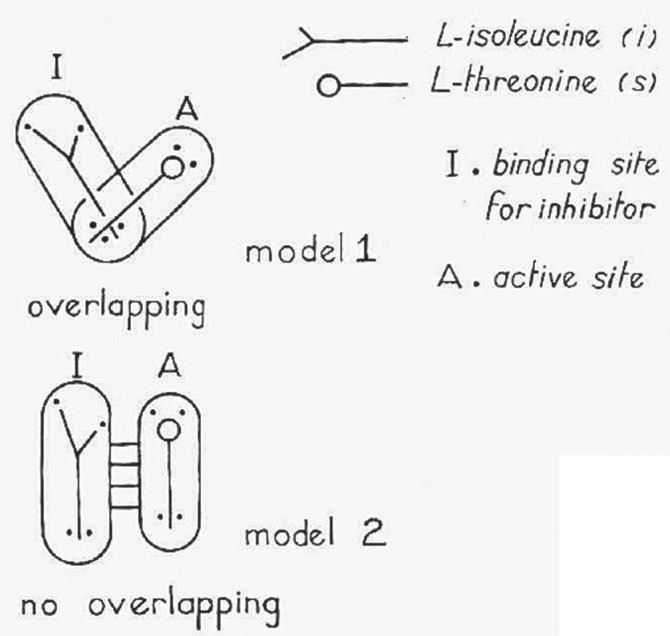
The generally accepted definition of allosteric interactions is that they constitute a mode of ligand–protein interaction radically different from classical molecular competition. Instead of mutual exclusion from a common binding site by steric hindrance, allosteric interactions are defined as indirect interactions between topographically distinct sites, mediated by a reversible alteration of the protein's molecular structure. The concept was first proposed to account for the inhibitory feedback mediated by the first enzyme in a bacterial biosynthetic pathway (l-threonine deaminase/aspartate transcarbamylase) where the feedback inhibitor (l-isoleucine/CTP) is not a steric analogue of the substrate (l-threonine/aspartate) [1–3]. As a PhD student in the laboratory of Jacques Monod, I struggled with l-threonine deaminase to find a way to dissociate regulatory interaction and catalytic activity in vitro. The data could not be accounted for by the classical Michaelis scheme of a competitive inhibition and it was suggested that substrate and regulatory effector bound at topographically distinct sites [1] (figure 1). The model of ‘no-overlapping’ sites was presented at the 1961 Cold Spring Harbor Symposium on Quantitative Biology [1], and the word allosteric was subsequently coined in the General Conclusions of the same meeting by Jacques Monod & François Jacob [2].
Figure 1.
First diagrammatic representation of a competitive vs allosteric interaction between ligands. This scheme was presented in [1] to account for the properties of the bacterial enzyme l-threonine deaminase at the 1961 Cold Spring Harbor Symposium on Quantitative Biology. The ‘no-overlapping’ model was named ‘allosteric’ by Monod & Jacob in the General Conclusions paper of the meeting.
The concept was expanded in 1963 [4] to explain properties of the few regulatory proteins then known, as well as Perutz's structural data on haemoglobin. An initially suggested mechanism underlying conformational change was inspired by Koshland's induced-fit theory [5] for the specificity of enzyme action, according to which the fit occurs ‘only after a change in shape of the enzyme molecule had been induced by the substrate’. Results obtained with l-threonine deaminase [1,6–12], in particular the effects of regulatory ligands on the cooperativity of substrate binding and their concomitant loss upon desensitization (noted independently with aspartate transcarbamylase [3,13]), soon led to a paradigmatic shift from instruction to selection. Rather than being instructed by the ligand, the conformational transition was proposed to pre-exist as an equilibrium among a few discrete structural states, totally independent of ligand structure or occupancy, and differentially stabilized by the various ligands. This conformational selection hypothesis is one of the key hallmarks of the 1965 Monod–Wyman–Changeux (MWC) model [14].
A second structural assumption of the model accounted for the observed cooperativities of substrate and regulatory ligand binding kinetics, by endowing the protein itself with a cooperative structure [14]. Regulatory proteins would be ‘oligomers’ comprising a small number of repeated units—protomers—and consequently possess at least one axis of symmetry, the conformation of each protomer being constrained by its association with the others [14]. In the absence of ligand the oligomers are, as already mentioned, assumed to naturally exist as a thermodynamic equilibrium of (at least) two discrete conformations, R (for relaxed) and T (for constrained, tendu in French), with different tertiary organization, inter-subunit bond energy, quaternary constraints, ligand binding affinity, and biological activity.
Lastly, ligands would shift the conformational equilibrium by stabilizing the oligomer conformation for which they have the highest affinity, thus mediating the important process of signal transduction [14]. An important parameter of the protein then becomes the intrinsic (ligand-free) equilibrium constant between R and T states Lo = To/Ro, termed the allosteric constant. In the model's formalization, ‘the microscopic dissociation constants, KoR and KoT, of the ligand for each state were assumed to be the same for all homologous sites in each of the two states’ regardless of ligand occupancy. The model therefore predicts both a ‘state function’ R describing the conformational equilibrium, and a ‘binding function’ Y, as distinct functions of ligand concentration [14]. Soon after the MWC model, Koshland et al. [15] proposed, in 1966, a sequential induced-fit mechanism of allosteric transition—known as the Koshland–Nemethy–Filmer (KNF) model—characterized by a progressive conformational change of the ligand-bound subunits within the oligomer. Unlike the MWC model, it excludes any change of protein conformation in the absence of ligand.
These research models triggered numerous experimental tests. Early on, Manfred Eigen and his group, measuring fast binding kinetics by T-jump relaxation, producing the first kinetic evidence supporting the MWC scheme [16]. A second confirmation was the demonstration [17] that ligand binding and conformational status as functions of ligand concentration do not superimpose, i.e. that, as the MWC model predicts, state function R differs from binding function Y.
Recent re-formulations of the MWC paradigm have emphasized the possible occurrence of multiple conformations in equilibrium and the notion of ‘population shifts’ by ligands within the energy landscape formalism (see [18–23]). Among many developments in the field during the past decades is the discovery of so-called ‘disordered’—or better, unordered—sequences in proteins [24]. There is evidence that in some systems, such as transcription factors, they facilitate the conformational changes that mediate long distance allosteric interactions [22] (see other articles in this issue).
Importantly, MWC (and KNF) models both formulate a static (equilibrium or steady-state) picture of allosteric behaviour. Understanding the phenomenon's dynamics requires structural studies at atomic resolution and complementary time-resolved analyses (including molecular dynamics) together with novel model systems and relevant technologies (see [18,20,25]). This recent development is presented in the last part of this paper.
The extension of the concept of allosteric interaction to intercellular communication, in particular neurotransmission in the nervous system [6–12,26], has opened a new field of research on brain chemistry and pharmacology (reviewed [27]). In this paper I shall focus on one typical allosteric machine that played a decisive role in the evolution of the domain: the nicotinic acetylcholine receptor (nAChR), the first receptor for a neurotransmitter to be chemically identified [28].
2. The nicotinic acetylcholine receptor
(a). Identification
Transmission of nerve signals at the synapse is an important physiological process where the temporal dimension of the response to the chemical signal is critical. Possible extension of the concept of allosteric interaction to ‘membrane phenomena involved in the recognition of (intercellular) communication signals and their transmission—synaptic transmission, for example’—was contemplated from the beginning [6–12]. Theoretical models were soon elaborated for membrane receptors composed of transmembrane oligomers [26,29] or even for large two-dimensional arrays of highly cooperative protein assemblies [26]. The lattice model was documented decades later, at the structural and functional level, in particular in bacterial chemoreceptors (reviewed [30]). Here I review the nAChR as a model of allosteric transmembrane oligomer together with its structure at atomic resolution and its molecular dynamics within the in vivo timescale of synaptic transmission.
Following Langley's (1905) proposal [31] of the pharmacological receptor concept it took about 65 years for the nAChR to be chemically identified through an elaborate process using fish electric organ (a very rich source of cholinergic synapses) and a snake venom toxin (a highly specific label of the nAChR) ([28,32–34], reviewed [34]). The upshot was that nAChRs are integral allosteric membrane proteins, of molecular mass about 290 kDa, that form oligomers comprising five identical or homologous subunits symmetrically arranged around a central ion channel, with a fivefold rotational symmetry axis perpendicular to the membrane (reviewed [34]) (figure 2). The primary structure of each subunit established through protein sequencing [37,38] and cDNA cloning and sequencing [39–41] reveals a large hydrophilic amino-terminal extracellular (EC) domain, a transmembrane (TM) domain comprising four hydrophobic segments (M1–M4) and a variable hydrophilic intracellular domain. The EC domain harbours two to five ACh binding sites located at the boundaries between subunits. These widely (approx. 40 Å) separated ACh binding sites are functionally linked to the 60 Å-distant, centrally located cationic ion channel delineated by the M2 α-helix (reviewed [34]) (figure 2). The gating of the ion channel by ACh takes place between topographically far distant sites. It is an unquestionable allosteric interaction. The nAChR protein thus possesses all the structural elements required to convert a chemical signal, typically a local rise of extracellular ACh concentration, into an electrical signal caused by the ion channel's opening [34]. It behaves as a typical, yet highly sophisticated, allosteric machine.
