Summary
The mitochondrial unfolded protein response (UPR mt), a cellular protective program that ensures proteostasis in the mitochondria, has recently emerged as a regulatory mechanism for adult stem cell maintenance that is conserved across tissues. Despite the emerging genetic evidence implicating the UPR mt in stem cell maintenance, the underlying molecular mechanism is unknown. While it has been speculated that the UPR mt is activated upon stem cell transition from quiescence to proliferation, the direct evidence is lacking. In this study, we devised three experimental approaches that enable us to monitor quiescent and proliferating hematopoietic stem cells (HSCs) and provided the direct evidence that the UPR mt is activated upon HSC transition from quiescence to proliferation, and more broadly, mitochondrial integrity is actively monitored at the restriction point to ensure metabolic fitness before stem cells are committed to proliferation.
Keywords: aging, hematopoietic stem cells, mitochondria, mitochondrial unfolded protein response, SIRT7, stem cell quiescence, stem cells
1. INTRODUCTION
Adult stem cells persist throughout the entire lifespan of an organism to repair tissue damage and maintain tissue homeostasis. Among their evolved adaptations are elaborate cellular protective programs that ensure stem cell integrity, tissue homeostasis, and organismal survival (Biteau, Hochmuth & Jasper, 2008; Brown et al., 2013; Ito et al., 2004; Rando, 2006; Renault et al., 2009; Rossi, Jamieson & Weissman, 2008; Rossi et al., 2007; Sahin & Depinho, 2010; Sperka, Wang & Rudolph, 2012; Walter et al., 2015). The mitochondrial unfolded protein response (UPRmt), a cellular protective program that ensures proteostasis in the mitochondria, has recently emerged as a regulatory mechanism for adult stem cell maintenance that is conserved across tissues (Berger et al., 2016; Mohrin et al., 2015; Zhang et al., 2016). This protective program is dysregulated during physiological aging, contributing to the functional deterioration of stem cells, tissue degeneration, and shortened organismal lifespan (Mohrin et al., 2015; Zhang et al., 2016). In addition to the UPRmt, deregulation of compensatory mitochondrial protective programs such as mitophagy and mitochondrial dynamics leads to compromised stem cells, further underscoring the importance of mitochondrial integrity in stem cell maintenance (Ho et al., 2017; Ito et al., 2016; Luchsinger, de Almeida, Corrigan, Mumau & Snoeck, 2016; Vannini et al., 2016).
Despite the emerging genetic evidence implicating the UPRmt in stem cell maintenance, the underlying molecular mechanism is unknown. The UPRmt is a nascent cellular pathway that is activated when cells experience mitochondrial protein folding stress and retrograde signaling from the mitochondria to the nucleus triggers transcriptional activation of nuclear‐encoded mitochondrial chaperones and proteases as well as repression of translation to reestablish proteostasis (Haynes, Fiorese & Lin, 2013; Haynes & Ron, 2010; Mohrin et al., 2015; Munch & Harper, 2016; Zhao et al., 2002). Primarily characterized in C. elegans, the UPRmt is activated during a developmental stage when there is a burst of mitochondrial biogenesis (Houtkooper et al., 2013; Lin et al., 2016; Merkwirth et al., 2016; Nargund, Pellegrino, Fiorese, Baker & Haynes, 2012; Pellegrino et al., 2014; Tian et al., 2016). It is therefore speculated that in stem cells, the UPRmt is activated under a physiological condition when mitochondrial biogenesis is induced. Adult stem cells frequently exit the cell cycle and are predominantly found in the quiescent (G0) state, where the number of mitochondria is low and glycolysis is the primary metabolic pathway to support energy production (Folmes, Dzeja, Nelson & Terzic, 2012; Takubo et al., 2013; Warr & Passegue, 2013; Yu et al., 2013). As stem cells transit from quiescence to proliferation, mitochondrial biogenesis is induced to enable metabolic reprogramming from glycolysis to oxidative phosphorylation to meet increasing energy demands. Because a major event during the transition from quiescence to proliferation is mitochondrial biogenesis, it raises the possibility that the UPRmt is activated during this transition. However, the direct evidence is lacking. In this study, we devised three experimental approaches that enable us to monitor quiescent and proliferating stem cells and directly test this hypothesis.
