Abstract
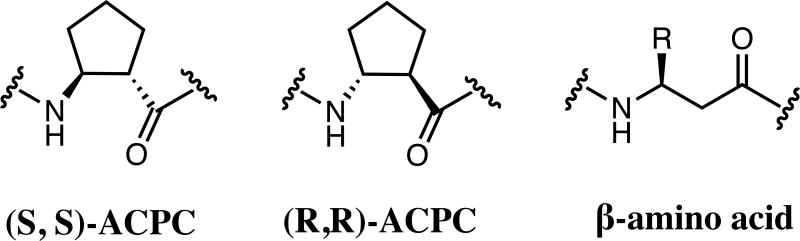
β-Amino acids have a backbone that is expanded by one carbon atom relative to α-amino acids, and β residues have been widely investigated as subunits in protein-like molecules that adopt discrete and predictable conformations. Two classes of β residue have been widely explored in the context of generating α-helix-like conformations: β3-amino acids, which are homologous to α-amino acids and bear a side chain on the backbone carbon adjacent to nitrogen; and residues constrained by a five-membered ring, such the one derived from trans-2-aminocyclopentanecarboxylic acid (ACPC). Substitution of α residues with their β3 homologues within an α-helix-forming sequence generally causes a decrease in conformational stability. Use of a ring-constrained β residue, however, can offset the destabilizing effect of α→β substitution. Here we extend the study of α→β substitutions, involving both β3 and ACPC residues, to short loops within a small tertiary motif. We start from previously reported variants of the Pin1 WW domain that contain a 2-, 3- or 4-residue β-hairpin loop, and we evaluate α→β replacements at each of the loop positions for each variant. By referral to the φ,ψ angles of the native structure, a stereochemically appropriate ACPC residue can be chosen. Use of such a logically chosen ACPC residue enhances conformational stability in several cases. The crystal structures of three β-containing Pin1 WW domain variants show that a native-like tertiary structure is maintained in each case.
Graphical abstract
Introduction
The diverse functions carried out by proteins usually depend on the folded conformations adopted by these macromolecules. Folding is dictated by α-amino acid sequence.[1] Natural proteins are constructed from 20 subunits, and in recent years there has been a growing effort to expand the realm of proteins and related macromolecules by broadening the set of subunits employed. Many studies of this type have involved unnatural side chains, with maintenance of a purely α residue backbone, but advances in chemical synthesis methods have begun to encourage exploration of subunits with unnatural backbones. These fundamental experiments should provide a foundation for reshaping natural polypeptides to display enhanced properties (e.g., resistance to proteolysis) or even entirely new functions. To date, the most systematically studied aspect of polypeptide backbone modification involves replacement of α-amino acid residues with β-amino acid residues in α-helix-forming segments. The backbone of a β-amino acid contains an "extra" carbon atom between carbonyl and nitrogen, relative to an α-amino acid. It has been demonstrated that an α-helix-like conformation can be maintained after insertion of up to 25–33% β residues, via α→β replacements that generate ααβ, αααβ or ααβαααβ backbone patterns.[2] These efforts have focused largely on two types of β-amino acid residues. One class includes direct homologues of the familiar α-amino acid residues, β3-amino acid residues, which bear a side chain on the backbone carbon adjacent to nitrogen (Scheme 1). Many protected β3-amino acids are commercially available, and they are readily incorporated via solid-phase synthesis. Another widely studied β-amino acid class contains a ring that limits conformational freedom. The constraint manifested in (S,S)-trans-2-aminocyclopentanecarboxylic acid (S,S-ACPC) (Scheme 1) is amenable to a right-handed α-helix-like secondary structure. Helix stability often declines as a result of α→β3 replacements, but β3→cyclic β replacements usually stabilize α/β-peptide helices.[2] Helical α/β-peptides can display useful activities, such as blocking pathogenic protein-protein interactions while resisting proteolytic degradation[3–5], or modulating the signaling profile, relative to a natural agonist, caused by activation of a cell-surface receptor.[6–8]
Scheme 1.
Chemical structures of both enantiomers of trans-ACPC and a generic β3-amino acid (R-group corresponds to side chain of analogous α-amino acid).
The studies described here are intended to expand our understanding of backbone modification via α→β replacement in terms of effects on folded structure and on conformational stability, relative to the corresponding all-α polypeptide. We focus on replacements in loop segments that connect two antiparallel β-strands, i.e., β-hairpin loops. Our experimental design builds from pioneering engineering studies by Kelly et al. in which a four-residue loop in the Pin1 WW domain was systematically contracted to a three-residue loop and a two- residue loop, without disruption of the overall tertiary structure of the domain.[9]
A few previous studies have examined non-natural subunits as replacements for dipeptide segments that form β-turns at the surface of a tertiary structure. A dibenzofuran-based unit developed by Kelly et al.[10] was evaluated as a β-turn mimic within several different protein contexts.[11–14] Alewood et al. replaced a β-turn within HIV-1 protease with the bicyclic turn dipeptide (BTD) unit, forming a variant of the enzyme that retains native-like activity and resistance to thermal inactivation.[15] The BTD unit has subsequently been used to replace β-turn-forming dipeptide segments in several other tertiary structures.[14,16] A β-turn mimic containing two sequential β-amino acid residues has been used to replace a β-turn-forming dipeptide segment in ribonuclease A; the resulting analogue retained native-like stability and catalytic activity.[17] D-Proline, which constrains the φ dihedral angle to +60° ± 20° due to restricted backbone rotation, has been used to stabilize type-I’ and II’ β-turns, which comprise residues with equivalent left-handed helical backbone geometries (typically Asn, Asp or Gly).[18–20] D-Proline replacements within protein β-turns do not always effectively reinforce native-like structure.[21]
