Abstract
Background:
The damaging effects of exposure to environmental toxicants differentially affect genetically distinct individuals, but the mechanisms contributing to these differences are poorly understood. Genetic variation affects the establishment of the gene regulatory landscape and thus gene expression, and we hypothesized that this contributes to the observed heterogeneity in individual responses to exogenous cellular insults.
Objectives:
We performed an in vivo study of how genetic variation and chromatin organization may dictate susceptibility to DNA damage, and influence the cellular response to such damage, caused by an environmental toxicant.
Materials and Methods:
We measured DNA damage, messenger RNA (mRNA) and microRNA (miRNA) expression, and genome-wide chromatin accessibility in lung tissue from two genetically divergent inbred mouse strains, C57BL/6J and CAST/EiJ, both in unexposed mice and in mice exposed to a model DNA-damaging chemical, 1,3-butadiene.
Results:
Our results showed that unexposed CAST/EiJ and C57BL/6J mice have very different chromatin organization and transcription profiles in the lung. Importantly, in unexposed CAST/EiJ mice, which acquired relatively less 1,3-butadiene-induced DNA damage, we observed increased transcription and a more accessible chromatin landscape around genes involved in detoxification pathways. Upon chemical exposure, chromatin was significantly remodeled in the lung of C57BL/6J mice, a strain that acquired higher levels of 1,3-butadiene–induced DNA damage, around the same genes, ultimately resembling the molecular profile of CAST/EiJ.
Conclusions:
These results suggest that strain-specific changes in chromatin and transcription in response to chemical exposure lead to a “compensation” for underlying genetic-driven interindividual differences in the baseline chromatin and transcriptional state. This work represents an example of how chemical and environmental exposures can be evaluated to better understand gene-by-environment interactions, and it demonstrates the important role of chromatin response in transcriptomic changes and, potentially, in deleterious effects of exposure. https://doi.org/10.1289/EHP1937
Introduction
Inter-individual genetic variation can have profound impacts on the metabolism of pharmaceutical drugs and environmental toxicants (Ma and Lu 2011; Pierce et al. 2012). The molecular consequences of chemical exposure can therefore also vary across individuals and populations and may be attributable to variation in the expression of key metabolic genes, in the immune response, and in the DNA damage response pathway. Emerging evidence also suggests that chemical-induced effects may be transmitted transgenerationally through epigenetic means (Nadeau 2009). Yet, the underlying mechanisms for how genetics, metabolism, gene expression, and gene regulation combinatorially dictate the response to chemical exposure both within and across individuals is poorly understood.
One such genotoxic chemical with variable damaging effects in genetically diverse individuals is 1,3-butadiene. 1,3-Butadiene is an industrial chemical that is primarily used in the production of synthetic rubbers and polymers (White 2007); it is a ubiquitous environmental pollutant, is present in both automobile exhaust and cigarette smoke, and is classified as carcinogenic to humans by the World Health Organization/International Association for Research on Cancer (WHO/IARC) (Baan et al. 2009). There have been four studies on the carcinogenicity of 1,3-butadiene exposure by inhalation in mice, all conducted in the same F1 hybrid mouse strain, B6C3F1 (IARC 2008). These studies showed that 1,3-butadiene induced tumors in multiple organs in this strain at exposure concentrations ranging from and durations of exposure from . Similar systematic carcinogenicity studies have not been performed in other mouse strains.
The carcinogenicity of 1,3-butadiene is mediated through the creation of highly reactive epoxide intermediates formed during 1,3-butadiene metabolism, which damage DNA through the formation of DNA adducts (Goggin et al. 2009; Swenberg et al. 2000a, 2000b). These DNA-reactive epoxide intermediates are initially processed through phase I metabolism (bioactivation) by cytochrome P450 oxidases and later conjugated and excreted through phase II metabolism (detoxification) by broad-specificity enzymes including glutathione S-transferases (Csanády et al. 1992).
In addition to DNA damage, 1,3-butadiene also causes alterations in gene expression, bulk DNA methylation, and bulk histone modifications in mouse lung and liver tissues, but not in kidney (Chappell et al. 2014; Koturbash et al. 2011b). Interestingly, these 1,3-butadiene–induced effects were found to vary across genetically divergent inbred mouse strains (Koturbash et al. 2011a). In particular, C57BL/6J (a Mus musculus domesticus subspecies and a classic laboratory strain) exhibited high levels of DNA adduct formation and changes in bulk histone modifications, whereas CAST/EiJ (a M. musculus castaneus subspecies and a wild-derived strain) exhibited relatively low levels of DNA adduct formation and bulk histone modifications. Because changes in adduct formation and bulk histone modifications do not specify which loci in the genome are affected, the mechanism behind these strain-specific differences is unknown.
Here, we sought to understand how genetic divergence influences the response to and consequences of chemical exposure. We therefore studied CAST/EiJ and C57BL/6J strain-specific differences in DNA adduct formation, messenger RNA (mRNA) expression, microRNA (miRNA) expression, and chromatin accessibility in lung tissue from mice exposed through inhalation to 1,3-butadiene.
Materials and Methods
Animals and 1,3-Butadiene Exposure
Male C57BL/6J and CAST/EiJ mice (Jackson Laboratory), approximately old at time of exposure, were housed in sterilized cages in a temperature-controlled () room with a light/dark cycle and were given ad libitum access to purified water and NIH-31 pelleted diet (Purina Mills). After of acclimation, the mice ( of age) were randomly allocated to a control group exposed to clean air or to an experimental group exposed to 1,3-butadiene for 6 h a day, Monday–Friday, across a period (and returned to their respective cages following each exposure). Immediately following the final exposure, mice were euthanized by exsanguination following deep nembutal ( intraperitoneal injection) anesthesia, and lungs were excised and snap-frozen immediately in liquid nitrogen and stored at for subsequent analyses. The animals were treated humanely and with regard for alleviation of suffering, and all procedures were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
The concentration of 1,3-butadiene in the exposure chamber was monitored twice a day: an hour after the beginning and an hour before the end of each exposure. An air sample was taken from the chamber and analyzed with a CP-3800 (Varian Inc.) gas chromatograph using a gas sample loop injector and a flame ionization detector, with a separation column to isolate the 1,3-butadiene from the air for integration of the response. Over the duration of the experiment, the average concentration of 1,3-butadiene in the exposure chamber was (see Figure S1). This concentration is within the range of concentrations that have been reported to cause tumors in the lung (among other tissues) of 1,3-butadiene–exposed B6C3F1 mice in chronic inhalation studies () (IARC 2008; Melnick and Sills 2001). Further, 1,3-butadiene has been shown to have a supralinear exposure–response curve in mice exposed to 1,3-butadiene at levels between 0 and (Melnick and Sills 2001), confirming that is a concentration that is expected to cause tumorigenesis and is therefore applicable for the study of potential mechanisms of 1,3-butadiene–induced tumorigenicity.
Determination of N7-Guanine Adduct Formation
Genomic DNA was isolated from flash-frozen mouse lung tissues using a Qiagen DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. Measurement of N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) was performed using liquid chromatography/positive ion electrospray ionization/tandem mass spectrometry () as described by Goggin et al. (2009), with minor modifications.
