The substrate-modulated, partial unwinding of transmembrane helices may facilitate the release of Na+ and substrate in LeuT.
Abstract
LeuT, a prokaryotic member of the neurotransmitter:sodium symporter (NSS) family, is an established structural model for mammalian NSS counterparts. We investigate the substrate translocation mechanism of LeuT by measuring the solution-phase structural dynamics of the transporter in distinct functional states by hydrogen/deuterium exchange mass spectrometry (HDX-MS). Our HDX-MS data pinpoint LeuT segments involved in substrate transport and reveal for the first time a comprehensive and detailed view of the dynamics associated with transition of the transporter between outward- and inward-facing configurations in a Na+- and K+-dependent manner. The results suggest that partial unwinding of transmembrane helices 1/5/6/7 drives LeuT from a substrate-bound, outward-facing occluded conformation toward an inward-facing open state. These hitherto unknown, large-scale conformational changes in functionally important transmembrane segments, observed for LeuT in detergent-solubilized form and when embedded in a native-like phospholipid bilayer, could be of physiological relevance for the translocation process.
INTRODUCTION
The neurotransmitter: sodium symporter (NSS) family includes prokaryotic and eukaryotic integral membrane proteins that harness the energy stored in the Na+ concentration gradient to transport solutes across the cell membrane (1). Mammalian NSS proteins play an important role in terminating the neurotransmission in the central nervous system by rapid uptake of neurotransmitters against their concentration gradient into the presynaptic neuron or neighboring glial cells (2). These secondary active transporters represent favorable targets for therapeutic drugs that act as potent transport inhibitors to increase neurotransmitter levels at the synaptic junction (1). Despite the importance of mammalian NSS proteins in neurobiology and pharmacology, the molecular mechanisms underlying the transport function have proven difficult to elucidate by traditional methods because the target proteins are unstable in detergent-solubilized form and are difficult to purify to the required quantity and purity.
The prokaryotic NSS protein LeuT from Aquifex aeolicus has emerged as an important structural model to investigate the structure-function relationship of mammalian NSS counterparts (3, 4). High-resolution crystal structures of LeuT in distinct functional states (5–8) have established a structural framework and given rise to mechanistic models depicting the substrate transport mechanism (9, 10). LeuT comprises 12 transmembrane segments (TMs) that are interconnected by relatively short extracellular and intracellular loops (ELs and ILs, respectively). The first TMs 1 to 5 share a similar structural repeat with the following TMs 6 to 10 but are inverted in the membrane plane (5). The specific arrangement of TM domains, also referred to as the “5+5” or LeuT fold (5), has been observed not only in other NSS proteins (11–13) but also in transporters without sequence homology to LeuT (14), suggesting the possibility of a conserved structural scaffold for many secondary active transporters (4). The primary binding site for the substrate (S1) and residues involved in coordination of two sodium ions (Na1 and Na2) is located approximately halfway across the membrane bilayer in the core of the transporter (5). The characteristic unwound regions of TMs 1 and 6 (providing both TMs with an a and a b section) fulfill a dual role in coordinating the sodium ions and forming interactions with the substrate molecule.
According to the widely embraced “alternating access” model (15), secondary active transporters isomerize between distinct functional states in a substrate-dependent manner. That is, the substrate binding site is alternatively exposed to either the intracellular or extracellular aqueous environment. X-ray crystallography provided structures of LeuT in “outward-facing open” (6, 7), “outward-facing occluded” (5), and “inward-facing open” (6) conformations and led to the identification of external and internal gating residues in LeuT and related transporters (16–18). On the basis of these structural snapshots, it has been hypothesized that local and large-scale structural rearrangements are required to regulate the molecular gates and the outward-to-inward transition of the transporter, respectively (6). Combined evidence from crystallographic, functional, and simulation studies suggest that the underlying allosteric couplings are essential for LeuT to function as a symporter (19). Several key aspects of the transport cycle, in particular the molecular mechanism related to the transition of LeuT to the inward-facing open state, have remained controversial and are the subject of extensive debate (4, 20, 21).
Here, we have studied the substrate translocation mechanism of LeuT by measuring the structural dynamics of the protein as a function of time and substrate/ion composition (leucine, Na+, K+, and Cs+) by local hydrogen/deuterium exchange mass spectrometry (HDX-MS). The exchange of hydrogen to deuterium (HDX) of backbone amides in a protein is dependent on the presence and stability of hydrogen bonds and thus provides a sensitive probe for higher-order structure and dynamics of the target protein in solution (22, 23). HDX-MS is a non-perturbing technique that allows the collection of structural dynamics data along the entire protein backbone in a coherent manner without the need for sequence alterations or changes to the covalent structure of the protein for labeling (24). Briefly, the target protein is diluted into deuterated buffer and labeled for various time intervals. The isotopic exchange reaction is quenched by lowering pH and temperature to approximately 2.5 and 0°C, respectively, and the protein is subsequently digested using an acid-stable protease (for example, pepsin). Chromatographic separation and mass analysis of these peptides, in turn, reveal the shift in mass over time (that is, deuterium uptake) of individual regions of the target protein, which is commonly referred to as local HDX analysis (25–27).
Our HDX-MS measurements provide detailed information about the regions in LeuT that undergo conformational changes upon binding biologically relevant ions and the substrate. Of special interest, the HDX data suggest that several helices undergo partial unwinding and that these unfolding events are dynamically coupled between individual helices that form the substrate binding site and the cytoplasmic gate. We envisage that partial unwinding of TM helices accompanies the outward-to-inward isomerization in LeuT and that the same concept might be relevant to structurally related transporters.
RESULTS
Pinpointing LeuT segments involved in substrate transport
For the HDX experiments, we used LeuT expressed in Escherichia coli C41 strain, purified and solubilized in 0.05% dodecyl-β-d-maltoside (DDM). The activity of LeuT was assessed by scintillation proximity assay (SPA), as described previously (28). Affinity for [3H]leucine and Na+ dependency were in agreement with previously published data for LeuT (fig. S1) (28). To examine the molecular mechanism underlying the transport function of LeuT, we tracked changes in deuterium uptake as a function of time and substrate/ion composition by local HDX-MS analyses. We reasoned that transporter segments, in which the protein backbone either becomes more dynamic or exhibits structural stabilization due to the isomerization of LeuT, would display increased and decreased deuterium uptake relative to a defined reference state, respectively. Online pepsin proteolysis yielded a total of 67 LeuT peptides covering 71% of the protein sequence (Fig. 1A) that complied with the requirements to monitor the time-resolved HDX (0.25 to 60 min) across different functional states of the transporter (Fig. 1B).
Fig. 1. Measuring the HDX in distinct LeuT regions by mass spectrometry.