The observation that both β3-amino acids and the cyclically-constrained subunit ACPC can serve as suitable replacements for α-amino acids within α-helices[2,22–23] led us to wonder whether these unnatural subunits might be tolerated as replacements in loops. We focused in particular on β-hairpin loops, that is, loops that connect adjacent strand segments that engage in antiparallel β-sheet interactions with one another, because this loop class has been very carefully studied and categorized by Thornton et al.[18,24–25] α-Amino acid residues in such loops tend to display backbone dihedral angles that place them in the αR, αL, γR or γL regions of a standard φ,ψ plot. A residue in the αR region has φ and ψ torsion angles that are consistent with participation in a right-handed α-helical conformation. The γR region is adjacent to the αR region in the φ,ψ plot. Many residues in natural proteins fall in the αR and γR regions. In contrast, relatively few residues fall in the αL or γL regions, which would be consistent with participation in a left-handed α-helix. However, Thornton et al. showed that residues displaying these unusual "left-handed" torsion angles are commonly observed at certain positions in β-hairpin loops.[25]
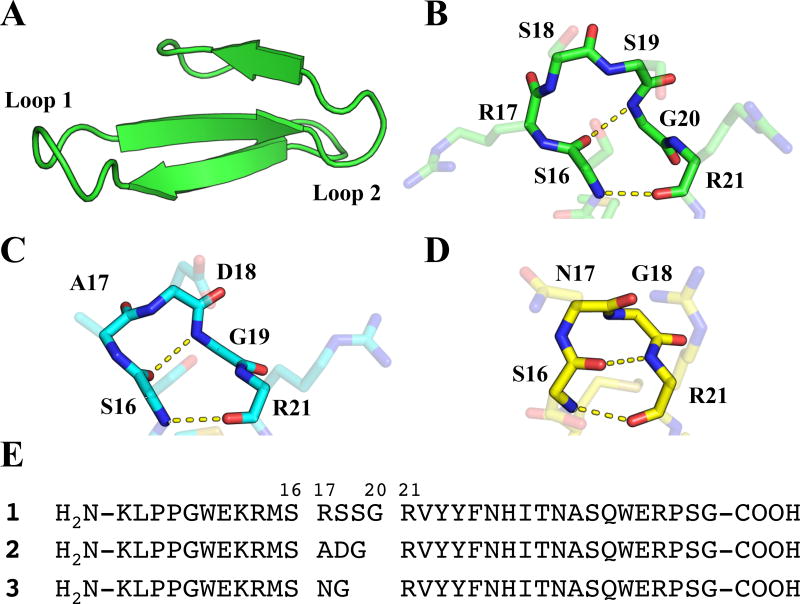
As the basis for our loop-replacement studies, we chose a set of three variants of the Pin1 WW domain described by Jäger et al. (Figure 1).[9] The WW domain tertiary motif is dominated by a three-stranded antiparallel β-sheet linked by two loop regions.[26] WW domains natively bind to proline-rich polypeptides[27–29], but have also been engineered to bind single-stranded DNA.[30] In the Pin1 WW domain, the first loop region (Loop 1) connects the first two β-strands and contains 4 residues, R17, S18, S19 and G20 (the β-sheet H-bond closest to the loop is formed between S16 NH and R21 carbonyl oxygen). Careful engineering enabled Jäger et al. to develop variants of the Pin1 WW domain containing a three-residue Loop 1 (A17, D18, G19) or a two-residue Loop 1 (N17, G18). High-resolution structural data for each variant revealed that the overall tertiary structure is maintained as Loop 1 is contracted. This set of WW domain variants allowed us to survey the impact of backbone modifications in a surface loop by replacing each α residue in each version of Loop 1 with the flexible β homologue (i.e., Arg replaced by β3-homoarginine (β3-hArg), Gly replaced by β-hGly, etc.). In a complementary set of studies, a cyclic β-residue (ACPC) was placed at each position of each variant of Loop 1. Based on results from α→β substitutions within α-helices, we hypothesized that β3-amino acid incorporation at loop sites would tend to destabilize the WW domain tertiary structure. We further predicted that ACPC replacements would be better tolerated than β3 replacements at most or all loop positions.[2] These hypotheses were tested by assessing tertiary structure stability via thermal denaturation. Additional insights were gained from the X-ray crystal structures we determined for three β-amino acid-substituted WW domain variants.
Figure 1.
(A) Structure of the isolated Pin1 WW domain, derived from PDB entry 4GWT. (B) View of Loop I within the Pin1 WW domain with backbone atoms (N, Cα, C, O) highlighted. Intramolecular hydrogen bonds correspond to a 4:6 β-hairpin classification. (C) View of the 3:5 β-hairpin from WW domain variant 2 (PDB: 2F21), with intramolecular hydrogen bonds shown. (D) View of the 2:2 β-hairpin from WW domain variant 3 (PDB: 1ZCN), with intramolecular hydrogen bonds shown. (E) Sequences of WW domain peptides 1, 2, and 3.
Results
Design of β-substituted WW domain variants
The α-peptide starting points for our α→β replacement studies include the native Pin1 WW domain (four-residue Loop 1) and the reported variants with three-residue or two-residue versions of Loop 1; these peptides are designated 1–3 (Figure 1e, Table 1). An initial set of nine β-containing variants was generated by systematically replacing each Loop 1 residue in 1–3 with the β3 homologue or β-hGly. A second set of variants was generated by replacing each Loop 1 residue in 1–3 with ACPC. For each β3 replacement, the configuration at the side chain-bearing carbon matches the configuration of the homologous L-α-amino acid residue. For ACPC replacements, however, the choice of configuration was informed by the backbone conformation of the α residue to be replaced. In the native Pin1 WW domain, the first three Loop 1 residues have backbone torsion angles that fall in the αR or γR region of the φ,ψ plot.[31–32] (S,S)-ACPC was employed at these sites in 1 because this absolute configuration is compatible with a right-handed α-helical conformation (Table 1). The fourth Loop 1 residue in 1, G20, displays φ,ψ angles in the γL region, and we evaluated both (S,S)-ACPC and (R,R)-ACPC at this site. In Pin1 WW domain variant 2, the first Loop 1 residue displays φ,ψ angles in the αR region, and the second Loop 1 residue displays φ,ψ angles in the γR region[9]; (S,S)-ACPC was the replacement at both of these sites. The third Loop 1 residue in 2, G19, displays φ,ψ angles in the γL region, and (R,R)-ACPC was used at this site. Both Loop 1 residues in 3 display torsion angles in the "left-handed" region of the φ,ψ plot[9], and (R,R)-ACPC was used in both cases.
Table 1.
