Summary
B-1a cells remain one of the most enigmatic lymphocyte subsets. In this review, we discuss recent advances in our understanding of the development of these cells and their regulation by the transcription factors Bhlhe41 and Arid3a as well as by the RNA-binding protein Lin28b. A large body of literature supports an instructive role of BCR signaling in B-1a cell development and linage commitment, which is initiated only after signaling from an autoreactive BCR. While both fetal and adult hematopoiesis can generate B-1a cells, the contribution of adult hematopoiesis to the B-1a cell compartment is low under physiological conditions. We discuss several models that can reconcile the instructive role of BCR signaling with this fetal bias in B-1a cell development.
Introduction
B-1 cells are an innate-like B lymphocyte subset that was discovered over 30 years ago [1]. These cells, which populate the peritoneal and pleural cavities, omentum and spleen, are believed to provide the first line of defense against pathogens and participate in the maintenance of tissue homeostasis [2]. While both fetal and adult precursors possess B-1 cell potential, B-1 cells are predominantly generated during fetal and neonatal life and can self-renew throughout the lifetime of the organism [3]. This is in stark contrast to conventional follicular (FO) and marginal zone (MZ) B cells (collectively called B-2 cells) that are constantly replenished from the adult bone marrow (BM). Unlike B-2 cells, B-1 cells can spontaneously differentiate into plasma cells that are believed to be the major source of secreted IgM (‘natural antibodies’) in unchallenged mice [2].
Most B-1 cells express the surface marker CD5 and are termed B-1a cells, while the minor subset of CD5− B-1 lymphocytes is referred to as B-1b cells. The developmental relationship and exact division of labor between these two cell types is not fully understood, but it was suggested that B-1a cells are the source of natural IgM in the steady state, while B-1b cells undergo T cell-independent responses to bacterial pathogens [4]. B-1a and B-1b cells share a common CD23− CD43+ B220lo cell surface phenotype [2], and CD5 remains the only marker that allows the distinction of the two cell types. Of note, CD5 expression follows a continuum from high to low rather than a discrete bimodal distribution. Comparison of gene expression profiles indicate that B-1a and B-1b cells are more similar to each other than to MZ or FO B cells (unpublished observations), suggesting a close relationship between the two subsets. The expression of CD5 is induced proportionally to the strength of T cell receptor (TCR) signaling during positive selection of T cells [5], where CD5 functions as a negative regulator of antigen receptor signaling [6]. Importantly, CD5 also negatively regulates B cell receptor (BCR) signaling in B-1a cells [7], suggesting that the gradient of CD5 expression reflects the different strengths of BCR signaling received by the cell. However, the CD5 expression level seems to be also developmentally regulated, as discussed below.
Instruction of B-1a cell lineage choice by BCR signaling
Many innate-like lymphoid lineages, such as NKT cells, some γδ T cell subsets and nonconventional αβ intraepithelial lymphocytes (IELs), are believed to be self-reactive [8]. Likewise, several lines of evidence suggest that B-1a cells are autoreactive [2], have a restricted BCR repertoire [9] and require a strong BCR signal for their differentiation [10]. Consequently, gene mutations that attenuate BCR signaling or co-stimulation pathways interfere with B-1a cell differentiation [11], whereas the loss of negative regulators of BCR signaling results in an expanded B-1a cell compartment [12–14]. A number of B-1a-specific BCRs, which recognize self carbohydrates and lipids either in their native or oxidized forms, have been identified to date [15]. It is believed that natural antibodies with these specificities can play a role in the maintenance of tissue homeostasis by clearing apoptotic cells and cellular debris, while simultaneously providing a first line of defense against pathogens that have similar epitopes in their membranes or cell walls. For example, a substantial fraction of the murine B•1a cells express the VH12/Vκ4 and VH11/Vκ14(Vκ9) BCRs [16, 17] that recognize phosphatidylcholine (PtC), which is present in the plasma membrane of host cells as well as in some pathogenic bacteria [18]. Transgenic expression of either of these BCR receptors promotes the development of B-1a cells at the expense of B-2 cells, revealing an important role of the BCR specificity in B-1a cell lineage choice [16, 17]. Direct evidence for the selection of B-1a cells on self-antigens is provided by a transgenic mouse model that expresses the heavy chain of a BCR recognizing the cell-surface protein Thy1 [19]. While this transgene efficiently induces the development of B-1a cells in Thy1-sufficient mice, B-1a cells are not generated on a Thy1-deficient background, clearly indicating that the positive selection of B-1a cells can be driven by self-antigen [19]. Importantly, a recently published ‘preview’ of data from Klaus Rajewsky’s lab indicates that mature FO B cells are still developmentally plastic and can be converted to the B-1 cell lineage by simply replacing a B-2 cell-specific BCR with a B-1a-typical BCR recognizing PtC [20]. Taken together, these results strongly suggest an instructive role of BCR signaling in B-1a cell lineage determination.