We tested this hypothesis in hematopoietic stem cells (HSCs), immunophenotypically defined as Lin−c‐Kit+Sca1+CD150+CD48−. About 90% of HSCs reside in a quiescent state under homeostatic conditions (Pietras, Warr & Passegue, 2011). We isolated HSCs from mouse bone marrow and stimulated them to exit quiescence ex vivo upon culture with cytokines. We first confirmed that HSCs stimulated with cytokines were actively proliferating (Figure 1a) and that mitochondrial mass was increased in HSCs upon proliferation (Figure 1b). Compared to freshly isolated quiescent HSCs, proliferating HSCs stimulated with cytokines exhibited increased expression of mitochondrial chaperones and proteases at the transcriptional level (Figure 1c). Because mitochondrial biogenesis upon HSC transition from quiescence to proliferation is regulated at the translational level mediated by mTOR (Chen et al., 2008; Gan et al., 2010; Gurumurthy et al., 2010; Morita et al., 2013; Nakada, Saunders & Morrison, 2010), increased expression of mitochondrial chaperones and proteases at the transcriptional level reflects de novo activation of the UPRmt.
Figure 1.
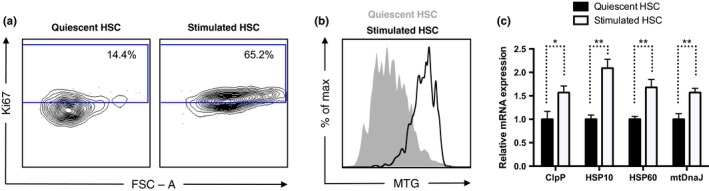
The UPR mt is activated upon HSC transition from quiescence to proliferation ex vivo. (a) Cell cycle analysis of HSCs using Ki‐67 staining showing increased proliferation in HSCs stimulated by ex vivo culture with cytokines compared to quiescent HSCs freshly isolated from mouse bone marrow. (b) MitoTracker Green staining showing increased mitochondrial mass in HSCs stimulated to proliferate via ex vivo culture with cytokines compared to quiescent HSCs freshly isolated from mouse bone marrow. (c) qPCR showing increased transcription of mitochondrial chaperones and proteases in HSCs stimulated to proliferate via ex vivo culture with cytokines compared to quiescent HSCs freshly isolated from mouse bone marrow. n = 3
We further validated these results by stimulating HSCs to exit quiescence in vivo upon transplantation. Compared to HSCs isolated from untransplanted mice, donor HSCs isolated from transplanted recipient mice 2 weeks post‐transplant were actively proliferating (Figure 2a), had increased mitochondrial mass (Figure 2b,c), and increased expression of mitochondrial chaperones and proteases at the transcriptional level (Figure 2d), indicative of activation of the UPRmt.
Figure 2.
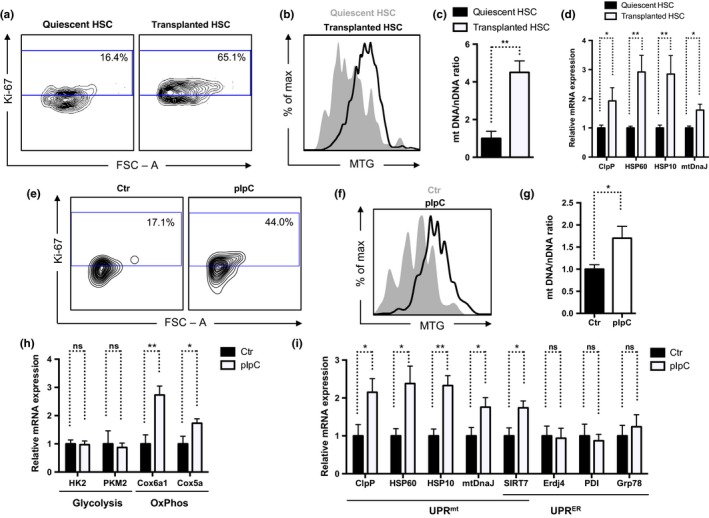
The UPR mt is activated upon HSC transition from quiescence to proliferation in vivo. (a) Cell cycle analysis of HSCs using Ki‐67 staining showing increased proliferation in HSCs stimulated by in vivo transplantation (2 week post‐transplantation) compared to quiescent HSCs freshly isolated from mouse bone marrow. (b, c) MitoTracker Green staining (b) and quantification of the mitochondrial to nuclear DNA ratio (c) showing increased mitochondrial mass in HSCs stimulated to proliferate via in vivo transplantation compared to quiescent HSCs freshly isolated from mouse bone marrow. n = 3. (d) qPCR showing increased transcription of mitochondrial chaperones and proteases in HSCs stimulated to proliferate via in vivo transplantation compared to quiescent HSCs freshly isolated from mouse bone marrow. n = 3. (e) Cell cycle analysis of HSCs using Ki‐67 staining showing increased proliferation in HSCs