Abstract
Background
The salmon louse, Lepeophtheirus salmonis, is an ectoparasitic copepod which feeds on the mucus, skin and blood of salmonid fish species. The parasite can persist on the surface of the fish without any effective control being exerted by the host immune system. Other ectoparasitic invertebrates produce compounds in their saliva, excretions and/or secretions which modulate the host immune responses allowing them to remain on or in the host during development. Similarly, compounds are produced in secretions of L. salmonis which are thought to be responsible for immunomodulation of the host responses as well as other aspects of crucial host-parasite interactions.
Methods
In this study we have identified and characterised the proteins in the excretory/secretory (E/S) products of L. salmonis using LC-ESI-MS/MS.
Results
In total 187 individual proteins were identified in the E/S collected from adult lice and pre-adult sea lice. Fifty-three proteins, including 13 serine-type endopeptidases, 1 peroxidase and 5 vitellogenin-like proteins were common to both adult and pre-adult E/S products. One hundred and seven proteins were identified in the adult E/S but not in the pre-adult E/S and these included serine and cysteine-type endopeptidases, vitellogenins, sphingomyelinase and calreticulin. A total of 27 proteins were identified in pre-adult E/S products but not in adult E/S.
Conclusions
The assigned functions of these E/S products and the potential roles they play in host-parasite interaction is discussed.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2885-6) contains supplementary material, which is available to authorized users.
Keywords: Lepeophtheirus salmonis, Excretory/secretory (E/S) products, Immunomodulators, Sea lice
Background
The sea louse or salmon louse, Lepeophtheirus salmonis (Krøyer, 1837) is an ectoparasitic copepod which feeds on the mucus, skin and blood of salmonids [1, 2]. There are eight developmental life stages from planktonic (2 naupliar stages) to infective copepodid through to chalimus (two male chalimus moults and two female chalimus moults), pre-adult (two moults) and adult [3]. Studies of the morphology of L. salmonis have revealed tegumental glands and labial (mouthpart) glands which are thought to be involved in secretion [4]. It has been reported that L. salmonis produces secretory compounds such as prostanoids and proteolytic enzymes which are involved in host immune-modulation and parasite virulence. The prostanoid prostaglandin E2 (PGE2) is present in secretory products produced by L. salmonis at levels similar to those observed in the saliva of terrestrial arthropods such as ticks [5] and the gene encoding prostaglandin E synthase (LsPGES2) has been identified and characterised in L. salmonis [6]. The effects of PGE2 have been well documented and these include causing renal vasodilation in rats [7], downregulation of cytokines IFN-γ and IL-2 in mouse T cell lines [8] and reducing adhesion of human lymphocytes to umbilical endothelial cells [9]. Prostaglandin E2 has also been shown to downregulate MHC I and II responses in lipopolysaccharide (LPS)-stimulated salmon head kidney cells including isolated leukocytes and cultured SHK-1 cells [10, 11].
Proteases are also secreted by L. salmonis [12–14] as well as other parasitic copepod species [15] and are thought to be important for feeding and immune evasion. Serine proteases such as trypsins, thought to be of L. salmonis origin, have been reported in salmon mucus [12] and in L. salmonis excretory/secretory (E/S) products [11]. Several of these trypsin-like serine proteases have been characterised and have increased mRNA transcript levels in the parasite midgut through all life stages as well as a suggested role in protein digestion [16].
Typically, secretions have been harvested from arthropods using bio-chemical neurotransmitters which induce salivation: tick saliva has been collected successfully following stimulation of salivation by dopamine and pilocarpine [17, 18] and this has led to the identification of numerous proteins using mass spectrometry [19]. Excretory/secretory compounds have also been collected from L. salmonis following dopamine stimulation either in solution or after application to the ventral surface of the cephalothorax [5]. The identification of proteins in both somatic and such E/S preparations from L. salmonis has been made possible through the generation of genomic and transcriptomic Expressed Sequence Tag (EST) datasets [20, 21]. Transcriptome assembly of RNAseq data from the closely related Caligus rogercresseyi has also led to a greater understanding into parasite protein functions and consequently has given insight to host-parasite interactions [22].
