Abstract
Microbial small RNAs (sRNAs) play essential roles against many stress conditions in cyanobacteria. However, little is known on their regulatory mechanisms on biofuels tolerance. In our previous sRNA analysis, a trans-encoded sRNA Nc117 was found involved in the tolerance to ethanol and 1-butanol in Synechocystis sp. PCC 6803. However, its functional mechanism is yet to be determined. In this study, functional characterization of sRNA Nc117 was performed. Briefly, the exact length of the trans-encoded sRNA Nc117 was determined to be 102 nucleotides using 3′ RACE, and the positive regulation of Nc117 on short chain alcohols tolerance was further confirmed. Then, computational target prediction and transcriptomic analysis were integrated to explore the potential targets of Nc117. A total of 119 up-regulated and 116 down-regulated genes were identified in nc117 overexpression strain compared with the wild type by comparative transcriptomic analysis, among which the upstream regions of five genes were overlapped with those predicted by computational target approach. Based on the phenotype analysis of gene deletion and overexpression strains under short chain alcohols stress, one gene slr0007 encoding D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase was determined as a potential target of Nc117, suggesting that the synthesis of LPS or S-layer glycoprotein may be responsible for the tolerance enhancement. As the first reported trans-encoded sRNA positively regulating biofuels tolerance in cyanobacteria, this study not only provided evidence for a new regulatory mechanism of trans-encoded sRNA in cyanobacteria, but also valuable information for rational construction of high-tolerant cyanobacterial chassis.
Keywords: trans-encoded sRNA, Synechocystis, tolerance, biofuels, regulation
Introduction
Biofuels have become a hot research area in recent decades due to their potential of replacing fossil (Atsumi et al., 2008). Cyanobacteria, as a group of autotrophic prokaryotes with the advantages of rapid growth, only consuming solar, water and CO2, hold great promise as an important alternative to generate biofuels (Angermayr et al., 2009). In recent years, cyanobacteria have been successfully engineered to synthesize various types of biofuels (Gao X. et al., 2016). For example, Lan et al. (2013) successfully incorporated biofuels biosynthetic pathways into cyanobacterial systems, and 404 mg/L production of 1-butanol was achieved in Synechococcus elongatus PCC 7942 (Lan et al., 2013). In addition, Gao Z. et al. (2016) reached 5.5 g/L production of ethanol by systematically optimizing metabolic pathway for ethanol production in engineered cyanobacteria (Gao Z. et al., 2016). However, there is still a big gap between the current productivity in cyanobacteria and other engineered bacteria, such as Escherichia coli and Clostridium, and the low tolerance to biofuels toxicity of cyanobacteria is regarded as one of the important limiting factors (Jin et al., 2014). It was found that the toxic effects caused by the product ethanol or isobutanol would result in significant cell growth retardation in Synechocystis sp. PCC 6803 (hereafter Synechocystis) and S. elongatus PCC 7942 (Atsumi et al., 2009; Dexter and Fu, 2009), thus limiting the potential of these biofuels production and the possibility of industrial application. In another study, it was reported that the addition of 0.20% (v/v) butanol caused a 50% growth decrease in Synechocystis (Tian et al., 2013), suggesting the great toxicity caused by biofuel butanol. Therefore, resistance mechanism of cyanobacteria to biofuels needs to be investigated, which is an important prerequisite for constructing highly biofuels-tolerant strains to improve the biofuels production in cyanobacteria. Recently, Kaczmarzyk et al. (2014) by overexpressing an RNA polymerase sigma factor, successfully increased butanol tolerance and lowered the intracellular concentration of reactive oxygen species in Synechocystis. Moreover, in our previous work, several regulatory genes (i.e., slr1037, sll0039, sll0794, and slr1860) related to alcohols tolerance were successfully identified (Song et al., 2014; Zhu et al., 2014; Gao et al., 2017). For the resistance mechanism to biofuels, based on several previous studies, cyanobacteria employed a combination of multiple resistance systems to adapt to biofuels stress or nutrient limited environments, including cell membrane tolerance mechanism, transport vector system, intracellular transformation and degradation, cell surface structure and morphology changes, as well as common pressure response tolerance mechanisms (Jie et al., 2012; Qiao et al., 2012, 2013; Huang et al., 2013; Tian et al., 2013; Xu et al., 2014). However, this brings great challenges to further improve tolerance by conventional sequential multi-gene modification approaches (Gao X. et al., 2016). As an alternative, “transcriptional engineering” for tolerance improvement (Anders and Huber, 2010), especially the sRNAs engineering that has the advantages such as rapid response, low metabolic burden and flexible and precise control, could be an applicable approach (Gaida et al., 2013).
Bacterial sRNAs between 50 and 300 nucleotides transcribed from the intergenic region, play important regulatory roles at the levels of post-transcriptional, regulating translation, decaying or protecting mRNA molecules, through base paring in bacterial cells (Johansen et al., 2008; McCullen et al., 2010). To some extent, deciphering sRNAs regulatory mechanism is still challenging due to their functional complexity. For example, sRNAs as a kind of regulatory molecules, mediate cellular responses to the environment stress, not by means of encoding protein products, but via the complementary pairs with mRNA molecules or via combining with some protein to influence molecular function of protein activity (Kopf and Hess, 2015). Base-pairing between targets of sRNA and mRNA may cause inhibition or activation of mRNA translation, or affect the stability of the target RNA by promoting or blocking cleavage of a ribonuclease (RNase) (Saramago et al., 2014). Besides, sRNA may interact with RNA chaperone protein Hfq to modify its activity and then regulate gene expression of a diversity of targets (Updegrove et al., 2016).
Although some of sRNAs have been systematically investigated in cyanobacteria, e.g., IsrR involved in iron depletion (Duhring et al., 2006), As1-Flv4 involved in inorganic carbon supply (Eisenhut et al., 2012), PsbA2R, PsbA3R (Sakurai et al., 2012), PsrR1 (Georg et al., 2014), and RblR (Hu et al., 2017) related to photosynthetic gene expression, and NsiR4 (Klahn et al., 2015) controlling nitrogen assimilation, several systematic sRNA studies suggested there were thousands of sRNAs in cyanobacteria (Mitschke et al., 2011; Kopf et al., 2014; Xu et al., 2014), and the majority of them were still functionally uncharacterized. It is noteworthy that based on our previous work, a trans-encoded sRNA Nc117 involved in tolerance to exogenous ethanol and 1-butanol was identified in Synechocystis (Pei et al., 2017). Due to the complexity of sRNA regulatory mechanism, details about this trans-encoded sRNA Nc117 involved in the biofuels tolerance are still unclear. To determine potential targets of Nc117 sRNA, in this study, a transcriptomic analysis combined with computational target prediction were utilized to explore its potential targets, and one target gene slr0007 encoding HddC positively regulated by Nc117, which participated in the tolerance to short chain alcohols in Synechocystis was successfully identified. As the first study of trans-sRNA involving biofuels tolerance in cyanobacteria, this study provided not only novel insights in regulatory mechanisms of sRNAs, but also a valuable target for biofuels tolerance enhancement in cyanobacteria.