B-1a cell development
While the role of BCR signaling in B-1a cell development is clearly documented, the exact developmental path of these cells is still hotly debated. It has been suggested that precursors prior to the appearance of definitive hematopoietic stem cells (HSCs) may have B-1a cell potential [21, 22], but the contribution of these progenitors to the B-1a cell pool still remains to be shown. It has also been proposed that HSCs, including those in fetal liver (FL), are unable to generate B-1a cells [23]. However, an elegant study using cellular barcoding to trace the progeny of HSCs clearly demonstrated that FL HSCs can efficiently give rise to B-1a cells and that a single FL HSC has both B-1a and B-2 cell potential [24]. Several studies of BM chimeras also demonstrate that B-1a cells can be generated from adult BM hematopoiesis [25, 26], although this process may be less efficient than for FL HSCs. While BM-derived B-1a cells seem to express lower levels of CD5 than their FL-derived counterparts [25, 27], they are otherwise phenotypically normal [25, 26] and frequently express the B-1a-specific VH12 BCR [27]. Of note, the lower CD5 expression likely explains why some studies conclude that B-1a cells cannot be generated from adult BM HSCs [24, 28]. Intriguingly, the low CD5 expression does not merely reflect attenuated BCR signaling due to a possible BCR repertoire change, as VH12/Vκ4 transgenic B-1a cells are normally CD5hi, but express lower CD5 levels in BM chimeras (unpublished observations).
While B-1a cell potential is clearly present in adult BM, several lines of evidence indicate that this potential is quite limited under physiological conditions. Early experiments demonstrated that in neonatal mice, which have been transiently depleted of B cells prior to reconstitution with adult peritoneal cells, the B-1a cell compartment remains largely of donor origin for several months after transplantation [3], demonstrating that endogenous BM hematopoiesis minimally contributes to the B-1a cell pool under these conditions. Moreover, recent fate mapping experiments with the HSC-restricted Pdzk1ip1-CreER line in tamoxifen-treated adult mice indicated that the contribution of adult hematopoiesis to the B-1a cell compartment is below 10% during a 46-week observation period [29].
While FL HSCs efficiently generate B-1a cells, cells with a B-1a cell surface phenotype cannot be detected in FL [27] and first emerge in the postnatal spleen only several days after birth [30]. These transitional B-1a cells (TrB-1a) express the early B cell marker CD93 and have an otherwise mature IgM+ CD5+ B220lo B-1a cell phenotype [30]. In addition, B-1-specified progenitors (Lin− IgM− CD93+ CD19+ B220neg-lo) have been described [31, 32] and, although they seem to be committed to the B cell lineage as indicated by the expression of Pax5 and its target gene Cd19 [33], they are thought to be functionally equivalent to the uncommitted pre-pro-B cells (Lin− IgM− CD93+ CD19− B220+ CD43+; referred to as B-2 progenitors) that give rise to B-2 cell development [31, 32]. The B-1-specified progenitors are more frequent in fetal and neonatal mice and, when sorted from fetal and adult BM, predominantly generate B-1 cells in injected mice [31]. These results are difficult to reconcile with the instructive role of BCR signaling discussed above. Our bioinformatic analysis of recently published RNA-seq data obtained from these B-1 progenitor populations [33] suggests a possible explanation for the apparent conundrum. All RNA-seq samples from the adult B-1 cell progenitors contained reads corresponding to V gene segments of immunoglobulin heavy- and light-chain genes, and two of the four samples additionally exhibited a strong enrichment of Ighv11-2 and Igkv14-126 sequences (Supplemental Figure 1A,B) that constitute the PtC-specific VH11/Vκ14(Vκ9) BCR, which is found in a large fraction of mature B-1a cells [9, 27]. Analysis of the sequence reads spanning the V(D)J junctions revealed the presence of productive Igh and Igk rearrangements and identified in-frame VH11 and Vκ14 CDR3 sequences (Supplemental Figure 1C,D), which have previously been described in the case of VH11 [9]. These analyses suggest that the sorted B-1 cell progenitor populations [33] contained at least some cells that expressed a fully rearranged B-1a-specific BCR and could thus expand upon cell transfer in recipient mice. Hence, we conclude that most experimental evidence is consistent with a scenario, in which commitment to the B-1a cell lineage takes place upon BCR expression and is first manifested by the appearance of TrB-1a cells in the postnatal spleen.
The results described above indicate that any model of B-1a cell development should take into account the following facts. First, the BCR plays an instructive role in the choice of the B-1a cell lineage. Second, both fetal and adult hematopoiesis have the potential to generate B-1a cells. Third, the actual contribution of adult hematopoiesis to the B-1a cell compartment is low in wild-type mice. Below we discuss three non-mutually exclusive hypotheses, which can explain these facts, together with recent advances in our understanding of the transcriptional regulation of B-1a cell development.
Distinct effects of autoreactive antigen receptors in fetal and adult lymphopoiesis
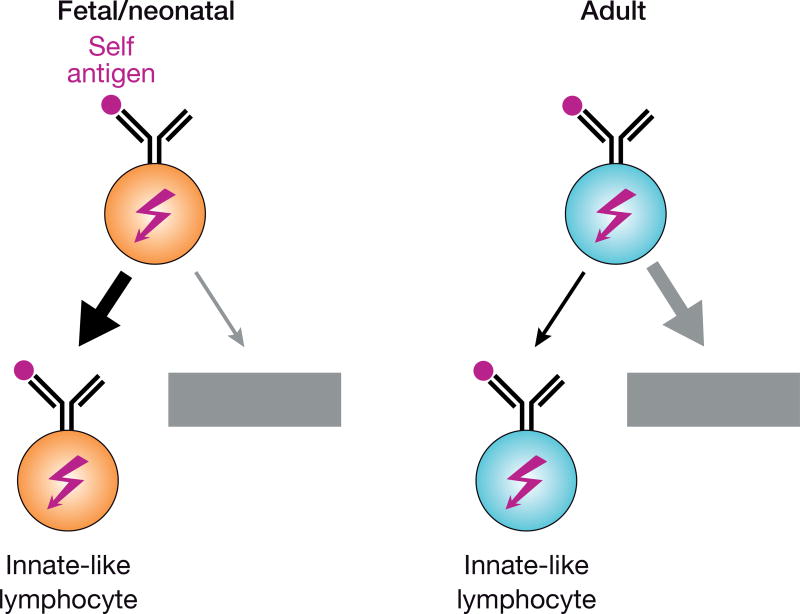
B-1a cells are not unique in their preferential fetal and neonatal origin as a number of innate-like T cell subsets are also generated early in ontogeny [34]. Such preferential production of innate-like lymphocytes early in life may be explained by a higher necessity for a first line of defense in an immunologically ‘naïve’ organism and may rely on a common mechanism. One shared feature of innate-like T cells and B-1a cells is their known or suspected self-reactivity. While autoreactive antigen receptors are thought to be largely eliminated from the repertoire, it is conceivable that T and B cells of fetal and neonatal origin may better tolerate strong antigen receptor signaling than their adult counterparts and may therefore be more efficiently diverted to innate-like lineages instead of being eliminated by tolerance mechanisms, such as negative selection (Figure 1; see also accompanying review by Joan Yuan). Indeed, this was directly demonstrated in an antigen-driven model of B-1 cell differentiation [35], where self-antigen diverts developing BCR-transgenic B cells to the B-1 lineage in chimeric mice generated by transplantation of FL cells, whereas the transgenic B cells are deleted in mice transplanted with BM progenitors [36].