Figure 2.
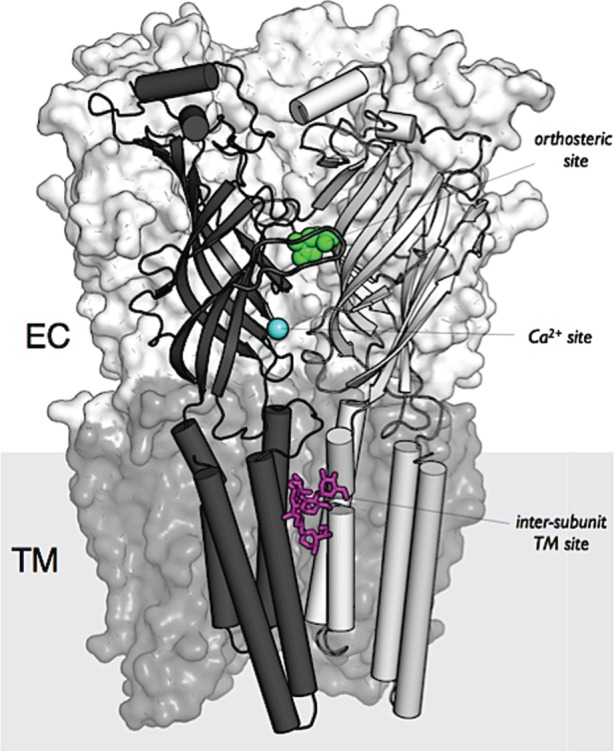
Structural organization of a pentameric ligand-gated ion channel of the nicotinic receptor family. Data from the crystal structure of GluCl [35] illustrating the pentameric organization, the distinction between the extracellular domain ECD and the transmembrane domain TM, the orthosteric site for glutamate (in green), the Ca2+ (cyan) and ivermectin (magenta) allosteric modulatory sites respectively in ECD and TM. Reproduced from Taly et al. [36, fig. 1].
Over the years the nAChR has become the ‘founding father’ of the superfamily of pentameric ligand-gated ion channels (pLGICs) which includes the 5-hydroxytryptamine receptor (5HT3R), the inhibitory anion-selective γ-aminobutyric acid type A (GABAAR)/glycine receptors (GlyR) and the invertebrate glutamate-gated chloride channel (GluCl) [34].
(b). In vivo dynamics of signal transmission
In the case of synaptic transmission, electrophysiological techniques made it possible, mid-twentieth century, to record the electric response of receptors to chemical transmitters on millisecond timescales [42,43]. At the neuromuscular junction, for instance, neurally released ACh elicits a fast (less than 0.2 ms) rise of postsynaptic potential followed by decay over a few milliseconds. The local ACh concentration in the synaptic cleft transiently rises (less than 1 ms) to 3 × 10−4 M over a background of 10−8 M [42,43]. In other words the physiological signal received by the nAChR is a transient millisecond pulse of ACh. The ionic response was interpreted, without structural evidence at the time, in terms of channel gating through an induced-fit mechanism: R + A ↔ RA ↔ R*A where RA is a closed, and R*A an open-channel state. Patch-clamp techniques revealed later that the kinetics of the postsynaptic response recorded at the cellular level represent the collective opening of a host of molecular channels, each individual opening having a square shape with risetime in the microsecond range and mean open duration of a few milliseconds [44]. These values set the time-range for the molecular dynamics studies presented in this review.
In addition to the fast channel gating process, Langley [31] had already noticed that prolonged application of the agonist nicotine blocks subsequent receptor responses, a process termed desensitization. To fit the electrophysiological data then available, Katz & Thesleff [45] proposed that ACh slowly (on a 10 ms−1 s timescale) stabilizes a new high-affinity closed (refractory), referred to as desensitized, state of the receptor. Subsequent electrophysiological and biochemical studies with nAChR-rich ‘excitable’ membrane fragments [46,47] and a fluorescent acetylcholine analogue (dansyl-C6-choline) allowed one to follow directly in vitro the binding kinetics of a nicotinic ligand and its conformational and ionic consequences without the need for in vivo electrophysiological recordings ([48–50], see also [51] with radiolabelled ligands). The data yielded the first millisecond-range in vitro demonstration of the allosteric transitions among several conformational states including : (i) a resting closed-channel R state stabilized by nicotinic antagonists; (ii) an active, fast, open-channel A state with low affinity for ACh and nicotinic agonists (kDa ACh: approx. 50–100 µM); and (iii) at least a fast (I) and a slow (D, desensitized, refractory) state with higher affinities for agonists (but also for antagonists) (kDa of I for ACh: approx. 1 µM; kDa of D for ACh: approx. 3–5 nM) ([48–51; see [52]). Contrary to an opinion widespread among pharmacologists, the highest-affinity states do not correspond to the active functional state of the receptor—far from it. On the other hand, the A state's low affinity for ACh allows binding and unbinding of ACh at millisecond rates and thus permits fast and repetitive transmission.
Moreover, in agreement with the MWC scheme, Jackson [53] observed spontaneous opening of muscle nAChR in the absence of ACh. A sizeable (approx. 20%) fraction of the receptor protein resides in the high-affinity D state in the total absence of ligand [48–50]. Recent analysis of the channel gating transition in terms of the MWC model further led to the conclusion that in vivo ‘diliganded and brief unliganded openings are generated by the same essential, global transition’ [54].
These findings rule out the induced-fit mechanism and validate the fundamental premise of the MWC model, namely that the basic activation mechanism involves a discrete R ↔ A conformational transition independent of ligand binding [48–54]. The nAChR protein behaves as a molecular machine that plays the role of ‘allosteric switch’ in the physiological timescale of synaptic transmission.
3. X-ray and electron microscopy high-resolution structures of pentameric receptors
An important step in the understanding of the allosteric mechanism mediating the nAChR signal transduction process within the timescale of synaptic transmission has been the availability of high-resolution structural data. They resulted in the first description of the allosteric transition of a LGIC and its allosteric modulation in structural terms.
Early electron micrographs of nAChR from fish electric organ [55,56] revealed ring-like pentameric particles (8–9 nm in diameter) with a hydrophilic core linked to a compact bundle. These nAChR images were the first ever of the structure of a neurotransmitter receptor. A step further was the X-ray structure of a water-soluble ACh binding protein (AChBP) from Lymnaea which displays, like the nAChR, a pentameric organization and approximately 30% sequence homology with the EC domain of nAChRs [57]. These structural data led to the development of atomic models for full-length nAChR [58] which were subsequently abandoned (see [36,59,60], for discussion) because, in particular, of altered conformational states and alignment errors.
The situation changed dramatically with the discovery in bacteria of DNA sequences homologous to eukaryotic nAChR [61]. One of such sequence from photosynthetic Gloeobacter violaceus [62] was cloned and expressed in eukaryotic cells. Electrophysiologically the protein behaves as a ligand-gated ion channel activated at acidic pH [62]. Purification and crystallization of that and a closely related protein led to the first full X-ray structures for a pentameric ligand-gated ion channel (pLGIC) in a closed-channel state (resolution 3.3 Å) from Erwinia chrysanthemi (ELIC) [63] and in an open-channel state (resolution 2.9 Å) from G. violaceus (GLIC) (figure 3) [64,65].
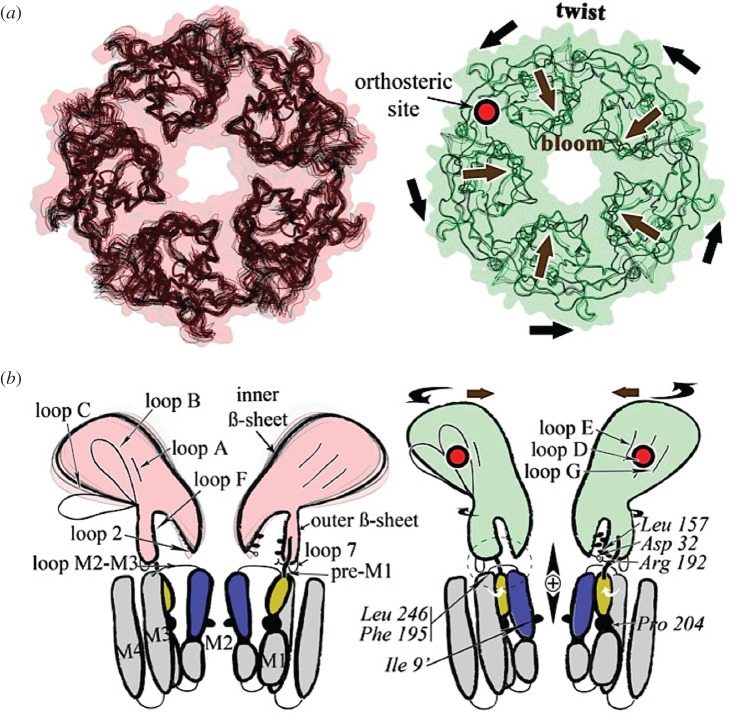
Figure 3.
