Abstract
In recent years, effective treatment of infections caused by Acinetobacter baumannii has become challenging due to the ability of the bacterium to acquire or up-regulate antimicrobial resistance determinants. Two component signal transduction systems are known to regulate expression of virulence factors including multidrug efflux pumps. Here, we investigated the role of the AdeRS two component signal transduction system in regulating the AdeAB efflux system, determined whether AdeA and/or AdeB can individually confer antimicrobial resistance, and explored the interplay between pentamidine resistance and growth conditions in A. baumannii ATCC 17978. Results identified that deletion of adeRS affected resistance towards chlorhexidine and 4’,6-diamidino-2-phenylindole dihydrochloride, two previously defined AdeABC substrates, and also identified an 8-fold decrease in resistance to pentamidine. Examination of ΔadeA, ΔadeB and ΔadeAB cells augmented results seen for ΔadeRS and identified a set of dicationic AdeAB substrates. RNA-sequencing of ΔadeRS revealed transcription of 290 genes were ≥2-fold altered compared to the wildtype. Pentamidine shock significantly increased adeA expression in the wildtype, but decreased it in ΔadeRS, implying that AdeRS activates adeAB transcription in ATCC 17978. Investigation under multiple growth conditions, including the use of Biolog phenotypic microarrays, revealed resistance to pentamidine in ATCC 17978 and mutants could be altered by bioavailability of iron or utilization of different carbon sources. In conclusion, the results of this study provide evidence that AdeAB in ATCC 17978 can confer intrinsic resistance to a subset of dicationic compounds and in particular, resistance to pentamidine can be significantly altered depending on the growth conditions.
Introduction
Acinetobacter baumannii causes a range of disease states including hospital-acquired pneumonia, blood stream, urinary, wound and bone infections, and is responsible for epidemic outbreaks of infection worldwide [1]. Such infections are often very difficult to treat due to the multidrug resistant (MDR) character of isolates displayed by this organism [2, 3]. In addition to the impressive propensity of the organism to acquire genetic elements carrying resistance determinants [2, 4, 5], up-regulation resulting in overproduction of resistance nodulation cell-division (RND) drug efflux systems through integration of insertion sequence elements or mutations in regulatory genes, has also been deemed a major contributor to the MDR phenotype [6–9]. The best studied RND efflux systems in A. baumannii include AdeABC [10], AdeFGH [11] and AdeIJK [12]. Of particular interest is the AdeABC system which affords resistance to diverse antibiotics, biocides and dyes [10, 13–15], and has gained attention due to its high incidence of over-expression across many MDR A. baumannii clinical isolates, primarily from incorporation of point mutations in the genes encoding its positive regulator, AdeRS [6, 8, 13, 16]. Typically RND pumps consist of three proteins that form a complex; the absence of any of these components renders the entire complex non-functional [17]. Interestingly, deletion of adeC in the A. baumannii strain BM4454 did not affect resistance towards two substrates of the pump suggesting that AdeAB can utilize an alternative outer membrane protein (OMP) to efflux antimicrobial compounds [13].
The genetic arrangement of the AdeABC system places adeABC in an operon that is divergently transcribed to the regulatory adeRS two component signal transduction system (TCSTS). Expression of adeABC occurs by binding of AdeR to a ten base-pair direct repeat motif found within the intercistronic region separating these operons [18, 19]. Many clinical A. baumannii isolates harbor different genetic arrangements of the adeRS and adeABC operons [20], and whether regulation via AdeRS is conserved in these strains is not completely understood.
With an increase in infections caused by MDR isolates across many bacterial species, including A. baumannii, understanding mechanisms of resistance and how resistance to and evasion of treatments has evolved over time has become a key research topic. Furthermore, determining the impact of expression of resistance determinants within the host environment and its effect on the efficacy of therapeutic treatments has gained attention. For example, when Pseudomonas aeruginosa is grown using L-glutamate as the sole carbon source, resistance to the related compounds polymyxin B and colistin increased ≥ 25- and 9-fold, respectively [21]. Other studies have shown that the bioavailability of cations such as iron can have a drastic effect towards the resistance of a number of antimicrobials across a range of pathogenic bacterial species [22]. Despite A. baumannii being recognized as a major human pathogen, these types of studies are limited for this organism.
This study aimed to determine the regulatory role of the AdeRS TCSTS in A. baumannii ATCC 17978, a clinical isolate which only encodes the adeAB subunits and identify whether AdeA and or AdeB alone can confer antimicrobial resistance in the ATCC 17978 background. Phenotypic characterization of a constructed panel of deletion strains identified that alterations to the adeRS and adeAB operons of ATCC 17978 reduced resistance to a subset of dicationic compounds, including pentamidine. As a recent study highlighted the effectiveness of pentamidine in combination therapy to treat infections caused by Gram-negative pathogens [23], we sought to further examine alternative mechanisms of pentamidine resistance in A. baumannii. The type of carbon source and availability of iron were identified as affecting pentamidine resistance, thereby revealing interconnectedness between metabolic and resistance strategies within this formidable pathogen.
Materials and methods
Bacterial strains, plasmids and growth conditions
A. baumannii ATCC 17978 [24] was obtained from the ATCC and is designated as wildtype (WT). Bacterial strains, plasmids and primers used in this study are summarized in S1 and S2 Tables, respectively. Bacterial strains were cultured using Luria-Bertani (LB) broth or LB agar plates, under aerobic conditions at 37°C, unless otherwise stated. Antibiotic concentrations used for selection were; ampicillin 100 mg/L, erythromycin (ERY) 25 mg/L, tetracycline 12 mg/L and gentamicin (GEN) 12.5 mg/L. All antimicrobial agents were purchased from Sigma with the exception of ampicillin which was purchased from AMRESCO.
M9 minimal medium agar plates were generated using a stock solution of 5 × M9 salts (200 mM Na2HPO4.7H2O, 110 mM KH2PO4, 43 mM NaCl and 93 mM NH4Cl) and subsequently diluted 1:5 and supplemented with 2 mM MgSO4, 0.1 mM CaCl2 and 0.4% (w/v) of various carbon sources on the day of use. M9 minimal medium was supplemented with 0.4% (w/v) of glucose, or succinic, fumaric, citric, α-ketoglutaric, pyruvic, glutamic or oxaloacetic acids. All carbon compounds were purchased from Sigma with the exception of citric and pyruvic acids which were purchased from Chem-Supply (Gillman, Australia). For solid media, J3 grade agar (Gelita, Australia) was used at a final concentration of 1%.
Iron-chelated and iron-rich conditions were achieved by the addition of dipyridyl (DIP) or FeSO4.7H2O (Scharlau) at final concentrations of 100 and 200 μM or 2.5 and 5 mM in Mueller-Hinton (MH) agar, respectively.
Antimicrobial susceptibility testing
The resistance profiles of WT and derivative strains were determined via broth microdilution methods [25] which were performed in duplicate with a minimum of three biological replicates using bacteria cultured in MH broth. Growth was monitored at 600 nm (OD600) using a FLUOstar Omega spectrometer (BMG Labtech, Germany) after overnight incubation at 37°C.
