Abstract
The 70 kDa heat-shock protein (Hsp70) is undoubtedly the most versatile of all molecular chaperones. Hsp70 is involved in numerous cellular protein folding processes, accompanying proteins throughout their lifespan from de novo folding at the ribosome to degradation at the proteasome, surveilling protein stability and functionality. Several properties of this ATP-dependent chaperone constitute the molecular basis for this versatility. With its substrate binding domain (SBD), Hsp70 transiently interacts with a short degenerative linear sequence motif found practically in all proteins and, in addition, with more folded protein conformers. Binding to polypeptides is tightly regulated by ATP binding and hydrolysis in the nucleotide binding domain, which is coupled to the SBD by an intricate allosteric mechanism. Hsp70 is regulated by a host of J-cochaperones, which act as targeting factors by regulating the ATPase activity of Hsp70 in synergism with the substrates themselves, and by several families of nucleotide exchange factors. In this review, I focus on the allosteric mechanism, which allows Hsp70s to interact with substrates with ultrahigh affinity through a non-equilibrium mode of action and summarize what mutagenesis and structural studies have taught us about the pathways and mechanics of interdomain communication.
This article is part of a discussion meeting issue ‘Allostery and molecular machines’.
Keywords: Hsp70, molecular chaperone, protein folding, interdomain communication
1. Hsp70: versatility of functions
The ATP-dependent 70 kDa heat-shock protein (Hsp70) chaperones are apparently universal and highly adaptable tools (figure 1a), being involved in a wide variety of cellular protein folding processes ranging from de novo folding to disaggregation and even disassembly of amyloid fibres, from protein translocation through membranous pores to protein degradation [4–6]. One reason for this versatility is certainly the ability of Hsp70s to bind short degenerative motifs within their substrate polypeptides, consisting of a core of five residues enriched in hydrophobic amino acids, flanked by positively charged residues [7]. Such sequence motifs are found on average every 30–40 residues in virtually all proteins. Interaction with such short motifs eliminates any size limitations for chaperone substrates. A second reason for the versatility of Hsp70 chaperones is their cooperation with J-proteins and nucleotide exchange factors, with additional cochaperones and with other chaperone systems [8]. J-proteins are modular multi-domain proteins characterized by the 75-residue J-domain, with which they interact with Hsp70s and stimulate their very low ATPase activity. J-proteins either bind to Hsp70 substrates themselves or are located within the cell where Hsp70 substrates appear, and target Hsp70s to their substrates. In the course of evolution, the number of J-proteins multiplied substantially from six in E. coli to 53 in humans [9] to 102 in Arabidopsis thaliana (determined by WU-Blast on the Arabidopsis thaliana genome database https://www.arabidopsis.org/). Hsp70s interact with four structurally unrelated families of nucleotide exchange factors: the GrpE proteins, the family of Bag proteins, the HspBP1 proteins and the Hsp110 s [10]. Bag-proteins are also multi-domain proteins, linking Hsp70s to diverse functions. Hsp70s in eukaryotic cells also interact with an anti-nucleotide exchange factor Hip [11], with Hop/Sti1, which connects Hsp70 to the Hsp90 chaperone system, and with the ubiquitin E3 ligase Chip [12,13], which links Hsp70 to the proteasome degradation system. In addition, Hsp70s cooperate in particular with Hsp90s in maturation and regulation of stability and activity of many native proteins [14], and with small Hsps and Hsp100 proteins in solubilization of amorphous protein aggregates [15]. A third reason for the versatility of Hsp70s is their intricate allosteric mechanism, which allows them to outrun competing folding reactions like aggregation and which will be the major topic of this review.
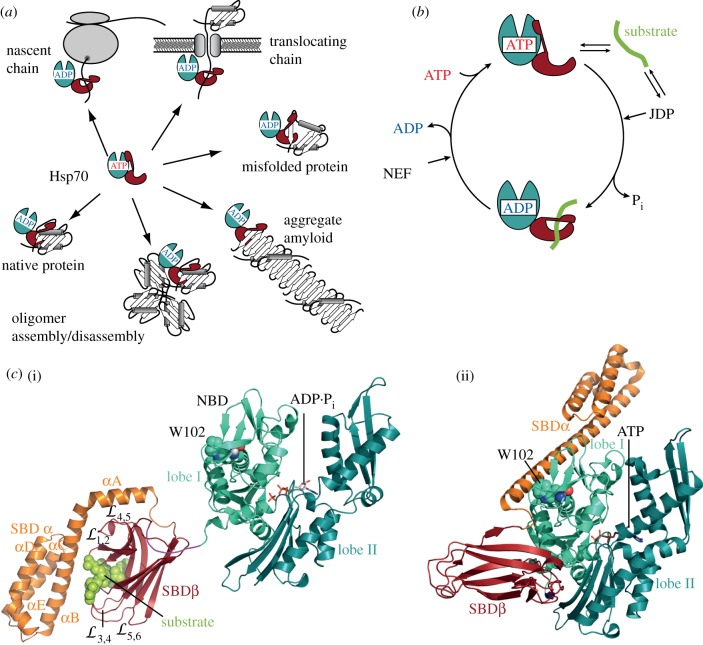
Figure 1.