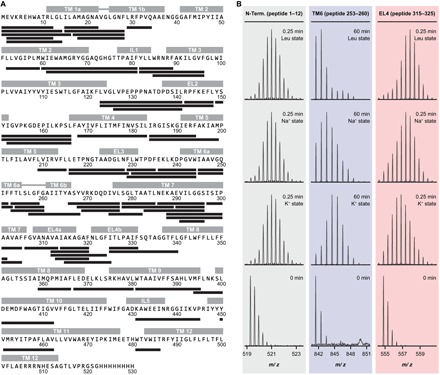
(A) Online pepsin proteolysis yielded a total of 67 LeuT peptides suitable for local HDX-MS analysis. The identified peptides are depicted as black bars and are aligned with the corresponding LeuT sequence. The peptides cover 71% of the protein sequence. Individual structural motifs in LeuT are indicated above the protein sequence in gray color. (B) Representative mass spectra for individual LeuT peptides that cover the N terminus (peptide 1–12), the substrate binding site in TM 6 (peptide 253–260), and EL4 (peptide 315–325). Mass spectra at the 0-min time point derive from unlabeled samples. The isotopic envelopes then shift to higher values on the m/z (mass-to-charge ratio) scale as a function of time (0.25 to 60 min) and ion/substrate composition (that is, K+, Na+, and Leu state) due to deuterium incorporation at the backbone amide position. Binding of Na+ or the combination of Na+ and leucine led to decreased HDX (structural stabilization) in the substrate binding site in TM 6 (peptide 253–260, blue panel) and to increased HDX (structural destabilization) in EL4 (peptide 315–325, red panel) relative to the K+ state. The exchange rate in the N terminus (peptide 1–12, gray panel) was not affected upon binding of Na+ and leucine.
We chose the purification conditions [200 mM KCl, 20 mM tris-HCl (pH 8.0), and 0.05% DDM] for the LeuT reference state and selectively shifted the conformational equilibrium of the transporter from a presumably more inward-facing open conformation (28) toward outward-oriented states of the transport cycle by varying the ion/substrate composition in a similar manner, as described previously (29–31). We thus measured the local HDX in LeuT as a function of time in K+ (200 mM KCl, inward-facing open K+ state), in Na+ (200 mM NaCl, outward-facing open Na+ state), and in the presence of 200 mM NaCl and varying concentrations of leucine (outward-facing occluded Leu state; see the Supplementary Materials).
We first studied the deuterium uptake of LeuT in the K+ state. The earliest measured time point most accurately samples the HDX of fast-exchanging, non–hydrogen-bonded amide hydrogens (22), and the observed deuterium uptake after 0.25 min of labeling correlated well with the expected higher-order structure of LeuT. We observed faster exchange rates in segments encompassing the more flexible loops (fig. S2). In particular, EL4a exchanged rapidly, demonstrating a pronounced dynamic behavior. In contrast, most peptides that cover individual TM helices showed limited deuterium incorporation, consistent with the respective backbone amide hydrogens being engaged in intramolecular hydrogen bonds. One exception is TM 4, which had an exchange rate similar to most loops. Strikingly, several LeuT peptides covering the intracellular halves of TMs 1a/5/7, the substrate binding site in TM 6, and EL4b displayed bimodal isotopic envelopes upon deuteration, demonstrating that these transporter regions coexist in a folded and an unfolded state, which interconvert in solution via slow and correlated unfolding/refolding motions. The structural interpretation of these unusual exchange kinetics (so-called EX1 kinetics) in LeuT is described in detail in a subsequent section.
HDX results for the K+ state then served as a baseline for determining the impact of Na+ and leucine binding on conformation and dynamics in LeuT. Distinct transporter segments including TMs 1a/1b/2/5/6a/6b/7 and interconnecting loops IL1/EL2/EL3/EL4b displayed perturbed HDX in the Na+- and leucine-bound states relative to the K+ state (Fig. 2, A and B). Mapping these differences in HDX onto available LeuT crystal structures revealed that regions exhibiting either structural stabilization or increased structural dynamics upon addition of Na+ or Na+/leucine were arranged in a symmetrical manner relative to the axis of the lipid membrane (Fig. 2, C and D). That is, structural motifs on the extracellular side became more dynamic, whereas substrate binding sites and LeuT segments on the intracellular side exhibited structural stabilization. Furthermore, differences in exchange seemed to be confined to the functionally important four-helix bundle domain (TMs 1/2/6/7) (32) and spatially neighboring transporter regions.
Fig. 2. Pinpointing LeuT segments involved in substrate transport.
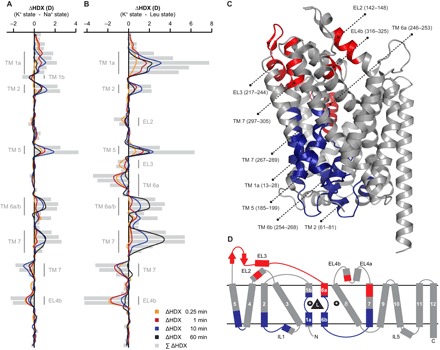
The average relative deuterium uptake of the Na+ state (A) or the Leu state (B) is subtracted from the average value of the K+ state for each peptide and time point. Individual LeuT peptides are plotted on the y axis in an ordered manner starting from the N to the C terminus. Differences in HDX (ΔHDX, colored lines) between the Na+- or Leu-bound state and the K+ state are plotted on the x axis. Positive and negative values indicate increased and decreased HDX in the K+ state, respectively. Values represent means of three independent measurements. Gray bars illustrate the sum of ΔHDX values for all sampled time points. Structural motifs in LeuT, for which we observed Na+- and/or leucine-induced changes in HDX, are annotated along the y axis. (C) Differences in HDX between the Leu state and the K+ state in (B) are mapped onto the LeuT crystal structure (pdb 2A65) displaying a substrate-bound, outward-facing occluded conformation. Red and blue colored regions indicate LeuT segments that became more dynamic (increased HDX) or exhibited structural stabilization (decreased HDX) in the Leu state, respectively. LeuT segments that were not affected upon Na+/substrate binding or for which no HDX data could be obtained are colored in gray. The same color scheme is applied to the topology map of LeuT in (D).
The Na+ and Leu states induced a similar HDX pattern along the protein backbone when analyzed relative to the results for the K+ state (compare Fig. 2A with Fig. 2B). This finding is consistent with the notion that both Na+ and the combination of Na+ and leucine shift the conformational ensemble of LeuT from an inward-facing toward an outward-facing transporter conformation. Despite perturbations in HDX being generally more pronounced for the Leu state, destabilization of EL3 and the TM 6a helix was only observed in the presence of the substrate and represented an apparent dissimilarity between the Na+ and the Leu state. The measured HDX values for the K+, Na+, and Leu states are summarized for each peptide as deuterium uptake plots in fig. S3. Surprisingly, except for the leucine binding site in TM 1a, varying the molar ratio between LeuT and leucine (see the Supplementary Materials) did not affect the exchange rate of individual transporter segments (fig. S4).
