Abstract
Poly(ADP-ribose) polymerases (PARPs) and sirtuins (SIRTs) are involved in the regulation of cell metabolism, transcription, and DNA repair. Alterations of these enzymes may play a crucial role in Alzheimer’s disease (AD). Our previous results indicated that amyloid beta (Aβ) peptides and inflammation led to activation of PARP1 and cell death. This study focused on a role of PARP1 in the regulation of gene expression for SIRTs and beta-amyloid precursor protein (βAPP) cleaving enzymes under Aβ42 oligomers (AβO) toxicity in pheochromocytoma cells (PC12) in culture. Moreover, the effect of endogenously liberated Aβ peptides in PC12 cells stably transfected with human gene for APP wild-type (APPwt) was analyzed. Our results demonstrated that AβO enhanced transcription of presenilins (Psen1 and Psen2), the crucial subunits of γ-secretase. Aβ peptides in APPwt cells activated expression of β-secretase (Bace1), Psen1, Psen2, and Parp1. The inhibitor of PARP1, PJ-34 in the presence of AβO upregulated transcription of α-secretase (Adam10), Psen1, and Psen2, but also Bace1. Concomitantly, PJ-34 enhanced mRNA level of nuclear Sirt1, Sirt6, mitochondrial Sirt4, and Parp3 in PC12 cells subjected to AβOs toxicity. Our data indicated that Aβ peptides through modulation of APP secretases may lead to a vicious metabolic circle, which could be responsible for maintaining Aβ at high level. PARP1 inhibition, besides activation of nuclear SIRTs and mitochondrial Sirt4 expression, enhanced transcription of enzyme(s) involved in βAPP metabolism, and this effect should be considered in its application against Aβ peptide toxicity.
Keywords: Poly(ADP-ribose) polymerases, Sirtuins, Amyloid, Alzheimer’s disease, APP, PARPs, SIRTs, Secretases, Neuroprotection
Introduction
Among all neurodegenerative disorders, Alzheimer’s disease (AD) is the most severe dementia that progressively and irreversibly impairs synaptic function and cognition [1–3]. Until now, the pathogenesis and pathomechanisms of AD is unknown, and no therapy to date has proven to be successful. For the last two decades, the amyloid hypothesis dominated in research field on AD [4–6].
Although the hypothesis of “pathogenic spread” of amyloid beta (Aβ) peptides in neurodegenerative disease has been widely suggested, recent studies have provided evidences for human transmission of amyloid β pathology and cerebral amyloid angiopathy [7, 8].
Aβ peptides are released from beta-amyloid precursor protein (βAPP) which is a membrane protein involved in regulation of synapse formation, neuronal growth, and repair [9, 10]. βAPP is metabolized by two separate pathways: non-amyloidogenic and amyloidogenic. In the non-amyloidogenic pathway, α- and γ-secretases are responsible for βAPP degradation to peptide p3 and αAPP. Moreover, several zinc metalloproteinases, including ADAM10, can mediate βAPP cleavage at α-secretase sites [11].
On the contrary, in the amyloidogenic pathway, βAPP is degraded at β-site by beta secretase (BACE1) and then by γ-secretase to yield Aβ peptides [12, 13]. γ-secretase is a tetrameric protein complex with presenilins (PSEN1 and PSEN2) as crucial components involved in processing at both α- and β- C-terminal fragment sites [14, 15]. The most common causes of familial AD are mutations of genes encoding presenilins 1 and 2 and β-secretase which lead to increased production of highly amyloidogenic Aβ42 isoform [16, 17].
Currently, among all Aβ forms oligomers (AβO) are suggested to be the most toxic and correlated with dementia [18]. Several studies have indicated that the most toxic are Aβ peptide dimers and probably trimers isolated from Alzheimer brains, and this molecular species may impair memory [19]. Independently of which types of Aβ oligomers are responsible for disrupting cognition, the question arises—what kind of molecular processes altered by Aβ peptides are responsible for dementia and cell loss and how to protect cells against Aβ toxicity?
It is generally accepted that oxidative stressors play a crucial role in pathomechanism of neurodegenerative diseases. Oxidative stress evoked by Aβ peptides may lead to disturbance of many key processes involved in regulation of cell viability and death [20, 21]. Mitochondria were recently indicated to be the target for APP that accumulates in the mitochondrial import channels and for Aβ that interacts with several proteins inside mitochondria [22–30]. Among several pro- and anti-oxidative enzymes, the most important stress response proteins are both NAD+ dependent enzymes families such as the following: sirtuins (SIRTs) and poly(ADP-ribose) polymerases (PARPs). These enzymes depending on stress conditions may exert neuroprotective effect or may lead to cells degeneration and death. The relationship between PARPs and SIRTs may play important role in regulation of cells’ fate [31–33].
SIRTs are a family of seven enzymes that are deacetylases [type III histone deacetylases (HDAC’s)], and some of them are ADP-ribosyltransferases. Three of them (SIRT3, SIRT4, SIRT5) primarily exist in mitochondria. The mitochondrial SIRTs can modulate the activity of antioxidative enzymes, and they can protect cell against oxidative stress by direct stimulation of expression and activity of Mn superoxide dismutase (SOD2) or indirectly by activation of glutathione peroxidase (GPx) and by enhancing mitochondrial electron transport activity [34, 35]. Mitochondrial SIRT3 and nuclear SIRT1 are the most well-studied SIRTs. They are also widely expressed in brain and other tissues that have high metabolic activity [36–39]. Sirtuins can interact with PARPs, and all these enzymes are involved in regulation of transcription, DNA repair, cell metabolism, survival, and death. PARPs are capable transfer and polymerization of ADP-ribose units using NAD+ molecules. PARP activity results in the attachment of linear or branched polymers of ADP-ribose to target proteins (heteromodification) or to PARP itself (automodification). PARP1 is responsible for over 90% of poly(ADP-ribosylation) in the brain [40–42]. Among the family, PARP1, PARP2, and PARP3 have been found to be induced by DNA strand breaks, and their role in DNA repair process has been reviewed [43, 44]. In AD and other neurodegenerative disorders, accumulation of Aβ in the central nervous system may induce neuronal cell death accompanied by overexpression of PARP1 [30, 45–50]. Overactivation of PARP1 may lead to the NAD+ depletion, followed by inhibition of SIRT1 activity. Another recent study revealed protective effects of PARP1 inhibitors on oxidative stress and mitochondrial integrity in an ex vivo model of AD induced by Aβ42 [51]. Moreover, in response to sustained stress, PARP1 can be overactivated and in consequence may alter the function of many transcription factors and other proteins including the following: p53, NF-κB, AIF, HIF-1, FOXO1, Bax, and Bcl-2 [48, 49, 52–54]. The last data of Martire et al. [55] demonstrated that PARP1 inhibition rescued metabolic dysfunction, bioenergetics impairment, and restored inhibition of pyruvate kinase 2 expression in animal and cellular models of AD. Despite intensive studies during the last decade, the role of PARP1 in the regulation of nuclear and mitochondrial SIRTs and enzymes responsible for APP metabolism is not fully understood and elucidated.