Small RNA and mRNA Sequencing and Data Processing
Total RNA was isolated from flash-frozen tissues using a Qiagen miRNeasy Kit according to the manufacturer’s protocol. RNA purity and integrity were evaluated using a Thermo Scientific Nanodrop 2000 and an Agilent 2100 Bioanalyzer, respectively. A minimum RNA integrity value of 7.0 was required for RNA samples to be used for library preparation and sequencing. Small RNA libraries were generated using an Illumina TruSeq Small RNA Sample Preparation Kit. Single-end () sequencing was performed (Illumina HiSeq 2500; see Excel Table S1). miRNAs were annotated and quantified using a bioinformatics analysis pipeline as described by Baran-Gale et al. (2013). Briefly, small RNA-seq reads were trimmed using cutAdapt to remove remnants of the 3′-adaptor sequence. A minimum of a 10-base overlap and a maximum of one base mismatch were allowed in the adapter sequence, and any reads separated by 65 nucleotides (nt) or fewer were merged. We utilized a tiered mapping approach that first determined exact match reads to the appropriate reference genomes [C57BL/6J: NCBI Build 37; CAST/EiJ: Build 37 pseudogenome (http://csbio.unc.edu/CCstatus/index.py?run=Pseudo)] using Bowtie (Langmead et al. 2009). The remaining sequences were aligned to regions with exact match reads allowing a maximum of one mismatch in the body or up to three mismatches at the 3′-end of the read (depending on the length of the read) using SHRimP2 (David et al. 2011).
Libraries for total RNA sequencing (RNA-seq) were prepared using the Illumina TruSeq Total RNA Sample Prep Kit (Illumina, Inc.) with ribosomal depletion. Paired-end () sequencing was performed (Illumina HiSeq 2500; see Excel Table S1). Reads were quality-filtered (score of in at least 90% of nucleotides), adapters removed, and aligned to appropriate reference genomes [C57BL/6: NCBI Build 37; CAST/EiJ: Build 37 pseudogenome (http://csbio.unc.edu/CCstatus/index.py?run=Pseudo)] using GSNAP software (Wu and Nacu 2010) and RefSeq splice site annotations. “Mod” files included with the CAST/EiJ pseudogenome that detail the basepair transformations were used to create the CAST/EiJ pseuodgenome from the C57BL/6J reference genome. RefSeq gene annotations were created for the CAST/EiJ genome by mapping C57BL/6J RefSeq annotations to the pseudogenome using a reversed mod file created from the mod file available for the pseudogenome. Certain types of genomic regions are known to induce artefactual signal caused by experimental or technical biases; thus, it has been suggested that these regions be masked or ignored (Carroll et al. 2014). Mouse blacklists were therefore created in a fashion consistent with ENCyclopedia Of DNA Elements (ENCODE) (ENCODE Project Consortium 2012) for the human genome, including problematic satellite repetitive elements (CENSAT, GSAT, MurSAT, and SYNREP, as defined by RepeatMasker), regions with sequence homology to mitochondrial DNA (NumtS), rRNA, and regions on the X chomosome (chrX) with strong sequence homology to the Y chromosome (chrY). A CAST/EiJ pseudogenome blacklist file was generated in the same way after generating RepeatMasker annotations and using BLAT to get the sequence homology pieces (NumtS, chrX/Y). Postalignment quantification of reads per kilobase million (RPKM) values was conducted using an in-house script with RefSeq gene annotations.
Assay for Transposase Accessible Chromatin (ATAC) Sequencing and Data Processing
Flash-frozen tissue was pulverized in liquid nitrogen using a BioPulverizer (BioSpec). This mechanical process breaks open the cells and allows even exposure of intact chromatin to transposase. Pulverized material was thawed in glycerol containing nuclear isolation buffer to stabilize nuclear structure (Masuda et al. 1991) and then filtered through Miracloth (Calbiochem) to remove large tissue debris. Nuclei were washed and directly used for treatment with Tn5 transposase. Single-end () sequencing was performed (Illumina HiSeq 2500; see Excel Table S1). Reads were quality-filtered (requiring a quality score in at least 90% of nucleotides), adapters removed, and aligned with GSNAP software (Wu and Nacu 2010; Zhang et al. 2012) to the appropriate reference genome. Postalignment blacklist filtering was performed as described for RNA-seq reads. Using the mod files described above, genomic coordinates for aligned ATAC-seq reads from CAST/EiJ samples were converted to the C57BL/6J mm9 reference genome coordinates, which enabled direct comparison of ATAC-seq data.
mRNA, miRNA, and Chromatin Accessibility Differential Analyses
Differentially expressed mRNAs and miRNAs were identified using the package DESeq2 v. 1.8.2 (Love et al. 2014) in R (version 3.2.3; R Development Core Team) with betaPrior set to “false.” For miRNAs, a minimum threshold of 10 reads in at least five mice from one strain was required.
For accessible regions, the union set of the top 50,000 peaks, as determined by F-seq (Boyle et al. 2008), from all samples was identified. The number of mapped reads within overlapping windows (peaks smaller than were expanded to ) within peaks was computed for each sample (323,243 windows). Differential chromatin accessibility was detected using the R package csaw v. 1.2.1 (Lun and Smyth 2016), which uses methods from the edgeR package (Robinson et al. 2010). Normalization factors were computed to correct for compositional bias between samples with the windowCounts function (with ) and the normOffsets function with default parameters. The function estimateDisp was used to estimate the dispersion, and a generalized linear model was fit using glmQLFit with . Following each test, significant neighboring windows [false discovery rate , ] were merged.
Identification of miRNA Regulatory Hubs
Candidate master miRNA regulators of mRNA expression among the 1,3-butadiene-altered genes were identified using miRhub as previously described (Baran-Gale et al. 2013), using the “nonnetwork” mode and requiring a predicted target site to be conserved only in the mouse. The miRhub algorithm uses a Monte Carlo simulation to determine if the predicted regulatory effect of each miRNA on a set of differentially expressed genes is significantly greater than what is expected by chance. Seed-based target predictions were used [genes and corresponding 3′-untranslated region (UTR) sequences were downloaded from http://www.targetscan.org (Lewis et al. 2005)], and each predicted miRNA–gene interaction was scored based on the strength of the seed match, the level of conservation of the target site, and the clustering of target sites within the 3′-UTR of the target gene. The set of differentially expressed genes used for the analysis consisted of those genes that were significantly differentially expressed () in 1,3-butadiene-treated mice relative to control mice, as determined by DESeq2.
Pathway Enrichment Analysis
Significant enrichment of biological pathways associated with mRNA and chromatin accessibility differences was determined using GSAASeqSP v. 1.2 (http://gsaa.unc.edu) (Xiong et al. 2012; Q Xiong et al. 2014) and the Reactome Pathway Database (Croft et al. 2014; Baderlab.org; updated 24 July 2015; Merico et al. 2010). The number of permutations was set to 2,000. Pathways with fewer than 15 genes or more than 500 genes were not included. For RNA-seq data, genes were ranked according to the significance of differential expression between groups. For ATAC-seq data, genomic regulatory regions were associated with genes using GREAT v. 3.0.0 (McLean et al. 2010), which were then ranked according to the significance of differential chromatin accessibility. These ranked gene lists and their associated weights {[, where FDR represents the FDR value for each gene, FC represents the fold change, and log is the natural log} were used as input to GSAASeqSP. To visualize significantly enriched reactome pathways () for the RNA-seq data, “makeDendrogram.py” (pyEnrichment: https://github.com/ofedrigo/pyEnrichment) and custom in-house R scripts were used to construct bubble plots using multidimensional scaling to calculate an optimal two-dimensional (2D) arrangement of pathways based on a distance matrix of between-pathway semantic scores. For each pathway, significance is represented by color, and occupancy (number of genes in the pathway) is represented by the size of the bubble. For all other enrichment analyses, significantly enriched pathways were determined using the runGSAhyper function of the R package Piano v. 1.10.2 (Väremo et al. 2013) along with the Reactome Pathway Database.
Motif Analysis
Enriched motifs in regions with significant differential chromatin accessibility () were identified using DREME (Bailey 2011) against a background set of all accessible regions. Significant hits (, where E represents a Bonferroni-corrected motif p-value) were selected, and these motifs were annotated using Tomtom (Gupta et al. 2007) and the HOCOMOCO v. 10 database (http://hocomoco11.autosome.ru/) (Kulakovskiy et al. 2016). Proximal regions [ to the nearest transcription start site (TSS)] were analyzed separately from distal regions.