Versatility of Hsp70s and functional cycle. (a) Hsp70s are able to interact with a wide variety of protein conformers, from completely unfolded polypeptides to native proteins and even amorphous protein aggregates and amyloid fibrils (modified from Schlecht et al. [1]). (b) Hsp70 chaperone cycle. Hsp70s exist in two conformational states, the ATP-bound state with low affinity for substrates but high substrate association and dissociation rates and the ADP-bound state with higher affinity for substrates but low substrate association and dissociation rates. J-domain proteins (JDP) regulate substrate binding by stimulating Hsp70's very low ATPase activity in synergism with substrates, and nucleotide exchange factors (NEF) regulate residence time of substrates on Hsp70s because nucleotide exchange is rate limiting for substrate release. (c) (i) Structure of the high-affinity ADP-bound conformation (PDB ID 2KHO; [2]) and (ii) the low-affinity ATP-bound conformation (PDB ID 4B9Q; [3]) of E. coli DnaK.
2. Evidence of allostery in Hsp70s
The first evidence for allostery in Hsp70s came from the observation that peptide substrates stimulate the weak ATPase activity of Hsp70s and that ATP binding decreased their affinity for substrates [16]. In addition to peptides, J-proteins were found to stimulate the ATPase activity [17] by acting in synergism with protein substrates [18–20]. The ATP-induced decrease in substrate affinity is due to approximately 100-fold increased peptide association rates and approximately 1000-fold increased peptide dissociation rates [21]. These changes in substrate affinity are accompanied by structural changes that are detected by partial proteolysis, tryptophan fluorescence, small angle X-ray scattering (SAXS), NMR and hydrogen exchange mass spectrometry. Upon tryptic digestion of the nucleotide-free and ADP-bound Hsp70, a prominent 44 kDa band appears, corresponding to the nucleotide binding domain (NBD), that is less pronounced when ATP-bound Hsp70 is digested [22–24]. Many Hsp70s have a single tryptophan in the NBD that reacts to ATP binding with a decrease in intensity and a blue shift of the emission maximum [24–27]. This change in tryptophan fluorescence is curiously not observed in the isolated NBD, but only if large parts of the SBD are present and therefore reports on conformational changes in the SBD [28,29]. SAXS measurements demonstrated that Hsp70s become more compact upon ATP binding as the radius of gyration decreases [30]. NMR and hydrogen exchange mass spectrometry also detect signatures of these conformational changes [31,32]. Based on these data and the structures of the individual domains [33,34], a functional cycle for Hsp70s was formulated (figure 1b): Hsp70 binds substrates in the ATP-bound state, which is characterized by high substrate association and dissociation rates but low affinity. ATP hydrolysis is very low in most Hsp70s with turnover numbers of 1 ATP per 20–30 min, but is stimulated in synergism by substrates and J-proteins, leading to the trapping of the substrate, because the ADP-bound state has low substrate exchange rates but higher affinity. Nucleotide exchange allows rebinding of ATP, and thus promotes substrate release. The non-equilibrium situation of this chaperone cycle leads to ultrahigh affinity [35].
A full understanding of all of the above-mentioned observations, however, only became possible with a structural model for full-length Hsp70 in the ATP-bound open conformation that was first hinted at by the structure of the homologous Hsp110 protein [36], before crystal structures of Hsp70 s finally revealed a detailed picture [3,37–40].
3. Hsp70 structure
NBD and the SBD were first crystalized in isolation [33,34] and also studied by NMR [41–44]. The N-terminal 45 kDa NBD consists of four subdomains (IA, IB, IIA, IIB) organized in two lobes (I and II) with a deep cleft between them, at the bottom of which ATP binds coordinated by residues from all four subdomains (figure 1c(i)). The 25 kDa SBD is built up of two subdomains, a β-sandwich subdomain (SBDβ) and an α-helical subdomain (SBDα). SBDβ is composed of two twisted, four-stranded β-sheets (strands 1, 2, 4, 5 and 3, 6, 7, 8) with upward protruding loops and the substrate binding cleft between strands 1 and 2 and strands 3 and 4. Between strands 3 and 4 is a deep hydrophobic pocket, tailored for a single large hydrophobic amino acid side chain. The substrate is further enclosed by two concentric pairs of loops forming a hydrophobic arch over the backbone of the co-crystallized peptide and contacting the peptide backbone via hydrogen bonds. The SBDα is composed of five α-helices (A–E), with helices A and B being packed tightly to the loops of the SBDβ, forming a lid over the substrate-binding cleft. NMR studies suggested that in the nucleotide-free and ADP-bound state SBD and NBD tumble largely independent of each other and are only tied together by the highly conserved flexible linker [2,31].