In this study, we investigated the effect of PARP1 inhibition on gene expression of enzymes responsible for stress response such as NAD+ dependent nuclear Sirt1, Sirt6, cytosolic Sirt2, and mitochondrial SIRTs (Sirt3, Sirt4, Sirt5) in Aβ toxicity. Moreover, for a better understanding of the role of poly(ADP-ribosylation) in βAPP metabolism, the effect of PARP inhibitor on gene expression of βAPP degrading enzymes was analyzed.
Materials and Methods
Preparation of Aβ1–42 Oligomers
Aβ 1–42 (rPeptide no. A-1163-2) oligomers were prepared according to Stine et al. [56], as described previously by Cieslik et al. [20]. Amyloid β was dissolved (5 mM Aβ 1–42) in anhydrous DMSO and then diluted to 100 μM concentration with ice-cold cell culture medium (phenol red-free Ham’s F-12; BioSource), immediately vortexing for 30 s, and then incubated at 4 °C for 24 h. The same procedure was used for preparation of Aβ scrambled 1-42 (Aβscr) (rPeptide no. A-1004-2). Conformation state of Aβ was confirmed using Thioflavin T (ThT) which is benzothiazole dye binding to amyloid fibrils. Electrophoretic analysis of used Aβ42 followed by silver staining was performed.
Cell Culture and Cell Treatment Protocol
Rat pheochromocytoma (PC12) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 5% heat-inactivated horse serum (HS), 2 mM L-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin in a 5% CO2 atmosphere at 37 °C.
Moreover, PC12 cells transfected with human gene for APP wild-type (APPwt) was used. As a control, we used cells transfected with an empty vector. These cells were a kind gift from Prof Dr. W. E. Mueller and Prof A. Eckert from the University of Frankfurt and Basel. Transfected PC12 cells APPwt and control were cultured in the same medium as mentioned before with addition of selective antibiotic 400 μg/ml G-418 sulfate. Cell treatment was performed in serum-free Neurobasal medium supplemented with B27 (Gibco), 1% penicillin/streptomycin, and 2 mM L-glutamine in order to stop proliferation of cells. Then, the PC12 cells were treated with 1 μM Aβ 1–42 oligomers (AβO) and (20 μM) PJ-34 for 24 h.
The procedure of cell transfection was described previously [57], and these APP transfected cells were used by us in several studies [26, 58–60]. Moreover, APP transfected cells were investigated under EM [61].
Determination of Cell Survival Using the MTT Test
Cell viability was evaluated by reduction of 2-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT) to formazan. After 24 h of treatment with the tested compounds, MTT (2.5 mg/ml) was added to the wells. The cells were incubated at 37 °C for 2 h. Then, the medium was removed, the formazan crystals were dissolved in 150 μL DMSO, and absorbance was measured at 595 nm [20].
Quantitative Real-Time PCR Assays
PC12 cells were washed twice with ice-cold PBS and suspended in 1 ml of TRI reagent (Sigma-Aldrich). RNA was isolated from the cell pellet according to the manufacturer’s protocol. Digestion of DNA contamination was performed by using DNase I according to the manufacturer’s protocol (Sigma-Aldrich). Reverse transcription was performed using a High Capacity cDNA Reverse Transcription Kit as described in the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). The level of mRNA for selected genes was analyzed using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. β-actin (Actb) was selected and used in all of the studies as a reference gene. Quantitative PCR was performed using an Applied Biosystems 7500 Real-Time PCR System. The following genes were analyzed: Sirt1 Rn01428096_m1, Sirt2 Rn01457502_m1, Sirt3 Rn01501410_m1, Sirt4 Rn01481485_m1, Sirt5 Rn01450559_m1, Sirt6 Rn01408249_m1, Parp1 Rn00565018_m1, Parp2 Rn01414610_m1, Parp3 Rn01447502_m1, Adam10 Rn01530753_m1, Bace1 Rn00569988_m1, Psen1 Rn00569763_m1, and Psen2 Rn00579412_m1. As a reference, we used Actb gene Rn00667869_m1. The relative level of mRNA was calculated using the ∆∆Ct method.
Statistical Analysis
All experiments were repeated at least three times and were performed either in triplicate or duplicate. The presented data are the means ± SEM. For statistical comparison, Student’s t test or one-way ANOVA followed by Newman-Keuls post hoc test were used. P values < 0.05 were considered statistically significant (*−p < 0.05; **−p < 0.01; ***−p < 0.001). The statistical analyses were performed by using Graph Pad Prism version 6.0 (Graph Pad Software, San Diego, CA, USA).
Results
In the present study, we have investigated the role of PARP1 in regulation/alterations of gene expression for stress responses proteins such as NAD+ dependent SIRTs and DNA bound PARPs in PC12 cells under Aβ toxicity. Moreover, the effect of PARP1 on gene expression of APP metabolizing enzymes which regulate Aβ concentration was analyzed. The effect of exogenous Aβ42 on the transcription of these enzymes was compared with the action of endogenously liberated Aβ peptides in PC12 cells with overexpressed human APP.
Description of Aβ42 Oligomers (AβO) and Their Role in Cell Viability
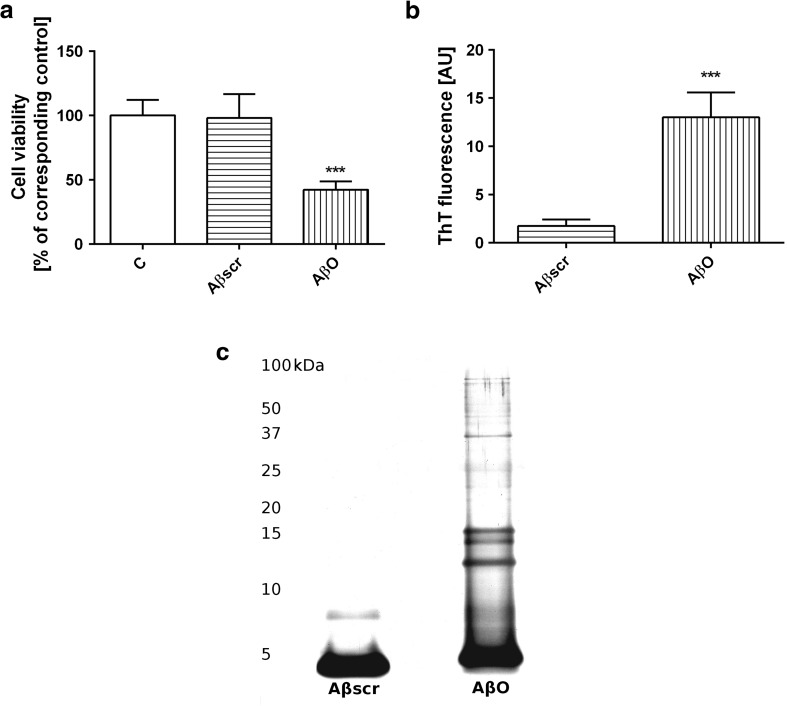
First of all, we have investigated whether Aβ42 in oligomeric form (1 μM) affects PC12 cells survival. Our data demonstrated that AβO decreased cell viability by about 60% compared to the control: non-treated cells and Aβ scrambled (Aβscr) peptide (Fig. 1a). We observed that Aβ42 oligomers enhanced ThT fluorescence, but such effect was not seen with Aβscr (Fig. 1b). Moreover, Aβ oligomers were separated using denaturing SDS-PAGE followed by silver staining. The results have shown the presence of Aβ oligomers, trimers, and tetramers and only small amount of polymers (Fig. 1c).
