Abstract
Study Objectives
Insomnia is a common sleep disorder that is associated with a range of adverse outcomes. Patients with insomnia exhibit hyperarousal in multiple domains, including an elevated metabolic rate, but specific metabolic molecular perturbations are unknown. Furthermore, objective clinical markers of insomnia are not available and current assessment of pathological extent relies on self-report. Here, we provide preliminary evidence that chronic insomnia is remarkably reflected in the periphery through detailed metabolic assessments.
Methods
Serum from confirmed patients with insomnia and matched good sleepers (n = 15 per group) was sampled at high temporal resolution (every 2 hr over 48 hr). Food intake was controlled by providing hourly isocaloric snacks, and sleep architecture was assessed by overnight polysomnography. Quantitative metabolic assessments were conducted using nuclear magnetic resonance spectroscopy.
Results
Global metabolic profiles differentiated patients with insomnia from healthy controls, with elevated amino acid and energy metabolites and reduced branched-chain amino acid catabolic products. Strikingly, branched-chain amino acid catabolism was found to be specifically altered during the night with ~10 per cent increased accumulation of glucose in insomnia patients. Rhythmicity analysis revealed 11 metabolites that cycled diurnally across both groups, with phase advances noted for acetone and delays for lactate and branched-chain amino acids and their products.
Conclusions
These preliminary observations suggest that insomnia is associated with quantitative metabolic dysregulation and supports the hyperarousal hypothesis. Furthermore, we posit that these changes lead to a state of metabolic desynchrony in insomnia that is involved in the pathophysiology of the disorder and/or mediates its impact on health outcomes.
Clinical Trials Registration
Keywords: insomnia, metabolism, hyperarousal, NMR spectroscopy, metabolomics
Statement of Significance
Insomnia is associated with significant public health burden, but the biological mechanisms by which insomnia affects health are unknown. Here, we provide preliminary evidence that chronic insomnia is associated with altered metabolism compared with good sleepers, with specific alterations in rhythmicity of anabolic vs catabolic metabolite levels in blood. In particular, glucose metabolism was found to be elevated in patients with insomnia. These results suggest that insomnia is associated with systemic metabolic dysregulation potentially involved in the pathophysiology of the disorder and/or impact on health outcomes.
Introduction
Chronic insomnia is estimated to be one of the top 10 causes of neuropsychiatric disability in the world by the National Institutes of Mental Health [1]. Insomnia currently affects 10%–15% of the population [2] and is associated with a number of negative sequelae including daytime fatigue, cognitive difficulties, impaired emotional regulation, and decreased quality of life [3]. Despite the public health impact of insomnia, very little is known about the underlying pathophysiology of the disorder or the mechanisms through which insomnia affects physical and mental health.
The metabolome offers a promising target for identifying these mechanisms for several reasons. First, there have been several studies documenting elevated metabolic parameters in patients with insomnia compared with good sleepers [4–6] although these studies did not seek to identify particular metabolic processes driving these differences. Brain metabolism studies by magnetic resonance suggested altered brain energetics and cell membrane dysregulation [7]. Second, it is likely that insomnia is associated with peripheral effects on physiology given that experimental sleep deprivation has been shown to affect peripheral metabolism [8]. Finally, metabolic byproducts are exchanged with the bloodstream to potentially reflect brain-related metabolism.
Comprehensive metabolic profiling of patients with insomnia and differential features from healthy controls is rare, and possibly absent. However, there is a recent surge of comprehensive metabolic profiling to investigate the effects of experimental sleep restriction or deprivation [8]. We have used mass spectrometry–based global profiling of blood serum in rodents and humans to demonstrate the presence of cross species biomarkers post partial sleep restriction [7]. Similarly, Van den Berg et al. have reported that plasma acylcarnitines are significantly affected following one night of sleep restriction [9].
We have recently performed a meta-analysis of sleep and circadian metabolomics studies and have shown that existing mass spec studies are over-represented in nonpolar species [8]. As a result, the goal of this study was to conduct metabolite profiling on blood from patients with chronic insomnia and matched good sleepers with high temporal resolution (sampling every 2 hr over 48 hr) in order to identify a metabolic signature of polar metabolites to reveal insights into the pathophysiology of this common sleep disorder. Individuals were required to be free of comorbidities and medications in order to determine whether insomnia is associated with metabolic differences in a relatively “pure” sample with fewer potential confounding factors. Our results suggest that in spite of similar sleep architecture to normal participants, the metabolic profiles are perturbed both globally through the day and temporally through the night sleep period. Specifically, we see preliminary evidence of a distinct signature of altered energy metabolism throughout the day and nighttime changes in branched-chain amino acid (BCAA) metabolism. Furthermore, a number of metabolites were rhythmic; however, there are phase differences between insomniacs and controls which lead to the hypothesis of metabolic desynchrony. We thus propose a future platform for investigating metabolic disorder phenotypes of patients with chronic insomnia.
Methods
Participants
Participants consisted of a group of with chronic insomnia disorder (n = 15) and age- and sex-matched good sleepers (n = 15). All participants were between the ages of 25 and 50 and had a BMI ≤ 29. Recruitment was from the general community and involved placement of advertisements in online classified posting and local newspapers and other media. Both patients with insomnia and controls were recruited through the same methods. Participants with insomnia met the following Research Diagnostic Criteria for primary insomnia: subjective complaint of difficulty initiating or maintaining sleep, waking up too early or nonrestorative sleep; daytime consequences as a result of the poor sleep; duration of at least 1 month; and sleep disturbance was not secondary to a medical or psychiatric condition based on the effects of a substance, as determined by clinical history. To exclude individuals with mild insomnia, insomnia had to occur on three or more nights per week for 3 months or longer. A 30 min criterion was used such that participants had to report taking 30 min or longer to fall asleep and/or spend 30 min awake during the night. To be considered a good sleeper, participants had to report no lifetime history of significant insomnia symptoms. A 15 min criterion was used such that good sleepers had to report taking 15 min or less to fall asleep and spend 15 min or less awake during the night over the past month. Exclusion criteria for both groups were as follows: medical or psychiatric comorbidities that could affect sleep (e.g. major depression or chronic pain) as assessed by a history and physical and by structured clinical interview with the SCID-IV; sleep disorders other than insomnia as determined by clinical history and screening polysomnography; engaging in shiftwork within the past 6 months; current use of any prescription medications of over the counter products; and women who were pregnant or lactating in the preceding 6 months. These eligibility requirements were chosen in order to have a relatively “clean” insomnia sample given that comorbidities, medications, and other factors would likely influence metabolism and confound the results of the study. Although this limits the generalizability of results, a priority was placed on internal rather than external validity in order to determine whether there is a metabolic signal worth further investigation.