stimulated by pIpC treatment (24 hr after treatment) compared to quiescent HSCs isolated from untreated mouse bone marrow. (f, g) MitoTracker Green staining (f) and quantification of the mitochondrial to nuclear DNA ratio (g) showing increased mitochondrial mass in HSCs stimulated to proliferate via pIpC compared to quiescent HSCs isolated from untreated mouse bone marrow. n = 3. (h, i) qPCR showing increased transcription of oxidative phosphorylation genes and mitochondrial chaperones and proteases, but not glycolysis genes and ER stress response genes in HSCs stimulated to proliferate via pIpC treatment compared to quiescent HSCs isolated from untreated mouse bone marrow. n = 3
An alternative approach to model HSC proliferation in vivo is to treat mice with polyinosinic:polycytidylic acid (pIpC), a synthetic double‐stranded RNA (dsRNA) mimetic that stimulates the multiple immune signaling pathways that are activated during a viral infection (Walter et al., 2015). Compared to HSCs isolated from untreated mice, HSCs isolated from mice 24 hr after the pIpC treatment showed increased proliferation (Figure 2e) and mitochondrial mass (Figure 2f,g), and induction of the expression of oxidative phosphorylation genes (Figure 2h) and mitochondrial chaperones and proteases (Figure 2i). In contrast, the expression of glycolysis genes (Figure 2h) and ER stress response genes (Figure 2i) was unchanged. These data are consistent with the notion that proliferative HSCs have increased mitochondrial number, experience a metabolic switch from glycolysis to oxidative phosphorylation, and induce the UPRmt to maintain the mitochondrial homeostasis.
Collectively, these results provide direct evidence that the UPRmt is activated upon HSC transition from quiescence to proliferation (Figures 1 and 2), and more broadly, mitochondrial integrity is actively monitored at the restriction point to ensure metabolic fitness before stem cells are committed to proliferation. Stem cell quiescence is a protective mechanism that prevents cell death and the depletion of the stem cell pool (Cheung & Rando, 2013). Consistent with the activation of the UPRmt at the transition from quiescence to proliferation, dysregulation of the UPRmt results in stem cell death, a reduced stem cell pool, and compromised stem cell self‐renewal (Berger et al., 2016; Mohrin et al., 2015; Zhang et al., 2016). Among the induction of the UPRmt is increased expression of SIRT7 (Figure 2i), which alleviates mitochondrial protein folding stress by repressing NRF1 activity and mitochondrial translation, reduces mitochondrial activity and proliferation, and gives cells more time to recover from stress (Mohrin et al., 2015). Failure to do so leads to HSC death. With aging, SIRT7 becomes inactivated, resulting in increased mitochondrial protein folding stress and functional decline (Mohrin et al., 2015). Identifying molecular regulators of the UPRmt in stem cells opens the door for novel therapeutic opportunities for improving stem cell maintenance, enhancing tissue regeneration, and extending lifespan and healthspan.
2. EXPERIMENTAL PROCEDURES
2.1. Mice
C57BL/6 mice were housed on a 12:12 hr light:dark cycle at 25°C. All animal procedures were in accordance with the animal care committee at the University of California, Berkeley.