The aim of this study was therefore to use these genomic and post-genomic resources to identify and characterize the immunododulatory role of proteins from L. salmonis E/S products from two different life stages. Proteins were identified from adult and pre-adult life stages using liquid chromatography-electrospray ionization/tandem mass spectrometry (LC-ESI-MS/MS).
Methods
Excretory/secretory material collection
Salmon lice were collected directly from farmed fish at the Atlantic salmon harvesting plant, Mallaig, Scotland and at the Marine Environment Research Laboratory (MERL), Machrihanish, Scotland. Lice were transported directly in sea water maintained at a temperature between 6–8 °C to the Moredun Research Institute, Scotland, where they were maintained at 6 °C in aerated sea water. For the collection of E/S material from adult stages of the parasite, 3 replicate preparations were made from 10 live adult female lice which had been placed into 10 ml of sterile saline (0.85% NaCl) and agitated for 3 h at 6 °C. For pre-adult stages, duplicate preparations of 120 pre-adult lice of mixed sex were placed into 10 ml of sterile saline and agitated for 3 h at 6 °C. Following incubation, the saline solution was harvested and trichloroacetic acid added to it to a final concentration of 10% to precipitate any proteins present. Precipitated material was centrifuged at 10, 000× g for 30 min, supernatants were discarded and the pellets allowed to air dry. The pellets were then re-suspended in 50 μl 0.1% Sarkosyl in dH2O.
The protein concentrations of each re-suspended pellet were determined using the bicinchoninic acid (BCA) protein assay microplate version according to the manufacturer’s guidelines (Thermo, Peirce, Paisley, Scotland).
SDS PAGE and LC-ESI-MS/MS
Ten microlitre samples containing between 0.1 and 1.9 mg/ml-1 of protein in each E/S preparation were denatured with Lithium Dodecyl Sample buffer (Invitrogen) for 5 min at 100 °C. The proteins were separated by electrophoresis using a 12% Bis-Tris NuPage gel (Invitrogen, Paisley, Scotland) at 150V for 90 min. Following electrophoresis, resolved proteins were visualised with colloidal Coomassie Blue (Simply Blue Safe Stain™, Invitrogen).
Gel lanes were excised and each sliced horizontally from top to bottom to yield a series of 25 equal gel slices of 2.5 mm deep. Each of the resulting gel slices were then subjected to standard in-gel destaining, reduction, alkylation and trypsinolysis procedures [23]. Digests were transferred to low-protein-binding high-performance liquid chromatography (HPLC) sample vials immediately prior to liquid chromatography using electrospray ionisation-tandem mass spectrometry (LC-ESI-MS/MS) analysis. Liquid chromatography was performed using an Ultimate 3000 nano-HPLC system (Dionex, Leeds, England) comprising a WPS-3000 well-plate micro auto sampler, a FLM-3000 flow manager and column compartment, a UVD-3000 UV detector, an LPG-3600 dual-gradient micro-pump and an SRD-3600 solvent rack controlled by Chromeleon™ chromatography software (Dionex). A micro-pump flow rate of 246 μl/min-1 was used in combination with a cap-flow splitter cartridge, affording a 1/82 flow split and a final flow rate of 3 μl/min-1 through a 5 cm × 200 μm ID (polystyrene-divinylbenzene) monolithic reversed phase column (Dionex) maintained at 50 °C. Samples of 4 μl were applied to the column by direct injection. Peptides were eluted by the application of a 15 min linear gradient from 8–45% solvent B (80% acetonitrile, 0.1% (v/v) formic acid) and directed through a 3 nl UV detector flow cell. LC was interfaced directly with a 3-D high capacity ion trap mass spectrometer (amaZon-ETD, Bruker Daltonics, Bremen, Germany) via a low-volume (50 μl/min-1 max) stainless steel nebuliser (Agilent, CA95051, United States) and ESI. Parameters for tandem MS analysis were based on those described previously [24].
