Abstract
Introduction
HER2 overexpression/amplification is identified in up to 40% of uterine serous carcinomas (USC) and 10% of ovarian serous carcinomas (OSC). However, clinical trials using various HER2-targeted agents failed to show significant responses. FDA-approved HER2 assays target only the protein's intracellular domain (ICD) and not the extracellular domain (ECD). Previous quantitative studies in breast cancer by our group have shown that ICD of HER2 is expressed in some cases that do not express the HER2 ECD. We measured HER2 ICD and ECD in USC and OSC samples, and determined their relationship with clinico-pathologic characteristics and survival.
Methods
We measured HER2 ICD and ECD levels in 2 cohorts of USC and OSC comprising 102 and 175 patients, respectively. HER2 antibodies targeting ICD (CB11) and ECD (SP3) were validated and standardized using the AQUA® method of quantitative immunofluorescence (QIF) and a previously reported HER2 standardization tissue microarray (TMA). Objective, population-based cut-points were used to stratify patients according to HER2 ICD/ECD status.
Results
In USC, 8% of patients with high HER2 ICD had low ECD levels (6/75 patients). In OSC, 42% of patients with high HER2 ICD had low ECD levels (29/69 patients). HER2 ICD/ECD status in USC and OSC was not significantly associated with major clinico-pathological features or survival.
Conclusion
Using objective, domain-specific HER2 measurement, 8% of USC and 42% of OSC patients with high HER2 ICD levels do not show uniform overexpression of the ECD. This may be related to the presence of p95 HER2, an oncogenic fragment generated by full protein cleavage or alternative initiation of translation. These observations raise the possibility that USC/OSCs expressing low ECD despite being HER2-positive by ICD measurement, may benefit from therapies directed against the intracellular domain (e.g. lapatinib or afatinib) alone or in combination with extracellular domain-directed drugs (e.g. trastuzumab, pertuzumab, T-DM1).
Keywords: HER2, Intracellular domain, Extracellular domain, Serous carcinoma, Quantitative immunofluorescence, Targeted therapy
1. Introduction
HER2 overexpression/amplification is observed in approximately 40% of uterine serous (USC) [1–3] and 20% of ovarian serous carcinomas (OSC) [4]. Current College of American Pathologists (CAP) guidelines for HER2 testing in breast and gastric cancer include chromogenic immunohistochemistry (IHC) and in situ hybridization (ISH) methods [5,6]. However, evaluation of HER2 IHC is subject to diverse methodological and interpretative challenges [7].
Despite high levels of HER2 present in these malignancies, 2 early phase clinical trials in patients with HER2-positive ovarian and endometrial tumors failed to show positive results and found objective responses in 7.3% and 0%, respectively [8,9]. Although methodological concerns have been raised [10], decreased response rates as compared to breast cancer may also be explained by the expression of oncogenic C-terminal fragments of HER2 (p95-HER2) potentially arising from protein cleavage or alternative initiation of translation [11,12]. Recent studies [13,14] using proprietary, quantitative methodology, found no correlation between p95 HER2 expression and HER2/CEP17 ratio (R2 = 0.0029) and a weak correlation between p95HER2 levels and total HER2 protein (R2 = 0.15), suggesting that traditional HER2 IHC and ISH screening will not identify this cases. Notably, such alternative HER2 forms cannot be targeted by trastuzumab, which binds the extracellular domain (ECD). Current U. S. Food and Drugs Administration (FDA)-approved antibody assays for HER2 detection target only the intracellular domain (ICD) and not the ECD of the protein, and therefore, are unable to distinguish these fragments from the full length protein.