Materials and Methods
Synechocystis Culture Conditions
For Synechocystis sp. PCC 6803 (ATCC 27184), WT and mutants were grown in BG11 medium (pH 7.5) under a light intensity of approximately 50 μmol photons m-2 s-1 in an illuminating incubator (HNY-211B Illuminating Shaker, Honour, China) at 130 rpm and 30°C with a starting cell density of OD630 = 0.04 (Pei et al., 2017). Cell density was measured with an ELx808 Absorbance Microplate Reader (BioTek, Winooski, VT, United States) at OD630. Five mL fresh cells at OD630 of 0.2 were collected by centrifugation at 3000 × g and 4°C and were then inoculated into 25 mL of BG11 liquid medium in a 100-mL flask. Ethanol 1.5% (v/v) and 1-butanol 0.25% (v/v) were added at the beginning of cultivation. 200 μL of culture was sampled and measured at OD630 every 24 h. Growth experiments were repeated at least three times to confirm growth patterns.
3′ RACE Analysis
Total RNA of Synechocystis was extracted under identical condition with transcriptome analysis (See below). Then RNA was added with a poly(A) tail using NEB E. coli poly(A) polymerase (New England Biolabs Inc., Ipswich, MA, United States). After that a specific primer containing oligo dT: 5′-CGTTGTAAAACGACGGCCAGTTTTTTTTTTTTTTTTTT-3′ and SuperScript® VILOTM cDNA Synthesis Kit were used to synthesize cDNA. At last, forward primer (5′-CGTCCAAACCTGAATAGATAATCCAT-3′) and reverse primer (5′-CGTTGTAAAACGACGGCCAG-3′) were used to amplify Nc117 containing 3′ end. The PCR product was purified and ligated into pTZ57R/T using InsTAclone PCR Cloning Kit (Thermo Fisher Scientific Inc., MA, United States) for sequencing analysis.
Mutants Construction
For construction of gene overexpression strains, E. coli DH5α was used for vectors construction and enrichment. Primers used in this study were listed in Table 1. Gene expression vector pJA2, kindly provided by Prof. Paul Hudson (KTH Royal Institute of Technology of Sweden) (Huang et al., 2010; Kaczmarzyk et al., 2014), was used to overexpress the sRNAs or target genes. All sRNAs or genes were cloned under the control of the psbA2 promoter. Briefly, the pJA2 backbone was amplified by PCR, treated with DpnI and digested with BamHI and XbaI to create cohesive ends. The sRNA or genes sequence was PCR-amplified using primers pJA2-sRNA/gene-F and pJA2-sRNA/gene-R and cloned into the BamHI/XbaI sites of pJA2, resulting in the recombinant plasmid pJA2-sRNA/gene. The plasmid was introduced into the WT by electro-transformation as previously described (Wang et al., 2016). Positive clones were grown on BG-11 agar plates with 10 μg/mL kanamycin and were confirmed by colony PCR analysis.
Table 1.
Primers used for mutants construction.
| Primer names | Primer sequences (5′ to 3′) |
|---|---|
| pJA2-nc117-F | TGCTCTAGAAACTTAAGAGCGAAGTAAGT |
| pJA2-nc117-R | CGCGGATCCGAAAATGGAAAGAAGACGCT |
| Δnc117-F1 | ACCCCCGATGATTTTGCCAT |
| Δnc117-R1 | CCAGTGGCTTCTGTTTCTATCAGCTTTCGCTCTTAAGTTCATGCC |
| Δnc117-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δnc117-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δnc117-F3 | CGATGAGTGGCAGGGCGGGGCGTAAATACTTTCCGAGCCCAGAAT |
| Δnc117-R3 | AATAGCTGTCGGCGATGGAG |
| pJA2-slr0007-F | TGCTCTAGAATGGCTCTTTCCCCCGCAGA |
| pJA2-slr0007-R | CGCGGATCCCTATTTGTCTAGGTCTTGAA |
| Δslr0007-F1 | CCCTGCCATTCAATCCGTCT |
| Δslr0007-R1 | CCAGTGGCTTCTGTTTCTATCAGCTAGAGTTGAAAAAGTAGAAAC |
| Δslr0007-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δslr0007-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δslr0007-F3 | CGATGAGTGGCAGGGCGGGGCGTAACTTAAAATGAGAAGCTAACT |
| Δslr0007-R3 | CAAGTTGATGCAGAGCGTGG |
| pJA2-slr2126-F | TGCTCTAGAATGTTTTTCATGCAAAATACTAAGT |
| pJA2-slr2126-R | CGCGGATCCTCACAAAGTTAGTTTTTGATCGAGA |
| Δslr2126-F1 | TTGTAATGGCCCCAGCTTGT |
| Δslr2126-R1 | AGCTGATAGAAACAGAAGCCACTGGGAATTAATAATATTCTTTGT |
| Δslr2126-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δslr2126-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δslr2126-F3 | CGATGAGTGGCAGGGCGGGGCGTAATACCCCCTACTACAAATCCC |
| Δslr2126-R3 | TCCGCTTCTTGGGACTGTTC |
| pJA2-sll1830-F | TGCTCTAGAATGATCAATCGTCAGGACCT |
| pJA2-sll1830-R | CGCGGATCCTCAGTAGCGTAAAACCAAGG |
| Δsll1830-F1 | CTTTCCATGGCCGCTAAACG |
| Δsll1830-R1 | CCAGTGGCTTCTGTTTCTATCAGCTGGCCATTGATTAAGTTAAGG |
| Δsll1830-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δsll1830-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δsll1830-F3 | CGATGAGTGGCAGGGCGGGGCGTAATTTGTCTTTACATAGGTCGA |
| Δsll1830-R3 | CCCGAATTTCTGTGCTCCCA |
| pJA2-slr2108-F | TGCTCTAGAATGGGAGTGGATGGGATGAC |
| pJA2-slr2108-R | CGCGGATCCTTAGGTCGTACATAAGTGCC |
| Δslr1501-F1 | GCATTGGGCAGTTGTAAGCC |
| Δslr1501-R1 | CCAGTGGCTTCTGTTTCTATCAGCTGACCCCTTGGGGAAAGTTTT |
| Δslr1501-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δslr1501-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δslr1501-F3 | CGATGAGTGGCAGGGCGGGGCGTAAAAAAGATAACTACCATGTTT |
| Δslr1501-R3 | ACCTAGTTCCATCACCCCGA |
| Δsll0784-F1 | TCGGCTTTTCCGGGCATAAT |
| Δsll0784-R1 | CCAGTGGCTTCTGTTTCTATCAGCTGTACAAAAATTTAAGGATTA |
| Δsll0784-F2 | AGCTGATAGAAACAGAAGCCACTGG |
| Δsll0784-R2 | TTACGCCCCGCCCTGCCACTCATCG |
| Δsll0784-F3 | CGATGAGTGGCAGGGCGGGGCGTAAAACGACTGGAATAGTTTCAG |
| Δsll0784-R3 | AAGGAAACTTCGCTTCTACATTGA |
| pJA2-up500-F | GTTCCGCGCACATTTCCCCGA |
For gene knockout and complementation mutants construction, a fusion PCR-based method was employed according to previous study (Wang et al., 2002). Briefly, for the gene target selected, three sets of primers were designed to amplify a linear DNA fragment containing the chloramphenicol resistance cassette (amplified from a plasmid pACYC184) with two flanking arms of DNA upstream and downstream of the target gene. The linear fused PCR amplicon was used directly for transformation into WT or WT-pJA2-nc117 strain by natural transformation. The chloramphenicol-resistant transformants were obtained and passed several times on fresh BG11 plates supplemented with 10 μg/mL chloramphenicol to achieve complete chromosome segregation (confirmed by PCR). The successful knockout mutants were confirmed by PCR and sequencing analysis. PCR primers for mutant construction were listed in Table 1.
