Abstract
Introduction
Oxygen from carbon dioxide, water or molecular oxygen, depending on the responsible enzyme, can lead to a large variety of metabolites through chemical modification.
Objectives
Pathway-specific labeling using isotopic molecular oxygen (18O2) makes it possible to determine the origin of oxygen atoms in metabolites and the presence of biosynthetic enzymes (e.g., oxygenases). In this study, we established the basis of 18O2-metabolome analysis.
Methods
18O2 labeled whole Medicago truncatula seedlings were prepared using 18O2-air and an economical sealed-glass bottle system. Metabolites were analyzed using high-accuracy and high-resolution mass spectrometry. Identification of the metabolite was confirmed by NMR following UHPLC–solid-phase extraction (SPE).
Results
A total of 511 peaks labeled by 18O2 from shoot and 343 peaks from root were annotated by untargeted metabolome analysis. Additionally, we identified a new flavonoid, apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside], that was labeled by 18O2. To the best of our knowledge, this is the first report of apigenin 4′-glucuronide in M. truncatula. Using MSn analysis, we estimated that 18O atoms were specifically incorporated in apigenin, the coumaroyl group, and glucuronic acid. For apigenin, an 18O atom was incorporated in the 4′-hydroxy group. Thus, non-specific incorporation of an 18O atom by recycling during one month of labeling is unlikely compared with the more specific oxygenase-catalyzing reaction.
Conclusion
Our finding indicated that 18O2 labeling was effective not only for the mining of unknown metabolites which were biosynthesized by oxygenase-related pathway but also for the identification of metabolites whose oxygen atoms were derived from oxygenase activity.
Electronic supplementary material
The online version of this article (10.1007/s11306-018-1364-6) contains supplementary material, which is available to authorized users.
Keywords: Stable-isotope, Untargeted metabolome analysis, Flavonoid, Medicago truncatula, Metabolite modification
Introduction
It is well known that plants biosynthesize a wide variety of metabolites. A statistical approach has estimated that there are over 1,060,000 metabolites in plant species (Afendi et al. 2012). Although these are often beneficial in both plant physiology and for drug development (Newman and Cragg 2012). only a small proportion of these metabolites (50,897 metabolites were recorded in KNApSAcK) have been identified (Nakamura et al. 2014). Therefore, it is highly probable that metabolome analyses will lead to the identification of the unknown metabolites, uncover unknown biological phenomena and lead to advancements in drug development.
For large-scale and high-throughput metabolite analyses, chromatography-coupled to mass spectrometry (MS) is a very powerful tool due to its ability to detect a wide range of metabolites (Nakabayashi and Saito 2013). However, MS merely provides information on the mass and intensity of a large number of metabolite-derived peaks for metabolite, and ‘metabolite annotation’ is a crucial aspect of MS-based metabolomics. Annotation is typically performed by comparing tandem mass spectrometry (MS/MS) spectra of unknowns with spectra of known metabolites (Sumner et al. 2007). However, this approach cannot be applied to many rare plant-specific metabolites, because corresponding authentic standards are not available in many cases. Moreover, unknown metabolites are often annotated as ‘unknown’, and the information is not effectively used for further estimation (Iijima et al. 2008). Currently, advanced MS instrumentation with high accuracy and resolution enables a standard compound-independent annotation strategy in which the elemental composition is estimated based on accurate mass values (Makarov and Scigelova 2010; Ohta et al. 2010). To avoid incorrect assignment of elemental compositions, the natural average abundance of stable isotopes has been used (Kind and Fiehn 2006, 2007).Using a different approach, Giavalisco et al. prepared Arabidopsis thaliana labeled with 13C, 15N, and 34S, and estimated the elemental composition using the number of stable isotopes incorporated in the compound as an index (Giavalisco et al. 2011). Therefore, use of stable isotopes is effective for the estimation of elemental composition.
13C, 15N, and 34S have also been used as a tag or tracer, and are sometimes enriched by feeding plants with 13CO2, 15N-, or 34S-inorganic salts during cultivation (Glaser et al. 2014; Harada et al. 2006; Huege et al. 2007; Nakabayashi et al. 2013). In addition, 18O also has potential roles in metabolite flux analysis and gene mining. In higher plants, oxygen atoms are incorporated into compounds from carbon dioxide, water, or molecular oxygen by each responsible enzyme. Among them, oxygenases (EC1.13 and EC1.14) are well known for transferring an oxygen atom from molecular oxygen, and not from carbon dioxide or water, into metabolites (Hayaishi et al. 1955; Mason et al. 1955). In 2004, Hecht et al. labeled hop (Humulus lupulus) with 18O2 (molecular oxygen) in vivo and analyzed the origin of the oxygen atoms of humulone and cohumulone by nuclear magnetic resonance (NMR) spectrometry (Hecht et al. 2004). The result indicated that oxygen atoms from 18O2 were distinguishable from unlabeled O2. Further, in vivo labeling with 18O2 can be effective for suggesting the existence of an oxygenase in a biosynthetic pathway. However, there are no reports that have applied 18O2 (molecular oxygen) for MS-based comprehensive metabolome analysis.