A recent study by our group showed that HER2 ICD and ECD are differentially expressed in HER2-positive breast malignancies [15]. Additionally, we found that benefit from trastuzumab treatment might be modulated by the ECD levels. Using a cohort of patients treated with chemotherapy and trastuzumab in the adjuvant setting, we showed that ECD, but not ICD measurement, was associated with increased 5-year disease-free survival (DFS). Moreover, while patients with low HER2 ICD levels did not have differences in survival with respect to cases with high ICD and low ECD expression, quantification of HER2 ECD in the ICD-high population did significantly stratify benefit from therapy. On the same line, a study including a metastatic cohort and quantitative p95 measurement [13] showed that estrogen receptor-positive patients with p95 levels above a pre-specified cut-point had worse progression-free and overall survival after trastuzumab treatment. Another study in the neoadjuvant setting using the same approach [14], p95HER2 levels were predictive of pathologic complete response in patients treated with trastuzumab or the combination of trastuzumab and lapatinib. In this study, we systematically examined HER2 ICD and ECD expression in human USC and OSC, using previously standardized assays, an established quantitative immunofluorescence method (QIF) and objective cut-points associated with response to trastuzumab in breast cancer. We assessed HER2 ICD/ECD levels and determined their associations with clinico-pathological characteristics and survival.
2. Methods
2.1. Patient cohort and tissue microarray construction
Two retrospective, stage I–IV uterine (USC) and ovarian high grade serous carcinoma (OSC) cohorts represented in tissue microarray (TMA) format, were used in this study (USC N = 102; OSC N = 175). Cases were collected between 1981 and 2014. Clinico-pathologic information from patients was obtained from clinical records and pathology reports, and it is summarized in Supplementary Table 1. Tissue specimens were included in a TMA as described [16]. Briefly, representative areas from primary tumors were selected in hematoxylin/eosin–stained preparations by a pathologist (D. C.) and 0.6 mm cores were obtained using a needle and arrayed in a recipient block. To increase representation and capture possible marker heterogeneity, 4 cores obtained from different areas of each tumor were included in the TMAs. Sections of the resultant TMA were cut and transferred to glass slides for histology processing and staining. Tissues were collected with specific consent or waived consent under the approved Yale Human Investigation committee protocol #9505008219.
2.2. Antibodies and immunofluorescent staining
Fresh TMA cuts were deparaffinized at 60 °C for 20 min, then incubated twice in xylene for 20 min. Antigen retrieval was performed with citrate buffer pH 6.0 at 97 °C for 20 min in a pressure-boiling container (PT Module, Lab Vision, Thermo Scientific, Waltham, MA, USA). Endogenous peroxidase activity was blocked with 2.5% hydroxyl peroxide in methanol for 30 min, followed by blocking with 0.3% bovine serum albumin in 0.1 mol/L of Tris-buffered saline for 30 min at room temperature. HER2 staining was carried out using U. S. Food and Drugs Administration (FDA)-approved companion diagnostic clone CB11 (mouse monoclonal antibody, Biocare Medical, Concord, CA, USA) against the intracellular domain (ICD) and clone SP3 (rabbit monoclonal antibody, Spring Biosciences, Pleasanton, CA, USA) against the extracellular domain (ECD) at an optimized titer (clone CB11: 10.4 µg/mL; clone SP3: 1:100), as previously reported by our group [15]. Slides were incubated overnight at 4 °C with primary antibodies and with cytokeratin at 1:100 dilution (polyclonal rabbit anticytokeratin, wide spectrum screening and monoclonal mouse antihuman cytokeratin clone AE1/AE3, Dako North America, Inc., Carpinteria, CA,USA). Sections were then incubated for 1 h at room temperature with Alexa 546-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Molecular Probes, Eugene, OR, USA) diluted 1:100 in mouse or rabbit EnVision amplification reagent (Dako). Cyanine 5 (Cy5) directly conjugated to tyramide (Perkin-Elmer, Waltham, MA, USA) at 1:50 dilution was used for target antibody detection. ProLong mounting medium (ProLong Gold; Molecular Probes) with 4,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei.
2.3. Fluorescent measurement and scoring
QIF was performed using the AQUA method [17–19]. Briefly, the QIF scores for HER2 CB11 and SP3 in the tumor compartment were calculated by dividing the target compartment pixel intensities by the area of cytokeratin positivity. QIF scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be comparable. All acquired TMA histospots were visually evaluated and cases with staining artifacts or <1% tumor (cytokeratin staining) were excluded from the analysis.
