SUMMARY
Beyond sample curation and basic pathologic characterization, the digitized H&E-stained images of TCGA samples remain underutilized. To highlight this resource, we present mappings of tumor-infiltrating lymphocytes (TILs) based on H&E images from 13 TCGA tumor types. These TIL maps are derived through computational staining using a convolutional neural network trained to classify patches of images. Affinity propagation revealed local spatial structure in TIL patterns and correlation with overall survival. TIL map structural patterns were grouped using standard histopathological parameters. These patterns are enriched in particular T cell subpopulations derived from molecular measures. TIL densities and spatial structure were differentially enriched among tumor types, immune subtypes, and tumor molecular subtypes, implying that spatial infiltrate state could reflect particular tumor cell aberration states. Obtaining spatial lymphocytic patterns linked to the rich genomic characterization of TCGA samples demonstrates one use for the TCGA image archives with insights into the tumor-immune microenvironment.
In Brief
Tumor-infiltrating lymphocytes (TILs) were identified from standard pathology cancer images by a deep-learning-derived “computational stain” developed by Saltz et al. They processed 5,202 digital images from 13 cancer types. Resulting TIL maps were correlated with TCGA molecular data, relating TIL content to survival, tumor subtypes, and immune profiles.
INTRODUCTION
Although studies in humans have shown that chronic inflammation can promote tumorigenesis (Trinchieri, 2012), the host immune system is equally capable of controlling tumor growth through the activation of adaptive and innate immune mechanisms (Galon et al., 2013). Such intratumoral processes are often referred to collectively as immunoediting, where this selective pressure can result in the emergence of tumor cells that escape immune surveillance and, ultimately, to tumor progression. At the same time, many observations suggest that high densities of tumor-infiltrating lymphocytes (TILs) correlate with favorable clinical outcomes (Mlecnik et al., 2011a) such as longer disease-free survival or improved overall survival (OS) in multiple cancer types (Angell and Galon, 2013). Recent studies further suggest that the importance of spatial context and the nature of cellular heterogeneity of the tumor microenvironment, in terms of the immune infiltrate involving the tumor center and/or invasive margin, can also correlate with cancer prognosis (Fridman et al., 2012). Prognostic factors, most notably the Immunoscore, that quantify such spatial TIL densities in different tumor regions have high prognostic value that can significantly supplement and sometimes even supersede the standard TNM classification and staging in certain settings(Galon et al., 2006; Broussard and Disis, 2011; Mlecnik et al., 2011b). Given this and the central role of immunotherapy treatments in contemporary cancer care, these assessments of tumor-associated lymphocytes are increasingly important both in the clinical assessment of pathology slides, as well as in translational research into the role of these lymphocytic populations.
Tissue diagnostic studies are carried out and interpreted by pathologists for virtually all cancer patients, and the overwhelming majority of these are stained with hematoxylin and eosin (H&E). The TCGA Pan Cancer Atlas dataset includes representative H&E diagnostic whole-slide images (WSIs) that enable spatial quantification and analysis of TILs and association with the wealth of molecular characterization conducted through the TCGA. Previously, this rich trove of imaging data has primarily been used solely to qualify samples for TCGA analysis and gleaning of some limited histopathologic parameters by expert pathologists. Using digital pathology and digitized whole-slide diagnostic tissue images, machine learning and deep learning approaches can create a “Computational Stain.” This allows identification and quantification of image features to formulate higher-order relationships that go beyond simple densities (e.g., of TILs) to explore quantitative assessments of lymphocyte clustering patterns, as well as characterization of the interrelationships between TILs and tumor regions. We apply this to the TCGA samples in a broad multi-cancer fashion. Only a few TCGA tumor types have been explored for TIL content based on feature extraction from histologic H&E images and in a more limited fashion (Rutledge et al., 2013; Cancer Genome Atlas Research Network, 2017).
Over the past 12 years, The Cancer Genome Atlas (TCGA) has profoundly illuminated the genomic landscape of human malignancy. More recently, it has been recognized that genomic data derived from bulk tumor samples, which include the tumor stromal, vascular, and immune compartments, as well as tumor cells, can provide detailed information about the tumor immune microenvironment. Molecular subtypes of ovarian, melanoma, and pancreatic cancer have been defined based on measures of immune infiltration (Cancer Genome Atlas Research Network, 2011; Cancer Genome Atlas Network, 2015; Bailey et al., 2016), and a number of other tumors show variation in immune gene expression by molecular subtype (Iglesia et al., 2014, 2016; Kardos et al., 2016). Recent publications (Charoentong et al., 2017; Li et al., 2016; Rooney et al., 2015) have presented comprehensive analyses of TCGA data on the basis of immune content response. A recent study (Thorsson et al., 2018) reports on a series of immunogenomic characterizations that include assessments such as total lymphocytic infiltrate, immune cell type fractions, immune gene expression signatures, HLA type and expression, neoantigen prediction, T cell and B cell repertoire, and viral RNA expression. From these base-level results, integrative analyses were performed to derive six immune subtypes, spanning tumor types and subtypes. The comprehensive pairing of clinical, sample, molecular tumor, and immune characterizations with H&E WSIs in the TCGA is a unique resource (Cooper et al., 2017) and offers the possibility of identifying relationships between computational staining of whole-slide images and other measures of immune response that may in turn inform research into immuno-oncological therapy. In this work, we characterize spatial patterns of TILs and present relationships between TIL patterns and immune subtypes, tumor types, immune cell fractions, and patient survival, illustrating the potential of this kind of analysis and the kinds of questions that can be explored. For example, through integration of spatial patterns with molecular TIL characterization, we found evidence for these patterns being enriched in particular T cell populations.
This study represents an important milestone in the use of digital-pathology-based quantification as we are able to present results relating spatial and molecular tumor immune characterizations for roughly 5,000 patients with 13 cancer types. TILs and spatial characterizations of TILs have shown significant value in diagnostic and prognostic settings, and the ability to quantify TILs from diagnostic tissue has proven to be demanding, expensive, challenging to scale, and beleaguered by subjectivity. Human review of diagnostic tissue is highly effective for traditional diagnosis but is qualitative and thus is prone to both inter- and intra- observer variability, particularly when attempting to quantify or reproducibly characterize feature-rich phenomena such as tumor-associated lymphocytic infiltrates. The spatial characterizations we present are high resolution, with TIL infiltration assessed in whole-slide images at a 50-micron resolution, and all TIL maps are available to the scientific community for further exploration. The recent FDA approval (FDA News Release, 2017) of whole-slide imaging for primary diagnostic use is leading to more widespread adoption of digital whole-slide imaging. It is widely expected that, within 5–10 years, the great majority of new pathology slides will be digitized, thus enabling the development and clinical adoption of various digital-pathology-based diagnostic and prognostic biomarkers that will likely provide decision support for traditional pathologic interpretation in the clinical setting.
RESULTS
Generating Maps of Tumor-Infiltrating Lymphocytes using Convolutional Neural Networks
In order to accurately generate maps of tumor-infiltrating lymphocytes (TIL Maps) from digitized H&E stained tissue specimens, we developed a comprehensive methodology and accompanying interactive tools. This methodology is termed Computational Staining and employs deep learning methods to analyze images and tools to incorporate expert feedback into the deep learning models. Such iterative feedback results in the improvement of the overall accuracy of TIL Maps. Key highlights and the validation strategy for Computational Staining are presented here, with further details provided in the Method Details.
Computational Staining uses convolutional neural networks (CNNs) to identify lymphocyte-infiltrated regions in digitized H&E stained tissue specimens. The CNN is a supervised deep learning method that has been successfully applied in a large number of image analysis problems (Cireşan et al., 2013; Huang et al., 2016; Xie et al., 2015a, 2015b; Wang et al., 2016; Sirinukunwattana et al., 2016; Bayramoglu and Heikkila, 2016; Su et al., 2015; Hou et al., 2016a; Murthy et al., 2017;Chen et al., 2017; Xu and Huang, 2016). A CNN first uses a set of training data to learn a classification (or predictive) model in the training phase. The resulting trained model is then used to classify new data elements in a prediction phase. Deep-learning-based automatic analysis methods generally require large annotated datasets. Many state-of-the-art methods employ semi-supervised training strategies to boost trained model performance using unlabeled data (Ranzato et al., 2006; Masci et al., 2011; Bayramoglu and Heikkila, 2016; Xu and Huang, 2016; Su et al., 2015). They (1) pre-train an autoencoder for unsupervised representation learning; (2) construct a CNN from the pretrained autoencoder; and (3) fine-tune the constructed CNN for supervised classification. One can train the unsupervised autoencoder on image patches with the object to be classified (e.g., nucleus) in the center of each patch (Hou et al., 2016a; Murthy et al., 2017) in order to capture the visual variance of the object more accurately. This method, however, requires a separate object detection step. Instead of tuning the detection and classification modules separately, recent studies (Graves and Jaitly, 2014; Ren et al., 2015; Redmon et al., 2016; Kokkinos 2017) have developed CNNs to perform these tasks in a unified but fully supervised pipeline.
Our methodology uses two CNNs: a lymphocyte infiltration classification CNN (lymphocyte CNN) and a necrosis segmentation CNN (necrosis CNN). The lymphocyte CNN categorizes tiny patches of an input image into those with lymphocyte infiltration and those without. It is a semi-supervised CNN, initialized by an unsupervised convolutional autoencoder (CAE). The necrosis CNN segments the regions of necrosis and is designed to eliminate false positives from necrotic regions where nuclei may have characteristics similar to those in lymphocyte-infiltrated regions. Details about the two CNNs are shown in Figure S1A and described in the Method Details.
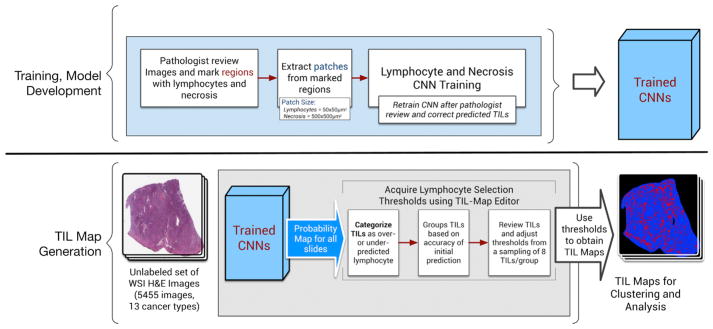
Figure 1 illustrates both the training and model development phase of our methodology (top half of the figure) and the use of the trained model to generate TIL Maps (bottom half of the figure). The CNN training and model development phase starts with expert pathologists reviewing a set of images and marking regions of lymphocytes and necrosis. The lymphocyte and necrosis regions are then subdivided into tiny patches to create the initial training dataset. Training with patches rather than with individual regions and cells is done for computational efficiency. The lymphocyte CNN is trained with 50 × 50 μm2 patches (equivalent to 100 × 100 square pixel patches in tissue images acquired at 20× magnification level) from WSIs. The necrosis CNN is trained with larger patches of size 500 × 500 μm2, as more contextual information results in superior prediction of patches being necrotic. The initial training step is followed by an iterative cycle of review and refinement steps to improve the prediction accuracy of the lymphocyte CNN. This prediction step generates a probability value of lymphocyte infiltration for each patch in the images. The patch-level predictions for an image are combined and represented to pathologists as a heatmap for review and visual editing using our TIL-Map editor tool. The pathologists refine the CNN predictions for an image by first adjusting the probability value threshold (which globally updates the labels of the patches in the image; if the probability value of a patch exceeds the adjusted threshold, the patch is labeled a TIL patch) and then manually editing the heatmap to correct prediction errors for individual or groups of patches. At the end of the editing step, the updated heatmaps are processed to augment the training dataset. The lymphocyte CNN is re-trained with the updated training dataset. This iterative process continues until adequate prediction accuracy is achieved, as determined by the pathologist feedback. The necrosis CNN was retrained only once in this study, because it achieved sufficient prediction accuracy. The training and re-training steps of both CNNs involve cross-validation to assess prediction performance and avoid overfitting (Hou et al., 2017). See the Method Details for an in-depth description of this process.