Interested participants meeting the basic criteria and who provided written informed consent for participation completed a screening visit, consisting of several self-reported questionnaires, a psychiatric interview (Structured Clinical Interview for DSM-IV) [10], a clinical sleep interview, and a physical examination, including height and weight. The Insomnia Severity Index (ISI) was used as a self-report estimate of sleep disturbance [11]. Individuals who met the inclusion criteria completed a 48 hr inpatient stay. Upon arrival, participants had an indwelling intravenous catheter placed and blood samples were then taken every 2 hr, for a total of 25 blood samples per participant. In order to maintain consistent caloric intake, participants ate small, isocaloric snacks every hour they were awake. Meals were prepared based on the participant’s height, weight, and body mass index. Participants were allowed to sleep each night according to their habitual sleep schedule. On one night, sleep was measured using overnight polysomnography.
Polysomnography
Standard polysomnographic (PSG) procedures were used to record the EEG, EOG, EMG, and EKG using an ambulatory system. Participants went to bed at their habitual bedtime. Electrode placements of FpZ, CZ, and OZ were used according to the International 10/20 system. Two EOG electrodes were placed, positioned 1 cm below and lateral to the outer canthus of the left eye and 1 cm above and lateral to the outer canthus of the right eye. Two surface EMG electrodes were taped onto the chin 2 cm apart. Additional leads were used to measure leg movements and breathing in order to rule out the presence of occult sleep disorders. Two electrodes were taped over the anterior tibialis muscle of each leg to detect leg movements during the night. Flexible Resp-EZ belts were placed around the abdomen and chest to measure breathing-related movements during the night. A nasal cannula was used to detect pressure and an oximeter probe placed on the finger to measure blood oxygen saturation. The criteria for defining sleep disorders were an apnea–hypopnea index greater than 15 events per hour for sleep apnea and a periodic limb movement index greater than 15 events per hour for periodic limb movements in sleep. Records were manually scored in 30 s epochs according to standard criteria. All records were scored by the same registered polysomnographic technician. PSG data were used to compute standard sleep architecture variables of the amount of each stage of sleep in terms of minutes and percentage of total sleep time. In addition, the following sleep continuity variables were computed: sleep latency (SL; time from lights out to the first epoch of any sleep stage), total sleep time (TST), wake after sleep onset (WASO; number of minutes spent awake between lights out and lights on), and sleep efficiency (SE; total sleep time divided by the total recording period).
Metabolomic assays
Metabolomics analysis of serum samples was carried out using nuclear magnetic resonance (NMR) spectroscopy as described previously [12]. This approach allows for rapid, unbiased, and quantitative metabolic profiles (fingerprints) to be acquired. The stability and reproducibility of the NMR assay allow for quantitative comparisons across the large number of samples analyzed in this study. Samples for NMR analysis were stored at −80°C, and subsequently thawed on ice with 250 μL serum removed and filtered using ultracentrifugation and buffered to pH 7.0 for analysis. One-dimensional proton NMR spectra were acquired using standard methods (NOESY pulse sequence) on a 700 MHz Bruker instrument equipped with a SampleJet sample changer. Samples from five participant pairs were acquired in analytical triplicates, whereas the samples from the remaining 10 pairs were acquired in analytical singlet, resulting in 1246 total spectral recordings. Additional two-dimensional NMR experiments were performed for the purpose of confirming chemical shift assignments, including homonuclear total correlation spectroscopy (2D 1H-1H TOCSY) and heteronuclear single quantum coherence spectroscopy (2D 1H-13C HSQC), using standard Bruker pulse programs. Raw NMR data were processed by spectral fitting using the targeted profiling method [12] by Chenomx Inc. to obtain quantitative metabolite information for all nighttime samples.
Data analysis
Individuals with insomnia and good sleeper controls were compared on demographic and both self-report and PSG sleep variables using paired sampled t-tests in order to account for the matched pair design based on age and sex matching.
Spectral preprocessing
All spectra were binned into 0.005 ppm bins throughout the spectral width. DSS, water, and urea signal regions were excluded from the binning process. The binned spectral intensity was normalized to total spectral integral of individual spectrum to generate the working data matrix. The data were mean centered followed by unit variance scaling and used for further multivariate analysis.
Multivariate data analysis
All multivariate data analyses were performed in Simca-P 14.0 (Umetrics AB, Umea, Sweden). Initially, principal component analysis (PCA), an unsupervised method, was used to obtain a global overview of the data from 1246 recorded spectra. PCA is also a useful tool to identify outlier samples in a multivariate data structure [13]. From the total data set of 1246 spectra, 59 outlier spectra were excluded (based on the 95% confidence interval) due to poor water suppression/baseline correction issues. As a result, 1187 spectra for supervised multivariate analysis remained. These samples were used for clustering using orthogonal partial least square—discriminant analysis (OPLS–DA). OPLS–DA is a supervised technique which allows for separation of between-class and within-class variability [13, 14]. OPLS–DA was performed in two sets. In set 1, the analytical triplicate samples were used and in set 2, the singlet samples were used. For each model, significant bins were selected by variable importance on projection (VIP) > 1.0; overlapped and direction conserved bins from set 1 and set 2 were considered for further analysis. The model fit was judged based on the cross-validation parameter Q2 and CV-ANOVA p-value. The cross-validation process employs an internal sevenfold validation. Briefly, 1/7th of the total samples are left out from the dataset and the model is fitted with the remaining samples. This model is then validated using the samples left out, thereby computing the Q2 as a measure of the predictive ability of the model. This process is repeated seven times such that each sample is used for prediction.
Initially, the bins representing spectral noise regions were excluded. Adjacent bins with opposing direction (a feature of spectral overlap) were also excluded. Spectral regions were assigned from the remaining bins. These regions were further investigated using Chenomx v 8.0 (Edmonton, Alberta, Canada) for metabolite assignment. The metabolites were assigned by targeted fitting of representative spectrum.
Targeted spectral profiling
In order to obtain a more quantitative temporal picture, nighttime samples (11 pm—7 am) from all participants were subjected to targeted spectral profiling using Chenomx v 8.0 to identify the concentration of the metabolites. Metabolites that were judged significant using multivariate analysis were profiled. Briefly, the processed spectral peaks were fitted using a pre-built metabolite library so that the residual signals could be minimized. A detailed description of this method can be found in the work of Weljie et al. [12].