2.4. Cut-point selection and statistical analysis
Joinpoint software [20] (version 4.04, National Cancer Institute) was used to obtain distribution-based cut-points in a previously characterized HER2 standardization TMA [15]. These cut-points were used to stratify HER2 CB11 and SP3 protein scores in low and high statuses. Protein levels were compared using linear regressions coefficients (R2). Patient characteristics were compared using χ2 test. Survival functions were compared using Kaplan-Meier estimates, and statistical significance was determined using the log-rank test. Disease-specific survival (DSS) data was available for all patients. Statistical analysis was carried out using GraphPad Prism v6.0 software (GraphPad Software, Inc., La Jolla, CA, USA) and JMP 11 software (SAS Institute, Cary, NC, USA). All P values were based on two-sided tests, and all values under 0.05 were considered statistically significant.
3. Results
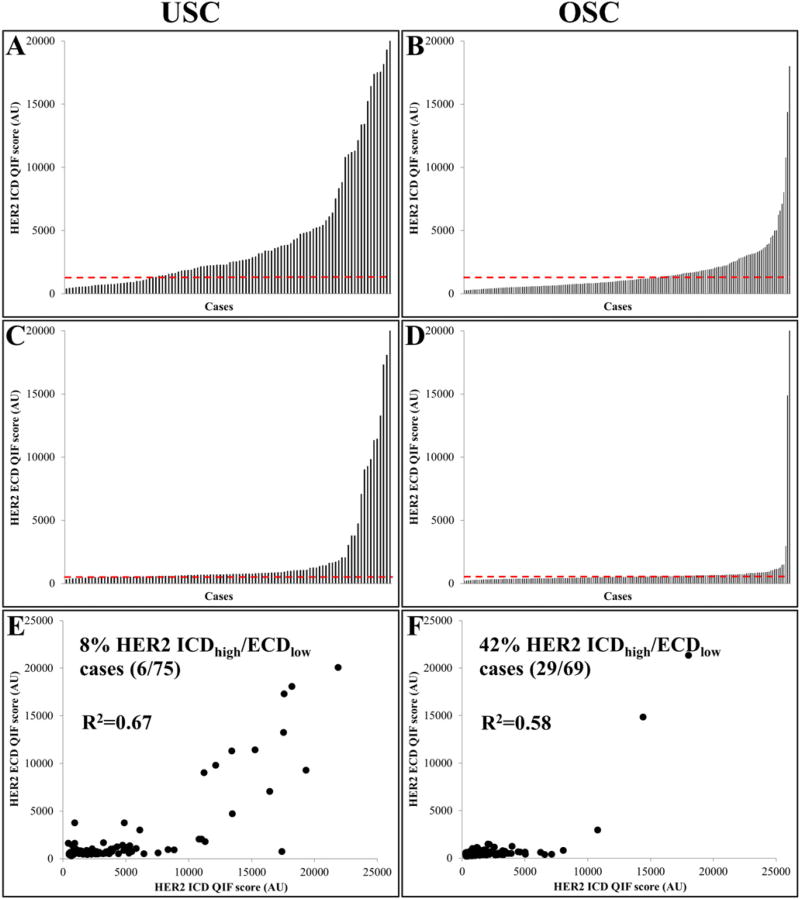
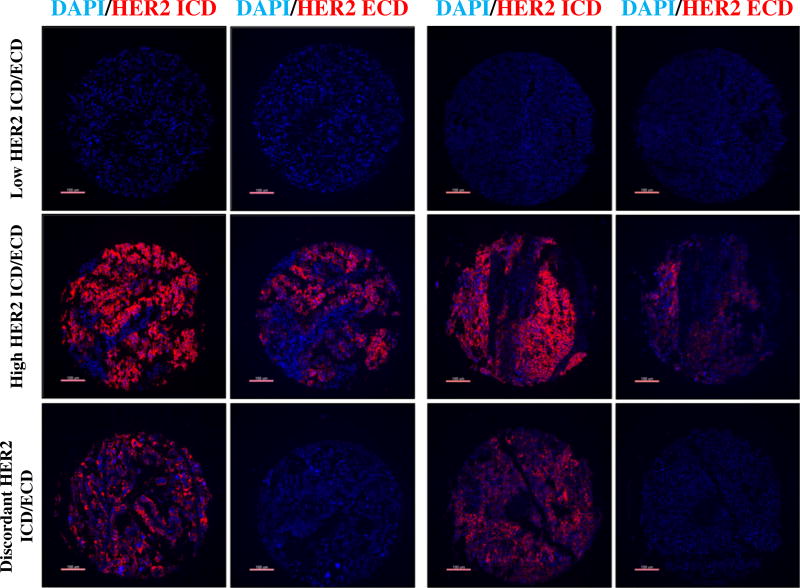
In uterine and ovarian serous carcinoma samples, HER2 showed a wide dynamic range of signal. In Fig. 1, bar charts depict the score distribution for HER2 ICD/ECD in USC and OSC (A, C and B, D, respectively). Overall, HER2 protein levels for both ICD and ECD were higher in USC than OSC. While the ICD showed a continuous range of scores in both USC and OSC, HER2 ECD presented a more bimodal distribution. Panels E and F show a positive relationship between scores from ECD and ICD scores in USC (R2 = 0.67) and OSC (R2 = 0.58), respectively. Using