Computational Prediction of sRNA Targets
After determination of the end of trans-encoded sRNA Nc117, target prediction was performed by IntaRNA software (Busch et al., 2008), and only the top 100 predictions obtained from IntaRNA with a free-energy cut-off of -15 kcal/mol were retained to remove potential false positive targets.
RNA Samples Collection and Library Preparation for Transcriptomic Analysis
The WT and Nc117 overexpression strains of Synechocystis were cultured under same condition as previous growth comparison, each sample with two biological replicates for RNA collection. Approximately 10 mg of cell pellets were frozen in liquid nitrogen immediately after centrifugation at 8000 × g for 10 min at 4°C, and cell walls were broken by liquid nitrogen mortar grinding. Total RNA extraction was achieved using a miRNeasy Mini Kit (Qiagen, CA, United States). Contaminating DNA in RNA samples was removed with DNase I according to the instructions for the miRNeasy Mini Kit (Qiagen, CA, United States). Total RNA of each sample was quantified and qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States), NanoDrop (Thermo Fisher Scientific Inc.) and 1% agarose gel. For each sample, 3 μg total RNA with RNA integrity number (RIN) value above 8 was used for following library preparation.
Next generation sequencing libraries were constructed according to the manufacturer’s protocol (NEBNext® UltraTM RNA Library Prep Kit for Illumina®). Briefly, rRNA was removed using a Ribo-Zero-rRNA Removal Kit. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA, United States). Then 3 μL USER Enzyme (NEB, United States) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed using Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 platform. Sequencing was carried out using a 2 × 150 bp paired-end (PE) configuration, while image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina, CA, United States) on the HiSeq instrument.
Transcriptomic Analysis
The raw mRNA sequence reads were pre-processed using a NGS QC Toolkit (v. 2.3) to remove low-quality bases with quality scores < 30 and adapter sequences (Patel and Jain, 2012). For paired-end Illumina reads, both pairs were removed if either pair mapped to rRNA sequences. Remaining reads were mapped to the Synechocystis genome which was downloaded from NCBI using Burrows-Wheeler Alignment tool software version (v. 0.7.5a) with default parameters (Li and Durbin, 2009). Raw counts of reads that uniquely mapped to each gene region were calculated by HTSeq (v. 0.6.1) (Anders et al., 2015). Then reads counts were normalized to the aligned FPKM (Fragments per kilobase of gene per million mapped fragments) to obtain the relative expression levels. Differential expression analysis between WT and nc117 overexpression strains (WT-pJA2-nc117) was performed using the DESeq2 software (Anders and Huber, 2010), which used a model based on the negative binomial distribution. The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with fold change > 1.2 and adjusted p-values < 0.05 were assigned as differentially expressed.
The metabolic pathway analysis of the genes was conducted according to KEGG Pathway Database. The significance of whether differently expressed genes were enriched in a given functional pathway or functional category was calculated by the Wallenius non-central hypergeometric test using the GOseq R package in which gene-length bias was corrected (Young et al., 2010).
qRT-PCR Validation
The RNA samples used in qRT-PCR were prepared from identical cultures for transcriptomic analysis above, and qRT-PCR analysis was performed as previously described (Wang et al., 2012). Quantification of mRNA expression was determined according to a standard process of qRT-PCR that used serial dilutions of known concentrations of chromosomal DNA as a template to construct a standard curve. A total of 20 genes were selected for validation and 16S rRNA was used as an internal control. Three technical replicates were analyzed for each sRNA. The data analysis was carried out using the StepOnePlus analytical software (Applied Biosystems, Foster City, CA, United States). Briefly, the amount of relative gene transcript was normalized by that of 16S rRNA in each sample, and the data presented were ratios of the amount of normalized transcripts in the treatment between the WT and mutant strains (All Primers were listed in Supplementary Table S1). Later, a Pearson correlation and statistically significant analysis with a set of transcripts between RNA-seq and qRT-PCR was conducted to ensure the reliability of RNA-seq by R software corr.test function.
Results
3′ End Determination of sRNA Nc117
The trans-encoded sRNA Nc117 was demonstrated to be involved in the tolerance to biofuels ethanol and 1-butanol in Synechocystis in our previous study (Pei et al., 2017). However, its exact length remained to be elucidated as our previous study employed the length of Nc117 for overexpression only basing on the RNA-seq data (Pei et al., 2017). Previously, Nc117 has also been reported by Mitschke et al. (2011) (named as Ncr1600), in which the 5′ end of Nc117 has been determined while its 3′ end remained unclear. Here in this study we further determined the 3′ end of Nc117 employing 3′ rapid amplification of cDNA end (3′ RACE) technology (Supplementary Figure S1). As shown in Figure 1A, a distinct signal with fragment length of approximately 110 nucleotides was detected for Nc117. After Sanger sequencing, we pinpointed the 3′ end of Ncl17 at 3,250,631 of the Synechocystis genome, which was 16 bp shorter than that reported in our previous study (Pei et al., 2017). Thus, the length of Nc117 was determined to be 102 nucleotides, which was located between slr0550 and a polycistron including slr0551, slr0552, and slr0553 encoding three hypothetical proteins, as depicted in Figure 1B. Interestingly, the TATA box motif was also found right before the TSS of Nc117 (Figure 1C), suggesting the reliability of our analysis.
FIGURE 1.
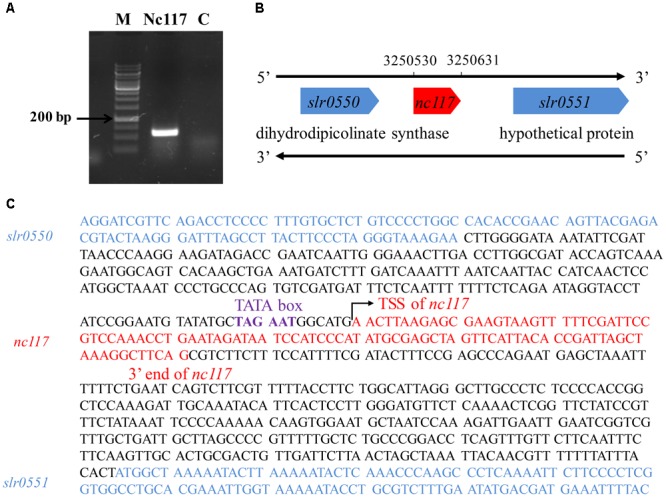
Determination of the origination and end of the sRNA Nc117. (A) 3′ ends of Nc117 orientation are determined by RACE. (B) Schematic presentation of Nc117. Number in the top DNA strands indicates the positions. (C) TSS and the 3′ end of Nc117 in the top DNA strands are indicated, respectively. The TATA box site is also indicated.