Figure 1. Distinct effects of autoreactive antigen receptors in fetal and adult lymphopoiesis.
Fetal and neonatal lymphocytes expressing autoreactive antigen receptors seem to tolerate strong signaling, which may promote their efficient diversion to innate-like T or B cell lineages [36]. In contrast, adult autoreactive lymphocytes are efficiently eliminated by tolerance mechanisms such as negative selection, receptor editing or anergy induction.
Role of premature Igk gene rearrangements in B-1a cell development
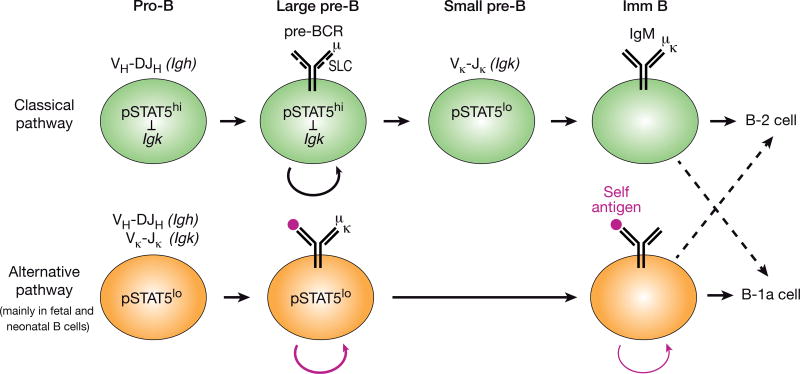
Another common theme in the differentiation of innate-like T and B-1a cells may be the timing of expression of a ‘complete’ antigen receptor. The majority of αβ T cells first rearrange the Tcrb locus in DN thymocytes followed by expression of the pre-TCR and subsequent differentiation to the DP thymocyte stage, where the Tcra locus is then recombined [37]. In contrast, γδ T lymphocytes, which are highly enriched for cells with innate-like properties, rearrange both the Trcg and Trcd loci in DN thymocytes, do not express the pre-TCR and are directly selected by signaling from the γδ TCR. Moreover, ‘premature’ expression of a pre-rearranged αβ TCR in transgenic mice leads to the development of cells with innate-like properties [38]. A small number of wild-type thymocytes is also thought to take this developmental route, and more cells do so in pre-TCR-deficient mice [39]. Akin to conventional αβ T cells, the majority of developing B cells first rearrange the immunoglobulin heavy-chain (Igh) locus in pro-B cells leading to expression of the pre-BCR and subsequent differentiation to small pre-B cells that rearrange the immunoglobulin light-chain (Igk or Igl) loci [37] (Figure 2). The sequential ordering of Igh and Igk recombination is enforced by IL-7 receptor (IL-7R) signaling through activation of the transcription factor STAT5. In pro-B cells, IL-7R signaling actively suppresses rearrangements at the Igk locus, as evidenced by a strong increase of Igk recombination in pro-B cells lacking IL-7Rα or STAT5 [40]. IL-7R signaling and its STAT5-mediated repression of the iEκ enhancer are lost during the subsequent pre-BCR transition, which activates Igk recombination in pre-B cells [41]. While B lymphopoiesis in the adult BM strictly depends on IL-7, it can take place in the FL in the absence of IL-7, consistent with the observation that B-1a cell differentiation is less dependent on this cytokine [42, 43]. Interestingly, a recent report revealed a strongly increased frequency of ‘premature’ Igk rearrangements in wild-type FL pro- B cells as a result of their reduced IL-7R/STAT5 activity [44] (Figure 2). Strikingly, the number of B-1a cells and their BCR repertoire are largely normal in pre-BCR-deficient mice, which are, however, severely deficient in B-1b and B-2 cells [44]. Hence, the simultaneous recombination of Igh and Igk loci in FL pro-B cells can directly generate a mature B cell receptor, thus bypassing the pre-BCR checkpoint [44] (Figure 2), which is consistent with a previous report demonstrating that a transgenic autoreactive BCR can efficiently replace the pre-BCR in early B lymphopoiesis [45]. In this context, it is important to note that pre-BCR signaling is known to counterselect against the generation of autoreactive BCRs [46], as potentially autoreactive heavy chains, such as VH11, may pair poorly with the surrogate light chains, thus preventing pre-BCR assembly [47]. Instead, the alternative pre-BCR-independent pathway of FL pro-B cells seems to promote the efficient generation of B-1a-specific autoreactive BCRs, which may explain the fetal bias of B-1a cell development in the context of instructive BCR signaling.