Materials and methods
Plant materials
The unlabeled and labeled Medicago truncatula, Jemalong A17, were prepared as described previously (Kera et al. 2014). Seeds were sterilized with 70% ethanol and 1% (v/v) sodium hypochlorite solution, and rinsed with sterile water. Four seeds were sown on an agar medium, prepared in a 500 mL sealed glass bottle, and were grown for 5 weeks under the following conditions: temperature, 25 °C and a day/night rhythm of 18 h/6 h. The medium contained 1 mM potassium phosphate monobasic, 30 µM boric acid, 11 µM manganese(II) chloride, 1 µM/zinc chloride, 1 mM potassium chloride, 1 mM molybdic acid (sodium salt), 3.2 µM cupric chloride, 84 nM cobalt chloride, 1 mM calcium chloride, 50 µM Fe-ethylenediaminetetraacetic acid (EDTA), 1 mM magnesium sulfate, 10 mM ammonium nitrate, 10 mM potassium nitrate, 0.5% glucose, 1× Gamborg’s Vitamin Solution (Sigma-Aldrich, USA), and 0.8% Bacto Agar (Becton Dickinson, USA). During cultivation, 1 L of 18O-labeled air was exchanged daily. The plants were carefully collected from the bottle and the roots were washed with sterile water to remove agar medium. The shoots and roots were then separated with a scalpel and immediately frozen using liquid nitrogen. The plants were labeled with 13C [13C6-glucose (ISOTEC, Korea) in medium and 13C-labeled air (388 ppm 13C–CO2 and balance air; Tatsuoka, Japan)], 15N [15N-ammonium nitrate (SI Science, Japan) and 10 mM 15N-potassium nitrate (SI Science, Japan)], 18O [18O-labeled air (21.7% 18O–O2, 0.0394% CO2, and balance N2; Tatsuoka, Japan)], and 34S [34S-magnesium sulfate (SI Science, Japan)]. Details of the sample preparation methods are also available in the database Metabolonote (http://metabolonote.kazusa.or.jp/SE37:/).
Analysis of the labeling efficiencies for 13C and 15N by sealed combustion method
The whole plants were lyophilized and ground by mortal. Samples were placed in quartz tubes that contained copper oxide. In the presence of Cu and Ag foil, the tubes were evacuated, sealed, and heated at 500 °C for 30 min and at 850 °C for 2 h, followed by cooling to room temperature. After combustion, the sample gas purified by cold trapping technique was analyzed by mass spectrometry RMI-2 (Hitachi, Japan).
Liquid chromatography–mass spectrometry (LC–MS) measurement
The analysis was performed as described previously (Kera et al. 2014). The metabolites, extracted with 80% methanol (Optima LC/MS; Thermo Fisher Scientific, USA) from approximately 0.25 mg of the lyophilized plants, were injected in a liquid chromatography (LC)–Orbitrap–MS [LC, Agilent 1100 series (Agilent, USA); LTQ–Orbitrap (Thermo Fisher Scientific, USA) or LC, Dionex Ultimate 3000 (Thermo Fisher Scientific, USA); Orbitrap Fusion (Thermo Fisher Scientific, USA)]. The metabolites were separated using a C18 reversed-phase column (TSKgel ODS-100V; 4.6 × 250 mm, 5 µm; TOSOH, Japan) with a flow rate of 500 µL min−1 and the following gradient: water + 0.1% formic acid/acetonitrile + 0.1% formic acid, 3/97 to 97/3% (0.0–45.0 min), 3/97% (45.1–50.0 min), and 3/97% (50.1–57.0 min). Full mass scans were recorded with high resolution (LTQ–Orbitrap, 60,000; Orbitrap Fusion, 120,000) and MS/MS were acquired on the five most intense ions from each full scan by ion trap or Orbitrap Fusion, covering a mass range from m/z 100.0–1500.0.
Data analysis
Data analysis was performed as described previously (Kera et al. 2014). All raw data files were converted into text files using MSGet (http://www.kazusa.or.jp/komics/software/MSGet), and peaks were extracted by PowerFT (http://www.kazusa.or.jp/komics/software/PowerGet/) (Sakurai et al. 2014). Unlabeled and blank data were aligned using PowerMatch (http://www.kazusa.or.jp/komics/software/PowerGet/) with manual curation, and valid peaks were selected after background correction. In the annotation process, the primary database search and the primary estimation of elemental composition were performed using PowerMatch and MFSearcher within a mass accuracy of 3 ppm (http://webs2.kazusa.or.jp/mfsearcher/) (Sakurai et al. 2013). Labeling data were further analysis by ShiftedIonsFinder (Kera et al. 2014).
To find the isotopic peaks in the labeled data and associate them with monoisotopic peaks from the unlabeled data, the labeled peak search was performed with the parameters as follows: Max fold, C = 100, N = 50, O = 50, S = 50; Mass difference = 3 ppm; and RT difference = 1. In flavonoid-like peak analysis, candidate peaks with chemical modification, such as glycosylation and acylation, were searched by comparing the peak list containing mass information of aglycone of typical flavonoids against the peak list from an unlabeled sample using ShiftedIonsFinder. Xylosylation (Xyl) (C5H8O4, m/z 132.04226), glucosylation (Glc) (C6H10O5, m/z 162.05282), rhamnosylation (Rha) (C6H10O4, m/z 146.05791), glucuronidation (GlcUA) (C6H8O6, m/z 176.03209), cinnamoylation (Cinnamoyl) (C9H6O1, m/z 130.04186), coumaroylation (Coumaroyl) (C9H6O2, m/z 146.03678), caffeoylation (Caffeoyl) (C9H6O3, m/z 162.03169), feruloylation (Feruloyl) (C10H8O3, m/z 176.04734), malonylation (Malonyl) (C3H2O3, m/z 89.0003939), and succinylation, (Succinyl) (C4H4O3, m/z 100.016044) were selected as modification groups. The settings were as follows: Max fold, Glc = 3, Rha = 3, GlcUA = 3, Cinnamoyl = 3, Coumaroyl = 3, Caffeoyl = 3, Feruloyl = 3, Malonyl = 3, Succinyl = 3; Mass difference = 3 ppm; RT difference = 60. The exported file from ShiftedIonsFinder was arranged by Excel.