Figure 1. Workflow for Training, Model Development, and Subsequent Generation of TIL Maps.
Top: for training and developing CNN models, a pathologist reviews images and marks regions with lymphocytes and necrosis. These training data are then broken down into patches that are then fed into a training stage to train CNNs for lymphocyte and necrosis detection. A pathologist periodically reviews the results for accuracy and corrects the prediction. This results in a pair of Trained CNNs. Bottom: these trained CNNs are then used on the full set of 5,455 images from 13 cancer types to generate TIL maps. During TIL map generation, a probability map for TILs is generated from each image. These probabilities are then reviewed and lymphocyte selection thresholds are established using a selective sampling strategy (further information in Method Details). These thresholds are then used to obtain the final TIL maps. See also Figure S1 and Tables S1 and S2.
The trained models are used on test datasets (bottom half of Figure 1). In this work, we applied our method to 5,455 diagnostic H&E WSIs from 13 TCGA tumor types in which lymphocytes are known to be present. See Additional Resources for listing and acronyms. We included uveal melanoma (UVM) as one of the 13 cancer types essentially as a type of negative control (Figure S3A), since it has the fewest immune cells among TCGA tumors (Thorsson et al., 2018). Tumor types were selected to represent a range of known positive involvement of lymphocytes and immunogenicity from literature and from molecular estimates of lymphocyte content. Each image was partitioned into patches of 50 × 50 μm2 and each patch was classified by the CNNs. TIL maps were successfully generated (see Figure S1C and Table S2) for 5,202 TCGA tumor images from 4,759 individual participants in the 13 tumor types. 253 images (4.6%) did not yield TIL maps because of low image quality or low prediction accuracy or because the images were duplicates (see Figure S1C).
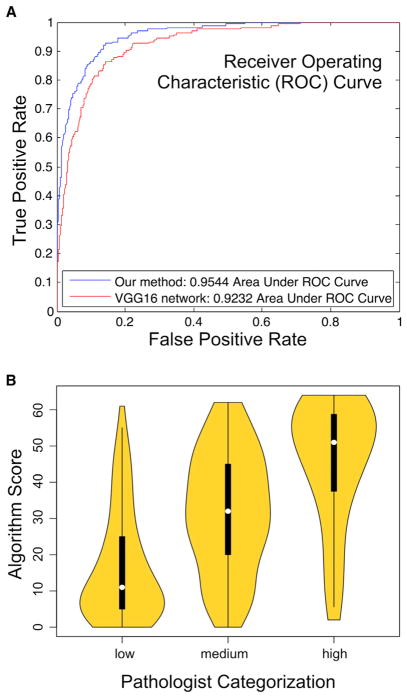
We assessed the performance of our approach in two complementary, yet orthogonal ways. The first assessment method, described in Zhao et al. (2017), compares performance prediction of our method with that of a popular and widely used CNN—called VGG16 (Simonyan and Zisserman, 2014)—using a set of WSIs from TCGA lung adenocarcinoma (LUAD) cases. The training set of the lymphocyte CNN consisted of 20,876 patches. Each patch usually contains 0 to 30 nuclei and was annotated by a pathologist as lymphocyte infiltrated or not lymphocyte infiltrated. The training set of the necrosis segmentation CNN consisted of 1,800 patches. Each patch was annotated with a necrosis region mask segmented by a pathologist. We sampled 2,480 patches to create the test dataset. The ROC curve shows that our approach slightly outperforms VGG16 by 3.1% with respect to the area under ROC curve (AUROC) metric (Figure 2A). We also performed direct comparison of TIL patch assignments by the Computational Staining pipeline with those by experienced pathologists by scoring 8 × 8 “super-patches” for TIL content. Three pathologists assessed 400 super-patches as having low, medium, or high TIL content, while machine-derived scores were assigned for the patch by counting TIL-positive patches (thus ranging from 0 to 64). Consistency was high among each of the pathologists (> 80%), as assessed by rescoring of 100 super-patches. As seen in Figure 2B, the median machine-derived score is quite distinct between the three ordinal bins. This is evidenced in strong correlation as assessed by the polyserial coefficient (Drasgow, 2014), designed for comparing ordinal with continuous values (0.36 with 95% CI [0.27,0.45], p value = 5.2 × 10−15, R package polycor).
Figure 2. Assessment of TIL Prediction.
(A) Receiver Operating Characteristic depicting performance of CNN. Applied to TCGA lung adenocarcinoma patches. The current method is compared with a popular CNN called VGG16 (see main text description).
(B) Comparison of TIL scores of super-patches between pathologists and computational stain. x axis: median scores from three pathologists assessing 400 super-patches as having low, medium, or high lymphocyte infiltrate. y axis: scores from computational staining, on a scale from 0 to 64.
Assessment and Correlates of TIL Spatial Fraction
Spatial Fraction of TILs
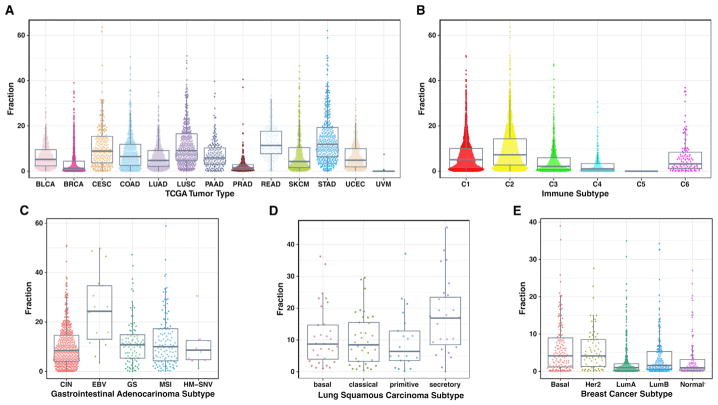
The spatial fraction of TILs was estimated as the fraction of TIL-positive patches among the total number of patches identified on the tissue sample. A wide range in spatial infiltrate is seen among the TCGA tumor types (Figure 3A and Table S1), with high infiltrates in gastric cancer (STAD) with a mean of 14.6%, rectal cancer (READ) at 13.0%, squamous cell carcinoma in the lung (LUSC) at 11.6%, while uveal melanoma (UVM) has only 1% estimated TIL fraction, consistent with its inclusion as negative control (Figure S3A). Wide differences are also seen grouping tumors by the nature of the immune response, according to a recent immune characterization of all TCGA tumors (Thorsson et al., 2018)(Figure 3B). The most immunologically active immune subtypes (e.g., C1, C2) tend to have the greatest spatial infiltration of lymphocytes. Within documented TCGA subtypes, which are typically characterized by specific molecular changes in tumor cells, strong differences are also seen (Figure S2A). EBV-positive gastric cancer is particularly rich in TILs, with an average of 25% of spatial regions infiltrated by TILs (Figure 3C). The lung squamous secretory subtype (Wilkerson et al., 2010) is also particularly rich in infiltrate (17%, Figure 3D) as is the mutation-rich POLE subtype of endometrial cancer. Among breast cancer tumors, the basal subtype has the greatest infiltrate (Figure 3E), consistent with what has been observed in other studies (Iglesia et al., 2014). Taken together, these data show that the nature of the infiltrate has strong ties to aspects of the tumor microenvironment and that the nature of the infiltrate may be reflective of particular molecular aberration states of tumor cells.
Figure 3. TIL Fraction by Tumor Category.
(A–E) Percent TIL fraction, the proportion of TIL-positive patches within a TIL map, is shown by various categorizations of TCGA tumor samples. Each plotted point represents a tumor sample for (A) 13 TCGA tumor types (4,612 cases), (B) six subtypes characterized by differences in the nature of the overall immune response (Thorsson et al., 2018) (C5 has very few samples here), (C) gastrointestinal adenocarcinoma subtypes, (D) lung squamous cell carcinoma subtypes, and (E) breast adenocarcinoma subtypes. See also Figure S2.
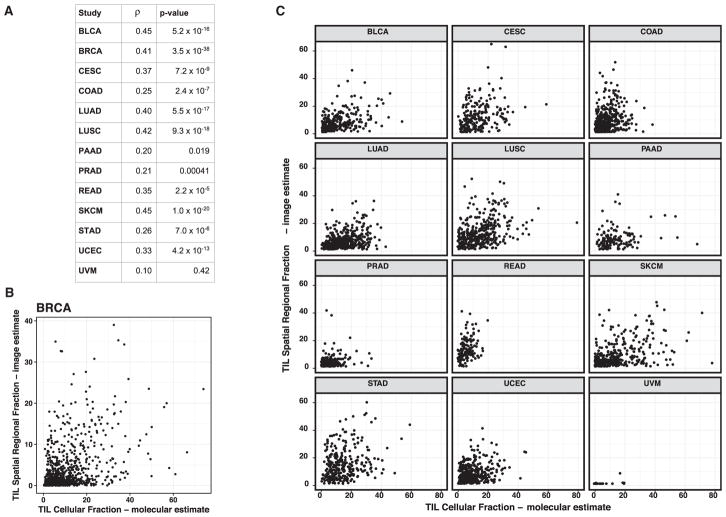
The spatial fraction of TILs was compared with molecular estimates of TIL content from molecular genomics assays (Thorsson et al., 2018). The molecular estimate of TIL fraction is obtained by multiplying an estimate of the overall leukocyte fraction, based on DNA methylation arrays, with an estimate of the lymphocyte proportion within the immune compartment obtained by applying CIBERSORT (Newman et al., 2015) to RNA sequencing data. Good, albeit imperfect, agreement is seen between the imaging and molecular estimates (Figure 4A), with Spearman correlation values ranging from 0.20 to 0.45 for the most part accompanied by highly significant p values, and with UVM, the negative control, showing no correlation. The reasons for the differences between the molecular estimates and spatial TIL fraction include: (1) molecular data are extracted from a fresh frozen tissue section in proximity to the formalin-fixed paraffin-embedded (FFPE) sample used to generate the diagnostic H&E image, but the exact spatial relation is unknown; (2) the molecular estimate is proportional to the number of lymphocytes, whereas the spatial fraction of TILs is estimated by tissue area; (3) the spatial analysis and TIL fraction are an assessment of lymphocyte-infiltrated tissue that can also include non-tumor regions on the diagnostic slides; and (4) the molecular quantification is obtained from frozen sections that are highly enriched for tumor as a criterion for project inclusion. We further examined the outlier cases (see Figures 4B and 4C) having high levels of discordance between molecular and spatial image-derived TIL estimates for several tumor types, including BRCA, SKCM, LUAD, LUSC, STAD, and READ. We determined that spatial TILs in non-tumor regions appeared to play a major explanatory role (Figures S3B and S3C). Attempts to exclude such areas by manual negative masking and/or CNN-based automation for tumor recognition will be included in future efforts in order to reduce the discordance between the molecular estimates from samples that are highly enriched for tumor and the spatial TIL estimates derived from diagnostic H&E images.