For disk diffusion assays, strains were grown to mid-log phase in MH broth, diluted to OD600 = 0.1 in fresh MH broth and 100 μL plated onto MH agar plates on which antimicrobial loaded filter disks were overlaid. Alternatively, when assessing zones of clearing using M9 minimal medium agar, strains were washed three times in phosphate buffered saline (PBS) and diluted to OD600 = 0.1 in fresh PBS. Tested antimicrobials were pentamidine isethionate, chlorhexidine dihydrochloride and 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) at the final concentrations of 125, 2.5 and 1.6 μg, respectively. After overnight incubation, callipers were used to measure the diffusion distances which were determined as half of the inhibition zone diameter, less the length of the disk. At least three independent experiments in duplicate were undertaken. Statistical significance was determined using the Student's t-test (two-tailed, unpaired) and P values <0.0005 were considered significant.
For plate dilution assays, strains were grown to mid-log phase in MH broth, washed three times in PBS and diluted to OD600 = 0.1 in fresh PBS. Serial dilutions (10-fold) were prepared of which 5 μl was spotted onto M9 minimal or MH agar containing various concentrations of pentamidine (32, 64, 128, 256 and 512 mg/L) and growth assessed after overnight incubation at 37°C. To test the effect of growth of different carbon sources, M9 minimal media was supplemented with 0.4% (w/v) of succinic, fumaric, citric, α-ketoglutaric or oxaloacetic acids.
Construction of A. baumannii ATCC 17978 deletion strains and genetic complementation
Two methods were adopted to generate specific gene deletions in A. baumannii ATCC 17978. The first method used to generate ΔadeRS utilized a sacB-based strategy [26], where genetic sequence flanking the region of interest and the ERY resistance cassette from pVA891 [27] were PCR amplified, purified and subsequently cloned into a modified pBluescript SK+ II vector that had the BamHI site removed by end-filling, generating pBl_adeRS. Confirmed clones were re-cloned into XbaI-digested pEX18Tc [28]. The resulting pEX18Tc_adeRS vector was introduced into ATCC 17978 cells via electroporation, as previously described [29]. Transformants were selected on LB agar supplemented with ERY. Counter-selection was undertaken on M9 minimal agar containing ERY and 5% sucrose. The second method used to generate ΔadeA, ΔadeB and ΔadeAB strains utilized the RecET recombinase system with modifications [30]. Briefly, 400–600 bp of sequence flanking the gene(s) were used as templates in a nested overlap extension PCR with the ERY resistance cassette from pVA891 [27]. Approximately, 3.5–5 μg of the purified linear PCR product was electroporated into ATCC 17978 cells harboring the vector pAT04 and recovered as previously described [30]. Recombinants were selected on LB agar supplemented with ERY. All mutants generated in this study were confirmed by PCR amplification and Sanger sequencing. Primers utilized to generate mutant strains are listed in S2 Table.
For genetic complementation of mutant strains, WT copies of adeRS and adeAB were cloned into pWH1266 [31] where transcription was driven by the tetracycline promoter. Resulting plasmids denoted pWH::adeRS and pWH::adeAB, respectively were confirmed by Sanger sequencing. The GEN resistance cassette from pUCGM [32] was PCR amplified and cloned into BamHI digested pWH::adeAB generating pWHgent::adeAB thus abrogating transcription from the pWH1266 tetracycline promoter. Plasmids were introduced into appropriate A. baumannii cells as previously described [33]. Primers used to generate complementation vectors are listed in S2 Table.
Cell treatments and RNA isolation
RNA was isolated from WT and ΔadeRS cells and Hiseq RNA transcriptome analysis performed following methodologies as outlined previously [34].
For pentamidine stress assays, WT and ΔadeRS strains were grown overnight in MH broth, sub-cultured 1:100 in fresh medium and grown to OD600 = 0.6; they were subsequently split into two 10 mL cultures. One 10 mL sample was treated with 7.8 mg/L of pentamidine (0.5 × MIC for ΔadeRS), whilst the other remained untreated. Cultures were grown for an additional 30 min before total RNA was extracted as outlined previously [34].
Bioinformatic analysis
Bioinformatic analysis of RNA-seq data was undertaken as described previously [34], with the modification that obtained reads were mapped to the recently re-sequenced A. baumannii ATCC 17978 genome (GenBank: CP012004). RNA-seq data have been deposited in the gene expression omnibus database, accession number GSE102711 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102711).
Quantitative Real-Time PCR
Pentamidine stress and RNA-seq validation experiments were achieved using a two-step qRT-PCR method. RNA samples were first purified as previously described [34], subsequently DNaseI treated (Promega) and then converted to cDNA using an iScriptTM cDNA synthesis kit (Biorad), following the manufacturer’s instructions. The cDNA generated was used as a template for qRT-PCR using the SYBR® Green JumpStart™ Taq readymix™ (Sigma) in a 20 μl final volume. Either a Rotor-Gene Q (Qiagen, Australia) or RG-3000 (Corbett Life Science, Australia) instrument was used for quantification of cDNA using the following protocol; 1 min at 95°C, followed by 40 cycles of 10 sec at 95°C, 15 sec at 57°C and 20 sec at 72°C. Melt curve analyses were undertaken to ensure only the desired amplicon was generated. Primers used (S2 Table) for amplification of cDNA transcripts were designed using NetPrimer (www.premiersoft.com). Transcriptional levels for RNA-seq validation experiments were corrected to GAPDH levels prior to normalization to the ATCC 17978 WT. For pentamidine stress experiments, transcriptional levels of adeA were corrected to 16S rDNA levels prior to being normalized to their respective untreated A. baumannii cultures. Transcriptional variations were calculated using the 2-ΔΔCT method [35]. Results for pentamidine stress experiments display the mean Log2 fold change (± SEM) of three biological replicates each undertaken in triplicate. Statistical analyses were performed by Student’s t-test, two-tailed, unpaired; ** = P <0.01 and *** = P <0.001.
Phenotypic microarray analysis
A. baumannii ΔadeRS cells were cultured on LB agar overnight at 37°C. A suspension of cells was made from a single colony in Biolog IF-0 inoculation fluid (Biolog, Inc.) to 85% transmittance and was subsequently diluted 1:200 in Biolog IF-0 containing dye A (Biolog, Inc.) and 0, 8, 16, 32 or 64 mg/L pentamidine. One hundred μL of each dilution was added to each well of the Biolog PM01 and PM02A MicroPlates and placed in an Omnilog automatic plate reader (Biolog, Inc.) for 72 h at 37°C. Color formation from the redox active dye (Biolog dye A) was monitored every 15 min. Data obtained from respiration of ΔadeRS under different pentamidine conditions were individually overlaid against the untreated control using the OmniLog File Management/Kinetic Analysis software v1.20.02, and analyzed using OmniLog Parametric Analysis software v1.20.02 (Biolog, Inc.).
Results
Generation of ΔadeRS and complemented strains
The adeRS operon of A. baumannii ATCC 17978 was disrupted through the introduction of an ERY resistance cassette to produce a ΔadeRS strain. Complementation of ΔadeRS was achieved via reintroduction of a WT copy of adeRS in trans on the pWH1266 shuttle vector, generating ΔadeRS pWH::adeRS and comparisons were made to cells carrying the empty vector control (ΔadeRS pWH1266). To ensure deletion of adeRS did not affect viability, growth was monitored by OD600 readings in MH broth over an 8 h period; results identified no significant perturbations in the growth rate in laboratory media compared to WT cells (data not shown).