previously validated cut-points, 8% of USC (6/75 cases) and 42% of OSC (29/69 cases) showed high ICD levels while expressing low ECD. As shown in Supplementary Table 2, HER2 ICD were tightly correlated across 4 tumor cores in USC and OSC indicating limited intra-tumor heterogeneity of HER2 in these malignancies (average inter-core R2 = 0.82 and 0.81, respectively). The same was true for ECD scores (average inter-core R2 = 0.84 and 0.92, respectively [Supplementary Table 3]). Fig. 2 illustrates representative USC and OSC cases with different HER2 ICD and ECD protein levels. Staining was observed in the tumor compartment, with membranous/cytoplasmic pattern. HER2 ICD/ECD status was not associated with major clinico-pathological characteristics in USC and OSC (Table 1).
Fig. 1.
HER2 intracellular (ICD) and extracellular (ECD) domain measurement in uterine (USC) and ovarian serous carcinomas (OSC). Bar plots show the distribution of scores forHER2 ICD/ECD in USC (A, C) and in OSC (B, D). Panels E and F depict ECD vs ICD scatterplots in USC and OSC, respectively. Red dotted line: median. QIF: quantitative immunofluorescence. AU: arbitrary units of fluorescence.
Fig. 2.
Domain-specific detection of HER2 in USC and OSC. Representative fluorescent microphotographs showing different patterns of HER2 ICD/ECD expression in human USC (left panels) and OSC (right panels). Blue channel: 4,6-diamidino-2-phenylindole; red channel: HER2 ICD/ECD. Bar: 100 µm.
Table 1.
Clinico-pathological characteristics according to HER2 ICD/ECD status in Yale USC and OSC cohorts.