Figure 4. Comparison of TIL Proportion from Imaging and Molecular Estimates.
(A) Spearman correlation coefficients and p values for comparison of TIL fraction from spatial estimates of TIL maps and molecular estimates of TIL fraction from processing of cancer genomics data using deconvolution methods (see main text).
(B) Each point represents a breast adenocarcinoma tumor sample, with the value of TIL fraction from TIL maps (x axis) and from molecular estimates (y axis).
(C) As in B for 12 additional TCGA tumor types. See also Figure S3 and the companion manuscript (Thorsson et al., 2018).
Automated Assessment of Local Structures in the TIL Infiltrate and Association with Molecular and Clinical Readouts
Local Spatial Structure of the Immune Infiltrate
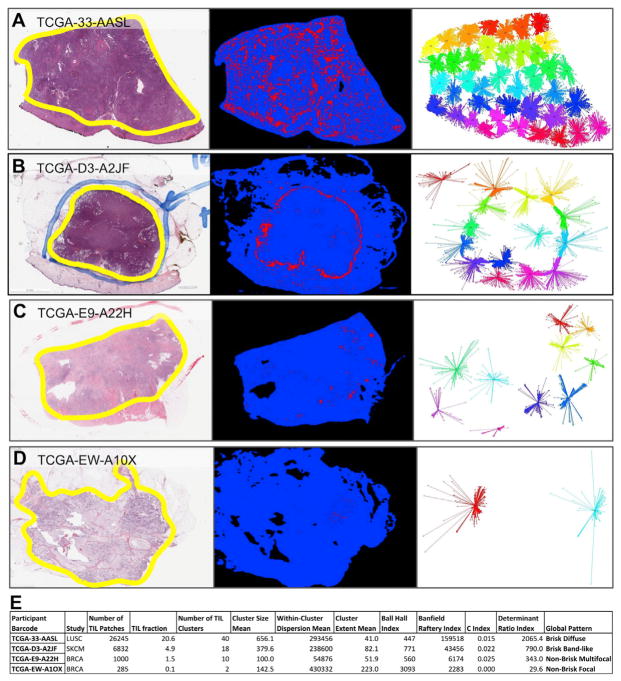
A unique feature of imaging data is the ability to go beyond total lymphocytic infiltrate load to the assessment of patterns of lymphocytic infiltration. To identify such patterns, we first used affinity propagation (Frey and Dueck, 2007) to find spatially connected and coherent regions (clusters) of TIL image patches (APCluster R package; Bodenhofer et al., 2011). Examples of H&E images, TIL maps, and clusters are shown in Figures 5A–5D for selected cases exemplifying sparse and dense lymphocyte infiltrates. For each slide, the resulting cluster pattern was characterized using measures for simple count and extent statistics but also by clustering indices, which assess more complex characteristics such as cluster shape. Summary measures include the number of clusters Ncluster, the mean number of TIL patches in the clusters NP, the mean of the within-cluster dispersion WCD, and the mean of cluster spatial extents CE (see Figure 5E, Method Details, and Table S1). In terms of TIL patch distances to a given cluster center, the dispersion is related to their variance, while spatial extent is akin to the maximal distance. Ncluster ranged from 2 to 46 over the entire cohort (4,480 cases, excluding non-tumor slides), with a median of 12, and the mean cluster membership was 293 TIL patches. We calculated the clustering indices of Ball and Hall (1965), Banfield and Raftery (1993), the C index, and the determinant ratio index, as implemented in the R package clusterCrit (see Method Details and Table S1). The Ball-Hall index is the mean of the dispersion through all of the clusters, equivalent to the mean of the squared distances of the points of the cluster with respect to its center. In our data, the Ball-Hall index is correlated (ρSpearman = 0.95) with the mean cluster extent, CE. The Banfield-Raftery index is the weighted sum of the logarithms of the mean cluster dispersion, which in our data correlates with Ncluster (ρSpearman = 0.95). We found similarity among several of the various scores (Figure S4A), including overall trending of some clustering indices to simpler measures such as Ncluster and TIL fraction. The C index is derived from pairwise distances and does not scale with any of the simpler measures. Values of these scores for the cases depicted in Figures 5A–5D are shown in Figure 5E. Clustering indices vary widely over slides, as illustrated in Figure 6A for the Ball-Hall index. Tumors with relatively high values of this index, such as BRCA and PRAD, are not among those with highest overall infiltrate (Figure 3A). Since the Ball-Hall index scales with approximately cluster extent, this implies that, in some of these tumor types of moderate infiltrate mass, TIL clusters of relatively large spatial extent are formed. In summary, this implies that, in some tumor types, local clustering of TILs may be a more distinctive feature than overall TIL infiltrate, in comparison with other tumor types.
Figure 5. Examples of TIL Map Structural Patterns.
(A–D) Four cases representing different degrees of lymphocyte infiltration. Each example is labeled by TCGA participant barcode and has the following three panels. Left: H&E diagnostic image at low magnification with tumor regions circled in yellow; middle: TIL map; red represents a positive TIL patch, blue represents a tissue region with no TIL patch, while black represents no tissue; right: diagrams of clusters of TIL patches derived from the affinity propagation clustering of the TIL patches. Line segments connect cluster members with a central representative for each cluster, and colors are arbitrarily assigned to aid visual separation of clusters.
(E) TIL map, cluster statistics, and global patterns for the four examples in A–D. Each column represents one way to characterize the TIL map, ranging from simple measures such as TIL count and density to more complex ones characterizing details of cluster properties and image patterns (see main text). See also Table S2.
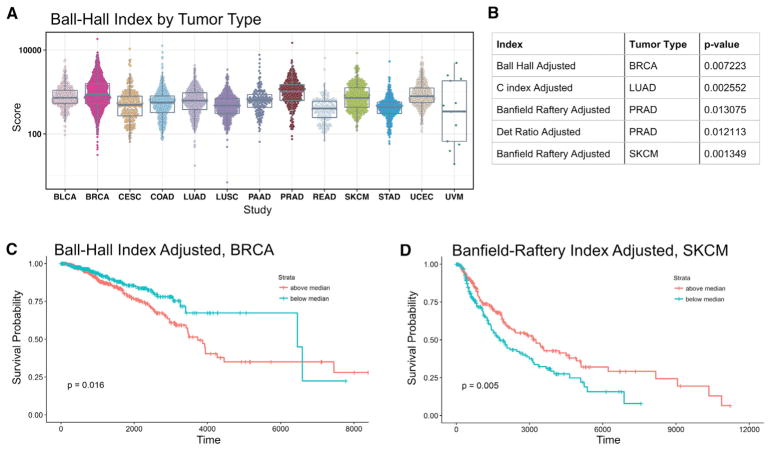
Figure 6. Associations of TIL Local Spatial Structure with Cancer Type and Survival.
Associations are shown with cluster indices, which summarize properties of clusters derived from affinity propagation clusters of the TIL map—properties that provide details on local structure beyond simple densities.
(A) Ball-Hall cluster indices for all slide images considered in the study. The Ball-Hall index is a particular clustering index, summarizing the mean, through all the clusters, of their mean dispersion and is equivalent to the mean of the squared distances of the points of the cluster with respect to its center. In our data, the Ball-Hall index is correlated (ρSpearman = 0.95) with the mean cluster extent, CE.
(B) Table of significant associations between TIL fraction-adjusted cluster indices and overall survival based on Cox regression, accounting for age and gender as additional clinical covariates.
(C) Overall survival for median-stratified TIL fraction-adjusted Ball-Hall index in breast cancer. Significance test p value is shown in the lower left.
(D) Same as C but for adjusted Banfield-Raftery index in skin cutaneous melanoma. The Banfield-Raftery index is the weighted sum of the logarithms of the mean cluster dispersion and, in our data, often correlates with the number of clusters. See also Figure S4.
Correlates of Local TIL Spatial Structure with Survival
We examined the extent to which TIL fraction might impact overall survival and the extent to which spatial characteristics of the tumor microenvironment—beyond overall densities—may provide additional predictive power of outcome. We used Cox regression, accounting for age and gender as additional clinical covariates to perform survival analysis. In order to mitigate possible problems in interpretation due to the inherent correlation between some clustering indices and the TIL densities, we used linear regression to obtain adjusted cluster indices by computing residuals with respect to TIL density (see Method Details). p values were obtained for four adjusted indices and 13 tumor types, which were then adjusted for multiple testing using the Benjamini-Hochberg procedure. Five associations between cluster index and outcome were significant (at p < 0.05) and are shown in Figure 6B. Interestingly, the various indices were significant across different tumor types. Examples of Kaplan-Meier curves for median-split clustering indices are shown in Figures 6C (BRCA) and 6D (SKCM). In SKCM, increased Banfield Raftery-index (“cluster count”) associates with superior survival, while in BRCA increased Ball-Hall index (“cluster extent”) associates with inferior survival, both adjusted for overall TIL density. Of interest, checkpoint inhibition immunotherapy has been successfully applied to melanoma, while breast cancer tumors have generally been unresponsive to checkpoint blockade therapy. The association of structure with survival, as evidenced by less favorable survival in tumors with elevated adjusted Ball-Hall index (“cluster extent”) could be worthy of further investigation as a stratification factor for patient tumors in clinical studies of response.
Characterization of Overall TIL Map Structural Patterns and Association with Molecular Estimates
We undertook further characterization of TIL spatial structure, looking beyond local spatial structures toward a global structure classification that reflects standard descriptions in current use by practicing pathologists. We incorporated qualitative and semiquantitative descriptions and scoring of the TIL map structural patterns in the combined intratumoral and peri-tumoral regions (collectively referred to as “tumor”) that are grossly defined by the corresponding H&E-stained whole-slide images.