Transcriptomic profiling of ΔadeRS
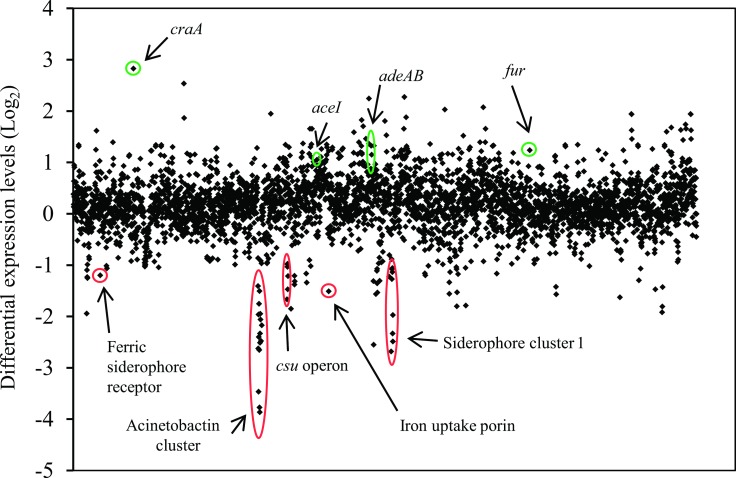
Transcriptome profiling by RNA-sequencing (RNA-seq) of RNA isolated from ΔadeRS compared to that from WT ATCC 17978 identified 290 differentially expressed (≥ 2-fold) genes; 210 up-regulated and 80 down-regulated (Fig 1 and S3 Table). RNA-seq results were confirmed by quantitative Real-Time PCR (qRT-PCR) for nine genes that displayed different levels of expression in ΔadeRS compared to WT; a good correlation between methods was observed (S1 Fig).
Fig 1. Global transcriptomic response differences of A. baumannii ATCC 17978 after deletion of adeRS.
Each diamond marker represents a predicted gene within the genome ordered according to locus tag along the X-axis and differential expression generated from normalized reads per kilobase mapped of WT against ΔadeRS are displayed on the Y-axis (Log2). Positive and negative Log2-values correlate to up- and down-regulated genes, respectively. Green and red circles highlight genes/gene clusters of interest that have been up- and down-regulated, respectively. See S3 Table for the full list of genes that were differentially expressed ≥ 1 Log2 fold.
Expression of a number of efflux proteins was affected by inactivation of adeRS. For example, the craA major facilitator superfamily transporter (ACX60_01760) shown to confer resistance to chloramphenicol and recently chlorhexidine efflux [36] was the highest up-regulated gene (7.1-fold; Fig 1) and adeA (ACX60_09125), adeB (ACX60_09130) and aceI (ACX60_07275) which encode the AdeAB and the AceI efflux pumps, were up-regulated by 2.5-, 1.43-, and 2-fold, respectively, following deletion of adeRS.
Expression of multiple genes involved in virulence were down-regulated in the ΔadeRS strain, including the type 1 pilus operon csuA/BABCDE (ACX60_06480–06505) (3.2- to 2-fold) and the siderophore-mediated iron-acquisition systems acinetobactin (ACX60_05590–05680) and siderophore 1 (ACX60_09665–09720) clusters (7.7- to 1.1-fold and 5.4- to 0.3-fold, respectively). In contrast, the ferric uptake regulator gene (fur) (ACX60_13910) was up-regulated 2.4-fold. Despite these alterations in expression levels of iron siderophore clusters and their regulator, no significant growth defects were identified when ΔadeRS was grown in the presence of 200 μM of DIP (data not shown).
Deletion of adeRS in ATCC 17978 reduced susceptibility to a limited number of antimicrobial agents
To assess if changes in expression of the adeAB and craA drug efflux genes identified in the transcriptome of the ΔadeRS derivative translated to an alteration in resistance profile, antibiogram analyses were undertaken. Resistance to a number of antibiotics including those that are known substrates of the CraA and AdeABC pumps were assessed. Surprisingly, despite craA being the highest up-regulated gene, no change in resistance to the primary substrate of CraA, chloramphenicol [36], was seen (data not shown). Previous studies examining the level of antimicrobial resistance conferred by AdeABC indicate that only when deletions are generated in strains which overexpress AdeABC is there a significant impact on the antibiogram [10, 13, 15, 16, 37], implying that AdeABC confers only minimal to no intrinsic resistance. This was supported in our analysis as the MIC for tetracycline, tigecycline, GEN, kanamycin, nalidixic acid, ampicillin, streptomycin and amikacin, all previously identified AdeABC substrates, remained unchanged, whilst resistance to norfloxacin and ciprofloxacin increased 2-fold (data not shown) and chlorhexidine decreased 2-fold for the ΔadeRS mutant compared to WT (Table 1).
Table 1. Antibiotic susceptibility of A. baumannii ATCC 17978, deletion mutants and complemented strains.
|
Strain |
MIC (mg/L)ab | ||
|---|---|---|---|
|
PENT |
DAPI |
CHX |
|
| WT | 125 | 4 | 8 |
| ΔadeA | 15.6 | 1 | 4 |
| ΔadeB | 15.6 | 1 | 4 |
| ΔadeABc | 15.6 | 1 | 4 |
| ΔadeRS d | 15.6 | 1 | 4 |
| ΔadeRS pWH::adeRS | 125 | 1 | 8 |
| ΔadeRS pWH1266 | 15.6 | 0.5 | 4/2 |
| ΔadeRS pWH::adeAB | 31.3 | 4 | 4 |
| ΔadeRS pWHgent::adeAB | 15.6 | 1 | 4 |
| ΔadeRS pWHgent | 7.8 | 0.5 | 4 |
| ΔadeA pWHgent::adeAB | 62.5 | 2 | 8/4 |
| ΔadeA pWHgent | 7.8 | 0.5 | 4 |
| ΔadeB pWHgent::adeAB | 62.5 | 4 | 4 |
| ΔadeB pWHgent | 7.8 | 1 | 2 |
| ΔadeAB pWHgent::adeAB | 62.5 | 4 | 4 |
| ΔadeAB pWHgent | 7.8 | 1 | 2 |
aPENT, pentamidine; DAPI, 4',6-diamidino-2-phenylindole; CHX, chlorhexidine.
bValues highlighted in bold indicate MIC values altered two-fold or greater compared to WT ATCC 17978.
cAntimicrobials that did not significantly differ from WT susceptibility levels included gentamicin, kanamycin, norfloxacin, ciprofloxacin, ampicillin, colistin, polymyxin B, dequalinium, tigecycline, triclosan and methyl viologen.
dAntimicrobials tested that did not significantly differ from WT susceptibility levels included rifampicin, benzylkonium, streptomycin, amikacin and novobiocin as well as antimicrobials listed above in ‘c’ with the exception of ciprofloxacin and norfloxacin.
A variety of additional compounds were tested, primarily focusing on substrates of other multidrug efflux pumps and/ or clinically relevant compounds, these included; colistin, polymyxin B, rifampicin, triclosan, novobiocin, benzalkonium, methyl viologen, pentamidine, DAPI and dequalinium. Of these, significant differences in the MICs of ΔadeRS versus WT were observed only for the diamidine compounds, pentamidine and DAPI, where an 8- and 4-fold reduction in resistance was identified, respectively (Table 1). Thus, taken together the ΔadeRS strain showed reduced resistance to the bisbiguanide chlorhexidine, and the diamidines pentamidine and DAPI which display structural similarities [38] (S2 Fig). Resistance to these substrates was partially or fully restored by complementation using ΔadeRS pWH::adeRS (Table 1).