Time course analysis by significance analysis on microarrays
Time-course analysis of nighttime metabolites was performed by significance analysis on microarrays (SAM) [15] using MeV 4.6. A slope-based method was employed. Briefly, SAM computes the signed area of two group time series data relative to one of the groups (baseline group) and a SAM score based on the signed areas. A positive SAM score would mean that the signed area is larger in group 2 (insomnia) and a negative score would mean the opposite.
The study was approved by the Institutional Review Board of the University of Pennsylvania and is listed on clinicaltrials.gov (NCT01957111).
Results
Recruitment of participants
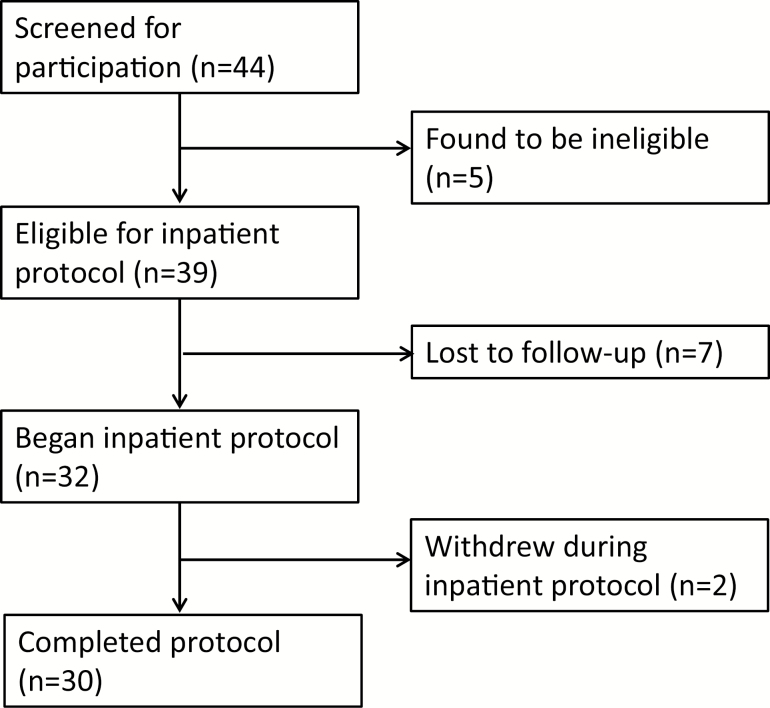
Forty-four people were screened for participation: five individuals were considered screen fails (two had BMIs over the cutoff, one medical concern precluding participation, one not meeting threshold for insomnia symptoms but not a good sleeper, one smoker), seven individuals were lost to follow-up before completing the hospital stay, and two individuals did not complete the full study protocol (see STROBE diagram in Figure 1). A total of 15 individuals with insomnia and 15 age- (± 5 years) and sex-matched good sleeper controls completed the study. The sample was largely non-Hispanic (87%) and identified as white (63%), with a smaller portion of participants identifying as Black/African American (20%) and Asian (10%), and 10 per cent preferred not to disclose their race. Each group had 10 females and 5 males; the insomnia group’s mean (SD) age was 37 (7.88) and the good sleeper group’s mean (SD) age was 35.93 (7.54). The insomnia group’s mean (SD) ISI was 15.14 (4.72) and the good sleeper group’s mean (SD) ISI was 1.73 (2.74). The only statistically significant group difference in PSG-assessed sleep was in the percentage of the night spent in REM, with the insomnia group having a slightly higher proportion compared with good sleepers (21.5% vs. 19.7%) (see Supplementary Table S1).
Figure 1.
STROBE diagram showing the flow of participants through the study.
Correlation of sleep parameters with metabolic profiles of insomnia and good sleepers
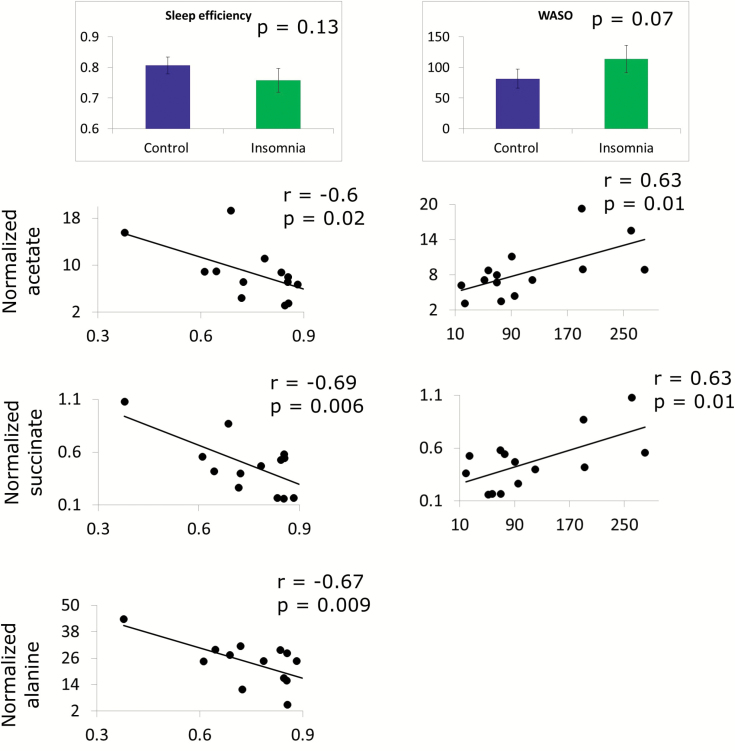
Among the PSG parameters, only sleep efficiency and WASO demonstrated statistically significant association with metabolic parameters. There were no significant group differences on the variables (p = 0.13 and 0.07 for sleep efficiency and WASO, respectively, Figure 2), but they were used as quantitative measures of insomnia severity. In order to understand if there is a quantitative relationship between these two parameters and peripheral metabolism, regression analysis was performed on 49 metabolites measured from first morning serum samples (7 am, post-PSG night) using quantitative NMR [12]. Each metabolite was compared with WASO and sleep efficiency using Pearson correlation separately for patients with insomnia and good sleepers (Figure 2). Significant correlations of acetate and succinate (negative with sleep efficiency, positive with WASO) and alanine (negative with sleep efficiency) were observed (Figure 2). Most of these correlations were weak/absent (|r| < 0.4, p > 0.1) in the good sleepers. Only acetate level of good sleepers was moderately correlated to sleep efficiency (r = 0.49, p = 0.09, data not shown); however, unlike insomnia group, the correlation was positive. Such observations suggest that the metabolic profile of patients with insomnia is closely related to the severity of objective sleep disturbance.