Results
Experimental Design
To compare transcriptional activity and chromatin accessibility between strains with or without 1,3-butadiene exposure (see “Methods”), we performed RNA-seq, small RNA-seq, and an assay for transposase-accessible chromatin (ATAC-seq) on lung tissue samples from two mouse strains, C57BL/6J [five control (i.e., exposed to filtered air in a chamber) and six 1,3-butadiene–exposed mice) and CAST/EiJ (seven control and six 1,3-butadiene–exposed mice). To confirm that the inhalational exposure system functioned as intended, we also quantified DNA adduct formation in the lungs of each strain following chemical exposure using liquid chromatography/positive ion electrospray ionization/tandem mass spectrometry ().
Strain-Specific DNA Damage Response to 1,3-Butadiene Exposure
Variation in 1,3-butadiene–induced DNA damage in the liver has been demonstrated between mouse strains, with higher levels of the DNA adduct N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) in the liver of exposed C57BL/6J mice than in exposed CAST/EiJ mice (Koturbash et al. 2011a). To test whether the lung exhibited a similar trend, we measured levels of THB-Gua in lung samples and found that adduct levels were also higher in the lung of exposed C57BL/6J mice than in exposed CAST/EiJ mice (Table 1).
Table 1.
Average level of N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) adducts in the lung of mice exposed to either clean air or 1,3-butadiene () for 10 days.
| Strain | Treatment | Number of lung samples | nucleotides |
|---|---|---|---|
| C57BL/6J | Control (clean air) | 5 | |
| 1,3-butadiene | 3 | *,† | |
| CAST/EiJ | Control (clean air) | 4 | |
| 1,3-butadiene | 6 | *,† |
Note: Adduct levels are represented by .
Indicates a significant difference () between the exposed and the control mice of the same strain.
Indicates a significant difference () between the exposed mice from each strain [based on one-way analysis of variance (ANOVA) with Tukey’s post hoc test].
Strain-Specific Baseline Gene Expression and Gene Regulation
To investigate the differential response to 1,3-butadiene exposure, we first compared mRNA expression (RNA-seq) in lung tissue from CAST/EiJ and C57BL/6J control mice, which were exposed to clean air, to determine whether there was variation in baseline molecular states that may have contributed to their response. We detected 10,250 genes that were differentially expressed between the two strains (), with 5,253 genes more highly expressed in CAST/EiJ and 4,997 genes more highly expressed in C57BL/6J (see Figure S2, Excel Table S2). To explore whether strain-specific differences in baseline gene expression profiles reflected changes in higher-level functional classes of genes, we performed pathway enrichment analysis using GSAASeqSP (Q Xiong et al. 2014) and the Reactome Pathway Database (Croft et al. 2014) (see Excel Table S3). We displayed the significantly altered pathways as a bubble plot, using a combination of distance measures and multidimensional scaling (Figure 1A, see “Methods”). Interestingly, phase II detoxification pathways (e.g., phase II conjugation) and DNA damage response pathways (e.g., p53-independent DNA damage response) were significantly enriched in genes more highly expressed in CAST/EiJ, whereas immune-related pathways [e.g., interferon (cytokine) signaling] were significantly enriched in genes more highly expressed in C57BL/6J (Figure 1A; see also Excel Table S3).
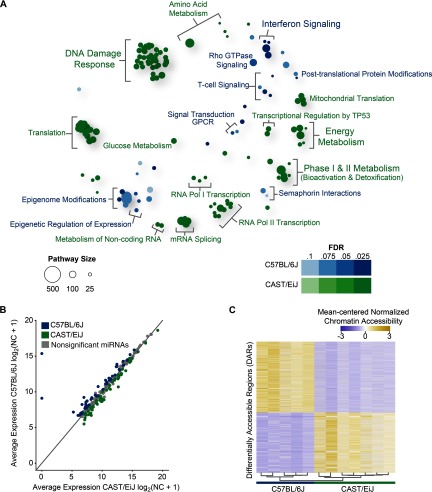
Figure 1.
Strain-specific transcriptome dynamics, microRNA (miRNA) profiles, and chromatin accessibility in the mouse lung without exposure to 1,3-butadiene. (A) Bubble plot showing enriched reactome pathways for differentially expressed genes more highly expressed in CAST/EiJ control mice compared with those more highly expressed in C57BL/6J control mice. For each pathway, significance is represented by shading, and the number of genes in each pathway is represented by bubble size. For clarity, groups of related pathways are labeled with a general descriptive term as opposed to individual pathway names. A false discovery rate was used to determine significance, but owing to the number of significant pathways, only the top 105 pathways in either direction are shown (see Excel Table S3 for a complete list). (B) Average expression of miRNAs in CAST/EiJ control (x-axis) compared with C57BL/6J control (y-axis). Expression values are based on normalized counts (NC). (C) Differentially accessible regions (DARs) identified using assay for transposase accessible chromatin (ATAC)-seq (, windows) exhibit distinct profiles in CAST/EiJ and C57BL/6J. Hierarchical clustering of samples is represented by a dendrogram. (D) Representative loci for DARs (asterisks) between CAST/EiJ control and C57BL/6J control.
Given the striking contrast between baseline CAST/EiJ and C57BL/6J gene expression in the lung, we further investigated whether miRNAs were likewise significantly different between strains. We detected 63 miRNAs that were more highly expressed in CAST/EiJ and 59 miRNAs that were more highly expressed in C57BL/6J (; Figure 1B; see also Excel Table S4). We then used miRHub (Baran-Gale et al. 2013) to determine whether any of these differentially expressed miRNAs were candidate master regulators or “regulatory hubs” of protein-coding genes that were differentially expressed between strains. Of the miRNAs that were more highly expressed in CAST/EiJ, we identified miR-210, miR-708, miR-3096-5p, and members of the miR-449a/c-5p family as regulatory hubs. In contrast, of the miRNAs that were more highly expressed in C57BL/6J, we identified miR-142-5p, miR-1264-3p, miR-350, and miR34b-5p as regulatory hubs. The up-regulation of miR-142-5p was particularly interesting because this miRNA is associated with a proinflammatory state and is critical to the pathogenesis of macrophage-mediated fibrosis in the lung (Su et al. 2015).
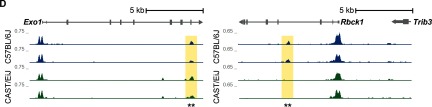
Next, we used ATAC-seq to investigate whether there was a similar modulation of chromatin accessibility. We observed dramatic strain-specific differences in the underlying chromatin landscape between CAST/EiJ and C57BL/6J. We identified 18,197 genomic regions with differential chromatin accessibility between strains (; see Excel Table S5), hereafter referred to as differentially accessible regions (DARs). Of these DARs, 8,365 exhibited greater accessibility in CAST/EiJ, whereas 9,832 exhibited greater accessibility in C57BL/6J (Figure 1C). For example, the Exo1 locus, which plays a critical role in the DNA damage response (Schaetzlein et al. 2007; Wei et al. 2003), contained a DAR with greater accessibility in CAST/EiJ (Figure 1D) and was associated with significantly increased expression in CAST/EiJ (see Excel Table S2). In contrast, a DAR in the Rbck1 locus, which encodes a protein with E3 ubiquitin ligase activity that is important in the activation of NF-κB proinflammatory signaling (Tian et al. 2007), exhibited greater accessibility in C57BL/6J (Figure 1D) and increased expression in C57BL/6J (see Excel Table S2).
As a consequence of the large number of DARs between strains, our pathway analysis identified few significantly enriched pathways in either strain at (see Excel Table S6). However, we did identify genes in interferon signaling as significantly enriched for regions more accessible in C57BL/6J relative to CAST/EiJ. We note that corresponding enrichment for this pathway was also found in the same direction based on differential gene expression (see Excel Table S3), providing evidence that interstrain differences in this immune system pathway were regulated at the chromatin level.