Figure 2. Generation of autoreactive BCRs by premature Igk rearrangements in fetal pro-B cells.
In the classical pathway of B cell development, pro-B cells experience strong IL-7R signaling, which activates the JAK/STAT pathway leading to active, phosphorylated (p) STAT5. Igk recombination is suppressed by pSTAT5 in IL-7R+ pro-B cells, which undergo VH-DJH recombination at the Igh locus, and is induced only after the pre-BCR transition in small IL-7R− pre-B cells containing low pSTAT5 levels. In contrast, IL-7R/pSTAT5 signaling is low already in fetal and neonatal pro-B cells, leading to efficient Igk rearrangements concomitant with Igh recombination, which directly generates mature BCRs. If an autoreactive BCR is generated, it is stimulated by self-antigen (red dots), which results in proliferative cell expansion and commitment to the B-1a cell lineage by bypassing the pre-BCR stage. The model of the alternative developmental pathway is based on recently published data [44].
Age-dependent BCR repertoire selection by clonal competition of B-1a cells
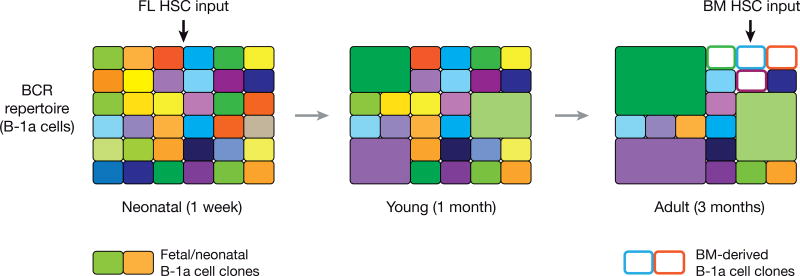
The clonal evolution of the B-1a BCR repertoire was recently analyzed by high-throughput sequencing of the complementarity-determining region 3 (CDR3) encoded by the V(D)J-rearranged Igh transcripts of splenic and peritoneal B-1a cells [9]. This elegant study revealed that the BCR repertoire of B-1a cells is continuously shaped by selection throughout the lifetime of a mouse. In neonatal life, the BCR repertoire is very diverse with even contribution of individual small B-1a cell clones to the overall diversity [9] (Figure 3). In contrast, the BCR repertoire of adult mice is dominated by a few large B-1a cell clones expressing certain BCRs, including those with PtC-binding specificity, which restricts the BCR repertoire of adult B-1a cells (Figure 3) [9]. Importantly, this oligoclonality is still observed under germ-free conditions and is thus not driven by microbiota-derived antigens, but likely results from competition of B-1a cell clones that have been selected on self-antigens [9].
Figure 3. Clonal evolution of the BCR repertoire of B-1a cells throughout adult life.
The schematic diagrams of the B-1a BCR repertoire in neonatal, young and adult mice are based on data obtained by high-throughput CDR3 sequencing of rearranged Igh transcripts [9]. Each rectangle represents a unique CDR3 sequence, and the size denotes the relative frequency (clone size) of an individual sequence. B-1a cell clones derived from FL HSCs are shown as filled colored rectangles, while B-1a cell clones originating from BM HSC are indicated by white rectangles.
These observations suggest a further explanation for the fetal bias of B-1a cell development in addition to the cell-intrinsic mechanisms described above. As the B-1a cell compartment of adult mice largely consists of cells of fetal and neonatal origin, which have undergone competition-based selection, it is conceivable that the low contribution of adult hematopoiesis to the B-1a cell pool can be explained in part by the failure of newly generated B-1a cells to efficiently compete with the already established B-1a cell clones (Figure 3). While this hypothesis requires experimental testing, it is consistent with published data. Neonatal mice, which are transiently depleted of their B cells, are known to readily restore their B-1a cell compartment, possibly by differentiation from endogenous HSCs [3]. However, endogenous hematopoiesis fails to contribute to the B-1a cell pool when the B cell depletion is accompanied by transfer of peritoneal B-1a cells from adult mice [3]. Hence, the presence of mature B-1a cells can interfere with the contribution of newly developing cells to the B-1a cell compartment, possibly through a competition-based mechanism. Interestingly, ~80% of the B-1a cell clones in the spleen of adult mice appear to be of postnatal origin, as they have N-nucleotide insertions in their CDR3 sequences [9] due to the postnatal expression of terminal deoxynucleotidyl transferase [48]. It is thus conceivable that the contribution of adult hematopoiesis to the B-1a cell pool is relatively high at the clonal level. However, the clone sizes of these B-1a cells may remain small due to inefficient competition with the established B-1a cell clones of fetal and neonatal origin, which is consistent with the observed low (< 10%) contribution of adult HSCs to the entire B-1a cell compartment [29].