Figure 2.
Correlation of insomnia-specific parameters with level of metabolites at 7 am in the morning. Only sleep efficiency and WASO showed some trend towards significance. The normalized levels of acetate, succinate, and alanine were significantly correlated with sleep efficiency and those of acetate and succinate were significantly correlated with WASO.
Global metabolic profile differentiates patients with insomnia from healthy controls
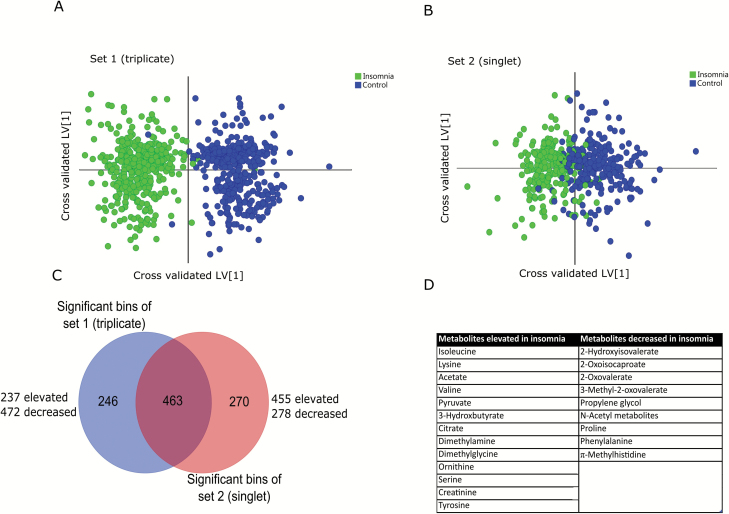
Our previous metabolomics work has demonstrated that significant variation can exist between measured batches of samples [7], and thus, the sample population was a priori divided into two subsets based on a matched-subjects design. The first subset was strategically designed with five patients with insomnia and five age- and sex-matched controls and metabolomic samples measured in analytical triplicate to assess analytical variance (total of 15 spectra/timepoint/group; 750 spectra total). In this subset—dubbed the triplicate subset—each objective time of day sampled was represented by six spectra per participant (i.e. three replicates each on days 1 and 2 at 8 am, etc.). The second subset—termed the singlet subset—consisted of the remaining 10 patients with insomnia and 10 matched controls measured across all time points over 48 hr (10 spectra/time point/group; 500 spectra total). Therefore, each objective time of day sampled was replicated across the 48 hr cycle. Outliers were detected using PCA as indicated before. OPLS–DA modeling was performed independently on the two subsets, resulting in clustering of insomnia and control samples from both (Figure 3A and B). The models were highly significant (Q2 = 0.84, CV-ANOVA p < 0.0001 for the triplicate subset and Q2 = 0.58, CV-ANOVA p < 0.0001 for the singlet subset, respectively). Significant bins from both models were selected based on variable importance on projection (VIP > 1.0) and they were overlapped across the two subsets (Figure 3C).
Figure 3.
Multivariate OPLS-DA analysis of insomnia and control samples over all collection time points. Cross-validated OPLS-DA scores plot showed significant clustering of insomnia and control samples over both triplicate (A) and singlet (B) sets. The bins were selected by overlap analysis (C) and metabolites were assigned by spectral profiling (D).
The overlapped bins were further pruned as detailed in Methods and the metabolites were assigned from the bins using Chenomx suite v 8.0 and 2-dimensional HSQC and TOCSY experiments. Metabolites significantly perturbed are listed in Figure 3. In aggregate, 13 metabolites were elevated and 9 metabolites were decreased in patients with insomnia across all times of day (Figure 3D). Loadings and VIP values are provided in Supplementary Table S2.
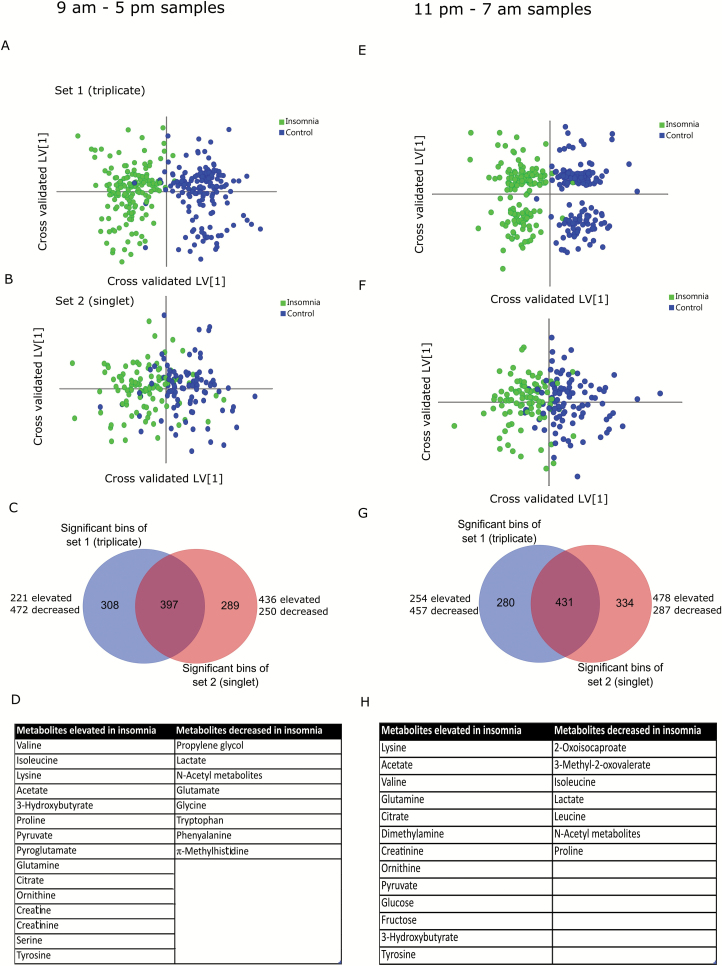
Time-of-day variations in metabolic profiles
To understand whether the overall metabolic profiles of patients with insomnia and good sleeper controls followed a diurnal pattern, the data were divided into discrete daytime (9 am–5 pm) or nighttime (11 pm–7 am) blocks, with the remaining time points excluded. OPLS-DA analysis indicated that insomnia and control individuals were strongly segregated during the daytime (Figure 4A and B, Q2 = 0.79, CV-ANOVA p < 0.0001 for triplicate and Q2 = 0.50, CV-ANOVA p < 0.0001 for singlet cohorts). Metabolites were assigned as before from the overlapped bins of two subsets (Figure 4C) and are listed in Figure 