UHPLC–ESI–QTOF–MS/MS measurement
The metabolites, extracted with 80% methanol were injected in a Waters ACQUITY UPLC (Waters, USA) coupled to a Bruker maXis Impact ESI–QTOF–MS system having a mass resolution of ∼ 40,000 (Bruker Daltonics, USA). The identification of the metabolite was confirmed by NMR following UHPLC–solid-phase extraction (SPE) using a Bruker/Spark Holland Prospekt II SPE system as described previously (Qiu et al. 2016). Details were provided in supplemental data 4.
Results and discussion
Establishment of an economical sealed-glass bottle system
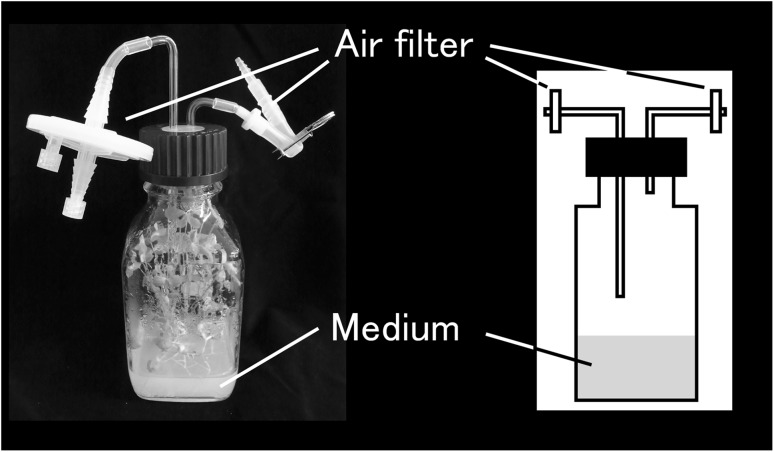
Although many stable isotopes, such as, 13C, 15N, and 34S, have been used for in vivo labeling, 18O (especially in its molecular oxygen form) has not been effectively used. A reported 13CO2 labeling procedure (Huege et al. 2007) is not suited for 18O2 labeling, because 18O-labeled air is too expensive to fill most growth chambers. Hecht et al. previously proposed an economical 18O2 labeling system to produce 18O-labeled hop (Hecht et al. 2004). In their system, several cones were enclosed in glass vessels filled with 20% 18O2 and 80% argon, and were incubated for 14 days without gas exchange. Therefore, we used a sealed-glass bottle with air filters (ADVANTEC, Japan) (Fig. 1) for gas exchange. Additionally, unlabeled and 13C-, 15N-, and 34S-labeled samples were prepared using the same setup, respectively. The labeling efficiencies for 13C-labeled and 15N-labeled samples were 59.6 and 78.8%, respectively, while those of unlabeled sample were 1.09 and 0.365%, respectively. These were not as high compared with those in previous reports, in which greater than 90% efficiency was achieved using a growth chamber (Giavalisco et al. 2011). In order to grow each labeling plants in equal condition, 13C-labeled air was exchanged daily in this study. However, continuous gas flow might improve it. For 15N-labeling, exchanging medium during long cultivation might effective. To enable exchange of medium, we plan to use liquid medium and modify Leonard jar (Leonard 1943) to keep sealed and exchange the gas in the future. Moreover, 78.8% of 15N labeling efficiency was not perfect but it seemed to be enough for further curation of the elemental composition. Because incorporation of 18O or 34S cannot be measured directly, the 80% methanol extraction of unlabeled, 18O-labeled, and 34S-labeled shoots were analyzed by LC–Orbitrap–MS using a mass accuracy of 3 ppm. The incorporation was manually confirmed in the mass spectra using Xcalibur (Thermo Fisher Scientific, USA) (data not shown). These results suggest that this labeling system works, although that there is still room for improvement in labeling efficiency.
Fig. 1.
The sealed-glass bottle system for economical labeling. The normal or labeled air was blown into the bottle through an air filter. After gas exchange, the flow channels of the filter were closed. The other labeled nutrients were supplied in the medium beforehand
Untargeted metabolite annotation of peaks able to be labeled by 18O2
For target analysis, whether a target metabolite is labeled by 18O2 or not, was investigated by searching for the calculated m/z values of 18O-labeled peaks in the 18O2 labeled sample. Here, this is calculated by adding the m/z difference between 16O and 18O of 2.0042462 to that of a target monoisotopic peak. However, this approach is not applicable to non-target analyses, because the monoisotopic m/z values of peaks that can be labeled by 18O2 are not assigned. One effective approach is to compare mass spectra from an unlabeled sample with those from a 18O2 labeled sample, and associate the peaks in the unlabeled sample with the peaks in the 18O2 labeled sample based on the specified m/z difference responsible for 18O-labeling. Therefore, the comprehensive search for peaks labeled by 18O2 can be accomplished in four steps: (1) preparation of an unlabeled peak list from unlabeled samples with tight intensity filter to reduce noise: (2) preparation of an 18O-labeled peak list from 18O2 labeled samples with a loose intensity filter to pick tiny peaks; (3) search for peaks labeled by 18O2 by association of the unlabeled peak list with the 18O-labeled peak list using ShiftedIonsFinder (Kera et al. 2014), and (4) curation of the search results by manually checking the mass spectra. Initially, a total of 3,417 peaks and 3,518 peaks were extracted as unlabeled peaks from M. truncatula shoots and roots, respectively. Because analysis using ShiftedIonsFinder requires manual curation of the mass spectra, we selected peaks labeled by 18O2 according to the search result of ShiftedIonsFinder and the existence of MS2 information for further annotation steps. Finally, a total of 511 peaks and 353 peaks were extracted as peaks labeled by 18O2 from M. truncatula shoots and roots, respectively (Supplement data 1, 2).