| Yale USC Cohort | Yale OSC Cohort | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| HER2 ICD status | HER2 ECD status | HER2 ICD status | HER2 ECD status | |||||||||
|
|
|
|
|
|||||||||
| ICD-low | ICD-high | P value | ECD-low | ECD-high | P value | ICD-low | ICD-high | P value | ECD-low | ECD-high | P value | |
| Age (years) | ||||||||||||
| <60 | 6 | 12 | 0.48 | 3 | 15 | 0.59 | 39 | 28 | 0.7 | 30 | 37 | 0.36 |
| ≥60 | 21 | 63 | 10 | 74 | 66 | 42 | 56 | 52 | ||||
| Stage | ||||||||||||
| Localized | 11 | 40 | 0.26 | 5 | 46 | 0.37 | 9 | 4 | 0.45 | 9 | 4 | 0.13 |
| Advanced | 16 | 35 | 8 | 43 | 94 | 66 | 76 | 84 | ||||
| Recurrence | ||||||||||||
| No | 22 | 60 | 0.87 | 10 | 72 | 0.74 | – | – | – | – | – | – |
| Yes | 5 | 15 | 3 | 17 | – | – | – | – | ||||
In univariate survival analysis, HER2 ICD and ECD levels did not stratify outcome in USC (log-rank P = 0.24 [Supplemental Fig. 1A] and P = 0.51 [Supplemental Fig. 1C]) or OSC (log-rank P = 0.07 [Supplemental Fig. 1B] and P = 0.65 [Supplemental Fig. 1D]. However, patients with high HER2 ICD levels exhibited a trend toward better disease-specific survival. The same pattern was observed when only advanced stage cases were tested for survival (Supplemental Fig. 2).
4. Discussion
Here, we report the measurement of HER2 ICD and ECD in uterine and ovarian serous carcinomas using QIF in formalin-fixed, paraffin-embedded tissues. Using objective methods and cut-points associated with response to adjuvant trastuzumab in breast cancer [15], we found that over 8% of USC and 42% of OSC express high levels of HER2 ICD, while having low levels of ECD. Additionally, we showed that USC have overall higher HER2 levels than OSC.
The loss of HER2 ECD has been previously studied in uterine serous malignancies. Todeschini et al. [21] found high levels of HER2 ECD in culture supernatants of HER2-positive USC cell lines. Also, they analyzed serum samples of patients harboring HER2-positive and negative USC, along with healthy donors. HER2 ECD levels did not differ between HER2-negative USC patients and healthy controls, but were significantly higher in HER2-positive USCs. A recent article by Growdon et al. [22] showed that 53% of high grade endometrial carcinomas with HER2 overexpression have high levels of p95-HER2. The authors used the proprietary, centralized VeraTag platform (Monogram Biosciences, San Francisco, CA. USA) and pre-specified cut-points. In accordance with our results, they did not find associations with stage or survival in these tumors.
Fewer data are available in ovarian cancer. A recent study by Montero et al. [23] showed that HER2 was the most frequently activated receptor tyrosine kinase in 16 ovarian cancer patient samples, 3 of which were serous carcinomas. In vitro, they found that lapatinib did not decrease cell proliferation in OVCAR8 (pure serous phenotype). Interestingly, this cell line was resistant to trastuzumab and pertuzumab, but not to T-DM1, while displaying high levels of pHER2 by immunoblot. Measurement of p95 was not available. Alternative mechanisms of primary resistance to trastuzumab, such as oncogenic PI3KCA mutations [24], alone or in conjunction with expression of C-terminus HER2 fragments might explain these results. T-DM1 might still be effective in this cell line, as the maytansinoid derivate can accumulate inside the cells after binding the full length HER2 protein.
Our study has a number of limitations. A major limitation is that it includes only retrospectively collected tissues with different treatments and variable follow up. A second issue is that the use of TMAs can underestimate or overestimate the levels of HER2 ICD/ECD due to tumor heterogeneity. We attempted to address this problem by testing 4 distant tumor cores and finding a high correlation between their scores. Ultimately, these observations will need to be validated on conventional whole tissue sections. Lastly, our study did not include a second set of USC/OSC cases for validation, patients treated with anti-HER2 therapies or response to treatment data. Presently, we wait for the results of ongoing trials NCT02491099 [A Phase II Evaluation of Afatinib in Patients With Persistent or Recurrent HER2-positive Uterine Serous Carcinoma (Afatinib)] and NCT01367002 (Evaluation of Carboplatin/Paclitaxel With and Without Trastuzumab (Herceptin) in Uterine Serous Cancer) and hope to test our hypothesis in these multicentric, prospective cohorts with HER2 therapies directed toward ICD and ECD, respectively.