α→β residue design strategy for loop residues in 1–3. a
| Variant (position) |
Backbone Geometry |
Acyclic β Replacement |
ACPC Replacement |
|---|---|---|---|
| 1(R17) | α/γR | β3R | [S,S] |
| 1(S18) | α/γR | β3S | [S,S] |
| 1(S19) | α/γR | β3S | [S,S] |
| 1(G20) | γL | βG | [R,R],[S,S] |
| 2(A17) | αR | β3A | [S,S] |
| 2(D18) | γR | β3D | [S,S] |
| 2(G19) | γL | βG | [R,R] |
| 3(N17) | αL | β3N | [R,R] |
| 3(G18) | γL | βG | [R,R] |
Loop 1 residue backbone conformations determined for reported structures of 1 (PDB 4GWT), 2 (PDB 2F21) and 3 (PDB 1ZCN), and β-amino acid replacements used at each sequence position. For sequence position G20 within variant 1, substitutions with both enantiomers of ACPC were evaluated. For variant 1, residues R17, S18 and S19 lie at the boundary between αR and γR regions of the Ramachandran plot. Backbone geometry assigned according to the convention described by Thornton et al.[25]
Tertiary structure stability
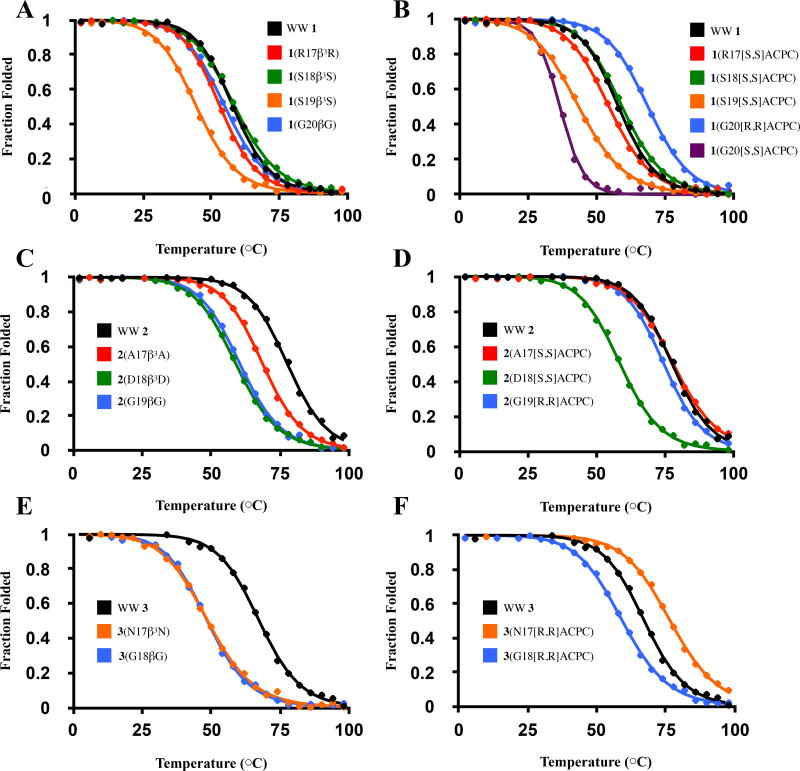
To assess the impact of α→β residue replacements on Pin1 WW domain conformational stability, we monitored folding via circular dichroism (CD) as a function of temperature for α-peptides 1–3 and each of the β-containing variants. Each polypeptide exhibited a CD maximum at 227 nm that is characteristic of the WW domain fold[33]; CD intensity at this wavelength was used to monitor temperature-dependent changes in folding. Thermal denaturation data were fitted to a two-state model to determine a melting temperature (TM) for each WW domain derivative (Figure 2). TM values in close agreement with reported values9 were obtained for 1, 2, and 3. To evaluate the impact of each α→β replacement on conformational stability relative to the appropriate all-α polypeptide, we report ΔTM values [TM(β replacement) – TM(parent)] (Table 2). To evaluate the impact of each flexible β→ACPC replacement on conformational stability, we report the quantity ΔTM(o) = [TM(ACPC) – TM(acyclic β)], which compares TM values corresponding to peptides containing ACPC- or β3-amino acid-substitution at a given position in Loop 1 (Table 2).
Figure 2.
Representative fitted thermal denaturation curves for all WW domain variants described in this study. Experimental data and best-fit lines are shown. For variants based on 1 (panels A, B), 2 (panels C, D) and 3 (panels E, F), thermal denaturation curves for β3 residue-containing and ACPC-containing variants based on each parent sequence are shown on adjacent plots.
Table 2.
Summary of thermal denaturation data.a
| Variant | TM (°C) | ΔTM (°C) | ΔTM(O) (°C) |
|---|---|---|---|
| WW 1 | 57.6 (± 0.6) | ||
| 1(R17β3R) | 53.4 (± 0.9) | −4.2 (± 1.1) | |
| 1(S18β3S) | 59.1 (± 0.5) | 1.5 (± 0.8) | |
| 1(S19β3S) | 44.4 (± 0.2) | −13.2 (± 0.6) | |
| 1(G20βG) | 54.7 (± 0.2) | −2.9 (± 0.6) | |
| 1(R17[S,S]ACPC) | 54.3 (± 0.3) | −3.3 (± 0.7) | 0.9 (± 0.9) |
| 1(S18[S,S]ACPC) | 59.3 (± 0.0) | 1.7 (± 0.6) | 0.2 (± 0.5) |
| 1(S19[S,S]ACPC) | 43.4 (± 0.6) | −14.2 (± 0.9) | −1.0 (± 0.6) |
| 1(G20[R,R]ACPC) | 68.5 (± 0.2) | 10.9 (± 0.6) | 13.8 (± 0.2) |
| 1(G20[S,S]ACPC) | 37.0 (± 0.4) | −20.6 (± 0.7) | −17.7 (± 0.4) |
| WW 2 | 77.5 (± 0.3) | ||
| 2(A17β3A) | 68.4 (± 0.2) | −9.1 (± 0.3) | |
| 2(D18β3D) | 58.2 (± 0.5) | −19.3 (± 0.6) | |
| 2(G19βG) | 60.2 (± 0.1) | −17.3 (± 0.3) | |
| 2(A17[S,S]ACPC) | 77.6 (± 0.3) | 0.1 (± 0.4) | 9.2 (± 0.3) |
| 2(D18[S,S]ACPC) | 58.9 (± 0.4) | −18.6 (± 0.5) | 0.7 (± 0.7) |
| 2(G19[R,R]ACPC) | 74.5 (± 0.2) | −3.0 (± 0.3) | 14.2 (± 0.2) |
| WW 3 | 67.4 (± 0.4) | ||
| 3(N17β3N) | 48.5 (± 0.3) | −18.9 (± 0.5) | |
| 3(G18βG) | 48.4 (± 0.4) | −19.0 (± 0.5) | |
| 3(N17[R,R]ACPC) | 77.0 (± 0.4) | 9.7 (± 0.5) | 28.5 (± 0.5) |
| 3(G18[R,R]ACPC) | 59.1 (± 0.2) | −8.2 (± 0.4) | 10.7 (± 0.4) |
Thermal denaturation data for all WW domain variants reported in this study. TM values are mean values of three replicates, with standard deviations shown in parentheses. For each substituted variant, ΔTM represents difference in TM relative to corresponding parent sequence. For ACPC-containing variants, ΔTM(O) represents difference in TM relative to variant containing β3 residue.
Substitution of R17, S18 or G20 in WW domain 1 with the flexible β homologue, to generate 1(R17β3R), 1(S18β3S) and 1(G20βG), had only a modest effect on TM. However, TM for 1(S19β3S) was dramatically decreased (~13 °C) relative to 1. Thus, the native Pin1 WW domain tertiary structure tolerates the insertion of an "extra" CH2 unit at several positions within Loop 1, but this insertion is quite destabilizing at S19. There is very little difference in terms of conformational stability between α→β3 replacement and α→ACPC replacement at R17, S18 or S19, which indicates that the structural fortification that we predicted for a preorganized cyclic β residue relative to a flexible β3 residue did not materialize. Replacement of G20 with a cyclic β residue caused divergent outcomes depending upon ACPC configuration. Incorporation of (S,S)-ACPC led to a substantial decline in conformational stability relative to 1 or 1(G20β), but incorporation of (R,R)-ACPC caused a significant increase in stability. Indeed, 1(G20[R,R]ACPC) was more stable than 1 itself and the most stable among all β-containing variants of 1.