Fig. 1.
The effect of Aβ oligomers on PC12 cells viability. The effect of oligomeric Aβ42 or scrambled Aβ at 1 μM concentration on PC12 cell viability after 24-h treatment (a). Thioflavin (ThT) fluorescence analysis of the Aβ42 scrambled and Aβ42 in oligomeric form (b). Proteins were subjected to denaturing SDS–PAGE followed by silver staining. Electrophoretic analysis of the Aβ42 scrambled and Aβ42 in oligomeric form (AβO) (c). For statistical comparison, Student’s t test or one-way analysis of variance (ANOVA) with Neuman-Keuls post hoc test was used. P values <0.05 were considered statistically significant (***−p < 0.001)
AβO and PARP-1 Inhibitor Alter Transcription of Sirtuins and DNA-Bound PARPs
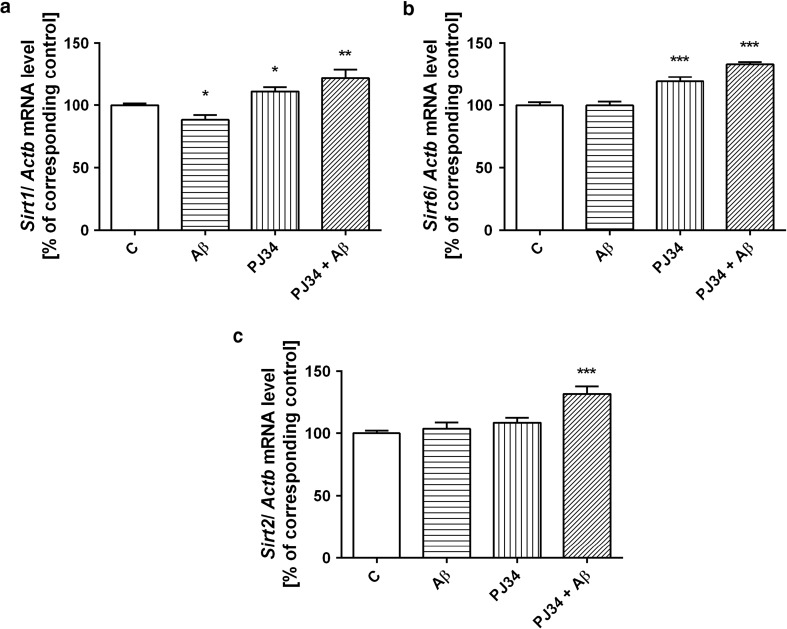
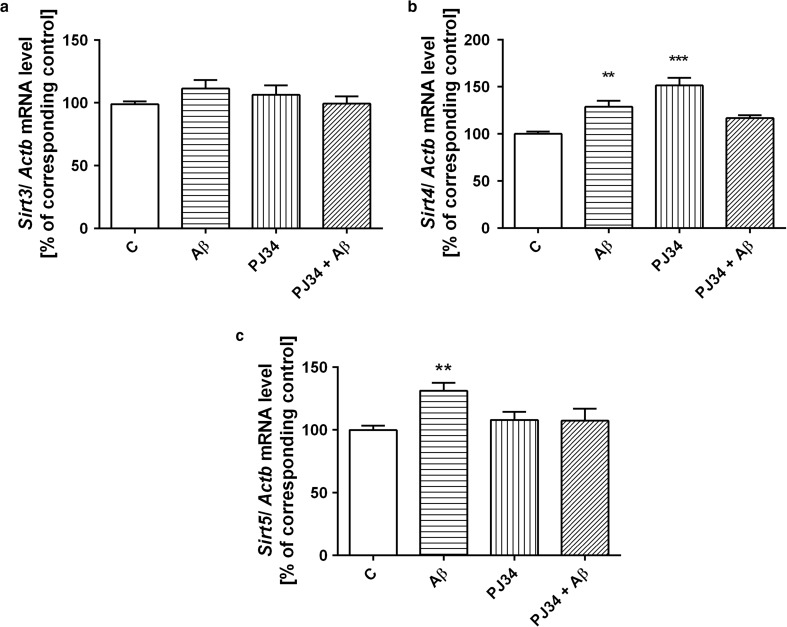
In the following experiments, the pharmacological inhibition of PARP1 (using specific inhibitor PJ-34) significantly activated transcription of Sirt1 and Sirt6 (Fig. 2a, b). AβO significantly decreased Sirt1 expression (Fig. 2a). AβO and PJ-34 acting separately had negligible stimulatory effect on cytosolic Sirt2 expression, but PJ-34 acting in the presence of AβO exerted significant activation on Sirt2 (Fig. 2c). AβOs influenced transcription of mitochondrial SIRTs with one exception of Sirt3 where negligible stimulatory effect was observed (Fig. 3a). AβOs significantly enhanced gene expression of mitochondrial Sirt4 and Sirt5 (Fig. 3b, c). Moreover, PJ-34 upregulated gene expression of Sirt4 (Fig. 3b). The further study demonstrated that AβOs had no significant effect on gene expression of Parp1 and other DNA bound PARPs (Table 1). Inhibition of PARP1 activity led to activation of gene expression of Parp3 in the absence and presence of AβO (Table 1).
Fig. 2.
The effect of oligomeric Aβ42 and PJ-34 treatment on gene expression of nuclear and cytosolic sirtuins. PC12 cells were incubated in the presence of oligomeric Aβ42 (AβO, 1 μM) and PJ-34 (20 μM) for 24 h (a–c). The levels of mRNA of nuclear Sirt1 (a), Sirt6 (b), and cytosolic Sirt2 (c) were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments with 3–4 replications. ***p < 0.001; **p < 0.01; *p < 0.05—the difference, that was statistically significant compared to the control cells, using one-way analysis of variance (ANOVA) with Neuman-Keuls post hoc test
Fig. 3.
The influence of PARP1 inhibitor PJ-34 and oligomeric Aβ42 treatment on gene expression of mitochondrial sirtuins. PC12 cells were incubated in the presence of oligomeric Aβ42 (AβO, 1 μM) and PJ-34 (20 μM) for 24 h (a–c). The levels of mRNA of mitochondrial sirtuins—Sirt3 (a), Sirt4 (b), and Sirt5 (c)—were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments with three replications. ***p < 0.001; **p < 0.01—the difference, that was statistically significant compared to the control cells, using one-way analysis of variance (ANOVA) with Neuman-Keuls post hoc test
Table 1.