Model of the gating mechanism mediated by the prokaryotic channel from Gloeobacter violaceus GLIC. Left, the pH7 closed state; right, the pH4 open state. (a) View from the top illustrating the twisting and blooming motion, indicated by black arrows. (b) View from the side schematically illustrating the transmission of the signal through the ECD–TM interface. Reproduced from Sauguet et al. [86, fig. 4].
Consistent with early models inferred from the primary sequence data of the nAChR [37–41], the EC domain carries the orthosteric ligand binding sites and folds into a highly conserved immunoglobulin-like β-sandwich; on the other hand, the TM domain consists of four α-helices organized as a well-conserved bundle (figure 2). The M2 helix lines, as expected, the channel walls [66–70] and is surrounded by a ring made of α-helices M1 and M3. The fourth TM α-helix, M4, is most peripheral and interacts extensively with the lipid bilayer (reviewed [34]).
Strikingly similar 3D X-ray and high-resolution electron microscopy (EM) structures have been reported with the anionic glutamate receptor from Caenorhabditis elegans (GluCl) [35], human GABAA β3R [71], mouse 5HT3R [72], fish glycine α1R [73] and α3R [74], and, last but not least, human nAChR α4β2 [75]. Also, the cytoplasmic domain, lacking in prokaryotic receptors, was revealed for the first time in 5HT3 receptors [72]. The available structural data beautifully illustrate that active sites and ion channel are far distant (at the protein scale), making the nAChR a typical allosteric membrane protein. They further show a remarkable conservation of 3D organization (down to the atomic level), illustrating the common phylogenetic origin of bacterial and brain pentameric receptors.
4. Structure of the ligand binding sites
A critical proposal of the allosteric scheme (figure 1) is that the interacting regulatory and biologically active sites are topographically distinct. The atomic structures of the orthosteric site and ion channel with the nAChR and its homologues definitively demonstrate this point together with the discovery of new allosteric modulatory sites.
(a). The orthosteric binding site
X-ray structures of the neurotransmitter—orthosteric ligand—binding pocket have confirmed its location in the EC domain at the interface between subunits. With or without ligand bound, the orthosteric pocket is found to be lined by amino acid side-chains from three main regions of the ‘principal’ subunit (loops A, B and C) and four from the ‘complementary’ subunit (loops D, E, F and G) ([62–65], reviewed [34]). Consistent with nAChR photolabelling and directed mutagenesis data from nAChR [76], the X-ray structure of AChBP shows that loops A (Tyr), B (Trp), C (two Tyr) and D (Trp) form an aromatic ‘box’ chelating the ammonium group of ACh with the tryptophan residue from loop B, establishing a direct cation–π interaction [77].
A similar organization is found in GluCl, GABAAR and α4β2 nAChR, with variations depending on the natural ligand ([35,71–75], reviewed [34]). Unexpectedly, the orthosteric ‘neurotransmitter’ site in pLGICs appears remarkably conserved from bacteria to humans.
(b). The ion channel binding site(s)
Channel blockers constitute an important category of nicotinic drugs that—like mecamylamine—are valuable therapeutic agents. They bind to the TM domain M2 of nAChR and prevent ion flux by sterically occluding the channel pore (reviewed [78–80]). High-resolution X-ray structures of nAChR and homologues ([35,62–65,71–75], reviewed [34]) confirm that binding sites for channel blockers are distributed throughout the transmembrane channel ([66–70], reviewed [34]) which consist of stacked pentameric rings of homologous amino acids. The data confirm the evolutionary stability of permeation and selectivity structure/function relationships in the TM domain from prokaryotes to eukaryotes.
(c). The allosteric modulatory site(s)
An important outcome of the research on the allosteric properties of nAChRs (and pLGIC in general) has been the discovery of diverse categories of modulatory sites, with positive (PAM) or negative (NAM) effects upon the signal transduction mechanism. These new modulatory sites are ‘allosteric’ in the sense that they are topographically distinct from the orthosteric sites and indirectly modulate the channel gating transition of the nAChR. They are distributed throughout every domain of the receptor molecule.
I review the main ones [79,80] (figure 2).
(i). Ca2+ site
Ca2+ potentiates most neuronal nAChRs [81,82] at the level of sites located in the EC domain at the subunit interfaces, below the orthosteric ACh site near the TM domain [83–86]. Homologues of these sites have been identified in prokaryotic ELIC [85] and in GLIC [86]. In agreement with the MWC model, in α7-nAChR, Ca2+ primarily affects the R-to-open A isomerization constant [83].
(ii). Transmembrane sites
A second class of allosteric modulatory sites lies at a rather original location within the TM domain. These sites accommodate pharmacological agents that regulate receptor activity (as PAMs or NAMs), such as the anthelmintic ivermectin [87], general anaesthetics (GAs) [88] or ethanol [89]. The X-ray structure of GLIC in complex with propofol or desflurane reveals a common site for GAs within the upper part of the TM domain of each subunit inside a cavity accessible to phospholipids from the lipid bilayer [88]. Structures of GluCl [35] and human GlyRα3 [90] bound to ivermectin show an ivermectin binding site located at the periphery of the upper part of the TM domain wedged by helices M3 and M1 at the subunit interface. Sites for ethanol in an ethanol-sensitized mutant of GLIC [89] appear closely related to the ivermectin site like the sites for several synthetic modulators of α7 nAChR such as PNU-120596 and LY 2087101 [91,92] and GA in GABAA receptors [93] (reviewed [79,80]).
(iii). Cytoplasmic domain sites
Allosteric modulatory sites are also present in the intracellular domain and may play important roles in clustering, stabilization, and modulation of receptor functions (reviewed [34]; [79]), in particular through the interaction with G-proteins and thus G-protein-coupled receptors [94].
In conclusion, several categories of allosteric modulatory sites, with positive (PAM) or negative (NAM) effects upon the signal transduction mechanism, have been discovered with nAChRs and closely related receptors by academic research groups [91,92,95–98] and companies (Abbott, Neuroresearch or Lilly), attesting of the rich diversity of allosteric modulators of potential pharmacological importance. These sites, topographically distinct from the orthosteric sites, are distributed throughout every domain of the receptor molecule. The nAChR is an allosteric machine under potential control by a broad diversity of chemical signals in addition to the neurotransmitter.
5. Interaction of AChR with the lipid membrane
The nAChR is an allosteric protein specialized in intercellular communication and integrated with the cell membrane. As a transmembrane protein, it exhibits a definite transverse polarity, with the neurotransmitter site facing the outside of the cell. The lipid bilayer forms the natural environment to the TM domain of the nAChR protein and may exert an allosteric modulatory role on receptor function. The precise composition of the membrane lipids [99] affects channel activity [49,50,100–102]. In the absence of anionic lipids and cholesterol, nAChRs appear to populate an ‘uncoupled’ conformation that binds agonists with resting-like low affinity but no longer undergoes the gating isomerization [60,103–108], supporting the view that binding of cholesterol to the TM domain affects the structure and/or dynamics of pLGICs. Computational analyses of the cryo-EM structure of Torpedo nAChR [108] and a homology model of the GABA receptor [109] reveal several cavities in the TM domain complementary to cholesterol and suggest that bound cholesterol at these sites stabilizes as a PAM the channel's open-pore conformation.
Lipids, free fatty acids, and steroids are known to allosterically modulate pLGICs and are thus likely natural ligand candidates for the GA sites [88,101–107]. The cholesterol binding site to the TM domain shows homologies with ivermectin binding to GluCl, cholesterol possibly acting as an endogenous allosteric modulator in eukaryotic pLGICs [103–109] (figure 4).
