SUMMARY
Helicobacter pylori is anomalous among non-nitrogen fixing bacteria in containing an incomplete NIF system for Fe-S cluster assembly comprising two essential proteins, NifS (cysteine desulfurase) and NifU (scaffold protein). Although nifU deletion strains cannot be obtained via the conventional gene replacement, a NifU-depleted strain was constructed and shown to be more sensitive to oxidative stress compared to wild-type (WT) strains. The hp1492 gene, encoding a putative Nfu-type Fe-S cluster carrier protein, was disrupted in three different H. pylori strains, indicating it is not essential. However, Δnfu strains have growth deficiency, are more sensitive to oxidative stress and are unable to colonize mouse stomachs. Moreover, Δnfu strains have lower aconitase activity, but higher hydrogenase activity than the WT. Recombinant Nfu was found to bind either one [2Fe-2S] or [4Fe-4S] cluster/dimer, based on analytical, UV-visible absorption/CD and resonance Raman studies. A bacterial two-hybrid system was used to ascertain interactions between Nfu, NifS, NifU, and each of 36 putative Fe-S-containing target proteins. Nfu, NifS, and NifU were found to interact with 15, 5 and 28 putative Fe-S proteins, respectively. The results indicate that Nfu, NifS and NifU play a major role in the biosynthesis and/or delivery of Fe-S clusters in H. pylori.
Graphical abstract
Helicobacter pylori is anomalous among non-nitrogen fixing bacteria in containing an incomplete NIF system for Fe-S cluster assembly, comprised of NifS, NifU and a Nfu-type protein. NifU-depleted strains and Δnfu mutants were constructed and characterized. Recombinant Nfu was found to bind either one [2Fe-2S] or [4Fe-4S] cluster/dimer, based on analytical, UV-visible absorption/CD and resonance Raman studies. A bacterial two-hybrid system was used to ascertain interactions between Nfu, NifS, NifU, and each of 36 putative Fe-S-containing target proteins.
INTRODUCTION
Iron-sulfur (Fe-S) clusters are ubiquitous in all domains of life and constitute essential protein cofactors with diverse functions ranging from electron transfer and catalysis to gene regulation and DNA repair. Maturation of Fe-S proteins is a multistep process involving assembly and transfer of intact Fe-S clusters. Three distinct bacterial systems for Fe-S cluster assembly have been described thus far: SUF (sulfur utilization factors), ISC (iron-sulfur cluster) and NIF (nitrogen fixation), the latter being used by nitrogen-fixing bacteria for the maturation of Fe-S proteins involved with nitrogenase biogenesis (reviewed in (Johnson et al., 2005)). Fe-S cluster assembly usually involves a cysteine desulfurase (SufS, IscS, and NifS) as the S donor and a source of Fe2+ ions (possibly from free Fe2+ pools in the cell) for assembly of Fe-S clusters on a scaffold protein (SufB, IscU, NifU) by an unknown mechanism. Intact Fe-S cluster transfer to recipient proteins occurs either directly or through the action of intermediate carrier or storage proteins, such as A-type carrier (ATC) proteins (IscA, SufA, NifIscA) (Vinella et al., 2009; Mapolelo et al., 2012), monothiol glutaredoxins (Bandyopadhyay et al., 2008a; Iwema et al., 2009; Shakamuri et al., 2012), and Nfu proteins (Nishio and Nakai, 2000; Bandyopadhyay et al., 2008b; Py et al., 2012; Gao et al., 2013). The latter proteins, also known as NifU-like proteins, all contain a domain with homology to the C-terminal domain of NifU (Jacobson et al., 1989), and bind [2Fe-2S] and/or [4Fe-4S] clusters at subunit interfaces via the conserved cysteines of the CXXC domain (Yabe et al., 2008; Gao et al., 2013).
Surprisingly, the gastric pathogen Helicobacter pylori relies exclusively on the NIF system, even though it does not fix nitrogen and does not have nitrogenase (Tomb et al., 1997; Olson et al., 2000). A previous study highlighted the versatility of the H. pylori NIF system, which can substitute for both SUF and ISC systems in E. coli (Tokumoto et al., 2004). Like other NIF-using organisms, the H. pylori NIF system is comprised of NifS and NifU, but does not contain the NifIscA ATC that is generally found in the nif operons of nitrogen fixing bacteria. Our lab previously showed that iron, oxidative stress and the Fur regulatory protein control nifS expression (Alamuri et al., 2006). Since nifS (hp0220 in strain 26695, (Tomb et al., 1997)) and nifU (hp0221) genes are part of the same operon (Sharma et al., 2010), it is reasonable to assume that nifU expression is also regulated by the same conditions and factors that govern nifS expression. However, this remains to be experimentally proven. In addition, previous results from our lab indicated that both nifU and nifS are essential genes in H. pylori, as we could not recover viable nifU or nifS mutants (Olson et al., 2000). In order to study the effect of a nifU deletion on the cell’s metabolism, we aimed at generating conditional nifU mutants. Their construction and characterization of are reported herein.
Besides NifU and NifS, H. pylori possess a NifS-like homolog (HP0405 in strain 26695). The hp0405 gene is not essential, since it has been successfully disrupted (for complementation purposes) in several studies done by our lab and by others (Benoit and Maier, 2003; Pereira and Hoover, 2005). Its role is yet unknown. In addition, H. pylori has a Nfu-type protein (HP1492 in strain 26695) that shares similarity with the C-terminal of NifU (Tokumoto et al., 2004). This is the only known type of Fe-S cluster carrier protein in H. pylori, which does not have any additional Nfu, ATC or monothiol glutaredoxin proteins. In the present study, recombinant H. pylori Nfu was expressed in E. coli, purified to homogeneity and shown to be capable of binding either [2Fe-2S] or [4Fe-4S] clusters using analytical and spectroscopic techniques. In addition, we successfully disrupted the hp1492/nfu gene in several wild-type strains, showing that it is not essential in H. pylori. Δnfu mutants were also characterized by studying their growth in presence of low or high O2 concentrations, assaying enzyme activities of select Fe-S-containing target proteins and assessing the capacity for Δnfu mutants to colonize mice. Another main goal of this study was to decipher the network of interactions between each of the three H. pylori NIF core components, i.e. NifS, NifU, and Nfu, and 36 other H. pylori proteins that are predicted to contain Fe-S clusters. The protein-protein interaction study was carried out in vivo, using a Bacterial Adenylate Cyclase Two-Hybrid (BACTH) system (Karimova et al., 1998). Results obtained with the BACTH system identified numerous interactions involving NifS, NifU or Nfu, ultimately leading to a better understanding of the H. pylori Nif-driven Fe-S maturation network.
RESULTS
Genome sequence analysis reveals 38 genes encoding putative Fe-S proteins
The list of putative Fe-S cluster-containing proteins was obtained by analyzing protein sequences (as inferred from DNA sequences) of 1,590 predicted open reading frames of H. pylori strain 26695 (Tomb et al., 1997). Various search engines and databases were used, including the Helicobacter pylori 26695 BioCyc database collection, GenBank, UniProt and STRING. H. pylori proteins with significant homology with previously characterized Fe-S proteins from other organisms were also included in the present study. We found 38 hypothetical Fe-S cluster-containing proteins, including NifU and Nfu (Table 1). Among them are 11 members of the radical S-adenosylmethionine (SAM) superfamily; proteins belonging to this group typically bind a [4Fe-4S] cluster using a conserved CX3CXφC motif, where φ is an aromatic residue (Grell et al., 2015). In addition, 9 proteins from this list are predicted to have [4Fe-4S]-binding ferredoxin-like domains, which harbor conserved CX2CX2CX3C(P) motifs. While the overwhelming majority of Fe-S target proteins are predicted to bind [4Fe-4S] clusters, a few are expected to coordinate [2Fe-2S] clusters, such as the fumarate reductase FrdB subunit (HP0191), the hypothetical protein HP0468, biotin synthase (HP1406) or components of the respiratory chain, like the ubiquinol cytochrome c oxidase (HP1540) or the NADH-Quinone oxidoreductase NQO3 subunit (HP1266).
Table 1.
H. pylori proteins predicted to contain Iron-Sulfur clusters.
| HP number* |
Protein ID |
Proposed function/Name | Putative [Fe-S] cluster # |
Putative ligands# |
|---|---|---|---|---|
| HP0117 | HP0117 | Hypothetical protein | [4Fe-4S] (SAM) | C33 C37 C40 |
| HP0132 | SdaA | L-serine dehydratase | [4Fe-4S] | C341 C383 C394 |
| HP0138 | HP0138 | Hypothetical, iron-sulfur cluster binding protein | 2[4Fe-4S] (ferredoxin) | C314 C318 C321 C325; C364 C368 C371 C375 |
| HP0142 | MutY | A/G-specific adenine glycosylase | [4Fe-4S] | C190 C196 C199 C205 |
| HP0191 | FrdB | Fumarate reductase, iron-sulfur subunit | [2Fe-2S], [3Fe-4S], [4Fe-4S] | C60 C65 C68 C80; C154 C157 C160 C221; C164 C211 C217 |
| HP0207 | ApbC, Mrp | Putative Fe-S carrier, ATPase | [4Fe-4S] | C274 C277 C366 |
| HP0221 | NifU | Scaffold protein NifU | [2Fe-2S], [4Fe-4S] | C161 C163 C196 C199; C54 C82 C131 |
| HP0269 | MiaB-like | tRNA-methylthiotransferase | 2[4Fe-4S] (SAM) | C10 C46 C78; C148 C152 C155 |
| HP0277 | HP0277 | Ferredoxin | 2[4Fe-4S] (ferredoxin) | C9 C13 C16 C20; C38 C41 C51 C55 |
| HP0285 | MiaB | tRNA-methylthiotransferase | 2[4Fe-4S] (SAM) | C11 C45 C74; C144 C148 C151 |
| HP0400 | IspH | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | [4Fe-4S] | C12 C92 C186 |
| HP0468 | HP0468 | Hypothetical protein | [2Fe-2S] | C235 C271 C307 C418 |
| HP0568 | HP0568 | Hypothetical, iron-sulfur cluster binding protein | [4Fe-4S] SPASM | Unknown |
| HP0585 | Nth | Endonuclease III | [4Fe-4S] | C194 C201 C205 C210 |
| HP0588 | OorD | 2-oxoglutarate oxidoreductase, δ subunit | 2[4Fe-4S] (ferredoxin) | C19 C22 C25 C29; C56 C59 C62 C66 |
| HP0625 | IspG | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | [4Fe-4S] | C264 C267 C299 E306 |
| HP0631 | HydA | Hydrogenase small subunit | 2[4Fe-4S], [3Fe-4S] | C86 C89 C185 C218; H256 C259 C284 C290; C299 C318 C321 |
| HP0654 | MqnE | Aminodeoxyfutalosine synthase | [4Fe-4S] (SAM) | C59 C63 C66 |
| HP0656 | MqnC | Dehypoxanthinylfutalosine cyclase | [4Fe-4S] (SAM) | C81 C85 C88 |
| HP0665 | HemN | O2-independent coproporphyrinogen-III oxidase | [4Fe-4S] (SAM) | C62 C66 C69 |
| HP0666 | GlpC | anaerobic Glycerol-3-phosphate dehydrogenase | 2[4Fe-4S] (ferredoxin) | C17 C20 C23 C27; C68 C71 C74 C78 |
| HP0734 | RimO | Ribosomal protein S12 methylthiotransferase | 2 [4Fe-4S] (SAM) | C16 C50 C82; C151 C155 C158 |
| HP0768 | MoaA | Molybdenum cofactor biosynthesis protein A | 2[4Fe-4S] (SAM) | C21 C25 C28; C249 C252 C266 |
| HP0779 | AcnB | Aconitase B subunit | [4Fe-4S] | C709 C767 C770 |
| HP0934 | QueE | 7-carboxy-7deazaguanine synthase | [4Fe-4S] (SAM) | C31 C35 C40 |
| HP1089 | AddB | ATP-dependent helicase-nuclease | [4Fe-4S] | C778 C781 C787 |
| HP1109 | PorD | Pyruvate-ferredoxin oxidoreductase, d subunit | 2[4Fe-4S] (ferredoxin) | C58 C61 C64 C68; C88 C91 C94 C98 |
| HP1222 | Dld | D-lactate dehydrogenase | 2[4Fe-4S] (ferredoxin) | C548 C551 C554 C558; C610 C613 C616 C620 |
| HP1226 | HemN | O2-independent coproporphyrinogen-III oxidase | [4Fe-4S] (SAM) | C17 C21 C24 |
| HP1261 | NuoB | NADH-Quinone oxidoreductase, subunit B | [4Fe-4S] | C32 C33 C97 C126 |
| HP1266 | Nqo3 | NADH-Quinone oxidoreductase, NQO3 subunit | 2[4Fe-4S], [2Fe-2S] | C34 C39 C45 C48; C143 C146 C149 C153; C203 C206 C209 C213 |
| HP1268 | Nqo9 | NADH-Quinone oxidoreductase, subunit I | 2 [4Fe-4S] (ferredoxin) | C82 C85 C88 C92; C121 C124 C127 C131 |
| HP1356 | NadA | Quinolinate synthase | [4Fe-4S] | C107 C193 C198 |
| HP1406 | BioB | Biotin synthase | [4Fe-4S](SAM), [2Fe-2S] | C17 C21 C24; C61 C96 C154 R221 |
| HP1428 | RlmN | Dual-specificity RNA methyltransferase | [4Fe-4S] (SAM) | C123 C127 C130 |
| HP1492 | Nfu | Nfu-like protein | [2Fe-2S], [4Fe-4S] | C49 C52 C79 |
| HP1508 | HP1508 | Ferredoxin | 2[4Fe-4S] (ferredoxin) | C249 C252 C255 C259; C273 C276 C279 C283 |
| HP1540 | FbcF | Ubiquinol cytochrome c oxidoreductase | [2Fe-2S] | C98 C104 C116 C118 |
HP number refers to strain 26695 (Tomb et al., 1997)
based on previously published studies on H. pylori proteins or sequence alignments with homologous proteins.