Confirmation of the Involvement of Nc117 in Short Chain Alcohols Tolerance
To further confirm the involvement of Nc117 in short chain alcohols tolerance, a series of engineered strains were constructed in the Synechocystis WT based on the exact length of Nc117 determined above, including nc117 overexpressing strain WT-pJA2-nc117, nc117 knockout strain WT-Δnc117, and the complementation strain WT-Δnc117/pJA2-nc117 by introducing the plasmid pJA2-nc117 into the WT-Δnc117 mutant, and then their phenotypes under short chain alcohols stress were comparatively monitored. Consistent with the previous phenotypic observation (Pei et al., 2017), the growth of WT-Δnc117 was slower than WT under 2.0% (v/v) ethanol or 0.25% (v/v) 1-butanol conditions, while the growth of WT-pJA2-nc117 was faster than the WT strain under the same conditions (Figure 2), indicating the direct involvement of Nc117 in short chain alcohols tolerance. In addition, the complementation of the sRNA nc117 in the WT-Δnc117 mutant was able to bring the short chain alcohols tolerance to the similar level as the WT, demonstrating that the sRNA Nc117 was indeed involved positively in short chain alcohols tolerance in Synechocystis.
FIGURE 2.
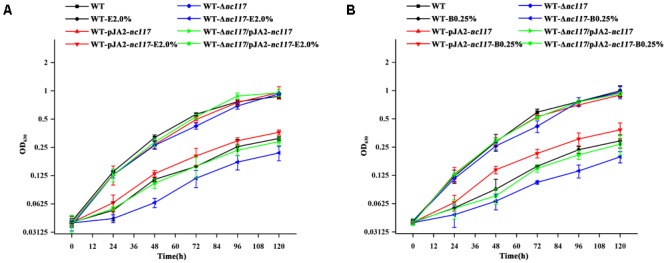
Growth curves of WT, nc117 overexpression, nc117 knockout and complementary strains under control and biofuels stress conditions. (A) WT, WT-pJA2-nc117, WT-Δnc117 and WT-Δnc117/pJA2-nc117 in normal BG11 medium with or without 2.0% (v/v) ethanol (E 2.0%). (B) WT, WT-pJA2-nc117, WT-Δnc117 and WT-Δnc117/pJA2-nc117 in normal BG11 medium with or without 0.25% (v/v) 1-butanol (B 0.25%). The error bar represents the calculated standard deviation of the measurements of three biological replicates.
Computational Target Prediction of Nc117 by IntaRNA
Based on the full length of the trans-encoded sRNA Nc117 determined in this study, target prediction was re-performed by IntaRNA software, and the top 100 predicted interaction targets were obtained (listed in Supplementary Table S2). Although the full sequence of Nc117 determined in this study had a few base pairs shifting compared to our previous analysis (Pei et al., 2017), more than half of the predicted target genes were identical to those reported in the previous study (Pei et al., 2017). According to the COG category classification, the functional pathways to which the predicted targets belonged included genes involved in replication, recombination and repair, cell wall/membrane/envelope biogenesis, amino acid transport and metabolism, and signal transduction mechanisms and so on (Supplementary Table S2). Besides, some genes predicted by IntaRNA have been reported involved in cyanobacterial tolerance to various kinds of environmental stresses previously. For example, slr0415 encoding Na(+)/H(+) antiporter was found related to salt stress response and internal pH regulation of the Synechocystis (Elanskaya et al., 2002), and the Δsll0272 mutant was sensitive to heat stress for growth in Synechocystis (Gao F. et al., 2016). Interestingly, most of the interaction areas were located in the central region of the sRNA Nc117 (Supplementary Figure S2), indicating possible conserved functional binding domain of sRNA Nc117. Accordingly, for the possible targets mRNA of Nc117, most of the interaction domains were close to or overlapped with the upstream region of the target genes, especially with the RBSs or the translational start codon (Supplementary Figure S3), which was consistent well with the sRNA interaction mechanism reported in bacteria (Georg et al., 2014).
Genome-Wide Identification of Potential Targets of Nc117 Based on Transcriptomic Analysis
Growth of nc117 overexpression strain WT-pJA2-nc117 was faster than the WT strain under short chain alcohols stress conditions (Figure 2). Then, RNA-sequencing (RNA-seq) based comparative transcriptomic analysis was conducted between WT and WT-pJA2-nc117 strains to investigate the target genes regulated by Nc117. After RNA-seq and data filtering, a total of 63 million clean reads were generated in four samples between WT and WT-pJA2-nc117 (each with two biological replicates). Subsequently, the qualified reads were mapped to the complete genome of Synechocystis, and all samples showed a mapping ratio greater than 96% (details in Supplementary Table S3). In addition, the Pearson correlation analysis between two biological replicates was greater than 0.99, indicating the reliability of our RNA-seq results (Figure 3A). To investigate target genes potentially regulated by sRNA Nc117, the differential expression profiling analysis was performed using DESeq2 software (Anders and Huber, 2010). Notably, a total of 235 genes were found differentially expressed in WT-pJA2-nc117 compared with WT (Supplementary Table S4). Among them, 119 and 116 genes were up- and down-regulated, respectively (Figure 3B). To investigate the reliability of RNA-seq results, a subset of 20 target genes were then randomly selected for qRT-PCR confirmation. As shown in Figure 3C, the Pearson correlation analysis between qRT-PCR and RNA-seq data showed a clear positive correlation with correlation coefficients greater than 0.8 and p-value less than 0.005, suggesting the high reliability of RNA-seq results.
FIGURE 3.
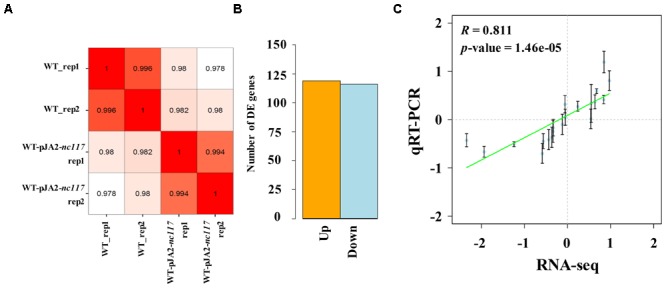
RNA-seq results analysis. (A) Pearson correlation analysis between all biological RNA-sample. (B) Differential expression analysis of RNA-seq. (C) Correlation between qRT-PCR and RNA-seq analyses for selected genes. For RNA-seq (horizontal coordinate), values represent log2 fold change of transcripts. For qRT-PCR (vertical coordinate), values represent the mean log2 fold change in transcripts. The error bar represents the standard deviation of three technical and three biological replicates. The Pearson correlation coefficients and their significances are indicated at the right lower corner of each plot.
Interestingly, among the differentially regulated genes, some genes were also associated with cyanobacterial tolerance to various kinds of environmental stresses. For example, slr0897 encoding β-1,3-1,4-glucanase functioning in salt stress tolerance in Synechocystis (Tamoi et al., 2007) was found up-regulated by 1.7-fold, while another gene slr1667 regulated by a cAMP receptor protein involved in the construction of cell surface against ultraviolet radiation (Yoshimura et al., 2002, 2010), was up-regulated by 2.22-fold. The scheme of pathway mediated by sRNA Nc117 revealed by RNA-seq results was shown in Figure 4. Among them, the most differentially regulated pathways based on RNA-seq transcriptomics analysis were TCSTS and ABC transporters (Figure 4). These two pathways were both reported to be involved in regulating resistance under stress conditions (Los et al., 2010; Foo et al., 2014). For example, Uchiyama et al. (2012) found that slr1214 encoding two-component response regulator PatA subfamily, may affect acid stress tolerance (Uchiyama et al., 2012). Although many differentially expressed genes were found encoding hypothetical proteins, the results showed that the up-regulated genes were significantly enriched in several functional categories, such as “photosynthesis”, “photosynthesis antenna proteins,” and “two-component system,” while most down-regulated genes were enriched in “pentose phosphate pathway” and “arginine and proline metabolism” (Supplementary Table S5).