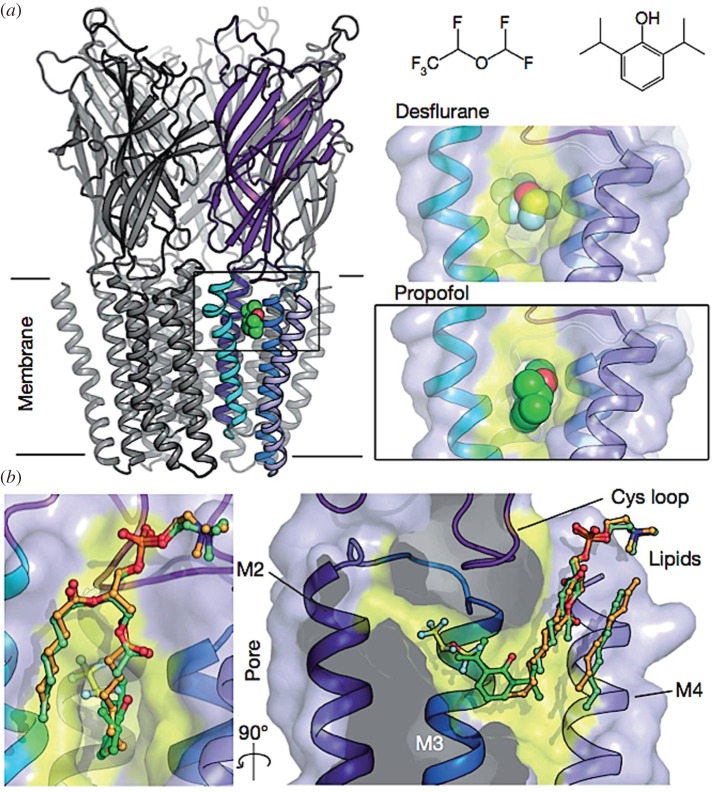
Figure 4.
X-ray structure of the allosteric site for general anaesthetics from the bacterium Gloeobacter violaceus. The general anaesthetic (propofol, desflurane) binding site is located within the transmembrane domain near the interface with the extracellular domain (a) and in close relationships with membrane lipids (b), which may behave as endogenous allosteric modulatory ligands. Reproduced from Nury et al. [88, fig. 1].
The lipid bilayer is the original physico-chemical medium that holds the receptor protein in place. But, diverse membrane lipids (including cholesterol) have been found to selectively modulate the activity of these allosteric machines. The transmembrane domain therefore becomes a strategic target for the design of pharmacological agents.
6. Molecular dynamics of nAChRs and homologues
As mentioned in the introduction, the MWC model (and the KNF model) explores allosteric transitions under the condition of thermodynamic equilibrium. The application of time-resolved analyses (including molecular dynamics) to the nAChR [25] together with relevant modelling approaches and technologies (see [18–23]) renew our understanding of the molecular mechanism involved in the gating process under conditions close to the actual physiological process.
(a). Early molecular dynamics studies
Even before high-resolution structural data became available, Taly et al. [25] developed by comparative analysis a 3D model of the α7-nAChR on which they performed the first, coarse-grained, molecular dynamics simulation of a pLGIC, applying the so-called ‘normal mode analysis.’ Approximating the surface of the conformational landscape, the analysis decomposes the receptor protein movements into discrete modes. Among the ten lowest-frequency modes, the first shows a structural reorganization described as concerted but opposite rotations of the upper (EC) and lower (TM) domains around the pore axis—a movement termed quaternary twist. The twist widens the ion channel and, by reshaping the subunits' interfaces in EC, opens or closes the agonist binding site(s) located there. These results were confirmed and extended on a new model of α7-nAChR based upon the crystal structures of ELIC [110] and GLIC [111] and then on GLIC with a 1 µs-long all-atom molecular dynamics simulation [112]. A parallel computational study was undertaken on nAChR [113] carrying pathological mutations associated with congenital myasthenia and autosomal dominant nocturnal frontal lobe epilepsy. These mutations constitutively stabilize the receptor in an active open (or closed) conformation. The mutant amino acids were found either at interfaces between subunits or, within subunits, between rigid domains, impeding the twist [113]. Taken together, these results suggest that quaternary twisting is a robust structural motion of the nAChR molecule accompanying ion channel opening.
(b). Molecular dynamics model of signal transduction
On the basis of structures for the GLIC open state, the ELIC ‘undefined’ closed state, and the GluCl open-channel form bound to the PAM ivermectin, Calimet et al. [114] carried out extensive all-atom molecular dynamics simulations over 0.2–0.5 µs. Removal of ivermectin causes the simulated trajectory to undergo a sequence of structural steps that couple agonist unbinding (from the EC domain) to ion-pore closing (in the TM domain). The simulation also shows that, in agreement with Taly et al. [25], a global twist initiates closure by facilitating the un-tilting of the pore-lining helices. The mechanistic scenario that emerges suggests that receptor twisting contributes to the activation process by ‘locking’ the ion channel in the open-pore form. A major outward tilting of the extracellular β-sandwiches further contributes to the allosteric communication between neurotransmitter binding site and ion pore. Crystallography of GLIC at both pH 7 (closed) and pH 4 (open) provided the first pair of gating endpoints in a single receptor [86] and the opportunity to generate a coarse-grained dynamic of the allosteric transition model [86, sup. mat.].
Taken together, the latest molecular dynamics ([34,115]), structural [86] and physiological [54,116] studies converge on a common detailed atomic model for the gating transition. According to the model, the stepwise process starts at the orthosteric binding site (loops A, B and C), propagates to the EC/TM interface (β1–β2 loop and Cys loop) via rigid-body rearrangements of the EC β-sandwiches, and moves on to the TM helices (M2, then M4 and M3) to finally open the gate. Two distinct sequential quaternary transitions take place: a radial concerted contraction or un-blooming of the EC domain, which opens the ion pore, followed by global concerted twisting to lock the channel in the active, open-channel state. Accordingly, gating of muscle nAChR is not a single-step ‘rigid’ event but proceeds through a concerted, stepwise, conformational sequence [34].
Structural and molecular dynamics data are consistent with the μs–ms kinetic analyses of single-channel recordings on muscle nAChR [54,116], which show that the nAChR isomerization is a well-defined sequence of protein domain motions that generate a propagated, Brownian, stepwise process ([54]; see also [116]). Moreover, the data are globally consistent with the MWC postulate that the conformational transition is ‘concerted’, i.e. conserves symmetry from start to end.
Finally, in addition to the closed resting state and the open active state, the structures of several fast (I) and slow (D) desensitized states—and uncoupled forms—have been identified with prokaryotic receptors. For example, the locally closed state [117] which shows an ‘active’ EC conformation with a ‘closed’ TM channel might be blocked in the middle of the stepwise R ↔ A transition [118] or alternatively be a fast desensitized I state [117]. The closed state of ELIC—often considered to be a resting state (see [114])—might then represent a slowly desensitized D state [34,86]. The ELIC structure has also been suggested to represent a ‘refractory’/’uncoupled’ state [60] (see [34] for discussion). The nAChR and homologues definitely exist in more than the two basic resting and active conformations of the minimal MWC model and this needs further investigation (see [34,119]).
(c). Allosteric modulation
Molecular dynamics is being applied to the analysis of the mode of action of allosteric modulators. Studies of the crystal structures of nAChR homologues GluCl and GLIC at pH 4 and pH 7 [34,86,88,120] are consistent with the view that allosteric modulators stabilize similar resting and active states to the orthosteric ligands. They further show that the gating transitions involve significant restructuring of subunit interfaces where sites for orthosteric ligands and allosteric modulators are primarily located [86]. This restructuring implements what was referred to as the ‘quaternary constraint’ in the original MWC model [14,34–36]. It includes a strong contraction of the orthosteric sites in the R → A transition [37,86] as a major change at the level of allosteric modulatory sites: the widening of the homologue of the Ca2+ pocket in the R state and the stabilization by positive allosteric modulators (ivermectin) of the untwisted (open-pore) configuration of the TM domain [86,117,118,120–123] (see [34,115]) (figure 5).
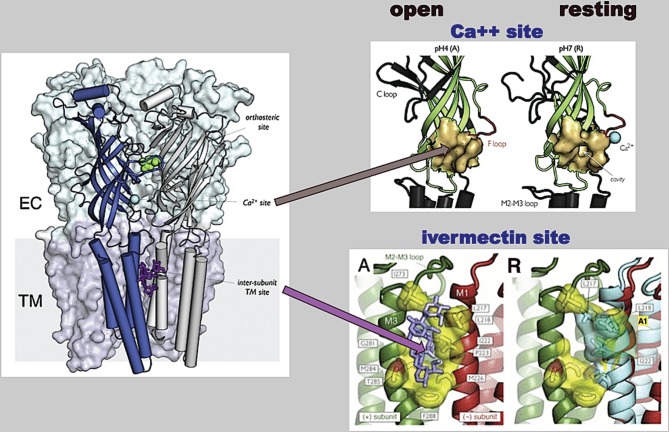
Figure 5.