Abreviations: SAM, S-Adenosyl Methionine ligand; SPASM domain: a domain found in [4Fe-4S] binding maturases of Subtilosin, PQQ, Anaerobic Sulfatases, and Mycofactocin (Haft and Basu, 2011).
Construction and characterization of conditional nifU mutants
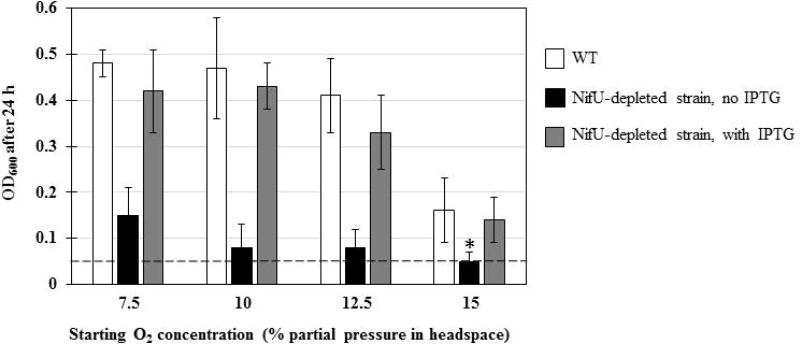
Previous attempts to disrupt the nifU locus in H. pylori were unsuccessful, suggesting the nifU gene is essential (Olson et al., 2000). In the current study, we generated conditional nifU mutants, first to confirm the essentiality of nifU and then to study the effects of its depletion on H. pylori’s growth and metabolism. Following a strategy previously described for other essential H. pylori genes (Boneca et al., 2008; Benoit and Maier, 2014), nifU (hp0221 in strain 26695) was first cloned under the control of an IPTG-inducible tac promoter in a E. coli-H. pylori shuttle plasmid conferring chloramphenicol (Cm) resistance (Table S1). After strain-specific DNA methylation, we attempted to introduce this plasmid in various H. pylori WT strains (26695, 43504, X47), however most strains were found to be refractory to transformation and we could only obtain Cmr mutants in strain 43504. This plasmid-harboring strain was used as parental strain to successfully disrupt the chromosomal nifU locus using an aphA3 cassette (kanamycin resistance), as confirmed by PCR (data not shown). These results confirm the essentiality of nifU, since the gene can only be disrupted when the plasmid-borne backup version is present. The conditional nifU mutant (NifU-depleted strain) was compared to the WT strain (43504) for its sensitivity to oxidative stress. NifU-depleted and WT strains were grown in BHI-βc liquid medium, with or without IPTG, under increasing (7.5 to 15%) O2 partial pressure and the growth yield (OD600) was scored after 24 h (Fig. 1). In the absence of IPTG, the NifU-depleted strain showed weak growth overall and was more sensitive to oxidative stress (no growth at 15% O2) than the WT, indicating that the nifU gene is needed for both growth and oxidative stress resistance in H. pylori. Interestingly, upon addition of 0.5 mM IPTG (to induce nifU expression from the plasmid), the NifU-depleted strain growth yield was similar to that of WT, under all starting O2 conditions tested (Fig. 1). Although we did not quantitate the amount of NifU synthesized in absence or presence of IPTG, our results suggest there is a direct correlation between IPTG-induced NifU levels, cell growth and resistance to oxygen stress.
Fig. 1. NifU-depleted strains are more sensitive to O2 stress.
WT strain 43504 and isogenic NifU-depleted mutant strains were grown on BA plates under microaerophilic conditions, resuspended in BHI-βc and used to inoculated 10 mL of BHI-βc, with or without supplemented 0.5 mM IPTG. IPTG was also added in WT cultures, as control. Cells were grown for 24 h at 37°C, 200 rpm shaking, in bottles with 150 mL head space filled with 10% H2, 5% CO2 and O2 concentrations as indicated (N2 as balance). Results represent 2 to 4 independent experiments (with bottles in duplicate) and are expressed as mean and standard deviation of OD600 after 24h. A dotted line indicates the initial OD600 at T0 (approximately 0.05). * indicates no growth (OD600 ≤ 0.05).
The hp1492 gene, encoding a Nfu-type protein, is not essential but needed for optimal growth
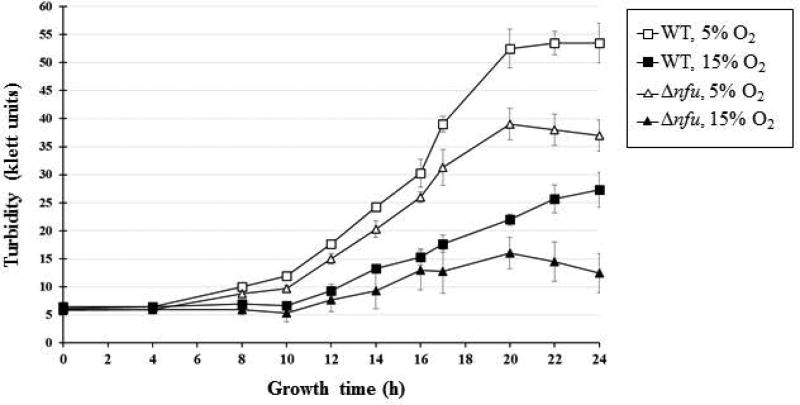
Sequence analysis of several available genomes indicates the presence of a gene (hp1492 in strain 2695 (Tomb et al., 1997)), encoding for an 89 amino-acid long peptide, that shares similarity with the C-terminus of the Fe-S scaffold protein NifU (Fig. 2). The presence of the gene in the H. pylori genome and the similarity of the protein to the C-terminus of NifU (referred to as a “Nfu-type” protein) were previously reported (Tokumoto et al., 2004); we refer to the H. pylori Nfu-type as “Nfu” in the present study. The nfu gene was successfully disrupted and replaced by a chloramphenicol resistance cassette in three different H. pylori parental strains, i.e. strains 26695, 43504 and X47 (data not shown), indicating that this gene is not essential in H. pylori, unlike nifU. This result is in agreement with a previous whole genome mutagenesis study from Salama et al., who were able to recover viable nfu::Tn7 transposon mutants in strain G27 (Salama et al., 2004). There were no apparent phenotypic differences (growth rate, colony size) between Δnfu and WT strains when cells were grown on plates under standard microaerophilic conditions (4–5 % O2). However, Δnfu mutant cells grown in liquid cultures under low (5 %) or high (15%) O2 partial pressure showed enfeebled growth compared to WT grown under the same conditions (Fig. 3). After 24h, Δnfu mutant cells achieved 70% and 45% of the WT growth yield under low and high O2 conditions, respectively, indicating the absence of Nfu had a significant impact on the cell’s metabolism and growth, especially under aerobic stress.
Fig. 2. The HP1492 (Nfu) protein sequence shares homology with the C-terminus domain of NifU.
The central domain of Nfu (HP1492, 89 amino acids) shares 34% identity and 55% similarity with the C-terminus part of NifU (HP0221, 321 amino acids), including a conserved CXXC motif (grey box) shown to be important for [2Fe-2S] as well as [4Fe-4S] coordination.
Fig. 3. X47 Δnfu mutants show growth deficiency.
H. pylori X47 (WT) cells and isogenic X47 Δnfu mutant cells were grown on BA agar under microaerophilic conditions, resuspended in BHI-βc and used to inoculate 10 mL of BHI-βc (starting OD600=0.05, which corresponds to approximately 5 Klett units). Cells were grown for 24 h at 37°C, 200 rpm shaking, in sealed bottles (with side arms) with 170 mL head space filled with 10% H2, 5% CO2 and either 5% or 15% O2 concentration, as indicated (N2 as balance). A Klett spectrophotometer was used to measure OD at various time points. Results are expressed as (mean and standard deviation) Klett units and represent data obtained from three bottles for each condition.
H. pylori Δnfu mutants have higher hydrogenase activity, but lower aconitase activity than WT
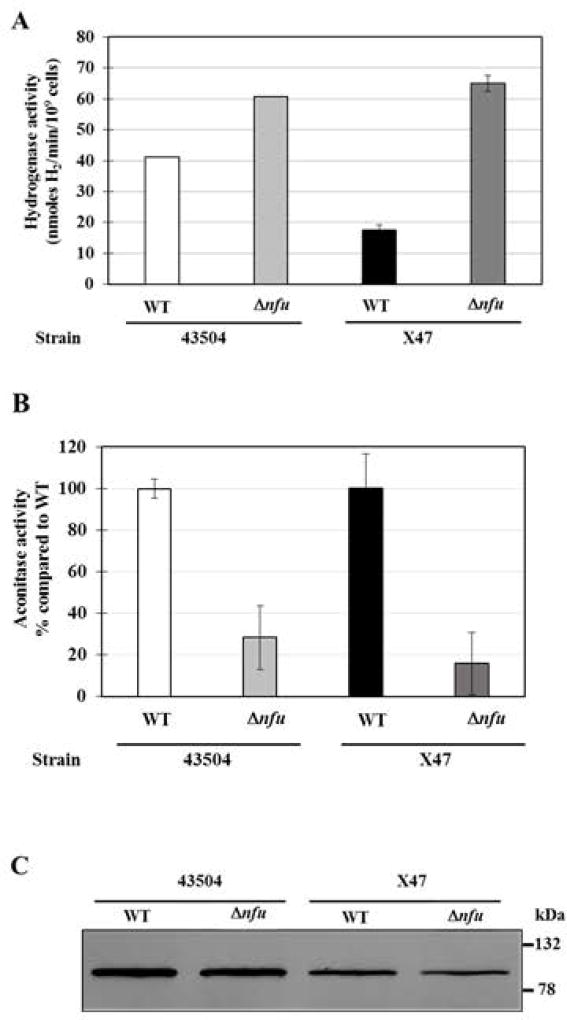
In order to determine whether Nfu is involved in the maturation of select Fe-S-containing proteins, we assayed hydrogenase and aconitase activities in Δnfu background strains. H. pylori possess only one hydrogenase, an (H2)-uptake type, heterotrimeric hydrogenase (HydABC, HP0631-HP0633 in strain 26695) and one aconitase (AcnB, HP0779 in strain 26695), and both enzymes are predicted to possess Fe-S clusters (Table 1) (Maier et al., 1996; Tomb et al., 1997). Whole cell hydrogenase assays were conducted to measure the H2-uptake activity of Δnfu mutants constructed in two independent background strains (Fig. 4A). While we anticipated a decrease or no change in hydrogenase activity in those mutants, surprisingly we found (1.5 to 4-fold) higher hydrogenase activity in both Δnfu mutants compared to their respective parental strains (Fig. 4A). While these results suggest that Nfu is not directly involved in hydrogenase Fe-S maturation in H. pylori, they are intriguing. These increased levels could reflect higher hydrogenase transcript levels due to higher Fe cellular content, since the hyd operon has been shown to be repressed by the Fur regulator under limiting iron concentrations (Ernst et al., 2005); however this hypothesis was not tested in the present study. In contrast to hydrogenase, aconitase activities in the two Δnfu mutants were significantly lower than WT strains (Fig.4B). The decrease in activity was not due to a decrease in aconitase enzyme, since immunoblots (using antiserum raised against E. coli AcnB) revealed comparable aconitase levels in H. pylori WT and Δnfu mutants (Fig. 4C). Therefore, the observed decreased aconitase activity in Δnfu mutants is probably due to a deficiency in Fe-S maturation and suggests a direct role for Nfu in delivering Fe-S clusters to the aconitase apo-protein.
Fig. 4. Hydrogenase activity, aconitase activity and aconitase levels in WT and Δnfu mutant strains.