The dramatic difference in conformational stability between the two diastereomers of 1(G20ACPC), ΔTM ~ 31°C, is attributed to the unusual backbone conformation of G20 in the Pin1 WW domain. The torsion angles of this glycine residue lie in the γL region of the φ,ψ plot. Extrapolation from the extensive structural data for α/β-peptides containing (S,S)-ACPC residues leads to the prediction that use of (R,R)-ACPC should enforce a backbone conformation compatible with that of a "left-handed" α residue conformation.
Substitution of each of the three Loop 1 residues in 2 with the homologous β residue was strongly destabilizing. ΔTM values ranged from −9.1 to −19.3 °C among 2(A17β3A), 2(D18β3D) and 2(G19βG). At two of the three loop positions, however, substitution with the appropriate enantiomer of ACPC provided variants (2(A17[S,S]ACPC) and 2(G19[R,R]ACPC)) that displayed a conformational stability comparable to that of the parent α-peptide. In contrast, 2(D18[S,S]ACPC) was strongly destabilized (ΔTM = −18.6 °C). This variant is nearly indistinguishable from the corresponding β3-containing analogue, 2(D18β3D), in terms of TM.
WW domain variant 3 contains a two-residue Loop 1 segment in which both residues occupy backbone torsion angles in the unusual αL or γL regions. Substitution of either sequence position with the analogous β3-residue was strongly destabilizing. For variant 3(N17β3N), this destabilization may arise from the inability of β3hAsn to mimic the distinctive conformational behavior observed for Asn residues in proteins. Asn can adopt φ,ψ angles in the αL/γL region of the Ramachandran plot, possibly because of favorable dipolar interactions between the side chain and the backbone[34]; however, it is unclear whether comparable conformational options are favorable for β hAsn. In contrast to the substantial destabilization resulting from α→β3 substitution at each position in the two-residue Loop 1 of 3, one α→ACPC substitution, to generate 3(N17[R,R]ACPC), is stabilizing relative to 3 (ΔTM = 9.7 °C). The other α→ACPC substitution, to generate 3(G18[R,R]ACPC), is destabilizing (ΔTM = −8.2 °C). At both loop positions in 3, β3→ACPC substitution is strongly stabilizing.
X-ray crystal structures of β-containing WW domain variants
To gain structural insights into the impact of α→β replacement in Loop 1 of 1, 2, and 3, we sought to crystallize each of our β-containing WW domain variants (Table 1). These efforts produced crystals suitable for diffraction measurements for three variants, 1(S18β3S), 1(S18[S,S]ACPC), and 3(N17[R,R]ACPC). Both 1(S18[S,S]ACPC) and 3(N17[R,R]ACPC) crystallized in space group P43212, while 1(S18β3S) crystallized in space group P6522. Initial attempts to crystallize 1(S18[S,S]ACPC) as a quasiracemate[35–37] with the enantiomer of parent WW domain 1 (i.e., the D-polypeptide) produced homochiral crystals of 1(S18[S,S]ACPC), leading us to abandon quasiracemic crystallization efforts.
Following X-ray data collection, we solved each structure via molecular replacement and refined the crystallographic models to resolution limits of 1.99 Å, 1.50 Å and 1.80 Å, for 1(S18β3S), 1(S18[S,S]ACPC), and 3(N17[R,R]ACPC), respectively (Table S1). Despite occupying two different space groups, all three structures manifest a crystal packing arrangement featuring an intermolecular contact in which Pro9 on one molecule is inserted into the substrate-binding cleft formed by Tyr23’ and Trp34’ on an adjacent, symmetry-related molecule (Figure S1). This packing contact, which is structurally analogous to binding of proline-containing substrates by the Pin1 WW domain[31], is found also in previously reported structures of the racemic Pin1 WW domain (PDB: 4GWT and 4GWV)[32]. That this contact is observed in structures of four distinct WW domain variants (one of these crystallized in two different racemic forms) points to the importance of this interaction mode both in partner recognition by WW domains and in facilitating crystallization of the Pin1 WW domain.
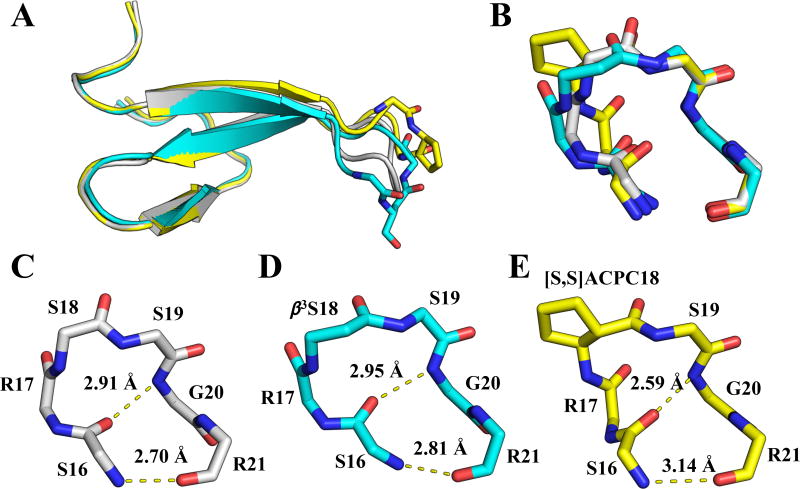
In all three X-ray structures reported here, the overall WW domain fold is unchanged by incorporation of a β-residue in Loop 1. However, differences in the conformation of Loop 1 are apparent between the all-α parent polypeptide and each β-residue-containing variant. Superposition of structures of 1, 1(S18β3S) and 1(S18[S,S]ACPC) shows global differences in the orientation of Loop 1 (Figure 3a). These differences may arise from packing interactions, which vary among the structures. Superposition of the isolated loops (S16 – R21) reveals distinct differences in the orientations of hydrogen bonding groups within each loop (Figure 3b). The substitution S18→β3S leads to a slight lengthening of both hydrogen bonds (N--O distance) within Loop 1 relative to the hydrogen bond lengths in structure 4GWT (Figure 3c–d). In contrast, the substitution S18→[S,S]ACPC leads to a significant shortening of one of these hydrogen bonds (by ~0.3 Å) and significant elongation of the other (by ~0.4 Å) (Figure 3c,e). In addition to being shortened by S18→[S,S]ACPC substitution, the Gly20(NH)→S16(CO) hydrogen bond is significantly more linear in this variant. Analysis of hydrogen bonds in organic crystals suggests that a linear arrangement of donor-H-acceptor is expected to be the most favorable hydrogen-bonding geometry.[38]
Figure 4.