In the ATP-bound open conformation, the SBD is dissociated into SBDβ and SBDα, and both SBD subdomains are docked onto two faces of the NBD (figure 1c(ii)). Closer comparison of the ADP-bound and ATP-bound structure revealed three important conformational differences. First, the two lobes of the NBD are rotated relative to each other (figure 2a) with the consequence that in the ATP-bound conformation the two γ-phosphate coordinating residues K70 and E171 are displaced relative to the post-hydrolysis state, suggesting a mechanism for catalysis (figure 2b see below). The rotation of the lobes also significantly remodels the surface to which the SBDβ is docked in the ATP-bound state and opens a lower crevice (figure 2c). Second, the interdomain linker is inserted into the lower crevice of the NBD. Third, the SBDβ docked onto the NBD has a much more open substrate-binding cleft, as the substrate enclosing loops are moved apart, but a more narrow substrate-binding pocket, as the distance between F426 and I 438, which line the hydrophobic pocket, is much smaller (figure 2d). Insertion of the substrate seems only possible if the two residues are moved apart. Either substrate binding induces such a conformational change that is then propagated to the NBD or the SBDβ is in a close-open conformational equilibrium and substrate binding fixes the conformation by inserting a large hydrophobic residue into the transiently opened hydrophobic pocket.
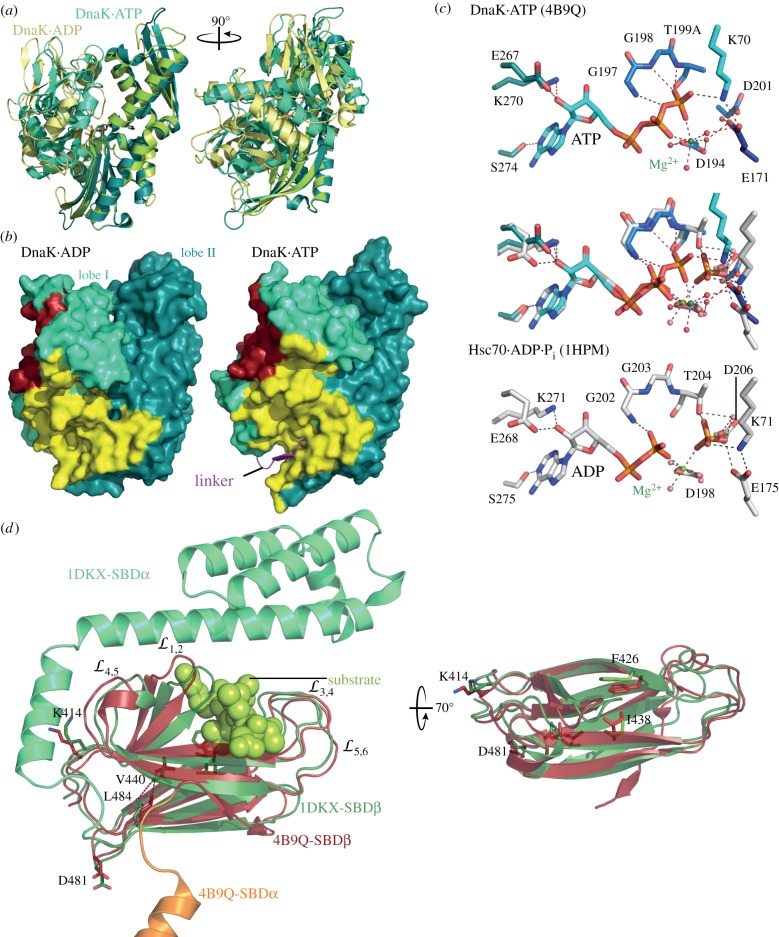
Figure 2.
ATP-induced conformational changes in NBD and SBD. (a) Overlay of the secondary structure representation of the ATP-bound NBD of E. coli DnaK (PDB ID 4B9Q; lobe I, greencyan; lobe II, deep teal) and a homology model of the NBD of E. coli DnaK on the structure of bovine Hsc70 (PDB ID 1HPM [45]; lobe I, yellow; lobe II, light green). (b) Surface representation of the NBD of E. coli DnaK in the ADP- (left) and ATP- (right) bound conformations. Coloured are lobe I (green cyan) and lobe II (deep teal), residues interacting in the ATP-bound open conformation with SBDβ (yellow) and SBDα (dark red), and the interdomain linker (magenta) inserted into the lower crevice of the NBD. (c) Conformational changes in the catalytic centre accompanying ATP hydrolysis. Upper panel, ATP and ATP-interacting residues in the structure of E. coli DnaK in the ATP-bound open conformation (PDB ID 4B9Q). Lower panel, ADP + inorganic phosphate and interacting residues in bovine Hsc70 (PCB ID 1HPM). Middle panel, overlay of upper and lower panel. Dashed lines indicate polar contacts. (d) ATP-induced conformational changes in the SBD. Overlay of the SBD of E. coli DnaK in the ATP-bound open conformation (PDB ID 4B9Q; SBDβ, dark red; SBDα, orange, truncated for space reasons) with the structure of the isolated SBD of E. coli DnaK (PDB ID 1DKX; [34]; SBDβ, dark green; SBDα, lime green). Indicated are the substrate (light green) enclosing loops (left panel) and residues lining the central hydrophobic substrate-binding pocket (right panel).