As seen in the recommendations from of the International TILs Working Group (Salgado et al., 2015), International Immuno-oncology Biomarkers Working Group (Hendry et al., 2017a, 2017b), and the prognostic descriptions used to characterize TILs in cutaneous melanoma (Crowson et al., 2006), pathologists classify patterns within the TIL maps in both the intratumoral and peritumoral regions. Correspondingly, patterns in the 5,202 TIL maps were visually assigned by a pathologist into one of five categories: “Brisk, diffuse” for diffusely infiltrative TILs scattered throughout at least 30% of the area of the tumor (1,856 cases); “Brisk, band-like” for immune responses forming band-like boundaries bordering the tumor at its periphery (1,185); “Non-brisk, multi-focal” for loosely scattered TILs present in less than 30% but more than 5% of the area of the tumor (1,083); “Non-brisk, focal” for TILs scattered throughout less than 5% but greater than 1% of the area of the tumor (874); and finally “None” in 143 cases where few TILs were present involving 1% or less of the area of the tumor (see Method Details). TIL maps with corresponding H&E images with insufficient or no grossly identifiable tumor at low magnification were designated as indeterminate (61). The examples in Figures 5A–5D are categorized as follows: Figure 5A, TCGA-33-AASL Brisk, diffuse pattern in a case of squamous cell carcinoma of the lung showing a relatively strong immune infiltrate within the tumor; Figure 5B, TCGA-D3-A2JF Brisk, band-like pattern in a case of cutaneous melanoma showing immune infiltrates forming boundaries bordering the tumor at its periphery and < 30% TILs in the intratumoral component; Figure 5C, TCGA-E9-A22H Non-brisk, multi-focal pattern in a case of invasive ductal carcinoma of the breast showing a weak immune response with loosely scattered TILs; Figure 5D, TCGA-EW-A1OX Non-brisk, focal pattern in a case of invasive ductal carcinoma of the breast showing a very weak immune response in a focal area (categories also listed in final column of Figure 5E).
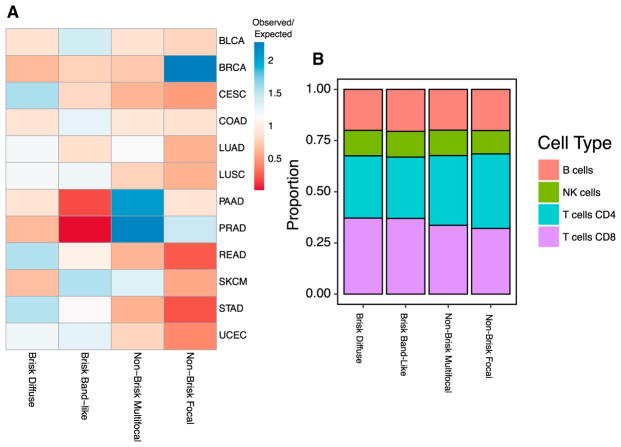
The TIL map global patterns are not distributed in an equal manner among TCGA tumor types. Figure 7A shows the ratio of observed counts over those expected randomly. BRCA is enriched in the “Non-brisk, focal” phenotype (374 observed; 166 expected; p value < 3 × 10−16, Fisher’s exact test, Benjamini-Hochberg adjusted). PAAD is enriched in the “Non-brisk, multi-focal” phenotype (70 observed; 36 expected; p = 8 × 10−8), as is PRAD (151; 70; p < 3 × 10−16). The “Brisk, band-like” phenotype is most enriched in SKCM (134; 86; 3 × 10−7) and very rare in PAAD (7; 37; 2 × 10−9) and PRAD, whereas “Brisk, diffuse” is more prevalent in STAD, READ, and CESC (p = 2 × 10−13, 4 × 10−6, and 3 × 10−10, respectively). Some TCGA subtypes also show enrichment in particular patterns (Figure S5A). For example, EBV-positive GI cancers are enriched in the “Brisk, diffuse” phenotype (14; 5; 6 × 10−3). Differences are also seen among immune subtypes (Figure S5B) defined in the TCGA pan-immune analysis (Thorsson et al., 2018), where the C4 subtype is enriched in the “Non-brisk, focal” (82; 44; 6 × 10−8). This is noteworthy, as subtype C4 is relatively richer in cells of the monocyte/macrophage lineage, which may play a role in sculpting the TME as evidenced in these patterns. Interestingly, the immune subtype C3, which tends to have good prognosis overall, has relatively few “Brisk band-like” structures (59; 162; < 3 × 10−16), perhaps reflective of the more moderate and tempered immune response or productive infiltration of lymphocytes into tumor regions. C2, which has relatively poor outcome, is somewhat richer in “Brisk” phenotypes, consistent with expectations that the relatively large degree of lymphocytic infiltrates are not adequately controlling tumor growth in this class of tumors. In summary, the global structural patterns show associations with distinct immune responses that can be either particular to subtypes, or shared across multiple tumor types, and may play a role in the determining the nature of the immune responses in the corresponding tumor microenvironments.
Figure 7. Association of Spatial Structural Patterns with Tumor Type and Cell Fractions.
(A) Each row corresponds to one of four spatial structure patterns, assigned in a manner consistent with the descriptions currently used to characterize the nature of the immune infiltrate in standard histopathological examinations, and each column is a TCGA tumor types. The values shown are the sample count for each tumor type and spatial structure pattern, divided by the counts expected by chance. The ratio of observed to expected co-membership counts is shown on a color scale, where the largest ratios are in red, values near unity as yellow, and blue represents fewer than expected counts.
(B) Estimates of the proportion of CD4, CD8, NK cells, and B cells were segregated by spatial structure patterns and averaged. Bars show the proportion within each structural pattern. These proportions are estimated using molecular data of the TCGA. See also Figure S5.
We also examined whether there was evidence of differences in the types of lymphocytes, such as signatures for CD4 T cells, CD8 T cells, B cells, and NK cells, represented in each phenotype. These cells cannot be distinguished by the H&E image analysis, but estimates of their proportions are available through analysis of the molecular data (Thorsson et al., 2018 and Method Details). Averaging these values within structural patterns, we see emerging relationships (Figures 7B and S5C), where “Brisk” phenotypes have a higher proportion of CD8 T cells than those seen in the “Non-Brisk” phenotypes (mean 13.2% versus 10.7%, p value < 2.2 × 10−16, Mann–Whitney–Wilcoxon test). Correspondingly, “Non-Brisk” phenotypes tend to have a slightly greater proportion of CD4 T cells (p = 0.03). Thus, by combining molecular estimates of cell proportion with structural analysis of imagining data, we see evidence that particular T cell subsets may play distinct roles in the formation of global structural patterns.
DISCUSSION
The scanned archival H&E archives of the TCGA are a rich but quite underutilized resource within this project. In effect, it is a largely ignored source of data that has only been manually and sporadically mined and awaits more systematic characterization using the variety of analytic tools and analyses currently available. These images have generally been used solely to ensure the correct diagnosis, and panels of expert pathologists also used the images to glean other variables such as mitotic activity, tumor grade, and histologic subtypes for some of the TCGA marker papers. The recently published sarcoma TCGA marker paper utilized automated feature extraction of nuclear properties for correlation with copy number load and genomic doubling (Cancer Genome Atlas Research Network, 2017). The cutaneous melanoma TCGA marker paper used a visual inspection of expert pathologists to assess the degree and pattern of lymphocytes in the frozen section images of the tissue going to the molecular platforms to correlate with other genomic and proteomic assessments of lymphocytic infiltrate and also directly with clinical outcome (Cancer Genome Atlas Network, 2015). This was a manual process done by expert pathologists, and there was no attempt at automation. The efforts presented in this present work represent an initial attempt to systematically employ automated image processing to assess lymphocytic infiltrates across multiple TCGA tumor types for correlation with genomic and epigenomic assessments of lymphocytic infiltrates, as well as clinical outcome. Our sincere hope is that this early attempt to exploit this remarkable TCGA resource of associated scanned histologic images will spur others to similar approaches.
We report a scalable and cost-effective methodology for computational staining to extract and characterize lymphocytes and lymphocytic infiltrates in intratumoral, peri-tumoral, and adjacent stromal regions. In comparing TIL fraction identified via molecular methods to TIL maps derived from digital image analyses of H&E images, we found good but certainly not perfect agreement. Several factors may be contributing. First, perfect agreement is not expected, since the estimates being compared are not of the same quantity or source. Indeed, the molecular estimates are analogous to cell count ratios, and the image fractions correspond to the proportion of spatial areas that contain TILs. Second, the exact spatial relation between the sample from which the molecular data is extracted (between the so-called frozen tissue top-section and bottom-section) and the diagnostic images from the FFPE examples used to generate the diagnostic H&E slides is not known. The TIL maps are derived from high-quality scanned diagnostic FFPE H&E slides from tissue samples in an adjacent or possibly a more distant portion of the tumor relative to where the top and bottom frozen sections are sampled. Unfortunately, the frozen section images are not of a quality that permits robust features extraction. Even though some degree of correlation is certainly expected since TIL status is often a property of the tumor as a whole, upon further evaluation, we observed regional differences in a subset of samples within the overall assessment. These differences are largely explained by the effect of spatial TILs in non-tumor regions in the diagnostic H&E images, which appeared quite different than the spatial TILs in the frozen section samples used for molecular TIL estimates.
Integrated analysis of TIL maps and molecular data reveals patterns and associations that can improve our understanding of the tumor microenvironment, and we illustrate some emerging relationships in this work. Both local patterns and overall structural patterns are differentially represented among tumor types, immune subtypes, and tumor molecular subtypes, the latter of which are typically driven by particular molecular alterations in the tumor cell compartment. This implies that the nature of spatial lymphocytic infiltrate state may be reflective of particular aberration states of tumor cells. In some tumor types (such as PAAD and PRAD), local clustering of TILs may be a more distinctive feature than overall TIL infiltrate, as compared with other tumor types. Structural patterns are further seen to be associated with survival, implying that the nature and effectiveness of immune response is encoded in patterns that may be assessable at the time of tumor diagnosis. For example, in breast cancer, less favorable survival in tumors with elevated adjusted Ball-Hall index (“cluster extent”) might be worth further investigation in terms of stratification of patient tumors in clinical studies of response. Overall structural patterns show associations with immune responses that are shared across multiple tumor types and may thus play a role in the determining the nature of those responses. For example, tumors with C2 immune subtypes, which tend to have relatively poor outcome, are somewhat richer in “Brisk” phenotypes, consistent with expectations that the relatively large degree of lymphocytic infiltrates are not adequately controlling tumor growth in these tissues. The immune subtype C3, which tends to have good prognosis overall, has fewer “Brisk band-like” structures, perhaps reflective of the more moderate and tempered immune response, or productive infiltration of lymphocytes into tumor regions. In contrast, tumors with the C4 immune subtype, which tends to be rich in cells of the monocyte/macrophage lineage, tend to have more “Non-brisk, focal” structures that may play a role in sculpting the TME as evidenced in these patterns. Finally, these patterns are enriched in particular T cell subpopulations as derived from molecular measures. For example, “Brisk” phenotypes have a higher proportion of CD8 T cells than those seen in the “Non-Brisk” phenotypes.
A number of factors can contribute to cancer patient outcome. In our analyses, we attempted to control for age and sex, but other factors such as tumor grade could affect the presence or function of tumor-infiltrating lymphocytes. Grade is more challenging to control for across tumor types, as some are not graded such as melanoma while others such as breast and prostate cancer have very different grading systems that are challenging to compare rigorously. We readily accept that tumor grade and potentially other factors could influence lymphocytic infiltrates in both degree and pattern.
These analyses and early results demonstrate the vast potential of combining analysis of spatial structure with advanced genomics and molecular assessment, as the TIL information is being provided in the context of tumor molecular data wide in detail and in scope. The TCGA molecular datasets and the characterizations performed on them through the work of the PanCancer Atlas consortium, including those on the tumor-immune interface and the tumor microenvironment, provide an extraordinarily rich source of correlative molecular information for our discovered TIL patterns.
