Abstract
A pyridoxal 5′-phosphate (PLP)-dependent enzyme, 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (S-adenosyl-l-Met methylthioadenosine-lyase, EC 4.4.1.14), catalyzes the conversion of S-adenosyl-l-methionine (AdoMet) to ACC. A tomato ACC synthase isozyme (LE-ACS2) with a deletion of 46 amino acids at the C terminus was chosen as the control enzyme for the study of the function of R286 in ACC synthase. R286 of the tomato ACC synthase was mutated to a leucine via site-directed mutagenesis. The ACC synthase mutant R286L was purified using a simplified two-step purification protocol. Circular dichroism (CD) analysis indicated that the overall three-dimensional structure of the mutant was indistinguishable from that of the control enzyme. Fluorescence spectroscopy revealed that the binding affinity of R286L ACC synthase for its cofactor PLP was reduced 20- to 25-fold compared with control. Kinetic analysis of R286L showed that this mutant ACC synthase had a significantly reduced turnover number (kcat) of 8.2 × 10−3 s−1 and an increased Km of 730 μm for AdoMet, leading to an 8,000-fold decrease in overall catalytic efficiency compared with the control enzyme. Thus, R286 of tomato ACC synthase is involved in binding both PLP and AdoMet.
The phytohormone ethylene participates in a variety of physiological processes during plant growth and development, including seed germination, ripening of climacteric fruit, sex expression, initiation of root hairs, gravitropism of stems, and abscission and senescence of flower petals and leaves (Yang and Hoffman, 1984; Abeles et al., 1992; Kende, 1993; Fluhr and Mattoo, 1996). Ethylene in higher plants is formed via the pathway of Met to S-adenosyl-l-Met (AdoMet) to 1-aminocyclopropane-1-carboxylic acid (ACC) and finally to C2H4 (ethylene; Adams and Yang, 1979). The committed step in the biosynthesis is the formation of ACC (Theologis, 1992; Kende, 1993; Fluhr and Mattoo, 1996), a reaction catalyzed by the pyridoxal 5′-phosphate (PLP)-dependent enzyme ACC synthase (S-adenosyl-l-Met methylthioadenosine-lyase, EC 4.4.1.14; Boller et al., 1979; Yu et al., 1979).
The proposed mechanism for the enzymatic generation of ACC involves the formation of a Schiff base between PLP and AdoMet, followed by an α, γ-elimination (Adams and Yang, 1979; Ramalingam et al., 1985; Yip et al., 1990). Earlier experiments suggested that K278 of the enzyme formed an internal Schiff base with PLP, because the amino acid residue was alkylated by the 2-aminobutyrate portion of AdoMet and formed a covalent bond with the coenzyme PLP in the presence of the reducing reagent NaB[3H]4 (Yip et al., 1990). Amino acid sequence alignment of synthases and Asp aminotransferases (AATases), both of which are members of subgroup I of aminotransferases (Mehta and Christen, 1994), have shown that 16 amino acids that are structurally or enzymatically important in AATases are all conserved in ACC synthases (Huang et al., 1991; White et al., 1994). Substitution of these highly conserved amino acids with alternative amino acids using either site-directed or random mutagenesis reduced or even abolished the enzyme activity (Tarun et al., 1998). Kinetic and/or spectroscopic analysis of several apple ACC synthase mutants with a point mutation at Y85, Y233, K273, or R407 strongly indicated the involvement of these conserved amino acids in PLP and substrate binding as well as in catalysis (Li and Mattoo, 1994; White et al., 1994).