The difference between ATP-bound and ADP-bound conformation allows the sketching of an allosteric mechanism to explain ATP-induced substrate release and substrate-triggered ATP hydrolysis. It seems straightforward to propose that ATP-induced rotation of the NBD lobes remodels the NBD surface to allow docking of the SBDβ and subsequent opening of the substrate-enclosing loops, accelerating substrate dissociation. The caveat with this simple sequence of events is that helix A of the SBDα needs to be removed first before the SBDβ can dock onto the NBD. How SBDβ-docking and SBDα dissociation from SBDβ is induced is still enigmatic. On the other side, to elucidate how substrates stimulate the ATPase activity, we first need to understand the catalytic mechanism of ATP hydrolysis by Hsp70s. Based on the overlay of the NBD in the ADP- and ATP-bound states (figure 2b), it was proposed that D201, E171 and T199 are the catalytic residues guiding a water molecule for the inline attack on the γ-phosphate and that K70 stabilizes the pentavalent transition state [3]. In addition, as will be discussed below, the differences between ATP- and ADP-bound conformation also suggested a pathway through which substrates trigger ATP hydrolysis [46].
4. Analysis of allostery by mutagenesis
Over the years, genetic screens [23,47–49] and analysis of the available structures of individual domains and of sequence conservation [1,20,46,50–53] revealed amino acid replacements in Hsp70s, mostly E. coli DnaK, that caused a defect in allostery (electronic supplementary material, table S1). A first attempt was performed by Gierasch and colleagues to analyse allostery more globally by comparing Hsp70s with their non-allosteric Hsp110 homologues using a statistical coupling analysis of co-evolving pairs of residues [54]. This analysis yielded approximately 20% of all residues in NBD (56 residues) and SBD (59 residues) as statistically significantly coupled. Regrettably, only two residues, one in each domain, were verified to affect allostery when replaced and the chosen residues were in the interface between NBD and SBDβ and suggested to interact with each other in a homology model. Therefore, whether all or a majority of the residues found in this approach have an impact on allostery remains to be shown.
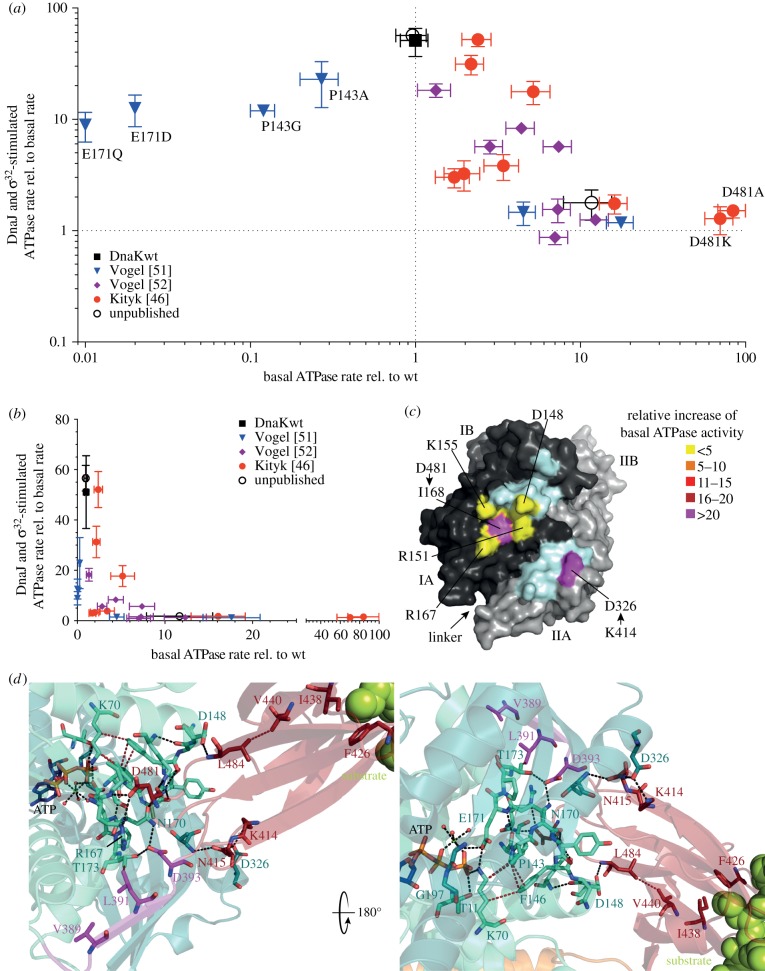
A global analysis of the phenotypes of amino acid replacement variants of E. coli DnaK that were characterized under comparable conditions sheds some light on interesting properties of the allosteric mechanism. Blotting the relative maximal stimulation of the ATPase activity by the simultaneous presence of the J-protein DnaJ and the protein substrate σ32 versus the basal ATPase rate relative to the ATPase rate of wild-type DnaK uncovers an inverse correlation between synergistic stimulation of the ATPase activity and basal ATPase rate (figure 3a,b). Notably, variants that are outside of this inverse correlation (blue triangles close to the y-axis in figure 3a,b) have replacements in or close to the catalytic centre (P143A, P143G, E171D and E171Q [51]), resulting in simultaneous reduction of basal and stimulated ATPase rates. Thus, this comparison suggests that the SBD represses the ATPase activity until substrate binding releases this repression. Such a hypothesis seems to be inconsistent with the fact that the isolated NBD (DnaK(2-385)) has an ATPase rate similar to the basal rate of full-length wild-type DnaK. However, the NBD including the interdomain linker (DnaK(2-393)) has a greatly elevated ATPase rate [31,52], suggesting that the linker is important for the stimulation of ATP hydrolysis and that the SBD prevents this. Of all DnaK variants in