4D. During the daytime, patients with insomnia showed elevated levels of 15 metabolites and decreased levels of 8 metabolites (Figure 4D) compared with good sleepers. These metabolites were used for pathway analysis using the metaboanalyst server (Metabanalyst v3.0). Significantly altered pathways (FDR < 0.05, pathway impact > 0) included arginine and proline metabolism, pyruvate metabolism, glycine, serine and threonine metabolism, and glycolysis/gluconeogenesis (Supplementary Figure S3). Similar analysis on the nighttime samples revealed significant clustering of the insomnia and control samples (Figure 4E and F, Q2 = 0.77, CV-ANOVA p < 0.0001 for triplicates and Q2 = 0.41, CV-ANOVA p < 0.0001 for singlets). Overlapped bins (Figure 4G) were assigned to metabolites (Figure 4H). During nighttime hours, patients with insomnia has elevated levels of 13 metabolites and decreased levels of 7 metabolites compared with good sleepers (Figure 4H). Pathway analysis using metaboanalyst server revealed significant alteration in BCAA metabolism, arginine and proline metabolism, pyruvate metabolism, and glycolysis/gluconeogenesis (Supplementary Figure S3). Interestingly, the BCAA metabolic pathway was specifically different in nighttime samples. Pyroglutamate, creatine, and serine were specifically increased during daytime in patients with insomnia, whereas dimethylamine, citrulline, glucose, and fructose were elevated at night. On the other hand, propylene glycol, glutamate, glycine, tryptophan, phenylalanine, and methylhistidine were decreased during daytime, and 2-oxoisocaproate and leucine were decreased during night. Isoleucine and proline were elevated during day and decreased during the nighttime in patients with insomnia. Loadings and VIP values from all the models described above are listed in Supplementary Table S2.
Figure 4.
Multivariate OPLS-DA analysis of insomnia and control samples over day (9 am–5 pm, A–D) and night (11 pm–7 am, E–H) samples. Cross-validated OPLS-DA scores plot showed significant clustering of insomnia and control samples over both triplicate (A/E) and singlet (B/D) sets. The bins were selected by overlap analysis (C/G) and metabolites were assigned by spectral profiling (D/H).
Time-dependent changes in metabolites significantly altered during nighttime
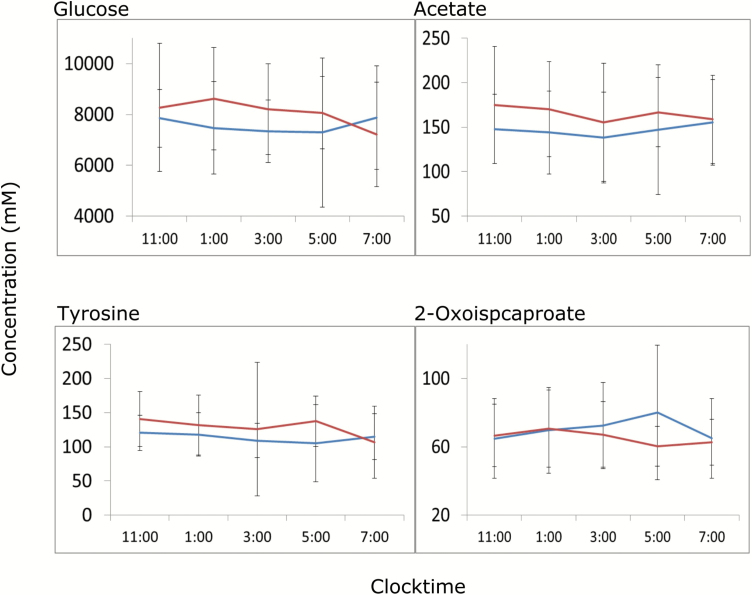
Metabolites that were differentially altered during the nighttime (11 pm–7 am) were further subjected to time course analysis using SAM. Individual metabolites were profiled from NMR spectra and time course samples from each individual were subjected to time course analysis. Only the second night was used to reduce possible acclimation effects. The analysis suggests a significant average elevation of glucose over the second night along with acetate and tyrosine, whereas leucine metabolite 2-oxoisocaproate decreased overnight in patients with insomnia (Figure 5). Separate AUC analysis on the metabolites suggested a 10%–12% elevation in overnight glucose in the patients with insomnia along with a 13 per cent elevation in acetate and 10 per cent depletion in 2-oxoisocaproate (Supplementary Table S4).
Figure 5.
Time series analysis of metabolites. Only the second night data are shown here. The samples were analyzed by two-group SAM for significantly different temporal trend of metabolites in insomnia (red) and control (blue) population. Metabolites with significant differences across control and insomnia population are presented.