The effect of Aβ42 oligomers and PARP1 inhibitor on the level of mRNA of DNA bound PARPs
| Gene | mRNA level of DNA bound PARPs/Actb [% of control] | ||
|---|---|---|---|
| Aβ | PJ-34 | Aβ + PJ-34 | |
| Parp1 | 120.6 ± 9.10 | 106.40 ± 2.75 | 115.10 ± 0.95 |
| Parp2 | 109.40 ± 5.31 | 100.40 ± 3.96 | 109.30 ± 6.23 |
| Parp3 | 98.94 ± 5.60 | 118.5 ± 0.70* | 130.00 ± 5.82* |
PC12 cells were incubated in the presence of oligomeric Aβ42 (AβO, 1 μM) and PJ-34 (20 μM) for 24 h. The levels of mRNA of DNA bound PARPs—Parp1, Parp2, and Parp3—were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments with three replications. *p < 0.05—the difference, that was statistically significant compared to the control cells, using one-way analysis of variance (ANOVA) with Neuman-Keuls post hoc test
AβO and PARP-1 Inhibitor Affect Genes Expression Involved in APP Metabolism
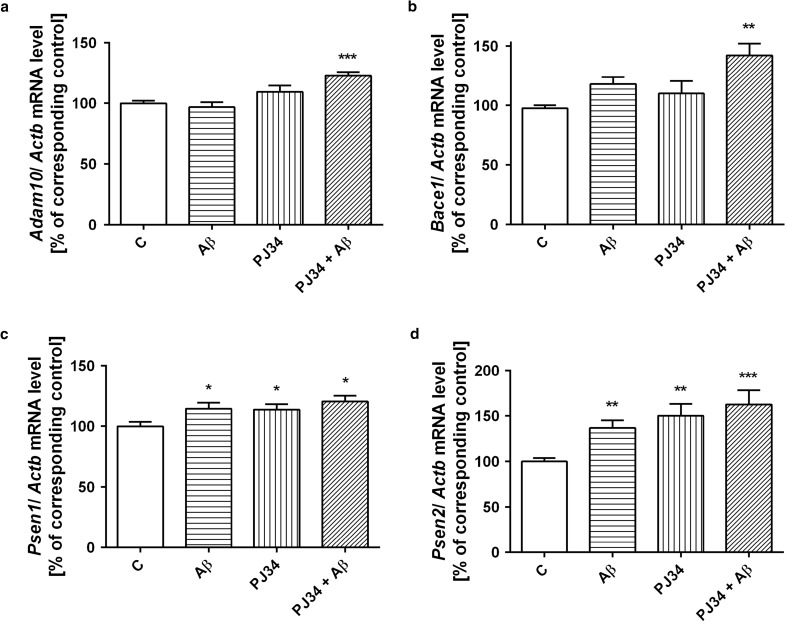
Then, question arises how AβO modulates transcription of enzymes responsible for APP processing in amyloidogenic and non-amyloidogenic pathways. In this experimental setting, AβOs exerted no effect on α-secretase (Adam10) expression and also on gene expression of β-secretase (Bace1) (Fig. 4a, b). PJ-34 significantly activated expression of Adam10 and Bace1 in the presence of AβO (Fig. 4a, b). Analysis of AβOs action on APP processing indicated the significant stimulatory effect of AβOs on gene expression of Psen1 and Psen2 (Fig. 4c, d). Moreover, PJ-34 had also stimulatory effect on Psen1/2 mRNA levels in the absence and presence of Aβ (Fig. 4c, d).
Fig. 4.
Expression levels of Adam10, Bace1, Psen1, and Psen2 genes in PC12 cells subjected to AβO42 (1 μM) and PJ-34 (20 μM) treatments (a–d). The levels of mRNA of secretases—Adam10 (a), Bace1 (b), Psen1 (c), and Psen2 (d)—were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments with three replications. ***p < 0.001; **p < 0.01, *p < 0.05—the difference, that was statistically significant compared to the control cells, using one-way analysis of variance (ANOVA) with Neuman-Keuls post hoc test
The Role of Endogenous Aβ Peptides on Transcription of APP Secretases and NAD+ Dependent Enzymes
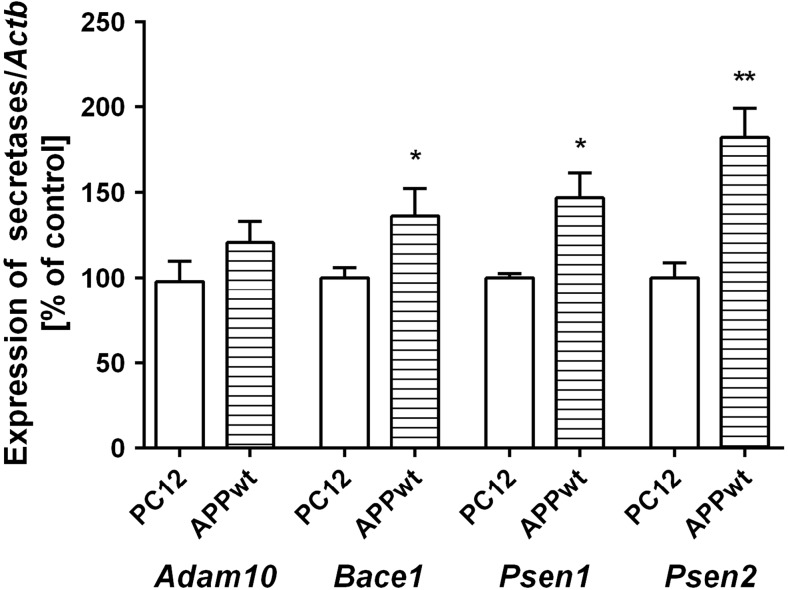
The effect of exogenous Aβ42 oligomers was then compared with the action of endogenously liberated Aβ peptides in APPwt cells. In this APP transfected cells, significant activation of gene expression of Bace1 and also of Psen1 and Psen2 was observed (Fig. 5). Although Parp1 expression was enhanced, transcription of other DNA bound PARPs and mitochondria SIRTs was not changed (Table 2). The relationship between described molecular events evoked by Aβ peptides, and the effect of PARP1 pharmacological inhibition is demonstrated on Fig. 6.
Fig. 5.
Expression levels of APP secretases in PC12 cells transfected with human gene for APP (APPwt) or with an empty vector. The levels of mRNA of secretases: Adam10, Bace1, Psen1, and Psen2 were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments. **p < 0.01; *p < 0.05—the difference, that was statistically significant compared to the control cells, using a Student’s t test
Table 2.
Gene expression and DNA bound PARPs and mitochondrial sirtuins in APPwt transfected cells
| Gene | PARPs and SIRTs/Actb mRNA level [% of control] |
|---|---|
| Parp1 | 158.6 ± 2.42* |
| Parp2 | 106.4 ± 1.27 |
| Parp3 | 91.4 ± 1.93 |
| Sirt3 | 120.6 ± 2.93 |
| Sirt4 | 74.43 ± 10.37 |
| Sirt5 | 58.78 ± 37.22 |
The levels of mRNA of DNA bound PARPs—Parp1, Parp2, Parp3—and mitochondria SIRTs—Sirt3, Sirt4, Sirt5—were analyzed via quantitative RT-PCR. The results of RT-PCR were normalized to Actb gene expression. Data represent the mean value ± SEM for three independent experiments. *p < 0.05—the difference, that was statistically significant compared to the control cells, using a Student’s t-test
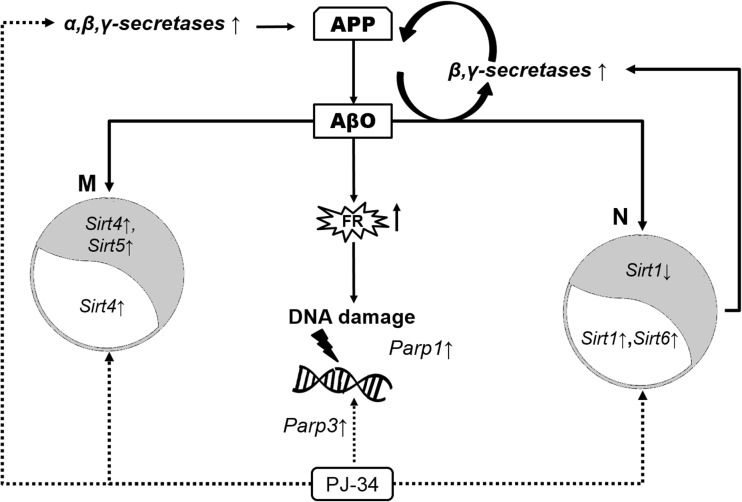
Fig. 6.