which residues that form direct contacts between NBD and SBDβ are replaced by non-interacting residues, two have a particular high basal ATPase rate, DnaK-D481A (increased 80-fold) and DnaK-K414I (increased 25-fold; figure 3c) [46]. Thus, the contacts between D481 and I168 and R167, and between K414 and D326 are particularly important for repressing the basal ATPase activity. The two residues at opposite ends of the NBD-interacting face of the SBDβ are ideally positioned to form a clamp contacting both NBD lobes after ATP-induced rotation and arresting of the NBD in a conformation unable to hydrolyse ATP [46]. Release of at least one of these contacts seems necessary to trigger ATP hydrolysis. N415 also forms contacts with D326, in addition to T221, but its contribution to repressing the ATPase activity seems to be limited as replacing it by glycine only increases the ATPase activity 1.6-fold [54].
Figure 3.
Allosteric network and consequences of amino acid replacements within this network. (a) Comparison of synergistic stimulation of the ATPase activity [kcat(mutant + DnaJ+σ32)/kcat(mutant)] with the basal ATPase [kcat(mutant)/kcat(wt)] of variants with single amino acid replacements coloured according to publication; DnaKwt, black. (b) Same as in (a) but on a linear scale. (c) Surface representations of the NBD of E. coli DnaK in the ATP-bound open conformation with lobe I coloured in dark grey, lobe II in light grey, and SBDβ interacting residues as indicated according to relative increase of basal ATPase activity [kcat(mutant)/kcat(wt)] (light cyan, residues not characterized). (d) Magnified view into the catalytic centre and the NBD-SBDβ interface of E. coli DnaK in the ATP-bound open conformation (PDB ID 4B9Q). Residues of the allosteric network are shown as sticks in atom colours with carbon in green cyan (NBD lobe I), deep teal (NBD lobe II), magenta (linker) and dark red (SBDβ). Black dashed lines indicate polar contacts (O-O or N-O-distance < 3.5 Å), brown dashed lines, hydrophobic contacts (C-C-distance < 4.2 Å). See also electronic supplementary material, table S1.
Locating all residues, the replacement of which cause defects in allosteric regulation, in the structure of the ATP-bound open conformation of DnaK reveals a network of interactions that seems to constitute the pathways of allostery (figure 3d). One pathway starting at the central hydrophobic residue of the substrate, which is inserted into the hydrophobic pocket between residues F426 and I438, transmits the signal of substrate binding to the catalytic centre through interacting residues V440, L484 and D148. V440 and L484 are displaced in an overlay between the SBDβ of the ATP-bound open conformation and the substrate-bound SBDβ (figure 2d). Replacing D148, V440 or L484 by alanine causes a complete loss of signal transmission from the substrate to the NBD as no stimulation of the ATPase activity by substrates and no synergistic stimulation by DnaJ and substrate are observed. By contrast, DnaK-D148A, V440A and L484A replacement variants have no defect in stimulation of the ATPase activity by DnaJ alone and no defects in substrate binding per se or ATP-induced substrate release. Based on these data and on the fact that SBDβ represses ATP hydrolysis through contacts between D481 and R167/I168 and between K414/N415-D326/T221, as mentioned above, it was proposed that substrate binding through V440, L484 and D148 triggers release of the SBDβ from the NBD, allowing the NBD lobes to rotate back into a position optimal for γ-phosphate cleavage. Such a hypothesis is consistent with a previous NMR study that found that SBDβ is released from the ATP-bound NBD in the presence of a peptide [55].