Database mining
Deconvoluted MS/MS data in .mgf (Mascot Generic Format) format were imported into ProteinScape™ V3.1 (Bruker Daltonics) proteomics data analysis software for downstream mining of a custom L. salmonis database. This custom database was constructed from all the “Lepeophtheirus salmonis” protein entries found in the NCBInr database as of January 2018 and comprised 38,092 sequences in total. Database searches were conducted utilising the Mascot™ V2.54.1 (Matrix Science) search engine. Mascot search parameters were set in accordance with published guidelines [25] and to this end, fixed (carbamidomethyl “C”) and variable (oxidation “M” and deamidation “N,Q”) modifications were selected along with peptide (MS) and secondary fragmentation (MS/MS) tolerance values of 0.5 Da whilst allowing for a single 13C isotope. Protein identifications obtained from each of the 25 individual gel slices per lane were compiled using the “protein list compilation” feature within Proteinscape [26]. From the compiled protein lists individual protein identifications were inspected manually and considered significant only if: (i) two peptides were matched for each protein; (ii) peptides were represented by a sequence coverage of > 5%; and (iii) each matched peptide contained an unbroken “b” or “y” ion series represented by a minimum of four contiguous amino acid residues.
Functional analysis
To investigate secretory involvement, signal peptide predictions were made from the translated cDNA sequence representing each protein using SignalP prediction software (CBS). Identified proteins were investigated using the Blast2Go software suite [27]. Sequences were blasted against the NCBInr database. Proteins were then assigned into functional groups by searching the InterPro databases and Gene ontology databases. Annotations from both searches were then merged. The Gene Ontology (GO) terms assigned to each protein were then used to construct pie charts based on Biological Process, Cellular Component and Molecular Function. The number of proteins and percentage were included with each GO term. Proteins with no assigned function or associated GO terms were allocated as “Other”.
Results
Protein profiles in E/S products
In total, 187 individual proteins were identified from E/S collected from adult and pre-adult sea lice. Adult E/S products displayed a wide spectrum of molecular weights with multiple discrete bands present between 24–220 kDa (Fig. 1a). The most dominant proteins with the greatest band intensity ranged between 170–212 kDa. A more diffuse protein gel profile was observed for the pre-adult E/S products with only a few discreet bands seen at 25, 35, 40 and 50 kDa with the dominant band at around 25 kDa (Fig. 1b).
Fig. 1.
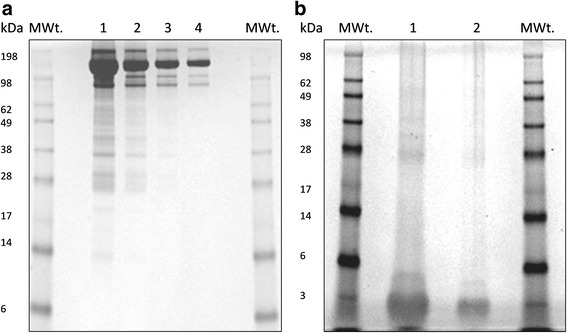
Lepeophtheirus salmonis excretory/secretory (E/S) products profiles on Coomassie Blue- stained polyacrylamide gels. a Lane MWt: molecular weight standards; Lane 1: undiluted adult female E/S products; Lane 2: 50% dilution of adult female E/S products; Lane 3: 25% dilution of adult female E/S products; Lane 4: 12.5% dilution of adult female E/S products. b Lane MWt: molecular weight standards; Lane 1: undiluted pre-adult E/S products; Lane 2: 50% dilution of pre-adult E/S products
Proteins common to adult and pre-adult E/S products
Fifty-three proteins were identified as being common to both adult and pre-adult E/S products (Additional file 1: Table S1). Nineteen of these 53 proteins were predicted to have a signal peptide among which were 6 serine type endopeptidases, 2 metalloendopeptidases, 1 peroxidase, 1 hydrolase, 1 hydratase, 1 protein binding, 2 vitellogenin-like proteins and 5 with no assigned function. In total 7 proteins had no assigned function or associated GO terms.