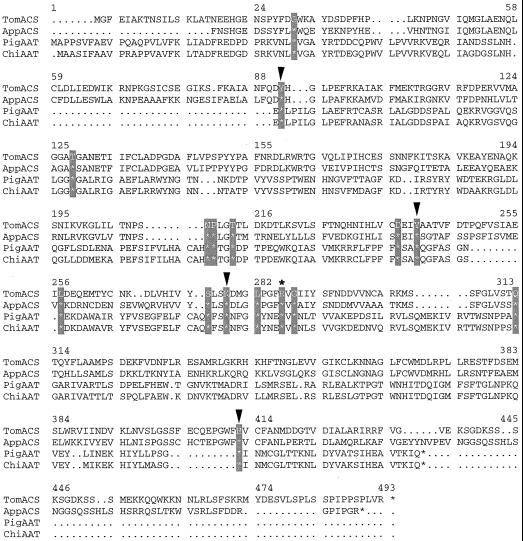
We selected R286 of the tomato ACC synthase, corresponding to R266 of the chicken heart mitochondrial AATase (Fig. 1), as the target for kinetic and biophysical study because it is one of the amino acids conserved among AATases and ACC synthases, and because its cognates have been strongly implicated in PLP binding for other PLP-dependent enzymes (Grishin et al., 1995; Osterman et al., 1997). Leu, which has a side chain size similar to Arg, was chosen as the substitute amino acid. We present results of circular dichroism (CD) spectrometry, fluorescence spectrometry, and kinetic properties of the resultant mutant enzyme, R286L. Our data strongly support the hypothesis that the conserved amino acid R286 has key roles in both PLP and AdoMet binding and in catalysis.
Figure 1.
Alignment of the amino acid sequences of AATase and ACC synthase. Amino acid sequences of tomato ACC synthase (TomACS), apple ACC synthase (AppACS), pig cytosolic AATase (PigAAT), and chicken mitochondrial AATase (ChiAAT) are compared. The 16 amino acid residues conserved among the AATase and ACC synthases are hatched. Arrowheads indicate the point mutations introduced into ACC synthase from previous studies (Li and Mattoo, 1994; White et al., 1994). The R286 of tomato ACC synthase mutated in this study is marked with an asterisk (*).
MATERIALS AND METHODS
Strains and Vectors
Bacterial strains used for subcloning were JM83 (F− araΔ [lac-proAB] rpsL [Strr] [φ80 δlacΔ{lac Z} M15] thi) from Biolab (Beverly, MA) and INVαF′ (F′ end A1 recA1 hsd R17 [rk−, mk+] sup E44 thi −1 gyr A96 rel A1*80lac ZΔM15Δ [lac ZYA-arg F] U169) from Invitrogen (Carlsbad, CA). The strain used for overexpressing ACC synthase was BL21 (DE3) pLysS: F− ompT, hsd SB (r−Bm−B), dcm, gal (DE3) pLysS, Cmr from Novagen (Madison, WI). The tomato ACC synthase gene (LE-ACS2) was amplified using PCR from a tomato-fruit-specific cDNA library and subcloned into the protein expression vector pET11d (Novagen), as previously described (Zhou et al., 1998).
Site-Directed Mutagenesis
Oligonucleotide primers for introducing R286L missense mutations into the ACC synthase were: R286LP1, 5′-T CAC ATC GTC TAC AGT CTT TCA AAA GAC ATG GGG TTA CCA GGA-3′; R286LP2, 5′-ATT AAC GAC ATC GTC GTT AAA AGA ATA TAT GAT TCC GAC TAA AAA TCC TGG TAA CCC CAT-3′.
Acc I and Tth111 I restriction sites (underlined) were incorporated into the primers. The bold, italic nucleotides AA represent the mutagenic nucleotides that convert R286 to L286. Both oligonucleotides have 15 bp of overlap at the 3′ ends. These were used to prime the synthesis of an 88-bp-long, double-stranded mutagenic DNA fragments encoding amino acid 271 to 299 in a PCR reaction containing 200 pmol of the primers and 1 unit of Taq DNA polymerase (CLONTECH Laboratories, Palo Alto, CA). The 88-bp PCR product was purified from a 2.5% (w/v) agarose gel and subcloned into a pCR2.1 vector. The PCR product was excised out of the PCR cloning vector by AccI and Tth111 I and inserted into the recombinant ACS expression plasmid pET11d*-del-1 (Li and Matto, 1994; Zhou et al., 1998), which had been predigested with AccI and Tth111 I, to generate pET11d*-ACS R286L. After confirmation of the existence of R286L mutation in the pET11d*-ACS recombinant using DNA sequencing, the plasmid was transformed into Escherichia coli BL21(DE3)pLysS.