Both AdeA and AdeB are required for intrinsic antimicrobial resistance in ATCC 17978
Since the 10-bp direct repeat where AdeR has been demonstrated to bind in other A. baumannii strains [18, 19] is also present in the ATCC 17978 adeA-adeR intercistronic region (data not shown), we hypothesized that the decreased resistance towards the subgroup of dicationic compounds seen in ΔadeRS resulted from changes in adeAB expression. To test this, deletion strains targeting adeAB, as well as individual adeA and adeB mutations were generated in ATCC 17978.
Using ΔadeAB, MIC analyses verified that deletion of the pump resulted in negligible changes in resistance to a subset of the antimicrobials tested for ΔadeRS (data not shown). However, an identical resistance pattern for the dicationic compounds as that afforded by ΔadeRS was observed (Table 1). Complementation of the inactivated genes partially restored resistance to all compounds, validating that AdeAB plays a direct role in resistance to these dicationic compounds (Table 1).
AdeRS is critical for increased expression of adeAB following pentamidine exposure
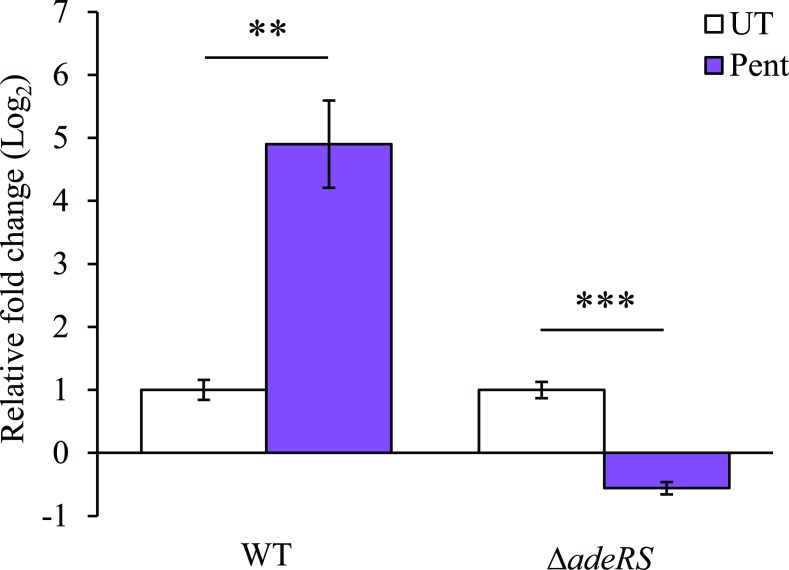
From MIC analysis of the ATCC 17978 derivatives, it was proposed that the presence of the AdeRS TCSTS increased adeAB expression consequently providing resistance to pentamidine, chlorhexidine and DAPI (Table 1). To confirm this, the level of adeA transcription was assessed by qRT-PCR of RNA isolated from WT and ΔadeRS strains after addition of a sub-inhibitory concentration of pentamidine. Transcription of the adeAB operon was significantly up-regulated in WT and down-regulated in ΔadeRS following pentamidine stress (Fig 2). Additionally, qRT-PCR was used to determine if adeS expression levels altered after pentamidine stress in WT cells. It was found that adeS expression increased less than <2-fold compared to untreated WT cells (data not shown). To phenotypically support the transcriptional evidence that adeRS initiates transcription of the ATCC 17978 adeAB operon, additional antibiograms were determined. Using the shuttle vector pWH1266, two clones were constructed; pWH::adeAB and pWHgent::adeAB, where the GEN resistance cartridge cloned in the latter vector inhibited transcription of adeAB from the tetracycline promoter naturally present in pWH1266. MIC analyses determined that the carriage of pWHgent::adeAB in ΔadeRS did not differ from results obtained for ΔadeRS. Conversely, ΔadeRS cells with pWH::adeAB displayed a 4- and 8-fold increase in pentamidine and DAPI resistance, respectively (Table 1). Collectively, these results suggest that expression of adeAB and subsequent resistance to the dicationic compounds in ATCC 17978 can only occur when AdeRS is present.
Fig 2. Increased expression of adeA following pentamidine stress is dependent on the presence of AdeRS in ATCC 17978.
Transcriptional levels of adeA (ACX60_09125) from WT and ΔadeRS were determined by qRT-PCR after 30 min shock with 7.8 mg/L of pentamidine (Pent) (0.5 × MIC of ΔadeRS) and corrected to untreated cells (UT) after normalization to 16S. Bars represent the mean fold change (Log2) of three biological replicates undertaken in triplicate, and error bars represent ± SEM. Statistical analyses were performed by Student’s t-test, two-tailed, unpaired; ** = P < 0.01 and *** = P < 0.001.
Carbon source utilization alters resistance to pentamidine
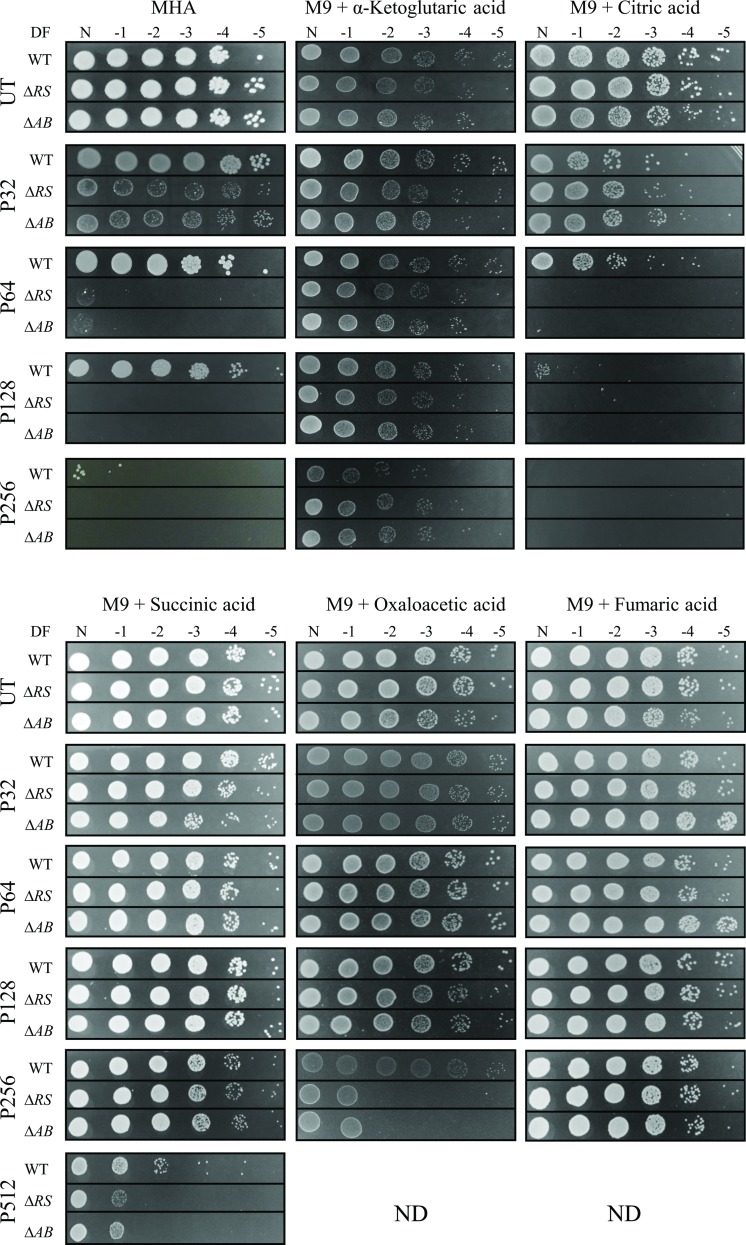
Pentamidine, a drug known to be effective in the treatment of fungal and protozoan infections has gained recent attention in a bacterial context. Pentamidine has shown synergy with Gram-positive antibiotics, potentiating their activity towards Gram-negative bacteria [23]. As such, pentamidine has been proposed to be utilized as an adjunct therapy for MDR bacteria, including A. baumannii [23]. To identify additional pentamidine resistance mechanisms employed by A. baumannii, growth in different media was assessed. Disk diffusion assays identified that in A. baumannii, resistance to pentamidine was affected by the carbon source provided in M9 minimal medium (S4 Table), whilst for chlorhexidine and DAPI this pattern was not conserved indicating a pentamidine-specific response (data not shown). To determine the MIC levels for pentamidine, plate dilution experiments were undertaken for WT, ΔadeRS and ΔadeAB strains provided with varied carbon sources (Fig 3). Growth of ΔadeRS and ΔadeAB cells on MH agar were significantly perturbed at 32 mg/L of pentamidine. This MIC drastically differs when succinic acid was utilized as the sole carbon source, as growth was maintained up to 512 mg/L of pentamidine for all strains tested (Fig 3). Fumaric, α-ketoglutaric and oxaloacetic acids also increased pentamidine resistance by 8-fold for ΔadeRS and ΔadeAB strains when compared to the MIC obtained for MH agar. Growth of ΔadeRS and ΔadeAB mutants was inhibited at a higher dilution factor in the presence of oxaloacetic acid compared to fumaric and α-ketoglutaric acids whilst growth using citrate as the sole carbon source negatively affected resistance to WT cells, decreasing resistance 2-fold.