The elemental compositions of peaks labeled by 18O2 were first predicted using MFSearcher (Sakurai et al. 2013). Because multiple elemental compositions are generally predicted for peaks with high m/z values, even if the deviation of m/z value is within 1 ppm, the predicted elemental compositions were manually curated according to a strategy in which the number of stable isotopes incorporated in the compound is used as an index (Giavalisco et al. 2011; Kera et al. 2014). As well as detecting 18O-labeled peaks, the corresponding 13C-, 15N-, and 34S-labeled peaks labeled by 18O2 were found from each labeled sample by comparing with unlabeled peak list using ShiftedIonsFinder. Considering that the labeling efficiency was not enough to determine the max number of each element, we excluded the elemental compositions in which the number of each element was less than that of detected. As a result, the single elemental composition increased from about 4% (shoot: 20 peaks, 3.9%; root: 14 peaks, 4.0%) to about 60% (shoot: 311 peaks, 60.9%; root: 222 peaks, 62.9%), while multiple elemental compositions decreased from about 95% (shoot: 489 peaks, 95.7%; root: 335 peaks, 94.9%) to about 38% (shoot: 193 peaks, 37.8%; root: 121 peaks, 34.3%) (Table 1). Among peaks having a single elemental composition, 195 peaks from shoot and 161 peaks from root were present in databases (see “Materials and methods”), suggesting that many secondary metabolites can be labeled with 18O2 by oxygenases (Table 2).
Table 1.
The status of estimating elemental composition within 3 ppm
| Status of elemental composition | Shoot | Root | ||||||
|---|---|---|---|---|---|---|---|---|
| Automatic estimation | Manual curation | Automatic estimation | Manual curation | |||||
| Number (peaks) | Rate (%) | Number (peaks) | Rate (%) | Number (peaks) | Rate (%) | Number (peaks) | Rate (%) | |
| Single | 20 | 3.9 | 311 | 60.9 | 14 | 4.0 | 222 | 62.9 |
| Multiple | 489 | 95.7 | 193 | 37.8 | 335 | 94.9 | 121 | 34.3 |
| No hits | 2 | 0.4 | 7 | 1.4 | 4 | 1.1 | 10 | 2.8 |
| Total | 511 | 100.0 | 511 | 100.0 | 353 | 100.0 | 353 | 100.0 |
Table 2.
The category of database search results for peaks having a single elemental composition
| Category | Shoot | Root | ||
|---|---|---|---|---|
| Number (peaks) | Rate (%) | Number (peaks) | Rate (%) | |
| Aminocarboxylic acid | 13 | 6.7 | 3 | 1.9 |
| Nucleoside | 2 | 1.0 | 0 | 0.0 |
| Organic acid | 2 | 1.0 | 1 | 0.6 |
| Peptide | 1 | 0.5 | 2 | 1.2 |
| Isoprenoid | 66 | 33.8 | 27 | 16.8 |
| Phenolic | 31 | 15.9 | 89 | 55.3 |
| Alkaloid | 2 | 1.0 | 1 | 0.6 |
| Other | 28 | 14.4 | 18 | 11.2 |
| Multiple | 48 | 24.6 | 19 | 11.8 |
| SEC (there are some DB hits, but unlikely) | 2 | 1.0 | 1 | 0.6 |
| Total | 195 | 100.0 | 161 | 100.0 |
Identification of 18O-labeled unknown flavonoids
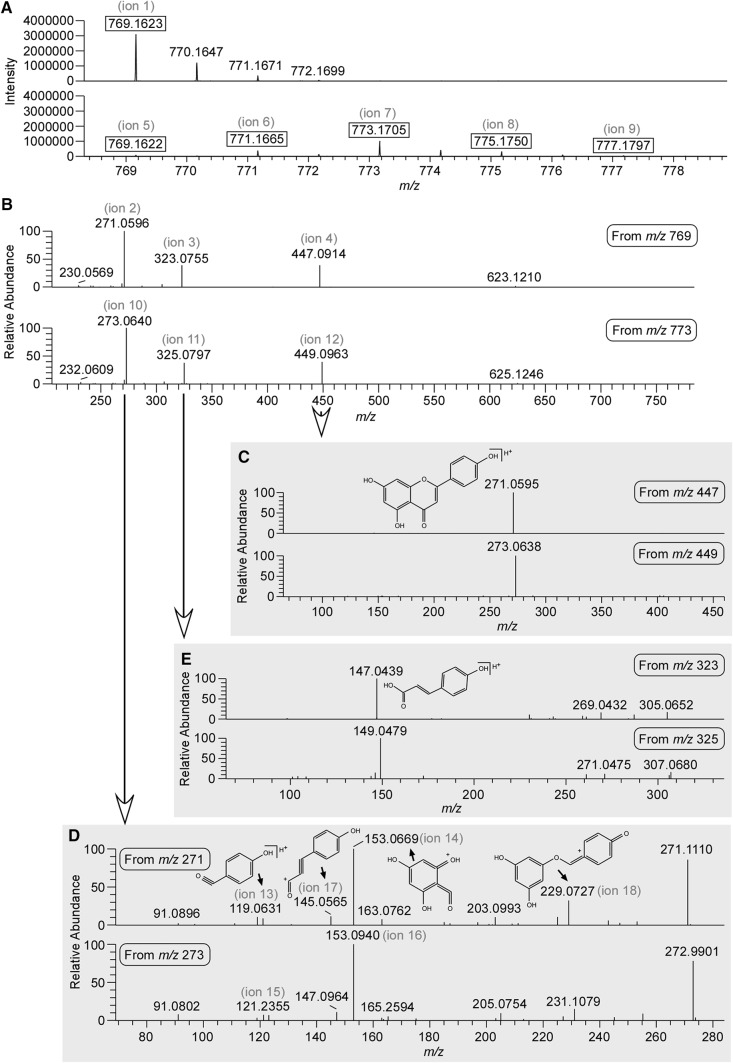
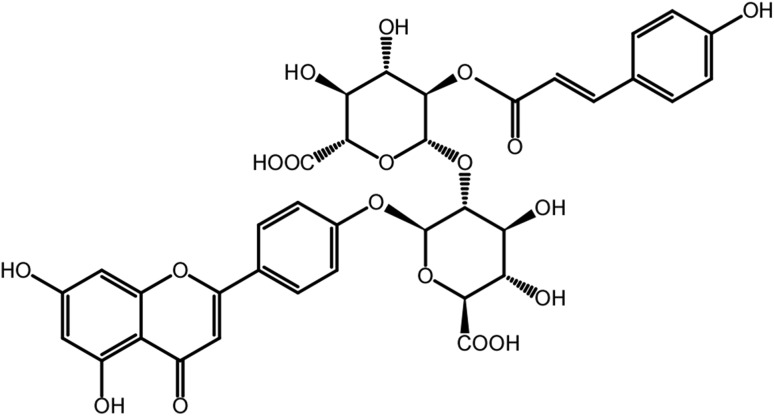