The clonal outgrowth of B-1a cells is reminiscent of the behavior of transformed cells, which suggests that B-1a cells may be prone to malignant transformation. Indeed, while the origin of B cell chronic lymphocytic leukemia (CLL) in humans remains unclear [49, 50], mouse models that recapitulate genetic aberrations or dysregulated gene expression characteristic of human CLL have shown that leukemia can originate from B-1a cells as evidenced by the expression of B-1a cell-specific BCRs by leukemia cells and by cell transfer experiments [51–55]. Strikingly, experimental overexpression of oncogenes or deletion of tumor suppressor genes is not strictly required to induce CLL development, as two transgenic mouse lines, each expressing a distinct B-1a-specific BCR, spontaneously develop CLL [55, 56]. Hence, the transgenic expression of specific autoreactive BCRs, which leads to the excessive generation of B-1a cells, can predispose to CLL in aged mice. As CLL develops in these mouse models at a late age (> 15 months), which is close to the normal lifespan of M. musculus, it appears that the benefit of B-1a cells to provide a first-line defense against pathogens [2] may outweigh the small risk of CLL development. This may, however, be a more serious problem for mammals with a longer lifespan, such as humans.
Transcriptional control of B-1a cell development
Our understanding of the transcriptional network that controls the identity and function of B-1a cells is still rudimentary. While some transcription factors, such as Ebf1 [57], Oct2 [58] and IκBNS [30], are important for both B-1a and B-2 cells, their ectopic expression (Ebf1) or loss (Oct2, IκBNS) more severely affect the development of B-1a cells. Here we discuss two transcriptional regulators, Arid3a and Bhlhe41, whose function is more restricted to B-1a cell development.
The Lin28b – Let-7 – Arid3a signaling axis has recently emerged as a key pathway controlling B-1a cell development, as discussed in detail in an accompanying review of this issue (review by Joan Yuan). The RNA-binding protein Lin28b is expressed in fetal but not adult hematopoiesis, and its ectopic expression in adult BM HSCs strongly enhances B-1a cell development, which indicates an important role for Lin28b in the developmental switch from fetal to adult hematopoiesis [24, 59, 60]. While Lin28b can bind to thousands of mRNAs [61], it functions as a negative regulator of the Let-7 microRNA family by binding to their precursor RNAs, which prevents microRNA processing [62]. Ectopic expression of Let-7b in FL pro-B cells interferes with the development of B-1a cells, indicating that the inhibition of Let-7 expression may be required for efficient B-1a cell generation [59]. A predicted target of negative regulation by the Let-7 microRNA is the mRNA encoding the transcription factor Arid3a [59]. Indeed, ectopic expression of Arid3a in adult pro-B cells enhances B-1a cell generation, while RNAi-mediated knockdown of its expression in FL pro-B cells interferes with B-1a cell differentiation [59]. Consistent with these findings, conditional inactivation of Arid3a results in a severe decrease of peritoneal B-1a cells [63]. Functional characterization of Arid3a by identifying critical downstream target genes will ultimately be required to fully understand the function of this pathway in controlling B-1a cell development.
In a search for regulators that are specifically expressed in mature B-1a cells [57], we recently identified the transcription factor Bhlhe41 (also known as Dec2 or Sharp1), which is highly and specifically expressed in postnatal and adult B-1 cells [27]. Consistent with this selective expression pattern, we identified an essential role for Bhlhe41, with a lesser contribution of its homolog Bhlhe40, in the differentiation and homeostasis of B-1a cells by analyzing Bhlhe41−/− and Bhlhe40−/− Bhlhe41−/− mice [27]. B-1a cells in these mutant mice are strongly reduced, which is already evident when the first IgM-expressing TrB-1a cells emerge in the postnatal spleen [27]. Mutant B-1a cells exhibit an abnormal cell surface phenotype and a drastically altered BCR repertoire, as exemplified by loss of the PtC-specific VH12/Vκ4 BCR [27]. Transgenic expression of a pre-rearranged VH12/Vκ4 BCR failed to rescue the mutant phenotype and revealed defective BCR signaling, enhanced proliferation and increased cell death of the mutant B-1a cells [27]. At the molecular level, Bhlhe41 directly represses the expression of cell cycle regulators and inhibitors of BCR signaling while promoting pro-survival signaling by the IL-5 receptor [27]. Hence, Bhlhe41 controls multiple aspects of B-1a cell biology by regulating the development, BCR repertoire and self-renewal of B-1a cells.
Evolutionary conservation of the B-1a cell lineage
The existence of a human B cell population, that is homologous to the peritoneal B-1a cells of the mouse, remains an area of hot debate. To date, such a human B cell type has not been unequivocally identified [2, 64–68]. Although some candidate B cell populations have been suggested [2, 64], they have been identified on the basis of cell surface markers and functional properties (such as spontaneous antibody secretion) that are not only specific for B-1a cells, but are also shared with activated B-2 cells [66, 68]. To our knowledge, the mouse is the only mammalian species, in which B-1a cells have unequivocally been identified. Even within the genus Mus, CD5+ B cells in the peritoneum appear to be restricted to the subspecies M. musculus domesticus, as these cells could not be detected in many other M. musculus subspecies [69]. Future research on the evolutionary conservation of the B-1a cell type should ideally take advantage of the published gene expression signature of murine B-1a cells [57, 70] rather than solely relying on cell surface markers and few functional properties.
Conclusions
Since their discovery more than 30 years ago, the innate-like B-1a cells have remained an enigmatic lymphocyte subset. Recent publications have shed new light on the development of these cells and their transcriptional regulation. We believe that the bulk of the literature is consistent with an instructive role of BCR signaling in B-1a cell lineage commitment and development. This process is more efficient in fetal than in adult B lymphopoiesis, possibly because fetal B lymphocytes better tolerate strong autoreactive BCR signaling, which may divert them to the B-1a cell lineage. Premature Igk rearrangements in FL pro-B cells efficiently generate autoreactive BCRs by a pre-BCR independent pathway, which may further contribute to the fetal bias of B-1a cell development. At the molecular level, the RNA-binding protein Lin28b as well as the transcription factors Arid3a and Bhlhe41 have been identified as key regulators of B-1a cell development, although their downstream transcriptional programs remain to be fully explored.