To identify transcription factors that may contribute to baseline expression differences between strains, we tested for enrichment of known transcription factor motifs separately in DARs either proximal () or distal to gene transcription start sites (see Figure S3). For DARs with increased accessibility in CAST/EiJ, we found significant enrichment of motifs for Hmga and Zfhx3, though interestingly, these factors were significantly up-regulated in C57BL/6J (see Excel Table S2). For DARs with greater accessibility in C57BL/6J, we found significant enrichment of motifs for Ets family, Sox family, and Sp1 (Specificity protein 1), with significantly higher expression of Ets1, several Sox genes, and Sp1 in C57BL/6J (see Excel Table S2). The ubiquitously expressed Sp1 has previously been linked to response to genotoxic chemicals (Boehme et al. 2011; Hsu et al. 2013; Magkoufopoulou et al. 2012) and is involved in the regulation of genes involved in many cellular processes, including cell proliferation, differentiation, and apoptosis, acting as both an activator and a repressor (Li and Davie 2010), perhaps through recruitment of acetyltransferases and deacetylases.
Collectively, these results demonstrate that at baseline, the molecular state in the lung was significantly different between CAST/EiJ and C57BL/6J mice in both gene expression and gene regulation. Further, these differences indicate there is variable activity in pathways that are critical for response to environmental toxicants and further suggest that baseline differences contribute to differences in susceptibility to DNA damage.
Effects of 1,3-Butadiene on Transcriptional Response and Chromatin in CAST/EiJ Mice
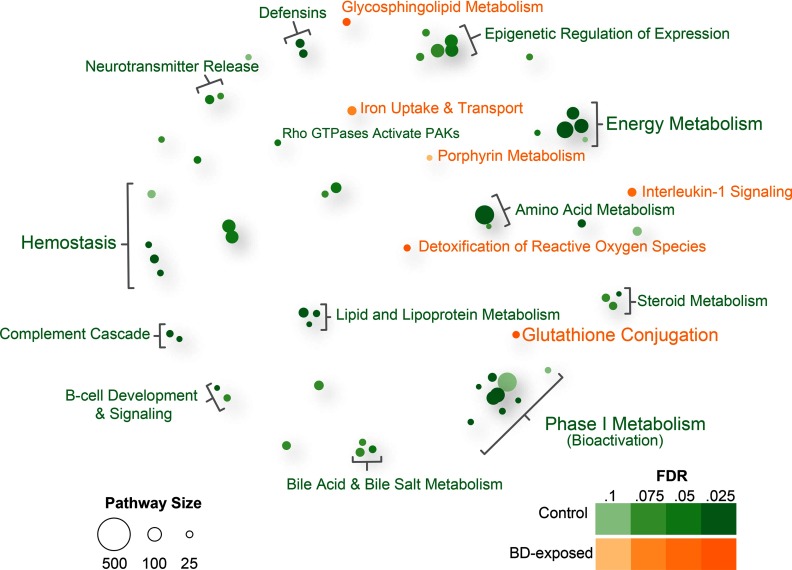
To evaluate strain-specific responses to 1,3-butadiene, we first assessed the effects of exposure on the lung transcriptome in CAST/EiJ mice, in which less DNA damage was observed than in C57BL/6J mice (Table 1), and we found that 1,3-butadiene induced relatively subtle changes, with only 113 genes up-regulated and 265 genes down-regulated () (Figure S4; see also Excel Table S2). Pathway enrichment analysis indicated the up-regulation of seven pathways (), the most significant of which was glutathione conjugation, a critical phase II metabolic pathway for the detoxification of exogenous xenobiotics such as 1,3-butadiene (Board and Menon 2013) (Figure 2; see also Excel Table S7). Down-regulated genes were enriched for a diverse set of pathways including those related to energy metabolism, hemostasis, and phase I metabolism (Figure 2; see also Excel Table S7). miRNA expression in CAST/EiJ mice was also largely unaffected by exposure, with only 10 miRNAs differentially expressed in exposed versus control mice (; see Excel Table S4). No miRNA regulatory hubs were detected. Further, 1,3-butadiene exposure had no significant effect on the chromatin landscape, resulting in no detected differentially accessible regions. Together, these findings show that the transcriptional response of CAST/EiJ mice to 1,3-butadiene exposure is intertwined with detoxification of the chemical, as indicated by the substantial shift from phase I bioactivation to phase II detoxification pathways. The lack of chromatin and miRNA changes indicates that baseline gene regulatory programs in the CAST/EiJ strain are poised to mount a robust response to efficiently detoxify chemicals such as 1,3-butadiene.
Figure 2.
Enriched reactome pathways for differentially expressed genes in lung tissue of 1,3-butadiene-exposed versus control CAST/EiJ mice. Bubble plot showing enriched reactome pathways for differentially expressed genes up-regulated in CAST/EiJ control or up-regulated in CAST/EiJ exposed to 1,3-butadiene (BD) at a false discovery rate . For each pathway, significance is represented by shading of the bubble, and the number of genes in each pathway is represented by bubble size. For clarity, groups of related pathways are labeled with a general descriptive term as opposed to individual pathway names (see Excel Table S7 for complete list). Neither microRNAs (miRNAs) nor differentially accessible regions (DARs) were differentially expressed in 1,3-butadiene versus control CAST/EiJ mice at an (data not shown).
Effect of 1,3-Butadiene on Transcriptional Responses and Chromatin in C57BL/6J Mice
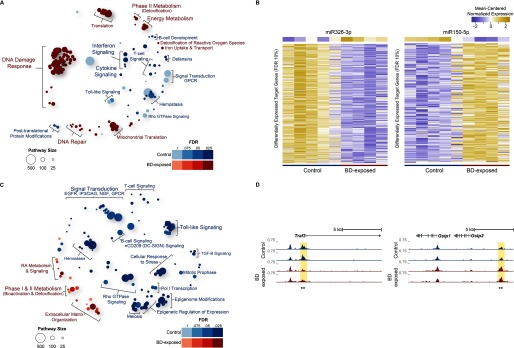
In contrast to the minimal response in CAST/EiJ mice, 1,3-butadiene exposure led to substantial changes in the lung of C57BL/6J mice, with 1,954 genes up-regulated and 2,162 genes down-regulated (; Figure S5; see also Excel Table S2). Up-regulated genes were significantly enriched in 266 pathways including DNA synthesis and replication, DNA damage response, and phase II detoxification pathways, including glutathione conjugation (Figure 3A; Excel Table S8). Down-regulated genes were enriched in 46 pathways, many related to immune signaling (e.g., interferon signaling, T-cell receptor signaling, and cytokine signaling in the immune system). miRNA expression was also significantly altered in C57BL/6J mice following 1,3-butadiene exposure, with 109 miRNAs differentially expressed between control and exposed groups (; see also Excel Table S4). Using miRhub, we identified miR-326-3p, up-regulated in response to exposure, as a significant regulatory hub (Figure 3B; ). miR-326-3p is an important regulator of cell proliferation and migration in lung cancer (Wang et al. 2016); it also promotes differentiation of interleukin 17–producing T helper cells (Du et al. 2009). We also identified miR-150-5p, down-regulated upon exposure, as a regulatory hub (Figure 3B; ). Notably, predicted mRNA targets of miR-150-5p whose expression was increased by 1,3-butadiene include the glucuronosyltransferases Ugt1a1, Ugt1a2, and Ugt1a5, which are involved in glucuronidation, a major detoxification reaction in phase II metabolism.
Figure 3.