A second pathway transmits the signal from the ATP binding pocket to the SBD causing the substantial conformational rearrangements. Replacement of the γ-phosphate, Mg2+ and K+-coordinating residues T11, K70, E171, D194 and D201 cause defects in allostery, in addition to defects in catalysis (electronic supplementary material, table S1) [23,51,56,57]. By contrast, replacement of T199, which also greatly reduces ATP hydrolysis activity, at least in DnaK and BiP, does not compromise allostery [56]. Thus, the residues T11, K70, E171, D194 and D201 sense γ-phosphate of ATP and induce conformational changes. The residues T11, K70 and E171 belong to NBD subdomain IA and D194 and D201 to IIA. When ATP binds T11, K70 and E171 have to move by 2.5, 2.1 and 1.7 Å, respectively, when compared with the ADP·Pi structure to be able to form hydrogen bonds with the γ-phosphate (figure 2c). This movement is transmitted from K70 to P143, known to be a critical switch in the NBD that determines the high-energy barrier for hydrolysis [51]. Replacement of P143 by alanine or glycine reduces the basal ATPase activity and abrogates allostery. P143 forms a stacking interaction with F146, which also forms van der Waals contacts to K70 (figure 3d). The F146A replacement reduces the ATP-induced peptide dissociation rates to 6% of the rates of DnaKwt, indicating that this residue is important for coupling ATP-sensing to opening of the substrate-binding cavity. Interestingly, F146 is not essential for signal transduction in the reverse direction, because the ATPase activity of DnaK-F146A is stimulated by substrate and DnaJ in synergism [46]. Another residue essential for signal transmission to the SBD is R151, which forms hydrogen bonds to the backbone carbonyls of A144, F146 and N147, thereby stabilizing the P143-A144-Y145-F146-N147-D148-loop, and to the side chain carbonyl of N170, which together with E171, P172 and T173 forms another constrained loop. In addition, R151 forms a hydrogen bond across the NBD-SBDβ-interface to the backbone carbonyl of D481. Thus, R151 is a central hub to sense ATP-induced displacements of K70 (through P143 and F146) and of E171 (through N170) and to sense the incoming signal from the substrate through D148, and may be instrumental for severing the D481-R167/I168 contact. Replacement of R151 by alanine and even lysine completely abrogates allostery in both directions [32,51]. The N170-E171-P172-T173-loop is further stabilized by H-bonds between N170/ND1 and T173/OG1 to D393, a residue of the universally conserved linker connecting NBD with SBD. The D393A replacement also abrogates allostery in both directions, suggesting that insertion of the linker into the lower crevice of the NBD is necessary to stabilize the NBD in the lobe-rotated conformation that allows SBDβ docking. The contacts between D393 and N170 and T173 also seem to be important for stimulation of the ATPase activity as an NBD fragment including the linker until D393 (DnaK(2-393)) has a 40-fold increased ATPase rate when compared with full-length wild-type DnaK [52], whereas the variant excluding D393 (DnaK(2-392)) has a 13-fold increased basal ATPase rate [31]. In addition to D393, the hydrophobic residues V389 and L391 contribute to the ability of the linker to stimulate the ATPase activity in this construct as DnaK(2-393)-V389A,L390A,L391A,L392A has no increased ATPase activity when compared with the linker-less NBD [52]. Of the four hydrophobic residues of the linker, only V389 and L391 are important for allostery [53].
Taken together, ATP binding leads to a displacement of T11, K70 and E171, which is transmitted through P143, F146 and R151 to stabilize the NBD lobes in the rotated conformation, allowing linker insertion into the lower crevice and SBDβ docking onto the NBD. The docked SBDβ is stabilized mainly by D481-R167/I168 and by K414/N145-D326/T221 contacts and represses the ATPase activity. Release of the SBDβ from the NBD is triggered by substrate through I438, V440, L484, D148 and R151, allowing at least partial back-rotation of the NBD lobes into a position optimal for γ-phosphate cleavage.
5. Outlook
Although mutant analysis has provided many insights into the allosteric network of Hsp70s, a number of questions remain. The described pathways of allosteric control are essential for functional interdomain communication. But are they also sufficient or are there still more branches to be discovered? Many residues in the NBD-SBDβ interface have not been tested for a role in interdomain communication. It is still not clear how contacts between NBD and SBDβ or between SBDα and SBDβ are broken and new methods may have to be developed for answering these questions. Also the dynamics of the conformational changes need to be explored to get a full picture of the intricate Hsp70 machine. Hsp70s in higher eukaryotes are heavily modified by post-translational modifications. Some of these modifications might also have an impact on the allosteric control. Finally, how DnaJ gears into this allosteric network is also currently unknown.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
The work of the author is funded by the Deutsche Forschungsgemeinschaft (MA 1278/4-3, SFB1036 TP9).