Fig 3. Resistance to pentamidine is modulated by carbon sources available in the growth medium.
Ten-fold serial dilutions of A. baumannii ATCC 17978 (WT), ΔadeRS (ΔRS) and ΔadeAB (ΔAB) cells were used to compare the concentration of pentamidine that inhibits growth in different media, Mueller-Hinton agar (MHA) was used as a comparative control. Images display serial 1:10 dilutions after overnight incubation at 37°C, where DF is abbreviated for dilution factor and N represents undiluted cells. Strains were grown in the absence of pentamidine (UT) or presence of 32, 64, 128, 256 and 512 mg/L of pentamidine (P32, P64, P128, P256, and P512, respectively). Carbon sources tested in M9 minimal medium were used at a final concentration of 0.4% (w/v). ND, not done due to precipitation of pentamidine once added into the molten medium. Figures are representative examples of results obtained.
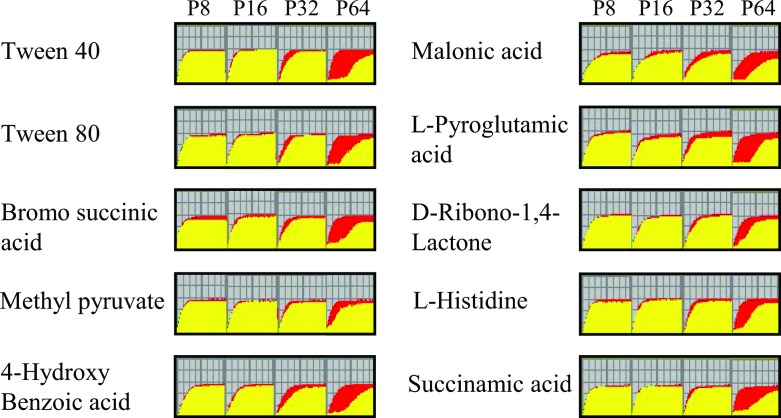
Biolog phenotypic arrays were undertaken to identify additional synergistic or antagonistic relationships between carbon sources and pentamidine resistance. Respiration of ΔadeRS cells for a total of 190 carbon compounds was assessed at various pentamidine concentrations (S3 Fig). From this, an additional ten compounds were identified that increased resistance to pentamidine in ΔadeRS at 64 mg/L (Fig 4). Despite minimal changes in pentamidine resistance in the presence of citric acid for ΔadeRS (Fig 3), this compound negatively affected respiration at lower pentamidine concentrations (S3 Fig). Surprisingly, succinic, α-ketoglutaric and fumaric acids failed to restore respiration at 64 mg/L of pentamidine for ΔadeRS (S3 Fig). By extending the incubation period for another 72 h, respiration in the presence of succinic or fumaric acids was restored to levels similar to the untreated control (data not shown). This may indicate that succinic and fumaric acids significantly lag in their ability to recover the cells from pentamidine in the IF-0 medium (Biolog Inc.) whilst α-ketoglutaric acid recovery is dependent on growth medium.
Fig 4. Kinetic response curves paralleling bacterial growth from Biolog PM01 and PM2A plates identify ten carbon sources that increase pentamidine resistance in ΔadeRS.
P8, P16, P32 and P64 represent kinetic response curves at 8, 16, 32, or 64 mg/L of pentamidine compared to the untreated control, respectively. Red curves represent respiration of untreated ΔadeRS, whilst respiratory activity which overlaps between the control and the sample in the different experimental conditions is represented in yellow. Only carbon compounds that promote at least 50% maximal respiration and induce a recovery response by 36 h are shown. See S3 Fig for respiration curves for all tested treatments.
Bioavailability of iron correlates with pentamidine resistance
Iron has been shown to influence pentamidine resistance in protozoan species [39], thus we assessed if its plays a similar role in A. baumannii. The addition of ferrous sulphate to the growth medium significantly reduced the zone of clearing from pentamidine in a dose-dependent manner up to a final concentration of 5 mM for WT and mutant strains (Table 2 and S4 Fig). Furthermore, chelation of iron using DIP resulted in WT cells becoming more susceptible to pentamidine compared to the untreated control (Table 2). This modified pentamidine susceptibility is iron specific as inclusion of other cations in the growth medium (zinc, copper, manganese, cobalt, nickel) did not significantly affect the zones of clearing (data not shown). Using inductively-coupled plasma mass spectrometry the internal iron concentration in WT and ΔadeRS cells was determined in the absence/presence of a sub-inhibitory concentration of pentamidine. Internal iron concentrations remained essentially unchanged in both strains under the different conditions tested (data not shown), indicating that iron/pentamidine interactions may occur outside of the cell and the observed response was not due to a reduced capacity to store iron.