H&E imaging is performed routinely in labs throughout the world as a component of tumor diagnostics. Methods for extracting information on TILs from H&E scanned images are potentially of enormous research validity and possible clinical applicability—hundreds of thousands of whole-slide images exist in public repositories, in hospital system databases, and many more will be generated for years to come. In a clinical setting, rapid and automated identification of the degree and nature of TIL infiltrate might be instrumental in determining whether options for immunotherapy should be explored or whether more detailed and costly immune diagnostics should be introduced. Indeed, our approach might also complement immunophenotyping data, and the patterns of immune infiltration assessed by pathologists are already widely employed in the standard clinical reports of primary melanomas as a prognostic factor. Applying methods like those we present here could also allow for very incisive research at very reasonable price points and levels of convenience. These kinds of analyses can only improve with more detailed molecular-marker-based assays such as immunohistochemistry, which are not currently applied in most standard clinical settings due to lack of clinical necessity. Since the TCGA cohorts often predate the broad clinical application of effective immunotherapy such as checkpoint inhibitors and contain little data regarding outcomes with such therapy, association of our TIL estimates and derived infiltration patterns await more appropriate datasets to test associations.
We believe our CNN-derived TIL mapping provides a reproducible and robust tool for the assessment of these lymphocytic infiltrates. The ability to assess this tumor feature is rapidly becoming vital to both clinical diagnosis and translational research for onco-immunologic cancer care. These results show that this approach correlates with molecular assessments of TILs generated by the molecular platforms of the TCGA and can also correlates with clinical outcome for certain tumor types. Importantly, this study shows the value of feature extraction from the information-rich resource of the scanned H&E image archive of the TCGA. This resource has not been exploited to the degree of the other TCGA molecular and clinical outcome resource and clearly not to the degree it can support. This present study demonstrates value that can be added by careful examination of this rich resource, and it is our sincere hope that others will soon explore the many facets of these imaging data.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Tumor-infiltrating lymphocyte maps | This paper | https://doi.org/10.7937/K9/TCIA.2018.Y75F9W1 |
| Software and Algorithms | ||
| Convolutional neural networks for TIL maps | This paper | https://doi.org/10.7937/K9/TCIA.2018.Y75F9W1 |
| QuIP | Saltz et al., 2017 | https://sbu-bmi.github.io/quip_distro/ |
| Theano | Theano Development Team, 2016 | http://deeplearning.net/software/theano/ |
| DeconvNet | Noh et al., 2015 | https://github.com/HyeonwooNoh/DeconvNet |
| CIBERSORT | Newman et al., 2015 | https://cibersort.stanford.edu/ |
| APCluster | Bodenhofer et al., 2011 | https://cran.r-project.org/web/packages/apcluster/index.html |
| clusterCrit | The Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/web/packages/clusterCrit/ |
| polycor | Drasgow, 2014 | https://cran.r-project.org/package=polycor |
| Other | ||
| Data (images, clinical and molecular) used in this study | National Cancer Institute Genomics Data Commons | https://gdc.cancer.gov/about-data/publications/tilmap |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Vésteinn Thorsson (Vesteinn.Thorsson@systemsbiology.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
A total of 4612 participants were included in this study. This study contained both males and females, with inclusions of genders dependent on tumor types. There were 2655 females and 1957 males. TCGA’s goal was to characterize adult human tumors; therefore, the vast majority of participants were over the age of 18. However, one participant under the age of 18 had tissue submitted prior to clinical data. Age was missing for 40 participants. The range of ages was 15–90 (maximum set to 90 for protection of human subjects) with a median age of diagnosis of 63 years of age. Institutional review boards at each tissue source site reviewed protocols and consent documentation and approved submission of cases to TCGA. Detailed clinical, pathologic and molecular characterization of these participants, as well as inclusion criteria and quality control procedures have been previously published for each of the individual TGCA cancer types.
Sample Inclusion Criteria
Surgical resection of biopsy bio-specimens were collected from patients that had not received prior treatment for their disease (ablation, chemotherapy, or radiotherapy). Institutional review boards at each tissue source site reviewed protocols and consent documentation and approved submission of cases to TCGA. Cases were staged according to the American Joint Committee on Cancer (AJCC). Each frozen primary tumor specimen had a companion normal tissue specimen (blood or blood components, including DNA extracted at the tissue source site). Adjacent tissue was submitted for some cases. Specimens were shipped overnight using a cryoport that maintained an average temperature of less than −180°C.
Pathology quality control was performed on each tumor and normal tissue (if available) specimen from either a frozen section slide prepared by the BCR or from a frozen section slide prepared by the Tissue Source Site (TSS). Hematoxylin and eosin (H&E) stained sections from each sample were subjected to independent pathology review to confirm that the tumor specimen was histologically consistent with the allowable hepatocellular carcinomas and the adjacent tissue specimen contained no tumor cells. Adjacent tissue with cirrhotic changes was not acceptable as a germline control, but was characterized if accompanied by DNA from a patient-matched blood specimen. The percent tumor nuclei, percent necrosis, and other pathology annotations were also assessed. Tumor samples with ≥ 60% tumor nuclei and ≤ 20% or less necrosis were submitted for nucleic acid extraction.
METHOD DETAILS
Image and Molecular Data Acquisition
Whole-slide tissue images were obtained from the public TCGA Data Portal (images are currently available from the Genomic Data Commons (GDC) Legacy Archive, following the deprecation of the TCGA Data Portal). Our study uses the diagnostic images, with some images from frozen tissue specimens used in the analysis of discrepancies with molecular estimates. The images were downloaded in the native image format, Aperio SVS files, in which they had been scanned. An SVS file stores an image in multiple resolutions, including the highest resolution the image data was captured; for example in an image that is acquired at a 40x magnification, each pixel is ~0.25 × 0.25 microns. An open source library called OpenSlide (http://openslide.org/formats/aperio/) was used to extract the highest resolution image data for our study. 5455 diagnostic slides were analyzed the 13 TCGA tumor types in the study.
Clinical and molecular data were obtained from processed and quality controlled files of the PanCancer Atlas consortium, available at (https://gdc.cancer.gov/about-data/publications/pancanatlas).
Convolutional Neural Networks for TIL Maps
Our overall methodology consists of two CNNs (a lymphocyte-infiltrated classification CNN (lymphocyte CNN) and a necrosis segmentation CNN), as well as mechanisms for capturing and incorporating feedback from pathologists, to evaluate and refine a generated Tumor-Infiltrating Lymphocyte (TIL) Map.
As is presented in the Results section, the lymphocyte CNN classifies image patches. Only foreground patches are processed and classified. To determine if a patch is a foreground patch, our analysis pipeline checks if the patch has enough tissue using the variance in Red, Green and Blue channels of the patch. A patch is labeled background and discarded if (σ(Red) + σ(Green) + σ(Blue))/3 < 18. The values of the Red, Green, and Blue channels range from 0 to 255. The threshold value of 18 was selected by adjusting it across several slides. We compute percent TIL values using only the foreground patches (i.e., patches with tissue). Note the set of patches with tissue includes TIL patches.
The lymphocyte CNN is a semi-supervised CNN, initialized by an unupervised Convolutional Autoencoder (CAE). The CNN and the CAE are designed to have relatively high resolution input such that one can recognize individual lymphocytes. We have chosen to apply unsupervised CAE pre-training because many studies have shown that it boosts the performance of the CNN, please refer to our technical report (Hou et al., 2017). Using the lung adenocarcinoma (LUAD) patches, we empirically showed that the CNN without pre-training achieved significantly lower area under the curve (AUC). The CAE encodes (compresses) an input image patch of 50 × 50 μm2 (100 × 100 square pixels, corresponding to 20x magnification) into several vectors of length 100, and then reconstruct the input image patch using these encoding vectors. We train the CAE in an unsupervised fashion, to minimize the pixel-wise image patch reconstruction error, with limited number of encoding vectors. By doing this, the CAE implicitly learns to encode the position, appearance and morphology etc. of nuclei, in the encoding vectors. Our guideline of designing the architecture of the CAE is that, each encoding vector, in the ideal case, should be capable of encoding one and only one nucleus. As a result, the CAE has 13 encoding layers and 3 pooling layers. The lymphocyte CNN is built based on the trained CAE: we discard the decoding (reconstruction) part of the CAE, and added several more layers on the encoding vectors. Therefore, our lymphocyte CNN is a 18-layer network with 14 convolutional layers, 3 pooling layers, and 1 fully connected layer (Zhao et al., 2017).
We use two different CNNs for classification of necrosis regions and TILs, because our experiments showed necrosis regions and lymphocytes are best recognized and classified at different image scales. The necrosis CNN model performs best with larger input tissue regions, whereas the lymphocyte CNN model achieves the best results with local, high-resolution image patches. The necrosis segmentation CNN is used to eliminate false positives from the lymphocyte CNN in necrotic regions. In these regions, nuclei may have characteristics similar to those in lymphocyte infiltrated regions. Because recognizing a region of 50 × 50 μm2 need contextual information in a larger region, we model this as a segmentation problem with larger input patches at a relatively lower resolution: 500 × 500 μm2 patches are extracted from the image and downsampled 3 times. The resulting patch is 333 × 333 pixels at 20x magnification. The necrosis segmentation CNN outputs pixel-wise segmentation results. We use DeconvNet (Noh et al., 2015) for this task because it is designed to predict pixel-wise class labels and handle structures and objects at multiple scales (which is more suitable for segmentation than patch-level classification) and it has been shown to achieve high prediction accuracy with several benchmark image datasets. We train DeconvNet to classify each pixel as inside or outside a necrosis region. The output of the necrosis segmentation CNN is resized to match the output resolution of the lymphocyte CNN. If over half of a 50x50 patch intersects with a necrotic region, the patch is classified as non-lymphocyte-infiltrated.
Convolutional Autoencoder Details
The Convolutional Autoencoder (CAE) contains one branch with a small number of low resolution, dense features maps, and a second branch with high resolution, but sparse feature maps The high resolution sparse feature maps are designed to capture foreground objects (e.g., cancer cell nuclei and lymphocytes) - these objects are sparsely distributed in the tissue and contain substantial high spatial frequency color and texture variability. The network learns foreground feature maps in a “crosswise sparse” manner: neurons across all feature maps are not activated (output zero) in most feature map locations. Only neurons in a few feature map locations can be activated. Since the non-activated neurons have no influence in the later decoding layers, the image foreground is reconstructed using only the non-zero responses in the foreground encoding feature maps. The low resolution dense feature maps are designed to encode background color and texture of the background. We first model the background (tissue, cytoplasm etc.) and then extract the foreground that contains nuclei.
The supervised CNN takes the unsupervised encoded features from the unsupervised CAE for classification. We initialize the parameters in these layers to be the same as the parameters in the CAE. We attach four 1x1 convolutional layers after the foreground encoding layer and two 3x3 convolutional layers after the background encoding layer. Each added layer has 320 convolutional filters. We then apply global average pooling on the two branches. The pooled features are then concatenated together, followed by a final classification layer with sigmoid activation function (Hou et al., 2017).