Purification of ACC Synthase
The R286L mutant protein (R286L) and the control enzyme (del-1 ACC synthase) were overexpressed and purified from E. coli BL21(DE3)pLysS (Studier, 1991) according to a previously described method (Zhou et al., 1998).
Determination of Protein Concentration
Protein was quantified using a protein micro-assay (Bio-Rad Laboratories, Hercules, CA) (Bradford, 1976) and a microplate reader (model MR5000, Dynatech, Chantilly, VA). For immunodetection of partially purified or purified ACC synthase, proteins were fractionated by SDS-PAGE on 12% (w/v) polyacrylamide gels and stained with 0.1% (w/v) Coomassie Brilliant Blue G-250. Protein markers used in SDS-PAGE were: phosphorylase b, 94 kD; BSA, 67 kD; ovalbumin, 43 kD; carbonic anhydrase, 30 kD; trypsin inhibitor, 20.1 kD; and α-lactal albumin, 14.4 kD. Western blotting was performed using a primary antibody raised against ACC synthase del-1 (Zhou et al., 1998) and an alkaline phosphatase-conjugated goat anti-rabbit IgG (H+L) secondary antibody according to the manufacturer's protocol (Qiagen USA, Valencia, CA).
Determination of Kinetic Parameters of ACC Synthase
Control and mutant (R286L) ACC synthase were subjected to gel filtration prior to biochemical and biophysical analysis. The column consisted of two individual 24-mL Superdex 200 columns linked in series and was pre-equilibrated with the gel filtration buffer containing 20 mm Tris-HCl, pH 7.5, 0.25 mm EDTA, 2 mm DTT, 0.15 m NaCl, and 1% (w/v) glycerol.
The ACC synthase assay was carried out in 500 μL of buffer containing 50 mm KH2PO4 (pH 9.5) and 5 mm DTT supplemented with various concentrations of AdoMet and PLP. The amounts of the enzyme used in the kinetic study were 0.15 μg for the control enzyme and 14 μg for R286L. The amount of ACC produced was measured according to the method of Lizada-Yang (1979). One unit of ACC synthase activity was defined as the formation of 1 nmol of ACC in 1 h at 30°C. At least five repetitions were performed for each data point. The steady-state kinetic parameters Km, kcat, and kcat/Km were determined according to the Michaelis-Menten equation using the Kaleidagraph program (Synergy Software, Reading, PA).
CD Spectrum Measurement
Protein samples were dissolved in a 0.5-mL quartz cuvette containing a buffer of 20 mm Tris-HCl (pH 7.5), 25 mm EDTA, 2 mm DTT, 150 mm NaCl, and 20% (w/v) glycerol, and were measured at room temperature using a spectropolarimeter (model J-715 CD, JASCO, Tokyo) under the following conditions: 0.1-cm cell length; 0.2-nm resolution; 2.0-nm band width; 20-mdeg sensitivity; 0.5-s response; 100 nm/min scanning speed; six accumulations. The protein concentration during CD spectropolarimeter analysis was 0.1 mg/mL. The CD spectra were plotted and analyzed (J-700 for Windows Standard Analysis, version 1.50.01, JASCO). The samples were measured in the absence or presence of PLP, AdoMet, or both, as indicated in the text.
Fluorescence Measurement
Fluorescence spectra were measured at either 25°C or 35°C on a fluorometer (LS50B, Perkin-Elmer, Foster City, CA). The excitation wavelength was 280 nm and the emission spectra covered wavelengths ranging from 300 to 400 nm. The bandwidths for both excitation and emission were 5 nm. The binding reaction between ACC synthase and PLP was carried out with a PLP concentration gradient ranging from 0.5 to 6 μm, a fixed ACC synthase concentration of 17 nm, and a buffer of 100 mm Tris/HCl (pH 7.5). Enzyme, AdoMet, and PLP were added to a 2-mL cuvette, followed by constant stirring during the measurement. Kd between the control or the mutant enzyme and PLP were calculated based on the following equation:
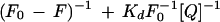 |
where F0 is the fluorescence intensity of protein without PLP or AdoMet. F is the fluorescence intensity of protein with PLP. Kd is the dissociation constant. [Q] is the concentration of PLP or AdoMet.