Dynamic reorganization of the allosteric modulatory sites from Ca2+ (ECD) (top) and for ivermectin (TM) (bottom) in the course of the gating transition. The comparison of GLIC pH4 (active) with GLIC pH7 (resting) reveals an important tertiary change in the EC subunits during activation, visualized by the crystal structure of GluCl with ivermectin bound [35] and contraction of the intersubunit TM binding site by the receptor's twisting. Reproduced from Cecchini & Changeux [34, figs 5 and 6].
From a drug design perspective, the ongoing structural and molecular dynamics studies suggest as logical targets both orthosteric and allosteric modulatory sites present on these multiple structurally distinct conformation(s)—rather than a single fixed binding site [34,48,86]. The consequences are important for the design of drugs with expected agonist versus antagonist properties. A new pharmacology is emerging.
7. Conclusion: allosteric receptors as the molecular machines of mind
High-resolution structural data from nAChR and pLGIC homologues, combined with molecular dynamics studies of the allosteric transition in pLGIC, have yielded a radically new description, in atomic detail, of the chemical-to-electric signal transduction mechanism present in the nervous system. The mechanism involves the propagation of a concerted, stepwise, conformational change on a microsecond physiological timescale. Parallel studies with the various nAChRs, ion channels, G-protein-coupled receptors, tyrosine kinase receptors, and even nuclear receptors (reviewed [20,27]) have significantly advanced the understanding of the ‘switching’ events mediated by these molecular machines, and highlighted important features common to the mechanisms involved [20,27,117,124–126].
This understanding has major practical consequences in the conception of new pharmacological agents at both orthosteric and allosteric modulatory sites in each of their conformational states [27,34,91]. Attesting to the growing interest in allosteric phenomena, the allosteric database lists 71 538 substances as allosteric modulators [127], among them 91 currently used as medicines. A new pharmacology is on the way.
The allosteric MWC model also predicts that altering the intrinsic, unliganded equilibrium between discrete conformational states, e.g. by gene mutation, may cause constitutive receptor activation (or inhibition) with resulting major human pathologies [20,27,124]. The Allosteric database lists 3350 allostery-related diseases [127].
The dynamics of human brain processes, including mental ones, are necessarily constrained by the timescale of the conformational transitions of the brain's building blocks, a fact demonstrated here with nicotinic receptors up to conscious processing [128]. The realization that many of these molecular machines have barely changed over 3 billion years, from bacteria to humans, raises an almost existential question: did such extraordinary structural and functional conservation impose upon human brain function a speed limitation in signal transmission originating as a bacterial clock? Is this why our brain networks propagate signals below the speed of sound as compared with computers operating at the speed of light?
These investigations further document and enrich what may be called a ‘chemical theory of higher brain functions’ [129–131]. Within this framework, all processes at the multiple levels of organization that span the human brain, from synapse to consciousness, rest upon a biochemical universe of allosteric transitions that mediate neuronal and interneuronal communications.
Acknowledgements
I thank Professor Paul deWeer and Henri Korn for careful language editing and editorial assistance and gratefully acknowledge the contribution of my past and present collaborators who contributed to this work. This paper was written in part at the Marine Biological Laboratory, Woods Hole, MA.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
Funding was provided by CNRS UMR 3571, Institut Pasteur, Paris F-75724, Communications Cellulaires, Collège de France, Paris F-75005, France and Human Brain Program SGA1 CDP6 grant. This project has also received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement no. 720270 (HBP SGA1).
References
- 1.Changeux J-P. 1961. The feedback control mechanisms of biosynthetic L-threonine deaminase by L-isoleucine. Cold Spring Harb. Symp. Quant. Biol. 26, 313–318. ( 10.1101/SQB.1961.026.01.037) [DOI] [PubMed] [Google Scholar]
- 2.Monod J, Jacob F. 1961. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26, 389–401. ( 10.1101/SQB.1961.026.01.048) [DOI] [PubMed] [Google Scholar]
- 3.Gerhart JC, Pardee AB. 1962. The enzymology of control by feedback inhibition. J. Biol. Chem. 237, 891–896. [PubMed] [Google Scholar]
- 4.Monod J, Changeux J-P, Jacob F. 1963. Allosteric proteins and cellular control systems. J. Mol. Biol. 6, 306–329. ( 10.1016/S0022-2836(63)80091-1) [DOI] [PubMed] [Google Scholar]
- 5.Koshland DE., Jr 1959. Enzyme flexibility and enzyme action. J. Cell Comp. Physiol. 54, 245–258. ( 10.1002/jcp.1030540420) [DOI] [PubMed] [Google Scholar]
- 6.Changeux J-P.1964. On the allosteric properties of biosynthetic l-threonine deaminase. PhD thesis, Institut Pasteur.
- 7.Changeux J-P. 1964. On the allosteric properties of L-threonine deaminase. I. Methods of studying biosynthetic L-threonine deaminase. Bull. Soc. Chim. Biol. 46, 927–946 (in French). [PubMed] [Google Scholar]
- 8.Changeux J-P. 1964. On the allosteric properties of biosynthetic L-threonine deaminase. II. Kinetics of action of biosynthetic L-threonine deaminase with respect to the natural substrate and inhibitor. Bull. Soc. Chim. Biol. 46, 947–961 (in French). [PubMed] [Google Scholar]
- 9.Changeux J-P. 1964. On the allosteric properties of L-threonine deaminase. 3. Interpretation of the inhibitory effect of L-isoleucine: steric hindrance or allosteric effect. Bull. Soc. Chim. Biol. 46, 1151–1173 (in French). [PubMed] [Google Scholar]
- 10.Changeux J-P. 1965. On the allosteric properties of L-threonine deaminase. 4. The desensitization phenomenon. Bull. Soc. Chim. Biol. 47, 113–139 (in French). [PubMed] [Google Scholar]
- 11.Changeux J-P. 1965. On the allosteric properties of L-threonine deaminase. 5. The allosteric transition. Bull. Soc. Chim. Biol. 47, 267–280 (in French). [PubMed] [Google Scholar]
- 12.Changeux J-P. 1965. On the allosteric properties of L-threonine deaminase. 6i. General discussion. Bull. Soc. Chim. Biol. 47, 281–300 (in French). [PubMed] [Google Scholar]
- 13.Gerhart JC, Pardee AB. 1964. Aspartate transcarbamylase, an enzyme designed for feedback inhibition. Fed. Proc. 23, 727–735. [PubMed] [Google Scholar]
- 14.Monod J, Wyman J, Changeux J-P. 1965. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118. ( 10.1016/S0022-2836(65)80285-6) [DOI] [PubMed] [Google Scholar]
- 15.Koshland DE Jr, Nemethy G, Filmer D. 1966. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385. ( 10.1021/bi00865a047) [DOI] [PubMed] [Google Scholar]
- 16.Kirschner K, Eigen M, Bittman R, Voigt B. 1965. The binding of nicotinamide-adenine dinucleotide to yeast d-glyceraldehyde-3-phosphate dehydrogenase: temperature-jump relaxation studies on the mechanism of an allosteric enzyme. Proc. Natl Acad. Sci. USA 56, 1661–1667. ( 10.1073/pnas.56.6.1661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Changeux J-P, Rubin M. 1968. Allosteric interactions in aspartate transcarbamylase. 3. Interpretation of experimental data in terms of the model of Monod, Wyman, and Changeux. Biochemistry 7, 553–561. ( 10.1021/bi00842a601) [DOI] [PubMed] [Google Scholar]
- 18.Cui Q, Karplus M. 2008. Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307. ( 10.1110/ps.03259908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh K, Sasai M. 2010. Entropic mechanism of large fluctuation in allosteric transition. Proc. Natl Acad. Sci. USA 107, 7775–7780. ( 10.1073/pnas.0912978107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Changeux J-P. 2013. 50 years of allosteric interactions: the twists and turns of the models. Nat. Rev. Mol. Cell Biol. 14, 819–829. ( 10.1038/nrm3695) [DOI] [PubMed] [Google Scholar]
- 21.Terada TP, Terada TP, Kimura T, Sasai M. 2013. Entropic mechanism of allosteric communication in conformational transitions of dihydrofolate reductase. J. Phys. Chem. B 117, 12 864–12 877. ( 10.1021/jp402071m) [DOI] [PubMed] [Google Scholar]