Adult female E/S products
In total, 107 proteins were identified which were unique to adult lice E/S products (Additional file 2: Table S2). Twenty nine of the 107 proteins were predicted to have a signal peptide and these included 2 serine type endopeptidases, 3 cysteine-type endopeptidases, 3 translocon subunits, 3 vitellogenins, 4 isomerases, 1 transferase, 1 hydrolase, 3 calcium binding proteins, 1 transmembrane emp24 protein 1 lysozyme, 1 sodium/calcium exchanger, 1 fibronectin, 1 heat-shock protein, 1 transforming growth factor, 1 endoplasmin and 2 uncharacterised proteins. The high molecular weight dominant bands observed in Fig. 1a correlate to the egg derived vitellogenin proteins (vitellogenin 1 and 2 and vitellogenin-like). The proteins in this group show the highest Proteinscape scores and sequence coverage and are represented within each replicated sample. Five proteins had no assigned function or associated GO terms.
Pre-adult E/S products
A total of 27 proteins were identified in pre-adult E/S products but not in adult E/S. Eight of the 27 proteins were predicted to have a signal peptide and these included 3 serine type endopeptidases, 2 cysteine type endopeptidases, 1 phosphatase, 1 acetylcholine receptor and 1 with no assigned function. (Additional file 3: Table S3). In total, 4 proteins had no assigned function or associated GO terms.
Assigned function of E/S products
Proteins common to the E/S products of both life stages (Fig. 2) or those that were detected only in adult stage (Fig. 3) or only in pre-adult stage (Fig. 4) were assigned to Biological Process, Cellular Component and Molecular Function. Proteins associated with proteolysis accounted for 34% of biological processes common to both life stages (Fig. 2a), 12% in adults only (Fig. 3a) and 13% in pre-adults only (Fig. 4a). Proteins involved in lipid transport were observed common to both stages at 11%, all being vitellogenin-like proteins and in adults only at 7% including vitellogenin 1, vitellogenin 2, vitellogenin-like and microsomal triglyceride transfer protein. No lipid transport proteins were observed only in pre-adults. Metabolic processes such as carbohydrate metabolism accounted for most proteins common to both stages, adult only and pre-adult only. Chitin metabolism was present only in pre-adults.
Fig. 2.
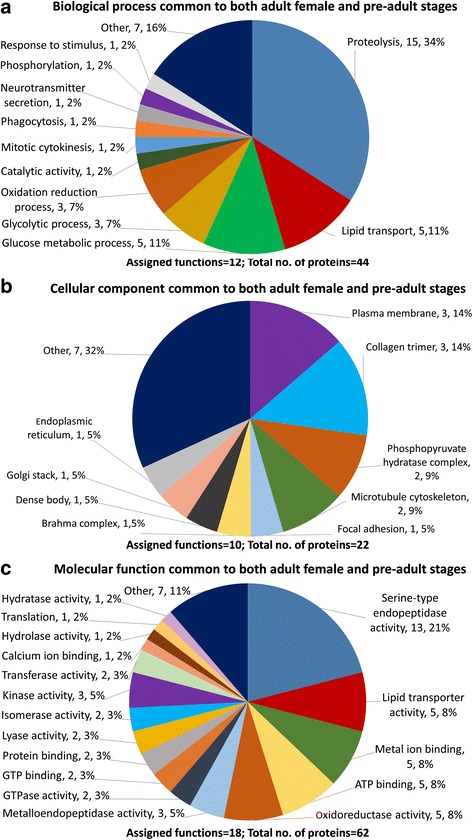
Lepeophtheirus salmonis excretory/secretory (E/S) products common to both adult female and pre-adult stages. a Biological Process. b Cellular Component. c Molecular Function
Fig. 3.