Metabolic desynchrony between patients with insomnia and healthy controls
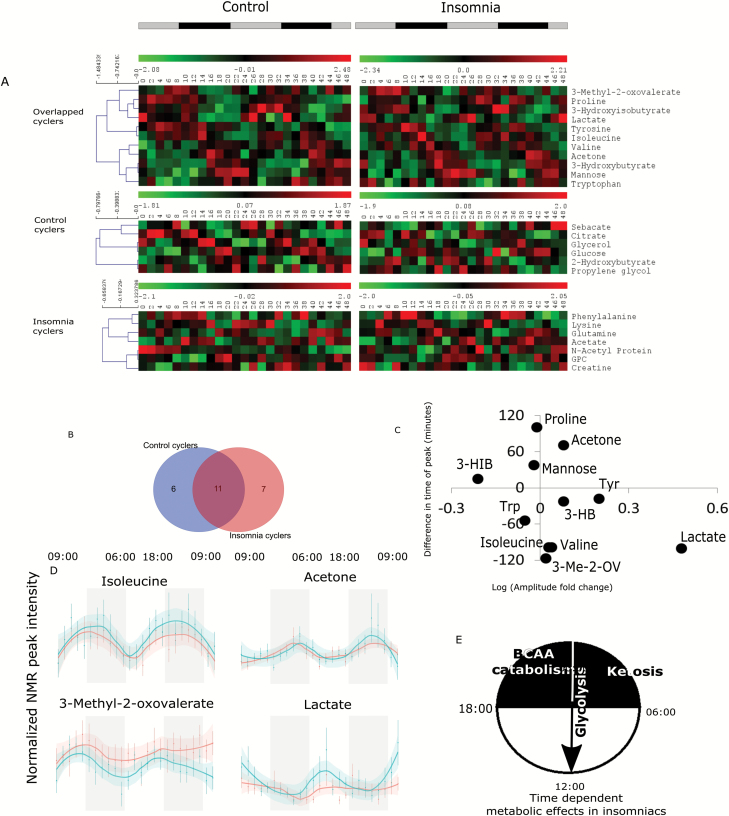
Recent studies have demonstrated that the metabolome and lipidome reflect diurnal rhythms in blood, urine, and saliva [16–18]. To understand how insomnia affects metabolic oscillations, all data from patients with insomnia and controls were independently subjected to JTK Cycle analysis for 24 hr periodic oscillation using Metacycle 2D [19]. A total of 17 metabolites (Figure 6A and B) were found to be oscillating in controls (p-value < 0.05, BH.Q < 0.2 for respective bins) (Figure 6A), whereas 18 metabolites were oscillating in patients with insomnia (Figure 6A and B). Eleven metabolites were found to be cycling in both groups (Figure 6A and B). Notably, distinct differences in oscillatory amplitude and phase were noted between insomnia and control groups for the common cyclers (Figure 6C). Specifically, lactate amplitude was increased in patients with insomnia with a concomitant decrease in phase. Valine, isoleucine, and 3-methyl-2-oxovalerate phases were earlier, whereas those of acetone and proline were later. 3-Hydroxyisobutyrate demonstrated suppressed amplitude. Control individual profiles revealed 6 metabolites that cycled only in this group, whereas 7 unique cyclers in participants with insomnia were observed (Figure 6A). Supplementary Figure S5 shows 48 hr timecourse plots of the metabolites demonstrating oscillations in both groups. Group differences in diurnal rhythms could be due to the insomnia group have delayed or advanced circadian phases relative to the good sleepers. Circadian markers were not assessed; however, there was no evidence of significant differences in habitual sleep/wake times between groups (mean [SD] bedtime was 11:14 [0:58] for the insomnia group and 10:55 [1:06] for the good sleepers).
Figure 6.
Analysis of metabolite rhythm in insomnia and control population using Metacycle 2D. Insomnia and control population was subjected to metabolite rhythm analysis separately. Control population showed 6 and insomnia population showed 7 unique cyclers, whereas 11 cyclers were common (A and B). Among the common cycling metabolites, many showed differences in fold change and amplitude (C). Representative time course plots are shown for four common metabolites (D, insomnia—blue, control—red). A proposed general metabolic state shift of insomniacs is shown over the diurnal day based on the time series analysis (E). 3-Me-2-OV = 3-methyl-2-oxovalerate; 3-HIB = 3-hydroxyisobutyrate; 3-HB = 3-hydroxybutyrate; Trp = tryptophan.
Discussion
Peripheral metabolic profiles of insomniacs support hyperarousal
This study is among the first to utilize comprehensive peripheral metabolic profiling to compare patients with insomnia and matched good sleeper controls. We used time course blood sampling of patients with insomnia and healthy controls and employed two subsets from the sample population in order to differentiate true biological and analytical variation. Using this design, we found preliminary evidence of clear metabolic differences in patients with insomnia, with divergent patterns exhibited during the day and night.
Insomnia was defined for this study as meeting diagnostic criteria for DSMIV Insomnia Disorder based on clinical interview by an experienced sleep disorders clinician (P.G.), but there was no requirement for a minimum severity on a quantitative measure such as the ISI. Although there were significant differences on the ISI between the insomnia and good sleeper groups, the mean for the insomnia group was only slightly above the standard cutoff of greater than 14 for moderate severity of insomnia [11]. The overall severity of insomnia for this sample was therefore not severe, which is supported by the lack of significant PSG differences between groups. This makes the results all the more striking and suggests that even milder insomnia is associated with robust metabolic effects in the periphery. Vgontzas and colleagues have made a compelling case that insomnia can be subdivided into cases of “objective insomnia,” in which there is PSG evidence of sleep duration less than 6 hr, and “subjective insomnia,” in which sleep duration is in a more normal range [20]. Objective insomnia is associated with greater biological severity across a range of measures and is thought to be a more severe phenotype. We would expect, had our sample had greater evidence of sleep disturbance on PSG, that the metabolic effects would be even larger. This also suggests that the metabolic differences between patients with insomnia and good sleepers are due to some aspect of insomnia that is distinct from objective sleep disturbance. For example, patients with insomnia have been found to have dysregulation of the HPA axis [21], and it may be that abnormal cortisol activity is partially responsible for these findings. This intriguing finding will need to be examined in future studies that seek to disentangle the effects of sleep disturbance compared with other biological aspects of insomnia.
Given the variability in insomnia severity in the sample, the association between metabolite profiles and quantitative measures of sleep disturbance was examined using PSG-defined sleep variables. Greater severity of sleep disturbance was associated with specific metabolites (acetate, alanine, and succinate), but only in the insomnia group, supporting the hypothesis that metabolic effects would be stronger in a more severe insomnia sample. Among these metabolites, nighttime acetate was also elevated in insomniacs, suggesting altered energy metabolism (Figures 4 and 5), potentially via elevated lipid breakdown.
Harper et al. have used 31P magnetic resonance spectroscopy (MRS) to show that phosophcreatine and phosphocholine levels in grey and white matter are differentially regulated in insomniacs with PSG-dependent differences, in support of the hyperarousal hypothesis, and increased energy demand in the brain [4, 5]. Our results in the periphery are consistent with this hypothesis, although it is not clear what might be driving this arousal. Changes in peripheral metabolites may cause central arousal, or vice versa.