- 22.Motlagh HN, Wrabl JO, Li J, Hilser VJ. 2014. The ensemble nature of allostery. Nature 508, 331–339. ( 10.1038/nature13001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai CJ, Nussinov R. 2014. A unified view of ‘how allostery works’. PLoS Comput. Biol. 10, e1003394 ( 10.1371/journal.pcbi.1003394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. 1998. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac. Symp. Biocomput. 1998, 473–484. [PubMed] [Google Scholar]
- 25.Taly A, Delarue M, Grutter T, Nilges M, Le Novère N, Corringer PJ, Changeux JP. 2005. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys. J. 88, 3954–3965. ( 10.1529/biophysj.104.050229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Changeux J-P, Thiéry J, Tung Y, Kittel C. 1967. On the cooperativity of biological membranes. Proc. Natl Acad. Sci. USA 57, 335–341. ( 10.1073/pnas.57.2.335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Changeux J-P, Christopoulos A. 2016. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell 166, 1084–1102. ( 10.1016/j.cell.2016.08.015) [DOI] [PubMed] [Google Scholar]
- 28.Changeux J-P, Kasai M, Lee CY. 1970. Use of a snake venom toxin to characterize the cholinergic receptor protein. Proc. Natl Acad. Sci. USA 67, 1241–1247. ( 10.1073/pnas.67.3.1241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlin A. 1967. On the application of ‘a plausible model’ of allosteric proteins to the receptor for acetylcholine. J. Theor. Biol. 16, 306–320. ( 10.1016/0022-5193(67)90011-2) [DOI] [PubMed] [Google Scholar]
- 30.Li M, Hazelbauer GL. 2014. Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl Acad. Sci. USA 111, 15 940–15 945. ( 10.1073/pnas.1415184111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langley JN. 1905. On the reaction of cells and of nerve-endings to certain poisons, chiefly as regards the reaction of striated muscle to nicotine and to curari. J. Physiol. 33, 374–413. ( 10.1113/jphysiol.1905.sp001128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miledi R, Molinoff P, Potter LT. 1971. Isolation of the cholinergic receptor protein of Torpedo electric tissue. Nature 229, 554–557. ( 10.1038/229554a0) [DOI] [PubMed] [Google Scholar]
- 33.Karlin A. 1993. Structure of nicotinic acetylcholine receptors. Curr. Opin. Neurobiol. 3, 299–309. ( 10.1016/0959-4388(93)90121-E) [DOI] [PubMed] [Google Scholar]
- 34.Cecchini M, Changeux J-P. 2015. The nicotinic acetylcholine receptor and its prokaryotic homologues: structure, conformational transitions & allosteric modulation. Neuropharmacology 96, 137–149. ( 10.1016/j.neuropharm.2014.12.006) [DOI] [PubMed] [Google Scholar]
- 35.Hibbs RE, Gouaux E. 2011. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60. ( 10.1038/nature10139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taly A, Hénin J, Changeux J-P, Cecchini M. 2014. Allosteric regulation of pentameric ligand-gated ion channels: an emerging mechanistic perspective. Channels 8, 1–11. ( 10.4161/chan.29444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devillers-Thiery A, Changeux JP, Paroutaud P, Strosberg AD. 1979. The amino-terminal sequence of the 40000 molecular weight subunit of the acetylcholine receptor protein from Torpedo marmorata. FEBS Lett. 104, 99–105. ( 10.1016/0014-5793(79)81092-3) [DOI] [PubMed] [Google Scholar]
- 38.Raftery MA, Hunkapiller MW, Strader CD, Hood LE. 1980. Acetylcholine receptor: complex of homologous subunits. Science 208, 1454–1456. ( 10.1126/science.7384786) [DOI] [PubMed] [Google Scholar]
- 39.Noda M, et al. 1982. Primary structure of α-subunit precursor of Torpedo californica acetylcholine receptor deduced from cDNA sequence. Nature 299, 793–797. ( 10.1038/299793a0) [DOI] [PubMed] [Google Scholar]
- 40.Ballivet M, Patrick J, Lee J, Heinemann S.. 1982. Molecular cloning of cDNA coding for the gamma subunit of Torpedo acetylcholine receptor. Proc. Natl Acad. Sci. USA 79, 4466–4470. ( 10.1073/pnas.79.14.4466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devillers-Thiery A, Giraudat J, Bentaboulet M, Changeux JP. 1983. Complete mRNA coding sequence of the acetylcholine binding alpha-subunit of Torpedo marmorata acetylcholine receptor: a model for the transmembrane organization of the polypeptide chain. Proc. Natl Acad. Sci. USA 80, 2067–7136. ( 10.1073/pnas.80.7.2067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katz B, Miledi R. 1973. The binding of acetylcholine to receptors and its removal from the synaptic cleft. J. Physiol. 231, 549–574. ( 10.1113/jphysiol.1973.sp010248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz B, Miledi R. 1977. Transmitter leakage from motor nerve endings. Proc. R. Soc. Lond. B 196, 59–72. ( 10.1098/rspb.1977.0029) [DOI] [PubMed] [Google Scholar]
- 44.Neher E, Sakmann B. 1976. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature 260, 799–802. ( 10.1038/260799a0) [DOI] [PubMed] [Google Scholar]
- 45.Katz B, Thesleff S. 1957. A study of the desensitization produced by acetylcholine at the motor end-plate. J. Physiol. 138, 63–80. ( 10.1113/jphysiol.1957.sp005838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasai M, Changeux J-P. 1971. In vitro excitation of purified membrane fragments by cholinergic agonists. J. Membr. Biol. 6, 1–80. ( 10.1007/BF01874112) [DOI] [PubMed] [Google Scholar]
- 47.Cohen JB, Weber M, Huchet M, Changeux J-P. 1972. Purification from Torpedo marmorata electric tissue of membrane fragments particularly rich in cholinergic receptor protein. FEBS Lett. 26, 43–47. ( 10.1016/0014-5793(72)80538-6) [DOI] [PubMed] [Google Scholar]
- 48.Heidmann T, Changeux J-P. 1979. Fast kinetic studies on the interaction of a fluorescent agonist with the membrane-bound acetylcholine receptor from Torpedo marmorata. Eur. J. Biochem. 94, 255–279 and 281–296 ( 10.1111/j.1432-1033.1979.tb12893.x) [DOI] [PubMed] [Google Scholar]
- 49.Heidmann T, Changeux J-P. 1980. Interaction of a fluorescent agonist with the membrane-bound acetylcholine receptor from Torpedo marmorata in the millisecond time range: resolution of an ‘intermediate’ conformational transition and evidence for positive cooperative effects. Biochem. Biophys. Res. Commun. 97, 889–896. ( 10.1016/0006-291X(80)91460-6) [DOI] [PubMed] [Google Scholar]
- 50.Heidmann T, Bernhardt J, Neumann E, Changeux J-P. 1980. Rapid kinetics of agonist binding and permeability response analyzed in parallel on acetylcholine receptor rich membranes from Torpedo marmorata. Biochemistry 22, 5452–5459. ( 10.1021/bi00292a029) [DOI] [PubMed] [Google Scholar]
- 51.Neubig RR, Boyd ND, Cohen JB. 1982. Conformations of Torpedo acetylcholine receptor associated with ion transport and desensitization. Biochemistry 21, 3460–3467. ( 10.1021/bi00257a032) [DOI] [PubMed] [Google Scholar]
- 52.Edelstein SJ, Schaad O, Henry E, Bertrand D, Changeux J-P. 1996. A kinetic mechanism for nicotinic acetylcholine receptors based on multiple allosteric transitions. Biol. Cybern. 75, 361–379. ( 10.1007/s004220050302) [DOI] [PubMed] [Google Scholar]
- 53.Jackson MB. 1984. Spontaneous openings of the acetylcholine receptor channel. Proc. Natl Acad. Sci. USA 81, 3901–3904. ( 10.1073/pnas.81.12.3901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auerbach A. 2012. Thinking in cycles: MWC is a good model for acetylcholine receptor-channels. J. Physiol. 590, 93–98. ( 10.1113/jphysiol.2011.214684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cartaud J, Benedetti EL, Cohen JB, Meunier JC, Changeux J-P. 1973. Presence of a lattice structure in membrane fragments rich in nicotinic receptor protein from the electric organ of Torpedo marmorata. FEBS Lett. 33, 109–113. ( 10.1016/0014-5793(73)80171-1) [DOI] [PubMed] [Google Scholar]
- 56.Brisson A, Unwin PN. 1985. Quaternary structure of the acetylcholine receptor. Nature 315, 474–477. ( 10.1038/315474a0) [DOI] [PubMed] [Google Scholar]
- 57.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. 2001. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276. ( 10.1038/35077011) [DOI] [PubMed] [Google Scholar]