FIGURE 4.
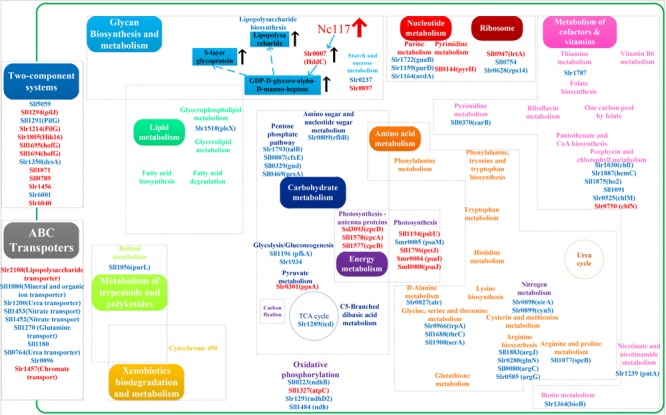
Scheme of global effects of sRNA Nc117 revealed by RNA-seq analysis. The metabolic features presented were deduced by RNA-seq analysis, and up-regulated genes (red) and down-regulated genes (blue) were labeled, respectively.
Identification of Direct Targets of Nc117 in Regulating Short Chain Alcohols Tolerance
Considering a large set of putative target genes of Nc117 identified by comparative transcriptomic analysis, an integrative analysis combining transcriptomic analysis and computational target prediction was performed to narrow down the possible target genes of Nc117 that might be responsible for the short chain alcohols tolerance. We first conducted a comparative analysis of the top ranked computational predicted targets and the upstream regions of significantly differently expressed genes in comparative transcriptomic analysis. Interestingly, the upstream regions of five genes, i.e., slr2108, slr0007, sll0784, sll1830, and slr1501 were identified overlapped. We then performed a qRT-PCR analysis for these five genes, and the results confirmed their differentially regulated expression in the nc117 overexpression strain revealed by the transcriptomic analysis (Table 2). Therefore, these genes were selected for further functional characterization. Briefly, all the selected five genes were individually overexpressed and knockout, and their phenotypes under short chain alcohols stress were comparatively monitored in shake flasks. The results showed that one strain, the slr0007 overexpressed strain, was found to be more resistant to ethanol and 1-butanol compared with WT (Figures 5A,B), and the knockout mutant Δslr0007 was more sensitive to ethanol compared with WT (Figure 5C), while there was no significant difference for the knockout mutants or overexpression strains of the other four genes compared with WT (data not shown). Furthermore, the complementation of slr0007 in the Δslr007 mutant restored the ethanol tolerance to the level of WT (Figure 5C), indicating that slr0007 was indeed involved in short chain alcohols tolerance positively regulated by Nc117.
Table 2.
Possible target genes identified by target prediction and RNA-seq.
| Gene | Fold change in RNA-seq analysis | Fold change in qRT-PCR analysis | Description |
|---|---|---|---|
| slr1501 | 0.30 | 0.77 ± 0.21 | Probable acetyltransferase |
| slr2108 | 1.46 | 1.27 ± 0.23 | Probable polysaccharide ABC transporter ATP binding subunit |
| sll1830 | 0.68 | 0.18 ± 0.15 | Unknown protein |
| slr0007 | 1.33 | 1.46 ± 0.20 | Probable sugar-phosphate nucleotidyltransferase |
| sll0784 | 0.42 | 0.43 ± 0.18 | Nitrilase |
FIGURE 5.
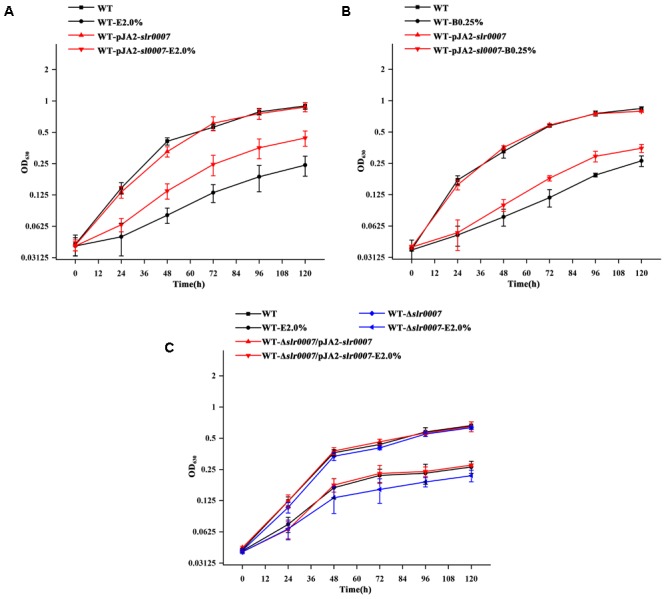
Growth curves of WT, overexpression, knockout and complementary strains under control and biofuels stress conditions. (A) WT, WT-pJA2-slr0007 in normal BG11 medium with or without 2.0% (v/v) ethanol (E 2.0%). (B) WT, WT-pJA2-slr0007 in normal BG11 medium with or without 0.25% (v/v) 1-butanol (B 0.25%). (C) WT, WT-Δslr0007 and WT-Δslr0007/pJA2-slr0007 in normal BG11 medium with or without 2.0% (v/v) ethanol (E 2.0%). The error bar represents the calculated standard deviation of the measurements of three biological replicates.
According to KEGG pathway database, slr0007 encodes the HddC involved in the LPS synthesis pathway, which might contribute to the short chain alcohols tolerance regulation (Leone et al., 2007). According to the comparative transcriptomic analysis above, the mRNA level of slr0007 was up-regulated 1.33-fold in the WT-pJA2-nc117 strain (Table 2). Concerning the mechanism of sRNA Nc117 in the regulation of slr0007, Nc117 may either enhance the de novo synthesis of slr0007 mRNA or suppress the degradation of slr0007 mRNA. To investigate whether the increased transcript accumulation of slr0007 in WT-pJA2-nc117 strain was due to transcriptional activation or post-transcriptional stabilization, a further bioinformatics analysis to predict the interaction between Nc117 and slr0007 together with its upstream region was firstly performed. As shown in Figure 6, the interaction region between Nc117 and slr0007 was predicted to be located in the upstream of slr0007 (intergenic region between slr0006 and slr0007). According to the previous RNA-seq analysis by Kopf et al. (2014), slr0006 and slr0007 composed an operon (Kopf et al., 2014), suggesting the interaction region between Nc117 and slr0007 might not be TSS or promoter region. However, according to a previous study by Pfeiffer et al. (2009), despite sRNAs could make base pair with the 5′ UTR of mRNA targets, some sRNAs such as MicC were shown to function with target mRNA by recognition of their mRNA molecule, guiding RNase E to cleave inside the coding sequence (Pfeiffer et al., 2009). Interestingly, our comparative transcriptomic results showed that the expression level of slr0006 was also 1.21-fold up-regulated in WT-pJA2-nc117 strain compared with WT. In addition, the AU content in the intergenic region between slr0006 and slr0007 was significantly higher than their coding sequence region, as shown in Figure 6, which is typical characteristic of a potential target cleavage site of RNase E in cyanobacteria (Horie et al., 2007). All these findings pointed to a hypothesis that the up-regulation of slr0006 and slr0007 in Nc117 overexpression strain was due to the protection role of Nc117 in protecting mRNA molecule of operon slr0006 and slr0007, although further evidences are still needed.