Partial unwinding of helices in LeuT
Studying protein dynamics in solution by HDX-MS allows the discovery of cooperative, local unfolding/refolding events along the protein backbone (27, 33, 34), in which the residence time of the unfolded state exceeds hundreds of milliseconds or more (35). These unfolding events are often interpreted as concerted conformational changes involving long-lived perturbations of the secondary structure within a domain. The corresponding peptide mass spectra are characterized by the time-dependent appearance of two distinct mass envelopes upon deuteration that are separated on the m/z scale. The mass envelope at lower m/z values (low-mass population) relates to the protein fraction that has not yet visited the unfolded state. Once multiple residues are simultaneously exposed to deuterated solvent through a local unfolding event, all the respective backbone amide hydrogens undergo correlated exchange (high-mass population) before the region is able to refold (that is, the chemical exchange rate greatly exceeds the rate of refolding). This nongradual increase in average mass, also referred to as the EX1 kinetic exchange regime, directly reports on the rate of unfolding or opening (kop) of the affected residues.
Analysis of the HDX-MS data on LeuT revealed a markedly high number of segments that exchanged according to such an EX1 kinetic regime. These segments were particularly concentrated on the intracellular side of the transporter (Fig. 3, A and B, orange colored segments). The characteristic bimodal isotopic envelopes were consistently observed in all biological replicates (n = 4) and for all studied conditions (K+, Na+, and Leu state). In control experiments using wild-type LeuT embedded in phospholipid bilayer nanodiscs (see the Supplementary Materials), we also detected EX1-type kinetics in the same transporter regions (fig. S5). However, because of the increased sample complexity of nanodisc-embedded LeuT [for example, the presence of phospholipids and nontargeted protein species including membrane scaffold protein (MSP) and porcine pepsin], we could not achieve the same high sequence coverage as for detergent-solubilized LeuT (fig. S6).
Fig. 3. Partial unwinding of individual helices in LeuT.

(A) LeuT segments that exchanged via an EX1 regime or that exhibited signs thereof are colored in orange and yellow in the LeuT crystal structure (pdb 2A65), respectively. The same color scheme is applied to the topology map of LeuT in (B). (C) Representative mass spectra for peptide 184–199, which covers the intracellular half of TM 5, for the K+, Na+, and Leu states and all sampled time points. The bimodal isotopic envelopes can be fitted to a low-mass (blue) and high-mass (red) population. Addition of Na+ or the combination of Na+ and leucine substantially decreased the rate of correlated exchange, that is, the conversion of the low-mass population to the high-mass population, relative to the K+ state. Na+- and leucine-induced stabilization of transmembrane helices was also evident for TMs 1a/6/7, with structural stabilization being consistently more pronounced in the Leu state than in the Na+ state (fig. S7).
For detergent-solubilized LeuT, we observed bimodal mass envelopes in peptides that cover the intracellular halves of TMs 1/5/7, the substrate binding site in TM 6, and EL4b. The correlated exchange in each of these structural motifs was monitored in at least two overlapping peptides and revealed the number of backbone amides that engage in a cooperative unfolding event. Four overlapping peptides covering residues 15–22, 15–28, 17–28, and 18–28 were used to examine the localized unfolding of TM 1. At the earliest measured time point (0.25 min), the average difference in HDX between the low- and high-mass population in peptide 15–28 was 5.6 ± 0.2 D, which corresponds to the correlated exchange of approximately nine backbone amide hydrogens when correcting for the measured partial loss of deuterium during HDX-MS analysis (back exchange; see the Supplementary Materials). It appears that amides undergoing correlated exchange are localized in the N-terminal half of TM 1 as fewer residues underwent correlated exchange in overlapping peptides 17–28 and 18–28 (relative to peptide 15–28). A similar effect is observed for the intracellular half of TM 5 (Fig. 3C). Deconvolution of the bimodal isotopic envelopes for peptides 184–194 and 184–199 revealed the involvement of five to six and seven backbone amides in a cooperative unfolding event, respectively. The increased correlated exchange in peptide 184–199 suggests that the unfolding event is centered on position 194 and neighboring residues of the TM 5 helix. In both TM 6 (peptides 253–259, 253–260, and 254–260) and EL4b (peptides 312–324 and 315–325), approximately four to five residues underwent correlated exchange. The location of the unfolding event in EL4b could be localized to residues 320–325 based on information from overlapping peptides. Finally, we observed distinct bimodal isotopic envelopes in LeuT segments corresponding to the intracellular half of TM 7 (peptides 275–282, 278–285, and 278–289). The average difference in HDX between the low- and high-mass population in peptide 278–289 was 5.7 ± 0.04 D, which corresponds to the concurrent exchange of approximately eight to nine backbone amide hydrogens in the TM 7 helix. Considering that the TM 7 helix starts at residue 276, the measured correlated exchange thus points toward a complete unwinding of the intracellular part of this TM.
We also found distinct signs of EX1 kinetics in the following LeuT segments: EL2 (peptides 123–148 and 142–148), the loop between EL3 and TM 6a (peptides 229–244, 230–244, and 236–244), TM 6a (peptides 245–252 and 245–253), the loop between TM 6b and TM 7 (peptides 266–277, 268–277, and 269–277), and the extracellular part of TM 7 (for example, peptide 297–305). However, although the exchange kinetics were compatible to an EX1 regime, the respective peptide mass spectra covering these individual transporter segments were not amenable to bimodal deconvolution because of various reasons such as insufficient separation of the two mass envelopes on the m/z scale or unfavorable unfolding kinetics. This is also partially illustrated in Fig. 3 (A and B), considering that transporter regions exhibiting signs of correlated exchange (yellow colored segments) are often located proximal in sequence to segments that exchanged according to an EX1 regime.
Strikingly, bimodal isotopic envelopes were solely observed in peptides covering transporter regions that were also conformationally affected upon Na+ and substrate binding (compare Fig. 2C with Fig. 3A). We thus reasoned that local unfolding events, such as the unwinding of transmembrane helices, are directly linked to the substrate translocation mechanism in LeuT and potentially represent the fundamental motions that enable the transporter to isomerize between distinct functional states in an ion- and substrate-dependent manner. The addition of Na+ or the combination of Na+ and leucine substantially lowered the rate of unfolding in TMs 1a/5/6/7 relative to the K+ state (Fig. 3C for TM 5 peptide 184–199 and fig. S7, A to C), with structural stabilization of transmembrane helices being consistently more pronounced in the Leu state than in the Na+ state. This trend was particularly apparent for peptides covering the substrate binding site in TM 6 and the intracellular half of TM 7 because binding of leucine to the transporter virtually abrogated the unwinding of these helices within the sampled time frame (fig. S7, B and C).
On the basis of time-resolved HDX data of regions undergoing correlated exchange (Fig. 4A), it is possible to approximate rate constants for the observed local unfolding events, that is, the rate constant of opening/unfolding (kop) and the half-life of the closed/folded state. We therefore plotted the relative abundance of the low-mass population in the K+ state for peptides amenable to bimodal deconvolution against labeling time (n = 3) and fitted the data points to an exponential decay function, as described previously (35). The results of the nonlinear regression for individual LeuT peptides covering the intracellular halves of TMs 1/5/7 (peptides 15–28, 184–199, and 278–285), the substrate binding site in TM 6 (peptide 253–260), and EL4b (peptide 315–325) are shown in Fig. 4B. The curve fits correlated well with experimental data (R > 0.85) and indicated that the rate of unwinding in individual helices differs by as much as two orders of magnitude (calculated kop values ranged from 0.06 to 0.0007 s−1; Fig. 4C). We further note that the determined kinetic parameters for the spatially neighboring TMs 1a and 5 were highly similar, suggesting the possibility that unwinding events in these LeuT helices are dynamically coupled.