(A) Overlay of structures of 1 (grey; PDB 4GWT), 1(S18β3S) (cyan) and 1(S18[S,S]ACPC) (yellow). (B) Close-up view of superimposed, isolated loops from (A). (C–E) 4:6 hydrogen-bonding arrangements in the three structures shown in (A) and (B), with distances between amide nitrogen and oxygen atoms reported in Å. For all residues except ACPC, only backbone atoms are shown.
The Loop 1 conformations differ somewhat between the crystal structures of 1(S18β3S) and 1(S18[S,S]ACPC) (Figure 3), but the β residues in these structures exhibit similar local conformations. β residue conformations are characterized by three backbone dihedral angles (φ, θ, and ψ) rather than the two angles within α residues (φ and ψ). Table 3 compares the β residue backbone dihedral angles observed in the three new structures with dihedral angle ranges previously observed[22] for β3 residues and cyclic β residues within α/β-peptides that adopt α-helix-like conformations. For 1(S18β3S) and 1(S18[S,S]ACPC), backbone dihedral angles of the β residues (β3-hSer and (S,S)-ACPC, respectively) are similar to one another. The φ and θ angles lie within or near the ranges reported for α-helix-mimetic β residues in both cases, and the ψ angle for (S,S)-ACPC in 1(S18[S,S]ACPC) lies near the α-helix-mimetic range as well. However, the ψ angle for β3-hSer in 1(S18β3S) lies well outside this range. For (R,R)-ACPC in 3(N17[R,R]ACPC), the θ and ψ torsion angles lie within or near the opposite ranges reported for α-helix-mimetic cyclic β residues, but the φ angle deviates considerably.
Table 3.
Comparison of β residue backbone dihedral angles.a
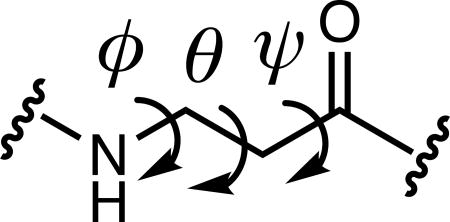
| |||
|---|---|---|---|
|
| |||
| ϕ (deg) | θ (deg) | ψ (deg) | |
| 1(S18β3S) | −126.0 | 73.4 | −149.4 |
| 1(S18[S,S]ACPC) | −100.1 | 89.3 | −129.3 |
| 3(N17[R,R]ACPC) | 49.0 | −92.3 | 126.4 |
| β3-residues22 | (−97 to −128) | (77 to 88) | (−96 to −116) |
| [S,S]ACPC/APC22 | (−105 to −133) | (78 to 104) | (−96 to −125) |
Backbone dihedral angles measured for β residues from reported structures of WW domain variants, compared to ranges of angles previously reported by Price et al.[22] for β3 residues as well as cyclic β residues [S,S]ACPC and its closely related nitrogen-containing analog [S,S]APC in a helical prototype. Angles derived from work of Price et al. are tabulated based on reported 95% confidence intervals, with limits shown here corresponding to extrema of reported ranges of angles (i.e., mean values plus or minus 95% confidence interval to produce maximum range for each angle). Note that hand of [R,R]ACPC (and sign of angles measured) is reversed relative to its enantiomer [S,S]ACPC.
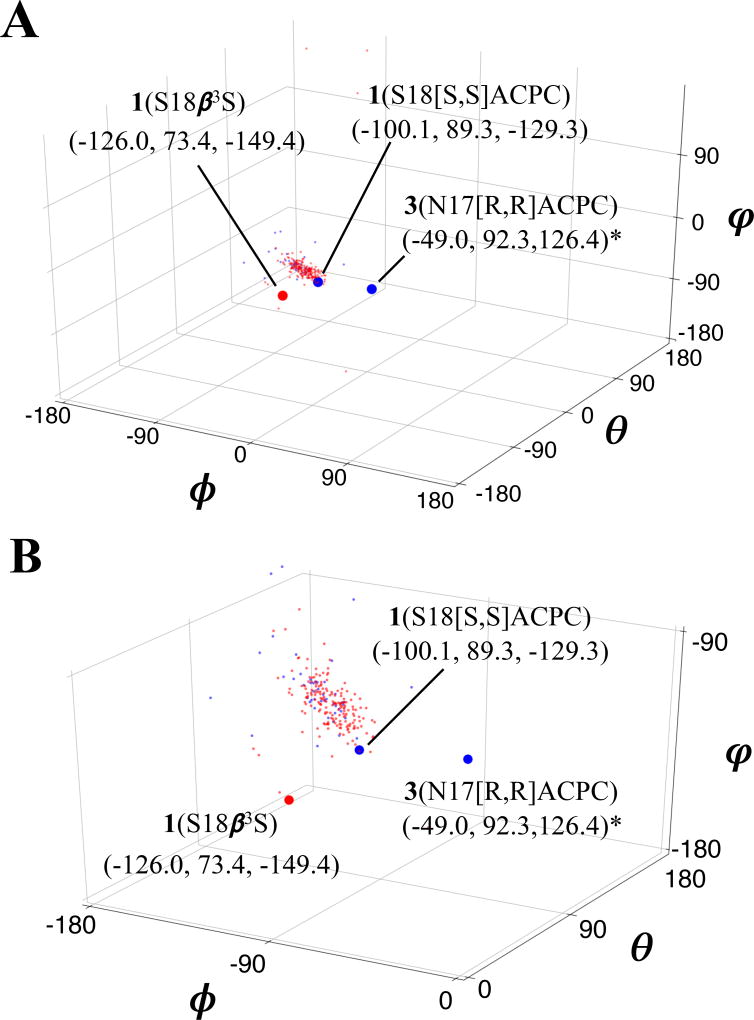
Comparison of β residue backbone torsion angles from the three structures reported here to a larger survey of [S,S]ACPC and β3-amino acid backbone angles from x-ray structures from the Protein Data Bank (PDB)[39] reveals differences that presumably arise from variability in the structural contexts of these β residue replacements. While β residues (both ACPC and β3 residues) represented in the PDB overwhelmingly appear in helical contexts, and are tightly clustered in terms of φ/θ/ψ (Figure 4), two of the three β residues in the structures we report (i.e., 1(S18β3S) and 3(N17[R,R]ACPC)) lie outside the distribution of “helix-like” backbone conformations. The same two residues lie outside the ranges of β residue backbone angles reported by Price et al.[22] It is unclear at this point whether the backbone torsion angles observed for these two β residues lie in a uniquely “turn-like” region of φ/θ/ψ-space, and further structural characterization of β residue substitutions in loop structures will be necessary to elucidate the relationship that may exist between parent loop structures and β residue-substituted variants thereof, represented in Ramachandran and φ/θ/ψ-space, respectively. However, it seems likely that successful β residue replacements in different loop structures will follow predictable sequences of backbone torsion angles, as seen for α-residues in loops and turns.[25]
Figure 5.