In summary, objective, domain-specific measurement of HER2 ICD and ECD in USC and OSC identifies populations that express low ECD despite being HER2-positive by ICD-targeted antibodies. These patients are unlikely to benefit from trastuzumab and other inhibitors binding the extracellular domain (e.g. pertuzumab, T-DM1), but may benefit from therapies that target the intracellular domain (e.g. lapatinib or afatinib) alone or in combination with other agents. Future studies will have to test the association between HER2 ICD/ECD status and response to targeted treatment.
Supplementary Material
HIGHLIGHTS.
Uterine serous carcinomas have higher HER2 levels than their ovarian counterpart.
Eight percent of USC have discordant levels of HER2 ICD/ECD.
Forty-two percent of OSC have discordant levels of HER2 ICD/ECD.
Acknowledgments
Dr. David L. Rimm has served as a consultant to Amgen, Applied Cellular Diagnostics, Avida Labs, Biocept, BMS, Cernostics, FivePrime, Genoptix/Novartis, Metamark Genetics, MDAgree, OptraScan, and Perkin Elmer. Cepheid, Genoptix, Gilead Sciences, Kolltan and OncoplexDx fund research in Dr. Rimm's lab.
The authors acknowledge Lori Charette and the Yale Pathology Tissue Services for production of the high-quality TMAs used in this study.
Footnotes
Disclosures
No potential conflicts of interest were disclosed by the rest of the authors.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2017.02.002.
References
- 1.Santin AD, Bellone S, Van Stedum S, Bushen W, De Las Casas LE, Korourian S, et al. Determination of HER2/neu status in uterine serous papillary carcinoma: comparative analysis of immunohistochemistry and fluorescence in situ hybridization. Gynecol. Oncol. 2005;98(1):24–30. doi: 10.1016/j.ygyno.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 2.Mentrikoski MJ, Stoler MH. HER2 immunohistochemistry significantly overestimates HER2 amplification in uterine papillary serous carcinomas. Am. J. Surg. Pathol. 2014;38(6):844–851. doi: 10.1097/PAS.0000000000000182. [DOI] [PubMed] [Google Scholar]
- 3.Buza N, Roque DM, Santin AD. HER2/neu in endometrial cancer: a promising therapeutic target with diagnostic challenges. Arch. Pathol. Lab. Med. 2014;138(3):343–350. doi: 10.5858/arpa.2012-0416-RA. [DOI] [PubMed] [Google Scholar]
- 4.Lassus H, Leminen A, Vayrynen A, Cheng G, Gustafsson JA, Isola J, et al. ERBB2 amplification is superior to protein expression status in predicting patient outcome in serous ovarian carcinoma. Gynecol. Oncol. 2004;92(1):31–39. doi: 10.1016/j.ygyno.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 6.Bartley AN, Christ J, Fitzgibbons PL, Hamilton SR, Kakar S, Shah MA, et al. Template for reporting results of HER2 (ERBB2) biomarker testing of specimens from patients with adenocarcinoma of the stomach or esophagogastric junction. Arch. Pathol. Lab. Med. 2015;139(5):618–620. doi: 10.5858/arpa.2014-0395-CP. [DOI] [PubMed] [Google Scholar]
- 7.Rimm DL. What brown cannot do for you. Nat. Biotechnol. 2006;24(8):914–916. doi: 10.1038/nbt0806-914. [DOI] [PubMed] [Google Scholar]
- 8.Bookman MA, Darcy KM, Clarke-Pearson D, Boothby RA, Horowitz IR. Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: a phase II trial of the Gynecologic Oncology Group. J. Clin. Oncol. 2003;21(2):283–290. doi: 10.1200/JCO.2003.10.104. [DOI] [PubMed] [Google Scholar]