Fig. 4. Extraction of kinetic measures for individual unfolding events in LeuT.

(A) Time-resolved HDX data (K+ state) for different LeuT peptides covering EL4, TMs 1a/5/7, and the substrate binding site in TM 6. The bimodal isotopic envelopes can be fitted to a low-mass (colored) and high-mass (gray) population. (B) The average relative abundance of the low-mass population is plotted against labeling time for each peptide in (A). Values represent means ± SD of three independent measurements. The data points for each peptide are fitted to an exponential decay function (see the Supplementary Materials). Note that each peptide is color-coded as specified in (A). (C) Tabular overview of the calculated kinetic parameters (that is, kop and the half-life of the low-mass population) for each LeuT peptide. The best-fit values are reported together with the values defining the 95% confidence interval (in square brackets). Notably, the calculated kop values for individual unfolding events in LeuT differed by as much as two orders of magnitude.
Impact of potassium on the conformational ensemble of LeuT
Recent experimental evidence (28) supports a critical role of K+ in the LeuT substrate transport mechanism. That is, K+ was shown to bind to LeuT and to promote an outward-closed/inward-facing configuration, thereby inhibiting the Na+-dependent binding of the substrate. This potentially facilitates transport by inhibiting substrate rebinding during the return step of the transporter. We therefore determined the local HDX in LeuT as a function of time and K+ concentration. As negative control, we used Cs+ (200 mM CsCl) to achieve similar ionic strength during labeling in the absence of K+. According to our previous data (28), Cs+ should not interact with LeuT under these conditions and thus allow assessment of the apo state compared to the K+-bound conformation. HDX-MS analysis was conducted in an identical manner as for the K+- and Na+/leucine-bound states (see the Supplementary Materials) and comprised the following conditions: Cs+ state (200 mM CsCl), K+ state (200 mM KCl), and K+High state (800 mM KCl).
On the basis of HDX-MS analysis, LeuT appeared to assume an outward-facing open conformation in the presence of 200 mM CsCl, which is similar to the Na+-bound conformation (compare Fig. 2A with Fig. 5A). We compared the obtained HDX results for LeuT in Cs+ with the K+ state (Fig. 5A) to assess how K+ affected the conformational equilibrium of LeuT. We observed perturbations in HDX for individual LeuT peptides that cover TMs 1a/5/7, the substrate binding site in TM 6, EL2, and EL4 (Fig. 5A). Structural motifs on the extracellular side became less dynamic (EL2 and EL4) in the presence of K+, whereas LeuT segments on the intracellular side (TM 1a and parts of TMs 5 and 7) exhibited structural destabilization. These changes in HDX indicate a K+-dependent closure of the transporter to the extracellular environment. Increasing the K+ concentration to 800 mM appeared to cause a further shift of the LeuT conformational ensemble toward an outward-closed/inward-facing conformation (Fig. 5B). Together, our HDX results for LeuT in the Cs+, K+, and K+High state overall support the notion of a potential role of K+ in the transport cycle, as K+ seemed to selectively shift the conformational ensemble of LeuT in a dose-dependent manner toward an outward-closed/inward-facing conformation.
Fig. 5. Impact of K+ on the conformational ensemble of LeuT.

(A) The average relative deuterium uptake of the K+ state is subtracted from the average value of the Cs+ state for each peptide and time point. Individual peptides are plotted on the y axis in an ordered manner starting from the N to the C terminus. Differences in HDX (ΔHDX, colored lines) between the two states are plotted on the x axis. Positive and negative values indicate increased and decreased HDX in the Cs+ state, respectively. Values represent means of three independent measurements. Gray bars illustrate the sum of ΔHDX values for all sampled time points. Structural motifs in LeuT, for which we observed K+-induced changes in HDX, are annotated along the y axis. (B) Same representation as in (A) for the comparison of the K+ and K+High states. Positive and negative values indicate increased and decreased HDX in the K+ state, respectively. Values represent means of three independent measurements. Notably, (A) and (B) are consistent with a concentration-dependent, K+-induced closure of LeuT to the extracellular environment.
DISCUSSION
Continuing crystallization efforts on LeuT (5–8) and other eukaryotic members of the NSS family (12, 13) have resulted in much needed insight into the structural architecture of this important class of transporters. It has become clear that NSS proteins of evolutionary distant species share a common structural fold and potentially operate via a preserved mechanism of action (4). However, the identified key conformational states of this transporter have not yet fully depicted the structural transitions required for substrate transport (4). For instance, it has remained unclear whether isomerization of LeuT to the inward-facing open conformation is primarily driven by a rotation of the four-helix bundle relative to the scaffold domain (TMs 3 to 5 and 8 to 10) with otherwise little additional conformational change (32) or rather relies on a more sophisticated helix bending mechanism (6). Biophysical techniques such as electron paramagnetic resonance (EPR) spectroscopy (30, 31) as well as various fluorescence resonance energy transfer (FRET)–based strategies (17, 28, 29, 36) have been applied to LeuT and demonstrated the highly dynamic nature of the transporter under different steady-state conditions. However, low-resolution biophysical techniques often require changes to the covalent structure of the target protein that might perturb its function. Furthermore, these techniques are often limited to detecting conformational changes only in specific regions of the protein. Here, we have probed the solution-phase dynamics of LeuT in a wild-type–like background by measuring the local HDX along the entire protein backbone as a function of time and ion/substrate composition.