2.2. Flow cytometry and cell sorting
Bone marrow cells were obtained by crushing the long bones with sterile PBS without calcium and magnesium supplemented with 2% FBS. Lineage staining contained a cocktail of biotinylated anti‐mouse antibodies to Mac‐1 (CD11b), Gr‐1 (Ly‐6G/C), Ter119 (Ly‐76), CD3, CD4, CD8a (Ly‐2), and B220 (CD45R) (BioLegend). For detection or sorting, we used streptavidin conjugated to APC‐Cy7, c‐Kit‐APC, Sca‐1‐Pacific blue, CD48‐FITC, and CD150‐PE (BioLegend). For congenic strain discrimination, anti‐CD45.1 PerCP and anti‐CD45.2 PE‐Cy7 antibodies (BioLegend) were used. For assessment of cell cycle analysis, Ki‐67 (BioLegend) staining was performed according to the manufacturer's recommendation after cell surface staining. The gates were drawn based on the fluorescence minus one (FMO) control. For mitochondrial mass, bone marrow cells were incubated with 100 nM MitoTracker Green (Invitrogen) for 30 min at 37°C in the dark after cell surface staining. All data were collected on a Fortessa (Becton Dickinson), and data analysis was performed with FlowJo (TreeStar). For cell sorting, lineage depletion or c‐kit enrichment was performed according to the manufacturer's instructions (Miltenyi Biotec). Cells were sorted using a Cytopeia INFLUX Sorter (Becton Dickinson). Antibody details are provided in Table S1.
To stimulate HSCs to exit quiescence, freshly isolated HSCs were cultured ex vivo in IMDM (Invitrogen) supplemented with 5% stem cell FBS (Stem Cell Technologies), 1% penicillin/streptomycin, sodium pyruvate, NEAA, l‐glutamine (Invitrogen), and cytokines (IL‐3 (10 ng/ml), GMCSF (10 ng/ml), SCF (25 ng/ml), IL‐11 (25 ng/ml), Flt3L (25 ng/ml), TPO (25 ng/ml) (PeproTech), and EPO (4 U/ml) (R&D)) for 48 hr. Alternatively, to stimulate HSCs to exit quiescence in vivo, 1 × 106 bone marrow cells were transplanted into lethally irradiated recipient mice. Two weeks post‐transplantation, donor HSCs were isolated via sorting. To induce in vivo cycling of HSCs, mice were injected intraperitoneally (i.p.) with 5 mg/kg polyinosinic:polycytidylic acid (Sigma) 24 hr prior to analysis.
2.3. mRNA analysis
RNA was isolated from cells using TRIzol reagent (Invitrogen). cDNA was generated using qScript™ cDNA SuperMix (Quanta Biosciences). Gene expression was determined by real‐time PCR using Eva qPCR SuperMix Kit (BioChain Institute) on an ABI StepOnePlus system. All data were normalized to β‐actin expression. PCR primer details are provided in Table S2.
2.4. mtDNA/nDNA
The mitochondrial DNA/nuclear DNA (mtDNA/nDNA) ratio was determined by isolating DNA from cells with TRIzol (Invitrogen), as described previously (Lai et al., 2008). The ratio of mtDNA/nDNA was calculated as previously described (Venegas et al., 2011).
2.5. Statistical analysis
The number of mice chosen for each experiment is based on the minimum number of mice necessary to have sufficient statistical power and is comparable to published literature for the same assays performed. Mice were randomized to groups, and analysis of mice and tissue samples was performed by investigators blinded to the treatment of the animals. Statistical analysis was performed with Excel (Microsoft). Means between two groups were compared with Student's t test. Error bars represent standard errors. In all corresponding figures, * represents p < .05, ** represents p < .01, *** represents p < .001, and ns represents p > .05.
Supporting information
ACKNOWLEDGMENTS
We thank H. Nolla, A. Valeros, and K. Heydari for cell sorting. This research was supported by NIH R01 AG040990 (D.C.), R01DK101885 (D.C.), National Institute of Food and Agriculture (D.C.), PackerWentz Endowment (D.C.), Chau Hoi Shuen Foundation (Chen), Siebel Stem Cell Institute (M.M., D.C.), NIH T32AG000266 (M.M.), NIH T32GM098218 (A.W.), and Glenn/AFAR Scholarship (H.L.).