Central energy metabolism is desynchronized in insomniacs
A significant strength of this study was the use of 48 hr blood sampling so that group comparisons could be made both globally and at particular times of day, as well as decreasing reliance on single time-point assessments that are subject to a number of confounding factors. Specifically, central carbon pathway (glycolysis/gluconeogenesis) metabolites were perturbed irrespective of time of day (Figures 3 and 4), pointing to a connection between insomnia and changes in energy metabolism. Indeed, decreased levels of lactate across the night and day along with increased glucose at night and pyruvate during the day (Figures 4 and 5) suggest specific, and chronic, changes in glycolysis/gluconeogenesis. Nighttime buildup of glucose in patients with insomnia compared with good sleepers (Figure 5) indicates that there is a decrease in nighttime glucose utilization. We should point out that such build-up/decay is not entirely free from diet-related effects. To that end, however, our data suggest that patients with insomnia and good sleepers may handle nutrient resources via significantly different modes. Metabolic oscillation analysis revealed that lactate levels began to increase at midnight and peaked around midday in insomniacs (Figure 6). Hyperarousal and hypermetabolism are hallmarks of insomnia and may be reflected by elevated nighttime glycolysis [23]; on the other hand, this suggests that there is an elevated bedtime catabolic activity. In general, sleep is considered to be important for anabolic processes [24], which seems to be affected by insomnia. However, irrespective of glycolytic activity, nighttime glucose build-up (Supplementary Table S4) in insomnia raises the possibility of prediabetic phenotypes. Epidemiological studies have found associations between chronic insomnia with incident diabetes [25]. These results are also reminiscent of circadian alignment studies which demonstrate that elevated glucose results from both time-of-day and circadian misalignment effects, affecting glucose tolerance [26].
Branched-chain amino acid catabolism is phase advanced in insomniacs
Recent research has unraveled potential underlying molecular mechanism of diabetes in addition to classic parameters such as blood glucose. For example, BCAAs have been implicated in development of diabetes and obesity [27, 28]. Interestingly, BCAA metabolism was perturbed only during the nighttime (Supplementary Figure S5). Specifically, branched-chain oxo-acids were decreased during nighttime in patients with insomnia (Figure 4H). Moreover, patients with insomnia also showed overall depletion of in leucine catabolic product 2-oxoisocaproate (Figure 5, Supplementary Table S4) during the second night compared with controls and an almost 1.5–2 hr peak offset of BCAAs and related metabolite 3-methyl-2-oxovalerate in oscillatory analysis (Figure 6). Together, this implies elevated and advanced nighttime BCAA catabolism can potentially hamper glucose oxidation leading to nighttime accumulation of glucose [21]. Therefore, bedtime metabolic activity is shifted towards catabolism in insomnia. Oishi et al. have created a mouse model of chronic sleep disturbance by exposure to psychophysiological stress that leads to increased sleep fragmentation and reduced circadian amplitude [29]. Animals with chronic sleep disturbance, compared with controls, had elevated levels of BCAAs in plasma at night, similar to what we observe in day time (Figure 4). Lim and colleagues examined the influence of dietary BCAA supplementation on sleep or wake disturbance in a mouse model of traumatic brain injury in which there was an impairment in both the ability to maintain sleep at night and wakefulness during the day, leading to greater state fragmentation [30]. BCAA supplementation led to reduced fragmentation and hence better sleep at night in part through improvement in the function of the orexin system, which is a critical component of the sleep/wake regulatory system [31]. Furthermore, it has been argued that BCAAs can lead to daytime fatigue because of their capacity to reduce central nervous system uptake of tryptophan and affect serotonin levels [22]. Interestingly, our data also suggest decreased daytime phenylalanine and tryptophan in the patients with insomnia (Figure 4). Given that serotonin plays a significant role in sleep/wake regulation, a similar mechanism at night could impair the ability initiate and maintain sleep. These disparate results suggest that BCAA metabolism has multiple interrelationships with the sleep/wake system and that insomnia may be associated with BCAA dysregulation. The role of BCAAs in the progression of glucose intolerance is also well documented [28]. Broadly, BCAA catabolism seems to be elevated and enter the tricarboxylic acid (TCA) cycle barring the glucose carbon, resulting in elevated glucose and glucose intolerance. We showed that altered nighttime BCAA catabolism and relative elevation in glucose concentration may be initial signs of glucose intolerance. Interestingly, acetate and succinate—the dethiolated forms of two entry points of BCAAs into TCA cycle—are positively correlated with WASO and sleep efficiency in patients with insomnia, indicating a phenotypic connection of molecular events. The hypothesis of potential circadian desynchrony in the insomnia group needs to take into consideration the fact that this study was not designed as a rigorous circadian protocol. No circadian phase markers were assessed, and there was no control of lighting levels or activity in the laboratory.
Biomarkers of insomnia compared with biomarkers of sleep deprivation/restriction
Insomnia is a chronic clinical disorder and is not directly equivalent to any acute experimental protocols such as recent reports describing metabolic effects of experimental sleep deprivation (SD)/restriction in healthy population [8], primarily because most human SD experiments are performed on otherwise healthy individuals who do not have any chronic conditions. Although SD experiments may be used to gauge acute effects of sleep disturbance, chronic insomnia models are not available. In spite of this important difference, some biomarker similarities are striking and may provide insight into common pathophysiological mechanisms for exploration. The most striking similarity is the elevation of glucose along with creatine post SD observed by Bell et al. [32]. Tryptophan and phenyalanine were found to be significantly altered in other studies [8]; however, the directionality does not match with our data. On the other hand, Davies et al. found that the rhythm of isoleucine remains conserved under regular and perturbed sleep [18]. Indeed, isoleucine was one of the conserved metabolites in our study. This pattern of conserved BCAA rhythm and elevated glucose post SD in multiple studies suggest that sleep loss may be generally related to altered glucose and BCAA metabolism.
A limitation of this study is the strict eligibility requirements, which required participants to be free of comorbidities and medications. This produced a relatively “clean” insomnia sample that may not be representative of the broader population of patients with insomnia. Although this limits the generalizability of results, a priority was placed on internal rather than external validity given that these other factors would likely also influence metabolism and act as confounder. For this initial foray into the metabolomics of insomnia, the goal was to determine whether there is a meaningful metabolic worth pursuing in future samples that are more representative.