As evident from Fig. 2C, binding of Na+ or the combination of Na+ and leucine stabilized the intracellular gating region, which implies a reorientation of LeuT toward a more outward-facing conformation, consistent with previous findings from both EPR (30, 31) and FRET spectroscopy studies (17, 28, 29, 36). Opening of LeuT to the extracellular environment was mainly facilitated by structural and dynamical changes in the four-helix bundle and neighboring transporter regions, emphasizing the central role of the bundle domain in substrate transport. In the context of the alternating access model, it furthermore appears that individual TM helices (for example, TMs 1/6/7) underwent hinge-like movements with pivot points located in the corresponding midsections. The increased differences in HDX (ΔHDX) upon leucine binding relative to the Na+-bound state (compare Fig. 2A with Fig. 2B) suggest that LeuT is trapped in a thermodynamically favorable outward-oriented conformation. Thus, the slowed exchange rates of intracellular structural motifs (for example, TM 1a and TM 5) in the Na+/leucine-bound state reflect the increased energy barriers associated with the structural rearrangements that facilitate isotopic exchange in the EX1 regime. Because LeuT predominantly adopts an inward-facing conformation in the K+ reference state (30), we envisaged finding a HDX-related, common denominator between the outward-oriented Na+ and Leu states, which would allow us to pinpoint LeuT segments involved in the outward-to-inward transition. On the basis of the crystal structure of LeuT in an inward-facing open conformation, it was inferred that inward-opening in LeuT requires substantial structural rearrangements including TMs 1/2/5/6/7 and EL4b (6). Comparison of the HDX-MS results obtained for the Na+ state (Fig. 2A) and Leu state (Fig. 2B) supports this notion. TMs 1a/2/5/6b/7 and EL4b were the only regions in LeuT displaying a consistent trend (that is, Na+- and leucine-induced stabilization or destabilization of the protein backbone) for both conditions when compared to the K+ reference state. Strikingly, the same transporter regions, with the exception of TM 2, exchanged according to an EX1 regime, signifying that the intracellular halves of TMs 1/5/7, the substrate binding site in TM 6, and EL4b exhibit unusually slow unfolding/refolding motions in solution. Distinct structural motifs in NSS proteins have previously been suggested to undergo ion- and substrate-dependent changes in secondary structure. For instance, partial unwinding of the TM 5 helix has been reported for the multihydrophobic amino acid transporter MhsT, a prokaryotic member of the NSS family from Bacillus halodurans. X-ray crystal structures of MhsT in an occluded inward-facing state (11) suggest that a conserved GlyX9Pro helix-breaking motif in TM 5 (equivalent to residues Gly190 to Pro200 in LeuT) enables the formation of a solvent pathway for intracellular release of the Na2 ion. Moreover, it has been shown that the higher-order structure in EL4 changes upon binding of monovalent metal ions to LeuT (28).
For a more detailed structural perspective, we mapped the five segments undergoing EX1 exchange (see Fig. 3) onto three crystal structures representing markedly different states in the LeuT transport cycle: a ligand-bound, outward-occluded state (pdb 2A65) (5); an apo inward-open state (pdb 3TT3) (6); and an apo outward-open return state (pdb 5JAE) (7). Inspection of these structures (fig. S8) suggested the implication of most segments in the transport mechanism, either because they exhibited marked conformational change (EL4b, TM 1a, and TM 5) as quantified by root mean square deviation (RMSD) measurements (table S1) or because they were directly in contact with the substrate binding site (TM 1a and TM 6a). For each segment, counting the number of hydrogen bonds lost or modified in the two most different structures indicated to which degree the known crystallographic record can account for the extent of unfolding revealed by EX1 exchange (summary of maximal variations in table S1 and detailed analysis in table S2). The largest variation was found in TM 5 segment 185–199 due to the obvious helix unwinding in structure 3TT3, and the number of hydrogen bonds lost was concomitant with the number of amide groups involved in EX1 exchange. However, it may be argued that the TM 5 conformation in 3TT3 results from direct contact with the antibody that stabilizes the inward-open state in the crystal (6) so that the observed unwinding event may be more similar to that reported for MhsT (11). For all other segments, the hydrogen bond variations among crystal structures cannot account for the magnitude of the observed EX1 exchange, which therefore provides novel evidence for concerted motions in LeuT not captured by available crystal structures. The most striking case is segment 279–289 of TM 7, in which eight to nine amide hydrogens exchange via the EX1 regime, demonstrating a concerted, slow unwinding of a substantial part of a TM helix, which has not previously been implicated in significant structural transitions during the transport cycle.
Further experimental evidence supporting the perception that the observed unwinding of TM helices in LeuT relates to the outward-to-inward transition of the transporter is provided by the fact that binding of Na+ or the combination of Na+ and leucine substantially decreased the unfolding rate of all corresponding TM domains (Fig. 3C and fig. S7, A to C). This observation is consistent with the concept that binding of Na+ and leucine stabilizes an inward-closed state in LeuT, thereby lowering the likelihood of intracellular gate opening (29). By pursuing a FRET-based imaging strategy, Zhao et al. (36) highlighted that the intracellular gating dynamics in LeuT are dampened in the presence of Na+ (about sevenfold) and that binding of leucine potentiates this effect. We observed a highly similar tendency with regard to the Na+- and substrate-induced stabilization of TM helices. Binding of leucine to LeuT virtually abrogated the unwinding of TMs 6 and 7 within the sampled time frame (fig. S7, B and C), demonstrating the ability of leucine to stabilize the Na+-bound complex in the absence of a Na+ concentration gradient. Similarly to TMs 1/5/6/7, we observed structural stabilization upon Na+/leucine binding in TMs 2 and 6b, yet these regions did not exchange via an EX1 regime. We speculate that TMs 2 and 6b may simply undergo inherently faster dynamics, which are not rate-limiting for the transition of LeuT between outward- and inward-facing conformations in a Na+- and substrate-dependent manner. The observation that higher concentrations of the substrate selectively decreased the exchange rate of TM 1a (fig. S4) may be due to direct interactions between leucine and the respective segment. One may speculate that conformational transitions between outward- and inward-facing LeuT states occur at a slower time scale than binding/rebinding of the substrate molecule. Alternatively, it could suggest that an additional low-affinity binding site for leucine, involving sites in TM 1a, is occupied at higher concentrations. Further experiments are needed to clarify this phenomenon.