(A) Hydrogenase activities. Whole cell hydrogenase assays were used to measure the H2-uptake activity of two independent Δnfu mutants and their parental strains. Results represent mean and standard deviation from 3 to 4 independent measurements and are expressed as nmoles of H2 used per min per 109 H. pylori cells. (B) Aconitase activities. Assays were carried out using cell-free extracts (CFE) from Δnfu and WT cells. Results shown represent means and standard deviations from three independent growth experiments, with assays done in triplicate. Activities are expressed as percentages compared to the parental strain.
(C) Aconitase immunoblot. Identical amounts of cell-free extracts (5 µg total protein) were loaded in each lane. Proteins were separated on a SDS-10% polyacrylamide gel, along with prestained mass standards (size are indicated on the right) and the proteins were then transferred onto a nitrocellulose membrane and subjected to immunoblotting using anti-E. coli AcnB antiserum. Densitometry analysis based on three independent immunoblots (including this one) revealed the following integrated density (pixels): 228,000 ± 17,000 for strain 43504, 186,000 ± 22,000 for 43504 Δnfu, 188,000 ± 56,000 for X47 and 160,000 ± 75,000 for X47 Δnfu.
Δnfu mutants are deficient in mouse stomach colonization
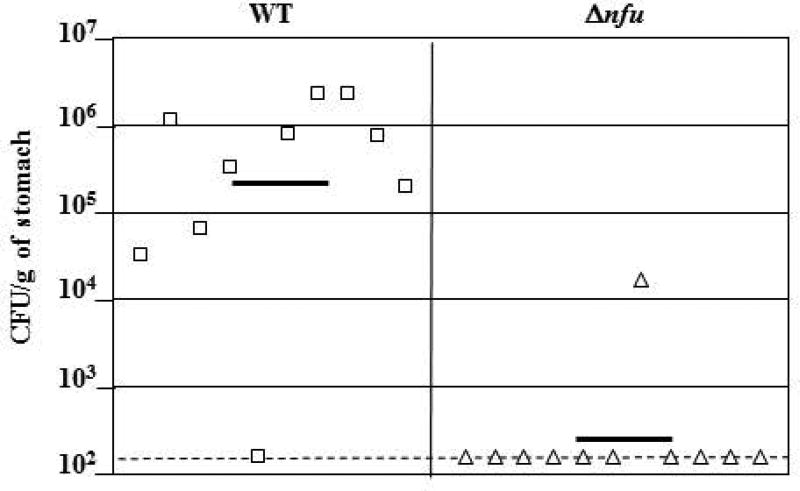
To further study the role of the nfu gene in vivo, we assessed the ability of the Δnfu mutant strain to colonize the gastric mucosa of mice. As described above, the Δnfu mutation was introduced in the mouse colonizing strain X47 (Kleanthous et al., 2001; Veyrier et al., 2013) and the mutant was orally given to C57BL female mice. After 3 weeks, mouse stomachs were harvested and colonization levels of the Δnfu mutant were compared to those of the WT strain X47 (Fig. 5). The WT strain was able to colonize 9 mice (n=10 total), with an average of 8.93 × 105 CFU recovered per g of colonized stomach. In contrast, the Δnfu deletion mutant was detected in only 1 mouse (n=11 total). These results indicate that the nfu gene plays a critical role in vivo, even though it is not required for survival when cells are grown under lab conditions.
Fig. 5. Mouse colonization by H. pylori X47 parent and Δnfu mutant strains.
Mice were inoculated with a dose of 1.5 × 108 viable cells. Colonization of the mouse stomachs was determined 3 weeks post inoculation. Mouse stomachs were homogenized and serial dilutions were plated. Data are presented as a scatter plot of numbers of CFU per gram of stomach (Log10 scale) as determined by plate counts. Each symbol represents the mean CFU count for one stomach (n=10 for WT, and n=11 for ∆nfu, respectively). Each horizontal bar represents the geometric mean of the colonization load for each group. The geometric mean for the ∆nfu mutant is significantly lower than for the wild-type strain (P<0.01, Student’s t-test). A dashed horizontal line shows the detection limit, which represents a count below 1.5 × 102 CFU per g of stomach.
Purified recombinant Nfu protein can coordinate both [2Fe-2S] and [4Fe-4S] clusters
The hp1492 ORF was cloned into an expression vector, and the recombinant Nfu protein was expressed in E. coli and FPLC-purified to near homogeneity (>95%). Upon purification, the Nfu protein was red in color, suggesting the presence of Fe-S centers, as previously described for recombinant H. pylori NifU (Olson et al., 2000). Under denaturing conditions, purified Nfu ran on a SDS 17.5% -PAGE gel with an apparent molecular mass of 11–12 kDa, consistent with its calculated theoretical mass (10.1 kDa). Under native conditions, Nfu formed higher molecular-mass complexes, suggesting oligomerization (data not shown). Size exclusion chromatography experiments revealed that Nfu eluted in three different fractions, with peaks (Ve) at 72.8, 77.8 and 87.7 mL, corresponding to approximately 41.4 kDa (tetrameric [Nfu]4), 25.1 kDa (dimeric [Nfu]2) and 9.3 kDa (monomeric Nfu), respectively. Fractions containing tetrameric and dimeric species were colored (red), while fractions containing monomeric Nfu were colorless.
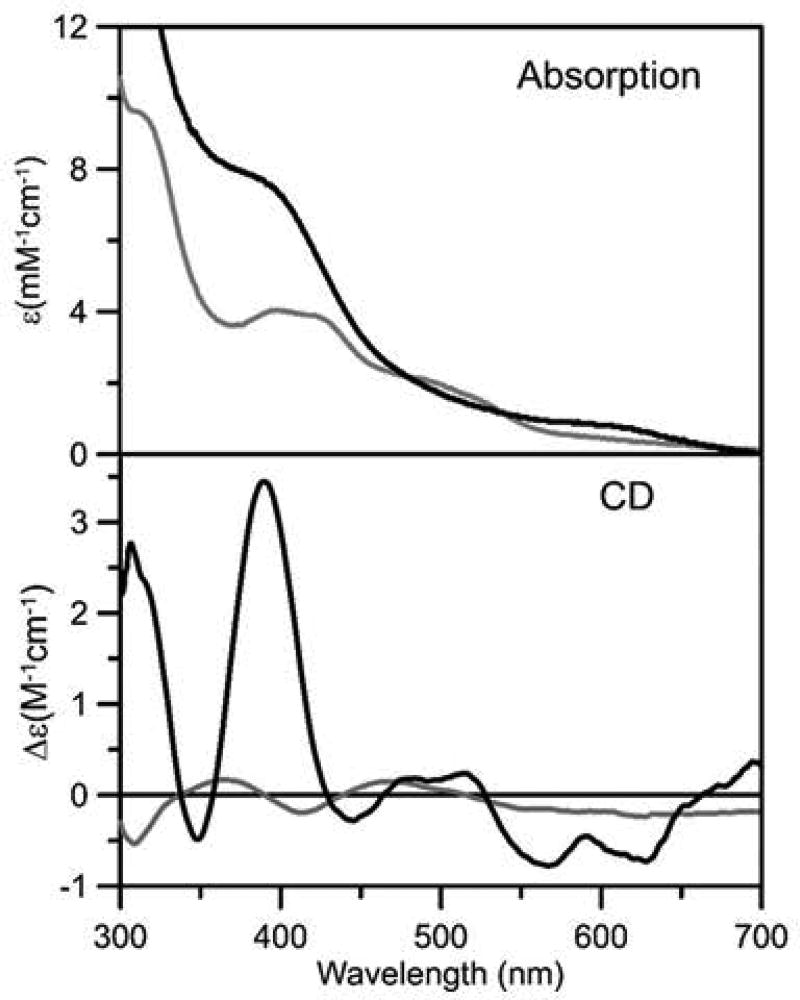
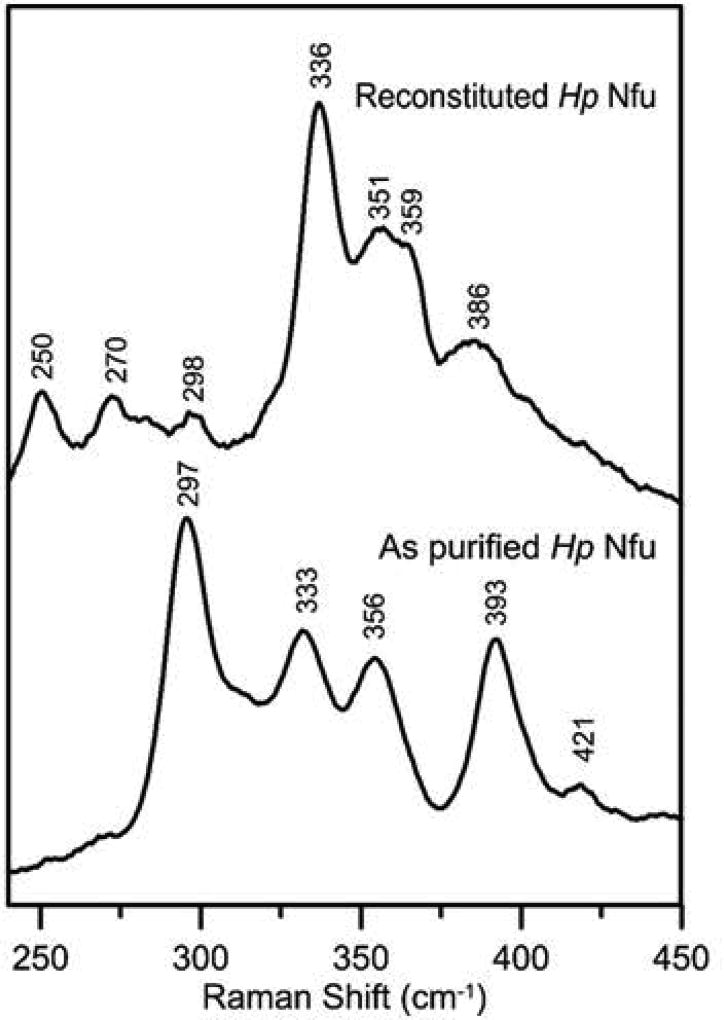
The Fe-S cluster type and stoichiometry in as purified recombinant Nfu and Nfu that was reconstituted in vitro in a cysteine desulfurase-mediated reaction were assessed using protein and iron analysis, in conjunction with UV/visible absorption and CD spectroscopy (Fig. 6), and resonance Raman spectroscopy (RR) (Fig. 7). As purified Nfu was found to contain 1.0 ±0.2 Fe/Nfu monomer and the UV-visible absorption/CD and RR spectra are very similar to those or the biological [2Fe-2S]2+ centers in general, and the previously characterized [2Fe-2S]2+ center in A. thaliana Nfu2 in particular (Gao et al., 2013). The absorption spectrum comprises broad S-to-Fe3+ charge transfer bands centered at 320, 420, 460, and 550 nm (Fig. 6). Moreover the ε420 value (4.0 mM−1cm−1) is indicative of approximately 0.5 [2Fe-2S]2+ cluster per Nfu monomer (ε420 values, based on [2Fe-2S]2+ cluster content, range from 7–11 mM−1cm−1 in characterized [2Fe-2S] cluster-containing proteins (Gao et al., 2013)), which is in accord with the analytical data that indicate one [2Fe-2S] cluster per Nfu dimer. The CD spectrum of [2Fe-2S]2+ centers is more sensitive to the protein environment than the absorption spectrum. Nevertheless, the CD spectrum of as purified H. pylori Nfu (Fig. 6) is very similar both in terms of form and intensity to that of the [2Fe-2S]2+ center in A. thaliana Nfu2 in the 300–500 nm region. The small difference in the 500–700-nm region likely reflect primary structure differences and the one [2Fe-2S] cluster per tetramer oligomeric structure of A. thaliana Nfu2 (Gao et al., 2013). The RR spectrum of the Fe-S center in H. pylori is uniquely indicative of a [2Fe-2S]2+ cluster (Fig. 7). Moreover, the RR spectra of the [2Fe-2S]2+ centers in H. pylori Nfu and A. thaliana Nfu2 are very similar, except for a 10 cm−1 downshift in the symmetric Fe-S(Cys) stretching normal mode in H. pylori Nfu (333 cm−1 compared with 343 cm−1 in A. thaliana Nfu2 (Gao et al., 2013), suggesting slightly weaker Fe-S(Cys) bonds in H. pylori Nfu. Both A. thaliana Nfu2 and H. pylori Nfu have anomalously high asymmetric Fe-S(Cys) stretching normal modes, 295 and 297 cm−1, respectively, compared to [2Fe-2S] cluster-containing ferredoxins with complete cysteinyl ligation, 281–291 cm−1. This is most likely a consequence of anomalous Fe-Sγ-Cβ-Cα dihedral angles due to ligation by two CXXC motifs at the subunit interface (Gao et al., 2013).