Table 2. Zones of clearing for A. baumannii ATCC 17978 and deletion mutants grown on Mueller-Hinton agar with the addition or chelation of iron after pentamidine exposure.
| Zone of clearing (mm)ab | |||||
|---|---|---|---|---|---|
| Strain | WT | ΔadeRS | ΔadeAB | ΔadeA | ΔadeB |
| Mediumc | |||||
| MHA control | 1.4 ± 0.3 | 6.9 ± 0.5 | 6.9 ± 0.3 | 6.9 ± 0.4 | 7.2 ± 0.4 |
| + 2.5 mM FeSO4 | 0.8 ± 0.1* | 5.4 ± 0.2* | 5.6 ± 0.3* | 5.4 ± 0.1*** | 5.6 ± 0.1*** |
| + 5 mM FeSO4 | 0.7 ± 0.2* | 2.5 ± 0.8*** | 2.8 ± 0.3*** | 3.2 ± 0.3*** | 2.9 ± 0.6*** |
| + 7.5 mM FeSO4 | NG | NG | NG | NG | NG |
| + 100 μM DIP | 1.7 ± 0.1 | 6.7 ± 0.2 | 6.9 ± 0.1 | 6.2 ± 0.2 | 6.6 ± 0.2 |
| + 200 μM DIP | 5 ± 0.3*** | 6.7 ± 0.4 | 6.6 ± 0.4 | 6.1 ± 0.1 | 6.4 ± 0.1 |
aAveraged values are displayed ± SD. Statistical analyses were performed by Student’s t-test, two-tailed, unpaired
* = P <0.0005 and
*** = P <0.000001.
bValues given in bold indicate a significant difference between a strain grown under iron-rich or iron-limited conditions versus its respective MHA control.
cMHA, Mueller-Hinton agar; FeSO4, ferrous sulphate; DIP, 2’,2’ dipyridyl; NG, no growth due to iron toxicity.
Discussion
The adeRS and adeABC operons of A. baumannii have gained considerable attention due to their role in regulating and conferring multidrug resistance, respectively [8, 13, 16, 40]. Multiple genetic arrangements of the adeABC operon in A. baumannii clinical isolates exist, where 35% of 116 diverse isolates lack the adeC OMP component [20]. This is also the case for the clinical isolate A. baumannii ATCC 17978, which was chosen for further analysis in this study. It is not uncommon for RND pumps to recruit an alternative OMP to form a functional complex [41–43]. This may also be the situation for AdeAB in A. baumannii ATCC 17978, since in an Escherichia coli background, adeAB can be co-expressed with adeK, an OMP belonging to the A. baumannii AdeIJK RND complex to confer resistance [44]. However, whether AdeB can work alone or function with alternative membrane fusion proteins such as adeI or adeF is not known. As the resistance profile of ΔadeA was indistinguishable from the ΔadeAB and ΔadeB strains, it provides confirmatory evidence that both adeA and adeB are required for efficient efflux to only a subset of dicationic compounds in ATCC 17978. Further assessments will be required to verify if this phenotype is maintained across other A. baumannii isolates and extends to other structurally-similar compounds.
It has been found previously, that the introduction of shuttle vectors expressing WT copies of genes in trans do not restore resistance profiles back to WT levels as also seen in this study [45–48]. Using qRT-PCR, adeS was expressed at low levels in WT cells even after pentamidine shock (data not shown). Complementation studies with adeRS thus could be influenced by copy number effects, which may perturb the native expression levels of these proteins within the cell. Additionally, the differing modes of action of the dicationic compounds may have influenced the MIC results, as fold-shifts in resistance were observed even when the empty pWH1266 vector was being expressed (Table 1).
The transcriptomic analysis of ATCC 17978 ΔadeRS revealed changes in gene expression were not limited to adeAB but included many genes, some of which have been shown to be important in A. baumannii pathogenesis [49, 50] and virulence in vivo [51]. Although this had been seen before in an adeRS mutant generated in the MDR A. baumannii isolate AYE [16], here the transcriptional changes were to a largely different subset of genes. Previously, deletion of adeRS in AYE resulted in decreased expression of adeABC by 128-, 91-, and 28-fold, respectively [16]. This decreased gene expression was expected as AYE naturally contains a point mutation in AdeS (producing an Ala 94 to Val substitution in AdeS) resulting in constitutive expression of adeABC and contributing to its MDR phenotype [2, 16]. Richmond et al., (2016) [16] proposed that the high similarities in phenotypic changes between their adeRS and adeB AYE deletion derivatives were largely due to downstream effects caused by reduced expression of adeABC. We cannot rule out the possibility that some of the changes in gene expression in our study have resulted from the slight increase seen in expression of adeAB, most likely caused by polar effects exerted by the ERY resistance cassette. However, our transcriptome data indicated adeAB were expressed at low levels and thus represent basal levels of expression; results consistent with ATCC 17978 being labelled as a ‘drug susceptible’ isolate [24].
Currently, the environmental signal(s) that interact with the periplasmic sensing domain of AdeS are not known. Antimicrobial compounds can directly stimulate autophosphorylation of specific TCSTS which in turn directly regulate the expression of genes providing resistance to that compound [52, 53]. It is unlikely that pentamidine is the environmental stimulus that is sensed by AdeS, as a number of conditions, including chlorhexidine shock, can also up-regulate adeAB expression in ATCC 17978 [54–57]. Instead, AdeS may respond to stimuli such as solutes that are excreted and accumulate in the periplasm when cells are subjected to various stressors including AdeAB substrates.
This study revealed unique interactions between pentamidine and a number of carbon sources which significantly alter the resistance profile to pentamidine for A. baumannii ATCC 17978 and derivatives. Biolog phenotypic arrays identified a number of succinic acid derivatives that also allowed respiration in the presence of a lethal concentration of pentamidine (Fig 4). Interestingly, when ΔadeRS utilized malonic acid as the sole carbon source, a potent inhibitor of the succinate dehydrogenase complex, cells were also able to respire at increased pentamidine concentrations (Fig 4). In Gram-negative bacteria, the mechanism of action for pentamidine is thought to primarily occur via binding to the lipid-A component of the outer membrane and not through inhibition of an intracellular target [23, 58]. Therefore, it seems unlikely that pentamidine interferes with enzymatic functions like that of succinate dehydrogenase, and instead the compounds which showed an increase in pentamidine resistance may contribute to either chelation of the compound or provide protective mechanisms that reduce binding to the lipid-A target. Interestingly, a 1956 study [59] identified that in the presence of α-ketoglutaric or glutamic acid, resistance to a lethal concentration of pentamidine could be achieved in E. coli, leading to the proposition that pentamidine interferes with the transaminase reaction in glutamic acid production. However, our study shows that glutamic acid does not affect pentamidine resistance (S4 Table), inferring that a different mechanism might be responsible for the increase in pentamidine resistance in A. baumannii.
Iron is an essential metal serving as a co-factor in numerous proteins involved in redox chemistry and electron transport. It is generally a limited resource for pathogenic bacteria such as A. baumannii where it is important for virulence and disease progression [6, 50]. Our studies showed that altering iron levels also had a marked effect on resistance to pentamidine. This is in agreement with the observation that citric and gluconic acids can act as iron-chelating agents [60, 61] and respiration activity in the presence of these compounds at the pentamidine concentrations tested were significantly altered (S3 Fig). Many clinically relevant antimicrobials have shown to be affected by the presence of iron, including compounds belonging to tetracycline, aminoglycoside and quinolone classes [22]. Activity of these compounds can be altered by numerous factors, including the formation of stable complexes which can affect drug efficacy or have unfavourable effects on patient health. Understanding the metabolic flux in A. baumannii and other MDR bacteria has the potential to lead to more effective therapeutic interventions thus reducing infection rates, knowledge critical to slow the progress towards our re-entry into a pre-antibiotic era.