Supplementary Material
Highlights.
The BCR specificity plays an instructive role in the B-1a cell lineage choice.
The clonal composition of B-1a cells is shaped throughout the lifetime of a mouse.
Premature Igk rearrangements in fetal pro-B cells can promote B-1a cell development.
Bhlhe41, Aridr3a and Lin28b emerged as key molecular regulators of B-1a cells.
Acknowledgments
We thank Lars Nitschke and Grace Liu for critical reading of the manuscript. This work was support by Boehringer Ingelheim, an ERC Advanced Grant (291740-LymphoControl; to M.B.) from the European Community’s Seventh Framework Program, the Austrian Science Fund (P28841; to T.K.) as well as the NIH grants R35GM122515 (to J.A.S.) and R21CA188968 (to J.A.S.). J.B.W. was previously supported by the T32 CA009161 training grant (Levy) and is currently supported by the 2T32 AI100853-06 (Reizis).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
* of special interest
** of outstanding interest
- 1.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The "Ly-1 B" cell subpopulation in normal immunodefective, and autoimmune mice. J. Exp. Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat. Rev. Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 3.Lalor PA, Herzenberg LA, Adams S, Stall AM. Feedback regulation of murine Ly-1 B cell development. Eur. J. Immunol. 1989;19:507–513. doi: 10.1002/eji.1830190315. [DOI] [PubMed] [Google Scholar]
- 4.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Müller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 7.Bikah G, Carey J, Ciallella JR, Tarakhovsky A, Bondada S. CD5-mediated negative regulation of antigen receptor-induced growth signals in B-1 B cells. Science. 1996;274:1906–1909. doi: 10.1126/science.274.5294.1906. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin TA, Hogquist KA, Jameson SC. The fourth way? Harnessing aggressive tendencies in the thymus. J. Immunol. 2004;173:6515–6520. doi: 10.4049/jimmunol.173.11.6515. [DOI] [PubMed] [Google Scholar]
- 9**.Yang Y, Wang C, Yang Q, Kantor AB, Chu H, Ghosn EE, Qin G, Mazmanian SK, Han J, Herzenberg LA. Distinct mechanisms define murine B cell lineage immunoglobulin heavy chain (IgH) repertoires. eLife. 2015;4:e09083. doi: 10.7554/eLife.09083. This high-throughput sequencing analysis demonstrates that the BCR repertoire of B-1a cells undergoes continuous evolution in adult mice, which result in a progressive restriction of the repertoire due to a dramatic expansion of individual B-1a cell clones with age. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K. B cell receptor signal strength determines B cell fate. Nat. Immunol. 2004;5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 11.Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu. Rev. Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- 12.Pan C, Baumgarth N, Parnes JR. CD72-deficient mice reveal nonredundant roles of CD72 in B cell development and activation. Immunity. 1999;11:495–506. doi: 10.1016/s1074-7613(00)80124-7. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, Winkler TH, Kneitz B, Crocker PR, Nitschke L. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat. Immunol. 2007;8:695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- 14.Pao LI, Lam KP, Henderson JM, Kutok JL, Alimzhanov M, Nitschke L, Thomas ML, Neel BG, Rajewsky K. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 2007;27:35–48. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Binder CJ. Natural IgM antibodies against oxidation-specific epitopes. J. Clin. Immunol. 2010;30:56–60. doi: 10.1007/s10875-010-9396-3. [DOI] [PubMed] [Google Scholar]
- 16.Arnold LW, Pennell CA, McCray SK, Clarke SH. Development of B-1 cells: segregation of phosphatidylcholine-specific B cells to the B-1 population occurs after immunoglobulin gene expression. J. Exp. Med. 1994;179:1585–1595. doi: 10.1084/jem.179.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chumley MJ, Dal Porto JM, Kawaguchi S, Cambier JC, Nemazee D, Hardy RR. A VH11Vκ9 B cell antigen receptor drives generation of CD5+ B cells both in vivo and in vitro. J. Immunol. 2000;164:4586–4593. doi: 10.4049/jimmunol.164.9.4586. [DOI] [PubMed] [Google Scholar]
- 18.Sohlenkamp C, López-Lara IM, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003;42:115–162. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- 19.Hayakawa K, Asano M, Shinton SA, Gui M, Allman D, Stewart CL, Silver J, Hardy RR. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 20.Rajewsky K. The Herzenberg lecture: how to make a B•1 cell? Ann. N.Y. Acad. Sci. 2015;1362:6–7. doi: 10.1111/nyas.12767. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Shelley WC, Seo W, Vemula S, Lin Y, Liu Y, Kapur R, Taniuchi I, Yoshimoto M. Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfβ for their development. Proc. Natl. Acad. Sci. USA. 2014;111:12151–12156. doi: 10.1073/pnas.1407370111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, Yoder MC. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. USA. 2011;108:1468–1473. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosn EEB, Waters J, Phillips M, Yamamoto R, Long BR, Yang Y, Gerstein R, Stoddart CA, Nakauchi H, Herzenberg LA. Fetal hematopoietic stem cell transplantation fails to fully regenerate the B-lymphocyte compartment. Stem Cell Reports. 2016;6:137–149. doi: 10.1016/j.stemcr.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Kristiansen TA, Jaensson Gyllenback E, Zriwil A, Bjorklund T, Daniel JA, Sitnicka E, Soneji S, Bryder D, Yuan J. Cellular barcoding links B-1a B cell potential to a fetal hematopoietic stem cell state at the single-cell level. Immunity. 2016;45:346–357. doi: 10.1016/j.immuni.2016.07.014. This elegant study using cellular barcoding to trace the progeny of HSCs demonstrates that a single fetal HSC can give rise to the development of B-1a and B-2 cells. [DOI] [PubMed] [Google Scholar]