As described in a previous report, ShiftedIonsFinder is useful for finding candidates of known and unknown flavonoids by calculating theoretical m/z values by adding modification groups to aglycone like a building block (Kera et al. 2014). Potential flavonoids were searched for within the 511 peaks of M. truncatula shoot with regard to some modification groups. As a result, 26 peaks were picked up as flavonoid candidates (Supplement data 3). As an example, we focused on peak No. 312 (ion 1: m/z 769.1623 in Fig. 2a), one of the flavonoid candidates whose mass was not matched to referenced databases. We first attempted to estimate its structure by a MS-based approach. This peak yielded three candidate elemental compositions, C47H29O9P1, C36H32O19, and C54H24O6 within 3 ppm, and 38 candidate flavonoids. In order to estimate its detailed structure, additional MSn analyses with high resolution and high mass accuracy using Orbitrap Fusion (Thermo Fisher Scientific, USA) were performed. At first, three major fragments (ion 2: m/z 271.0596; ion 3: m/z 323.0756; and ion 4: m/z 447.0916) were detected by MS2 analysis from the parent monoisotopic ion (Fig. 2b). In positive electrospray ionization-mode fragmentation of flavonoid glycosides, it is well known that after cleavage of glycosidic bonds between the glycoside group and aglycone (Cuyckens and Claeys 2004), the product ion for a flavonoid aglycone results. It is strongly suggested that ion 2 at m/z 271.0596 is the aglycone. Among the 38 candidate flavonoids suggested by ShiftedIonsFinder, the structure having such an aglycone was [Aglycone + 2 GlcUA + Coumaroyl + H]+ (Table 3). This estimation indicates that ion 3 (m/z 323.0756) was [GlcUA + Coumaroyl + H]+, and ion 4 (m/z 447.0916) was [Aglycone + GlcUA + Coumaroyl + H]+. In fact, MS3 analysis of ion 3 and ion 4 supported the estimation, and the fragmentation pattern of ion 2 agreed with that of apigenin using m/zCloud (https://www.m/zcloud.org/; Thermo Fisher Scientific, USA). Taken together, this unknown peak No. 312 (m/z 769.1624) was annotated as [Apigenin] + 2 GlcUA + Coumaroyl + [H]+, and the elemental composition was C36H32O19. The structure was finally identified as apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside] using a UHPLC–MS–SPE–NMR system (Fig. 3, Supplement data 4) (Sumner et al. 2014, 2015). To date, many types of apigenin glucuronides have been reported from M. truncatula (Kowalska et al. 2007; Marczak et al. 2010) and Medicago sativa (Stochmal et al. 2001a, b). Apigenin 7-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside] has already been reported (Marczak et al. 2010). Interestingly, glucuronidation at the 4′ position of apigenin was found in M. sativa, but not in M. truncatula. Additionally, we also identified peak No. 335 (m/z 799.1730) as apigenin 4′-O-[2′-O-feruloyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside], which was found in M. sativa. This result is the first report of apigenin 4′-glucuronides present in M. truncatula. There were other peaks with the same mass values (m/z 769.1624 and m/z 799.1730) at different retention times in our data, suggesting the presence of structural isomers in M. truncatula.
Fig. 2.
MSn analysis of peak no. 312, apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside], by LC–orbitrap-fusion. a The full scan of unlabeled (above) and 18O-labeled sample (bottom) by Orbitrap. b MS2 analysis of peak no. 312 (above) and ion 7 (bottom) by Orbitrap. c MS3 analysis of ion 4 (above) and ion 12 (bottom) by Orbitrap. d MS3 analysis of ion 2 (above) and ion 10 (bottom) by ion trap. e MS3 analysis of ion 3 (above) and ion 11 (bottom) by Orbitrap
Table 3.