FIGURE 6.
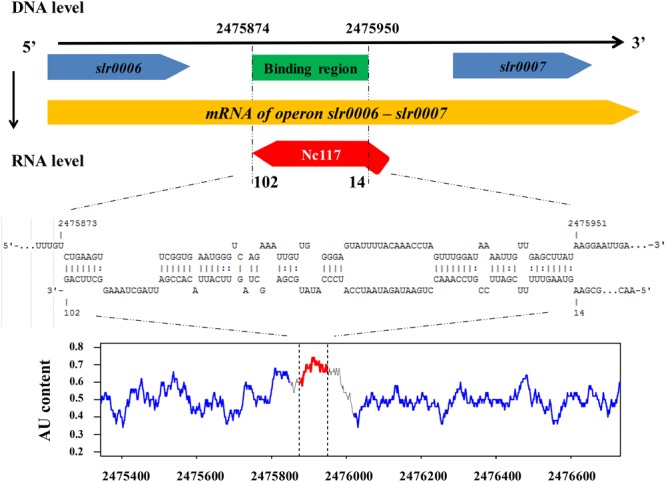
Interaction model of Nc117 and the target gene. The upper part shows the location and interact region between Nc117 and slr0006-slr0007 operon in DNA level and RNA level. The lower part indicated the AU content (box in 50 bp) of slr0006-slr0007 operon, red represents interact region and blue represents gene slr0006 (left) and slr0007 (right) coding sequence.
Discussion
As regulatory elements to deal with stress conditions in prokaryotes, sRNAs have received considerable attention in recent years. To understand the regulation mechanism of sRNAs, the primary targets should first be identified. So far, a series of methods for determining sRNAs target genes were developed (Vogel and Wagner, 2007). Among them, computational target prediction was demonstrated to be one of the effective approaches (Busch et al., 2008). In this study, the computational target prediction was first employed to explore potential targets of Nc117, and the top 100 potential targets were obtained. Although only 5 targets were demonstrated to be overlapped with those identified by the following RNA-seq transcriptomic analysis, considering the possibility that Nc117 might be involved in other functions besides short chain alcohols resistance, we can’t fully exclude some of these predicted targets as potential targets of Nc117, which remained to be investigated. Meanwhile, RNA-seq based comparative transcriptomic analysis of nc117 overexpression strain WT-pJA2-nc117 and WT revealed that 119 and 116 genes were up- and down-regulated, respectively, although most of these regulated genes may be indirect. By functional characterization of overexpression and knockout of the overlapped 5 potential targets, only slr0007 was demonstrated to be the potential target of Nc117 in regulating short chain alcohols tolerance (Figure 5). For the other four genes (slr2108, sll0784, sll1830, and slr1501), the similar phenotype in alcohols resistance was not observed as slr0007, which might be due to several possible reasons: (i) the significance of involvement in alcohols tolerance: these genes may be involved in alcohols tolerance, but we failed to identify them under the tested conditions; (ii) the changes of these genes may be secondary responses of genes regulated by Nc117, or be involved in other functions instead of short chain alcohols resistance that are still yet to be investigated; (iii) the false positive from the deficiency of computational target prediction and RNA-seq transcriptomic analysis. To further get more reliable target genes, more analysis such as comparative proteomics analysis can be employed.
The gene slr0007 was demonstrated to be the potential target of Nc117 in regulating short chain alcohols tolerance. According to an early conclusion by Cuccui et al. (2012), most Gram-negative bacteria tended to have higher organic solvent tolerance than Gram-positive bacteria, and it was speculated that the extra outer membrane structure unique to Gram-negative bacteria may be one of the key reasons for tolerance differences. LPS, as a main component of the outer membrane of Gram-negative bacteria, helped maintain the integrity of the outer membrane structure and was the first barrier to resist external environmental stress (Snyder et al., 2009). It prevents not only the dissolution of complement, antimicrobial peptide and detergent, but also the penetration of harmful substances to some extent (Trautner and Vermaas, 2013). According to Park et al. (2017), the carbohydrate glycero-manno-heptose and its derivatives were crucial parts of LPS in most Gram-negative bacteria, and they existed widely in core oligosaccharide region, O antigens and even glycan chains of S-layer glycoproteins. The target identified in this study, HddC encoded by slr0007 was one of the essential enzymes for the synthesis of carbohydrate glycero-manno-heptose (Cuccui et al., 2012). In addition, HddC may be related to the synthesis of surface layer glycoprotein, as an early study by Trautner and Vermaas (2013) showed that deletion of S-layer glycoprotein in Synechocystis led to low survival ability when exposed to lysozyme treatment or under low osmotic pressure, as well as the protective effect of the S-layer glycoprotein on the cyanobacterium (Trautner and Vermaas, 2013). In this respect, our results pointed to the proposed regulation mechanism of Nc117 that it may directly regulate synthesis of LPS or S-layer glycoprotein, correspondingly affecting tolerance to short chain alcohols. On the other hand, trans-encoded sRNAs usually regulate mRNAs by short, imperfect base pairing interactions. Some of these sRNAs interaction base pairs are at or near the 5′ UTR of their targets, and other family members interaction base pair may be at more distant sites (Storz et al., 2011). They have positive or negative regulatory effects on the expression of the corresponding genes by influencing translation or changing the stability of mRNA. Although it was hypothesized that the up-regulation of slr0006 and slr0007 in Nc117 overexpression strain was due to the protection role of Nc117 in protecting mRNA molecule of operon slr0006 and slr0007, further evidences are also still needed. Nevertheless, the identification of the involvement of slr0007 in alcohols tolerance helped to better understand the tolerance regulation mechanism to alcohols in cyanobacteria, and could be used to further rationally design and construct more tolerant cyanobacterial chassis, for example, by gene mutation to gain a protein with improved activity or other synthetic biology approaches, in the future.
Conclusion
sRNAs are important regulatory elements for coping with stress in prokaryotes. In this study, functional characterization of Nc117 in regulating biofuels tolerance in Synechocystis was conducted. By employing computational target prediction, the top 100 target genes were obtained. In addition, by comparative transcriptomic RNA-seq analysis, a total of 119 up-regulated and 116 down-regulated genes were identified. Integration of both analysis allowed us narrow down the possible targets to five for which the upstream regions of 5 genes were overlapped with those identified by computational target prediction approach. Notably, further functional characterization using overexpression and knockout strains showed that one target gene slr0007 was directly regulated by Nc117 involved in short chain alcohols tolerance. The results suggested that Nc117 was involved in short chain alcohols tolerance by regulating slr0007 and synthesis of LPS or S-layer glycoprotein. This study not only provided a novel regulatory mechanism of trans-encoded sRNA in cyanobacteria, but also valuable information for high-tolerant chassis construction in photosynthetic cyanobacteria.