RESULTS
Generation and Purification of Mutant ACC Synthase R286L
The control enzyme in the study, del-1, was generated by the removal of 46 amino acids from the C terminus of tomato ACC synthase (LE-ACS2), and displayed a 4-fold reduction in catalytic activity compared with the wild-type enzyme (Li and Mattoo, 1994). Mutant enzyme ACC synthase R286L was constructed by inserting an 88-bp, double-stranded PCR product, which replaced the Arg codon with a Leu codon at the 286 position, into the recombinant ACS expression plasmid pET11d*-del-1.
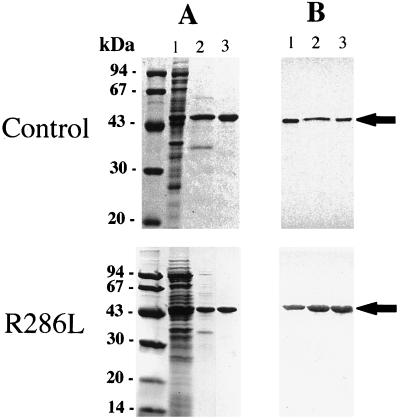
Both the control and R286L enzymes were purified according to procedures recently established in this laboratory (Huxtable et al., 1998; Zhou et al., 1998). About 16 mg each of the control and mutant enzyme was isolated from 2 L of induced E. coli cells through the two-step column purification procedure. Their purity was estimated to be at least 96% by densitometric scanning of the purified enzyme fractionated on SDS-PAGE gels and stained with Coomassie Brilliant Blue (Fig. 2). Western-blot analysis showed that: (a) R286L was purified to a similar homogeneity to that of the control enzyme, (b) the molecular masses of the control ACC synthase and R286L were the same, and (c) the size of 47 kD was consistent with the value deduced from the cDNA inserts (Fig. 2).
Figure 2.
SDS-PAGE and immunoblot analysis of control and mutant ACC synthase. A, SDS-PAGE of protein samples from different stages of purification. Lane 1, Soluble cell extract; lane 2, protein sample eluted from DEAE expanded bed; lane 3, protein sample eluted from the hydroxylapatite column. B, The immunoblot was color-developed with the primary antibody raised against tomato del-1 ACC synthase, followed by the secondary antibody conjugated with alkaline phosphatase. ACC synthase is marked by arrows.
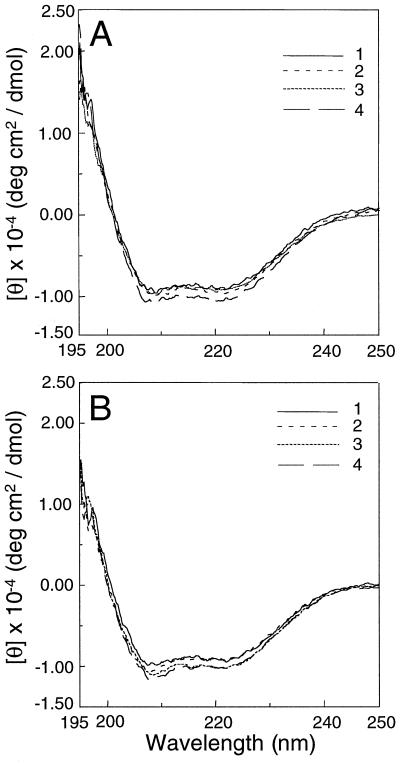
CD Spectra of R286L
The CD spectra of the purified control enzyme and R286L were measured. It was apparent that the CD spectra of R286L alone (Fig. 3B, curve 1), with the substrate AdoMet (Fig. 3B, curve 2), with co-factor PLP (Fig. 3B, curve 3), or with both AdoMet and PLP (Fig. 3B, curve 4) were nearly identical to those of the control enzyme (Fig. 3A). These data indicate that the R286L mutation did not cause a major change in the overall secondary structures of the enzyme. It also indicates that the CD spectra of both the control ACC synthase and R286L were not significantly altered by the binding of the cofactor or the substrate alone or in combination. (Fig. 3).