CNN Training and Testing Details
We train our CAE on the unlabeled dataset, minimizing the pixel-wise root mean squared error between the input images and the reconstructed images. No regularization loss is deployed. We use stochastic gradient descent with batch size 32, learning rate 0.03 and momentum 0.9, and train the network until convergence (6 epochs).
For the lymphocyte CNN (constructed from the CAE) training, we use stochastic gradient descent with batch size 100, learning rate 0.001, and momentum 0.985. We train the CNN until convergence (64 epochs) and divide the learning rate by 10 at the 20th, 32th, and 52th epoch. We use sigmoid as the nonlinearity function in the last layer and log-likelihood as the loss function. No regularization loss is deployed. We apply three types of data augmentation. First, the input images are randomly cropped from a larger image. Second, the colors of the input images are randomly perturbed. Third, we randomly rotate and mirror the input images. We trained the CAE and CNN on a single Tesla K40 GPU. During testing phase, we augmented the test patch 24 times and averaged the prediction results. The CAE and CNN used the Theano library (http://deeplearning.net/software/theano/).
CNN-VGG Comparison Experiment Details
We fine-tuned the VGG 16-layer network which was pre-trained on ImageNet. Fine-tuning the VGG16 network has been shown to be robust for pathology image classification (Xu et.al. 2015; Hou et al., 2016b). We used stochastic gradient descent with batch size 32, learning rate 0.0001, and momentum 0.985. We trained the lymphocyte CNN until convergence (32 epochs). We used the same loss function and data augmentation method used for the proposed CNN. To match the input size of the VGG16 network, we re-sized the input patches from 100 × 100 pixels to 224 × 224 pixels. Same as the proposed CNN, during testing phase, we augment the test patch 24 times and average the prediction results.
Iterative Model Training and Data Labeling
We have implemented an iterative workflow as depicted in Figure S1 in order to train the CNN models. First, an unsupervised image analysis of WSIs is executed to initialize a CNN model. This model is refined in an iterative process in which CNN predictions are reviewed, corrected and refined by expert pathologists and the CNN model is re-trained with the updated data in order to improve its classification performance. After a training phase, the CNN model is applied to patches in the test set. For each test patch, the lymphocyte CNN produces a probability of the patch being a lymphocyte-infiltrated patch. The label of the patch is decided by simple thresholding; if the probability value is above a predefined threshold, the patch is classified as lymphocyte-infiltrated.
Training a fully supervised CNN requires a large number of training instances with ground truth labels. Masci et al. (Masci et al., 2011) have shown that utilizing unlabeled instances can boost the performance of a CNN. Drawing from those findings, we first trained an unsupervised Convolutional Auto-Encoder (CAE) to learn the representation of nuclei and lymphocytes in histopathology images and initialize the lymphocyte CNN (Zhao et al., 2017). In this way, the initial lymphocyte CNN model captures the appearance of histopathology images without supervised training. We initialized the weights of the necrosis segmentation CNN randomly following the DeconvNet approach. We then trained the CNNs with labeled images. The training phases of the CNNs involve a cross-validation step to assess prediction performance and avoid overfitting (Hou et al., 2017).
Review and Refinement of CNN Predictions
We developed a web application, called the TIL-Map editor, to support the review and refinement by the pathologists of the tumor-infiltrating lymphocyte patch predictions and the segmentation of necrotic regions. The TIL-Map editor extends caMicroscope (Sharma et al., 2014) interface to enable the visualization of patch-level classification labels as a heatmap overlay on a WSI. It is distributed as part of a suite of tools called QuIP - Quantitative Imaging for Pathology (Saltz et al., 2017). QuIP is an open-source software system which consists of a suite of integrated data services and web-based user applications designed for the management and analysis of whole-slide tissue images and indexing and exploration of image features. When using the TIL-Map editor, a user can interactively visualize, pan, and zoom-in/out of the whole-slide tissue image and interactively pan and zoom around the image, in a manner similar to various online mapping systems. It display the TIL-Maps, as polygonal overlays that appear over the H&E image. The intermediate and final TIL Maps are stored in the QuIP FeatureDB, which manages and indexes both the image metadata and the TIL classification results. Figure 2B shows an example heatmap along with the TIL-Map editor.
Each patch in a WSI is represented as a rectangle and associated with a classification label and the probability value computed by the CNN. This information is stored as a data element (document) in FeatureDB and indexed to speed up queries by the TIL-Map editor to retrieve and display subsets of patches. After classification results for a set of WSIs have been loaded to the database, a pathologist can use a web browser to view and update the classification results. The pathologist would use the TIL-Map editor to examine an image, query FeatureDB to retrieve patches visible within the view point and zoom level and display them as a two-color heatmap. The pathologist can edit the heatmap using the “Lymphocyte Sensitivity,” “Necrosis Specificity,” “Smoothness” sliders in a panel. These slides allow the pathologist to change the threshold value which determines if a patch should be classified as lymphocyte-infiltrated or not. For finer-grain editing of individual patches or sets of patches, the pathologist can use the “Markup Edit” function to markup specific patches and label them as lymphocyte-infiltrated or not-lymphocyte-infiltrated. The pathologist can then save the updated patch labels to the database. The updated patch labels are used to retrain the CNN. Changes to the heatmap are only visible to the user him/herself: multiple users can work independently selecting lymphocyte sensitivity and making finer-grain editing in the same slide without knowing each other’s editing choices.
In this work, a team of three pathologists from Stony Brook Medicine and MD Anderson Cancer Center reviewed and refined 10 to 20 WSIs in each cancer type using the TIL-Map editor. Each image was assigned to two pathologists. Each pathologist separately adjusted the “Lymphocyte Sensitivity,” “Necrosis Specificity,” “Smoothness” thresholds and manually edited regions in the images using the “Markup Edit” tool in order to generate an accurate patch-level classification in the entire image. Depending on the pathologists consensus, if retraining was needed, the pathologists collaboratively generated a consensus lymphocyte heatmap for each image. Data from these consensus heatmaps was input back to the lymphocyte CNN in a training step to further improve its performance.
Determining Lymphocyte Selection Thresholds
The trained lymphocyte and necrosis CNNs was applied to 5455 diagnostic slides available for the 13 TCGA tumor types in the study. We then determined selection thresholds based on overall probability estimates for each slide to correct for possible slide-specific bias, in which the CNN was seen to systematically over or under predicts lymphocytes depending on the overall characteristics of the whole slide. The process of determining the lymphocyte selection thresholds is shown in Figure S1. The first step is to classify each slide into categories that reflect whether there is systematic over or under prediction of lymphocytes. To do this. for each slide, ten patches were sampled from 10 ranges of the lymphocyte CNN’s scores (0.10–0.20, 0.20–0.25, 0.25–0.30, 0.30–0.40, 0.40–0.50, 0.60–0.70, 0.70–0.80, 0.80–0.90, 0.90–1.00). Three pathologists labeled them as lymphocyte infiltrated or not. Based on the number of labeled lymphocyte/non-lymphocyte patches, each slide was categorized into 1 of 7 groups: Groups A-G, based corresponding to 0,1,2,3–7,8,9, and 10 positive patches respectively. The second step is to select a threshold in each group. In each group, we randomly selected 8 slides and manually adjusted thresholds for each of them using our visual TIL-Map editor. The threshold of all slides in one group was set to be the average threshold selected for the eight slides sampled in that group. Note that if we categorize the slides into more number of groups, then we have to manually select thresholds for more slides, since per group, a meaningful averaged threshold requires a minimum number of selected thresholds. On the other hand, if we categorize the slides into fewer groups, the intra-group variance of possible slide-specific biases might be too large. Therefore, we select seven as the number of groups, striking a balance between efficiency and effectiveness.
Subsequent to processing as described above, incomplete TIL maps or those with failed predictions were removed, and for LUAD additional manual review was performed to remove TIL maps derived from poor slides, such as those that were out-of-focus or only partially visible. This resulted in 5202 TIL maps (see Figure S1C, Table S2) for further analysis and distribution. For a number of TCGA cases, multiple diagnostics slides are available, distinguished by TCGA slide ID barcode suffixes DX1, DX2, …, DX13. All cases have a DX1 diagnostic slide; hence these slides and corresponding TIL maps were used in subsequent correlative analyses. The 5202 slide-derived TIL maps correspond to 4759 TCGA participants and slide IDs with suffix DX1.
Molecular Data Estimates of Immune Response
We used estimates of tumor and immune characteristics derived and made available in (Thorsson et al., 2018). The estimate of TIL fraction by genomics measurements is obtained as described therein, by multiplying overall leukocyte fraction derived from DNA methylation with an aggregated proportion of immune-cell fractions within the immune compartment estimated using CIBERSORT (Newman et al., 2015). The lymphocyte fraction is an aggregation of CIBERSORT estimates of naive and memory B cells, naive, resting and activated memory CD4 T cells, follicular helper T cells, T regulatory cells, gamma-delta T cells, CD8 T cells, activated and resting NK cells and plasma cells. To compare with these data with TIL estimates from images, participant and slide barcodes were restricted to those satisfying the inclusion criteria of the TCGA PanCancer Atlas and Immune Response Working Group. Of the 4705 cases with characterized TIL map clusters and patterns (see below), 4612 were thus available for molecular data integration and comparison (Table S1, see also Figure S1C, Table S2).
Local Spatial Structure of Immune Infiltrate
We used the APCluster R package (Bodenhofer et al., 2011) to apply the affinity propagation algorithm to obtain local TIL cluster patterns. The affinity propagation approach (Frey and Dueck, 2007) simultaneously considers all data points as potential exemplars (i.e., the centers of clusters) from among possible data points. Treating each data point as a node in a network, it recursively transmits real-valued messages along edges of the network until it finds a good set of exemplars and corresponding clusters. We define the similarities between data points (TIL patches) as the negative square Euclidean distance between them. Aside from the similarity matrix itself, the most important input parameter is the so-called “input preference” which can be interpreted as the tendency of a data sample to become an exemplar. The function apcluster in the package contains an argument q that allows setting the ‘input preference’ parameter to a certain quantile of the input similarities: resulting in the median for q = 0.5 and in the minimum for q = 0. To select this parameter, we generated synthetic data points in a plane comprising two distinct Gaussian clouds of points. Using the synthetic data, we observed that q = 0 was best able to cluster these points into two clusters, and used this value for identifying TIL clusters. Of the 5202 TIL maps, 5144 clustering results were generated (see Figure S1C, Table S2), with the remainder failing to complete clustering runs in time or failing due to memory errors, mostly in slides with numerous TILs.