Clinical implications of the study
Currently, there are no objective biomarkers of insomnia in clinical use and the availability of such a tool would be of tremendous value to sleep medicine. These data provide a proof of principle that metabolic profiling can be used to identify a biological “signature” of insomnia. This biomarker signature will need to be validated in future studies, especially given the relatively small sample size reported here. A validated signature would have potential use as a diagnostic tool for insomnia and for monitoring of treatment outcome. Recently, the work of Irwin and colleagues has shown that insomnia is associated with inflammatory biomarkers of disease risk and that these indices improve following nonpharmacologic treatment [22, 33]. The addition of metabolic profiling adds to the biomarkers identified in these studies and may lead to more comprehensive biomarker panels that could be integrated into routine clinical care. Second, the identification of metabolic processes that are dysregulated in insomnia may shed light on the pathophysiology of the disorder and the mechanisms through which it negatively affects physical and mental health, both of which are not understood. This study was not designed to assess whether the metabolic effects were a cause or a consequence of insomnia but future work in this area can begin to delineate the direction of causation and may lead to novel approaches to prevention and treatment of insomnia. Finally, the availability of biomarkers of insomnia would facilitate the identification of subtypes. It is widely assumed that insomnia can result from myriad biological processes, much as fever is a common endpoint of many mechanisms. Efforts to delineate diagnostic subtypes of insomnia-based self-report and clinical measures have failed to demonstrate sufficient reliability for clinical use [34]. Ultimately, a thorough understanding of insomnia will require a better understanding of the different pathways to insomnia so that prevention and treatment can be tailored to the unique constellation of factors present for each patient.
Supplementary Material
Supplementary material is available at SLEEP online.
Funding
Supported in part by a research grant from the Investigator-Initiated Studies Program of Merck, Sharp & Dohme Corp. The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000003. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
Notes
Conflict of interest statement. The authors have no competing financial or nonfinancial interests related to the work presented in this manuscript.
References
- 1. Organization WH. The Global Burden of Disease: 2004 Update. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 2. Roth T, et al. Prevalence and perceived health associated with insomnia based on DSM-IV-TR; International Statistical Classification of Diseases and Related Health Problems, Tenth Revision; and Research Diagnostic Criteria/International Classification of Sleep Disorders, Second Edition criteria: results from the America Insomnia Survey. Biol psychiatry. 2011;69(6):592–600. [DOI] [PubMed] [Google Scholar]
- 3. Benca RM. Consequences of insomnia and its therapies. J Clin Psychiatry. 2001;62Suppl 10:33–38. [PubMed] [Google Scholar]
- 4. Harper DG, et al. Energetic and cell membrane metabolic products in patients with primary insomnia: a 31-phosphorus magnetic resonance spectroscopy study at 4 tesla. Sleep. 2013;36(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonnet MH, et al. 24-Hour metabolic rate in insomniacs and matched normal sleepers. Sleep. 1995;18(7):581–588. [DOI] [PubMed] [Google Scholar]
- 6. Bonnet MH, et al. Physiological activation in patients with Sleep State Misperception. Psychosom Med. 1997;59(5):533–540. [DOI] [PubMed] [Google Scholar]
- 7. Weljie AM, et al. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112(8):2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhoades SD, et al. Time is ripe: maturation of metabolomics in chronobiology. Curr Opin Biotechnol. 2017;43:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van den Berg R, et al. A single night of sleep curtailment increases plasma acylcarnitines: novel insights in the relationship between sleep and insulin resistance. Arch Biochem Biophys. 2016;589:145–151. [DOI] [PubMed] [Google Scholar]
- 10. First M, et al. Structured Clinical Interview for Axis I DSM-IV-TR Disorders. New York: New York State Psychiatric Institute; 2001. [Google Scholar]
- 11. Bastien CH, et al. Validation of the Insomnia Severity Index as an outcome measure for insomnia research. Sleep Med. 2001;2(4):297–307. [DOI] [PubMed] [Google Scholar]
- 12. Weljie AM, et al. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78(13):4430–4442. [DOI] [PubMed] [Google Scholar]
- 13. Madsen R, et al. Chemometrics in metabolomics–a review in human disease diagnosis. Anal Chim Acta. 2010;659(1-2):23–33. [DOI] [PubMed] [Google Scholar]
- 14. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemom. 2002;16(3):119–128. [Google Scholar]
- 15. Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dallmann R, et al. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109(7):2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chua EC, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2013;110(35):14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davies SK, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111(29):10761–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu G, et al. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32(21):3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vgontzas AN, et al. Insomnia with objective short sleep duration: the most biologically severe phenotype of the disorder. Sleep Med Rev. 2013;17(4):241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vgontzas AN, et al. Sleep deprivation effects on the activity of the hypothalamic-pituitary-adrenal and growth axes: potential clinical implications. Clin Endocrinol (Oxf). 1999;51(2):205–215. [DOI] [PubMed] [Google Scholar]
- 22. Carroll JE, et al. Improved sleep quality in older adults with insomnia reduces biomarkers of disease risk: pilot results from a randomized controlled comparative efficacy trial. Psychoneuroendocrinology. 2015;55:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nofzinger EA, et al. Functional neuroimaging evidence for hyperarousal in insomnia. Am J Psychiatry. 2004;161(11):2126–2128. [DOI] [PubMed] [Google Scholar]
- 24. Borbély AA, et al. The two-process model of sleep regulation: a reappraisal. J Sleep Res. 2016;25(2):131–143. [DOI] [PubMed] [Google Scholar]
- 25. Vgontzas AN, et al. Insomnia with objective short sleep duration is associated with type 2 diabetes: a population-based study. Diabetes Care. 2009;32(11):1980–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morris CJ, et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci U S A. 2015;112(17):E2225–E223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. White PJ, et al. Branched-chain amino acid restriction in Zucker-fatty rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Mol Metab. 2016;5(7):538–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oishi K, et al. Disruption of behavioral circadian rhythms induced by psychophysiological stress affects plasma free amino acid profiles without affecting peripheral clock gene expression in mice. Biochem Biophys Res Commun. 2014;450(1):880–884. [DOI] [PubMed] [Google Scholar]
- 30. Lim MM, et al. Dietary therapy mitigates persistent wake deficits caused by mild traumatic brain injury. Sci Transl Med. 2013;5(215):215ra173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brisbare-Roch C, et al. Promotion of sleep by targeting the orexin system in rats, dogs and humans. Nat Med. 2007;13(2):150–155. [DOI] [PubMed] [Google Scholar]
- 32. Bell LN, et al. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Irwin MR, et al. Tai chi, cellular inflammation, and transcriptome dynamics in breast cancer survivors with insomnia: a randomized controlled trial. J Natl Cancer Inst Monogr. 2014;2014(50):295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edinger JD, et al. Testing the reliability and validity of DSM-IV-TR and ICSD-2 insomnia diagnoses. Results of a multitrait-multimethod analysis. Arch Gen Psychiatry. 2011;68(10):992–1002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.