Mohrin M, Widjaja A, Liu Y, Luo H, Chen D. The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell. 2018;17:e12756 10.1111/acel.12756
REFERENCES
- Berger, E. , Rath, E. , Yuan, D. , Waldschmitt, N. , Khaloian, S. , Allgauer, M. , … Haller, D. (2016). Mitochondrial function controls intestinal epithelial stemness and proliferation. Nature Communications, 7, 13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau, B. , Hochmuth, C. E. , & Jasper, H. (2008). JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell, 3, 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. , Xie, S. , Qiu, X. , Mohrin, M. , Shin, J. , Liu, Y. , … Chen, D. (2013). SIRT3 reverses aging‐associated degeneration. Cell Reports, 3, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Liu, Y. , Liu, R. , Ikenoue, T. , Guan, K. L. , Liu, Y. , & Zheng, P. (2008). TSC‐mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. Journal of Experimental Medicine, 205, 2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, T. H. , & Rando, T. A. (2013). Molecular regulation of stem cell quiescence. Nature Reviews Molecular Cell Biology, 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes, C. D. , Dzeja, P. P. , Nelson, T. J. , & Terzic, A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell, 11, 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, B. , Hu, J. , Jiang, S. , Liu, Y. , Sahin, E. , Zhuang, L. , … Depinho, R. A. (2010). Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature, 468, 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy, S. , Xie, S. Z. , Alagesan, B. , Kim, J. , Yusuf, R. Z. , Saez, B. , … Bardeesy, N. (2010). The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature, 468, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, C. M. , Fiorese, C. J. , & Lin, Y. F. (2013). Evaluating and responding to mitochondrial dysfunction: The mitochondrial unfolded‐protein response and beyond. Trends in Cell Biology, 23, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, C. M. , & Ron, D. (2010). The mitochondrial UPR ‐ protecting organelle protein homeostasis. Journal of Cell Science, 123, 3849–3855. [DOI] [PubMed] [Google Scholar]
- Ho, T. T. , Warr, M. R. , Adelman, E. R. , Lansinger, O. M. , Flach, J. , Verovskaya, E. V. , … Passegue, E. (2017). Autophagy maintains the metabolism and function of young and old stem cells. Nature, 543, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper, R. H. , Mouchiroud, L. , Ryu, D. , Moullan, N. , Katsyuba, E. , Knott, G. , … Auwerx, J. (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature, 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K. , Hirao, A. , Arai, F. , Matsuoka, S. , Takubo, K. , Hamaguchi, I. , … Suda, T. (2004). Regulation of oxidative stress by ATM is required for self‐renewal of haematopoietic stem cells. Nature, 431, 997–1002. [DOI] [PubMed] [Google Scholar]
- Ito, K. , Turcotte, R. , Cui, J. , Zimmerman, S. E. , Pinho, S. , Mizoguchi, T. , … Ito, K. (2016). Self‐renewal of a purified Tie2 + hematopoietic stem cell population relies on mitochondrial clearance. Science, 354, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, L. , Leone, T. C. , Zechner, C. , Schaeffer, P. J. , Kelly, S. M. , Flanagan, D. P. , Medeiros, D. M. , Kovacs, A. , & Kelly, D. P. (2008). Transcriptional coactivators PGC‐1α and PGC‐lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev., 22, 1948–1961. 10.1101/gad.1661708pmid:18628400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. F. , Schulz, A. M. , Pellegrino, M. W. , Lu, Y. , Shaham, S. , & Haynes, C. M. (2016). Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature, 533, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger, L. L. , de Almeida, M. J. , Corrigan, D. J. , Mumau, M. , & Snoeck, H. W. (2016). Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature, 529, 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth, C. , Jovaisaite, V. , Durieux, J. , Matilainen, O. , Jordan, S. D. , Quiros, P. M. , … Dillin, A. (2016). Two conserved histone demethylases regulate mitochondrial stress‐induced longevity. Cell, 165, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin, M. , Shin, J. , Liu, Y. , Brown, K. , Luo, H. , Xi, Y. , … Chen, D. (2015). Stem cell aging. A mitochondrial UPR‐mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science, 347, 1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, M. , Gravel, S. P. , Chenard, V. , Sikstrom, K. , Zheng, L. , Alain, T. , … Sonenberg, N. (2013). mTORC1 controls mitochondrial activity and biogenesis through 4E‐BP‐dependent translational regulation. Cell Metabolism, 18, 698–711. [DOI] [PubMed] [Google Scholar]
