Abstract
Summary
Although the genome-wide association study (GWAS) is a powerful method to identify disease-associated variants, it does not directly address the biological mechanisms underlying such genetic association signals. Here, we present PGA, a Perl- and Java-based program for post-GWAS analysis that predicts likely disease genes given a list of GWAS-reported variants. Designed with a command line interface, PGA incorporates genomic and eQTL data in identifying disease gene candidates and uses gene network and ontology data to score them based upon the strength of their relationship to the disease in question.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
In the past several years, genome-wide association studies (GWAS) have been successfully applied to various human complex diseases leading to the identification of a large number of disease-associated genetic loci. Interpreting these results, however, remains elusive as GWAS only detect statistical associations—not functional signals—among a subset of all variants and most associated SNPs are non-coding—either intronic or intergenic. To uncover the biological mechanisms underlying disease association signals, it is necessary to identify genes potentially affected by the reported variants as possible sources of these signals.
For lack of a better approach, in current GWAS the genes closest to or in the vicinity of disease-associated variants are used as the causal genes. This method cannot effectively handle variants found in gene deserts or in close proximity to multiple genes and also overlooks the possibility that risk variants may be contained in regulatory elements and therefore affect distant genes.
Here, we present a Perl- and Java-based application with a command line interface for post-GWAS analysis. It integrates both gene network and annotation data with GWAS signals to predict disease causal genes and assign them evidence-based scores. By considering associations between regulatory elements and promoters, our program can predict disease genes both proximal and distal to GWAS signals and regulatory elements (e.g. enhancers) that could harbor non-coding causal SNPs.
2 Materials and methods
Following the framework of our recent post-GWAS analysis of schizophrenia (Lin et al., 2016), this application performs two distinct operations: it identifies candidate disease genes given a set of GWAS-reported variants, and it scores these candidate genes to prioritize those most likely to be the sources of the disease-association signals.
Identifying risk regions. As GWAS only detect statistical associations from a pre-select subset of variants, it is necessary to identify all unexamined variants that are in strong linkage disequilibrium (LD) with the GWAS-reported variants as potential alternative sources of the disease-association signals. Using VCFtools (Danecek et al., 2011) and a 1000 Genomes Project (1KG) reference panel (Genomes Project et al., 2012), we calculate the LD between each GWAS-reported variant and every 1KG variant within a 400-kb range. An LD block is then formed from all within-range SNPs with > 0.5 and indexed by the corresponding GWAS variant. We merge all overlapping or adjacent (within 250 kb) LD blocks to form genomic risk regions and then use them to identify both proximal and distal risk gene candidates.
Identifying risk gene candidates. Proximal risk gene candidates are genes that—after extending their ranges by 20 kb on each end—overlap with these genomic risk regions. Distal risk gene candidates are genes affected by an expression quantitative trait locus (eQTL) or transcriptional regulatory element (TRE) containing any of the variants found in strong LD with GWAS-reported variants in an LD block (including the GWAS-reported variant itself). In order to incorporate this gene regulatory information, we collected lists of eQTL and TREs with their target genes from ENCODE (Thurman et al., 2012; Wang et al., 2018), FANTOM5 (Andersson et al., 2014; Wang et al., 2018), and GTEx (Lonsdale et al., 2013) data.
Scoring risk gene candidates. Variants associated with a particular disease may implicate a large number of disease gene candidates—particularly when distal gene candidates are considered as well—and it is therefore useful to prioritize these candidate genes. Our application employs a statistical method to score the disease-relatedness of risk gene candidates using predictive features derived from gene networks and annotation based on a set of training genes that are known to play a role in the etiology of the disease.
Given this set of known disease genes D and the set of known genes G (from GENCODE v19), we obtain the set of background genes B = G – D. Then, from the set of known disease genes D we extract our predictive features: the frequent combinations of the Gene Ontology (GO) terms associated with the genes in D and of the neighbors of genes in D in the genome-scale human protein-protein interaction network that we employ (Li et al., 2017). GO terms of genes in D include both annotated GO terms and their ancestor GO terms along the path of the ‘is a’ relationship in the gene ontology structure. Our application uses the FP-growth algorithm for frequent itemset mining (Han et al., 2000) with a support value of We limit the predictive features to 3-itemsets to avoid redundancy and intensive computation.