We proceeded to extract relevant kinetic measures for individual unfolding reactions observed in TMs 1/5/6/7 and EL4 (Fig. 4). We thus studied and established the spatiotemporal relationship between individual unfolding events in LeuT. The calculated kop values indicate that isomerization of LeuT between conformational states relies on an ordered series of local structural changes involving the unwinding of helices. On the basis of the presented HDX data and previously published research findings, we hypothesize that substrate transport in LeuT occurs according to the following model (Fig. 6): LeuT predominantly adopts an outward-facing open conformation under apo state conditions. The obtained HDX data for LeuT in the Cs+ state and recent FRET measurements (28) support this view. The S1 binding pocket is accessible to the extracellular aqueous solution such that Na+ ions can bind to the Na1 and Na2 sites. Notably, we observe, in agreement with other studies (29, 36), that the outward-facing open conformation is further stabilized upon formation of the Na+-bound complex. Subsequent binding of a substrate molecule prompts closure of the extracellular gate, giving rise to an outward-facing occluded LeuT conformation. Structural rearrangements in EL4 (calculated kop: 0.06 s−1) ensure that this loop meets the optimal length requirements to act as an “extracellular lid” (5) in LeuT and simultaneously promote the forward transition in the transport cycle. Computational simulations on the bacterial hydantoin transporter Mhp1 (37), a secondary active transporter bearing the LeuT fold, support this notion. We assume, based on our HDX measurements, that the substrate-bound, outward-facing occluded state represents the thermodynamically most favorable LeuT conformation in the transport cycle. However, unwinding of the intracellular half of TM 5 may allow the formation of a solvent pathway for intracellular release of the Na2 ion, in a similar manner as described for MhsT (11). Concurrent unwinding of the TM 1a helix, which is involved in the coordination of the Na2 ion through backbone interactions at Gly20 and Val23 (5), further facilitates Na+ translocation. We reason that the structural transitions for Na2 release are dynamically coupled, and find that the calculated kop values for TMs 1a and 5 (0.03 and 0.02 s−1, respectively) are in excellent agreement with the previously reported rate constant of conformational change at the inner gate (~0.02 s−1) (29). The highly conserved proline kink in TM 5 and the unwound region in TM 1 may act as hinges, thereby preventing the propagation of structural transitions at the inner gate to the extracellular side of the transporter. Abstraction of the Na2 ion is associated with considerable destabilization of the substrate-bound complex and potentially induces a population shift toward an inward-facing occluded conformation compatible with the LeuT state observed by site-directed fluorescence quenching spectroscopy (17). At this point in the transport cycle, LeuT is primed for intracellular gate opening and substrate release. Our HDX measurements imply that this final isomerization step involves the observed unwinding of both the intracellular half of TM 7 (calculated kop: 0.003 s−1) and the substrate binding site in TM 6 (calculated kop: 0.0007 s−1). In the outward-facing occluded crystal structure (pdb 2A65) (5), TM 7 shields the Na1 ion and the substrate binding site in TM 6 from solvent access from the intracellular side. Consequently, the concerted and large-scale conformational rearrangements in TM 7 and the substrate binding site in TM 6 may represent fundamental motions that enable LeuT to release its cargo toward the intracellular side. Compared to all other regions exhibiting EX1 kinetics, we further note that the unfolding events in TMs 6 and 7 occur at the slowest rates, implicating that these structural transitions are related to substrate release as the rate-limiting step in the transport cycle. In excellent agreement, we find that the determined kop rate constant for unfolding of the substrate binding site in TM 6 (0.0007 s−1) is highly similar to the substrate turnover rate of LeuT in proteoliposomes (kcat ~0.0003 s−1) (38). It is tempting to speculate that the rate of unfolding in the identified EX1 segments as well as the substrate turnover rate of LeuT would be substantially increased under native conditions in the hyperthermophilic A. aeolicus bacterium, which commonly grows at a temperature of approximately 95°C. We envisage that helical interruptions in TM 6 (Ser256 to Gly260) and TM 7 (Gly294) (5) may allow structural transitions in TMs 6 and 7 at the inner gate with minimal perturbation to the higher-order structure on the extracellular side of LeuT. Finally, our HDX measurements support the suggestion that K+ modulates the conformational ensemble of LeuT in a dose-dependent manner by favoring a more outward-closed/inward-facing transporter conformation (28). The functional role and the underlying molecular mechanism of K+-induced conformational change in LeuT, however, remain to be further elucidated.
Fig. 6. Proposed substrate transport mechanism in LeuT.

(1) Under apo state conditions, LeuT preferentially assumes an outward-facing open conformation with relatively high Gibbs free energy. (2) HDX is decreased upon Na+ binding, suggesting stabilization of the protein backbone in basically the same transporter conformation as for apo state, presumably the outward-facing open. (3) According to crystal structures (5), binding of a transportable hydrophobic amino acid (for example, leucine) prompts occlusion of the extracellular vestibule. EL4 and the water-mediated salt bridge between Arg30 and Asp404 prevent water access to the substrate binding site. On the basis of HDX data, it appears that the substrate-bound, outward-facing occluded state represents the thermodynamically most favorable LeuT conformation in the transport cycle. (4) As shown for MhsT (11) and supported by the results herein, the concurrent unwinding of the intracellular halves of TMs 1 and 5 creates a solvent pathway for intracellular release of the Na2 ion, which triggers transition of LeuT to an inward-facing occluded transporter conformation (5). (6) The rates for the observed EX1 kinetics suggest that the release of both the substrate molecule and the Na1 ion is facilitated by a partial unwinding of the intracellular part of TM 7, thereby enabling subsequent isomerization of LeuT to an inward-facing open state (7). Notably, binding of intracellular K+ to LeuT (not indicated in the diagram) potentially prevents substrate rebinding as well as isomerization of LeuT from an inward-facing open (7) to an outward-facing open conformation (1) through a poorly understood return mechanism.
Very recently, Adhikary et al. (39) have reported on the local HDX behavior of LeuT reconstituted into phospholipid-containing bilayer nanodiscs using MS. We have independently performed control experiments on LeuT reconstituted into phospholipid-containing nanodiscs to validate our HDX results for detergent-solubilized LeuT. We observe that individual regions of nanodisc-embedded, wild-type LeuT undergo EX1 exchange with the characteristic and time-dependent interconversion of the low- and high-mass envelopes (see fig. S5). Consequently, our mechanistic inferences (Fig. 6) are supported by HDX-MS measurements of both detergent-solubilized LeuT and LeuT embedded in a native-like phospholipid environment.
In summary, the possibility to study the conformational dynamics of LeuT along the entire protein backbone under different steady-state conditions allowed us to explore the extensive allosteric regulation in LeuT in a coherent and spatiotemporally resolved manner, which revealed novel mechanistic features inherent to the substrate transport mechanism. We propose that isomerization of LeuT relies on the partial unwinding of TMs 1a/5/7 and the substrate binding site in TM 6. Furthermore, we reason that the release of the substrate molecule and both Na+ ions is accomplished by these conformational rearrangements in TMs 1/5/6a/7. In Fig. 6, we provide a rational work hypothesis for the transport mechanism of LeuT by combining our findings from solution-phase HDX measurements with the established structural and functional framework for LeuT and related transporters. Considering that the proposed substrate transport mechanism for LeuT is facilitated by its higher-order structure, we envisage that other transporters bearing the conserved LeuT fold may operate via a similar mechanism of action. Finally, our results show that the HDX-MS technique represents a unique and useful approach to probe the elusive dynamics of transporters. Specifically, the technique appears to hold an unrecognized potential for identification and characterization of slow (that is, millisecond-to-second time scale) fundamental motions that facilitate ion- and substrate-dependent alternating access.
MATERIALS AND METHODS
Chemicals and reagents
All chemicals and reagents were purchased from Sigma-Aldrich and were of the highest grade commercially available unless stated otherwise.
Expression and purification of LeuT
Wild-type LeuT was overexpressed in E. coli C41(DE3) transformed with the pET16b vector encoding C-terminally His-tagged protein (expression plasmid was provided by E. Gouaux, Vollum Institute, Portland, OR). After membrane isolation and solubilization in 1% (w/v) DDM, LeuT was purified in buffer A [20 mM tris-HCl (pH 8.0), 200 mM KCl, 0.05% (w/v) DDM, and 20% (v/v) glycerol] using nickel affinity chromatography, essentially as described previously (28). A brief description of the experimental procedure can be found in the Supplementary Materials.
Expression and purification of membrane scaffold protein MSP1D1
MSP1D1 comprising a polyhistidine tag was overexpressed in E. coli BL21(DE3) cells and purified essentially as described previously (40). A brief description of the experimental procedure can be found in the Supplementary Materials.