References
- 1.Schlecht R, Erbse AH, Bukau B, Mayer MP. 2011. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351. ( 10.1038/nsmb.2006) [DOI] [PubMed] [Google Scholar]
- 2.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. 2009. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl Acad. Sci. USA. 106, 8471–8476. ( 10.1073/pnas.0903503106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kityk R, Kopp J, Sinning I, Mayer MP. 2012. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell. 48, 863–874. ( 10.1016/j.molcel.2012.09.023) [DOI] [PubMed] [Google Scholar]
- 4.Deuerling E, Bukau B. 2004. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit. Rev. Biochem. Mol. Biol. 39, 261–277. ( 10.1080/10409230490892496) [DOI] [PubMed] [Google Scholar]
- 5.Nillegoda NB, Bukau B. 2015. Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front. Mol. Biosci. 2, 57 ( 10.3389/fmolb.2015.00057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meimaridou E, Gooljar SB, Chapple JP. 2009. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J. Mol. Endocrinol. 42, 1–9. ( 10.1677/JME-08-0116) [DOI] [PubMed] [Google Scholar]
- 7.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507. ( 10.1093/emboj/16.7.1501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuiderweg ERP, Hightower LE, Gestwicki JE.. 2017. The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones 22, 173–189. ( 10.1007/s12192-017-0776-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. ( 10.1038/nrm2941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracher A, Bracher A, Verghese J, Verghese J. 2015. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2, 10 ( 10.3389/fmolb.2015.00010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Hartl FU, Bracher A. 2013. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 20, 929–935. ( 10.1038/nsmb.2608) [DOI] [PubMed] [Google Scholar]
- 12.Qian S-B, McDonough H, Boellmann F, Cyr DM, Patterson C. 2006. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440, 551–555. ( 10.1038/nature04600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stankiewicz M, Nikolay R, Rybin V, Mayer MP. 2010. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 277, 3353–3367. ( 10.1111/j.1742-4658.2010.07737.x) [DOI] [PubMed] [Google Scholar]
- 14.Wegele H, Müller L, Buchner J. 2004. Hsp70 and Hsp90--a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44. ( 10.1007/s10254-003-0021-1) [DOI] [PubMed] [Google Scholar]
- 15.Mogk A, Kummer E, Bukau B. 2015. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2, 22 ( 10.3389/fmolb.2015.00022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn GC, Chappell TG, Rothman JE. 1989. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390. ( 10.1126/science.2756425) [DOI] [PubMed] [Google Scholar]
- 17.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl Acad. Sci. USA 88, 2874–2878. ( 10.1073/pnas.88.7.2874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karzai AW, McMacken R. 1996. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11 236–11 246. ( 10.1074/jbc.271.19.11236) [DOI] [PubMed] [Google Scholar]
- 19.Barouch W, Prasad K, Greene L, Eisenberg E. 1997. Auxilin-induced interaction of the molecular chaperone Hsc70 with clathrin baskets. Biochemistry 36, 4303–4308. ( 10.1021/bi962727z) [DOI] [PubMed] [Google Scholar]
- 20.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl Acad. Sci. USA 96, 5452–5457. ( 10.1073/pnas.96.10.5452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid D, Baici A, Gehring H, Christen P. 1994. Kinetics of molecular chaperone action. Science 263, 971–973. ( 10.1126/science.8310296) [DOI] [PubMed] [Google Scholar]
- 22.Kassenbrock CK, Kelly RB. 1989. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. Eur. Mol. Biol. Organ. 8, 1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei J, Gaut JR, Hendershot LM. 1995. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270, 26 677–26 682. ( 10.1074/jbc.270.44.26677) [DOI] [PubMed] [Google Scholar]
- 24.Buchberger A, Theyssen H, Schröder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. 1995. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270, 16 903–16 910. ( 10.1074/jbc.270.28.16903) [DOI] [PubMed] [Google Scholar]
- 25.Palleros D, Reid K, McCarty J, Walker G. 1992. DnaK, hsp73, and their molten globules. Two different ways heat shock proteins respond to …. J. Biol. Chem. 267, 5279–5285. [PubMed] [Google Scholar]
- 26.Banecki B, Zylicz M, Bertoli E, Tanfani F.. 1992. Structural and functional relationships in DnaK and DnaK756 heat-shock proteins from …. J. Biol. Chem. 267, 25051–25058. [PubMed] [Google Scholar]
- 27.Ha JH, McKay DB. 1995. Kinetics of nucleotide-induced changes in the tryptophan fluorescence of the molecular chaperone Hsc70 and its subfragments suggest the ATP-induced conformational change follows initial ATP binding. Biochemistry 34, 11 635–11 644. ( 10.1021/bi00036a040) [DOI] [PubMed] [Google Scholar]
- 28.Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J. 1996. The second step of ATP binding to DnaK induces peptide release. J. Mol. Biol. 263, 657–670. ( 10.1006/jmbi.1996.0606) [DOI] [PubMed] [Google Scholar]
- 29.Moro F, Fernández V, Muga A.. 2003. Interdomain interaction through helices A and B of DnaK peptide binding domain. FEBS Lett. 533, 119–123. [DOI] [PubMed] [Google Scholar]
- 30.Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. 1995. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry 34, 12 095–12 106. ( 10.1021/bi00038a002) [DOI] [PubMed] [Google Scholar]