Enriched reactome pathways for differentially expressed genes in lung tissue of 1,3-butadiene–exposed versus control C57BL/6J mice. (A) Bubble plot showing enriched reactome pathways for differentially expressed genes up-regulated in C57BL/6J control mice or up-regulated in C57BL/6J mice exposed to 1,3-butadiene (BD) at a false discovery rate . For each pathway, significance is represented by bubble shading, and the number of genes in each pathway is represented by bubble size. For clarity, groups of related pathways are labeled with a general descriptive term as opposed to individual pathway names. The top 105 pathways in either direction are shown (see Excel Table S8 for complete list). (B) miR-326-3p is a significant regulatory hub (miRHub, ) and is up-regulated in C57BL/6J in response to 1,3-butadiene (BD) exposure. The predicted messenger RNA (mRNA) targets of miR-326-3p are down-regulated in response to exposure. miR-150-5p is also a significant regulatory hub (miRHub, ) but was down-regulated in C57BL/6J in response to exposure. The predicted mRNA targets of miR-150-5p were subsequently up-regulated following exposure. (C) Bubble plot showing enriched reactome pathways for differentially accessible regions (DARs) up-regulated in C57BL/6J control or up-regulated in C57BL/6J exposed mice at an . For each pathway, significance is represented by bubble shading, and the number of genes in each pathway is represented by bubble size. For clarity, groups of related pathways are labeled with a general descriptive term as opposed to individual pathway names. Only the top 105 pathways in either direction are shown (see Excel Table S9 for complete list). (D) Representative loci for DARs (asterisks) between C57BL/6J control and C57BL/6J exposed mice.
Chromatin accessibility also changed dramatically, with 2,634 DARs identified between control and exposed C57BL/6J mice (; see also Excel Table S5). Of these, 1,533 DARs exhibited greater accessibility in control mice and were significantly enriched for immune and signaling-related pathways (Figure 3C), as shown at the Traf3 locus, which regulates pathways involved in activation of the immune response (Figure 3D) (Häcker et al. 2011). In contrast, 1,101 DARs exhibited greater accessibility in exposed mice and were enriched for both phase I bioactivation and phase II detoxification pathways (Figure 3C), such as within the glutathione S-transferase Gstp2 locus, a key enzyme in glutathione conjugation (Figure 3D) (Board and Menon 2013). Both sets of DARs were analyzed for transcription factor motif enrichment in regions proximal and distal to gene transcription start sites (see Figure S6). For DARs with increased accessibility following exposure, we observed significant enrichment of motifs for Mafk/Bach1, Dlx3 and Dbp. In contrast, in DARs with decreased accessibility following exposure, we detected enrichment of motifs for Egr4 and Sp1. In addition, Sp1 showed significantly decreased expression upon exposure (; see also Excel Table S2). This result suggests that Sp1 not only contributes to baseline differences in chromatin organization between strains (see Figure S3) but may also play a key role in the strain-specific chromatin remodeling response to exposure in C57BL/6J mice.
Together, results from the two strains indicate that the transcriptional and chromatin response of the C57BL/6J strain to 1,3-butadiene exposure is far more pronounced than that of the CAST/EiJ strain and that it centers on the down-regulation of pathways related to the immune system and on the up-regulation of pathways related to the detoxification of 1,3-butadiene and the DNA damage response.
Strain-Specific Differences in Sp1-Mediated Regulation of Transcriptional States in Response to 1,3-Butadiene
Our motif analyses identified Sp1 as enriched in DARs found in the comparison between control mice of each strain, as well as in 1,3-butdiene exposure–related DARs in C57BL/6J mice. We sought to better understand the dynamics and potential functional associations of these regions. We identified 2,188 proximal and 149 distal DARs that were significantly more accessible in C57BL/6J than in CAST/EiJ and that also contained an Sp1 motif. These C57BL/6J-enriched DARs, on average, showed decreased accessibility in C57BL/6J lung upon 1,3-butadiene exposure in proximal regions (Figure 4A), and to a lesser extent in distal regions (see Figure S7), whereas there was no change in the same regions in the lung of CAST/EiJ mice upon exposure. In addition, as noted above, Sp1 itself was significantly more highly expressed in C57BL/6J than in CAST/EiJ at baseline, and expression was significantly decreased in C57BL/6J upon exposure (see Excel Table S2), in concordance with the chromatin accessibility measures. Together, these suggest that Sp1 has a more prominent regulatory role in C57BL/6J that became repressed when these mice were exposed to 1,3-butadiene.
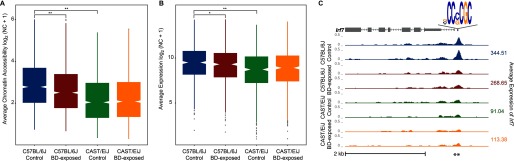
Figure 4.
Accessibility of Sp1 motif-containing differentially accessible regions (DARs) and expression of associated genes according to 1,3-butadiene (BD) exposure and strain. (A) Differences in chromatin accessibility in proximal regions containing the Sp1 motif in the lung of control and 1,3-butadiene-exposed mice of both strains (independent t-test, *, **) (B) Differences in messenger RNA (mRNA) level of genes associated with these regions in the same tissues (independent t-test, *, **). (C) Difference in mRNA level of the immune response pathway gene Irf7, the locus for which contains an Sp1 motif, following 1,3-butadiene exposure in C57BL/6J and CAST/EiJ mice.
To better assess the downstream effects of Sp1 regulation, we analyzed expression levels of putative target genes near DARs containing an Sp1 motif. For 719 genes linked to one of the 2,188 proximal DARs, we found that on average, these genes showed significantly increased expression in C57BL/6J relative to CAST/EiJ in control mice, and they showed decreased expression in C57BL/6J and no change in CAST/EiJ upon 1,3-butadiene exposure (Figure 4B). These 719 genes are enriched in immune response pathways, such as interferon signaling, as demonstrated by the interferon regulatory factor 7 (Irf7) locus (Figure 4C; see also Excel Table S9). Similarly, the 67 genes linked to one of the 149 distal DARs were expressed more highly in C57BL/6J at baseline and decreased in expression on exposure in only C57BL/6J mice (see Figure S6). Together, these findings suggest that Sp1 plays an important role in establishing the proinflammatory immune system state seen in clean air–exposed C57BL/6J (but not CAST/EiJ) that may contribute to increased DNA damage; they also suggest that the removal of Sp1 by exposure to 1,3-butadiene is associated with a decrease in these immune-related pathways.
Comparative Analysis of Strain-Specific, Exposure-Induced Pathway Enrichment
The above Sp1 results suggest that 1,3-butadiene exposure may reprogram the C57BL/6J transcriptome and chromatin landscape such that they become more like those of CAST/EiJ. To investigate this more broadly, we analyzed expression patterns across all genes and found that the majority of genes for which expression was affected by 1,3-butadiene exposure in C57BL/6J mice were directionally altered to more closely resemble expression levels in CAST/EiJ mice (Figure 5A; see also Excel Table S2). Nearly all of these genes could be divided into four groups. The first group contained 1,757 genes that were down-regulated in C57BL/6J upon exposure, thereby approximating the expression profile of CAST/EiJ (Figure 5A, Group 1), and was enriched for genes in the immune and hemostasis-related pathways (see Excel Table S10). The second group, containing 1,534 genes up-regulated in exposed C57BL/6J mice to better resemble CAST/EiJ, was enriched for genes in the phase II detoxification and DNA damage response pathways (Figure 5A, Group 2; see also Excel Table S10). These pathway enrichments were not surprising given our previous analysis of the C57BL/6J response to exposure (Figure 3A). In addition, we observed sets of 376 (Group 3) and 345 (Group 4) differentially expressed genes in exposed C57BL/6J mice in which expression in C57BL/6J diverged in direction compared with expression in CAST/EiJ. These genes were also enriched for pathways related to detoxification of 1,3-butadiene but included Phase I bioactivation pathways, which were absent in Group 2 (Figure 5A).