Interactions between Aβ oligomers and PARP1 inhibitor (PJ-34) on genes expression of sirtuins, DNA bound PARPs, and APP metabolizing enzymes. Effect of Aβ in mitochondria (M) and nucleus (N) is demonstrated on the grey field
Discussion
Our data demonstrate for the first time that PARP1 pharmacological inhibition by PJ-34 significantly enhances the expression of nuclear Sirt1 and Sirt6 in control cells and in Aβ42 toxicity. It was previously shown that in mammalian cells subjected to oxidative stress, SIRT1 and SIRT6 are responsible for DNA double-strand breaks repair [62–64]. Moreover, Sirt6 knockout mice exhibit premature aging phenotype and disturbed base excision repair (BER). In our experimental conditions, activation of Sirt1 and Sirt6 expression suggests the significant involvement of these both nuclear SIRTs in oxidative stress evoked by Aβ toxicity and by PARP1 inhibition. The inhibition of PARP1 activates also gene expression of cytosolic Sirt2 and mitochondrial Sirt4, however, remains without effect on Sirt3 and Sirt5. Transcription of mitochondrial Sirt4 and Sirt5 is significantly enhanced by AβO. These both SIRTs could be responsible for the mitochondrial proteins modifications by several processes as mono ADP-ribosylation, succinylation, and sumolynation which may subsequently affect mitochondrial protein function. PARP inhibitor decreases Sirt4 and Sirt5 transcription elevated by AβO toxicity.
Moreover, our results indicated that PARP1 inhibition and AβOs lead to activation of genes coding enzymes responsible for APP metabolism. PARP1 pharmacological inhibition enhanced mRNA level of Psen1 and Psen2 crucial subunits of γ-secretase involved in degradation of APP. PARP1 inhibition in the presence of AβOs enhanced gene expression of α-secretase and in the same experimental condition exerted significant stimulatory effect on transcription of β-secretase. Endogenously liberated Aβ peptides in βAPP transfected cells were found to exert stimulatory effects on transcription of PSENs and also β-secretase.
These results suggest that the vicious toxic cycle between Aβ and βAPP metabolism may be responsible for high levels of neurotoxic C99 fragment and Aβ peptides in cells. Our previous results showed that oxidative/genotoxic stress evoked by Aβ, inflammation processes, and brain ischemia led to activation of PARP1 and accumulation of PAR [52, 65–68]. In this study, we have observed that Aβ enhanced significantly gene expression of Parp1 in APPwt cells. Inhibitors of PARP are therefore suggested to be promising in protection of cells against neurodegeneration. They have stimulatory effect on gene expression of Sirt1, Sirt6, and non-amyloidogenic pathway which may exert cytoprotective effect. On the other hand, however, their stimulatory effect on gene expression of Bace1 should be taken under consideration. In this experimental setting using AβO, the expression of Parp1 was only slightly elevated. However, pharmacological inhibition of PARP1 activity evoked significant enhancement of gene expression of Parp3. Recent studies reported a particular role of PARP3 in cellular response to DNA double-strand breaks. It was described that PARP3 shares high degree of structural similarities with PARP1 and PARP2. This findings may suggest that under inhibition of PARP1, PARP3 could be responsible for the maintenance of genomic integrity, through interaction with Polycomb group proteins involved in BER/SSBR DNA repair and non-homologous endjoining (NHEJ) [69, 70].
It was previously documented that massive oxidative stress and DNA damage may lead to overactivation of DNA bound PARP(s) and to NAD+ reduction. Consequently, it may decrease activity of SIRT1, which is one of the positive regulators of α-secretase, a crucial enzyme in non-amyloidogenic processing of APP [71]. Concomitantly with these events, amyloidogenic pathway of APP processing may be strengthened with excessive production and accumulation of neurotoxic Aβ [72]. Inhibition of β- and γ-secretase or activation of α-secretase was suggested as promising targets in therapy of AD. Unfortunately, this type of therapeutic strategy is till now not successful [73, 74]. Some of the inhibitors and activators have entered human clinical trials, but only those targeted SIRT1 seem to be promising. Recent data about beneficial action of SIRTs focused mainly on SIRT1. Several direct and indirect activators were considered for further studies. However, the therapeutic strategy based on these compounds is till now debatable [75]. It was observed that PARP2 deficiency increased Sirt1 transcription [76]. Also, PARP1 inhibitors were proposed to be the best activators of SIRTs. Recently, Bai et al. [77] demonstrated that PARP1 inhibition increased mitochondria metabolism through SIRT1 activation. Their data showed that PARP1 inhibition had strong effect on NAD+ level and other metabolic processes through modulation of SIRT1 activity which in turn enhanced mitochondria content and function. In this way, PARP1 inhibition may exert protection against metabolic, and probably also against neurodegenerative disorders. Our data indicated significant effect of PARP1 pharmacological inhibition on Sirt1, Sirt6, and Sirt4 and Parp3 expression levels. However, the data of Lapucci et al. [78] showed that mitochondria homeostasis is compromised in cell lines exposed to PARP1 pharmacological inhibitors or small interfering RNA. The last data emphasized the relevance of PARP1 inhibitors in therapy of mitochondrial respiratory disorders [79–82]. Molecular interaction between PARPs and SIRTs and the role of PARP1 in neurodegenerative diseases is very complex and till now not fully elucidated [31–33, 49]. Moreover, recently Szczesny et al. [83] suggested the opposing roles of mitochondrial and nuclear PARP1 in regulation of mitochondria and nuclear DNA integrity and in regulation of mitochondria function. Until now, little is known about relationship between PARPs and mitochondrial SIRTs including SIRT4. In contrast to SIRT1 and SIRT3 which possess mainly deacetylase activity, SIRT4 and SIRT5 exhibit low deacetylase activity. SIRT4 mainly performs protein mono-ADP-ribosylation and regulates ATP homeostasis and mitochondrial biogenesis [84]. SIRT5, another mitochondrial sirtuin, is capable to carry out demalonylation, desuccinylation, and glutarylation of mitochondrial proteins with almost 1000 times higher catalytic activity than deacetylation [85, 86]. SIRT5 can also modulate mitochondrial physiology and promotes respiration and ATP synthesis via desuccinylation [87]. Moreover, SIRT5 is a regulator of lysine malonylation [88]. SIRT5 also desuccinylates and activates Cu/Zn superoxide dismutase (SOD1) independent of deacetylation to eliminate reactive oxygen species (ROS) [89]. In this study, we show that short-term action of Aβ oligomers lead to activation of gene expression for Sirt4 and Sirt5. However, endogenously liberated Aβ peptides in APPwt cells had no effects on mitochondrial SIRTs transcription, suggesting transient stimulatory effect of exogenous AβO. It was demonstrated previously that modulation of SIRT1 could be beneficial in AD via activation of non-amyloidogenic processing of βAPP [90]. Our results demonstrated that PARP1 inhibition activated Sirt1 expression in the presence of AβO and upregulated gene expression of α-secretase (Adam10). Recently, Sirt1 was also shown to decrease Bace1 expression levels and β-amyloid generation in cells culture [91]. In vitro studies on human cell lines and rat primary cortical neurons have shown that γ-secretase is also regulated by SIRTs [92].