- 31.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. 2007. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 26, 27–39. ( 10.1016/j.molcel.2007.02.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rist W, Graf C, Bukau B, Mayer MP. 2006. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J. Biol. Chem. 281, 16 493–16 501. ( 10.1074/jbc.M600847200) [DOI] [PubMed] [Google Scholar]
- 33.Flaherty KM, DeLuca-Flaherty C, McKay DB. 1990. Three-dimensional structure of the ATPase fragment of a 70 K heat-shock cognate protein. Nature 346, 623–628. ( 10.1038/346623a0) [DOI] [PubMed] [Google Scholar]
- 34.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614. ( 10.1126/science.272.5268.1606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Los Rios P, Barducci A. 2014. Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. Elife 3, e02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Hendrickson WA. 2007. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120. ( 10.1016/j.cell.2007.08.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi R, et al. 2013. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907. ( 10.1038/nsmb.2583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Nune M, Zong Y, Zhou L, Liu Q. 2015. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23, 2191–2203. ( 10.1016/j.str.2015.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gumiero A, et al. 2016. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nat. Commun. 7, 13563 ( 10.1038/ncomms13563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Zong Y, Su J, Li H, Zhu H, Columbus L, Zhou L, Liu Q. 2017. Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat. Commun. 8, 1201 ( 10.1038/s41467-017-01310-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Kurochkin AV, Pang Y, Hu W, Flynn GC, Zuiderweg ER. 1998. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry 37, 7929–7940. ( 10.1021/bi9800855) [DOI] [PubMed] [Google Scholar]
- 42.Morshauser RC, Hu W, Wang H, Pang Y, Flynn GC, Zuiderweg ER. 1999. High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 289, 1387–1403. ( 10.1006/jmbi.1999.2776) [DOI] [PubMed] [Google Scholar]
- 43.Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, Gierasch LM. 2000. Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303. ( 10.1038/74062) [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zuiderweg ERP. 2004. The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc. Natl Acad. Sci. USA 101, 10 272–10 277. ( 10.1073/pnas.0401313101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilbanks SM, McKay DB. 1995. How potassium affects the activity of the molecular chaperone Hsc70. II. Potassium binds specifically in the ATPase active site. J. Biol. Chem. 270, 2251–2257. ( 10.1074/jbc.270.5.2251) [DOI] [PubMed] [Google Scholar]
- 46.Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. 2015. Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308 ( 10.1038/ncomms9308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burkholder WF, Panagiotidis CA, Silverstein SJ, Cegielska A, Gottesman ME, Gaitanaris GA. 1994. Isolation and characterization of an Escherichia coli DnaK mutant with impaired ATPase activity. J. Mol. Biol. 242, 364–377. ( 10.1006/jmbi.1994.1587) [DOI] [PubMed] [Google Scholar]
- 48.Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. 1998. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl Acad. Sci. USA 95, 15 223–15 228. ( 10.1073/pnas.95.26.15223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery DL, Morimoto RI, Gierasch LM. 1999. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286, 915–932. ( 10.1006/jmbi.1998.2514) [DOI] [PubMed] [Google Scholar]
- 50.Gässler CS, Buchberger A, Laufen T, Mayer MP, Schröder H, Valencia A, Bukau B. 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl Acad. Sci. USA 95, 15 229–15 234. ( 10.1073/pnas.95.26.15229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel M, Bukau B, Mayer MP. 2006. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21, 359–367. ( 10.1016/j.molcel.2005.12.017) [DOI] [PubMed] [Google Scholar]
- 52.Vogel M, Mayer MP, Bukau B. 2006. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 281, 38 705–38 711. ( 10.1074/jbc.M609020200) [DOI] [PubMed] [Google Scholar]
- 53.Kumar DP, Vorvis C, Sarbeng EB, Cabra Ledesma VC, Willis JE, Liu Q. 2011. The four hydrophobic residues on the Hsp70 inter-domain linker have two distinct roles. J. Mol. Biol. 411, 1099–1113. ( 10.1016/j.jmb.2011.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smock RG, Rivoire O, Russ WP, Swain JF, Leibler S, Ranganathan R, Gierasch LM. 2010. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol. Syst. Biol. 6, 414 ( 10.1038/msb.2010.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuravleva A, Clerico EM, Gierasch LM. 2012. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307. ( 10.1016/j.cell.2012.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barthel TK, Zhang J, Walker GC. 2001. ATPase-defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP-induced conformational change and peptide release. J. Bacteriol. 183, 5482–5490. ( 10.1128/JB.183.19.5482-5490.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson ER, McKay DB. 1999. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochemistry 38, 10 823–10 830. ( 10.1021/bi990816g) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.