Fig. 6. UV-visible absorption and CD spectra of as purified (gray line) and reconstituted (black line) recombinant H. pylori Nfu.
Spectra were recorded in 1 mm cuvettes with samples that were approximately 0.2 mM in Nfu monomer. ε and Δε values are expressed per Nfu monomer.
Fig. 7. Resonance Raman of as purified and reconstituted recombinant H. pylori Nfu.
Spectra were recorded at 17 K using 457.9-nm laser excitation. Each spectrum is the sum of 100 scans, with each scan involving counting photons for 1 s every 0.5 cm−1 with 7 cm−1 spectral resolution. Bands resulting from the frozen buffer solution have been subtracted from both spectra.
Completely different UV-visible absorption/CD and RR spectra that are indicative of a [4Fe-4S]2+ cluster were observed after in vitro cysteine desulfurase-mediated chemical reconstitution of apo H. pylori Nfu (Fig. 6 and 7). Protein and iron determinations indicated 2.1 ±0.3 Fe/Nfu monomer. Coupled with the ε400 value (7.7 mM−1cm−1) for the broad shoulder at ~400 nm that dominates the absorption spectra of all [4Fe-4S]2+ centers, this indicates one [4Fe-4S] cluster per Nfu dimer. The CD spectrum is more intense than most biological [4Fe-4S]2+ centers, but is very similar in form and intensity to the CD spectrum of reconstituted A. thaliana Nfu2 which was shown to exclusively contain [4Fe-4S]2+ clusters based on Mössbauer spectroscopy (Gao et al., 2013). The RR spectrum is also uniquely indicative of a biological [4Fe-4S]2+ center (Fig. 7) and can be assigned to the Fe-S stretching normal modes of an all cysteinyl-ligated [4Fe-4S]2+ cluster by direct analogy with the published assignments (Czernuszewicz et al., 1987). Hence H. pylori Nfu, can bind either one [2Fe-2S]2+ or one [4Fe-4S]2+ cluster per Nfu dimer. However, in contrast to [2Fe-2S] cluster-bound H. pylori Nfu, the [4Fe-4S] cluster-bound form is readily degraded upon exposure to air.
Use of BACTH system reveals a network of protein interactions involving H. pylori Fe-S cluster maturation proteins
To investigate protein-protein interactions between NifS, NifU or Nfu and putative Fe-S clusters-containing proteins, we used a bacterial (E. coli) adenylate cyclase-based two-hybrid system (Karimova et al., 1998). Briefly, interaction between plasmid-encoded peptides T18 and T25 is needed to turn on the adenylate cyclase in E. coli cya- mutants; production of cAMP (from the adenylate cyclase) leads to activation of cAMP-CRP dependent operons, such as lac or mal operons. The (protein-protein) interaction-dependent activation of the aforementioned operons can be monitored on screening media, such as Mac Conkey supplemented with maltose (MC-Mal) and LB supplemented with X-Gal (LB-X-GaL), or on a selection medium, such as M63 with Maltose (M63-Mal).
We cloned nifS (hp0220) in plasmid pKT25, thus generating a T25-NifS fusion. We cloned nifU (hp0221), or nfu (hp1492) in plasmids pKT25 and pKNT25, resulting in T25-NifU, T25-Nfu, NifU-T25 and Nfu-T25 fusion proteins, respectively. The same 3 genes nifS, nifU and nfu, as well as 36 genes encoding for putative Fe-S-containing proteins, were individually cloned in plasmid pUT18C, generating T18-target fusion proteins. Next, E. coli cya mutants (BTH101, Table S1) were co-transformed with both pK(N)T25 and pUT18C derivatives, yielding 240 different combinations, including vector (pKT25) only controls. For initial screening, cells were spotted on MC-Mal and LB-X-Gal (Fig. S1). Plates were incubated for 48 h at 30°C under aerobic conditions. Strong interactions between H. pylori proteins yielded red and blue colonies on MC-Mal and LB-X-Gal, respectively (Fig. S1). Since all combinations that included pKNT25-Nfu plasmid (encoding for Nfu-T25) appeared positive on both MC-Mal and LB-X-Gal, even in presence of the vector-only pUT18C negative control, those were labelled as false positive and therefore excluded from the study (data not shown). For the remaining 200 clones, all negative controls were indeed “negative” and there was high correlation between results obtained on both chromogenic media, e.g. clones turning red on MC-Mal were blue on LB-X-Gal (Fig. S1). These preliminary screening experiments revealed that each of the three “bait” proteins (NifS, NifU or Nfu) was able to interact with itself. In addition, NifS-NifU and NifU-Nfu complexes were also identified (Fig. S1). The majority of strong interactions involved NifU rather than Nfu. A few interactions also involved NifS.
While both MC and LB-X-Gal yielded promising results, neither screening medium was sensitive enough to detect weak interactions, or to discriminate between weak interactions and negative controls even after prolonged incubation times. Therefore, we aimed at improving characterization of protein-protein interactions by monitoring growth of all co-transformed E. coli cells in M63-Mal minimal medium. In this semi-quantitative experiment, the duration and/or strength of protein-protein interactions correlates to the amount of cAMP produced by the Cya enzyme, which in turns leads to activation of the mal operon, allowing E. coli to use maltose and grow in the minimal medium. Hence, E. coli cells were incubated in 96-well microplates under aerobic conditions and the growth (OD595) was recorded after 72 h at 30°C (Table 2). Growth was scored as follows: OD595 ≤ 0.05 (white boxes), no growth e.g. no detectable interaction, including all vector-only negative controls; 0.05<OD595<0.1 (orange boxes), weak interactions; 0.1≤ OD595<0.2 (blue boxes), intermediate interactions; OD595≥0.2 (green boxes), strong interactions, including the Zip-Zip positive control. Overall, there was good correlation between cell growth in M63 (96-well microplates) and colonies’ phenotype on either MC-Mal or LB-X-Gal plates. Indeed, E. coli cells with intermediate or strong levels of interactions grew well in M63-maltose (Table 2) and appeared red or blue on the respective chromogenic media (Fig. S1 and S2). M63-Mal growth experiments confirmed that NifS, NifU and Nfu could interact with themselves (dimerization) and also indicated each protein could interact with either of the two other components (NifS-NifU, NifS-Nfu and NifU-Nfu complexes). NifS-Nfu and NifU-Nfu interactions were confirmed with additional T18 and T25 fusions (Fig. S2). M63-Mal growth experiments also revealed additional interactions between NifU, Nfu and putative Fe-S-containing proteins (Table 2 and Table S3). Almost all the target proteins that interacted with Nfu were also found to interact with NifU, with the exception of HP0277 (ferredoxin), which was shown to make complexes with either NifS or Nfu, but not with NifU. Preliminary experiments with T18-AcnB (HP0779) suggested AcnB only interacts with (T25-)Nfu, but not with (T25-) NifS or NifU (Table 2 and Fig. S1) however complementary BACTH screening and growth experiments with T25-AcnB or AcnB-T25 and T18-Nfu, Nfu-T18, T18-NifU or T18-NifS revealed that AcnB can also interact with NifU as well as with NifS (Fig. S2 and Table S3).
Table 2.
Growth of co-transformed E. coli in M63-maltose minimal medium.
| Growth (OD595) in M63 after 72h at 30°C (Mean ± SD, n=3–5) | |||||||
|---|---|---|---|---|---|---|---|
| FUSION PROTEIN 1 (in pUT18C) | FUSION PROTEIN 2 (in pKT25 or pKNT25) | ||||||
| # | Fusion protein 1 |
H. pylori Protein ID |
T25 only (control) |
T25-HP0220 (NifS) |
T25-HP0221 (NifU) |
HP0221-T25 (NifU) |
T25-HP1492 (Nfu) |
| T18 only | None (Neg. Cont.) | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.04 ± 0.01 | |
| 1 | T18-HP0117 | HP0117 | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.04 ± 0.01 | 0.36 ± 0.04 | 0.09 ± 0.04 |
| 2 | T18-HP0132 | SdaA | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.21 ± 0.05 | 0.14 ± 0.05 | 0.27 ± 0.03 |
| 3 | T18-HP0138 | HP0138 | 0.02 ± 0.02 | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| 4 | T18-HP0142 | MutY | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.05 ± 0.03 | 0.44 ± 0.11 | 0.04 ± 0.02 |
| 5 | T18-HP0191 | FrdB | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.02 ± 0.03 | 0.04 ± 0.02 | 0.04 ± 0.02 |
| 6 | T18-HP0207 | Mrp, ApbC | 0.04 ± 0.01 | 0.23 ± 0.07 | 0.14 ± 0.04 | 0.13 ± 0.05 | 0.21 ± 0.01 |
| 7 | T18-HP0220 | NifS | 0.02 ± 0.02 | 0.29 ± 0.02 | 0.13 ± 0.04 | 0.11 ± 0.04 | 0.22 ± 0.02 |
| 8 | T18-HP0221 | NifU | 0.02 ± 0.01 | 0.12 ± 0.07 | 0.37 ± 0.03 | 0.04 ± 0.01 | 0.08 ± 0.01 |
| 9 | T18-HP0269 | MiaB-like | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.05 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 |
| 10 | T18-HP0277 | HP0277 | 0.04 ± 0.01 | 0.30 ± 0.09 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.40 ± 0.04 |
| 11 | T18-HP0285 | MiaB | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.25 ± 0.07 | 0.21 ± 0.04 |
| 12 | T18-HP0400 | IspH | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.11 ± 0.02 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| 13 | T18-HP0468 | HP0468 | 0.03 ± 0.01 | 0.05 ± 0.03 | 0.04 ± 0.01 | 0.29 ± 0.04 | 0.04 ± 0.02 |
| 14 | T18-HP0568 | HP0568 | 0.04 ± 0.01 | 0.05 ± 0.01 | 0.13 ± 0.06 | 0.36 ± 0.04 | 0.14 ± 0.02 |
| 15 | T18-HP0585 | Nth | 0.02 ± 0.02 | 0.03 ± 0.01 | 0.21 ± 0.05 | 0.15 ± 0.07 | 0.06 ± 0.01 |
| 16 | T18-HP0588 | OorD | 0.03 ± 0.02 | 0.05 ± 0.03 | 0.15 ± 0.01 | 0.14 ± 0.05 | 0.18 ± 0.07 |
| 17 | T18-HP0625 | IspG | 0.05 ± 0.02 | 0.04 ± 0.01 | 0.11 ± 0.03 | 0.37 ± 0.01 | 0.03 ± 0.01 |
| 18 | T18-HP0631 | HydA | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.08 ± 0.03 | 0.24 ± 0.04 | 0.04 ± 0.01 |