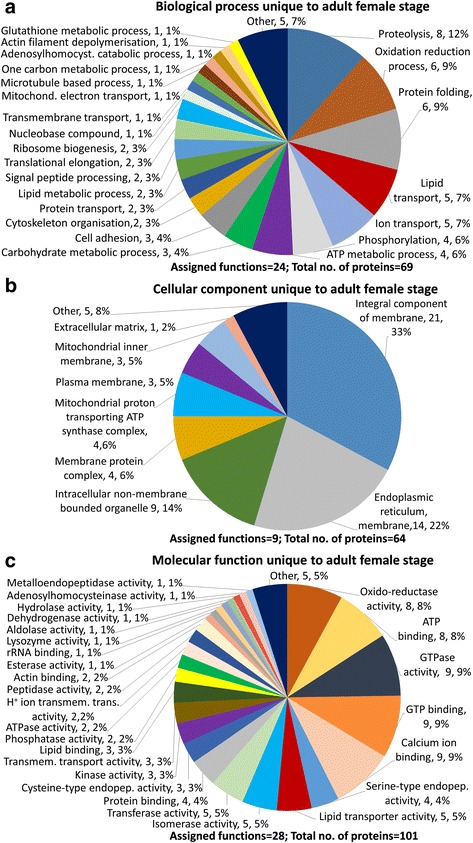
Lepeophtheirus salmonis excretory/secretory (E/S) products unique to adult female stage. a Biological Process. b Cellular Component. c Molecular Function
Fig. 4.
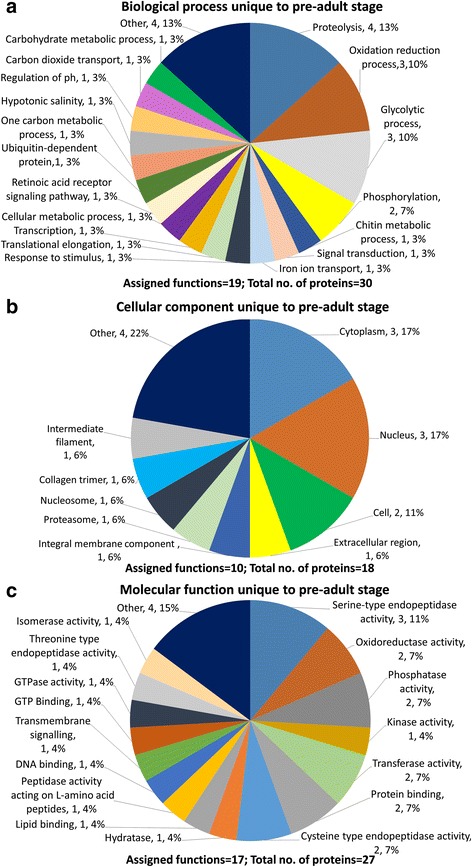
Lepeophtheirus salmonis excretory/secretory (E/S) products unique to pre-adult stage. a Biological Process. b Cellular Component. c Molecular Function
Proteins assigned to have cellular component consisted mostly of membrane proteins common to both life stages (Fig. 2b), adults only (Fig. 3b), and pre-adults only (Fig. 4b). A large proportion of proteins were assigned as other and had no assigned function or associated GO terms.
Proteins assigned to have molecular function, presented a wide range of enzymes observed in both life stages including metallopeptidases, reductases, transferases, kinases, GTPases, isomerases, lyases, hydrolases and hydratases. Serine type endopeptidases common to both stages, adult only and pre-adult only, constituted 21%, 4% and 11%, respectively (Figs. 2c, 3c, 4c). Cysteine endopeptidases were observed in adult only and pre-adult only stages at 3% and 7%, respectively. One threonine peptidase was detected in pre-adult E/S products only. Proteins associated with oxidoreductase activity were observed in 8%, 8% and 7% common to both stages, adult only and pre-adult only, respectively. Enzymes unique to adult only E/S products were dehydrogenase, aldolase, esterase, lysozyme and adenosylhomocysteinase. Protein, GTP, DNA, calcium and ion-binding proteins were among those proteins common to both stages, in adult and pre-adult only samples.