- 9.Fleming GF, Sill MW, Darcy KM, McMeekin DS, Thigpen JT, Adler LM, et al. Phase II trial of trastuzumab in women with advanced or recurrent, HER2-positive endometrial carcinoma: a Gynecologic Oncology Group study. Gynecol. Oncol. 2010;116(1):15–20. doi: 10.1016/j.ygyno.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santin AD. Letter to the editor referring to the manuscript entitled: “Phase II trial of trastuzumab in women with advanced or recurrent HER-positive endometrial carcinoma: a Gynecologic Oncology Group study” recently reported by Fleming et al., (Gynecol Oncol. 116;15–20;2010) Gynecol. Oncol. 2010;118(1):95–96. doi: 10.1016/j.ygyno.2010.01.043. (author reply 6–7) [DOI] [PubMed] [Google Scholar]
- 11.Anido J, Scaltriti M, Bech Serra JJ, Santiago Josefat B, Todo FR, Baselga J, et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. EMBO J. 2006;25(13):3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J. Natl. Cancer Inst. 2007;99(8):628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 13.Duchnowska R, Sperinde J, Chenna A, Haddad M, Paquet A, Lie Y, et al. Quantitative measurements of tumoral p95HER2 protein expression in metastatic breast cancer patients treated with trastuzumab: independent validation of the p95HER2 clinical cutoff. Clin. Cancer Res. 2014;20(10):2805–2813. doi: 10.1158/1078-0432.CCR-13-2782. [DOI] [PubMed] [Google Scholar]
- 14.Scaltriti M, Nuciforo P, Bradbury I, Sperinde J, Agbor-Tarh D, Campbell C, et al. High HER2 expression correlates with response to the combination of lapatinib and trastuzumab. Clin. Cancer Res. 2015;21(3):569–576. doi: 10.1158/1078-0432.CCR-14-1824. [DOI] [PubMed] [Google Scholar]
- 15.Carvajal-Hausdorf DE, Schalper KA, Pusztai L, Psyrri A, Kalogeras KT, Kotoula V, et al. Measurement of domain-specific HER2 (ERBB2) expression may classify benefit from trastuzumab in breast cancer. J. Natl. Cancer Inst. 2015;107(8) doi: 10.1093/jnci/djv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat. Clin. Pract. Oncol. 2004;1(2):104–111. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- 17.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 18.Neumeister VM, Anagnostou V, Siddiqui S, England AM, Zarrella ER, Vassilakopoulou M, et al. Quantitative assessment of effect of preanalytic cold ischemic time on protein expression in breast cancer tissues. J. Natl. Cancer Inst. 2012;104(23):1815–1824. doi: 10.1093/jnci/djs438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab. Investig. 2014;94(1):107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Fay MP, Feuer EJ, Midthune DN. Permutation tests for joinpoint regression with applications to cancer rates. Stat. Med. 2000;19(3):335–351. doi: 10.1002/(sici)1097-0258(20000215)19:3<335::aid-sim336>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Todeschini P, Cocco E, Bellone S, Varughese J, Lin K, Carrara L, et al. Her2/neu extracellular domain shedding in uterine serous carcinoma: implications for immunotherapy with trastuzumab. Br. J. Cancer. 2011;105(8):1176–1182. doi: 10.1038/bjc.2011.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Growdon WB, Groeneweg J, Byron V, DiGloria C, Borger DR, Tambouret R, et al. HER2 over-expressing high grade endometrial cancer expresses high levels of p95HER2 variant. Gynecol. Oncol. 2015;137(1):160–166. doi: 10.1016/j.ygyno.2015.01.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montero JC, Garcia-Alonso S, Ocana A, Pandiella A. Identification of therapeutic targets in ovarian cancer through active tyrosine kinase profiling. Oncotarget. 2015;6(30):30057–30071. doi: 10.18632/oncotarget.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black JD, Lopez S, Cocco E, Bellone S, Altwerger G, Schwab CL, et al. PIK3CA oncogenic mutations represent a major mechanism of resistance to trastuzumab in HER2/neu overexpressing uterine serous carcinomas. Br. J. Cancer. 2015;113(11):1641. doi: 10.1038/bjc.2015.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.