- 25.Holodick NE, Repetny K, Zhong X, Rothstein TL. Adult BM generates CD5+ B1 cells containing abundant N•region additions. Eur. J. Immunol. 2009;39:2383–2394. doi: 10.1002/eji.200838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Düber S, Hafner M, Krey M, Lienenklaus S, Roy B, Hobeika E, Reth M, Buch T, Waisman A, Kretschmer K. Induction of B-cell development in adult mice reveals the ability of bone marrow to produce B-1a cells. Blood. 2009;114:4960–4967. doi: 10.1182/blood-2009-04-218156. [DOI] [PubMed] [Google Scholar]
- 27**.Kreslavsky T, Vilagos B, Tagoh H, Poliakova DK, Schwickert TA, Wöhner M, Jaritz M, Weiss S, Taneja R, Rossner MJ, et al. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat. Immunol. 2017;18:442–455. doi: 10.1038/ni.3694. This study reveals that the B-1 cell-specific transcription factor Bhlhe41 controls multiple aspects of B-1a cell biology by regulating the development, BCR repertoire and self-renewal of B-1a cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa K, Hardy RR, Herzenberg LA, Herzenberg LA. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J. Exp. Med. 1985;161:1554–1568. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Sawai CM, Babovic S, Upadhaya S, Knapp DJ, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity. 2016;45:597–609. doi: 10.1016/j.immuni.2016.08.007. This fate-mapping study shows that adult HSCs contribute to the B-1a cell compartment at a low (< 10%) frequency in mice under steady-state conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen GK, Àdori M, Khoenkhoen S, Dosenovic P, Beutler B, Karlsson Hedestam GB. B-1a transitional cells are phenotypically distinct and are lacking in mice deficient in IκBNS. Proc. Natl. Acad. Sci. USA. 2014;111:E4119–E4126. doi: 10.1073/pnas.1415866111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat. Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 32.Montecino-Rodriguez E, Dorshkind K. B-1 B cell development in the fetus and adult. Immunity. 2012;36:13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montecino-Rodriguez E, Fice M, Casero D, Berent-Maoz B, Barber CL, Dorshkind K. Distinct genetic networks orchestrate the emergence of specific waves of fetal and adult B-1 and B-2 development. Immunity. 2016;45:527–539. doi: 10.1016/j.immuni.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vermijlen D, Prinz I. Ontogeny of innate T Lymphocytes – some innate lymphocytes are more nnate than others. Front Immunol. 2014;5:486. doi: 10.3389/fimmu.2014.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferry H, Jones M, Vaux DJ, Roberts ISD, Cornall RJ. The cellular location of self-antigen determines the positive and negative delection of autoreactive B cells. J. Exp. Med. 2003;198:1415–1425. doi: 10.1084/jem.20030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferry H, Crockford TL, Leung JCH, Cornall RJ. Signals from a self-antigen induce positive selection in early B cell ontogeny but are tolerogenic in adults. J. Immunol. 2006;176:7402–7411. doi: 10.4049/jimmunol.176.12.7402. [DOI] [PubMed] [Google Scholar]
- 37.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 38.Kreslavsky T, Gleimer M, von Boehmer H. αβ versus γδ lineage choice at the first TCR-controlled checkpoint. Curr. Opin. Immunol. 2010;22:185–192. doi: 10.1016/j.coi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aifantis I, Bassing CH, Garbe AI, Sawai K, Alt FW, von Boehmer H. The Eδ delta enhancer controls the generation of CD4− CD8− αβTCR-expressing T cells that can give rise to different lineages of αβ T cells. J. Exp. Med. 2006;203:1543–1550. doi: 10.1084/jem.20051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hesslein DG, Yang SY, Schatz DG. Origins of peripheral B cells in IL-7 receptor-deficient mice. Mol. Immunol. 2006;43:326–334. doi: 10.1016/j.molimm.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Erlandsson L, Licence S, Gaspal F, Bell S, Lane P, Corcoran AE, Mårtensson IL. Impaired B•1 and B•2 B cell development and atypical splenic B cell structures in IL•7 receptor•deficient mice. Eur. J. Immunol. 2004;34:3595–3603. doi: 10.1002/eji.200425217. [DOI] [PubMed] [Google Scholar]
- 44**.Wong JB, Hewitt SL, Heltemes-Harris LM, Mandal M, Johnson K, Rajewsky K, Koralov SB, Clark MR, Farrar MA, Skok JA. B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage. bioRxiv, submitted online Nov. 13, 2017; doi: http://dx.doi.org/10.1101/214908. This study demonstrates that FL pro-B cells undergo premature Igk rearrangements at a high frequency concomitant with Igh rearrangements, which may directly generate autoreactive BCR that bypass the pre-BCR checkpoint. Notably, the number of B-1a cells and their BCR repertoire are largely normal in pre-BCR-deficient mice, suggesting that this alternative developmental pathway may contribute to the fetal bias of B-1a cell development.