Discussion
Proteins secreted by salmon lice have long been thought to be important for the success of the parasite in evading the host immune response [12, 28]. Here, we have identified a suite of proteins excreted or secreted by adult and pre-adult salmon lice L. salmonis which may aid in understanding the interactions between the salmon louse and its host S. salar. In the present study, 21% of the 62 proteins, assigned to molecular function that we identified as being common to adult and pre-adult lice were serine peptidases, specifically trypsins and 5% were metallopeptidases. In addition, a serine collagenase was represented amongst these “common to both stages” proteinases which potentially facilitate feeding and digestion of host tissues, specifically for the breakdown of connective tissue and in this case mucus, skin and blood digestion. Metallopeptidases have been identified in the saliva of ticks (Ixodes scapularis) where they have effects on the ability of the tick to transmit pathogens by reducing adherence of polymorphonuclear leukocytes to the spirochete Borrelia burgdorferi, the causative agent of Lyme’s disease [29]. Other potential immunomodulatory proteins were the anti-oxidant family proteins, thioredoxin 2 and thiol peroxiredoxin isoform X3. These proteins may play a role in immune modulation by protecting against host reactive oxygen species as has been shown in helminth parasites [30]. Salmon louse E/S products have previously been shown to have immunomodulatory properties, for example causing decreased expression of IL-1β and MHC I in salmon head kidney macrophages [5].
Arginine kinase, which is solely expressed in invertebrates, is also present in L. salmonis (Additional file 1: Table S1) and in ectoparasite species, and is thought to play a key role in energy metabolism facilitated by buffering levels of intracellular adenosine triphosphate in muscle tissue [31]. Arginine kinase is a significant shellfish allergen [32] and has recently also been identified as an important allergen of house dust mites [33].
The hydratase protein, ganglioside GM2 activator-like protein observed in both life stages, contains an ML domain homologous to that found in Der p2 which is a major allergen in the house dust mite Dermatophagoides pteronyssinus [34]. In the hard tick Ixodes ricinus, the expression of the transcript encoding a Der p2 homologue is substantially increased during feeding and the encoded protein has been shown to have potential immunomodulatory properties on the host [35].
The processes involved in collecting and analysing E/S proteins, such as live lice collection and culture, genomic resource for the identification of specific peptides and bioinformatics to understand structure and function make the type of study reported here technically challenging. Fast et al. [11] reported low secreted protein yields from mixed life stage, dopamine induced L. salmonis with mostly low molecular weight proteins observed by SDS-PAGE. In our study presented here, the protein band profiles of E/S products observed from adult lice and pre-adult lice were markedly different. Protein gel profiles of pre-adult E/S products, show a very discreet group of bands between 20–55 kDa in size amongst which 11% of the identifiable proteins were serine type endopeptidases. Serine enodpeptidases, specifically trypsins, from cattle warble fly (Hypoderma lineatum) larvae have been associated with complement C3 degradation in cattle [36], demonstrating a possible immunomodulatory role for these molecules. Experiments carried out by Firth et al. [12] suggest that serine endopeptidases are secreted by L. salmonis in the presence of salmon mucus and, given the relatively high number of serine peptidases identified here in the pre-adult stages of the parasite, these proteins may be of key importance to the success of the louse in evading the host immune system before going into the final stage in the parasite life-cycle.