Scintillation proximity assay
The activity of purified LeuT was tested by assessing its binding affinity for [3H]leucine and Na+ dependency by SPA, as described previously (28). A brief description of the experimental procedure can be found in the Supplementary Materials.
Hydrogen/deuterium exchange
We induced different functional states of LeuT by varying the ion and substrate composition in the sample solution in a similar manner as described previously (29–31). Except for the Leu state at nonsaturating conditions, different functional states were induced by adding an excess of the respective ion and substrate, and the sample solution was equilibrated for at least 30 min at 25°C before labeling with deuterium oxide. The concentration of tris-HCl [20 mM (pH 8.0)] and DDM [0.05% (w/v)] was kept constant for all samples throughout the labeling workflow. The corresponding sample preparation is described in more detail in the Supplementary Materials. The sample preparation procedure for nanodisc-embedded LeuT is entirely described in the Supplementary Materials.
Subsequent to the equilibration with ions and the substrate, HDX was initiated by diluting LeuT 10-fold in deuterated buffer, and the samples were labeled at a constant temperature of 25°C for the indicated time intervals (0.25 to 60 min). All labeling buffers contained 20 mM tris-HCl (pH 8.0) and 0.05% (w/v) DDM. The labeling buffers were supplemented with the corresponding salt needed to maintain equilibration conditions. Ice-cold quench buffer [220 mM phosphate buffer (pH 2.3) and 6 M urea] was added in equal volume to the sample solution at the indicated time points to inhibit the isotopic exchange reaction. Quenched protein samples were immediately frozen and stored at −80°C until further use. LeuT samples, in which the ion and substrate composition matched the purification conditions and in which LeuT was labeled for 72 hours, served as equilibrium-labeled control samples. The equilibrium-labeled control samples could not be prepared using a chemical denaturant due to protein aggregation. In turn, we observed that the deuterium uptake for the majority of identified LeuT peptides did not further increase from 60 min of labeling to 72 hours. Therefore, we used a 72-hour sample as a simulated equilibrium-labeled control, noting that deuteration for some peptides from TMs appeared to be incomplete even after such extended incubation times.
Liquid chromatography and MS
To obtain local HDX information, approximately 80 pmol of labeled LeuT was loaded onto a refrigerated (0°C) ultraperformance liquid chromatographic system (nanoACQUITY HDX technology, Waters) coupled to a hybrid Q-ToF Synapt G2 mass spectrometer (Waters). Online proteolysis was accomplished at pH 2.5, 20°C, and a constant flow rate of 200 μl/min by using an in-house packed pepsin column (IDEX) containing immobilized pepsin on agarose resin beads (Thermo Scientific Pierce). The use of rhizopuspepsin (also known as protease XVIII) as well as nepenthesin I and nepenthesin II from tropical pitcher plants was unfavorable when compared to the traditional pepsin digestion and did not result in increased or complementary sequence coverage for LeuT. The resulting peptide mixture was separated by reversed-phase liquid chromatography (see the Supplementary Materials for details) and eluted into a Synapt G2 mass spectrometer, which was interfaced with an electrospray ionization source and operated in positive ionization mode. Human Glu-Fibrinopeptide B (Sigma-Aldrich) served as an internal standard and was acquired throughout the analysis. To minimize spectral overlap, ions were further separated in the gas phase using ion mobility. The ion mobility cell was operated at a wave velocity of 600 m/s, a wave height of 40 V, and a constant nitrogen gas flow of 90 ml/min. Peptides were identified by data-independent MS/MS analysis (MSE) of LeuT (approximately 100 pmol per injection), with the ion mobility cell turned off using collision-induced dissociation and subsequent database searching in the PLGS 3.0 software.
Peptide identification criteria and HDX data analysis
Peptide hits were filtered according to fragmentation quality (minimum fragmentation products per amino acid: 0.2), mass accuracy (maximum MH+ error: 10 parts per million), and reproducibility (peptide identification in 50% of MSE runs) before their integration into HDX analysis. HDX-MS data were processed in the DynamX 3.0 software, and all peptide assignments were manually verified. Noisy and overlapping HDX data were discarded from HDX analysis. Back exchange (BE) was calculated for each peptide on the basis of average values of at least three independent measurements according to the following equation
where m90% is the measured average mass of the peptide for the equilibrium-labeled control, m0% is the average mass of the unlabeled peptide, and mmax is the theoretical average mass of the equilibrium-labeled peptide. We considered that the N-terminal residue and all proline residues in a given peptide do not contribute to the measured deuterium uptake. The overall average deuterium back exchange level in the local HDX-MS setup was calculated to be 38 ± 9%. We thereby only considered LeuT peptides that did not display a measurable difference in HDX between the 60-min time point and the equilibrium-labeled control sample.
Peptides exhibiting bimodal isotopic envelopes upon deuteration were further analyzed using HX-Express 2.0. A brief description of the analysis procedure for peptides displaying EX1 kinetics can be found in the Supplementary Materials.
Supplementary Material
Acknowledgments
We would like to thank J. S. Mortensen and L. Rosenquist from the Molecular Neuropharmacology and Genetics Laboratory, University of Copenhagen, for their help with the SPA measurements and the purification of LeuT. We thank I. R. Möller from the Department of Pharmacy, University of Copenhagen, for fruitful scientific discussions. We acknowledge the Protein Modeling Facility (PMF) of the Swiss Institute of Bioinformatics for structural bioinformatics. Funding: This work was made possible through support from the Danish Council for Independent Research (0602-02740B to K.D.R. and 0602-02100B and 4183-00581 to C.J.L.) and by the University of Copenhagen bioSYNergy Center of Excellence (C.J.L.). Modeling work by M.A.C. was supported by resources from the PMF, University of Lausanne, Switzerland. Author contributions: K.D.R. and C.J.L. conceived and supervised the work. P.S.M. and K.G. produced recombinant LeuT. P.S.M. performed the HDX-MS and SPA experiments. P.S.M. and K.D.R. analyzed the HDX-MS data. C.J.L., K.D.R., and U.G. contributed with equipment, reagents, and knowhow. M.A.C. performed the structural analyses. P.S.M., C.J.L., and K.D.R. prepared the manuscript, and all authors read and commented on it. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar6179/DC1
Supplementary Materials and Methods
fig. S1. Na+-dependent binding of [3H]leucine to purified LeuT.
fig. S2. Correlation between the measured HDX and secondary structure elements in LeuT.
fig. S3. Deuterium uptake plots for detergent-solubilized LeuT.
fig. S4. Impact of different leucine concentrations on local HDX rates in LeuT.
fig. S5. Partial unwinding of individual helices in LeuT reconstituted into phospholipid-containing bilayer nanodiscs.
fig. S6. Sequence coverage map for LeuT reconstituted into phospholipid-containing bilayer nanodiscs.
fig. S7. Na+- and substrate-induced stabilization of TM helices.
fig. S8. Compared conformations of EX1 segments in crystal structures.
table S1. RMSD and hydrogen bond variations between three x-ray structures of LeuT for the EX1 segments.
table S2. Detailed hydrogen bond analysis in three x-ray structures of LeuT for backbone amide groups in the EX1 segments.