Global (A) and zoomed-in (B) views of β3-residue (red; 185 shown) and ACPC (blue; 46 shown) backbone geometry in φ/θ/ψ space, from previously-reported structures (see Supporting Information for relevant PDB accession codes) and three WW domain x-ray structures reported here. Backbone angles corresponding to the new WW domain structures are shown as large red or blue circles. *Note that in both plots, signs of backbone dihedral angles for [R,R]ACPC in structure of 3(N17[R,R]ACPC) have been reversed to facilitate comparison with angles derived from “right-handed” residues.
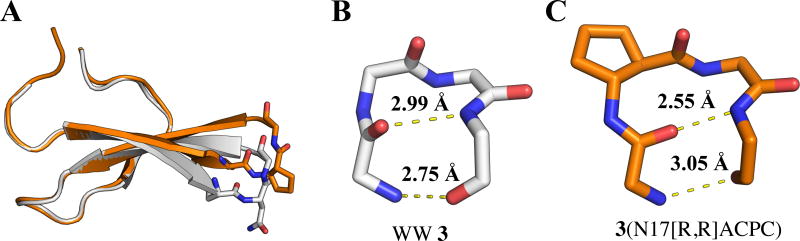
Among the β residue backbone torsion angles summarized in Table 3, the φ value for (R,R)-ACPC in 3(N17[R,R]ACPC) is the most dramatic outlier relative to torsion angles observed for β residues in α-helix-like conformations. This φ torsion angle orients the carbonyl of the preceding residue (Ser16) toward the interior of the β-hairpin. We compared the conformation of Loop 1 in PDB entry 1ZCN, which contains 3 crystallized in the context of the full Pin1 enzyme, with the conformation of the substituted loop from the structure of 3(N17[R,R]ACPC) (Figure 5). The substitution of ACPC into the two-residue loop of 3 leads to significant shortening of one hydrogen bond (R21(NH)→S16(CO); by 0.4 Å) and significant lengthening of another (S16(NH)→R21(CO), by 0.3 Å). A related pair of hydrogen bond length changes was observed for 1(S18[S,S]ACPC) relative to 1 itself. In 3(N17[R,R]ACPC), the shortened hydrogen bond is significantly more linear than in the structure of 3 (PDB: 1ZCN). In contrast, the lengthened hydrogen bond found in 3(N17[R,R]ACPC) is distorted away from a linear donor-H-acceptor arrangement.
Figure 6.
(A) Overlay of structures of 3 (grey; PDB 1ZCN) and 3(N17[R,R]ACPC) (orange). β-hairpin hydrogen bond distances (O—N) in 3 (B) and 3(N17[R,R]ACPC) (C) shown in Å.
DISCUSSION
The set of tertiary structures we employed as starting points has allowed us to evaluate the impact of systematic α→β replacement across a series of homologous loops that varies incrementally in length. Our findings reveal trends that should inform future efforts to incorporate β-amino acid residues into surface loops of target proteins. The extent to which these trends represent robust design guidelines will not be clear, however, until comparable loop-replacement studies have been conducted in the context of other tertiary folding motifs.
The impact of α→β3 replacement (which includes Gly→βhGly replacement, for the purposes of this analysis) differs between the native Pin1 WW domain (1) and analogues 2 and 3, in which the progressively shorter loops presumably generate higher local order. For the four-residue loop in 1, replacement of the original α residue with the homologous β residue at R17, S18 or G20 has relatively little impact on conformational stability (Table 2, Figure 2). In contrast, α→β3 replacement at any of the three Loop 1 positions in 2 or either of the Loop 1 positions in 3 is highly destabilizing, according to thermal denaturation data. Each of these backbone modifications places an additional CH2 unit into the loop, relative to the original all-α loop. Conformational destabilization that results from this backbone lengthening could arise from an increased entropic cost for folding, which presumably requires restriction of rotation about the "extra" backbone bond in a β-containing loop relative to the homologous all-α loop. An alternative hypothesis is that the added CH2 in the β-containing loop raises the enthalpic cost of folding because achieving the proper tertiary structure requires development of strain within the backbone-modified loop.[40] These two destabilizing mechanisms could operate in tandem. α→β3 replacements have previously been observed to cause destabilization of other small tertiary structures including the B1 domain of protein G (GB1)[41] and the villin headpiece subdomain (VHP)[37].
In light of the tertiary structural destabilization that is common for α→β3 replacements, it is noteworthy that such replacements have little effect on stability at three of the four Loop 1 positions in the native Pin1 WW domain. This behavior may arise because Loop 1 appears to be highly dynamic within the WW domain tertiary structure.[32,42] If the local mobility of this segment changes little between the unfolded and folded states, then introducing additional flexibility via α→β3 substitution would not necessarily increase the entropic cost of folding. The observation that α→β3 substitution at S19 causes a pronounced destabilization, however, shows that even in a segment that remains flexible after folding, some positions may not accommodate backbone modification without significant energetic cost. The diminished stability of the 1(S19β3S) tertiary structure relative to that of 1 may indicate that S19 plays a role in orienting the backbone NH of G20 for hydrogen bonding to the carbonyl of S16. This hydrogen bond is seen in the crystal structure of 1[31–32] as well as in our new crystal structures of 1(S18β3S) and 1(S18[S,S]ACPC) (Figure 4).
The variable loop lengths in our set of poly-α-peptide starting molecules, 1–3, provide multiple opportunities to compare the effects of flexible replacements (β3 homologue or βhGly) and constrained replacements (ACPC) in terms of tertiary structure stability. Flexible vs. constrained β residue comparisons in several systems involving individual helices suggested indirectly that β3→cyclic β replacements stabilize an α-helix-like conformation.[2,22] For helices with the right-handed twist that is favored by L-α-amino acid residues, the most favorable β residue constraint observed to date is that manifested by (S,S)-ACPC. Multiple crystal structures show that this type of constraint is compatible with the backbone φ, θ and ψ torsion angles displayed by β3 residues in α-helix-like conformations. However, β3 vs. cyclic β residue comparisons in the context of specific tertiary structures have revealed more complex behavior. For α-helical segments within both GB1 and VHP, some β3→cyclic β replacements enhance tertiary stability while others have little effect on tertiary stability or are mildly destabilizing.[37,41] The results we report for β3→cyclic β replacements within loop segments of Pin1 WW domain-derived polypeptides constitute a significant expansion of the knowledge data base related to β-amino acid substitutions outside α-helical contexts.