Thirty-eight structural candidates of peak No. 312 (m/z 769.1624) by ShiftedIonsFinder
| Ave. mass (detected) | Ave. mass (actual) | Name | Class | Ioniz.Mode | Xyl (C5H8O4): lag (times) | Rha (C6H10O4): lag (times) | Glc (C6H10O5): lag (times) | GlcUA (C6H8O6): lag (times) | Cinnamoyl (C9H6O1): lag (times) | Coumaroyl (C9H6O2) : lag (times) | Caffeoyl (C9H6O3): lag (times) | Feruloyl (C10H8O3): lag (times) | Malonyl (C3H2O3): lag (times) | Succinyl (C4H4O3): lag (times) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 255.0652 | 254.0579 | Chrysin (C15H10O4) | Flavone | [M + H]+ | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| 255.0652 | 254.0579 | Daidzein (C15H10O4) | Isoflavone | [M + H]+ | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| 271.0601 | 270.0528 | Sulphuretin (C15H10O5) | Aurone | [M + H]+ | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| 271.0601 | 270.0528 | Apigenin (C15H10O5) | Flavone | [M + H]+ | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| 271.0601 | 270.0528 | Genistein (C15H10O5) | Isoflavone | [M + H]+ | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| 271.0601 | 271.0606 | Pelargonidin (C15H11O5) | Anthocyanidin | [M]+ | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| 273.0757 | 272.0685 | "Naringenin chalcone, butein (C15H12O5)" | Chalcone | [M + H]+ | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 273.0757 | 272.0685 | Naringenin (C15H12O5) | Flavanone | [M + H]+ | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 275.0914 | 274.0841 | Phloretin (C15H14O5) | Dihydrochalcone | [M + H]+ | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 |
| 287.0550 | 286.0477 | Aureusidin (C15H10O6) | Aurone | [M + H]+ | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| 287.0550 | 286.0477 | Luteolin (C15H10O6) | Flavone | [M + H]+ | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| 287.0550 | 286.0477 | Kaempferol (C15H10O6) | Flavonol | [M + H]+ | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| 287.0550 | 287.0556 | Cyanidin (C15H11O6) | Anthocyanidin | [M]+ | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| 289.0707 | 288.0634 | Pentahydroxychalcone (C15H12O6) | Chalcone | [M + H]+ | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Pentahydroxychalcone (C15H12O6) | Chalcone | [M + H]+ | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Pentahydroxychalcone (C15H12O6) | Chalcone | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| 289.0707 | 288.0634 | Pentahydroxychalcone (C15H12O6) | Chalcone | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 289.0707 | 288.0634 | Eriodictyol (C15H12O6) | Flavanone | [M + H]+ | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Eriodictyol (C15H12O6) | Flavanone | [M + H]+ | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Eriodictyol (C15H12O6) | Flavanone | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| 289.0707 | 288.0634 | Eriodictyol (C15H12O6) | Flavanone | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 289.0707 | 288.0634 | Dihydrokaempferol (C15H12O6) | Flavanonol | [M + H]+ | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Dihydrokaempferol (C15H12O6) | Flavanonol | [M + H]+ | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 289.0707 | 288.0634 | Dihydrokaempferol (C15H12O6) | Flavanonol | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| 289.0707 | 288.0634 | Dihydrokaempferol (C15H12O6) | Flavanonol | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 291.0863 | 290.0790 | Leucopelargonidin (C15H14O6) | Leucoanthocyanidin | [M + H]+ | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 |
| 291.0863 | 290.0790 | Catechin (C15H14O6) | Flavanol | [M + H]+ | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 |
| 303.0863 | 302.0790 | Hesperetin (C16H14O6) | Flavanone | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 |
| 305.0656 | 304.0583 | Dihydroquercetin (C15H12O7) | Flavanonol | [M + H]+ | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 |
| 305.0656 | 304.0583 | Dihydroquercetin (C15H12O7) | Flavanonol | [M + H]+ | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| 305.0656 | 304.0583 | Dihydroquercetin (C15H12O7) | Flavanonol | [M + H]+ | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| 307.0812 | 306.0740 | Leucocyanidin (C15H14O7) | Leucoanthocyanidin | [M + H]+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 307.0812 | 306.0740 | Leucocyanidin (C15H14O7) | Leucoanthocyanidin | [M + H]+ | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 |
| 307.0812 | 306.0740 | Gallocatechin (C15H14O7) | Flavanol | [M + H]+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 307.0812 | 306.0740 | Gallocatechin (C15H14O7) | Flavanol | [M + H]+ | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 |
| 321.0605 | 320.0532 | Dihydromyricetin (C15H12O8) | Flavanonol | [M + H]+ | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 |
| 321.0605 | 320.0532 | Dihydromyricetin (C15H12O8) | Flavanonol | [M + H]+ | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| 323.0761 | 322.0689 | Leucodelphinidin (C15H14O8) | Leucoanthocyanidin | [M + H]+ | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
Lag (times) means the number of each possible modification group (Kera et al. 2014)
Fig. 3.
The structure of apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside]. It was identified using a UHPLC–MS–SPE–NMR system and the detail information was shown in supplemental data 4
Discussion of 18O-labeled positions in the metabolite