Figure 3.
The CD spectra of control (A) and R286L (B) ACC synthase. Buffer 1 (curve 1) contains 20 mm Tris-HCl (pH 7.5), 20% (w/v) glycerol, 25 mm EDTA, 2 mm DTT, and 150 mm NaCl; buffer 2 (curve 2) is buffer 1 supplemented with 100 μm AdoMet; buffer 3 (curve 3) is buffer 1 supplemented with 10 μm PLP; and buffer 4 (curve 4) is buffer 1 supplemented with both 100 μm AdoMet and 10 μm PLP. The measurements were done at room temperature.
Kinetic Parameters of the ACC Synthase Mutant
The control enzyme was assayed for its activity at various pH values ranging from 6.0 to 11. The peak specific activity of the control ACC synthase was found to be 830,000 units/mg at pH 9.5. The specific activities of the control and mutant enzymes assayed in 5,000 μm PLP were nearly identical to the specific activities in 50 or 500 μm PLP (data not shown). Consequently, 500 μm PLP was used in the standard enzyme assay to compensate for the reduced binding affinity of R286L for PLP.
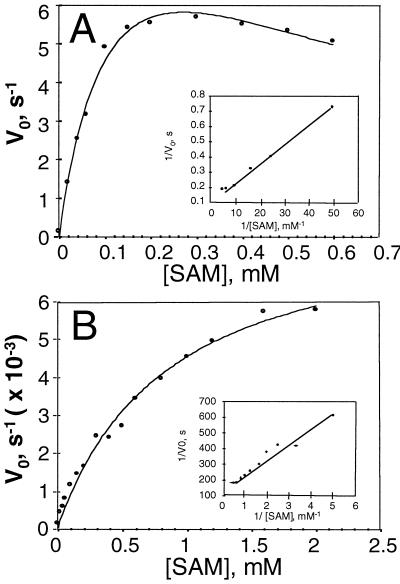
As reported previously (Li and Mattoo, 1994), the catalysis of the control enzyme de1-1 ACC synthase differs from the wild-type tomato ACC synthase, and is inhibited at substrate concentrations of 0.3 mm or above (Fig. 4A). R286L, on the other hand, similar to the wild-type enzyme, exhibited a typical Michaelis-Menten kinetic behavior even when the substrate concentration reached 2.0 mm (Fig. 4B). The biochemical parameters of the two enzymes calculated from Lineweaver-Burk plots are summarized in Table I. The control enzyme had a Km of 120 μm for AdoMet and a kcat of 9.8 s−1, whereas the mutant R286L had a Km of 730 μm and a kcat of 8.2 × 10−3 s−1. The overall catalytic efficiency of the mutant expressed in the form of kcat/Km was 8 × 103 times lower than that of the control.
Figure 4.
Kinetic parameters of control (A) and R286L (B) ACC synthase. Enzyme activity was measured in a reaction buffer of 500 μL containing 50 mm KH2PO4 (pH 9.5), 5 mm DTT, and 500 μm PLP. The amount of enzyme used in each assay mixture was 0.15 μg for the control and 14 μg for R286L. Each point represents the average of five replicates. One unit of ACC synthase activity is defined as 1 nmol of ACC formed per hour at 30°C.
Table I.
Kinetic and equilibrium constants of control and R286L ACC synthase
| Enzyme | Km SAM | kcat | kcat/Km SAM | KD PLP | |
|---|---|---|---|---|---|
| 25°C | 35°C | ||||
| mm | s−1 | m−1 s−1 | μm | ||
| Control (del-1) | 0.12a | 9.79 | 8.15 × 104 | 0.53b | 0.63b |
| R286L | 0.75a | 8.20 × 10−3 | 1.09 × 10 | 1.08 × 10b | 1.61 × 10b |
| Control/R286L | 0.16 | 1.20 × 103 | 7.49 × 103 | 4.9 × 10−2 | 3.9 × 10−2 |
Kinetic data were collected at pH 9.5 with a fixed PLP concentration of 500 μm and a variable AdoMet concentration from 20 to 2,000 μm.