Conclusions
Overall, this is the first study which has demonstrated that the AdeAB system in A. baumannii ATCC 17978 provides intrinsic resistance to a subset of dicationic compounds, and efflux of these compounds via AdeAB is directly regulated by the AdeRS TCSTS. RNA-seq identified that deletion of adeRS produced significant changes in the transcriptome. These results support the notion that strain specific variations are apparent [16]. We have provided evidence that in ATCC 17978, AdeRS is directly responsible for the activation of adeAB gene expression, as ΔadeRS failed to increase expression upon subjection to one of the pumps newly identified intrinsic substrates, pentamidine. It will be of interest to assess whether these dicationic compounds also extend as substrates towards AdeAB(C) pumps present in other A. baumannii isolates and if expression levels of adeAB(C) upon exposure to these substrates and other potential stressors also occur. This information may help to identify the stimulus that activates the AdeRS TCSTS. We have also demonstrated for the first time that for pentamidine to exert its antibacterial effect in A. baumannii, a dependence on the availability of iron is required, and that growth in the presence of selected carbon sources has a profound effect on its resistance.
Supporting information
The transcriptomic results obtained by RNA-sequencing were validated by qRT-PCR analysis. The level of nine genes that displayed differential expression or remained essentially unchanged between ΔadeRS and WT ATCC 17978 were chosen for comparison. Expression levels for qRT-PCR experiments were corrected to those obtained for GAPDH (ACX60_05065) prior to normalisation against WT ATCC 17978 transcriptional levels. Grey and black bars represent values obtained from RNA-seq and qRT-PCR results, respectively. Differential expression between ΔadeRS and WT ATCC 17978 are given in Log2-values.
(TIF)
Compounds include (a) pentamidine, (b) DAPI and (c) chlorhexidine. For pentamidine and chlorhexidine, the cationic nitrogenous groups are separated by a long carbon chain, forming symmetrical compounds, whereas, DAPI lacks this long linker and is asymmetric.
(TIF)
Respiration of ATCC 17978 ΔadeRS cells in the presence of pentamidine (8, 16, 32, or 64 mg/L) against untreated control cells are shown for (a) Biolog PM01 plates and (b) Biolog PM02A plates. Respiration activity for both plates were monitored in IF-0 (Biolog, Inc.) liquid medium for 72 h at 37°C. The curve in each well represents the colour intensity of a redox-active dye (y axis) over time (x axis: 72 h). Respiration of ΔadeRS cells are shown in red (control), green (under different concentrations of pentamidine), and yellow (depicts the regions of respiratory overlap). Black numbered squares represent carbon sources which decreased resistance to pentamidine (1, D-gluconic acid; 2, citric acid).
(TIF)
Resistance to pentamidine was assessed by disc diffusion assays in Mueller-Hinton agar (MHA) for ATCC 17978 and ΔadeRS, ΔadeA, ΔadeB and ΔadeAB deletion derivatives. Zones of clearing were compared to iron rich conditions from the addition of ferrous sulphate (FeSO4) at the final concentrations of 2.5 and 5 mM and iron-chelated conditions obtained by the addition of 2’,2’ dipyridyl (DIP) at the final concentrations of 100 and 200 μM in MHA. Images displayed are a representative of the typical results obtained.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Professor Bryan Davies from the Department of Molecular Biosciences at the University of Texas for providing the plasmid pAT04.
Data Availability
Relevant data are within the paper and its Supporting Information files. Additionally, RNA-seq data have been deposited in the gene expression omnibus database, accession number GSE102711.
Funding Statement
This work was supported by the Australian National Health and Medical Research Council (Project Grant 1047509) and a Flinders Medical Research Foundation Grant to MHB and UHS. FGA was supported by AJ and IM Naylon and Playford Trust Ph.D. Scholarships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–82. doi: 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2(1):e7 doi: 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(9):3569–77. doi: 10.1128/AAC.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell GA, Hamidian M, Hall RM. IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere. 2016;1(3):e00117–16. doi: 10.1128/mSphere.00117-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother. 2014;69(10):2625–8. doi: 10.1093/jac/dku188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright MS, Jacobs MR, Bonomo RA, Adams MD. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. MBio. 2017;8(2):e02193–16. doi: 10.1128/mBio.02193-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saranathan R, Pagal S, Sawant AR, Tomar A, Madhangi M, Sah S, et al. Disruption of TetR type regulator adeN by mobile genetic element confers elevated virulence in Acinetobacter baumannii. Virulence. 2017:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013;57(7):2989–95. doi: 10.1128/AAC.02556-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne S, Courvalin P, Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55(3):947–53. doi: 10.1128/AAC.01388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–80. doi: 10.1128/AAC.45.12.3375-3380.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(10):4389–93. doi: 10.1128/AAC.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52(2):557–62. doi: 10.1128/AAC.00732-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48(9):3298–304. doi: 10.1128/AAC.48.9.3298-3304.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(6):2065–9. doi: 10.1128/AAC.01198-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother. 2010;65(2):228–32. doi: 10.1093/jac/dkp427 [DOI] [PubMed] [Google Scholar]
- 16.Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, et al. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio. 2016;7(2):e00430–16. doi: 10.1128/mBio.00430-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794(5):769–81. doi: 10.1016/j.bbapap.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang TY, Huang BJ, Sun JR, Perng CL, Chan MC, Yu CP, et al. AdeR protein regulates adeABC expression by binding to a direct-repeat motif in the intercistronic spacer. Microbiol Res. 2016;183:60–7. doi: 10.1016/j.micres.2015.11.010 [DOI] [PubMed] [Google Scholar]
- 19.Wen YO, Z., Yu Y, Zhou X, Pei YD, B., Higgins P, Zheng F. Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region Nucleic Acids Res. 2017;45(16):9773–87. doi: 10.1093/nar/gkx624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemec A, Maixnerova M, van der Reijden TJ, van den Broek PJ, Dijkshoorn L. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J Antimicrob Chemother. 2007;60(3):483–9. doi: 10.1093/jac/dkm231 [DOI] [PubMed] [Google Scholar]
- 21.Conrad RS, Wulf RG, Clay DL. Effects of carbon sources on antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1979;15(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezraty B, Barras F. The 'liaisons dangereuses' between iron and antibiotics. FEMS Microbiol Rev. 2016;40(3):418–35. doi: 10.1093/femsre/fuw004 [DOI] [PubMed] [Google Scholar]
- 23.Stokes JM, MacNair CR, Ilyas B, French S, Cote JP, Bouwman C, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. 2017;2:17028 doi: 10.1038/nmicrobiol.2017.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21(5):601–14. doi: 10.1101/gad.1510307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–75. doi: 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 26.Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109(1):137–41. [DOI] [PubMed] [Google Scholar]
- 27.Macrina FL, Evans RP, Tobian JA, Hartley DL, Clewell DB, Jones KR. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25(1):145–50. [DOI] [PubMed] [Google Scholar]
- 28.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77–86. [DOI] [PubMed] [Google Scholar]