- 58.Unwin N. 2013. Nicotinic acetylcholine receptor and the structural basis of neuromuscular transmission: insights from Torpedo postsynaptic membranes. Q Rev. Biophys. 46, 283–322. ( 10.1017/S0033583513000061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paas Y, Gibor G, Grailhe R, Savatier-Duclert N, Dufresne V, Sunesen M, de Carvalho LP, Changeux J-P, Attali B. 2005. Pore conformations and gating mechanism of a Cys-loop receptor. Proc. Natl Acad. Sci. USA 102, 15 877–15 882. ( 10.1073/pnas.0507599102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.daCosta CJ, Dey L, Therien JP, Baenziger JE. 2013. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat. Chem. Biol. 9, 701–707. ( 10.1038/nchembio.1338) [DOI] [PubMed] [Google Scholar]
- 61.Tasneem A, Iyer LM, Jakobsson E, Aravind L. 2005. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6, R4 ( 10.1186/gb-2004-6-1-r4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bocquet N, Prado de Carvalho L, Cartaud J, Neyton J, Le Poupon C, Taly A, Grutter T, Changeux J-P, Corringer PJ. 2007. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119. ( 10.1038/nature05371) [DOI] [PubMed] [Google Scholar]
- 63.Hilf RJ, Dutzler R. 2008. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379. ( 10.1038/nature06717) [DOI] [PubMed] [Google Scholar]
- 64.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux J-P, Delarue M, Corringer PJ. 2009. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111 ( 10.1038/nature07462) [DOI] [PubMed] [Google Scholar]
- 65.Hilf RJ, Dutzler R. 2009. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115 ( 10.1038/nature07461) [DOI] [PubMed] [Google Scholar]
- 66.Giraudat J, Dennis M, Heidmann T, Chang JY, Changeux J-P. 1986. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: serine-262 of the delta subunit is labeled by [3H] chlorpromazine. Proc. Natl Acad. Sci. USA 83, 2719–2723. ( 10.1073/pnas.83.8.2719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giraudat J, Dennis M, Heidmann T, Haumont PY, Lederer F, Changeux J-P. 1987. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: [3H]chlorpromazine labels homologous residues in the beta and delta chains. Biochemistry 26, 2410–2418. ( 10.1021/bi00383a003) [DOI] [PubMed] [Google Scholar]
- 68.Hucho F, Oberthür W, Lottspeich F. 1986. The ion channel of the nicotinic acetylcholine receptor is formed by the homologous helices M II of the receptor subunits. FEBS Lett. 205, 137–142. ( 10.1016/0014-5793(86)80881-X) [DOI] [PubMed] [Google Scholar]
- 69.Imoto K, et al. 1988. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature 335, 645–648. ( 10.1038/335645a0) [DOI] [PubMed] [Google Scholar]
- 70.Leonard RJ, Labarca CG, Charnet P, Davidson N, Lester HA. 1988. Evidence that the M2 membrane-spanning region lines the ion channel pore of the nicotinic receptor. Science 242, 1578–1581. ( 10.1126/science.2462281) [DOI] [PubMed] [Google Scholar]
- 71.Miller PS, Aricescu AR. 2014. Crystal structure of a human GABAA receptor. Nature 512, 270–275. ( 10.1038/nature13293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hassaine G, et al. 2014. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281. ( 10.1038/nature13552) [DOI] [PubMed] [Google Scholar]
- 73.Du J, Lü W, Wu S, Cheng Y, Gouaux E. 2015. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229. ( 10.1038/nature14853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. 2015. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature 526, 277–280. ( 10.1038/nature14972) [DOI] [PubMed] [Google Scholar]
- 75.Morales-Perez CL, Noviello CM, Hibbs RE. 2016. X-ray structure of the human α4β2 nicotinic receptor. Nature 538, 411–415. ( 10.1038/nature19785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Changeux J-P. 1990. Functional architecture and dynamics of the nicotinic acetylcholine receptor: an allosteric ligand-gated ion channel. In Fidia research foundation neuroscience award lectures, vol. 4 (eds Changeux JP, Llinas R, Purves D, Bloom F), pp. 17–168. New-York, NY: Raven Press. [Google Scholar]
- 77.Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. 1998. From ab initio quantum mechanics to molecular neurobiology: a cation–π binding site in the nicotinic receptor. Proc. Natl Acad. Sci. USA 95, 12 088–12 093. ( 10.1073/pnas.95.21.12088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neher E, Marty A, Fenwick E. 1983. Ionic channels for signal transmission and propagation. Prog. Brain Res. 58, 39–48. ( 10.1016/S0079-6123(08)60005-9) [DOI] [PubMed] [Google Scholar]
- 79.Changeux J-P. 2013. The concept of allosteric modulation: an overview. Drug Discov. Today Technol. 10, e223–e228. ( 10.1016/j.ddtec.2012.07.007) [DOI] [PubMed] [Google Scholar]
- 80.Sauguet L, Shahsavar A, Delarue M. 2015. Crystallographic studies of pharmacological sites in pentameric ligand-gated ion channels. Biochim. Biophys. Acta 1850, 511–523. ( 10.1016/j.bbagen.2014.05.007) [DOI] [PubMed] [Google Scholar]
- 81.Mulle C, Léna C, Changeux J-P. 1992. Potentiation of nicotinic receptor response by external calcium in rat central neurons. Neuron 8, 937–945. ( 10.1016/0896-6273(92)90208-U) [DOI] [PubMed] [Google Scholar]
- 82.Vernino S, Amador M, Luetje CW, Patrick J, Dani JA. 1992. Calcium modulation and high calcium permeability of neuronal nicotinic acetylcholine receptors. Neuron 8, 127–134. ( 10.1016/0896-6273(92)90114-S) [DOI] [PubMed] [Google Scholar]
- 83.Galzi JL, Bertrand S, Corringer PJ, Changeux J-P, Bertrand D. 1996. Identification of calcium binding sites that regulate potentiation of a neuronal nicotinic acetylcholine receptor. EMBO J. 15, 5824–5832. [PMC free article] [PubMed] [Google Scholar]
- 84.Le Novere N, Grutter T, Changeux J-P. 2002. Models of the extracellular domain of the nicotinic receptors and of agonist and Ca2+ binding sites. Proc. Natl Acad. Sci. USA 99, 3210–3215. ( 10.1073/pnas.042699699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zimmermann I, Dutzler R. 2011. Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 9, e1001101 ( 10.1371/journal.pbio.1001101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sauguet L, et al. 2014. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl Acad. Sci. USA 111, 966–971. ( 10.1073/pnas.1314997111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krause RM, Buisson B, Bertrand S, Corringer PJ, Galzi JL, Changeux J-P, Bertrand D. 1998. Ivermectin: a positive allosteric effector of the α7 neuronal nicotinic acetylcholine receptor. Mol. Pharmacol. 53, 283–294. ( 10.1124/mol.53.2.283) [DOI] [PubMed] [Google Scholar]
- 88.Nury H, et al. 2011. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature 469, 428–431. ( 10.1038/nature09647) [DOI] [PubMed] [Google Scholar]
- 89.Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, Harris RA, Delarue M. 2013. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat. Commun. 4, 1697 ( 10.1038/ncomms2682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang X, Chen H, Shaffer PL. 2017. Crystal structures of human GlyRα3 bound to ivermectin. Structure 25, 945–950e2. ( 10.1016/j.str.2017.04.007) [DOI] [PubMed] [Google Scholar]
- 91.Bertrand D, Gopalakrishnan M. 2007. Allosteric modulation of nicotinic acetylcholine receptors. Biochem. Pharmacol. 74, 1155–1163. ( 10.1016/j.bcp.2007.07.011) [DOI] [PubMed] [Google Scholar]
- 92.Forman SA, Chiara DC, Miller KW. 2015. Anesthetics target interfacial transmembrane sites in nicotinic acetylcholine receptors. Neuropharmacology 96, 169–177. ( 10.1016/j.neuropharm.2014.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olsen R. 2014. Analysis of γ-aminobutyric acid GABA type A receptor subtypes using isosteric and allosteric ligands. Neurochem. Res. 39, 1924–1941. ( 10.1007/s11064-014-1382-3) [DOI] [PubMed] [Google Scholar]
- 94.Kabbani N, Woll MP, Levenson R, Lindstrom JM, Changeux J-P. 2007. Intracellular complexes of the β2 subunit of the nicotinic acetylcholine receptor in brain identified by proteomics. Proc. Natl Acad. Sci. USA 104, 20 570–20 575. ( 10.1073/pnas.0710314104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alcaino C, Musgaard M, Minguez T, Mazzaferro S, Faundez M, Iturriaga-Vasquez P, Biggin PC, Bermudez I.. 2017. Role of the Cys loop and transmembrane domain in the allosteric modulation of α4β2 nicotinic acetylcholine receptors. J. Biol. Chem. 292, 551–562. ( 10.1074/jbc.M116.751206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gill JK, Chatzidaki A, Ursu D, Sher E, Millar NS.. 2013. Contrasting properties of α7-selective orthosteric and allosteric agonists examined on native nicotinic acetylcholine receptors. PLoS ONE 8, e55047 ( 10.1371/journal.pone.0055047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thomsen MS, Arvaniti M, Jensen MM, Shulepko MA, Dolgikh DA, Pinborg LH, Härtig W, Lyukmanova EN, Mikkelsen JD. 2016. Lynx1 and Aβ1-42 bind competitively to multiple nicotinic acetylcholine receptor subtypes. Neurobiol. Aging 46, 13–21. ( 10.1016/j.neurobiolaging.2016.06.009) [DOI] [PubMed] [Google Scholar]