The first three of the four Loop 1 residues in the native Pin1 WW domain display local conformations in or near the αR region of the Ramachandran plot. Therefore, we used (S,S)-ACPC as the replacement for R17, S18 and S19. In each case, the resulting Pin1 WW domain variant manifested conformational stability very similar to that of the analogue containing a flexible β3 residue, according to thermal denaturation data. At the fourth Loop 1 position, G20, (S,S)-ACPC proved to be highly destabilizing, but (R,R)-ACPC was highly stabilizing. This stereochemical preference can be rationalized based on the crystal structure of 1, which shows G20 in the γL region of the φ,ψ plot. This comparison of the two diastereomeric derivatives of 1 bearing an ACPC residue in position 20 led us to focus on (R,R)-ACPC for replacement of G19 in 2, N17 of 3 and G18 in 3, because the crystal structures of 2 and 3 shows the two Gly residues to display γL backbone torsion angles and the Asn residue to display αL torsion angles.
For two of the three Loop 1 residues in 2, introduction of the ACPC residue with the appropriate configuration enhances stability relative to the β3 homologue or βhGly at that position. At position 18, in contrast, substitution with β3-hAsp or (S,S)-ACPC leads to a similar outcome. For both of the Loop 1 residues in 3, introduction of (R,R)-ACPC improves stability relative to β3-hAsn or β-hGly. In the former case, the ACPC-containing polypeptide displays a substantially elevated TM relative to 3.
CONCLUSIONS
Past studies of α-helical peptides and small tertiary structures have shown that α→β3 substitutions generally lead to a decrease in conformational stability, but that the original secondary or tertiary structure is often retained. The results of our α→β3 substitution studies in two-residue and three-residue variants of the Pin1 WW domain are consistent with these precedents. Comparable modifications of the native four-residue Loop 1 of the Pin1 WW domain, however, indicate that a flexible segment[32,42] can accommodate α→β3 substitutions at some positions without significant loss of conformational stability.
All previous studies of α→ACPC replacement have employed the (S,S) enantiomer, because these replacements have occurred in α-helix-forming segments (αR region of φ,ψ plot). Among our starting structures, however, several Loop 1 residues display backbone torsion angles in the unusual αL or γL region. Our comparison of the two ACPC enantiomers as replacements for G20 in 1 suggests that (R,R)-ACPC is the more appropriate replacement at such positions. Results from α→β replacements at all nine Loop 1 positions among 1–3 indicate that a variant containing the appropriate ACPC enantiomer can be either comparable in stability to the variant containing a β3 replacement at the same site in Loop 1, or significantly more stable than the β3 variant (five of nine cases). In two cases, α→(R,R)-ACPC replacement generates a polypeptide with significantly enhanced thermal stability relative to the all-α polypeptide. It is not obvious from our data how to predict which loop positions will be indifferent to flexible β vs. constrained β substitution, and which will give rise to conformational stabilization for constrained β relative to flexible β substitution. However, it is noteworthy that three of the four indifferent loop sites in our set occur in the flexible Loop 1 of the native Pin1 WW domain (1), and we speculate that flexible loops will in general be more prone to such flexible vs. constrained β indifference relative to rigid loops.
The trends we observe should provide a basis for implementing α→β replacements in the loops of other tertiary structures without compromising the folding pattern or conformational stability. This type of modification might provide protein analogues that display diminished susceptibility to proteolysis and thereby improved performance in vivo. The results obtained with the native Pin1 WW domain are particularly interesting in this regard. Loop 1 in this variant retains considerable flexibility upon WW domain folding, and α→β3 substitutions are well tolerated at three of four Loop 1 positions. Flexible loops can be sites for enzymatic cleavage of proteins[43], and a single β residue is expected to confer significant resistance to proteolysis for a few peptide bonds on either side of the substitution site. Protein loops are commonly found at protein-protein interaction interfaces[44], and β residue substitutions within these contexts might prove useful for modulating signaling pathways.[6–8] Recent results suggest that it may ultimately be possible to generate protein analogues with occasional α→β3 substitution via ribosomal synthesis[45–47], which raises the prospect of employing targeted replacements in flexible loops to generate metabolically stabilized variants of full-fledged proteins. Future studies could also evaluate whether either enantiomer of the cyclic β residue cis-ACPC[48], which has not been explored as a replacement for α-amino acids in proteins, can be accommodated by or even stabilize protein secondary structures such as loops or β-turns.
EXPERIMENTAL SECTION
Solid-phase peptide synthesis
All WW domain variants were prepared via solid-phase synthesis employing an Fmoc protecting group strategy. WW domain polypeptides were purified by reversed-phase high-performance liquid chromatography (HPLC), and polypeptide purity and identity were assessed by analytical HPLC and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry, respectively. Full details on WW domain synthesis, purification and characterization can be found in the Supporting Information.
Thermal denaturation measurements
Circular dichroism data were collected on an Aviv model 420 spectropolarimeter, using solutions of WW domain variants (~40 µM) in 10 mM sodium phosphate buffer (pH 7). For each WW domain variant, the circular dichroism signal at a 227 nm maximum[33] was monitored as a function of temperature over a range of 2 – 98 °C, and melting temperatures (TM) were obtained as unfolding transition midpoints by fitting data to a two-state model. For each WW domain variant, an average TM value and standard deviation were obtained from individual fits to three replicate measurements. Full details on thermal denaturation experiments can be found in the Supporting Information.
WW domain variant crystallization
Crystals of 1(S18β3S), 1(S18[S,S]ACPC), and 3(N17[R,R]ACPC) were grown via hanging-drop vapor diffusion experiments using sparse-matrix screens available from Hampton Research (Index and Crystal Screen II). For 1(S18β3S) and 1(S18[S,S]ACPC), crystals were transferred to precipitant solutions supplemented with glycerol prior to vitrification in liquid N2. For 3(N17[R,R]ACPC), crystals were transferred to Paratone-N prior to vitrification. For 1(S18[S,S]ACPC) and 3(N17[R,R]ACPC), diffraction data were collected using synchrotron radiation, while for 1(S18β3S), diffraction data were collected using a sealed-tube Mo X-ray source (Bruker). Full details on WW domain variant crystallization, X-ray data collection and processing can be found in the Supporting Information.
WW domain variant structure solution and refinement
X-ray structures of 1(S18β3S), 1(S18[S,S]ACPC), and 3(N17[R,R]ACPC) were solved via molecular replacement in Phaser[49] and refined via maximum likelihood methods in Refmac5[50]. Full details on WW domain X-ray structural solution and model refinements can be found in the Supporting Information. X-ray crystallographic model coordinates and structure factors have been deposited in the PDB[39] under accession codes 5VTK (1(S18β3S)), 5VTJ (1(S18[S,S]ACPC)), and 5VTI (3(N17[R,R]ACPC)).