- 29.Dorsey CW, Tomaras AP, Actis LA. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl Environ Microbiol. 2002;68(12):6353–60. doi: 10.1128/AEM.68.12.6353-6360.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, et al. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio. 2014;5(4):e01313–14. doi: 10.1128/mBio.01313-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87(1):45–51. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer HD. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 1993;15(5):831–4. [PubMed] [Google Scholar]
- 33.Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LD, Paulsen IT, Brown MH. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun. 2013;81(7):2574–83. doi: 10.1128/IAI.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giles SK, Stroeher UH, Eijkelkamp BA, Brown MH. Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 2015;15:116 doi: 10.1186/s12866-015-0440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 36.Li L, Hassan KA, Brown MH, Paulsen IT. Rapid multiplexed phenotypic screening identifies drug resistance functions for three novel efflux pumps in Acinetobacter baumannii. J Antimicrob Chemother. 2016;71(5):1223–32. doi: 10.1093/jac/dkv460 [DOI] [PubMed] [Google Scholar]
- 37.Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, De E, et al. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio. 2015;6(2):e00309–15. doi: 10.1128/mBio.00309-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown MH, Skurray RA. Staphylococcal multidrug efflux protein QacA. J Mol Microbiol Biotechnol. 2001;3(2):163–70. [PubMed] [Google Scholar]
- 39.Wong IL, Chow LM. The role of Leishmania enriettii multidrug resistance protein 1 (LeMDR1) in mediating drug resistance is iron-dependent. Mol Biochem Parasitol. 2006;150(2):278–87. doi: 10.1016/j.molbiopara.2006.08.014 [DOI] [PubMed] [Google Scholar]
- 40.Nowak J, Schneiders T, Seifert H, Higgins PG. The Asp20-to-Asn substitution in the response regulator AdeR leads to enhanced efflux activity of AdeB in Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(2):1085–90. doi: 10.1128/AAC.02413-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuanchuen R, Narasaki CT, Schweizer HP. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J Bacteriol. 2002;184(18):5036–44. doi: 10.1128/JB.184.18.5036-5044.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43(2):415–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178(19):5803–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara E, Nikaido H. Properties of AdeABC and AdeIJK efflux systems of Acinetobacter baumanii compared with those of AcrAB-TolC system of Escherichia coli. Antimicrob Agents Chemother. 2014;58:7250–7. doi: 10.1128/AAC.03728-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Quan J, Yang Y, Ji J, Liu L, Fu Y, et al. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 2016;35(8):1371–5. doi: 10.1007/s10096-016-2674-0 [DOI] [PubMed] [Google Scholar]
- 46.Liou ML, Soo PC, Ling SR, Kuo HY, Tang CY, Chang KC. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect. 2014;47(4):275–81. doi: 10.1016/j.jmii.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 47.Saroj SD, Clemmer KM, Bonomo RA, Rather PN. Novel mechanism for fluoroquinolone resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56(9):4955–7. doi: 10.1128/AAC.00739-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Zhou Z, He F, Ruan Z, Jiang Y, Hua X, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One. 2018;13(2):e0192288 doi: 10.1371/journal.pone.0192288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149(Pt 12):3473–84. doi: 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- 50.Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80(3):1015–24. doi: 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray GL, Tsyganov K, Kostoulias XP, Bulach DM, Powell D, Creek DJ, et al. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis. 2017;215(suppl_1):S52–7. doi: 10.1093/infdis/jiw529 [DOI] [PubMed] [Google Scholar]
- 52.Koteva K, Hong HJ, Wang XD, Nazi I, Hughes D, Naldrett MJ, et al. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat Chem Biol. 2010;6(5):327–9. doi: 10.1038/nchembio.350 [DOI] [PubMed] [Google Scholar]
- 53.Li L, Wang Q, Zhang H, Yang M, Khan MI, Zhou X. Sensor histidine kinase is a β-lactam receptor and induces resistance to β-lactam antibiotics. Proc Natl Acad Sci USA. 2016;113(6):1648–53. doi: 10.1073/pnas.1520300113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, et al. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci USA. 2013;110(50):20254–9. doi: 10.1073/pnas.1317052110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12:126 doi: 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin MF, Lin YY, Lan CY. The role of the two-component system BaeSR in disposing chemicals through regulating transporter systems in Acinetobacter baumannii. PLoS One. 2015;10(7):e0132843 doi: 10.1371/journal.pone.0132843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6(4):e1000834 doi: 10.1371/journal.ppat.1000834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David SA, Bechtel B, Annaiah C, Mathan VI, Balaram P. Interaction of cationic amphiphilic drugs with lipid A: implications for development of endotoxin antagonists. Biochim Biophys Acta. 1994;1212(2):167–75. [DOI] [PubMed] [Google Scholar]
- 59.Amos H, Vollmayer E. Effect of pentamidine on the growth of Escherichia coli. J Bacteriol. 1957;73(2):172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamm RE, Shull CM Jr., Grant DM. Citrate complexes with iron(II) and iron(III). J Am Chem Soc 1953;76:2111–4. [Google Scholar]
- 61.Pecsok RL, Sandera J. The gluconate complexes. II. The ferric-gluconate system. J Am Chem Soc. 1955;77(6):1489–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The transcriptomic results obtained by RNA-sequencing were validated by qRT-PCR analysis. The level of nine genes that displayed differential expression or remained essentially unchanged between ΔadeRS and WT ATCC 17978 were chosen for comparison. Expression levels for qRT-PCR experiments were corrected to those obtained for GAPDH (ACX60_05065) prior to normalisation against WT ATCC 17978 transcriptional levels. Grey and black bars represent values obtained from RNA-seq and qRT-PCR results, respectively. Differential expression between ΔadeRS and WT ATCC 17978 are given in Log2-values.
(TIF)
Compounds include (a) pentamidine, (b) DAPI and (c) chlorhexidine. For pentamidine and chlorhexidine, the cationic nitrogenous groups are separated by a long carbon chain, forming symmetrical compounds, whereas, DAPI lacks this long linker and is asymmetric.
(TIF)
Respiration of ATCC 17978 ΔadeRS cells in the presence of pentamidine (8, 16, 32, or 64 mg/L) against untreated control cells are shown for (a) Biolog PM01 plates and (b) Biolog PM02A plates. Respiration activity for both plates were monitored in IF-0 (Biolog, Inc.) liquid medium for 72 h at 37°C. The curve in each well represents the colour intensity of a redox-active dye (y axis) over time (x axis: 72 h). Respiration of ΔadeRS cells are shown in red (control), green (under different concentrations of pentamidine), and yellow (depicts the regions of respiratory overlap). Black numbered squares represent carbon sources which decreased resistance to pentamidine (1, D-gluconic acid; 2, citric acid).
(TIF)
Resistance to pentamidine was assessed by disc diffusion assays in Mueller-Hinton agar (MHA) for ATCC 17978 and ΔadeRS, ΔadeA, ΔadeB and ΔadeAB deletion derivatives. Zones of clearing were compared to iron rich conditions from the addition of ferrous sulphate (FeSO4) at the final concentrations of 2.5 and 5 mM and iron-chelated conditions obtained by the addition of 2’,2’ dipyridyl (DIP) at the final concentrations of 100 and 200 μM in MHA. Images displayed are a representative of the typical results obtained.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Relevant data are within the paper and its Supporting Information files. Additionally, RNA-seq data have been deposited in the gene expression omnibus database, accession number GSE102711.