An important aim of this study is to estimate the 18O-labeled position in metabolites with MS-based information. For peak No. 312 (m/z 769.1624, C36H32O19, apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside]), five corresponding ions (ion 5: m/z 769.1622; ion 6: m/z 771.1665; ion 7: m/z 773.1705; ion 8: m/z 775.1750; and ion 9: m/z 777.1797) were detected, and the intensity of ion 7 was highest in the 18O-labeled sample (Fig. 2a). Although we have not investigate whether the 18O abundance ratio changed during cultivation, that of peak No. 312 from several five weeks old plants was similar. This indicates that the labeled molecule containing two 18O atoms was more abundant, and also that there are two major oxygenase-catalyzed steps in the biosynthetic pathway of apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside]. In order to estimate the 18O-labeled position, MSn analysis for the monoisotopic ion and ion 7: m/z 773.1705 was performed using Orbitrap Fusion. In MS2 analysis of ion 7: m/z 773.1705, the fragmentation pattern was similar to that of the monoisotopic ion, containing three major fragments (ion 10: m/z 273.0640; ion 11: m/z 325.0797; and ion 12: m/z 449.0963) (Fig. 2b). Comparison with the mass of each responsible ion indicated that apigenin had one 18O atom, [GlcUA + Coumaroyl] (2′-O-coumaroyl-glucuronopyranoside) had one 18O atom, and [Apigenin + GlcUA] (apigenin 4′-O-glucuronopyranoside) had one 18O atom, respectively. Here, one of the two 18O atoms was thought to be incorporated in apigenin. Detection of the major fragment of m/z 273.0638 from ion 12: m/z 449.0963 in MS3 analysis supports this estimation (Fig. 2c). In some, but not all, cases, MSn analysis can indicate a more confined 18O-labeled position. In MS3 analysis of apigenin (ion 2), two characteristic fragments (ion 13: m/z 119.0631 and ion 14: m/z 153.0669) are known retro-Diels–Alder fragment ions (Fig. 2d). Focusing on the responsible ions (ion 15: m/z 121.2355 and ion 16: m/z 153.0940) in MS3 analysis of ion 10, the 18O responsible mass shift was detected between ion 13 and ion 15, but not between ion 14 and ion 16 (Fig. 2d). In addition to ion 13, other fragment ions containing a 4′-hydroxy group (ion 17: m/z 145.0565 and ion 18: m/z 229.0727) also showed the 18O mass shift, whereas ion 16 contains all four oxygen atoms except the 4′-hydroxy group. Hence, the 18O atom was specifically incorporated into the 4′-hydroxy group of apigenin. According to the apigenin biosynthetic pathway, the hydroxy groups at positions 1, 4, 5, and 7 are derived from malonyl CoA, which is synthesized by an oxygenase-independent pathway, and the 4′-hydroxy group is derived from p-coumaroyl CoA by cinnamic acid 4-hydroxylase (C4H), which is a cytochrome P450 monooxygenase (Saito et al. 2013). Thus, specific incorporation into the 4′-hydroxy group is reasonable and indicates that non-specific incorporation of an 18O atom by recycling during one month of labeling is unlikely compared with the oxygenase-catalyzing reaction. On the other hand, by MS3 analysis of ion 3: m/z 323.0756 and ion 11: m/z 325.0797, one major fragment (m/z 147.0439 [Coumaroyl]) was detected from ion 3, and one major fragment (m/z 149.0479) was detected from ion 11, indicating that one 18O atom in ion 11 was derived from [Coumaroyl] (Fig. 2e). We could not determine the detailed labeling position by MSn analysis, but an 18O atom was also thought to be incorporated at the C-4 position of [Coumaroyl] by C4H. Finally, tiny ion 8: m/z 775.1750 and ion 9: m/z 777.1797 were responsible for the incorporation in [GlcUA] (data not shown). Since UDP-glucuronic acid is biosynthesized in plants through both a UDP-glucose 6-dehydrogenase-catalyzing reaction, in which H2O is used as an oxygen source, and a myo-inositol oxygenase-catalyzing reaction, in which O2 is used (Roberts 1971), these weak ions might reflect the biosynthetic pathway in M. truncatula. Consequently, these data well supported that ion 7 containing two 18O atoms from [Coumaroyl] was more abundant.
Concluding remarks
We have studied pathway-specific metabolome analysis using 18O2-labeled M. truncatula. This economical 18O2 labeling system enables whole plant labeling for over 1 month with 18O2-air. From untargeted analysis data, we succeeded in the identification of an unknown 18O-labeled flavonoid, apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside]. By MSn analysis of this flavonoid in unlabeled and 18O2-labeled samples, we estimated that 18O atoms were specifically incorporated in apigenin, the coumaroyl group, and glucuronic acid. For apigenin, an 18O atom was specifically incorporated in the 4′-hydroxy group. This approach enables the determination of not only the presence of oxygenase in the biosynthetic pathway but also that the identification of oxygen atoms in metabolites derived from oxygenases. Therefore, pathway-specific metabolome analysis can be a key for linking metabolite information to enzymatic information.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Strategic International Collaborative Research Program (SICORP), JST, and Japan Advanced Plant Science Network. We would also like to acknowledge support from the USA National Science Foundation Award #1139489 which was a joint initiative with the Japanese Science and Technology Agency Metabolomics for a Low Carbon Society program, USA NSF RCN Award#1340058, and USA NSF MRI #1126719. We would like to thank Dr. Nozomu Sakurai, Mr. Manabu Yamada, Dr. Kazuto Mannen for supporting data analysis.
Author contributions
KK designed the study, and wrote the initial draft of the manuscript. DDF contributed to UHPLC–ESI–QTOF–MS/MS analysis and assisted in the preparation of the manuscript. DJW contributed to NMR analysis and identification of apigenin 4′-O-[2′-O-coumaroyl-glucuronopyranosyl-(1–2)-O-glucuronopyranoside]. YN contributed to LC–MS analysis and data analysis. NS contributed to establish an economical sealed-glass bottle system. TA and YO contributed to data analysis and untargeted metabolite annotation. LWS and HS devised the project and the main conceptual ideas. All authors critically discussed and reviewed the final manuscript.
Funding
This study was funded by Strategic International Collaborative Research Program (SICORP) of JST and NSF as shown in acknowledgement.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11306-018-1364-6) contains supplementary material, which is available to authorized users.