The dissociation constants for the ACC synthase-PLP complex were determined based on data obtained from the fluorescence spectroscopic measurements at pH 7.5.
Determination of the Binding Affinity of ACC Synthase for PLP
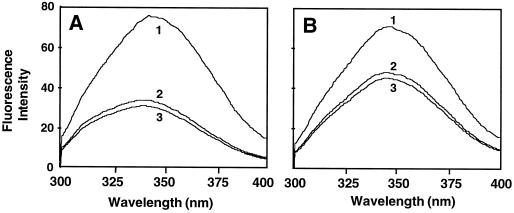
We used fluorescence titration to determine the binding affinities of both the control and the mutant enzyme for PLP. The fluorescence spectra of the control ACC synthase (Fig. 5A) and R286L (Fig. 5B) were examined in the absence of PLP (curve 1), in the presence of PLP (curve 2), and in the presence of PLP and AdoMet (curve 3). Both enzymes exhibited maximal fluorescence intensity around 345 nm in the absence of PLP (Fig. 5). The addition of PLP dramatically reduced the fluorescence intensity of R286L (Fig. 5B). The quenching effect of PLP with the control enzyme was even more pronounced for the control enzyme, and PLP caused a 5-nm blue shift in the fluorescence spectra of the control enzyme (Fig. 5A).
Figure 5.
Fluorescence spectra of control (A) and R286L (B) ACC synthase. Protein samples were dissolved in 10 mm Tris/HCl buffer (pH 7.5) to a final concentration of 17 nm. Spectra were obtained at a scan rate of 1 nm s−1. Spectrum 1, Protein sample alone; spectrum 2, protein plus PLP; spectrum 3, protein plus both PLP and AdoMet.
The dissociation constants of the control ACC synthase and the mutant, determined using Equation 1, are listed in Table I. The dissociation constant of the control enzyme was 20-fold less at 25°C and 26-fold less at 35°C than that of R286L. The increase in the dissociation constants for both enzymes at the higher temperature indicated that the binding of PLP to the enzymes was exothermic. The effect of the substrate AdoMet on the formation of the ACC synthase-PLP complex for both the control and R286L enzymes was negligible, because the addition of 25 μm AdoMet to the ACC synthase-PLP complex did not change significantly the fluorescence intensities of either enzyme (Fig. 5).
DISCUSSION
Del-1 ACC synthase was chosen as the control enzyme in this study because E. coli consistently expresses large quantities of soluble enzyme that can be purified easily for biochemical and biophysical analyses. Our previous study indicated that the C terminus of the enzyme was dispensable with regard to enzymatic activity (Li and Mattoo, 1994). This observation has been confirmed by a recent random mutagenesis study that failed to generate any inactive mutants following mutations involving the last 56 amino acids at the C terminus of tomato ACC synthase (LE-ACS2; Tarun et al., 1998). The control enzyme had a Km of 0.12 mm and a kcat of 9.8 s−1 under the assay conditions specified in “Materials and Methods.” This represents a higher catalytic efficiency of the enzyme than previously reported (Km = 0.28 mm and kcat = 5.84 s−1; Li and Mattoo, 1994), which likely is due to the higher pH (9.5) used in the reaction buffer in the present study. Effects of pH on steady-state kinetic parameters were found in a study of apple ACC synthase (Li and Mattoo, 1994).
Substitution of R286 with Leu did not disrupt the gross secondary structure of the enzyme (Fig. 3), but led to a lower affinity for both PLP and AdoMet, and to a drastic reduction in catalysis (Table I). Our results are consistent with those from the random mutagenesis study, in which the replacement of R286 by Val, Thr, Ile, and Ala converted the enzyme into class I mutants that retained normal levels of protein expression but lost 95% to 100% of their catalytic activity (Tarun et al., 1998).