In contrast, the adult lice E/S products contained a wide spectrum of proteins from 20–220 kDa in size. Accordingly, the protein yield was higher with a broader diversity of protein types identified. This was despite using much greater numbers of pre-adult lice (n = 120) for collection of secretions than adult lice (n = 10). The differential protein profile between stages may reflect the importance of these proteins for feeding. On becoming sexually mature, adult lice appear to feed on blood more readily than pre-adult lice [1, 2]. In our study, we identified 3 cysteine proteases including Cathepsin L suggesting an important facilitatory role in increased feeding in this stage. A Cathepsin L has been shown to be vital in blood meal digestion in hard ticks (I. ricinus) [37] and a Fasciola hepatica cathepsin L has also been shown to have immunomodulatory properties by suppressing ovine T-lymphocyte proliferation and CD4 expression during parasitism by liver flukes [38].
It is worth noting that in previous studies, prostaglandins, thought to be important immune modulators, were observed in both excretory/secretory and secretory only sample collections [5]. However, prostaglandin synthase E2, although observed here, did not meet stipulated confidence criteria sufficiently to merit inclusion in the catalogue of identified ES proteins. Eichner et al. [6] recently reported that knockdown of the gene encoding PGE2 synthase by RNAi did not adversely affect parasite survival or the ability to infect the host and that the encoded protein was most likely involved in muscle contraction in planktonic stages and reproduction in adult females. Thus, the role of PGE2 in salmon louse immune-modulatory mechanisms remains to be determined.
Conclusions
The proteins identified in this study give an insight into the complexity of L. salmonis secretions, from serine proteases and redoxins which may be involved in host immune evasion, to metallopeptidases and cysteine peptidases such as cathepsin L thought to be involved in feeding. Utilizing the most up to date genomic resources currently available for L. salmonis, we can confidently identify a significant portion of the entire louse E/S protein complement. The biological functions of the E/S proteins identified here facilitate a better understanding of the ways in which the sea louse parasite interacts with its host environment.
Additional files
Table S1. E/S products common to both adult female and pre-adult stages. Data includes accession number, protein name, Proteinscape scores, no. of matched peptides, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 32 kb)
Table S2. E/S products unique to adult female stage. Data includes accession number, protein name, Proteinscape scores, no. of matched peptides, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 48 kb)
Table S3. E/S products unique to pre-adult stage. Data includes accession number, protein name, Proteinscape scores, no. of peptides matched, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 18 kb)
Acknowledgements
The authors would like to acknowledge the contributions of Erin Manson of the Moredun Proteomics Facility for expertise with sample processing for LC-ESI-MS/MS and staff of the MERL facility, Machrihanish for expertise in culture and provision of salmon lice.
Funding
Funding for this study was provided by the Technology Strategy Board (Innovate UK), Project No.101077 and Zoetis Animal Health.
Availability of data and materials
The datasets which support the conclusions are included within the article and its additional files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009391.
Abbreviations
- E/S
excretory/secretory
- LPS
lipopolysaccharide
- LC-ESI-MS/MS
liquid chromatography-electrospray ionisation-tandem mass spectrometry
- MERL
Marine Environment Research Laboratory
- NaCl
Sodium chloride
- NCIBnr
National Centre for Biotechnology Information non-redundant
- HPLC
high-performance liquid chromatography
- GO
Gene Ontology
- BCA
bicinchoninic acid
- kDa
kilodalton
- EST
expressed sequence tag
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PGE2
prostaglandin synthase E2
- MHC
major histocompatibility complex
- IFN- γ
interferon gamma
- IL-2
interleukin 2
- GTP
guanidine triphosphate
- RNAseq
RNA sequencing
- RNAi
RNA interference
Authors’ contributions
SH and DK designed and planned the experiments. SH, SJM, CM, HM and WR were involved with sample collection. SH and KM carried out the laboratory experiments and analysed the data. SH and AJN wrote the manuscript. KM, SJM, NFI, SA, RR, PS and JB revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval
The project was ethically approved by the University of Stirling ethics committee. Salmon lice were collected from donor fish using methods in accordance with The Home Office (Scientific Procedures Act) 1986.
Consent for publication
Consent for publication was approved by the funders.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2885-6) contains supplementary material, which is available to authorized users.
Contributor Information
Scott Hamilton, Email: scott.hamilton@moredun.ac.uk.