| 19 | T18-HP0654 | MqnE | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.09 ± 0.04 | 0.33 ± 0.05 | 0.04 ± 0.01 |
| 20 | T18-HP0656 | MqnC | 0.01 ± 0.01 | 0.05 ± 0.01 | 0.13 ± 0.02 | 0.26 ± 0.04 | 0.04 ± 0.01 |
| 21 | T18-HP0665 | HemN | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.07 ± 0.04 | 0.06 ± 0.01 | 0.05 ± 0.03 |
| 22 | T18-HP0666 | GlpC | 0.02 ± 0.03 | 0.03 ± 0.02 | 0.48 ± 0.08 | 0.11 ± 0.02 | 0.03 ± 0.03 |
| 23 | T18-HP0734 | RimO | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.40 ± 0.02 | 0.48 ± 0.01 | 0.12 ± 0.07 |
| 24 | T18-HP0768 | MoaA | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.03 ± 0.02 | 0.21 ± 0.04 | 0.05 ± 0.01 |
| 25 | T18-HP0779 | AcnB | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.17 ± 0.04 |
| 26 | T18-HP0934 | QueE | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.03 ± 0.02 | 0.13 ± 0.05 | 0.05 ± 0.01 |
| 27 | T18-HP1089 | AddB | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.26 ± 0.04 | 0.29 ± 0.01 | 0.04 ± 0.01 |
| 28 | T18-HP1109 | PorD | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.04 ± 0.03 | 0.05 ± 0.02 | 0.05 ± 0.01 |
| 29 | T18-HP1222 | Dld | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.03 |
| 30 | T18-HP1226 | HemN | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.03 ± 0.01 |
| 31 | T18-HP1261 | NuoB | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.05 ± 0.02 |
| 32 | T18-HP1266 | Nqo3 | 0.04 ± 0.03 | 0.16 ± 0.08 | 0.32 ± 0.09 | 0.04 ± 0.01 | 0.24 ± 0.03 |
| 33 | T18-HP1268 | Nqo9 | 0.02 ± 0.02 | 0.03 ± 0.02 | 0.33 ± 0.02 | 0.22 ± 0.14 | 0.18 ± 0.05 |
| 34 | T18-HP1356 | NadA | 0.03 ± 0.02 | 0.04 ± 0.03 | 0.02 ± 0.01 | 0.08 ± 0.02 | 0.03 ± 0.03 |
| 35 | T18-HP1406 | BioB | 0.03 ± 0.02 | 0.02 ± 0.02 | 0.03 ± 0.03 | 0.26 ± 0.05 | 0.04 ± 0.02 |
| 36 | T18-HP1428 | RlmN | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.15 ± 0.02 | 0.27 ± 0.07 | 0.08 ± 0.01 |
| 37 | T18-HP1492 | Nfu | 0.02 ± 0.01 | 0.32 ± 0.06 | 0.13 ± 0.02 | 0.29 ± 0.01 | 0.39 ± 0.04 |
| 38 | T18-HP1508 | HP1508 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.03 | 0.01 ± 0.01 | 0.03 ± 0.02 |
| 39 | T18-HP1540 | FbcF | 0.05 ± 0.01 | 0.05 ± 0.02 | 0.24 ± 0.04 | 0.44 ± 0.06 | 0.17 ± 0.10 |
| T18-Zip/T25-Zip: Positive Control | 0.23 ± 0.04 | Interaction & Color key | No detectable | Moderate | |||
| Weak | Strong | ||||||
Size exclusion chromatography (SEC) confirms NifS-NifU interactions
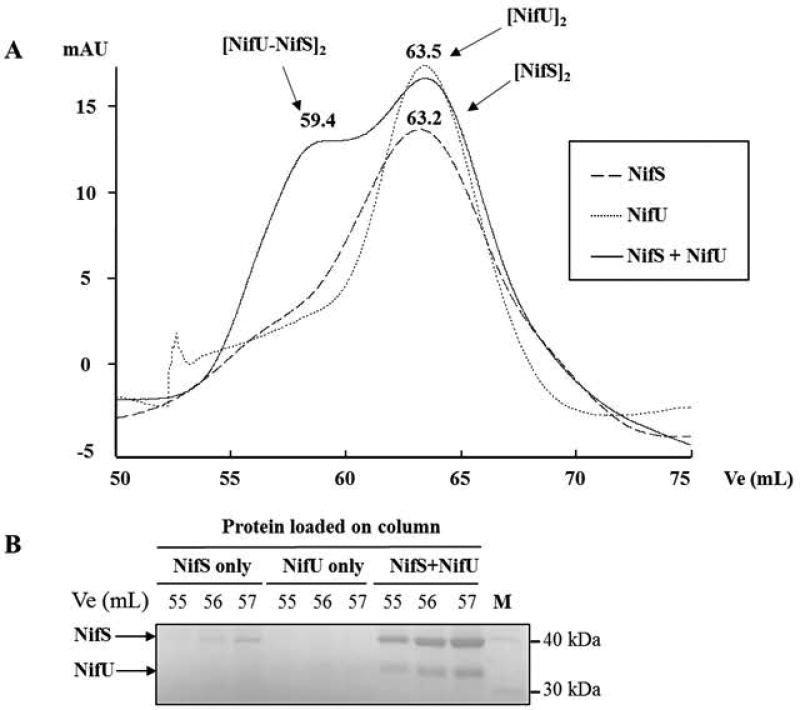
In order to confirm interactions between the core components of the NIF system (NifS, NifU, Nfu), as well as with AcnB (HP0779), each protein was injected separately or after incubation with other proteins. The Ve/Vo ratio was determined for each complex and used to calculate corresponding molecular masses, using a calibration curve. This strategy revealed that both NifS and NifU preferentially form homodimers in solution when run separately (Fig. 8, panel A). When both proteins were incubated and injected together, a new peak appeared (Ve=59.4 mL), indicating the presence of a complex with a calculated mass of 157 kDa, in good agreement with the formation of a [NifS-NifU]2 hetero-tetramer complex. We analyzed the three same fractions (Ve=55, 56, 57 mL) from each run by SDS-PAGE (Fig. 8, panel B): when injected separately, NifS was barely detected, and NifU was not at all detected; in contrast, when injected together, both NifS and NifU were clearly visualized in these fractions, confirming the presence of the [NifS-NifU]2 hetero-tetramer. SEC was also used to confirm other putative complexes (as suggested by BACTH) such as NifS-Nfu, NifU-Nfu or AcnB-Nfu complexes, however we were unable to isolate such complexes using this method.
Fig. 8. Size exclusion chromatography analysis of NifS-NifU protein complexes.
Purified NifS and NifU were run separately, then together, on a HiLoad 16/60 Superdex 75, and 1 mL fractions were collected and analyzed by SDS-PAGE. Elution volumes (Ve) were recorded for each protein complex, Ve/Vo ratio were calculated and a standard curve with molecular weight markers was used to determine molecular weights for each complex. (A) Chromatograms showing NifS only, NifU only and NifS and NifU together. Ve (mL) and the proposed protein oligomeric state are shown above each peak. (B) Fractions corresponding to the same Ve (55, 56, 57 mL) for each experiment were analyzed on SDS-PAGE. NifS (42.4 kDa) and NifU (36.3 kDa) are indicated by an arrow.
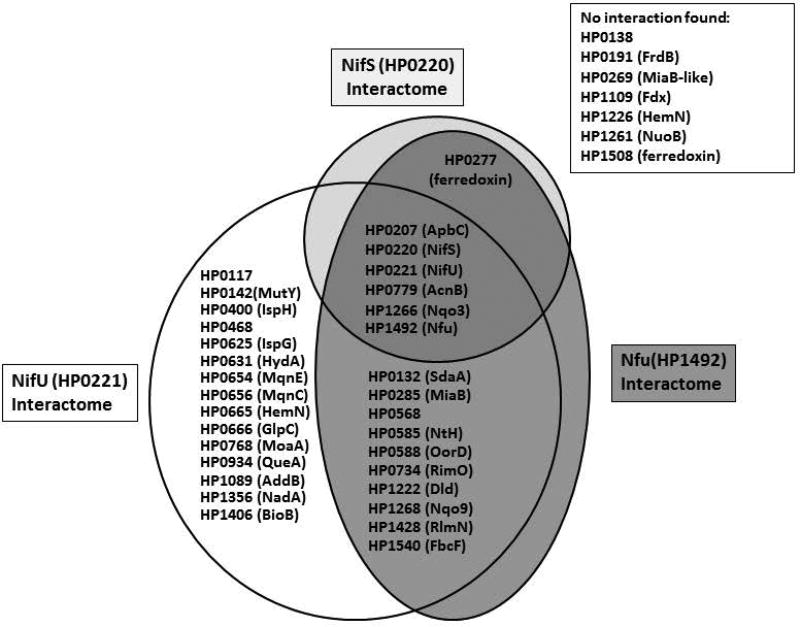
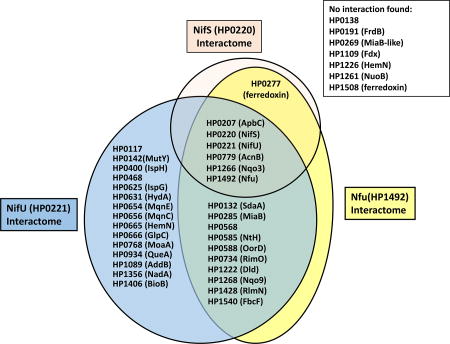
Taken together, BACTH and SEC results suggest NifU interacted with 29 proteins, including itself, NifS and 27 putative Fe-S cluster-containing proteins. Likewise, Nfu was found to interact with 16 proteins, comprising itself, NifS and 14 putative Fe-S cluster-containing proteins. NifS interacted with itself and 5 other proteins. Protein-protein interactions between NifS, NiU, Nfu and each of the putative Fe-S cluster-binding proteins are summarized in Fig. 9.
Fig. 9. Summary of protein-protein interactions between H. pylori NifS, NifU, Nfu and putative Fe-S recipient proteins.
Based on BACTH (screening and selection) results, proteins found to interact with either NifS, NifU or/and Nfu are shown in respective interaction balloons. Proteins shown on the right were not found to interact with any of the three cluster-maturation bait proteins.
DISCUSSION
While most bacteria possess multiple independent systems (ISC, NIF, SUF) to synthesize and transfer Fe-S clusters, the ε-proteobacterium H. pylori appears to be unique: not only does it exclusively use one system, the NIF system, but it uses proteins that are the hallmarks of nitrogen-fixing bacteria, a group H. pylori does not belong to. Other members of the ε-proteobacteria, such as Campylobacter jejuni and Wolinella succinogenes, also have NIF as their unique Fe-S maturation system. Two genes/proteins had previously been identified as being members of the Fe-S cluster synthesis system in H. pylori: NifS, the cysteine desulfurase and S-donor, and NifU, the scaffold protein, both previously studied in our lab (Olson et al., 2000). Both NifS and NifU appeared to be essential, as previous attempts to construct nifS or nifU mutants failed (Olson et al., 2000). The essentiality of both genes was expected, since the NIF system is the only Fe-S maturation system known in H. pylori and Fe-S clusters are required for key proteins to perform their metabolic function. In order to study the role of nifU in H. pylori, we generated conditional nifU mutants. This was achieved by first introducing a plasmid-borne, IPTG-inducible copy of nifU in a WT strain, prior to inactivating the chromosomal nifU allele. This strategy has been successfully used by our lab and others to target genes encoding essential proteins in H. pylori, such as penicillin-binding proteins 1 and 3 (Boneca et al., 2008), or Twin Arginine Translocase C (Benoit and Maier, 2014). When grown under increasing O2 concentrations (and without supplemented IPTG), the NifU-depleted strain grew poorly under most O2 concentrations and it did not not at all under 15% O2 concentration. This phenotype was not unexpected. Indeed, Fe-S clusters in NifU (leaky expression in the absence of IPTG) and other Fe-S proteins are likely to become unstable under high O2 concentration and this instability would lead to an insufficient supply of Fe-S clusters. Besides, a number of Fe-S cluster-containing enzymes are essential, including some involved in aerobic respiration; for instance three proteins, NuoB (HP1261), Nqo3 (HP1266) and Nqo9 (HP1268) are part of the multiprotein NADH:ubiquinone respiration complex, while a fourth (FbcF, HP1540), an ubiquinol:cytochrome c oxidoreductase subunit, is also needed for respiration (and previously shown to be essential in H. pylori) (Benoit and Maier, 2014). In the present study, Nqo3, Nqo9 and FbcF were all found to interact with NifU in our BACTH assay (Table 2). Upon addition of IPTG in the growth medium, the NifU-depleted strain grew better and was less sensitive to O2-generated oxidative stress, suggesting that higher NifU levels helped H. pylori grow, respire O2 and cope with oxidative stress. Further studies with this NifU-depleted strain will be needed to understand better the role of NifU in vivo.
Besides NifU and NifS, H. pylori possess a small Nfu-like protein that shares some homology with the C-terminus domain of NifU, including a conserved CXXC domain (Tokumoto et al., 2004). Proteins containing the Nfu-domain have been characterized in several organisms, including prokaryotes such as E. coli (Angelini et al., 2008), the cyanobacterium Synechococcus (Balasubramanian et al., 2006) and Azotobacter vinelandii (Bandyopadhyay et al., 2008b), as well as in eukaryotes such as Saccharomyces cerevisiae (Schilke et al., 1999), Arabidopsis thaliana (Touraine et al., 2004; Yabe et al., 2004; Yabe et al., 2008) or Homo sapiens (Tong et al., 2003). Mutations in nfu-type genes can have severe consequences: for instance, disruption of nfu is lethal for the cyanobacterium Synechococcus (Balasubramanian et al., 2006), and a single point mutation in the human nfu1 gene leads to a fatal disease known as multiple mitochondrial dysfunctional syndrome (MMDS1)(Ahting et al., 2015). The H. pylori nfu gene had not been characterized before, therefore it was not clear which impact the disruption of nfu would have on the gastric pathogen’s metabolism and viability.