References
- Afendi FM, Okada T, Yamazaki M, et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant and Cell Physiology. 2012;53(2):e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Cuyckens F, Claeys M. Mass spectrometry in the structural analysis of flavonoids. Journal of Mass Spectrometry. 2004;39(1):1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, et al. Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labelling, in combination with high-resolution mass spectrometry. The Plant Journal. 2011;68(2):364–376. doi: 10.1111/j.1365-313X.2011.04682.x. [DOI] [PubMed] [Google Scholar]
- Glaser K, Kanawati B, Kubo T, Schmitt-Kopplin P, Grill E. Exploring the Arabidopsis sulfur metabolome. The Plant Journal. 2014;77(1):31–45. doi: 10.1111/tpj.12359. [DOI] [PubMed] [Google Scholar]
- Harada K, Fukusaki E, Bamba T, Sato F, Kobayashi A. In vivo 15N-enrichment of metabolites in suspension cultured cells and its application to metabolomics. Biotechnology Progress. 2006;22(4):1003–1011. doi: 10.1021/bp060139z. [DOI] [PubMed] [Google Scholar]
- Hayaishi O, Katagiri M, Rothberg S. Mechanism of the pyrocatechase reaction. Journal of the American Chemical Society. 1955;77(20):5450–5451. doi: 10.1021/ja01625a095. [DOI] [Google Scholar]
- Hecht S, Kammhuber K, Reiner J, Bacher A, Eisenreich W. Biosynthetic experiments with tall plants under field conditions. 18O2 incorporation into humulone from Humulus lupulus. Phytochemistry. 2004;65(8):1057–1060. doi: 10.1016/j.phytochem.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Huege J, Sulpice R, Gibon Y, Lisec J, Koehl K, Kopka J. GC-EI-TOF-MS analysis of in vivo carbon-partitioning into soluble metabolite pools of higher plants by monitoring isotope dilution after 13CO2 labelling. Phytochemistry. 2007;68(16–18):2258–2272. doi: 10.1016/j.phytochem.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, et al. Metabolite annotations based on the integration of mass spectral information. The Plant Journal. 2008;54(5):949–962. doi: 10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kera K, Ogata Y, Ara T, et al. ShiftedIonsFinder: A standalone Java tool for finding peaks with specified mass differences by comparing mass spectra of isotope-labeled and unlabeled data sets. Plant Biotechnology. 2014;31(3):269–274. doi: 10.5511/plantbiotechnology.14.0609c. [DOI] [Google Scholar]
- Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska I, Stochmal A, Kapusta I, et al. Flavonoids from barrel medic (Medicago truncatula) aerial parts. Journal of Agricultural and Food Chemistry. 2007;55(7):2645–2652. doi: 10.1021/jf063635b. [DOI] [PubMed] [Google Scholar]
- Leonard LT. A simple assembly for use in the testing of cultures of rhizobia. Journal of Bacteriology. 1943;45(6):523–527. doi: 10.1128/jb.45.6.523-527.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov A, Scigelova M. Coupling liquid chromatography to Orbitrap mass spectrometry. Journal of Chromatography A. 2010;1217(25):3938–3945. doi: 10.1016/j.chroma.2010.02.022. [DOI] [PubMed] [Google Scholar]
- Marczak L, Stobiecki M, Jasinski M, Oleszek W, Kachlicki P. Fragmentation pathways of acylated flavonoid diglucuronides from leaves of Medicago truncatula. Phytochemical Analysis. 2010;21(3):224–233. doi: 10.1002/pca.1189. [DOI] [PubMed] [Google Scholar]
- Mason H, Fowlks W, Peterson E. Oxygen transfer and electron transport by the phenolase complex 1. Journal of the American Chemical Society. 1955;77(10):2914–2915. doi: 10.1021/ja01615a088. [DOI] [Google Scholar]
- Nakabayashi R, Saito K. Metabolomics for unknown plant metabolites. Analytical and Bioanalytical Chemistry. 2013;405(15):5005–5011. doi: 10.1007/s00216-013-6869-2. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Sawada Y, Yamada Y, et al. Combination of liquid chromatography-Fourier transform ion cyclotron resonance-mass spectrometry with 13C-labeling for chemical assignment of sulfur-containing metabolites in onion bulbs. Analytical Chemistry. 2013;85(3):1310–1315. doi: 10.1021/ac302733c. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Afendi FM, Parvin AK, et al. KNApSAcK metabolite activity database for retrieving the relationships between metabolites and biological activities. Plant and Cell Physiology. 2014;55(1):e7. doi: 10.1093/pcp/pct176. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. Journal of Natural Products. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta D, Kanaya S, Suzuki H. Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Current Opinion in Biotechnology. 2010;21(1):35–44. doi: 10.1016/j.copbio.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Qiu F, Fine DD, Wherritt DJ, Lei Z, Sumner LW. PlantMAT: A metabolomics tool for predicting the specialized metabolic potential of a system and for large-scale metabolite identifications. Analytical Chemistry. 2016;88(23):11373–11383. doi: 10.1021/acs.analchem.6b00906. [DOI] [PubMed] [Google Scholar]
- Roberts RM. The formation of uridine diphosphate-glucuronic acid in plants. Uridine diphosphate-glucuronic acid pyrophosphorylase from barley seedlings. The Journal of Biological Chemistry. 1971;246(16):4995–5002. [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, et al. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiology and Biochemistry. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Enomoto M, et al. Tools and databases of the KOMICS web portal for preprocessing, mining, and dissemination of metabolomics data. BioMed Research International. 2014;2014:1–11. doi: 10.1155/2014/194812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Kanaya S, et al. An application of a relational database system for high-throughput prediction of elemental compositions from accurate mass values. Bioinformatics. 2013;29(2):290–291. doi: 10.1093/bioinformatics/bts660. [DOI] [PubMed] [Google Scholar]
- Stochmal A, Piacente S, Pizza C, De Riccardis F, Leitz R, Oleszek W. Alfalfa (Medicago sativa L.) flavonoids. 1. Apigenin and luteolin glycosides from aerial parts. Journal of Agricultural and Food Chemistry. 2001;49(2):753–758. doi: 10.1021/jf000876p. [DOI] [PubMed] [Google Scholar]
- Stochmal A, Simonet AM, Macias FA, et al. Acylated apigenin glycosides from alfalfa (Medicago sativa L.) var Artal. Phytochemistry. 2001;57(8):1223–1226. doi: 10.1016/S0031-9422(01)00204-7. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner, L. W., Lei, Z., Fine, D., et al. (2014). Plant metabolomics research highlight: Prof. Lloyd W. Sumner combines UHPLC-MS-SPE-NMR and Prof. Kazuki Saito employs FT-ICR-MS in novel strategies to identify unknown plant metabolites. Bruker Application Note # LCMS-85.
- Sumner LW, Lei Z, Nikolau BJ, Saito K. Modern plant metabolomics: Advanced natural product gene discoveries, improved technologies, and future prospects. Natural Product Reports. 2015;32(2):212–229. doi: 10.1039/C4NP00072B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.