Cluster characterization was made using simple measures of counts and membership and cluster indices from the R package clusterCrit by Bernard Desgraupes. The Ball-Hall, Banfield-Raftery, C Index, and Determinant Ration indices are detailed in the package documentation.
| Variable | Definition or Reference |
|---|---|
| Number of TIL Patches | TIL patch count |
| TIL fraction | (TIL patch count)/(Total number of available patches on tissue slice) |
| Number of TIL Clusters | Number of clusters, from affinity propagation clustering |
| Cluster Size Mean | Mean of the cluster membership counts |
| Cluster Size Standard Deviation | Standard deviation of the cluster membership counts |
| Within-Cluster Dispersion Mean | Mean of the values of WGSSk, the within-cluster dispersion (see below) |
| Within-Cluster Dispersion Standard Deviation | Standard deviation of the values of WGSSk |
| Cluster Extent Mean | Mean of the maximum distances to clusters exemplars. The cluster examplar is the most representative TIL patch for the cluster, as defined in the affinity propagation method |
| Cluster Extent Standard Deviation | Standard deviation of the maximum distances to exemplars |
| Ball Hall Index | Ball and Hall (1965). Available at: http://www.dtic.mil/docs/citations/AD0699616 |
| Banfield Raftery Index | Banfield and Raftery (1993) |
| C Index | Hubert and Schultz (1976) |
| Determinant Ratio Index | Scott and Symons (1971) |
| Ball Hall Index - TIL count adjusted | ‘Adjusted’ refers to the residual of the corresponding index after regression against %TIL density |
| Banfield Raftery Index - TIL count adjusted | ‘Adjusted’ refers to the residual of the corresponding index after regression against %TIL density |
| C Index - TIL count adjusted | ‘Adjusted’ refers to the residual of the corresponding index after regression against %TIL density |
| Determinant Ratio Index - TIL count adjusted | ‘Adjusted’ refers to the residual of the corresponding index after regression against %TIL density |
In the above, WGSSk is a within-cluster dispersion which is the sum of the squared distances between the observations and the barycenter of the cluster (see https://CRAN.R-project.org/package=clusterCrit) for details. To compute the adjusted indices, linear regression was used to model the relationship between the clustering index and the %TIL density. The regression residual was used as the adjusted index. Cluster characteristic were generated for all 5144 slides with cluster results (4705 with DX1 suffix)(see Figure S1C, Table S2) and adjusted indices for 4509 cases.
Assessment of TIL Map Structural Patterns
In order to perform a comprehensive assessment of the TIL map structural patterns, the collection of 5202 H&E images (see above, 4759 with DX1 suffix) and the corresponding TIL maps were visually inspected to ensure that each H&E image had a correctly matched TIL map, after which, a subset of 500 H&E images and corresponding TIL maps were closely inspected at higher power magnification (100x to 200x) in 30–50 fields to ensure that the lymphocyte-detection algorithm was performing as intended and not mistakenly identifying tumor cells as lymphocytes across the various tumor types as a quality-control measure. We further employed H&E images and corresponding TIL maps from cases of uveal melanoma as negative controls because melanoma tumor cells and melanotic pigment can be a difficult challenge for the lymphocyte-detection algorithm.
After the negative controls were verified and quality measures were satisfactorily addressed, TIL maps (total N = 4455) were assessed in a two part fashion by a qualitative description and a semiquantitative score based on visual inspection with respect to the tumor region only, which is determined by histopathologic evaluation at low-power magnification (40x) of the corresponding H&E diagnostic whole-slide image. The tumor region represents the combined intra-tumoral and peri-tumoral regions and excludes the adjacent non-tumor regions.
The qualitative description characterizes the nature of the immune infiltrate with respect to the gross spatial distribution of the TILs in only the tumor region with terms like “Focal” (localized), “Multi-focal” (loosely scattered), “Diffuse” (spread out over a large area), and “Band-like” (well-defined boundaries bordering the tumor at its periphery). The semiquantitative scoring evaluates the relative strength of the immune response terms like “None,” “Non-brisk” (minimal to mild partial immune response), and “Brisk” (moderate to strong immune response).
Taken together, “Non-brisk, focal” is indicative of a “very weak” but minimally present immune response with a low density of TILs in a localized area of the tumor, whereas “Non-brisk, multi-focal” is indicative of a weak partial immune response with loosely scattered TILs in a few areas of the tumor. However, “Brisk, diffuse” represents a moderate to strong immune response with a relatively dense and spread out pattern of TILs across > 30% of the tumor even if there are band-like boundaries bordering the tumor at its periphery. The “Brisk, band-like” description was reserved for cases where the TIL map patterns showed relatively organized structures that appear as boundaries bordering the tumor at its periphery and < 30% TILs in the intra-tumoral component. “None” was selected in cases where few TILs were present in less than 1% of the area of the tumor and “Indeterminate” was used if there was insufficient or no grossly identifiable tumor in the H&E image at low-power with the corresponding TIL map regardless of pattern and semiquantitative distribution of TILs.
Summary Table of Criteria Used to Characterize TIL Map Structural Patterns
| Category | Immune Response | Qualitative Pattern | Proportion of Tumor composed of Lymphocytes |
|---|---|---|---|
| Indeterminate | Insufficient and/or no tumor in the H&E image at low-power | Not applicable | Not applicable |
| None | No response | No pattern | <1% TILs |
| Non-brisk, focal | Very Weak (minimal) | Localized | <5% TILs |
| Non-brisk, multi-focal | Weak (mild) | Loosely scattered foci | >5%–30% TILs |
| Brisk, diffuse | Moderate to Strong | Diffuse and dense infiltrate | >30% TILs in the intra-tumoral component* |
| Brisk, band-like | Not applicable | Infiltrate bordering the tumor at its periphery | <30% TILs in the intra-tumoral component* |
If the TIL map patterns revealed both diffuse and band-like immune responses, the predominant pattern was characterized and the difference between “Brisk, diffuse” and “Brisk, band-like” was based on whether the relative distribution of TILs in the intra-tumoral component appeared to be greater than or less than 30%, respectively.
DATA AND SOFTWARE AVAILABILITY
The original H&E stained whole-slide images used in this work can be downloaded from the Genomic Data Commons. All TCGA molecular data can be obtained from the Genomic Data Commons, as well as derived data matrices of the PanCancer Atlas. Integration with immune signatures of the TCGA immune response working group is available through CRI iAtlas web resource. Links to these data resources can be found at the accompanying publication manuscript page (https://gdc.cancer.gov/about-data/publications/tilmap).
The analysis codes used in this work is version controlled has also been containerized and made available as a Docker image. The QuIP software for iterative refinement of CNN prediction results is also available. The training datasets for the CNN models and the TIL maps generated in this study are also available for download. These different software resources as well as the TIL maps are available on the Cancer Imaging Archive, at: https://doi.org/10.7937/K9/TCIA.2018.Y75F9W1
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical details of all experiments are reported in the text, figure legends and figures, including statistical analysis performed, statistical significance and counts.
Supplementary Material
Highlights.
Deep learning based computational stain for staining tumorinfiltrating lymphocytes (TILs)
TIL patterns generated from 4,759 TCGA subjects (5,202 H& slides), 13 cancer types
Computationally stained TILs correlate with pathologist eye and molecular estimates
TIL patterns linked to tumor and immune molecular features, cancer type, and outcome
Acknowledgments
We are grateful to all the patients and families who contributed to this study. Funding from the Cancer Research Institute is gratefully acknowledged, as is support from National Cancer Institute (NCI) through U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845,U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, P30 CA016672, U24CA180924, U24CA210950, U24CA215109, NCI Contract HHSN261201400007C, and Leidos Biomedical Contract 14X138. A.U.K.R. and P.S were supported by CCSG Bioinformatics Shared Resource P30 CA01667, ITCR U24 Supplement 1U24CA199461-01, a gift from Agilent technologies, CPRIT RP150578, and a Research Scholar Grant from the American Cancer Society (RSG-16-005-01). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation XSEDE Science Gateways program under grant ACI-1548562 allocation TG-ASC130023. The authors would like to thank Stony Brook Research Computing and Cyberinfrastructure and the Institute for Advanced Computational Science at Stony Brook University for access to the high-performance LIred and SeaWulf computing systems, the latter of which was supported by National Science Foundation grant (#1531492).
TCGA Tumor Types Used in this Study
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD
Colon adenocarcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- PAAD
Pancreatic adenocarcinoma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SKCM
Skin Cutaneous Melanoma
- STAD
Stomach adenocarcinoma
- UCEC
Uterine Corpus Endometrial Carcinoma
- UVM
Uveal Melanoma
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.086.
AUTHOR CONTRIBUTIONS
Conceptualization, J.H.S., V.T., A.J.L., T.K., I.S.; Methodology, J.H.S., V.T., A.S., A.J.L., A.U.K.R., I.S., T.Z., D.S., V.N., P.S., T.K., L.H., R.G.; Investigation, J.H.S., V.T., A.S., A.J.L., A.U.K.R., I.S., J.V.A., R.B., T.Z., D.S., V.N., P.S., T.K., L.H., R.G.; Writing – Original Draft, J.H.S., V.T., A.S., A.J.L., A.U.K.R., K.R.S., D.S., V.N., T.K., L.H., R.G.; Writing – Review & Editing, J.H.S., V.T., A.S., A.J.L., K.R.S., D.S., T.K., L.H., R.G.; Supervision, J.H.S., V.T., A.J.L., K.R.S., D.S., T.K.; Visualization, V.T., A.S., A.U.K.R., T.Z., P.S., L.H., R.G.; Data Curation, V.T., A.S., I.S., A.J.L., V.N., T.K., L.H., R.G.; Software, A.S., V.N., P.S., T.K., L.H.; Formal Analysis; J.H.S., V.T., A.U.K.R., V.N., P.S., T.K., L.H.
DECLARATION OF INTERESTS
Michael Seiler, Peter G. Smith, Ping Zhu, Silvia Buonamici, and Lihua Yu are employees of H3 Biomedicine, Inc. Parts of this work are the subject of a patent application: WO2017040526 titled “Splice variants associated with neo-morphic sf3b1 mutants.” Shouyoung Peng, Anant A. Agrawal, James Palacino, and Teng Teng are employees of H3 Biomedicine, Inc. Andrew D. Cherniack, Ashton C. Berger, and Galen F. Gao receive research support from Bayer Pharmaceuticals. Gordon B. Mills serves on the External Scientific Review Board of Astrazeneca. Anil Sood is on the Scientific Advisory Board for Kiyatec and is a shareholder in BioPath. Jonathan S. Serody receives funding from Merck, Inc. Kyle R. Covington is an employee of Castle Biosciences, Inc. Preethi H. Gunaratne is founder, CSO, and shareholder of NextmiRNA Therapeutics. Christina Yau is a part-time employee/consultant at NantOmics. Franz X. Schaub is an employee and shareholder of SEngine Precision Medicine, Inc. Carla Grandori is an employee, founder, and shareholder of SEngine Precision Medicine, Inc. Robert N. Eisenman is a member of the Scientific Advisory Boards and shareholder of Shenogen Pharma and Kronos Bio. Daniel J. Weisenberger is a consultant for Zymo Research Corporation. Joshua M. Stuart is the founder of Five3 Genomics and shareholder of NantOmics. Marc T. Goodman receives research support from Merck, Inc. Andrew J. Gentles is a consultant for Cibermed. Charles M. Perou is an equity stock holder, consultant, and Board of Directors member of BioClassifier and GeneCentric Diagnostics and is also listed as an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays. Matthew Meyerson receives research support from Bayer Pharmaceuticals; is an equity holder in, consultant for, and Scientific Advisory Board chair for OrigiMed; and is an inventor of a patent for EGFR mutation diagnosis in lung cancer, licensed to LabCorp. Eduard Porta-Pardo is an inventor of a patent for domainXplorer. Han Liang is a shareholder and scientific advisor of Precision Scientific and Eagle Nebula. Da Yang is an inventor on a pending patent application describing the use of antisense oligonucleotides against specific lncRNA sequence as diagnostic and therapeutic tools. Yonghong Xiao was an employee and shareholder of TESARO, Inc. Bin Feng is an employee and shareholder of TESARO, Inc. Carter Van Waes received research funding for the study of IAP inhibitor ASTX660 through a Cooperative Agreement between NIDCD, NIH, and Astex Pharmaceuticals. Raunaq Malhotra is an employee and shareholder of Seven Bridges, Inc. Peter W. Laird serves on the Scientific Advisory Board for AnchorDx. Joel Tepper is a consultant at EMD Serono. Kenneth Wang serves on the Advisory Board for Boston Scientific, Microtech, and Olympus. Andrea Califano is a founder, shareholder, and advisory board member of DarwinHealth, Inc. and a shareholder and advisory board member of Tempus, Inc. Toni K. Choueiri serves as needed on advisory boards for Bristol-Myers Squibb, Merck, and Roche. Lawrence Kwong receives research support from Array BioPharma. Sharon E. Plon is a member of the Scientific Advisory Board for Baylor Genetics Laboratory. Beth Y. Karlan serves on the Advisory Board of Invitae.