Additional Information
The raw mRNA sequence data of Synechocystis are deposited in the SRA database of NCBI with accession numbers SRP130885.
Author Contributions
GP, LC, and WZ conceived the study. GP executed the sRNA and transcriptome bioinformatic analysis. YB, GP, and ZC carried out the mutant construction, phenotypic analysis, qRT-PCR experiments, and determination of RNA half-lives. TS and GP performed the 3′ RACE. YB, GP, TS, LC, and WZ designed and revised this paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- 3′ RACE
3′ rapid amplification of complementary DNA end
- cDNA
complementary DNA
- HddC
D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- qRT-PCR
quantitative real-time polymerase chain reaction
- RBS
ribosomal binding site
- RNA-seq
RNA sequencing
- sRNA
small RNA
- TCSTS
two-component signal transduction systems
- TSS
transcriptional starting site
- UTR
untranslated region
- WT
wild type
Footnotes
Funding. The research was supported by grants from National Basic Research Program of China (National “973” Program, Project No. 2014CB745101), the National Natural Science Foundation of China (NSFC) (No. 31470217, 21621004, 31770100, 31170043, 31270086, and 31370115), and the Tianjin Municipal Science and Technology Commission (No. 15JCZDJC32500).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00863/full#supplementary-material
3′ end determination of sRNA Nc117 by 3′ RACE and previous 5′ origination location.
Predicted interaction region in sRNA.
Predicted interaction region in mRNA.
Primers used for qRT-PCR validation.
Targets information for Nc117 based on IntaRNA prediction.
RNA-seq mapping statistics of different samples.
RNA-seq analysis of wild type Synechocystis and nc117 overexpression strain.
Pathway enrichment analysis for differential expressed genes.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S. A., Hellingwerf K. J., Lindblad P., De Mattos M. J. (2009). Energy biotechnology with cyanobacteria. Curr. Opin. Biotechnol. 20 257–263. 10.1016/j.copbio.2009.05.011 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Cann A. F., Connor M. R., Shen C. R., Smith K. M., Brynildsen M. P., et al. (2008). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10 305–311. 10.1016/j.ymben.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Higashide W., Liao J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27 1177–1180. 10.1038/nbt.1586 [DOI] [PubMed] [Google Scholar]
- Busch A., Richter A. S., Backofen R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccui J., Milne T. S., Harmer N., George A. J., Harding S. V., Dean R. E., et al. (2012). Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect. Immun. 80 1209–1221. 10.1128/IAI.05805-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J., Fu P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2 857–864. 10.1039/b811937f [DOI] [Google Scholar]
- Duhring U., Axmann I. M., Hess W. R., Wilde A. (2006). An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U.S.A. 103 7054–7058. 10.1073/pnas.0600927103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., Georg J., Klahn S., Sakurai I., Mustila H., Zhang P., et al. (2012). The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J. Biol. Chem. 287 33153–33162. 10.1074/jbc.M112.391755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanskaya I. V., Karandashova I. V., Bogachev A. V., Hagemann M. (2002). Functional analysis of the Na+/H+ antiporter encoding genes of the cyanobacterium Synechocystis PCC 6803. Biochem. Biokhimiia 67 432–440. 10.1023/A:1015281906254 [DOI] [PubMed] [Google Scholar]
- Foo J. L., Jensen H. M., Dahl R. H., George K., Keasling J. D., Lee T. S., et al. (2014). Improving microbial biogasoline production in Escherichia coli using tolerance engineering. mBio 5:e01932. 10.1128/mBio.01932-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida S. M., Al-Hinai M. A., Indurthi D. C., Nicolaou S. A., Papoutsakis E. T. (2013). Synthetic tolerance: three noncoding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Res. 41 8726–8737. 10.1093/nar/gkt651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Zhao J., Wang X., Qin S., Wei L., Ma W. (2016). NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. Strain PCC 6803. Plant Physiol. 170 752–760. 10.1104/pp.15.01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Sun T., Pei G., Chen L., Zhang W. (2016). Cyanobacterial chassis engineering for enhancing production of biofuels and chemicals. Appl. Microbiol. Biotechnol. 100 3401–3413. 10.1007/s00253-016-7374-2 [DOI] [PubMed] [Google Scholar]
- Gao Z., Zhao H., Li Z., Tan X., Lu X. (2016). Correction: photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 9 1113–1113. 10.1039/C5EE90067K [DOI] [Google Scholar]
- Gao X., Sun T., Wu L., Chen L., Zhang W. (2017). Co-overexpression of response regulator genes slr1037 and sll0039 improves tolerance of Synechocystis sp. PCC 6803 to 1-butanol. Bioresour. Technol. 245 1476–1483. 10.1016/j.biortech.2017.04.112 [DOI] [PubMed] [Google Scholar]
- Georg J., Dienst D., Schurgers N., Wallner T., Kopp D., Stazic D., et al. (2014). The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26 3661–3679. 10.1105/tpc.114.129767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y., Ito Y., Ono M., Moriwaki N., Kato H., Hamakubo Y., et al. (2007). Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol. Genet. Genomics 278 331–346. 10.1007/s00438-007-0254-9 [DOI] [PubMed] [Google Scholar]
- Hu J., Li T., Xu W., Zhan J., Chen H., He C., et al. (2017). Small antisense RNA RblR positively regulates RuBisCo in Synechocystis sp. PCC 6803. Front. Microbiol. 8:231. 10.3389/fmicb.2017.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. H., Camsund D., Lindblad P., Heidorn T. (2010). Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 38 2577–2593. 10.1093/nar/gkq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chen L., Te R., Qiao J., Wang J., Zhang W. (2013). Complementary iTRAQ proteomics and RNA-seq transcriptomics reveal multiple levels of regulation in response to nitrogen starvation in Synechocystis sp. PCC 6803. Mol. Biosyst. 9 2565–2574. 10.1039/c3mb70188c [DOI] [PubMed] [Google Scholar]
- Jie L., Lei C., Wang J., Qiao J., Zhang W. (2012). Proteomic analysis reveals resistance mechanism against biofuel hexane in Synechocystis sp. PCC 6803. Biotechnol. Biofuels 5 1–17. 10.1186/1754-6834-5-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Chen L., Wang J., Zhang W. (2014). Engineering biofuel tolerance in non-native producing microorganisms. Biotechnol. Adv. 32 541–548. 10.1016/j.biotechadv.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Johansen J., Eriksen M., Kallipolitis B., Valentin-Hansen P. (2008). Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and sigmaE-dependent CyaR-ompX regulatory case. J. Mol. Biol. 383 1–9. 10.1016/j.jmb.2008.06.058 [DOI] [PubMed] [Google Scholar]
- Kaczmarzyk D., Anfelt J., Sarnegrim A., Hudson E. P. (2014). Overexpression of sigma factor SigB improves temperature and butanol tolerance of Synechocystis sp. PCC6803. J. Biotechnol. 182-183 54–60. 10.1016/j.jbiotec.2014.04.017 [DOI] [PubMed] [Google Scholar]