After feature extraction, we assign a score to each predictive feature f based on the frequency of its association with genes in D and B:
in which FDis the frequency with which f occurs in D and ND is the number of genes in D. FB and NBare corresponding values in B. Next, for each candidate disease gene, we identify all the predictive features with which it is associated and assign it the highest score of these features. In the event that a risk gene candidate is a training gene as well, the score Sf of each predictive feature it contains must be adjusted:
As network and annotation scores are treated separately, each gene has two different scores that are combined to produce a final gene score:
in which Sf(n) and Sf(a) are the network and annotation-based scores, respectively, and α is a coefficient controlling the relative weights of these two scores on the final gene score. α = 0.4 yields the best predictive power according to our evaluation (data not shown). Every candidate gene is excluded from gene sets B and D when scored to avoid biased scoring.
Notably, our gene scoring method is limited by information available about known disease genes and is based on the hypothesis that novel disease genes will be involved in the same pathways and mechanisms as known disease genes.
Evaluating scores. Our application produces a score threshold to indicate that candidate genes with scores greater than this threshold should be considered as putative causal disease genes. To evaluate the prediction precision of a score threshold, we use disease training genes as the positive gene set and random background genes as negative gene sets. The threshold is the value that achieves a prediction precision ≥ 0.8.
Application output and efficiency. PGA produces a number of output files: a list of scored disease gene candidates based on the scoring method outlined above, a list of risk regions indexed by associated variants and their linked genes (with scores and markings indicating proximal or distal), and several intermediate files such as the regulatory information for tracing the links of distal genes to GWAS signals and lists of network and annotation-based predictive features that can be reused with the same training gene set in order to avoid redundant computation. The first operation performed by the application, generating risk gene candidates, requires between 15 s and 90 s per input variant, dependent on the size of the 1KG reference panel in use. The next operation, scoring these candidate genes, can take less than one to several hours, depending on the size of the network/annotation data and their relationship with the training gene set used to generate predictive features. As the frequent itemset mining takes the majority of the time at this step, using pre-generated predictive feature sets significantly reduces the time used to score candidate genes.
Custom annotation data. PGA automatically uses built-in loci-gene regulatory information from ENCODE, FANTOM5 and GTEx eQTL data to identify distal risk gene candidates. It can also incorporate additional loci-gene regulatory information provided by users to potentially uncover more risk gene candidates.
3 Application
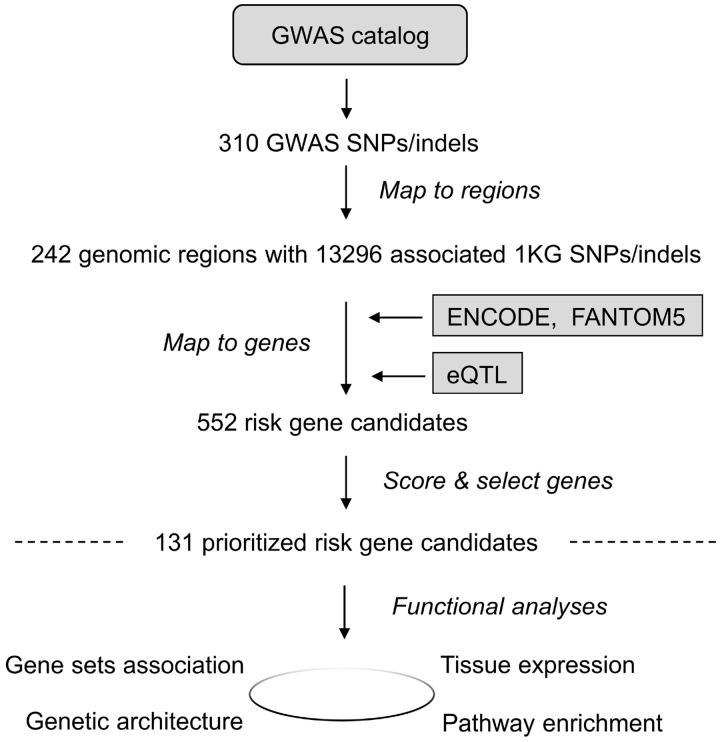
PGA can identify putative risk genes proximal or distal to GWAS signals. In a case study of Alzheimer’s disease (AD), we first collected the top 50 AD risk genes from MalaCards (Rappaport et al., 2017) as training genes (Supplementary Table S1) and 310 AD-associated SNPs from the GWAS Catalog (MacArthur et al., 2017) as variant input (Supplementary Table S2). Given these two types of input, PGA identified 242 risk genomic regions and 552 connected candidate genes (Fig. 1 and Supplementary Table S3), of which 131 were scored high and thus predicted as putative AD risk genes (Supplementary Table S4). In the subsequent GO term and pathway analysis of these genes (Supplementary Tables S5 and S6), the most significantly enriched biological process (BP) GO term, ‘Negative regulation of beta-amyloid formation,’ is directly related to the pathogenesis of AD (Sadigh-Eteghad et al., 2015). Among the many over-represented inflammatory signaling pathways, ‘Signaling by Interleukins’ and ‘Cytokine Signaling in Immune system’ have both been suggested to play a role in the pathology and progression of AD (Weisman et al., 2006).