Summarizing, our results indicate that inhibition of PARP1 through the modulation of transcription of SIRTs and APP-cleaving enzymes may have significant implication on cellular processes and cell fate. In consequence, it may be important in therapy of neurodegenerative disorders.
Compliance with Ethical Standards
Funding
This study was supported by the National Science Centre; Grant No: 2013/09/B/NZ3/01350.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Koffie RM, Meyer-Luehmann M, Hashimoto T, Adams KW, Mielke ML, Garcia-Alloza M, Micheva KD, Smith SJ et al (2009) Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc Natl Acad Sci U S A 106(10):4012–4017. doi:10.1073/pnas.0811698106 [DOI] [PMC free article] [PubMed]
- 2.Leon R, Garcia AG, Marco-Contelles J. Recent advances in the multitarget-directed ligands approach for the treatment of Alzheimer’s disease. Med Res Rev. 2013;33(1):139–189. doi: 10.1002/med.20248. [DOI] [PubMed] [Google Scholar]
- 3.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 4.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8(1):79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ (1991) Amyloid protein and Alzheimer’s disease. Scientific American 265 (5):68-71, 74-66, 78 [DOI] [PubMed]
- 6.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 7.Walsh DM, Selkoe DJ. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat Rev Neurosci. 2016;17(4):251–260. doi: 10.1038/nrn.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaunmuktane Z, Mead S, Ellis M, Wadsworth JD, Nicoll AJ, Kenny J, Launchbury F, Linehan J et al (2015) Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature 525(7568):247–250. doi:10.1038/nature15369 [DOI] [PubMed]
- 9.Nicolas M, Hassan BA. Amyloid precursor protein and neural development. Development (Cambridge, England) 2014;141(13):2543–2548. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H, Koo EH. Biology and pathophysiology of the amyloid precursor protein. Mol Neurodegener. 2011;6(1):27. doi: 10.1186/1750-1326-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allinson TMJ, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein α-secretases. J Neurosci Res. 2003;74(3):342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4(3):233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 13.Laird FM, Cai H, Savonenko AV, Farah MH, He K, Melnikova T, Wen H, Chiang HC et al (2005) BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J Neurosci 25(50):11693–11709. doi:10.1523/jneurosci.2766-05.2005 [DOI] [PMC free article] [PubMed]
- 14.Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB, Saurat N, McGlade A et al (2015) APP metabolism regulates tau proteostasis in human cerebral cortex neurons. Cell Rep 11(5):689–696. doi:10.1016/j.celrep.2015.03.068 [DOI] [PMC free article] [PubMed]
- 15.Yan R, Vassar R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. The Lancet Neurology. 2014;13(3):319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398(6727):513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Bhattacharjee S, Jones BM, Hill JM, Clement C, Sambamurti K, Dua P, Lukiw WJ (2015) Beta-amyloid precursor protein (βAPP) processing in Alzheimer’s disease (AD) and age-related macular degeneration (AMD). Mol Neurobiol 52(1):533–544. doi:10.1007/s12035-014-8886-3 [DOI] [PMC free article] [PubMed]
- 18.Larson ME, Lesne SE (2012) Soluble Aβ oligomer production and toxicity. J Neurochem 120(Suppl 1):125–139. doi:10.1111/j.1471-4159.2011.07478.x [DOI] [PMC free article] [PubMed]
- 19.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA et al (2008) Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med 14(8):837–842. doi:10.1038/nm1782 [DOI] [PMC free article] [PubMed]
- 20.Cieslik M, Czapski GA, Strosznajder JB (2015) The molecular mechanism of amyloid β42 peptide toxicity: the role of sphingosine kinase-1 and mitochondrial Sirtuins. PLoS One 10(9):e0137193. doi:10.1371/journal.pone.0137193 [DOI] [PMC free article] [PubMed]
- 21.Butterfield DA, Swomley AM, Sultana R. Amyloid β-peptide (1–42)-induced oxidative stress in Alzheimer disease: importance in disease pathogenesis and progression. Antioxid Redox Signal. 2013;19(8):823–835. doi: 10.1089/ars.2012.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer’s amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol. 2003;161(1):41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calkins MJ, Reddy PH. Amyloid beta impairs mitochondrial anterograde transport and degenerates synapses in Alzheimer’s disease neurons. Biochim Biophys Acta. 2011;1812(4):507–513. doi: 10.1016/j.bbadis.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckert A, Hauptmann S, Scherping I, Meinhardt J, Rhein V, Drose S, Brandt U, Fandrich M et al (2008) Oligomeric and fibrillar species of β-amyloid (Aβ42) both impair mitochondrial function in P301L tau transgenic mice. Journal of molecular medicine (Berlin, Germany) 86(11):1255–1267. doi:10.1007/s00109-008-0391-6 [DOI] [PubMed]
- 25.Kaminsky YG, Tikhonova LA, Kosenko EA. Critical analysis of Alzheimer’s amyloid-beta toxicity to mitochondria. Frontiers in bioscience (Landmark edition) 2015;20:173–197. doi: 10.2741/4304. [DOI] [PubMed] [Google Scholar]
- 26.Kazmierczak A, Strosznajder JB, Adamczyk A. Alpha-Synuclein enhances secretion and toxicity of amyloid beta peptides in PC12 cells. Neurochem Int. 2008;53(6–8):263–269. doi: 10.1016/j.neuint.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Pagani L, Eckert A. Amyloid-Beta interaction with mitochondria. Int J Alzheimers Dis. 2011;2011:925050. doi: 10.4061/2011/925050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlov PF, Wiehager B, Sakai J, Frykman S, Behbahani H, Winblad B, Ankarcrona M. Mitochondrial gamma-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25(1):78–88. doi: 10.1096/fj.10-157230. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt K, Grimm A, Kazmierczak A, Strosznajder JB, Gotz J, Eckert A. Insights into mitochondrial dysfunction: aging, amyloid-beta, and tau-a deleterious trio. Antioxid Redox Signal. 2012;16(12):1456–1466. doi: 10.1089/ars.2011.4400. [DOI] [PubMed] [Google Scholar]
- 30.Strosznajder JB, Czapski GA, Adamczyk A, Strosznajder RP. Poly(ADP-ribose) polymerase-1 in amyloid beta toxicity and Alzheimer’s disease. Mol Neurobiol. 2012;46(1):78–84. doi: 10.1007/s12035-012-8258-9. [DOI] [PubMed] [Google Scholar]
- 31.Jęśko H, Strosznajder RP. Sirtuins and their interactions with transcription factors and poly(ADP-ribose) polymerases. Folia Neuropathol. 2016;3:212–233. doi: 10.5114/fn.2016.62531. [DOI] [PubMed] [Google Scholar]