- Klahn S., Schaal C., Georg J., Baumgartner D., Knippen G., Hagemann M., et al. (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U.S.A. 112 E6243–E6252. 10.1073/pnas.1508412112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Hess W. R. (2015). Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 39 301–315. 10.1093/femsre/fuv017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Klahn S., Scholz I., Matthiessen J. K., Hess W. R., Voss B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 21 527–539. 10.1093/dnares/dsu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E. I., Ro S. Y., Liao J. C. (2013). Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energy Environ. Sci. 6 2672–2681. 10.1039/c3ee41405a [DOI] [Google Scholar]
- Leone S., Silipo A., L Nazarenko E., Lanzetta R., Parrilli M., Molinaro A. (2007). Molecular structure of endotoxins from Gram-negative marine bacteria: an update. Mar. Drugs 5 85–112. 10.3390/md503085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los D. A., Zorina A., Sinetova M., Kryazhov S., Mironov K., Zinchenko V. V. (2010). Stress sensors and signal transducers in cyanobacteria. Sensors 10 2386–2415. 10.3390/s100302386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen C. A., Benhammou J. N., Majdalani N., Gottesman S. (2010). Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J. Bacteriol. 192 5559–5571. 10.1128/JB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J., Georg J., Scholz I., Sharma C. M., Dienst D., Bantscheff J., et al. (2011). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108 2124–2129. 10.1073/pnas.1015154108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee D., Seo E. K., Ryu J. S., Shin D. H. (2017). General assay for enzymes in the heptose biosynthesis pathways using electrospray ionization mass spectrometry. Appl. Microbiol. Biotechnol. 101 4521–4532. 10.1007/s00253-017-8148-1 [DOI] [PubMed] [Google Scholar]
- Patel R. K., Jain M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei G., Sun T., Chen S., Chen L., Zhang W. (2017). Systematic and functional identification of small non-coding RNAs associated with exogenous biofuel stress in cyanobacterium Synechocystis sp. PCC 6803. Biotechnol. Biofuels 10:57. 10.1186/s13068-017-0743-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V., Papenfort K., Lucchini S., Hinton J. C., Vogel J. (2009). Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16 840–846. 10.1038/nsmb.1631 [DOI] [PubMed] [Google Scholar]
- Qiao J., Huang S., Te R., Wang J., Chen L., Zhang W. (2013). Integrated proteomic and transcriptomic analysis reveals novel genes and regulatory mechanisms involved in salt stress responses in Synechocystis sp. PCC 6803. Appl. Microbiol. Biotechnol. 97 8253–8264. 10.1007/s00253-013-5139-8 [DOI] [PubMed] [Google Scholar]
- Qiao J., Wang J., Chen L., Tian X., Huang S., Ren X., et al. (2012). Quantitative iTRAQ LC-MS/MS proteomics reveals metabolic responses to biofuel ethanol in cyanobacterial Synechocystis sp. PCC 6803. J. Proteome Res. 11 5286–5300. 10.1021/pr300504w [DOI] [PubMed] [Google Scholar]
- Sakurai I., Stazic D., Eisenhut M., Vuorio E., Steglich C., Hess W. R., et al. (2012). Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 160 1000–1010. 10.1104/pp.112.202127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramago M., Barria C., Dos Santos R. F., Silva I. J., Pobre V., Domingues S., et al. (2014). The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 18 105–115. 10.1016/j.mib.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Snyder D. S., Brahamsha B., Azadi P., Palenik B. (2009). Structure of compositionally simple lipopolysaccharide from marine synechococcus. J. Bacteriol. 191 5499–5509. 10.1128/JB.00121-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Chen L., Wang J., Lu Y., Jiang W., Zhang W. (2014). A transcriptional regulator Sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803. Mol. Cell. Proteom. 13 3519–3532. 10.1074/mcp.M113.035675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Vogel J., Wassarman K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43 880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi M., Kurotaki H., Fukamizo T. (2007). Beta-1,4-glucanase-like protein from the cyanobacterium Synechocystis PCC6803 is a beta-1,3-1,4-glucanase and functions in salt stress tolerance. Biochem. J. 405 139–146. 10.1042/BJ20070171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Chen L., Wang J., Qiao J., Zhang W. (2013). Quantitative proteomics reveals dynamic responses of Synechocystis sp. PCC 6803 to next-generation biofuel butanol. J. Proteomics 78 326–345. 10.1016/j.jprot.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Trautner C., Vermaas W. F. (2013). The sll1951 gene encodes the surface layer protein of Synechocystis sp. strain PCC 6803. J. Bacteriol. 195 5370–5380. 10.1128/JB.00615-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama J., Asakura R., Kimura M., Moriyama A., Tahara H., Kobayashi Y., et al. (2012). Slr0967 and Sll0939 induced by the SphR response regulator in Synechocystis sp. PCC 6803 are essential for growth under acid stress conditions. Biochim. Biophys. Acta 1817 1270–1276. 10.1016/j.bbabio.2012.03.028 [DOI] [PubMed] [Google Scholar]
- Updegrove T. B., Zhang A., Storz G. (2016). Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30 133–138. 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Wagner E. G. (2007). Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 10 262–270. 10.1016/j.mib.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Wang H., Postier B., Burnap R. (2002). Optimization of fusion PCR for in vitro construction of gene knockout fragments. Biotechniques 33 26 28 30. [DOI] [PubMed] [Google Scholar]
- Wang J., Chen L., Huang S., Liu J., Ren X., Tian X., et al. (2012). RNA-seq based identification and mutant validation of gene targets related to ethanol resistance in cyanobacterial Synechocystis sp. PCC 6803. Biotechnol. Biofuels 5:89. 10.1186/1754-6834-5-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun T., Gao X., Shi M., Wu L., Chen L., et al. (2016). Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 34 60–70. 10.1016/j.ymben.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Xu W., Chen H., He C. L., Wang Q. (2014). Deep sequencing-based identification of small regulatory RNAs in Synechocystis sp. PCC 6803. PLoS One 9:e92711. 10.1371/journal.pone.0092711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H., Kaneko Y., Ehira S., Yoshihara S., Ikeuchi M., Ohmori M. (2010). CccS and CccP are involved in construction of cell surface components in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 51 1163–1172. 10.1093/pcp/pcq081 [DOI] [PubMed] [Google Scholar]
- Yoshimura H., Yanagisawa S., Kanehisa M., Ohmori M. (2002). Screening for the target gene of cyanobacterial cAMP receptor protein SYCRP1. Mol. Microbiol. 43 843–853. 10.1046/j.1365-2958.2002.02790.x [DOI] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Pei G., Niu X., Shi M., Zhang M., Chen L., et al. (2014). Metabolomic analysis reveals functional overlapping of three signal transduction proteins in regulating ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Mol. Biosyst. 11 770–782. 10.1039/c4mb00651h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3′ end determination of sRNA Nc117 by 3′ RACE and previous 5′ origination location.
Predicted interaction region in sRNA.
Predicted interaction region in mRNA.
Primers used for qRT-PCR validation.
Targets information for Nc117 based on IntaRNA prediction.
RNA-seq mapping statistics of different samples.
RNA-seq analysis of wild type Synechocystis and nc117 overexpression strain.
Pathway enrichment analysis for differential expressed genes.