Fig. 1.
The flowchart of the integrated post-GWAS study of Alzheimer’s disease. 131 putative risk genes were identified from 310 GWAS reported SNPs for AD
We also examined the effectiveness of PGA in putative disease risk gene identification. In an AD risk genomic region in 22q13.2 (Supplementary Fig. S1A), PGA linked five candidate genes to the AD-associated SNP (rs7364180). Although this SNP is located in an intron of CCDC134, PGA identified a high scoring gene in the risk region, SREBF2, which has been implicated in the pathology of AD (Barbero-Camps et al., 2013). In another case, PGA linked five candidate genes to three AD-associated SNPs (rs9877502, rs61174035 and rs10937470) in an AD risk genomic region in 3q28 (Supplementary Fig. S1B). Among them, three are proximal candidate genes that have low scores. PGA identified a high-scoring distal gene, IL1RAP, as a possible AD risk gene underlying the disease association signals of these SNPs. Interestingly, IL1RAP was implicated as a novel causal gene for AD in a recent GWAS study (Ramanan et al., 2015). The details of regulatory information of high scoring genes are shown in Supplementary Table S7.
Indeed, a systematic performance evaluation of PGA’s results indicated that it is more effective than other methods at prioritizing risk genes in AD GWAS (Supplementary Fig. S2). The superior performance of PGA is due to the fact that it integrates different types of data (Supplementary Table S8) allowing it to uncover plausible risk genes implicated by GWAS signals that might be missed by other methods.
Funding
This work was supported by NIH grants R01 HG008153 from the National Human Genome Research Institute and R01 AG057909 from the National Institute on Aging to Z.D.Z. This work was also supported by NIH grant U01 MH101720 from the National Institute of Mental Health to the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome.
Conflict of Interest: none declared.
Supplementary Material
References
- Andersson R. et al. (2014) An atlas of active enhancers across human cell types and tissues. Nature, 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero-Camps E. et al. (2013) APP/PS1 mice overexpressing SREBP-2 exhibit combined Abeta accumulation and tau pathology underlying Alzheimer’s disease. Human Mol. Genet., 22, 3460–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C. et al. (2012) An integrated map of genetic variation from 1, 092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. et al. (2000) Mining frequent patterns without candidate generation. In: SIGMOD ‘00 Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data Association for Computing Machinery (ACM), Dallas, Texas, USA. [Google Scholar]
- Li T.B. et al. (2017) A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods, 14, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.R. et al. (2016) Integrated post-GWAS analysis sheds new light on the disease mechanisms of schizophrenia. Genetics, 204, 1587–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J. et al. (2013) The genotype-tissue expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J. et al. (2017) The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res., 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V.K. et al. (2015) GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain, 138, 3076–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport N. et al. (2017) MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res., 45, D877–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadigh-Eteghad S. et al. (2015) Amyloid-beta: a crucial factor in Alzheimer’s disease. Med. Princ. Pract., 24, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R.E. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2018) HEDD: Human Enhancer Disease Database. Nucleic Acids Res., 46, D113–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman D. et al. (2006) Interleukins, inflammation, and mechanisms of Alzheimer’s disease. Vitam. Horm., 74, 505–530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.