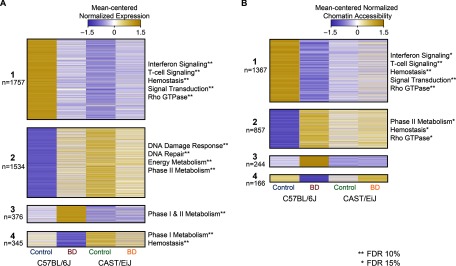
Figure 5.
Mean-centered normalized gene expression and chromatin assembly for enriched pathways according to exposure and strain. (A) Heat map of mean-centered normalized expression for genes that were differentially expressed in C57BL/6J mice following 1,3-butadiene (BD) exposure across all strains and conditions. Genes are divided into four groups based on their overall pattern of expression (see Excel Table S2 for genes in each group). For clarity, significantly enriched pathways within each group are represented by a general descriptive term (asterisks denote the significance cut-off for each pathway as indicated on the figure) as opposed to individual pathway names (see Excel Table S10 for a complete list of pathways). (B) Heat map of mean-centered normalized chromatin accessibility for regions that were differentially accessible in C57BL/6J mice following exposure across all strains and conditions. Regions are divided into four groups based on their overall pattern of accessibility (see Excel Table S5 for regions in each group). For clarity, significantly enriched pathways within each group are represented by a general descriptive term as opposed to individual pathway names (see Excel Table S10 for a complete list). FDR, false discovery rate.
The chromatin landscape of C57BL/6J was also remodeled following 1,3-butadiene exposure to more closely mirror that of CAST/EiJ (Figure 5B). DARs identified in C57BL/6J between control and exposed mice showed the same four patterns of change as those occurring with gene expression, with the first group similarly enriched for immune and hemostasis-related pathways and the second group enriched for phase II detoxification pathways (Figure 5B, Groups 1 and 2; see also Excel Table S10).
Taken together, these results suggest that CAST/EiJ mice are better poised to process the DNA-reactive metabolic intermediates of 1,3-butadiene on the molecular level relative to C57BL/6J mice. In fact, upon exposure, C57BL/6J gene expression and regulation profiles seem to migrate toward what appears to be a more optimal cellular state that efficiently mitigates the damaging effects of this chemical. This finding suggests a model (Figure 6) in which differences in susceptibility to DNA damage by environmental toxicants are largely dependent on the baseline activity of key genes and the corresponding chromatin environment. When expression and chromatin profiles are highly divergent from those necessary to efficiently combat these chemicals, DNA will incur increased damage as the necessary cellular reprogramming occurs.
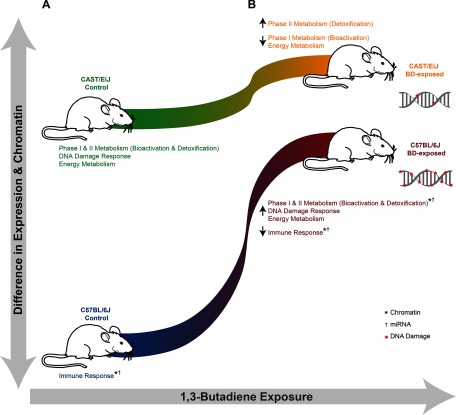
Figure 6.
Conceptual model of strain-specific differences in gene expression and chromatin state in the lung in response to 1,3-butadiene (BD) exposure. (A) Gene expression and chromatin accessibility of detoxification pathways, as well as those related to energy metabolism and the DNA damage response, in the lung of unexposed CAST/EiJ relative to the lung of unexposed C57BL/6J. (B) Changes in C57BL/6J detoxification-related gene expression and regulation profiles after exposure to 1,3-butadiene result in a molecular state that resembles that of CAST/EiJ. miRNA, microRNA.
Discussion
On a population level, environmental toxicants have serious health and economic consequences. Similar to complex diseases, the burden at the individual level can vary widely, and it has been difficult to determine the source of this variability. At a molecular level, genetic background plays a critical role in dictating an individual’s response to the environment and his or her susceptibility to disease. Yet, how this genetic variation intersects with the gene regulatory landscape to modify an individual’s ability to effectively metabolize environmental toxicants, as well as their ability to repair DNA damage resulting from toxicant exposure, remains an unexplored question. Our study represents a novel investigation into the genome-wide impact of both genetic background and exposure to the environmental toxicant 1,3-butadiene on the transcriptional and gene regulatory landscape in the mouse lung. Although 1,3-butadiene is a well-studied, classic model toxicant, little is known about the complexity of individual variation in response to exposure or to genotoxic carcinogens in general. Our study represents a significant step forward in our understanding of this complexity at a genome-wide scale. Importantly, this work represents a new paradigm for investigating the effects of chemical or environmental exposures on the transcriptional and regulatory landscapes in genetically diverse individuals.
Our results reveal striking differences in the underlying chromatin and transcriptional landscape between two genetically divergent mouse strains, CAST/EiJ and C57BL/6J, and provide strong evidence that susceptibility to the damaging effects of 1,3-butadiene exposure is associated with the baseline molecular state in the lung. In particular, we found that the baseline activities of cytochrome P450 and glutathione-S-transferase genes, key players in the bioactivation and detoxification of 1,3-butadiene, respectively, are increased in CAST/EiJ relative to C57BL/6J. The increased baseline activity of detoxification pathways, as well as those related to energy metabolism and the DNA damage response, may better position CAST/EiJ mice to more effectively metabolize 1,3-butadiene upon exposure and quickly repair chemically induced DNA damage, resulting in a reduced number of DNA adducts in this strain. Relatedly, a higher level of expression of inflammatory response genes in C57BL/6J lung than in CAST/EiJ lung in clean air-exposed mice could be related to DNA damage levels because increases in inflammation from exogenous agents have been shown to increase DNA damage (Kiraly et al. 2015).
In contrast, our results indicate that C57BL/6J mice have more active immune system–related processes than CAST/EiJ at baseline, a finding that is consistent with prior studies. In a study of respiratory infection by Influenza A and Severe Acute Respiratory Coronavirus (SARS-CoV), C57BL/6J mice were shown to incur a reduced viral load, demonstrate less weight loss, and display fewer differentially expressed genes upon infection compared with CAST/EiJ mice (H Xiong et al. 2014). Further, a separate study of SARS-CoV infection in multiple mouse strains found that the CAST/EiJ strain was extremely susceptible to infection (Gralinski et al. 2015). Therefore, whereas the CAST/EiJ strain may be at an advantage in its ability to respond to environmental toxicant exposure, the C57BL/6J strain may be better poised to fight infection. Future studies in a greater diversity of mice may also clarify whether activity levels in detoxification and immune-related response are generally anticorrelated.
Upon exposure to 1,3-butadiene, distinct chromatin and transcriptional responses were observed in a strain-specific manner. Notably, CAST/EiJ mice exhibited a relatively minor response to 1,3-butadiene exposure, with no overt changes in DNA damage response or repair pathways. This finding suggests that the level of DNA damage in CAST/EiJ is not high enough to induce DNA damage repair activity and may be due to more efficient recognition of DNA damage and the increased expression of detoxification genes in CAST/EiJ relative to C57BL/6J at baseline.
In contrast, exposure to 1,3-butadiene elicited a profound chromatin and transcriptional remodeling response in C57BL/6J, primarily characterized by a down-regulation of genes with immune-related functions and an up-regulation of genes involved in detoxification, DNA damage response, and DNA repair pathways. The down-regulation of immune-related pathways was particularly interesting because immunosuppression has been related to carcinogenesis (Pardoll 2015) and has been reported in humans and animals exposed to cigarette smoke (Mehta et al. 2008), pesticides (Banerjee et al. 1996), and mycotoxins (Capriotti et al. 2012), all of which have genotoxic potential. Further, suppression of T-lymphocyte production has been reported in mice that were exposed to 1,3-butadiene (Thurmond et al. 1986), and down-regulation of regulatory T cells has also been shown to be associated with indoor exposure to the genotoxic chemical benzene (Herberth et al. 2014).