In the present study, we disrupted nfu in three different parental strains, showing that it is dispensable for H. pylori, under conditions we used. We hypothesized that deletion of the nfu gene would impact the maturation of select Fe-S enzymes, such as aconitase or hydrogenase, as described in other microorganisms (Bandyopadhyay et al., 2008b; Py et al., 2012; Mashruwala et al., 2015). Therefore, we assayed aconitase and hydrogenase activity in two independent Δnfu mutants. Compared to their parental strains, aconitase activity in those mutants was sharply decreased (by 70 to 80 %). Similar results (i.e. significant decrease in aconitase activity) were also reported for A. vinelandii nfuA mutants, S. aureus nfu mutants and E. coli nfuA mutants, respectively, and these phenotypes were generally attributed to an aconitase [4Fe-4S] cluster maturation deficiency by the authors of these studies (Bandyopadhyay et al., 2008b; Py et al., 2012; Mashruwala et al., 2015). Besides, each of the three aforementioned Nfu-type proteins was shown to interact with and transfer [4Fe-4S] clusters to apo-aconitase (Bandyopadhyay et al., 2008b; Py et al., 2012; Mashruwala et al., 2015). In the present study, using BACTH, we showed that AcnB (HP0779) can interact with Nfu, as well as with NifS and NifU (see Sup. Fig. S2). H. pylori aconitase is a bifunctional enzyme that can act as post-transcriptional regulator (Austin and Maier, 2013; Austin et al., 2015), in addition to its well-known contribution to the tricarboxylic acid (TCA) cycle (Pitson et al., 1999). Indeed, previous results from our lab suggest AcnB is a pleiotropic regulator in H. pylori, controlling genes involved in peptidoglycan modification, flagella synthesis, nickel homeostasis or oxidative stress resistance (Austin and Maier, 2013; Austin et al., 2015). Since our results suggest that AcnB depends on Nfu for its Fe-S maturation, and because the Fe-S status of AcnB dictates its cellular function (regulation vs TCA), we hypothesize Nfu could also be considered a broad-function regulator in H. pylori; more work is needed to experimentally test this hypothesis.
Regarding hydrogenase activity, we found that H. pylori Δnfu mutants had higher hydrogenase activity (compared to WT), an unexpected phenotype we hypothetically attribute to higher cellular pools of free Fe in the absence of the Nfu Fe-S carrier protein. Indeed, the hydABCDE operon encoding for hydrogenase was previously shown to be repressed by the apo (iron-free) form of the Fur regulatory protein (Ernst et al., 2005). Therefore, increased Fe levels in Δnfu mutants would lead to de-repression, increased hyd transcript levels and increased hydrogenase activity. However, this remains to be experimentally proven. This result (higher hydrogenase activity in mutants) also suggests that Nfu is not directly involved in the maturation (Fe-S delivery) of hydrogenase. Such a lack of interaction was (indirectly) confirmed by our BACTH screening: we found that NifU-but not Nfu-was capable of interacting with the hydrogenase Fe-S subunit HydA, therefore suggesting that NifU is involved in hydrogenase Fe-S maturation while Nfu is not.
In addition, the Δnfu deletion was associated with deficient growth in liquid medium, similar to what we observed for the NifU-depleted strain. Both the growth rate and the growth yield (after 24 h) were reduced in the Δnfu mutant (compared to wild-type) when cells were grown under low (5%) and the phenotype was even more obvious when cells were grown at high (15%) O2 partial pressure. As suggested by our results, Nfu interacts with 14 Fe-S clusters containing proteins, including essential proteins involved in cell metabolism and respiration. Therefore it is not surprising to Nfu is needed for optimal growth and aerobic respiration in H. pylori. The Δnfu deletion was also associated with severe mouse colonization deficiency, indicating that Nfu plays a critical role in H. pylori. These results are in agreement with a recent study from Mashruwala et al., who showed that a Staphylococcus aureus nfu mutant is more susceptible to oxidative killing by human polymorphonuclear leukocytes and displays decreased tissue colonization in mice (Mashruwala et al., 2015).
Using UV/visible absorption/CD and resonance Raman spectroscopy, we showed that H. pylori Nfu (expressed as recombinant protein in E. coli) can accommodate both [2Fe-2S] (as purified) and [4Fe-4S] clusters upon reconstitution. At this stage, it is not clear whether the [2Fe-2S] cluster is physiologically relevant, i.e. it can accommodate [2Fe-2S] clusters and transfer them to putative [2Fe-2S]-containing recipient proteins, such as fumarate reductase (HP0191) or the Rieske protein (HP1540) (Table 1). Alternatively, the presence of the [2Fe-2S] cluster as purified could be the result of heterologous expression in E. coli, which contains the ISC and SUF systems for Fe-S cluster assembly. While some prokaryotic proteins with Nfu domains, like E. coli NfuA or A. vinlandii NfuA were reported to bind only [4Fe-4S] clusters (Bandyopadhyay et al., 2008b; Py et al., 2012), A. thaliana Nfu-type Nfu2 was shown to accommodate both [2Fe-2S] and [4Fe-4S] clusters; in addition, the protein is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins (Gao et al., 2013). The Nfu domains of all three proteins share 30 to 35% identity with H. pylori Nfu. More work will be needed to determine the nature of the Fe-S clusters in H. pylori Nfu in vivo.
To investigate protein-protein interactions between Fe-S clusters-containing H. pylori proteins, we used a bacterial adenylate cyclase-based, two-hybrid system (Karimova et al., 1998). BACTH results unambiguously showed that the three core components of the Nif system, NifS, NifU, and Nfu can individually interact together to make heterodimeric (NifS-NifU, Nifs-Nfu and NifU-Nfu) complexes. The NifS-NifU complex was confirmed by SEC studies. Based on these studies, NifS and NifU are able to form a hetero-tetrameric complex. NifS and NifU were expected to interact, since (i) they are the two main known components of the NIF maturation system, with clearly assigned roles (S-donor for NifS, scaffold protein for NifU, (Olson et al., 2000; Johnson et al., 2005)) and (ii) a NifS-NifU interaction has been shown in a yeast two-hybrid (Y2H) proteome-wide screening of the closely related ε-proteobacterium C. jejuni (Parrish et al., 2007). The fact that Nfu can interact with NifU is interesting and suggests that Nfu can act as a Fe-S carrier protein, transferring NifU-assembled Fe-S clusters to recipient proteins that cannot get their clusters directly from NifU. However, the existence of a NifS/Nfu interaction (as suggested by BACTH results) is intriguing since it raises the possibility that Nfu could receive sulfur from NifS. Therefore, Nfu could potentially be a stand-alone scaffold protein, playing a role similar to NifU in H. pylori. However, there are at least two alternative explanations. First, the apo form of Nfu could play a potential regulatory role in releasing sulfide from the cysteine persulfide on NifS by dithiol/disulfide exchange reactivity involving the CXXC motif, as suggested by the work of Cowan and coworkers on a human Nfu protein (Liu and Cowan, 2007; Liu et al., 2009). Second, Nfu could accept clusters from the NifS/NifU complex by binding to both NifS and NifU. This raises the possibility of a Nfu/NifS/NifU multi-component platform, on which Fe-S-recipient proteins would bind to get their Fe-S clusters. In E. coli, IscS (NifS homolog) has been shown to form complexes not only with IscU (NifU homolog) but also with several other partners, such as TusA (Shi et al., 2010), ferredoxin (Yan et al., 2013), bacterial frataxin CyaY (Prischi et al., 2010) and the ancillary protein IscX (Kim et al., 2014; di Maio et al., 2017). Likewise, a model involving a ternary complex IscS-IscU-CyaY has been recently proposed (di Maio et al. 2017). Clearly more work is required to understand the significance of the NifS/Nfu interaction in H. pylori.
Sequence analysis of all (1,590) predicted open reading frames of H. pylori strain 26695 suggested 36 putative Fe-S cluster-coordinating proteins based on their primary amino acid sequence, besides NifU and Nfu (Table 1 and 2). Therefore the BACTH system was also used to screen for interactions between NifS, NifU, Nfu and those 36 proteins. Two proteome-wide studies of H. pylori protein-protein interactions using Y2H technology have been reported (Rain et al., 2001; Hauser et al., 2014). In the first one, neither NifS nor NifU were use as baits in the study (Rain et al., 2001). When Nfu was used as bait, the study identified 6 proteins (HP0172, HP0351, HP0464, HP1041, HP1286, HP1533) as potential targets (Rain et al., 2001); however based on their sequence and/or known functions, none of them appears to be a member of the H. pylori Fe-S protein network. The second, more recent proteome-wide Y2H study did not include NifU or Nfu as baits, and only identified 3 putative partners for NifS (HP0069, HP1262, HP1536); again, none of these proteins appears to be a putative Fe-S protein. In our study, we found that 15 proteins out of the 34 putative Fe-S cluster-coordinating proteins could interact with NifU, 11 others with NifU or Nfu, and one (HP1266; Nqo3) with each of the 3 donor proteins (NifS, NifU, Nfu) (Fig. 8). We also found that one protein (HP0277) appears to interact with NifS or Nfu, but not with NifU. HP0277 encodes for a ferredoxin-type protein. However, while most ferredoxin-like proteins harbor one or two conserved CX2CX2CX3C(P) motifs in their sequence (a hallmark of [4Fe-4S] cluster binding), HP0277 has an unusual CX2CX9CX3CP domain.
Seven proteins were not found to interact with any of the three proteins (NifS, NifU, Nfu) used as baits in the BACTH system (Fig. 9). These are: HP0138 (hypothetical protein); HP0191 (fumarate reductase β subunit); HP0269 (MiaB-like protein)l HP1109 (pyruvate ferredoxin oxidoreductase, δ subunit); HP1226 (HemN, O2-independent coproporphyrinogen-III oxidase); HP1261 (NuoB, NADH-Quinone oxidoreductase subunit B); and HP1508 (ferredoxin-like). Nothing is known so far about the role and the structure of the HP0138 protein, therefore it is hard to speculate on whether the protein can accommodate Fe-S clusters, if any, and where it would get them from. As for the fumarate reductase FrdB subunit, it is expected to contain several diverse Fe-S clusters: indeed, FrdB homologs have been shown to coordinate one [2Fe-2S], one [4Fe-4S] and one [3Fe-4S] cluster (Morningstar et al., 1985). In addition, the presence of Fe-S clusters in recombinant H. pylori FrdB has been experimentally confirmed (Mileni et al., 2006). HP0269 is a MiaB-like protein, homologous to HP0285, shown in the present study to interact with both NifU and Nfu. The δ subunit of pyruvate ferredoxin oxidoreductase (HP1109) and the ferredoxin-like HP1508 are both strong candidates for [4Fe-4S] binding, since both possess two conserved CX2CX2CX3C(P) motifs. HP1226 is a homolog of HP0665 (both are annotated as “HemN”), the latter found to interact with NifU in the present study. Finally, NuoB (HP1261), one of the NADH-quinone respiratory complex subunit, failed to turn on the BACTH system. Based on its sequence and the fact that two other NADH-quinone oxidoreductase subunits, HP1266 (Nqo3) and HP1268 (Nqo9) interact strongly with NifU and Nfu in the present study, NuoB was expected to interact with at least one of the Fe-S scaffold or carrier, but it did not. Therefore, we believe that the negative phenotype observed for the six aforementioned proteins is probably more imputable to limits of the screening system (BACTH) rather than to an actual lack of interaction between these proteins and components of the Nif maturation system. For instance, the C- or N- (T18- or T25-) fusion could disrupt the interacting interface. We cannot rule out such interactions, and further work will be needed to determine if any or several of these six proteins belong to the NIF-mediated network.
In conclusion, we have shown H. pylori Nfu can bind [2Fe-2S] or [4Fe-4S] clusters with a stoichiometry of one cluster per dimer. The combined use of bacterial two-hybrid technology and targeted mutagenesis has shed light on the respective role of NifU and Nfu in H. pylori. The existence of protein-protein interactions between the Fe-S scaffold protein NifU or the Fe-S carrier protein Nfu and any of the putative Fe-S cluster containing protein, as shown by BACTH herein, strongly suggests (but does not demonstrate per se) Fe-S transfer from the former(s) to the latter(s). Formal Fe-S transfer between Fe-S donors and recipients will still have to be demonstrated on a case-by-case basis.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
E. coli and H. pylori strains and plasmids used in this study are listed in Table S1. Genomic DNA from H. pylori strain 26695 was used as template for all PCR amplifications. All plasmids (including all BACTH plasmids) were sequenced at the Georgia Genomics Facility, University of Georgia, Athens, GA.
Growth conditions
E. coli cells were grown aerobically in Luria-Bertani (LB) medium or plates at 37°C, unless indicated otherwise. For BACTH screening, we used MacConkey (MC) plates (BD-Difco # 281810), supplemented with 1% glucose-free maltose (Mal, BD-Difco #216830), or LB plates supplemented with 20 µg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal, Fisher). Both MC-Mal and LB-X-Gal were supplemented with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG, GoldBio), 0.1 mM FeCl3, 0.5 mM L-cysteine, 100 µg/mL ampicillin (Amp) and 30 µg/mL kanamycin (Kan). For selection, we used the following Fe-enriched M63 minimal medium: (NH4)2SO4 (2 g/L), KH2PO4 (13.6 g/L), Thiamine B1 (1 mg/L), 1 mM MgSO4, FeSO4.7H2O (5 mg/L), pH 7. This medium was supplemented with 0.4% Mal, 0.5 mM IPTG, 50 µg/mL Amp and 30 µg/mL Kan. All BACTH growth experiments were carried out at 30°C in aerobiosis (exposed to air).