- 32.Jęśko H, Wencel P, Strosznajder RP, Strosznajder JB (2016) Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem Res:1–15 [DOI] [PMC free article] [PubMed]
- 33.Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome Integrity. 2013;4:6–6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kincaid B, Bossy-Wetzel E. Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci. 2013;5:48. doi: 10.3389/fnagi.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280(14):13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 36.Dittenhafer-Reed KE, Richards AL, Fan J, Smallegan MJ, Fotuhi Siahpirani A, Kemmerer ZA, Prolla TA, Roy S et al (2015) SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metab 21(4):637–646. doi:10.1016/j.cmet.2015.03.007 [DOI] [PMC free article] [PubMed]
- 37.Mahlknecht U, Voelter-Mahlknecht S. Genomic organization and localization of the NAD-dependent histone deacetylase gene sirtuin 3 (Sirt3) in the mouse. Int J Oncol. 2011;38(3):813–822. doi: 10.3892/ijo.2010.872. [DOI] [PubMed] [Google Scholar]
- 38.Matsushima S, Sadoshima J. The role of sirtuins in cardiac disease. Am J Phys Heart Circ Phys. 2015;309(9):H1375–H1389. doi: 10.1152/ajpheart.00053.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava S, Baxa U, Niu G, Chen X, Veech RL. A ketogenic diet increases brown adipose tissue mitochondrial proteins and UCP1 levels in mice. IUBMB Life. 2013;65(1):58–66. doi: 10.1002/iub.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 41.Banasik M, Stedeford T, Strosznajder RP. Natural inhibitors of poly(ADP-ribose) polymerase-1. Mol Neurobiol. 2012;46(1):55–63. doi: 10.1007/s12035-012-8257-x. [DOI] [PubMed] [Google Scholar]
- 42.Pieper AA, Blackshaw S, Clements EE, Brat DJ, Krug DK, White AJ, Pinto-Garcia P, Favit A et al (2000) Poly(ADP-ribosyl) ation basally activated by DNA strand breaks reflects glutamate–nitric oxide neurotransmission. Proc Natl Acad Sci U S A 97(4):1845–1850 [DOI] [PMC free article] [PubMed]
- 43.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329(1):18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105(8):1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abeti R, Abramov AY, Duchen MR. Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain. 2011;134(Pt 6):1658–1672. doi: 10.1093/brain/awr104. [DOI] [PubMed] [Google Scholar]
- 46.Adamczyk A, Czapski GA, Jesko H, Strosznajder RP. Non a beta component of Alzheimer’s disease amyloid and amyloid beta peptides evoked poly(ADP-ribose) polymerase-dependent release of apoptosis-inducing factor from rat brain mitochondria. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2005;56(Suppl 2):5–13. [PubMed] [Google Scholar]
- 47.Love S, Barber R, Wilcock GK. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer’s disease. Brain. 1999;122(Pt 2):247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- 48.Martire S, Fuso A, Rotili D, Tempera I, Giordano C, De Zottis I, Muzi A, Vernole P et al (2013) PARP-1 modulates amyloid beta peptide-induced neuronal damage. PLoS One 8(9):e72169. doi:10.1371/journal.pone.0072169 [DOI] [PMC free article] [PubMed]
- 49.Martire S, Mosca L, d'Erme M. PARP-1 involvement in neurodegeneration: a focus on Alzheimer’s and Parkinson’s diseases. Mech Ageing Dev. 2015;146-148:53–64. doi: 10.1016/j.mad.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Strosznajder JB, Jesko H, Strosznajder RP. Age-related alteration of poly(ADP-ribose) polymerase activity in different parts of the brain. Acta Biochim Pol. 2000;47(2):331–337. [PubMed] [Google Scholar]
- 51.Bayrakdar ET, Armagan G, Uyanikgil Y, Kanit L, Koylu E, Yalcin A. Ex vivo protective effects of nicotinamide and 3-aminobenzamide on rat synaptosomes treated with Abeta(1–42) Cell Biochem Funct. 2014;32(7):557–564. doi: 10.1002/cbf.3049. [DOI] [PubMed] [Google Scholar]
- 52.Strosznajder RP, Czubowicz K, Jesko H, Strosznajder JB. Poly(ADP-ribose) metabolism in brain and its role in ischemia pathology. Mol Neurobiol. 2010;41(2–3):187–196. doi: 10.1007/s12035-010-8124-6. [DOI] [PubMed] [Google Scholar]
- 53.Kraus WL, Hottiger MO. PARP-1 and gene regulation: progress and puzzles. Mol Asp Med. 2013;34(6):1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Wesierska-Gadek J, Schmid G. Poly(ADP-ribose) polymerase-1 regulates the stability of the wild-type p53 protein. Cellular & molecular biology letters. 2001;6(2):117–140. [PubMed] [Google Scholar]
- 55.Martire S, Fuso A, Mosca L, Forte E, Correani V, Fontana M, Scarpa S, Maras B et al (2016) Bioenergetic impairment in animal and cellular models of Alzheimer’s disease: PARP-1 inhibition rescues metabolic dysfunctions. J Alzheimers Dis 54(1):307–324. doi:10.3233/JAD-151040 [DOI] [PubMed]
- 56.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278(13):11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 57.Eckert A, Steiner B, Marques C, Leutz S, Romig H, Haass C, Muller WE. Elevated vulnerability to oxidative stress-induced cell death and activation of caspase-3 by the Swedish amyloid precursor protein mutation. J Neurosci Res. 2001;64(2):183–192. doi: 10.1002/jnr.1064. [DOI] [PubMed] [Google Scholar]
- 58.Keil U, Bonert A, Marques CA, Scherping I, Weyermann J, Strosznajder JB, Muller-Spahn F, Haass C et al (2004) Amyloid beta-induced changes in nitric oxide production and mitochondrial activity lead to apoptosis. J Biol Chem 279(48):50310–50320. doi:10.1074/jbc.M405600200 [DOI] [PubMed]
- 59.Gassowska M, Cieslik M, Wilkaniec A, Strosznajder JB. Sphingosine kinases/sphingosine-1-phosphate and death Signalling in APP-transfected cells. Neurochem Res. 2014;39(4):645–652. doi: 10.1007/s11064-014-1240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chalimoniuk M, Stolecka A, Cakala M, Hauptmann S, Schulz K, Lipka U, Leuner K, Eckert A et al (2007) Amyloid beta enhances cytosolic phospholipase A2 level and arachidonic acid release via nitric oxide in APP-transfected PC12 cells. Acta Biochim Pol 54(3):611–623 [PubMed]
- 61.Pajak B, Songin M, Strosznajder JB, Gajkowska B. Alzheimer’s disease genetic mutation evokes ultrastructural alterations: correlation to an intracellular Abeta deposition and the level of GSK-3beta-P(Y216) phosphorylated form. Neurotoxicology. 2009;30(4):581–588. doi: 10.1016/j.neuro.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chalkiadaki A, Guarente L (2015) The multifaceted functions of sirtuins in cancer. Nat Rev Cancer 15 (10):608–624. doi:10.1038/nrc3985 [DOI] [PubMed]