In addition to an overall down-regulation of immune-related pathways in C57BL/6J following exposure to 1,3-butadiene, we also observed a significant up-regulation of pathways related to the detoxification of 1,3-butadiene along with the large-scale chromatin rearrangements necessary to facilitate this response. This result provides further evidence that, unlike CAST/EiJ, C57BL/6J is not poised at baseline to effectively cope with 1,3-butadiene exposure and therefore must dramatically reorganize its transcriptional and regulatory landscape. Because these molecular changes take time, C57BL/6J incurs increased damage, as indicated by direct measurements of THB-Gua adducts in the lung and by the observed increased activity of pathways related to recognition, response, and repair of DNA damage.
Our results indicate that the up-regulation of immune response pathways in C57BL/6J relative to CAST/EiJ at baseline and the down-regulation of these pathways in C57BL/6J in response to 1,3-butadiene exposure are modulated by the transcription factor Sp1. Specifically, our results revealed that Sp1 is more highly expressed and that regions containing Sp1 motifs, which are significantly enriched for immune response pathways, are more accessible in C57BL/6J than in CAST/EiJ at baseline. Following exposure, Sp1 expression decreases, and regions containing Sp1 motifs are remodeled to become less accessible in C57BL/6J, thereby more closely resembling the molecular state in CAST/EiJ.
Exposure to cadmium, a toxic heavy metal, has also been shown to down-regulate Sp1 in rat lung epithelial cells (Watkin et al. 2003) and in primary mouse kidney cells (Tabatabai et al. 2005). Other genotoxic chemicals have also been reported to affect expression of Sp1 in HepG2 liver cells (Boehme et al. 2011; Magkoufopoulou et al. 2012). However, prior studies have not evaluated the role of Sp1 in modulating the transcriptional and chromatin landscape in response to toxicant exposure.
The changes that were observed in response to 1,3-butadiene exposure in C57BL/6J mice resulted in transcriptomic and chromatin profiles that more closely resembled those of CAST/EiJ mice. Collectively, our results indicate that the transcriptome and gene regulatory landscape in CAST/EiJ is better poised to process the DNA-reactive metabolic intermediates of 1,3-butadiene than that in C57BL/6J. This was particularly evident in the significant up-regulation of both detoxification pathways and pathways related to energy metabolism and the DNA damage response in CAST/EiJ relative to C57BL/6J at baseline. Interestingly, upon exposure, C57BL/6J gene expression and regulation profiles were directionally altered to more closely resemble the molecular state of CAST/EiJ. These results suggest that the baseline activity of key genes and the corresponding chromatin environment together dictate strain-specific differences in susceptibility to chemically induced DNA damage.
Limitations and Future Directions
The results from our study raise important questions to explore in future investigations regarding the role of genetic divergence in mediating susceptibility to chemically induced DNA damage. In particular, larger-scale studies in genetically diverse populations are needed to identify specific genetic variants that contribute to interindividual differences in the baseline activity of key genes and the corresponding chromatin landscape. Including a larger panel of genetically diverse inbred mouse strains will provide a better understanding of the mechanisms of how genetic variation dictates the response to and consequences of toxicant exposure. Assessing the level of DNA damage at a range of time points will help us better understand how quickly these molecular changes occur, if and when adaptation in the transcriptomic or chromatin response occurs after prolonged exposure, and how that adaptation (or the lack thereof) may affect the amount of damage incurred. Importantly, determining the persistence of these changes after an initial exposure will reveal whether there exists an epigenetic memory that may reduce the effects of future exposures.
Conclusions
Overall, the results of our investigation highlight the differences in response to genotoxic chemical exposure between two genetically divergent mouse strains across the transcriptome and gene regulatory landscape. Our results emphasize the importance of considering genetic differences in model organisms that may influence metabolism and other underlying mechanisms of toxicity, as well as that of including epigenetic end points in hazard assessments of known and potential environmental carcinogens. More broadly, these results suggest that interindividual variations in tolerance to disease susceptibility from exposures may largely depend on how prepared at a molecular level target tissues are to efficiently mitigate the damaging effects of specific environmental agents.
Supplemental Material
Supplemental Material
Acknowledgments
We would like to thank O. Fedrigo at the Duke sequencing core for providing advice on assay for transposase accessible chromatin (ATAC)-seq library sequencing; we also thank the staff at the UNC High Throughput Sequencing Facility for providing sequencing support.
All RNA-seq, small RNA-seq, and ATAC-seq data sets generated during this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession ID GSE86623 or are included in this published article (and/or the supplemental materials).
This research was funded, in part, by a grant from the National Institutes of Health (R01 ES023195).
References
- Baan R, Grosse Y, Straif K, Secretan B, El Ghissassi F, Bouvard V, et al. 2009. A review of human carcinogens–Part F: Chemical agents and related occupations. Lancet Oncol 10(12):1143–1144, PMID: 19998521, 10.1016/S1470-2045(09)70358-4. [DOI] [PubMed] [Google Scholar]
- Baderlab. Enrichment Map Gene Sets (mouse gene sets). Updated 24 July 2015 http://download.baderlab.org/EM_Genesets/July_24_2015/Mouse/symbol/Pathways/Mouse_Reactome_July_24_2015_symbol.gmt [accessed 24 August 2015]. [Google Scholar]
- Bailey TL. 2011. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27(12):1653–1659, PMID: 21543442, 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee BD, Koner BC, Ray A. 1996. Immunotoxicity of pesticides: Perspectives and trends. Indian J Exp Biol 34(8):723–733, PMID: 8979476. [PubMed] [Google Scholar]
- Baran-Gale J, Fannin EE, Kurtz CL, Sethupathy P. 2013. Beta cell 5'-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One 8(9):e73240, PMID: 24039891, 10.1371/journal.pone.0073240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board PG, Menon D. 2013. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim Biophys Acta 1830(5):3267–3288, PMID: 23201197, 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Boehme K, Dietz Y, Hewitt P, Mueller SO. 2011. Genomic profiling uncovers a molecular pattern for toxicological characterization of mutagens and promutagens in vitro. Toxicol Sci 122(1):185–197, PMID: 21527772, 10.1093/toxsci/kfr090. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. 2008. F-Seq: A feature density estimator for high-throughput sequence tags. Bioinformatics 24(21):2537–2538, PMID: 18784119, 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti AL, Caruso G, Cavaliere C, Foglia P, Samperi R, Laganà A. 2012. Multiclass mycotoxin analysis in food, environmental and biological matrices with chromatography/mass spectrometry. Mass Spectrom Rev 31(4):466–503, PMID: 22065561, 10.1002/mas.20351. [DOI] [PubMed] [Google Scholar]
- Carroll TS, Ziwei L, Salama R, Stark R, de Santiago I. 2014. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 5:75, PMID: 24782889, 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell G, Kobets T, O’Brien B, Tretyakova N, Sangaraju D, Kosyk O, et al. 2014. Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice. Toxicol Sci 142(2):375–384, PMID: 25237060, 10.1093/toxsci/kfu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. 2014. The reactome pathway knowledgebase. Nucleic Acids Res 42(Database issue):D472–D477, PMID: 24243840, 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanády GA, Guengerich FP, Bond JA. 1992. Comparison of the biotransformation of 1,3-butadiene and its metabolite, butadiene monoepoxide, by hepatic and pulmonary tissues from humans, rats and mice. Carcinogenesis 13(7):1143–1153, PMID: 1638680. [DOI] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27(7):1011–1012, PMID: 21278192, 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57–74, PMID: 22955616, 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. 2009. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 10(12):1252–1259, PMID: 19838199, 10.1289/ehp.1205266. [DOI] [PubMed] [Google Scholar]