H. pylori was routinely grown on Brucella agar plates supplemented with 10% defibrinated sheep blood (BA plates), at 37°C under microaerophilic conditions (5% CO2, 4% O2 and 91% N2). Chloramphenicol (Cm, 25 µg/ml for Δnfu mutants, 8 µg/ml for ΔnifU conditional mutants), Kan (30 µg/ml) or IPTG (1 mM) was added as needed. For mouse colonization experiments, BA was supplemented with amphotericin B (10 µg/ml), vancomycin (10 µg/ml) and bacitracin (50 µg/ml). All antibiotics were purchased from Sigma. For liquid cultures, Brain-Heart Infusion (BHI) supplemented with 0.4% β-cyclodextrin (Sigma) was used (BHI-βc). Briefly, sealed 165 mL bottles with a 20 mL side arm containing 10 mL of BHI-βc were flushed with anaerobic mixture gas (5% CO2, 10 % H2 and 85% N2) for 10 min. O2 (5% or 15% partial pressure) was added to the bottles. 0.5 mM IPTG was added as needed. H. pylori cells grown on BA or BA-IPTG for less than 24 h were resuspended in BHI-βc, standardized to the same OD600, and inoculated. Cells were grown at 37°C under constant shaking (200 rpm). Cell growth (as shown by turbidity) was monitored for 24 h using a Klett spectrophotometer.
Construction of H. pylori Δnfu mutants
A ∆nfu deletion-insertion strain containing a cat cassette replacing the coding sequence for nfu (hp1492) was constructed using a splicing-by-overlap-extension (SOE) PCR method, as follows. H. pylori WT 26695 (Table S1) genomic DNA was used as a template for polymerase chain reaction (PCR) to amplify fragments of DNA flanking hp1492. Primers ΔNfu-1 and ΔNfu-2 (Table S2) were used to amplify a 450 bp-long DNA sequence located upstream of hp1492. Primers ΔNfu-3 and ΔNfu-4 were used to amplify a 470 bp-long sequence located downstream of hp1492. The final SOE amplification step included both PCR products, a 0.8 kb-long cat cassette (Wang and Taylor, 1990) and primers ΔNfu-1 and ΔNfu-4. The resulting 1640 bp-long PCR product was introduced by natural transformation into various H. pylori parental strains (X47, 43504 and 26695) and cells were plated on BA supplemented with chloramphenicol. Δnfu::cat mutants appeared after 3 to 5 days. The concomitant deletion of nfu and the insertion of cat was confirmed by PCR, using genomic DNA from mutants as template.
Construction of conditional nifU mutants
Conditional ΔnifU mutants were constructed in two steps. Briefly, a 1 kb-long PCR product containing the nifU ORF was amplified using primers NifU-NdeI and NifU-BamHI (Table S2). After digestion with NdeI and BamHI, the PCR product was ligated into similarly digested plasmid pILL2150 (Boneca et al., 2008) to generate plasmid pILL-nifU. In this plasmid, the nifU gene is under the control of a (IPTG-inducible) Ptac promoter. Plasmid pILL-nifU was methylated in presence of S-adenosylmethionine (New England Biolabs, Ipswich, MA) and cell-free extracts from H. pylori wild-type strain 43504, as described by Boneca et al. (Boneca et al., 2008). Methylated plasmid pILL-nifU was then introduced into strain 43504, generating chloramphenicol-resistant strain SLB1355 (Table S1). This plasmid-containing strain was used as host strain to generate ΔnifU conditional mutants (second step). Briefly, a ΔnifU::aphA3 construct was generated by SOE-PCR as follows. Primers ΔnifU-1 and ΔnifU-2 (Table S2) were used to amplify a 510 bp-long DNA sequence located upstream of nifU (hp0221). Primers ΔNifu-3 and ΔNifu-4 were used to amplify a 480 bp-long sequence located downstream of hp0221. The final amplification step included each purified PCR product, a 960 bp-long aphA3 cassette conferring kanamycin resistance (Taylor et al., 1988), and primers ΔnifU-1 and ΔnifU-4. The final 1960 bp-long PCR product containing ΔnifU::aphA3 was introduced by natural transformation into strain SLB1355. Mutants appeared 3 to 5 days after transformation, on plates supplemented with Kan (20 µg/ml) as well as Cm (8 µg/ml) and 1 mM IPTG. The concomitant deletion of nifU and the insertion of aphA3 (mutant strain SLB1372) was confirmed by PCR, using genomic DNA from mutants as template.
Expression and purification of NifS, NifU and Nfu (HP1492)
NifS, NifU and Nfu were expressed as recombinant proteins using E. coli BL21 RIL as host strain. The cloning of nifS (hp0220) or nifU (hp0221) in expression vector pET21b was previously reported (Olson et al., 2000). For Nfu expression, primers Nfu-NdeI and Nfu-XhoI were used to amplify a 270 bp DNA sequence containing the whole hp1492 (nfu) ORF (with its stop codon), as well as to incorporate a 5' NdeI and a 3' XhoI restriction site, respectively. The PCR product was digested with NdeI and XhoI, gel-purified and cloned into similarly digested pET21b plasmid, generating pET-Nfu. E. coli RIL strains harboring either plasmid pET-NifS, pET-NifU or pET-Nfu were grown at 37°C in 800 mL LB supplemented with 0.1 mM FeCl3, 1 mM L-cysteine, ampicillin (100 mg/L) and chloramphenicol (30 mg/L) to an OD600 of 0.3–05. Cultures were cooled at 25°C, protein expression was induced by adding 0.25 mM IPTG in the medium and leaving the cells for 3 to 4 h at 25°C. Cells were harvested by centrifugation (15,000 × g, 20 min, 4°C) and subsequent steps were performed at 4°C. Purification of NifU and NifS was done under reducing, semi-anaerobic conditions, as previously reported (Olson et al., 2000). Purification of Nfu was carried out as follows. Briefly, cells were washed with 200 ml of 50 mM Tris-HCl, pH 7.5, with 25 mM NaCl and 1 mM dithiothreitol (DTT) (buffer A) and resuspended in 5 ml of the same buffer. Phenylmethylsulfonyl fluoride was added to a final concentration of 0.5 mM. Bacteria were lysed by three passages through a cold French pressure cell at 18,000 lb/in2, cell debris were removed by centrifugation at 15,000 × g, and the supernatant was subjected to ultracentrifugation at 100,000 × g for 2 h. The membrane-free supernatant was applied to a 5-ml Q Sepharose anion exchange column (GE healthcare, Piscataway, NJ) and a linear gradient with buffer B (buffer A with 1 M NaCl) was used to purify the protein. Unexpectedly, Nfu did not bind to the Q column under these conditions (despite having a pI of 6.57) and was eluted in the unbound fraction. Fractions of interest were pooled and concentrated using an YM-10 cutoff Centricon device (Millipore, Billerica, MA) to a final volume of 1 ml. The protein was further purified on a size exclusion column (HiLoad 16/60 Superdex 75, GE Healthcare) giving nearly homogeneous protein (>95% purity). Samples of interest were pooled, concentrated again on 10 kDa YMCO Centricon and the protein concentration was determined with the BCA protein kit (Thermo Fisher Pierce, Rockford, IL, USA). Upon purification, both purified NifU and Nfu proteins were red/brown in color, suggesting the presence of Fe-S centers, while purified NifS had a yellow hue, in agreement with the expected presence of the pyridoxal phosphate cofactor, as previously described(Olson et al., 2000). Fe concentrations were determined colorimetrically with bathophenanthroline under reducing conditions, after digesting proteins with KMnO4/HCl, as described by Fish (Fish, 1988).
Size exclusion chromatography (SEC) studies
SEC was used to study protein-protein interactions between purified AcnB, Nfu, NifS and NifU. Each purified protein was loaded either individually or after incubation with another protein (15 min at room temperature) onto a size exclusion column (HiLoad 16/60 Superdex 75). Buffer A (see above) was used at 0.5 ml/min and the elution volume (Ve) for each protein complex was recorded. Fractions of interest were subjected to SDS-PAGE using a Mini-Protean II apparatus (Bio-Rad, Hercules, CA, USA), according to the method of Laemmli (Laemmli, 1970). Chromatograms were integrated and analyzed using Unicorn 5.01 software. Molecular weights were determined by running a standard molecular weight determination kit (Sigma, MW-GF-70). The kit included the following compound and proteins: blue dextran (2,000 kDa), thyroglobulin (669 kDa), apoferritin (443 kDa), BSA (66.2 kDa), Carbonic anhydrase (29 kDa) and cytochrome C (12.4 kDa). The equation used to calculate molecular masses of protein complexes was as follows: Log10 (Mol. Mass) = -1.7274 (Ve/Vo) + 4.7754.
Fe-S cluster reconstitution and spectroscopic characterization of Nfu
Apo H. pylori Nfu was prepared by incubating as-purified samples with 50-fold excess of EDTA and 20-fold excess of potassium ferricyanide for 30 min, followed by anaerobic purification using gel filtration to remove residual iron and sulfide inside a glove box under Ar (< 2 ppm O2). Reconstitution of Fe-S clusters on apo H. pylori Nfu was conducted by incubating 0.2−0.3 mM apo Nfu with 10–20-fold excess of ferrous ammonium sulfate (FAS), 10−20-fold excess of L-cysteine, and a catalytic amount of E. coli IscS under strictly anaerobic conditions. Cluster-bound Nfu was repurified anaerobically using gel filtration and concentrated using YM3 Centricon ultrafiltration. Preparation of the apo protein, cluster reconstitution, repurification, and all sample handling procedures were carried out in a glove box under Ar (< 2 ppm O2).
UV-visible absorption spectra were recorded using septum-sealed 1-mm or 1-cm quartz cuvettes at room temperature, using a Shimadzu UV-3101 PC scanning spectrophotometer. Circular dichroism (CD) spectra were recorded with the same cuvettes using a JASCO J-715 spectropolarimeter (Jasco, Easton, MD). For low temperature resonance Raman (RR) spectra, samples were concentrated to ~2 mM in [2Fe-2S] or [4Fe-4S] clusters and frozen as droplets on an O-ring-sealed gold-plated copper sample holder and mounted to the coldfinger of a Displex Model CSA-202E closed-cycle refrigerator (Air Products, Allentown, PA) at 17 K. The RR spectra were acquired using a Ramanor U1000 spectrometer (Instruments SA, Edison, NJ) coupled with a Sabre argon laser (Coherent, Santa Clara, CA).
Hydrogenase assays
Cells were grown on BA plates, harvested and resuspended in phosphate buffered saline (PBS). Cell density (OD600) was measured and hydrogen uptake in whole cells was followed using a previously described amperometric method (Maier et al., 1996). Activities are reported as nmoles of H2 used per min per 109 cells, and represent 3 to 4 independent measurements.
Aconitase assays
Aconitase assays were done as previously described (Skovran and Downs, 2000), with modifications. Briefly H. pylori WT and Δnfu mutant cells were grown for less than 48 h on BA plates, harvested, washed and broken (by sonication) in ice-cold Tris 100 mM, 20 mM citrate, pH 8. Supernatant (cell-free extract, CFE) was isolated and assays were performed immediately by mixing 50 µg to 150 µg of CFE total protein with 20 mM DL-trisodium isocitrate (in 10 mM Tris, pH 8). Aconitase activity was assayed at room temperature by recording the increase in absorbance at 240 nm (formation of cis-aconitate, ε240= 3,600 M−1) using a Beckman DU640B spectrophotometer. Protein concentration was determined using the BCA kit. Aconitase activity ranged from 80 to 380 nmoles of cis-aconitate formed per min per mg of total protein for WT strain 43504 and 60 to 120 nmoles of cis-aconitate formed per min per mg of total protein for WT strain X47. Results shown represent 3 independent growth experiments, with assays done in triplicate. Results are reported as (mean and standard deviation) percentages activity compared to WT.
Aconitase immunoblotting
Equal amounts of CFE total protein (5 µg) from WT and Δnfu mutants were subjected to SDS-10% PAGE using a Mini-Protean II apparatus (Bio-Rad, Hercules, CA, USA), according to the method of Laemmli (Laemmli, 1970) and transferred to a nitrocellulose membrane (0.2 µm pore size; Bio-Rad). The membrane was blocked by incubation in 20 mM Tris-HCl (pH 7.6) 137 mM NaCl buffer (Tris buffer saline, TBS) supplemented with 3% gelatin (J.T. Baker, Center Valley, PA, USA). This was followed by a 1 h incubation along with a 1:2,000 dilution of anti-E. coli AcnB (rabbit polyclonal) antiserum, in TBS buffer with 0.1% Tween 20 (TTBS), 1% gelatin. The membrane was washed with TTBS and then incubated for 1 h with the secondary antibody (goat anti-rabbit immunoglobulin G coupled with alkaline phosphatase, Bio-Rad) diluted 1:2,000 in TTBS-1% gelatin. The membrane was again washed with TTBS buffer. Bound antibodies were detected following addition of the chromogenic reagents nitro blue tetrazolium (0.25 mg/mL) and 5-bromo-4-chloro-3-indolyl phosphate (0.125 mg/mL) (Sigma, St Louis, MO, USA) in Tris-HCl 10 mM, pH 9.5, NaCl 150 mM. We performed three immunoblottings (independent growth experiments) and used Image J (Schneider et al., 2012) to estimate the density (pixels) of each AcnB-specific band.