Supplementary Material
Acknowledgments
This work was supported in part by the NIH (R01 GM061238 to SHG and R01 GM100346 to Michael G. Thomas and KTF). DEM was supported in part by a Biophysics Training Grant (T32 GM08293), and DFK was supported in part by a Biotechnology Training Grant (T32 GM00839). Use of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC0206CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
References
- 1.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Horne WS, Price JL, Gellman SH. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar M–I, Purcell AW, Devi R, Lew R, Rossjohn J, Smith AI, Perlmutter P. Org. Biomol. Chem. 2007;5:2884–2890. doi: 10.1039/b708507a. [DOI] [PubMed] [Google Scholar]
- 4.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson LM, Mortenson DE, Yun HG, Horne WS, Ketas TJ, Lu M, Moore JP, Gellman SH. J. Am. Chem. Soc. 2012;134:7317–7320. doi: 10.1021/ja302428d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boersma MD, Haase HS, Peterson-Kaufman KJ, Lee EF, Clarke OB, Colman PM, Smith BJ, Horne WS, Fairlie WD, Gellman SH. J. Am. Chem. Soc. 2012;134:315–323. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheloha RW, Maeda A, Dean T, Gardella TJ, Gellman SH. Nat. Biotech. 2014;32:653–655. doi: 10.1038/nbt.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hager MV, Johnson LM, Wootten D, Sexton PM, Gellman SH. J. Am. Chem. Soc. 2016;138:14970–14979. doi: 10.1021/jacs.6b08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jäger M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz H, Espina JR, Kelly JW. J. Am. Chem. Soc. 1992;114:8316–8318. [Google Scholar]
- 11.Jean F, Buisine E, Melnyk O, Drobecq H, Odaert B, Hugues M, Lippens G, Tartar A. J. Am. Chem. Soc. 1998;120:6076–6083. [Google Scholar]
- 12.Odaert B, Jean F, Boutillon C, Buisine E, Melnyk O, Tartar A, Lippens G. Protein Sci. 1999;8:2773–2783. doi: 10.1110/ps.8.12.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaul R, Angeles AR, Jäger M, Powers ET, Kelly JW. J. Am. Chem. Soc. 2001;123:5206–5212. doi: 10.1021/ja0102890. [DOI] [PubMed] [Google Scholar]
- 14.Fuller AA, Du D, Liu F, Davoren JE, Bhabha G, Kroon G, Case DA, Dyson HJ, Powers ET, Wipf P, Gruebele M, Kelly JW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca M, Kent SBH, Alewood PF. Protein Sci. 1993;2:1085–1091. doi: 10.1002/pro.5560020702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viles JH, Patel SU, Mitchell JBO, Moody CM, Justice DE, Uppenbrink J, Doyle PM, Harris CJ, Sadler PJ, Thornton JM. J. Mol. Biol. 1998;279:973–986. doi: 10.1006/jmbi.1998.1764. [DOI] [PubMed] [Google Scholar]
- 17.Arnold U, Hinderaker MP, Nilsson BL, Huck BR, Gellman SH, Raines RT. J. Am. Chem. Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson EG, Thornton JM. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karle IL, Awasthi SK, Balaram P. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8189–8193. doi: 10.1073/pnas.93.16.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanger HE, Gellman SH. J. Am. Chem. Soc. 1998;120:4236–4237. [Google Scholar]
- 21.George KL, Horne WS. J Am. Chem. Soc. 2017;139:7931–7938. doi: 10.1021/jacs.7b03114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price JL, Horne WS, Gellman SH. J. Am. Chem. Soc. 2010;132:12378–12387. doi: 10.1021/ja103543s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson LM, Gellman SH. Methods Enzymol. 2013;523:407–429. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sibanda BL, Thornton JM. Nature. 1985;316:170–174. doi: 10.1038/316170a0. [DOI] [PubMed] [Google Scholar]
- 25.Sibanda BL, Blundell TL, Thornton JM. J. Mol. Biol. 1989;206:759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 26.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen HI, Sudol M. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 29.Lu P–J, Zhou XZ, Shen M, Lu KP. Science. 1999;283:1325. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 30.Stewart AL, Park JH, Waters ML. Biochemistry. 2011;50:2575–2584. doi: 10.1021/bi101116a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Nat. Struct. Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 32.Mortenson DE, Kreitler DF, Yun HG, Gellman SH, Forest KT. Acta Crystallogr. 2013;D69:2506–2512. doi: 10.1107/S090744491302444X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jäger M, Dendle M, Fuller AA, Kelly JW. Protein Sci. 2007;16:2306–2313. doi: 10.1110/ps.072904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deane CM, Allen FH, Taylor R, Blundell TL. Protein Eng. 1999;12:1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 35.Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SBH. J. Am. Chem. Soc. 2008;130:9695–9701. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortenson DE, Satyshur KA, Guzei IA, Forest KT, Gellman SH. J. Am. Chem. Soc. 2012;134:2473–2476. doi: 10.1021/ja210045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreitler DF, Mortenson DE, Forest KT, Gellman SH. J. Am. Chem. Soc. 2016;138:6498–6505. doi: 10.1021/jacs.6b01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J Donohue. J. Phys. Chem. 1952;56:502–510. [Google Scholar]
- 39.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinert ZE, Horne WS. Chem. Sci. 2014;5:3325–3330. doi: 10.1039/C4SC01094A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinert ZE, Lengyel GA, Horne WS. J. Am. Chem. Soc. 2013;135:12528–12531. doi: 10.1021/ja405422v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalski JA, Liu K, Kelly JW. Biopolymers. 2002;63:111–121. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 43.Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM, Riedl SJ, Salvesen GS. Nat. Struct. Mol. Biol. 2009;16:1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavenonis J, Sheneman BA, Siegert TR, Eshelman MR, Kritzer JA. Nat. Chem. Biol. 2014;10:716–722. doi: 10.1038/nchembio.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maini R, Chowdhury SR, Dedkova LM, Roy B, Daskalova SM, Paul R, Chen S, Hecht SM. Biochemistry. 2015;54:3694–3706. doi: 10.1021/acs.biochem.5b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujino T, Goto Y, Suga H, Murakami H. J. Am. Chem. Soc. 2016;138:1962–1969. doi: 10.1021/jacs.5b12482. [DOI] [PubMed] [Google Scholar]
- 47.Czekster CM, Robertson WE, Walker AS, Söll D, Schepartz A. J. Am. Chem. Soc. 2016;138:5194–5197. doi: 10.1021/jacs.6b01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinek TA, Tóth GA, Vass E, Hollósi M, Fülöp F. Angew. Chem. Int. Ed. 2002;41:1718–1721. doi: 10.1002/1521-3773(20020517)41:10<1718::aid-anie1718>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.