- Goggin M, Swenberg JA, Walker VE, Tretyakova N. 2009. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res 69(6):2479–2486, PMID: 19276346, 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski LE, Ferris MT, Aylor DL, Whitmore AC, Green R, Frieman MB, et al. 2015. Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet 11(10):e1005504, PMID: 26452100, 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8(2):R24, PMID: 17324271, 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker H, Tseng PH, Karin M. 2011. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol 11(7):457–468, PMID: 21660053, 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- Herberth G, Bauer M, Gasch M, Hinz D, Röder S, Olek S, et al. 2014. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol 133(2):543–550, PMID: 23978443, 10.1016/j.jaci.2013.06.036. [DOI] [PubMed] [Google Scholar]
- Hsu T, Huang KM, Tsai HT, Sung ST, Ho TN. 2013. Cadmium(Cd)-induced oxidative stress down-regulates the gene expression of DNA mismatch recognition proteins MutS homolog 2 (MSH2) and MSH6 in zebrafish (Danio rerio) embryos. Aquat Toxicol 126:9–16, PMID: 23143036, 10.1016/j.aquatox.2012.09.020. [DOI] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer). 2008. IARC monographs on the evaluation of carcinogenic risks to humans. Volume 97. 1,3-butadiene, ethylene oxide and vinyl halides (vinyl fluoride, vinyl chloride and vinyl bromide). IARC Monogr Eval Carcinog Risks Hum 97:3–471, PMID: 10925842. [PMC free article] [PubMed] [Google Scholar]
- Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. 2015. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS Genet 11(2):e1004901, PMID: 25647331, 10.1371/journal.pgen.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Swenberg JA, et al. 2011a. Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol Sci 122(2):448–456, PMID: 21602187, 10.1093/toxsci/kfr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Tryndyak V, et al. 2011b. Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ Health Perspect 119(5):635–640, PMID: 21147608, 10.1289/ehp.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, et al. 2016. Hocomoco: Expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res 44(D1):D116–D125, PMID: 26586801, 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25, PMID: 19261174, 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20, PMID: 15652477, 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li L, Davie JR. 2010. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat 192(5):275–283, PMID: 20810260, 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550, PMID: 25516281, 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Smyth GK. 2016. csaw: A Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44(5):e45, PMID: 26578583, 10.1093/nar/gkv1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Lu AY. 2011. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev 63(2):437–459, PMID: 21436344, 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- Magkoufopoulou C, Claessen SM, Tsamou M, Jennen DG, Kleinjans JC, van Delft JH. 2012. A transcriptomics-based in vitro assay for predicting chemical genotoxicity in vivo. Carcinogenesis 33(7):1421–1429, PMID: 22623647, 10.1093/carcin/bgs182. [DOI] [PubMed] [Google Scholar]
- Masuda K, Takahashi S, Nomura K, Inoue M. 1991. A simple procedure for the isolation of pure nuclei from carrot embryos in synchronized cultures. Plant Cell Rep 10(6–7):329–333, PMID: 24221667, 10.1007/BF00193152. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28(5):495–501, PMID: 20436461, 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta H, Nazzal K, Sadikot RT. 2008. Cigarette smoking and innate immunity. Inflamm Res 57(11):497–503, PMID: 19109742, 10.1007/s00011-008-8078-6. [DOI] [PubMed] [Google Scholar]
- Melnick RL, Sills RC. 2001. Comparative carcinogenicity of 1,3-butadiene, isoprene, and chloroprene in rats and mice. Chem Biol Interact 135-136:27–42, PMID: 11397379, 10.1016/S0009-2797(01)00213-7. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5(11):e13984, PMID: 21085593, 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH. 2009. Transgenerational genetic effects on phenotypic variation and disease risk. Hum Mol Genet 18(R2):R202–R210, PMID: 19808797, 10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D. 2015. Cancer and the immune system: Basic concepts and targets for intervention. Semin Oncol 42(4):523–538, PMID: 26320058, 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, et al. 2012. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet 8(2):e1002522, PMID: 22383894, 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140, PMID: 19910308, 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, et al. 2007. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell 130(5):863–877, PMID: 17803909, 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Zhao Q, He C, Huang D, Liu J, Chen F, et al. 2015. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun 6:8523, PMID: 26436920, 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Christova-Gueorguieva NI, Upton PB, Ranasinghe A, Scheller N, Wu KY, et al. 2000a. “1,3-Butadiene: Cancer, Mutations, and Adducts. Part V. Hemoglobin Adducts as Biomarkers of Butadiene Exposure and Metabolism.” Cambridge, MA:Health Effects Institute. [PubMed] [Google Scholar]
- Swenberg JA, Ham AJ, Koc H, Morinello E, Ranasinghe A, Tretyakova N, et al. 2000b. DNA adducts: Effects of low exposure to ethylene oxide, vinyl chloride and butadiene. Mutat Res 464(1):77–86, PMID: 10633179, 10.1016/S1383-5718(99)00168-0. [DOI] [PubMed] [Google Scholar]
- Tabatabai NM, Blumenthal SS, Petering DH. 2005. Adverse effect of cadmium on binding of transcription factor Sp1 to the GC-rich regions of the mouse sodium-glucose cotransporter 1, SGLT1, promoter. Toxicology 207(3):369–382, PMID: 15664265, 10.1016/j.tox.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Thurmond LM, Lauer LD, House RV, Stillman WS, Irons RD, Steinhagen WH, et al. 1986. Effect of short-term inhalation exposure to 1,3-butadiene on murine immune functions. Toxicol Appl Pharmacol 86(2):170–179, PMID: 3787617, 10.1016/0041-008X(86)90047-5. [DOI] [PubMed] [Google Scholar]
- Tian Y, Zhang Y, Zhong B, Wang YY, Diao FC, Wang RP, et al. 2007. RBCK1 negatively regulates tumor necrosis factor- and interleukin-1-triggered NF-kappaB activation by targeting TAB2/3 for degradation. J Biol Chem 282(23):16776–16782, PMID: 17449468, 10.1074/jbc.M701913200. [DOI] [PubMed] [Google Scholar]
- Väremo L, Nielsen J, Nookaew I. 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41(8):4378–4391, PMID: 23444143, 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chen X, Xu T, Xia R, Han L, Chen W, et al. 2016. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res 6(2):173–186, PMID: 27186394. [PMC free article] [PubMed] [Google Scholar]
- Watkin RD, Nawrot T, Potts RJ, Hart BA. 2003. Mechanisms regulating the cadmium-mediated suppression of Sp1 transcription factor activity in alveolar epithelial cells. Toxicology 184(2–3):157–178, PMID: 12499119, 10.1016/S0300-483X(02)00577-2. [DOI] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, et al. 2003. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev 17(5):603–614, PMID: 12629043, 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WC. 2007. Butadiene production process overview. Chem Biol Interact 166(1–3):10–14, PMID: 17324391, 10.1016/j.cbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881, PMID: 20147302, 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Morrison J, Ferris MT, Gralinski LE, Whitmore AC, Green R, et al. 2014. Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression. G3 (Bethesda) 4(8):1429–1444, PMID: 24902603, 10.1534/g3.114.011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS. 2012. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res 22(2):386–397, PMID: 21940837, 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Mukherjee S, Furey TS. 2014. GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Sci Rep 4:6347, PMID: 25213199, 10.1038/srep06347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu Y, Schnable JC, Zeng Z, Freeling M, Crawford GE, et al. 2012. High-resolution mapping of open chromatin in the rice genome. Genome Res 22(1):151–162, PMID: 22110044, 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.