REFERENCES AND NOTES
- 1.Kristensen A. S., Andersen J., Jørgensen T. N., Sørensen L., Eriksen J., Loland C. J., Strømgaard K., Gether U., SLC6 neurotransmitter transporters: Structure, function, and regulation. Pharmacol. Rev. 63, 585–640 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Torres G. E., Gainetdinov R. R., Caron M. G., Plasma membrane monoamine transporters: Structure, regulation and function. Nat. Rev. Neurosci. 4, 13–25 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Krishnamurthy H., Piscitelli C. L., Gouaux E., Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loland C. J., The use of LeuT as a model in elucidating binding sites for substrates and inhibitors in neurotransmitter transporters. Biochim. Biophys. Acta 1850, 500–510 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E., Crystal structure of a bacterial homologue of Na+/Cl–-dependent neurotransmitter transporters. Nature 437, 215–223 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy H., Gouaux E., X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 481, 469–474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malinauskaite L., Said S., Sahin C., Grouleff J., Shahsavar A., Bjerregaard H., Noer P., Severinsen K., Boesen T., Schiøtt B., Sinning S., Nissen P., A conserved leucine occupies the empty substrate site of LeuT in the Na+-free return state. Nat. Commun. 7, 11673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Elferich J., Gouaux E., Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat. Struct. Mol. Biol. 19, 212–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Focke P. J., Wang X., Larsson H. P., Neurotransmitter transporters: Structure meets function. Structure 21, 694–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E., A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinauskaite L., Quick M., Reinhard L., Lyons J. A., Yano H., Javitch J. A., Nissen P., A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat. Struct. Mol. Biol. 21, 1006–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penmatsa A., Wang K. H., Gouaux E., X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503, 85–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman J. A., Green E. M., Gouaux E., X-ray structures and mechanism of the human serotonin transporter. Nature 532, 334–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer P. L., Goehring A., Shankaranarayanan A., Gouaux E., Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jardetzky O., Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966). [DOI] [PubMed] [Google Scholar]
- 16.Beuming T., Kniazeff J., Bergmann M. L., Shi L., Gracia L., Raniszewska K., Newman A. H., Javitch J. A., Weinstein H., Gether U., Loland C. J., The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat. Neurosci. 11, 780–789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billesbølle C. B., Krüger M. B., Shi L., Quick M., Li Z., Stolzenberg S., Kniazeff J., Gotfryd K., Mortensen J. S., Javitch J. A., Weinstein H., Loland C. J., Gether U., Substrate-induced unlocking of the inner gate determines the catalytic efficiency of a neurotransmitter:sodium symporter. J. Biol. Chem. 290, 26725–26738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kniazeff J., Shi L., Loland C. J., Javitch J. A., Weinstein H., Gether U., An intracellular interaction network regulates conformational transitions in the dopamine transporter. J. Biol. Chem. 283, 17691–17701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeVine M. V., Cuendet M. A., Khelashvili G., Weinstein H., Allosteric mechanisms of molecular machines at the membrane: Transport by sodium-coupled symporters. Chem. Rev. 116, 6552–6587 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Piscitelli C. L., Krishnamurthy H., Gouaux E., Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature 468, 1129–1132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi L. L., Quick M., Zhao Y., Weinstein H., Javitch J. A., The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skinner J. J., Lim W. K., Bédard S., Black B. E., Englander S. W., Protein dynamics viewed by hydrogen exchange. Protein Sci. 21, 996–1005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skinner J. J., Lim W. K., Bédard S., Black B. E., Englander S. W., Protein hydrogen exchange: Testing current models. Protein Sci. 21, 987–995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Smith D. L., Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engen J. R., Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 81, 7870–7875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.P. F. Jensen, K. D. Rand, Hydrogen exchange: A sensitive analytical window into protein conformation and dynamics, in Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods, and Applications, D. D. Weis, Ed. (Wiley, 2016). [Google Scholar]
- 27.Konermann L., Pan J., Liu Y.-H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Billesbølle C. B., Mortensen J. S., Sohail A., Schmidt S. G., Shi L., Sitte H. H., Gether U., Loland C. J., Transition metal ion FRET uncovers K+ regulation of a neurotransmitter/sodium symporter. Nat. Commun. 7, 12755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y., Terry D., Shi L., Weinstein H., Blanchard S. C., Javitch J. A., Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465, 188–193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claxton D. P., Quick M., Shi L., de Carvalho F. D., Weinstein H., Javitch J. A., McHaourab H. S., Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 17, 822–829 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazmier K., Sharma S., Quick M., Islam S. M., Roux B., Weinstein H., Javitch J. A., Mchaourab H. S., Conformational dynamics of ligand-dependent alternating access in LeuT. Nat. Struct. Mol. Biol. 21, 472–479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrest L. R., Zhang Y.-W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G., Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferraro D. M., Lazo N., Robertson A. D., EX1 hydrogen exchange and protein folding. Biochemistry 43, 587–594 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Weis D. D., Wales T. E., Engen J. R., Hotchko M., Ten Eyck L. F., Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 17, 1498–1509 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Trelle M. B., Madsen J. B., Andreasen P. A., Jørgensen T. J. D., Local transient unfolding of native state PAI-1 associated with serpin metastability. Angew. Chem. Int. Ed. Engl. 53, 9751–9754 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y., Terry D. S., Shi L., Quick M., Weinstein H., Blanchard S. C., Javitch J. A., Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature 474, 109–113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H. D., Zhu F., Conformational changes in two inter-helical loops of Mhp1 membrane transporter. PLOS ONE 10, e0133388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh S. K., Yamashita A., Gouaux E., Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448, 952–956 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Adhikary S., Deredge D. J., Nagarajan A., Forrest L. R., Wintrode P. L., Singh S. K., Conformational dynamics of a neurotransmitter:sodium symporter in a lipid bilayer. Proc. Natl. Acad. Sci. U.S.A. 114, E1786–E1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasr M. L., Singh S. K., Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay. Biochemistry 53, 4–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar6179/DC1
Supplementary Materials and Methods
fig. S1. Na+-dependent binding of [3H]leucine to purified LeuT.
fig. S2. Correlation between the measured HDX and secondary structure elements in LeuT.
fig. S3. Deuterium uptake plots for detergent-solubilized LeuT.
fig. S4. Impact of different leucine concentrations on local HDX rates in LeuT.
fig. S5. Partial unwinding of individual helices in LeuT reconstituted into phospholipid-containing bilayer nanodiscs.
fig. S6. Sequence coverage map for LeuT reconstituted into phospholipid-containing bilayer nanodiscs.
fig. S7. Na+- and substrate-induced stabilization of TM helices.
fig. S8. Compared conformations of EX1 segments in crystal structures.
table S1. RMSD and hydrogen bond variations between three x-ray structures of LeuT for the EX1 segments.
table S2. Detailed hydrogen bond analysis in three x-ray structures of LeuT for backbone amide groups in the EX1 segments.