References
- Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25:261–267. doi: 10.1016/j.coi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJC, Quinn MC, et al. Australian Pancreatic Cancer Genome Initiative. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- Ball GH, Hall DJ. Technical Report April 1965 prepared for the Information Sciences Branch of the Office of Naval Research. Stanford Research Institute - Clearinghouse for Federal Scientific and Technical Information; 1965. ISODATA, a novel method of data analysis and pattern classification; pp. 2–50. [Google Scholar]
- Banfield JD, Raftery AE. Model-Based Gaussian and Non-Gaussian Clustering. Biometrics. 1993;49:803–821. [Google Scholar]
- Bayramoglu N, Heikkila J. Transfer learning for cell nuclei classification in histopathology images. In: Hua G, Jégou H, editors. Computer Vision – ECCV 2016 Workshops. Springer; 2016. pp. 532–539. Lecture Notes in Computer Science. [Google Scholar]
- Bodenhofer U, Kothmeier A, Hochreiter S. APCluster: an R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–2464. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- Broussard EK, Disis ML. TNM staging in colorectal cancer: T is for T cell and M is for memory. J Clin Oncol. 2011;29:601–603. doi: 10.1200/JCO.2010.32.9078. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171:950–965e28. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Chen H, Qi X, Yu L, Dou Q, Qin J, Heng PA. DCAN: Deep contour-aware networks for object instance segmentation from histology images. Med Image Anal. 2017;36:135–146. doi: 10.1016/j.media.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention –MICCAI 2013. Vol. 8150. Springer; 2013. pp. 411–418. Lecture Notes in Computer Science. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Demicco EG, Saltz JH, Powell RT, Rao A, Lazar AJ. PanCancer insights from The Cancer Genome Atlas: the pathologist’s perspective. J Pathol. 2017 doi: 10.1002/path.5028. Published online December 30, 2017. https://doi.org/10.1002/path.5028. [DOI] [PMC free article] [PubMed]
- Crowson AN, Magro CM, Mihm MC. Prognosticators of melanoma, the melanoma report, and the sentinel lymph node. Mod Pathol. 2006;19(Suppl 2):S71–S87. doi: 10.1038/modpathol.3800517. [DOI] [PubMed] [Google Scholar]
- Drasgow F. Polychoric and Polyserial Correlations. In: Balakrishnan N, Colton T, Everitt B, Piegorsch W, Ruggeri F, Teugels JL, editors. Wiley StatsRef: Statistics Reference. John Wiley & Sons; 2014. pp. 68–74. [Google Scholar]
- FDA News Release. FDA allows marketing of first whole slide imaging system for digital pathology. 2017 https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm552742.htm.
- Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Graves A, Jaitly N. Towards end-to-end speech recognition with recurrent neural networks. JMLR; Proceedings of the 31st International Conference on Machine Learning; 2014. pp. 1764–1772. [Google Scholar]
- Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv Anat Pathol. 2017a;24:235–251. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol. 2017b;24:311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Samaras D, Kurc T, Gao Y, Davis JE, Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. Computer Vision and Pattern Recognition. 2016a doi: 10.1109/CVPR.2016.266. arXiv:1504.07947v5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Singh K, Samaras D, Kurc TM, Gao Y, Seidman RJ, Saltz JH. 2016 New York Scientific Data Summit (NYSDS) IEEE; 2016b. Automatic histopathology image analysis with CNNs; pp. 1–6. [Google Scholar]
- Hou L, Nguyen V, Samaras D, Kurc TM, Gao Y, Zhao T, Saltz JH. Sparse Autoencoder for Unsupervised Nucleus Detection and Representation in Histopathology Images. Computer Vision and Pattern Recognition. 2017 doi: 10.1016/j.patcog.2018.09.007. arXiv:1704.00406v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, Weinberger KQ, van der Maaten L. Densely connected convolutional networks. Computer Vision and Pattern Recognition. 2016 arXiv:1608.06993v5. [Google Scholar]
- Hubert L, Schultz J. Quadratic Assignment as a General Data-analysis Strategy. British Journal of Mathematical and Statistical Psychology. 1976;29:190–241. [Google Scholar]
- Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, Serody JS. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20:3818–3829. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst. 2016;108:djw144. doi: 10.1093/jnci/djw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos J, Chai S, Mose LE, Selitsky SR, Krishnan B, Saito R, Iglesia MD, Milowsky MI, Parker JS, Kim WY, Vincent BG. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight. 2016;1:e85902. doi: 10.1172/jci.insight.85902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinos I. Ubernet: Training a universal convolutional neural network for low-,mid-, and high-level vision using diverse datasets and limited memory. Computer Vision and Pattern Recognition. 2017 arXiv:1609.02132v1. [Google Scholar]
- Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masci J, Meier U, Cireşan D, Schmidhuber J. Stacked Convolutional Auto-Encoders for Hierarchical Feature Extraction. In: Honkela T, Duch W, Girolami M, Kaski S, editors. Artificial Neural Networks and Machine Learning – ICANN 2011. Springer; 2011. pp. 52–59. [Google Scholar]
- Mlecnik B, Bindea G, Pagès F, Galon J. Tumor immunosurveillance in human cancers. Cancer Metastasis Rev. 2011a;30:5–12. doi: 10.1007/s10555-011-9270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pagès F, Galon J. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011b;29:610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- Murthy V, Hou L, Samaras D, Kurc TM, Saltz JH. Center-focusing multitask CNN with injected features for classification of glioma nuclear images. Winter Conference on Applications of Computer Vision (WACV) - Computer Vision and Pattern Recognition; 2017. arXiv:1612.06825v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh H, Hong S, Han B. Learning Deconvolution Network for Semantic Segmentation. 2015 IEEE International Conference on Computer Vision (ICCV) - Computer Vision and Pattern Recognition; 2015. pp. 1520–1528. arXiv:1505.04366v1. [Google Scholar]
- Ranzato M, Poultney C, Chopra S, LeCun Y. Efficient learning of sparse representations with an energy-based model. Proceedings of the 19th International Conference on Neural Information Processing Systems; 2006. pp. 1137–1144. [Google Scholar]
- Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: Unified, real-time object detection. Computer Vision and Pattern Recognition. 2016 arXiv:1506.02640v5. [Google Scholar]
- Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards real-time object detection with region proposal networks. Computer Vision and Pattern Recognition. 2015 doi: 10.1109/TPAMI.2016.2577031. arXiv:1506.01497v3. [DOI] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge WC, Kong J, Gao J, Gutman DA, Cooper LAD, Appin C, Park Y, Scarpace L, Mikkelsen T, Cohen ML, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. 2013;19:4951–4960. doi: 10.1158/1078-0432.CCR-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, et al. International TILs Working Group 2014. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz J, Sharma A, Iyer G, Bremer E, Wang F, Jasniewski A, DiPrima T, Almeida JS, Gao Y, Zhao T, et al. A Containerized Software System for Generation, Management, and Exploration of Features from Whole Slide Tissue Images. Cancer Res. 2017;77:e79–e82. doi: 10.1158/0008-5472.CAN-17-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AJ, Symons MJ. Clustering Methods Based on Likelihood Ratio Criteria. Biometrics. 1971;27:387–397. [Google Scholar]
- Sharma A, Kazerouni A, Saghar N, Commean P, Tarbox L, Prior F. Framework for Data Management and Visualization of The National Lung Screening Trial Pathology Images. Pathology Informatics Summit 2014 (J Pathol Inform) 2014:S30–S31. [Google Scholar]
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Computer Vision and Pattern Recognition. 2014 arXiv:1409.1556v6. [Google Scholar]
- Sirinukunwattana K, Ahmed Raza SE, Tsang Yee-Wah, Snead DR, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35:1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- Su H, Xing F, Kong X, Xie Y, Zhang S, Yang L. Robust cell detection and segmentation in histopathological images using sparse reconstruction and stacked denoising autoencoders. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Vol. 9351. Springer; 2015. pp. 383–390. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theano Development Team. Theano: A Python framework for fast computation of mathematical expressions. Symbolic Computation; Learning; Mathematical Software. 2016 arXiv:1605.02688v1. [Google Scholar]
- Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Yang T-HO, Porta-Pardo E, Gao G, Plaisier CL, Eddy JA, et al. The Immune Landscape of Cancer. Immunity. 2018:48. https://doi.org/10.1016/j.immuni.2018.03.023.
- Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- Wang S, Yao J, Xu Z, Huang J. Subtype cell detection with an accelerated deep convolution neural network. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Vol. 9901. Springer; 2016. pp. 640–648. Lecture Notes in Computer Science. [Google Scholar]
- Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kong X, Xing F, Liu F, Su H, Yang L. Deep voting: A robust approach toward nucleus localization in microscopy images. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Vol. 9351. Springer; 2015a. pp. 374–382. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xing F, Kong X, Su H, Yang L. Beyond classification: structured regression for robust cell detection using convolutional neural network. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention –MICCAI 2015. Vol. 9351. Springer; 2015b. pp. 358–365. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang J. Detecting 10,000 cells in one second. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Vol. 9901. Springer; 2016. pp. 676–684. Lecture Notes in Computer Science. [Google Scholar]
- Xu Y, Jia Z, Ai Y, Zhang F, Lai M, Chang EIC. Deep convolutional activation features for large scale brain tumor histopathology image classification and segmentation. 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2015. pp. 947–951. [Google Scholar]
- Zhao T, Hou L, Nguyen V, Gao Y, Samaras D, Kurc TM, Saltz JH. Using machine methods to score tumor-infiltrating lymphocytes in lung cancer. USCAP 2017 Annual Meeting. Proffered papers, Section A.2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.