- Munch, C. , & Harper, J. W. (2016). Mitochondrial unfolded protein response controls matrix pre‐RNA processing and translation. Nature, 534, 710–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, D. , Saunders, T. L. , & Morrison, S. J. (2010). Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature, 468, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund, A. M. , Pellegrino, M. W. , Fiorese, C. J. , Baker, B. M. , & Haynes, C. M. (2012). Mitochondrial import efficiency of ATFS‐1 regulates mitochondrial UPR activation. Science, 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino, M. W. , Nargund, A. M. , Kirienko, N. V. , Gillis, R. , Fiorese, C. J. , & Haynes, C. M. (2014). Mitochondrial UPR‐regulated innate immunity provides resistance to pathogen infection. Nature, 516, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E. M. , Warr, M. R. , & Passegue, E. (2011). Cell cycle regulation in hematopoietic stem cells. The Journal of Cell Biology, 195, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T. A. (2006). Stem cells, ageing and the quest for immortality. Nature, 441, 1080–1086. [DOI] [PubMed] [Google Scholar]
- Renault, V. M. , Rafalski, V. A. , Morgan, A. A. , Salih, D. A. , Brett, J. O. , Webb, A. E. , … Brunet, A. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell, 5, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, D. J. , Bryder, D. , Seita, J. , Nussenzweig, A. , Hoeijmakers, J. , & Weissman, I. L. (2007). Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature, 447, 725–729. [DOI] [PubMed] [Google Scholar]
- Rossi, D. J. , Jamieson, C. H. , & Weissman, I. L. (2008). Stems cells and the pathways to aging and cancer. Cell, 132, 681–696. [DOI] [PubMed] [Google Scholar]
- Sahin, E. , & Depinho, R. A. (2010). Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature, 464, 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka, T. , Wang, J. , & Rudolph, K. L. (2012). DNA damage checkpoints in stem cells, ageing and cancer. Nature Reviews Molecular Cell Biology, 13, 579–590. [DOI] [PubMed] [Google Scholar]
- Takubo, K. , Nagamatsu, G. , Kobayashi, C. I. , Nakamura‐Ishizu, A. , Kobayashi, H. , Ikeda, E. , … Suda, T. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell, 12, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y. , Garcia, G. , Bian, Q. , Steffen, K. K. , Joe, L. , Wolff, S. , … Dillin, A. (2016). Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell, 165, 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini, N. , Girotra, M. , Naveiras, O. , Nikitin, G. , Campos, V. , Giger, S. , … Lutolf, M. P. (2016). Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nature Communications, 7, 13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas, V. , Wang, J. , Dimmock, D. , & Wong, L. J. (2011). Real‐time quantitative PCR analysis of mitochondrial DNA content. Curr. Protoc. Hum. Genet., Chapter 19, Unit 19.7. 10.1002/0471142905.hg1907s68 [DOI] [PubMed] [Google Scholar]
- Walter, D. , Lier, A. , Geiselhart, A. , Thalheimer, F. B. , Huntscha, S. , Sobotta, M. C. , … Milsom, M. D. (2015). Exit from dormancy provokes DNA‐damage‐induced attrition in haematopoietic stem cells. Nature, 520, 549–552. [DOI] [PubMed] [Google Scholar]
- Warr, M. R. , & Passegue, E. (2013). Metabolic makeover for HSCs. Cell Stem Cell, 12, 1–3. [DOI] [PubMed] [Google Scholar]
- Yu, W. M. , Liu, X. , Shen, J. , Jovanovic, O. , Pohl, E. E. , Gerson, S. L. , … Qu, C. K. (2013). Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell, 12, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Ryu, D. , Wu, Y. , Gariani, K. , Wang, X. , Luan, P. , … Auwerx, J. (2016). NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science, 352, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Wang, J. , Levichkin, I. V. , Stasinopoulos, S. , Ryan, M. T. , & Hoogenraad, N. J. (2002). A mitochondrial specific stress response in mammalian cells. The EMBO Journal, 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