- 45.Eschbach C, Bach MP, Fidler I, Pelanda R, Köhler F, Rajewsky K, Jumaa H. Efficient generation of B lymphocytes by recognition of self•antigens. Eur. J. Immunol. 2011;41:2397–2403. doi: 10.1002/eji.201041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keenan RA, De Riva A, Corleis B, Hepburn L, Licence S, Winkler TH, Martensson IL. Censoring of autoreactive B cell development by the pre-B cell receptor. Science. 2008;321:696–699. doi: 10.1126/science.1157533. [DOI] [PubMed] [Google Scholar]
- 47.Wasserman R, Li Y-S, Shinton SA, Carmack CE, Manser T, Wiest DL, Hayakawa K, Hardy RR. A novel mechanism for B cell repertoire maturation based on response by B cell precursors to pre-B receptor assembly. J. Exp. Med. 1998;187:259–264. doi: 10.1084/jem.187.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y-S, Hayakawa K, Hardy RR. The regulated expression of B lineage-associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2011;117:1781–1791. doi: 10.1182/blood-2010-07-155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Kipps TJ. The pathogenesis of chronic lymphocytic leukemia. Annu. Rev. Pathol. 2014;9:103–118. doi: 10.1146/annurev-pathol-020712-163955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan X-J, Albesiano E, Zanesi N, Yancopoulos S, Sawyer A, Romano E, Petlickovski A, Efremov DG, Croce CM, Chiorazzi N. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2006;103:11713–11718. doi: 10.1073/pnas.0604564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Lia M, Carette A, Tang H, Shen Q, Mo T, Bhagat G, Dalla-Favera R, Klein U. Functional dissection of the chromosome 13q14 tumor-suppressor locus using transgenic mouse lines. Blood. 2012;119:2981–2990. doi: 10.1182/blood-2011-09-381814. [DOI] [PubMed] [Google Scholar]
- 54.Simonetti G, Bertilaccio MTS, Ghia P, Klein U. Mouse models in the study of chronic lymphocytic leukemia pathogenesis and therapy. Blood. 2014;124:1010–1019. doi: 10.1182/blood-2014-05-577122. [DOI] [PubMed] [Google Scholar]
- 55*.Hayakawa K, Formica AM, Brill-Dashoff J, Shinton SA, Ichikawa D, Zhou Y, Morse HC, Hardy RR. Early generated B1 B cells with restricted BCRs become chronic lymphocytic leukemia with continued c-Myc and low Bmf expression. J. Exp. Med. 2016;213:3007–3024. doi: 10.1084/jem.20160712. This study demonstrates that B-1a cells give rise to CLL in the Eµ-TCL1 transgenic model and that transgenic mice expressing a B-1a-specific BCR can spontaneously develop CLL at an old age. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayakawa K, Formica AM, Colombo MJ, Shinton SA, Brill-Dashoff J, Morse HC, Iii, Li YS, Hardy RR. Loss of a chromosomal region with synteny to human 13q14 occurs in mouse chronic lymphocytic leukemia that originates from early-generated B-1 B cells. Leukemia. 2016;30:1510–1519. doi: 10.1038/leu.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J, Bilic I, Minnich M, Axelsson E, Jaritz M, et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 2012;209:775–792. doi: 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emslie D, D'Costa K, Hasbold J, Metcalf D, Takatsu K, Hodgkin PO, Corcoran LM. Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor alpha chain expression on activated B cells. J Exp Med. 2008;205:409–421. doi: 10.1084/jem.20072049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Zhou Y, Li Y-S, Bandi SR, Tang L, Shinton SA, Hayakawa K, Hardy RR. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J. Exp. Med. 2015;212:569–580. doi: 10.1084/jem.20141510. This study implicates the transcription factor Arid3a as a positive regulator of B-1a cell development as part of the Lin28b – Let-7 – Arid3a signaling axis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graf R, Munschauer M, Mastrobuoni G, Mayr F, Heinemann U, Kempa S, Rajewsky N, Landthaler M. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 2013;10:1146–1159. doi: 10.4161/rna.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Habir K, Aeinehband S, Wermeling F, Malin S. A role for the transcription factor Arid3a in mouse B2 lymphocyte expansion and peritoneal B1a generation. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70−. J. Exp. Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells are CD3−: A reply to “A human equivalent of mouse B-1 cells?” and “The nature of circulating CD27+CD43+ B cells”. J. Exp. Med. 2011;208:2566–2569. doi: 10.1084/jem.20111761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Descatoire M, Weill J-C, Reynaud C-A, Weller S. A human equivalent of mouse B-1 cells? J. Exp. Med. 2011;208:2563–2564. doi: 10.1084/jem.20112232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Andres M, Grosserichter-Wagener C, Teodosio C, van Dongen JJM, Orfao A, van Zelm MC. The nature of circulating CD27+ CD43+ B cells. J. Exp. Med. 2011;208:2565–2566. doi: 10.1084/jem.20112203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Covens K, Verbinnen B, Geukens N, Meyts I, Schuit F, Van Lommel L, Jacquemin M, Bossuyt X. Characterization of proposed human B-1 cells reveals pre-plasmablast phenotype. Blood. 2013;121:5176–5183. doi: 10.1182/blood-2012-12-471953. [DOI] [PubMed] [Google Scholar]
- 69.Thiriot A, Drapier A-M, Vieira P, Fitting C, Cavaillon J-M, Cazenave P-A, Rueff-Juy D. The Bw cells, a novel B cell population conserved in the whole genus Mus. J. Immunol. 2007;179:6568–6578. doi: 10.4049/jimmunol.179.10.6568. [DOI] [PubMed] [Google Scholar]
- 70.Mabbott NA, Gray D. Identification of co-expressed gene signatures in mouse B1, marginal zone and B2 B-cell populations. Immunology. 2014;141:79–95. doi: 10.1111/imm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.