The biochemical parameter kcat remained almost unchanged over a wide range of PLP concentrations (50–5,000 μm, data not shown), and the PLP binding constant K (= 1/Kd) for the control ACC synthase was much larger than kcat (Table I), suggesting that PLP binding is not a rate-limiting step. The dramatic decrease in the binding affinity for PLP in the mutant enzyme, as revealed by fluorescence spectroscopy data (Table I), strongly suggests that R286 is a principal candidate for PLP binding. Its role in the formation of the ACC synthase-PLP complex is probably comparable to that of the corresponding R266 of AATase, forming a hydrogen bond with the phosphate oxygens OP2 and OP4 of PLP (McPhalen et al., 1992).
The ionic bridge across the −NH2+ group of Arg and the phosphate moiety is understood to be a critical factor in the interaction between PLP and a number of PLP-dependent enzymes, including eukaryotic Orn decarboxylases (Grishin et al., 1995), bacterial diaminopimelate decarboxylases (Grishin et al., 1995), Arg decarboxylase (Grishin et al., 1995), Asp aminotransferase (Kirsch et al., 1984), and d-amino acid aminotransferase (Sugio et al., 1995). Other structural features responsible for the interaction include a Gly-rich loop and β/α-barrel. When R244, a catalytic amino acid involved in the binding of a phosphate group, is converted to a Leu at the active site of E. coli S-adenosylmethionine synthetase (Reczkowski et al., 1998), a 7- to 10-fold increase in Km and a 1,000-fold decrease in kcat was observed, a very similar alteration of the steady-state kinetic parameters reported here for R286L.
In addition to its involvement in PLP binding, R286 probably serves additional functions in ACC formation. In the control enzyme, substitution of R286 with Leu, an amino acid residue that has the same number of carbons as Arg, caused a nearly 1.2- × 103-fold reduction in kcat, suggesting that R286 participates in substrate catalysis. Whether R286 is a part of the charge relay system in proton abstraction or if it functions to stabilize the transition stage of ACC synthase similar to the function of Y70 in AATase (Inoue et al., 1991) remains to be investigated. Finally, the significant decrease in the binding affinity for AdoMet (Table I) may be due to the loss of the correct orientation of PLP in the active site, which may be required for the formation of external aldimine between PLP and AdoMet in the active site.
ACKNOWLEDGMENTS
The authors thank Drs. Susan Huxtable, Heng Ming Ke, Ming Jie Zhang, and Jun Wang for their constructive suggestions.
Footnotes
This work was supported by the Reseach Grant Council of Hong Kong (grant no. HKUST649/96M) and by the Biotechnology Research Institute (grant no. BRI–96–III–3).
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. New York: Academic Press; 1992. [Google Scholar]
- Adams DO, Yang SF. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci USA. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Herner RC, Kende H. Assay for and enzymatic formation of an ethylene precursor, 1-aminocyclopropane-1-carboxylic acid [Tomatoes] Planta. 1979;145:293–303. doi: 10.1007/BF00454455. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Fluhr R, Mattoo AK. Ethylene-biosynthesis and perception. Crit Rev Plant Sci. 1996;15:479–523. [Google Scholar]
- Grishin NV, Phillips MA, Goldsmith EJ. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Prot Sci. 1995;4:1291–1304. doi: 10.1002/pro.5560040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Parks JE, Rottman WH, Theologis A. Two genes encoding 1-aminocyclopropane-1-carboxylate synthase in zucchini (Cucurbita pepo) are clustered and similar but differentially regulated. Proc Natl Acad Sci USA. 1991;88:7021–7025. doi: 10.1073/pnas.88.16.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxtable S, Zhou H, Wong S, Li N. Renaturation of 1-aminocyclopropane-1-carboxylate synthase expressed in Escherichia coli in the form of inclusion bodies into a dimeric and catalytically active enzyme. Prot Exp Purif. 1998;12:305–314. doi: 10.1006/prep.1997.0847. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kuramitsu S, Okamoto A, Hirotsu K, Higuchi T, Kagamiyama H. Site-directed mutagenesis of Escherichia coli aspartate aminotransferase: role of Tyr70 in the catalytic processes. Biochemistry. 1991;30:7796–7801. doi: 10.1021/bi00245a019. [DOI] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H, Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984;174:497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- Li N, Mattoo AK. Deletion of the carboxyl-terminal region of 1-aminocyclopropane-1-carboxylic acid synthase, a key protein in the biosynthesis of ethylene, results in catalytically hyperactive, monomeric enzyme. J Biol Chem. 1994;269:6908–6917. [PubMed] [Google Scholar]
- Lizada C, Yang SF. A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal Biochem. 1979;100:140–145. doi: 10.1016/0003-2697(79)90123-4. [DOI] [PubMed] [Google Scholar]
- McPhalen CA, Vincent MG, Jansonius JN. X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase. J Mol Biol. 1992;225:495–517. doi: 10.1016/0022-2836(92)90935-d. [DOI] [PubMed] [Google Scholar]
- Mehta PK, Christen P. Homology of 1-aminocyclopropane-1-carboxylate synthase, 8-amino-7-oxononanoate synthase, 2-amino-6-caprolactam racemase, 2,2-dialkylglycine decarboxylase, glutamate-1-semialdehyde 2,1-aminomutase and isopenicillin-N-epimerase with aminotransferases. Biochem Biophys Res Commun. 1994;198:138–143. doi: 10.1006/bbrc.1994.1020. [DOI] [PubMed] [Google Scholar]
- Osterman AL, Brooks HB, Rizo J, Phillips MA. Role of Arg-277 in the binding of pyridoxal 5′-phosphate to Trypanosoma brucei ornithine decarboxylase. Biochemistry. 1997;36:4558–4567. doi: 10.1021/bi962916h. [DOI] [PubMed] [Google Scholar]
- Ramalingam K, Lee KM, Woodard RW, Bleecker AB, Kende H. Stereochemical course of the reaction catalyzed by the pyridoxal phosphate-dependent enzyme 1-aminocyclopropane-1-carboxylate synthase. Proc Natl Acad Sci USA. 1985;82:7820–7824. doi: 10.1073/pnas.82.23.7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczkowski RS, Taylor JC, Markham GD. The active-site arginine of S-adenosylmethionine synthetase orients the reaction intermediate. Biochemistry. 1998;37:13499–13506. doi: 10.1021/bi9811011. [DOI] [PubMed] [Google Scholar]
- Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- Sugio S, Petsko GA, Manning JM, Soda K, Ringe D. Crystal structure of a d-amino acid aminotransferase: how the protein controls stereoselectivity. Biochemistry. 1995;34:9661–9669. doi: 10.1021/bi00030a002. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Lee JS, Theologis A. Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: a key enzyme in ethylene biosynthesis. Proc Natl Acad Sci USA. 1998;95:9796–9801. doi: 10.1073/pnas.95.17.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A. One rotten apple spoils the whole bushel: the role of ethylene in fruit ripening. Cell. 1992;70:181–184. doi: 10.1016/0092-8674(92)90093-r. [DOI] [PubMed] [Google Scholar]
- White MF, Vasquez J, Yang SF, Kirsch JF. Expression of apple 1-aminocyclopropane-1-carboxylate synthase in Escherichia coli: kinetic characterization of wild-type and active-site mutant forms. Proc Natl Acad Sci USA. 1994;91:12428–12432. doi: 10.1073/pnas.91.26.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yip WK, Dong JG, Kenny JW, Thompson GA, Yang SF. Characterization and sequencing of the active site of 1-aminocyclopropane-1-carboxylate synthase. Proc Natl Acad Sci USA. 1990;87:7930–7934. doi: 10.1073/pnas.87.20.7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YB, Adams DO, Yang SF. 1-Aminocyclopropane-1-carboxylate synthase, a key enzyme in ethylene biosynthesis. Archiv Biochem Biophys. 1979;198:280–286. doi: 10.1016/0003-9861(79)90420-x. [DOI] [PubMed] [Google Scholar]
- Zhou HQ, Huxtable S, Xin H, Li N. Enhanced high-level expression of soluble 1-aminocyclopropane-1-carboxylate synthase and rapid purification by expanded-bed adsorption. Protein Expr Purif. 1998;14:178–184. doi: 10.1006/prep.1998.0923. [DOI] [PubMed] [Google Scholar]