Mouse colonization experiments
All procedures were approved by the Institutional Animal Care and Use Committee of the University of Georgia. H. pylori X47 (mouse-adapted, parental strain) and X47 Δnfu mutant strains were grown for 24 hours on BA plates, harvested and resuspended in sterile PBS buffer (pH 7.4) to a final OD600 of 2. Five to six week-old females C57BL/6 mice (Charles River Labs) were infected via oral gavage with 0.15 mL of bacterial suspension (approximately 1.5 × 108 H. pylori cells per mouse). Mice were sacrificed by CO2 asphyxiation and cervical dislocation three weeks post inoculation. Stomachs were quickly removed, weighed, and gently homogenized in 5 mL sterile PBS using Dounce hand homogenizer. Dilutions were made in sterile PBS and plated (0.1 mL) in duplicate on plates supplemented with amphotericin B, bacitracin and vancomycin. Plates were incubated for 5 to 7 days at 37°C in a 4% O2 partial pressure atmosphere. Colonies were then counted and the data are expressed as CFU per gram (g) of mouse stomach. The detection limit of the assay is 150 CFU per g of stomach. Student’s t-test was used to assess significance by comparing geometrical means of colonization capacity between strains.
Genome sequence analysis
Fe-S cluster-containing proteins were predicted by analyzing all open reading frames of strain 26695 and using the following databases and prediction tools: Helicobacter pylori 26695 Biocyc database collection (www.helicobacter.biocyc.org), Genbank (www.ncbi.nlm.nih.gov), Uniprot (www.uniprot.org), EMBL-EBI (www.ebi.ac.uk) and STRING (www.string-db.org).
Bacterial Adenylate Cyclase Two Hybrid (BACTH)
A BACTH-based kit (Euromedex, France) was used to study protein-protein interactions between the three donor proteins, NifS, NifU and Nfu and each of the 36 putative Fe-S-containing target proteins. Each open reading frame (ORF) was PCR-amplified (without start and stop codons) using primers designed to introduce a XbaI restriction site on the 5’ end (PstI for hp0117) and a KpnI restriction site on the 3’ end, respectively (Table S2). Next, each PCR product was digested with XbaI (PstI for hp0117) and KpnI and ligated into similarly digested pUT18C plasmid to generate in-frame gene fusions (Table S1). In addition, nifS, nifU and nfu were also cloned in plasmids pUT18, pKT25 or pKNT25 (Table S1). Ligation mixtures were introduced into E. coli TOP10 and transformants were selected on LB plates supplemented with 100 µg/mL Amp (for pUT18C derivatives) or LB plates supplemented with 30 µg/mL Kan (for pKT25 or pKNT25 derivatives). Recombinant plasmids were verified by restriction profiles and DNA sequencing. Finally, E. coli BTH101 (cya mutant, Euromedex) cells were co-transformed with a combination of one pUT18C derivative and one pK(N)T25 derivative; co-transformants were selected on LB plates supplemented with both Kan and Amp. Individual colonies were picked and grown overnight at 30°C in LB supplemented with both antibiotics. This cell suspension was used as inoculum for all subsequent screening and growth experiments. Interactions between Cya T18- and Cya T25-fusion proteins were analyzed using three complementary methods: 1) screening on MC-Mal plates: clones positive for protein-protein interaction turned red; 2) screening on LB-X-Gal plates: clones positive for protein-protein interaction turned blue; and 3) selection in M63-Mal liquid medium: protein-protein interaction is required for E. coli cya mutants to grow on this medium. In addition to pUT18C-only negative controls, a negative control (pKT25 vector only) was included with each (pUT18C derivative) combination. A positive control (pUT18C-zip with pKT25-zip, provided with the kit) was also included in each experiment. For MC-Mal or LB-X-Gal plates, 1 µL of LB-grown cells was spotted and plates were incubated at 30°C for 48 h under aerobic conditions. For 96-well plates, 2 µL of LB-grown cells were used to inoculate 200 µL of M63 minimal medium (1:100) and plates were incubated at 30°C for 72 h, after which A595 was recorded (Biotek Synergy Mx, Winooski, VT). Each plate included a blank (2 µL of LB as inoculum). Results shown are the average and SD of blank-subtracted A595 from 3 to 5 independent growth experiments.
Supplementary Material
Acknowledgments
This work was supported by the Georgia Research Foundation (RJM) and NIH R37GM62524 (MKJ). We thank Susan Maier for technical assistance with the mouse colonization experiment and Marie-Anaïs Benoit for help with BACTH experiments. We are grateful to Dr. Vincent Starai and his lab (Department of Microbiology, University of Georgia) for the use of the Biotek 96-well microplate reader. We thank Dr. Diana Downs (Department of Microbiology, University of Georgia) for the gift of anti-E. coli AcnB antiserum.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahting U, Mayr JA, Vanlander AV, Hardy SA, Santra S, Makowski C, et al. Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front Genet. 2015;6:123. doi: 10.3389/fgene.2015.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P, Mehta N, Burk A, Maier RJ. Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur. J Bacteriol. 2006;188(14):5325–5330. doi: 10.1128/JB.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283(20):14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- Austin CM, Maier RJ. Aconitase-mediated posttranscriptional regulation of Helicobacter pylori peptidoglycan deacetylase. J Bacteriol. 2013;195(23):5316–5322. doi: 10.1128/JB.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CM, Wang G, Maier RJ. Aconitase Functions as a Pleiotropic Posttranscriptional Regulator in Helicobacter pylori. J Bacteriol. 2015;197(19):3076–3086. doi: 10.1128/JB.00529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Shen G, Bryant DA, Golbeck JH. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 2006;188(9):3182–3191. doi: 10.1128/JB.188.9.3182-3191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008a;27(7):1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008b;283(20):14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185(16):4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit SL, Maier RJ. Twin-arginine translocation system in Helicobacter pylori: TatC, but not TatB, is essential for viability. MBio. 2014;5(1):e01016–01013. doi: 10.1128/mBio.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca IG, Ecobichon C, Chaput C, Mathieu A, Guadagnini S, Prevost MC, et al. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 2008;74(7):2095–2102. doi: 10.1128/AEM.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czernuszewicz RS, Macor KA, Johnson MK, Gewirth A, Spiro TG. Vibrational mode structure and symmetry in proteins and analogues containing Fe4S4 clusters: Resonance Raman evidence for different degrees. J Am Chem Soc. 1987;109:7178–7187. [Google Scholar]
- di Maio D, Chandramouli B, Yan R, Brancato G, Pastore A. Understanding the role of dynamics in the iron sulfur cluster molecular machine. Biochim Biophys Acta. 2017;1861(1 Pt A):3154–3163. doi: 10.1016/j.bbagen.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, et al. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology. 2005;151(Pt 2):533–546. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- Gao H, Subramanian S, Couturier J, Naik SG, Kim SK, Leustek T, et al. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry. 2013;52(38):6633–6645. doi: 10.1021/bi4007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell TA, Goldman PJ, Drennan CL. SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J Biol Chem. 2015;290(7):3964–3971. doi: 10.1074/jbc.R114.581249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Basu MK. Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J Bacteriol. 2011;193(11):2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R, Ceol A, Rajagopala SV, Mosca R, Siszler G, Wermke N, et al. A second-generation protein-protein interaction network of Helicobacter pylori. Mol Cell Proteomics. 2014;13(5):1318–1329. doi: 10.1074/mcp.O113.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48(26):6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219(1–2):49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95(10):5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Bothe JR, Frederick RO, Holder JC, Markley JL. Role of IscX in iron-sulfur cluster biogenesis in Escherichia coli. J Am Chem Soc. 2014;136(22):7933–7942. doi: 10.1021/ja501260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous H, Tibbitts TJ, Gray HL, Myers GA, Lee CK, Ermak TH, Monath TP. Sterilizing immunity against experimental Helicobacter pylori infection is challenge-strain dependent. Vaccine. 2001;19(32):4883–4895. doi: 10.1016/s0264-410x(01)00248-1. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cowan JA. Iron sulfur cluster biosynthesis. Human NFU mediates sulfide delivery to ISU in the final step of [2Fe-2S] cluster assembly. Chem Commun (Camb) 2007;(30):3192–3194. doi: 10.1039/b704928e. [DOI] [PubMed] [Google Scholar]
- Liu Y, Qi W, Cowan JA. Iron-sulfur cluster biosynthesis: functional characterization of the N- and C-terminal domains of human NFU. Biochemistry. 2009;48(5):973–980. doi: 10.1021/bi801645z. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Fu C, Gilbert J, Moshiri F, Olson J, Plaut AG. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141(1):71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Spectroscopic and functional characterization of iron-sulfur cluster-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry. 2012;51(41):8071–8084. doi: 10.1021/bi3006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, et al. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol. 2015;95(3):383–409. doi: 10.1111/mmi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileni M, MacMillan F, Tziatzios C, Zwicker K, Haas AH, Mantele W, et al. Heterologous production in Wolinella succinogenes and characterization of the quinol:fumarate reductase enzymes from Helicobacter pylori and Campylobacter jejuni. Biochem J. 2006;395(1):191–201. doi: 10.1042/BJ20051675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morningstar JE, Johnson MK, Cecchini G, Ackrell BA, Kearney EB. The high potential iron-sulfur center in Escherichia coli fumarate reductase is a three-iron cluster. J Biol Chem. 1985;260(25):13631–13638. [PubMed] [Google Scholar]
- Nishio K, Nakai M. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J Biol Chem. 2000;275(30):22615–22618. doi: 10.1074/jbc.C000279200. [DOI] [PubMed] [Google Scholar]
- Olson JW, Agar JN, Johnson MK, Maier RJ. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry. 2000;39(51):16213–16219. doi: 10.1021/bi001744s. [DOI] [PubMed] [Google Scholar]
- Parrish JR, Yu J, Liu G, Hines JA, Chan JE, Mangiola BA, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8(7):R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Hoover TR. Stable accumulation of sigma54 in Helicobacter pylori requires the novel protein HP0958. J Bacteriol. 2005;187(13):4463–4469. doi: 10.1128/JB.187.13.4463-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson SM, Mendz GL, Srinivasan S, Hazell SL. The tricarboxylic acid cycle of Helicobacter pylori. Eur J Biochem. 1999;260(1):258–267. doi: 10.1046/j.1432-1327.1999.00153.x. [DOI] [PubMed] [Google Scholar]
- Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, et al. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat Commun. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, et al. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol Microbiol. 2012;86(1):155–171. doi: 10.1111/j.1365-2958.2012.08181.x. [DOI] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409(6817):211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186(23):7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96(18):10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134(37):15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, et al. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8(4):e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovran E, Downs DM. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar typhimurium: implications for thiamine synthesis. J Bacteriol. 2000;182(14):3896–3903. doi: 10.1128/jb.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DE, Yan W, Ng LK, Manavathu EK, Courvalin P. Genetic characterization of kanamycin resistance in Campylobacter coli. Ann Inst Pasteur Microbiol. 1988;139(6):665–676. doi: 10.1016/0769-2609(88)90071-3. [DOI] [PubMed] [Google Scholar]
- Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem. 2004;136(2):199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Tong WH, Jameson GN, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci U S A. 2003;100(17):9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine B, Boutin JP, Marion-Poll A, Briat JF, Peltier G, Lobreaux S. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 2004;40(1):101–111. doi: 10.1111/j.1365-313X.2004.02189.x. [DOI] [PubMed] [Google Scholar]
- Veyrier FJ, Ecobichon C, Boneca IG. Draft genome sequence of strain X47-2AL, a feline Helicobacter pylori isolate. Genome Announc. 2013;1(6) doi: 10.1128/genomeA.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5(5):e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94(1):23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M. The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell. 2004;16(4):993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Yamashita E, Kikuchi A, Morimoto K, Nakagawa A, Tsukihara T, Nakai M. Structural analysis of Arabidopsis CnfU protein: an iron-sulfur cluster biosynthetic scaffold in chloroplasts. J Mol Biol. 2008;381(1):160–173. doi: 10.1016/j.jmb.2008.05.072. [DOI] [PubMed] [Google Scholar]
- Yan R, Konarev PV, Iannuzzi C, Adinolfi S, Roche B, Kelly G, et al. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J Biol Chem. 2013;288(34):24777–24787. doi: 10.1074/jbc.M113.480327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.