- 98.Bagdas D, et al. 2016. The α7 nicotinic receptor dual allosteric agonist and positive allosteric modulator GAT107 reverses nociception in mouse models of inflammatory and neuropathic pain. Br. J. Pharmacol. 173, 2506–2520. ( 10.1111/bph.13528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Popot JL, Demel RA, Sobel A, Van Deenen LL, Changeux J-P. 1978. Interaction of the acetylcholine (nicotinic) receptor protein from Torpedo marmorata electric organ with monolayers of pure lipids. Eur. J. Biochem. 85, 27–42. ( 10.1111/j.1432-1033.1978.tb12209.x) [DOI] [PubMed] [Google Scholar]
- 100.Popot JL, Cartaud J, Changeux J-P. 1981. Reconstitution of a functional acetylcholine receptor. Incorporation into artificial lipid vesicles and pharmacology of the agonist-controlled permeability changes. Eur. J. Biochem. 118, 203–214. ( 10.1111/j.1432-1033.1981.tb06388.x) [DOI] [PubMed] [Google Scholar]
- 101.Ochoa EL, Dalziel AW, McNamee MG. 1983. Reconstitution of acetylcholine receptor function in lipid vesicles of defined composition. Biochim. Biophys. Acta 727, 151–162. ( 10.1016/0005-2736(83)90379-6) [DOI] [PubMed] [Google Scholar]
- 102.Martinez KL, Gohon Y, Corringer PJ, Tribet C, Mérola F, Changeux J-P, Popot JL. 2002. Allosteric transitions of Torpedo acetylcholine receptor in lipids, detergent and amphipols: molecular interactions vs. physical constraints. FEBS Lett. 528, 251–256. ( 10.1016/S0014-5793(02)03306-9) [DOI] [PubMed] [Google Scholar]
- 103.daCosta CJ, Baenziger JE. 2009. A lipid-dependent uncoupled conformation of the acetylcholine receptor. J. Biol. Chem. 284, 17 819–17 825. ( 10.1074/jbc.M900030200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marsh D, Barrantes FJ. 1978. Immobilized lipid in acetylcholine receptor-rich membranes from Torpedo marmorata. Proc. Natl Acad. Sci. USA 75, 4329–4333. ( 10.1073/pnas.75.9.4329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jones OT, McNamee MG. 1988. Annular and nonannular binding sites for cholesterol associated with the nicotinic acetylcholine receptor. Biochemistry 27, 2364–2374. ( 10.1021/bi00407a018) [DOI] [PubMed] [Google Scholar]
- 106.Addona GH, Sandermann H Jr, Kloczewiak MA, Husain SS, Miller KW.. 1998. Where does cholesterol act during activation of the nicotinic acetylcholine receptor? Biochim. Biophys. Acta 1370, 299–309. ( 10.1016/S0005-2736(97)00280-0) [DOI] [PubMed] [Google Scholar]
- 107.Di Scala C, Mazzarino M, Yahi N, Varini K, Garmy N, Fantini J, Chahinian H. 2017. Anandamide-ceramide interactions in a membrane environment: molecular dynamic simulations data. Data Brief 14, 163–167. ( 10.1016/j.dib.2017.07.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brannigan G, Hénin J, Law R, Eckenhoff R, Klein ML. 2008. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl Acad. Sci. USA 105, 14 418–14 423. ( 10.1073/pnas.0803029105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hénin J, Salari R, Murlidaran S, Brannigan G. 2014. A predicted binding site for cholesterol on the GABAA receptor. Biophys. J. 106, 1938–1949. ( 10.1016/j.bpj.2014.03.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng X, Ivanov I, Wang H, Sine SM, McCammon JA. 2009. Molecular dynamics simulations of ELIC-a prokaryotic homologue of the nicotinic acetylcholine receptor. Biophys. J. 96, 4502–4513. ( 10.1016/j.bpj.2009.03.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheng X, Lu B, Grant B, Law RJ, McCammon JA. 2006. Channel opening motion of α7 nicotinic acetylcholine receptor as suggested by normal mode analysis. J. Mol. Biol. 355, 310–324. ( 10.1016/j.jmb.2005.10.039) [DOI] [PubMed] [Google Scholar]
- 112.Nury H, Poitevin F, Van Renterghem C, Changeux J-P, Corringer PJ, Delarue M, Baaden M. 2010. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc. Natl Acad. Sci. USA 107, 6275–6280. ( 10.1073/pnas.1001832107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taly A, Corringer PJ, Grutter T, Prado de Carvalho L, Karplus M, Changeux J-P. 2006. Implications of the quaternary twist allosteric model for the physiology and pathology of nicotinic acetylcholine receptors. Proc. Natl Acad. Sci. USA 103, 16 965–16 970. ( 10.1073/pnas.0607477103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Calimet N, Simoes M, Changeux JP, Karplus M, Taly A, Cecchini M. 2013. A gating mechanism of pentameric ligand-gated ion channels. Proc. Natl Acad. Sci. USA 110, E3987–E3996. ( 10.1073/pnas.1313785110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin NE, Malik S, Calimet N, Changeux J-P, Cecchini M. 2017. Un-gating and allosteric modulation of a pentameric ligand-gated ion channel captured by molecular dynamics. PLoS Comput. Biol. 13, e1005784 ( 10.1371/journal.pcbi.1005784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng W, Auerbach A. 2011. Decrypting the sequence of structural events during the gating transition of pentameric ligand-gated ion channels based on an interpolated elastic network model. PLoS Comput. Biol. 7, e1001046 ( 10.1371/journal.pcbi.1001046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prevost MS, Sauguet L, Nury H, Van Renterghem C, Huon C, Poitevin F, Baaden M, Delarue M, Corringer PJ. 2012. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat. Struct. Mol. Biol. 19, 642–649. ( 10.1038/nsmb.2307) [DOI] [PubMed] [Google Scholar]
- 118.Sivilotti L, Colquhoun D. 2016. In praise of single channel kinetics. J. Gen. Physiol. 148, 79–88. ( 10.1085/jgp.201611649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cerdan A, Martin NE, Cecchini M. Submitted. The physiologically active state of the glycine receptor captured by molecular dynamics. [DOI] [PubMed]
- 120.Althoff T, Hibbs RE, Banerjee S, Gouaux E. 2014. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337. ( 10.1038/nature13669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Velisetty P, Chalamalasetti SV, Chakrapani S. 2012. Conformational transitions underlying pore opening and desensitization in membrane-embedded Gloeobacter violaceus ligand-gated ion channel (GLIC). J. Biol. Chem. 287, 36 864–36 872. ( 10.1074/jbc.M112.401067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gonzalez-Gutierrez G, Lukk T, Agarwal V, Papke D, Nair SK, Grosman C. 2012. Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl Acad. Sci. USA 109, 6331–6336. ( 10.1073/pnas.1119268109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.daCosta CJ, Baenziger JE. 2013. Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure 21, 1271–1283. ( 10.1016/j.str.2013.06.019) [DOI] [PubMed] [Google Scholar]
- 124.Christopoulos A, et al. 2014. International Union of Basic and Clinical Pharmacology. XC. Multisite pharmacology: recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol. Rev. 66, 918–947. ( 10.1124/pr.114.008862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marzen S, Garcia HG, Phillips R. 2013. Statistical mechanics of Monod–Wyman–Changeux (MWC) models. J. Mol. Biol. 425, 1433–1460. ( 10.1016/j.jmb.2013.03.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yan L, Ravasio R, Brito C, Wyart M. 2017. Principles for optimal cooperativity in allosteric materials arXiv 1708.01820. (https://arxiv.org/abs/1708.01820)
- 127.Huang Z, et al. 2011. ASD: a comprehensive database of allosteric proteins and modulators. Nucleic Acids Res. 39, 663–669. ( 10.1093/nar/gkq1022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koukouli F, Rooy M, Changeux J-P, Maskos U. 2016. Nicotinic receptors in mouse prefrontal cortex modulate ultraslow fluctuations related to conscious processing. Proc. Natl Acad. Sci. USA 113, 14 823–14 828. ( 10.1073/pnas.1614417113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Changeux J-P. 1983. L’homme neuronal. Paris: Fayard. Neuronal man (transl. L Garey). New York, NY: Pantheon.
- 130.Changeux J-P. 1985. Neuronal man. (Transl. by L Garey). New York, NY: Pantheon.
- 131.Changeux J-P. 2013. The concept of allosteric interaction and its consequences for the chemistry of the brain. J. Biol. Chem. 288, 26 969–26 986. ( 10.1074/jbc.X113.503375) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.