- 64.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323(5917):1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cieslik M, Pyszko J, Strosznajder JB. Docosahexaenoic acid and tetracyclines as promising neuroprotective compounds with poly(ADP-ribose) polymerase inhibitory activities for oxidative/genotoxic stress treatment. Neurochem Int. 2013;62(5):626–636. doi: 10.1016/j.neuint.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 66.Czapski GA, Adamczyk A, Strosznajder RP, Strosznajder JB. Expression and activity of PARP family members in the hippocampus during systemic inflammation: their role in the regulation of prooxidative genes. Neurochem Int. 2013;62(5):664–673. doi: 10.1016/j.neuint.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 67.Strosznajder RP, Gadamski R, Czapski GA, Jesko H, Strosznajder JB. Poly(ADP-ribose) polymerase during reperfusion after transient forebrain ischemia: its role in brain edema and cell death. J Mol Neurosci. 2003;20(1):61–72. doi: 10.1385/JMN:20:1:61. [DOI] [PubMed] [Google Scholar]
- 68.Strosznajder R, Gadamski R, Walski M. Inhibition of poly(ADP-ribose) polymerase activity protects hippocampal cells against morphological and ultrastructural alteration evoked by ischemia-reperfusion injury. Folia Neuropathol. 2005;43(3):156–165. [PubMed] [Google Scholar]
- 69.Boehler C, Dantzer F. PARP-3, a DNA-dependent PARP with emerging roles in double-strand break repair and mitotic progression. Cell Cycle. 2011;10(7):1023–1024. doi: 10.4161/cc.10.7.15169. [DOI] [PubMed] [Google Scholar]
- 70.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou J-M, Bresson A, Sanglier-Cianferani S, Smith S et al (2011) Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A 108(7):2783–2788. doi:10.1073/pnas.1016574108 [DOI] [PMC free article] [PubMed]
- 71.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD(+): an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonda DJ, Lee HG, Camins A, Pallas M, Casadesus G, Smith MA, Zhu X. The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. The Lancet Neurology. 2011;10(3):275–279. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hardy J, Bogdanovic N, Winblad B, Portelius E, Andreasen N, Cedazo-Minguez A, Zetterberg H. Pathways to Alzheimer’s disease. J Intern Med. 2014;275(3):296–303. doi: 10.1111/joim.12192. [DOI] [PubMed] [Google Scholar]
- 74.Iqbal K, Bolognin S, Kazim SF. A novel pharmacologic therapeutic approach to Alzheimer disease and cognitive aging. Alzheimer's & Dementia: The Journal of the Alzheimer's Association. 2014;10(4):P175. doi: 10.1016/j.jalz.2014.04.189. [DOI] [Google Scholar]
- 75.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M et al (2010) SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem 285(11):8340–8351. doi:10.1074/jbc.M109.088682 [DOI] [PMC free article] [PubMed]
- 76.Bai P, Canto C, Brunyánszki A, Huber A, Szántó M, Cen Y, Yamamoto H, Houten SM et al (2011) PARP-2 regulates SIRT1 expression and whole body energy expenditure. Cell Metab 13(4):450–460. doi:10.1016/j.cmet.2011.03.013 [DOI] [PMC free article] [PubMed]
- 77.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A et al (2011) PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13(4):461–468. doi:10.1016/j.cmet.2011.03.004 [DOI] [PMC free article] [PubMed]
- 78.Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol Pharmacol. 2011;79(6):932–940. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- 79.Felici R, Cavone L, Lapucci A, Guasti D, Bani D, Chiarugi A. PARP inhibition delays progression of mitochondrial encephalopathy in mice. Neurotherapeutics. 2014;11(3):651–664. doi: 10.1007/s13311-014-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Felici R, Lapucci A, Cavone L, Pratesi S, Berlinguer-Palmini R, Chiarugi A. Pharmacological NAD-boosting strategies improve mitochondrial homeostasis in human complex I-mutant fibroblasts. Mol Pharmacol. 2015;87(6):965–971. doi: 10.1124/mol.114.097204. [DOI] [PubMed] [Google Scholar]
- 81.Pirinen E, Canto C, Jo YS, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L et al (2014) Pharmacological inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 19(6):1034–1041. doi:10.1016/j.cmet.2014.04.002 [DOI] [PMC free article] [PubMed]
- 82.Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve Anthony A, Li W, Leoni V, Schon Eric A et al (2014) NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab 19(6):1042–1049. doi:10.1016/j.cmet.2014.04.001 [DOI] [PMC free article] [PubMed]
- 83.Szczesny B, Brunyanszki A, Olah G, Mitra S, Szabo C. Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: implications for the regulation of mitochondrial function. Nucleic Acids Res. 2014;42(21):13161–13173. doi: 10.1093/nar/gku1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E, Kolthur-Seetharam U (2013) SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging 5 (11):835-849. Doi:10.18632/aging.100616 [DOI] [PMC free article] [PubMed]
- 85.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J et al (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334(6057):806–809. doi:10.1126/science.1207861 [DOI] [PMC free article] [PubMed]
- 86.Yang X, Liu B, Zhu W, Luo J. SIRT5, functions in cellular metabolism with a multiple enzymatic activities. Sci China Life Sci. 2015;58(9):912–914. doi: 10.1007/s11427-015-4902-8. [DOI] [PubMed] [Google Scholar]
- 87.Buler M, Aatsinki SM, Izzi V, Uusimaa J, Hakkola J. SIRT5 is under the control of PGC-1alpha and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28(7):3225–3237. doi: 10.1096/fj.13-245241. [DOI] [PubMed] [Google Scholar]
- 88.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M et al (2015) SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell 59(2):321–332. doi:10.1016/j.molcel.2015.05.022 [DOI] [PMC free article] [PubMed]
- 89.Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH et al (2013) SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun 441(1):191–195. doi:10.1016/j.bbrc.2013.10.033 [DOI] [PubMed]
- 90.Theendakara V, Patent A, Peters Libeu CA, Philpot B, Flores S, Descamps O, Poksay KS, Zhang Q et al (2013) Neuroprotective Sirtuin ratio reversed by ApoE4. Proc Natl Acad Sci U S A 110(45):18303–18308. doi:10.1073/pnas.1314145110 [DOI] [PMC free article] [PubMed]
- 91.Marwarha G, Raza S, Meiers C, Ghribi O. Leptin attenuates BACE1 expression and amyloid-beta genesis via the activation of SIRT1 signaling pathway. Biochim Biophys Acta. 2014;1842(9):1587–1595. doi: 10.1016/j.bbadis.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Wu F, Schweizer C, Rudinskiy N, Taylor DM, Kazantsev A, Luthi-Carter R, Fraering PC. Novel gamma-secretase inhibitors uncover a common nucleotide-binding site in JAK3, SIRT2, and PS1. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24(7):2464–2474. doi: 10.1096/fj.09-148031. [DOI] [PubMed] [Google Scholar]