Kevin McLean, Email: kevin.mclean@moredun.ac.uk.
Sean J. Monaghan, Email: s.j.monaghan@stir.ac.uk
Carol McNair, Email: carolmmcnair@yahoo.co.uk.
Neil F. Inglis, Email: neil.inglis@moredun.ac.uk
Hazel McDonald, Email: h.c.mcdonald@stir.ac.uk.
Sandra Adams, Email: aa2@stir.ac.uk.
Randolph Richards, Email: r.h.richards@stir.ac.uk.
William Roy, Email: william.roy@stir.ac.uk.
Patrick Smith, Email: patrick.tethysaquaculture@gmail.com.
James Bron, Email: j.e.bron@stir.ac.uk.
Alasdair J. Nisbet, Email: alasdair.nisbet@moredun.ac.uk
David Knox, Email: davepknox@gmail.com.
References
- 1.Brandal PO, Egidius E, Romslo I. Host blood: a major food component for the parasitic copepod Lepeophtheirus salmonis Kroyer, 1838 (Crustacea: Caligidae) Norw J Zool. 1976;24:341–343. [Google Scholar]
- 2.Pike AW, Wadsworth SL. Sealice on salmonids: their biology and control. Adv Parasitol. 1999;44:233–237. doi: 10.1016/S0065-308X(08)60233-X. [DOI] [PubMed] [Google Scholar]
- 3.Hamre LA, Eichner C, Caipang CMA, Dalvin ST, Bron JE, Nilsen F, et al. The salmon louse Lepeophtheirus salmonis (Copepoda: Caligidae) life cycle has only two chalimus stages. PLoS One. 2013;8:e73539. doi: 10.1371/journal.pone.0073539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overgard AC, Hamre LA, Harasimczuk E, Dalvin S, Nilsen F, Grotmol S. Exocrine glands of Lepeophtheirus salmonis (Copepoda: Caligidae): distribution, developmental appearance, and site of secretion. J Morphol. 2016;277:1616–1630. doi: 10.1002/jmor.20611. [DOI] [PubMed] [Google Scholar]
- 5.Fast MD, Ross NW, Craft CA, Locke SJ, MacKinnon SL, Johnson SC. Lepeophtheirus salmonis: characterization of prostaglandin E(2) in secretory products of the salmon louse by RP-HPLC and mass spectrometry. Exp Parasitol. 2004;107:5–13. doi: 10.1016/j.exppara.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Eichner C, Overgard AC, Nilsen F, Dalvin S. Molecular characterization and knock-down of salmon louse (Lepeophtheirus salmonis) prostaglandin E synthase. Exp Parasitol. 2015;159:79–93. doi: 10.1016/j.exppara.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Haylor J, Towers J. Renal vasodilator activity of prostaglandin E2 in the rat anaesthetized with pentobarbitone. Br J Pharmacol. 1982;76:131–137. doi: 10.1111/j.1476-5381.1982.tb09198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betz M, Fox BS. Prostaglandin E2 inhibits production of Th1 lymphokines but not of Th2 lymphokines. J Immunol. 1991;146:108–113. [PubMed] [Google Scholar]
- 9.To SST, Schrieber L. Effect of leukotriene B4 and prostaglandin E2 on the adhesion of lymphocytes to endothelial cells. Clin Exp Immunol. 1990;81:160–165. doi: 10.1111/j.1365-2249.1990.tb05308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fast MD, Ross NW, Johnson SC. Prostaglandin E(2) modulation of gene expression in an Atlantic salmon (Salmo salar) macrophage-like cell line (SHK-1) Dev Comp Immunol. 2005;29:951–963. doi: 10.1016/j.dci.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Fast MD, Johnson SC, Eddy TD, Pinto D, Ross NW. Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 2007;29:179–189. doi: 10.1111/j.1365-3024.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 12.Firth KJ, Johnson SC, Ross NW. Characterization of proteases in the skin mucus of Atlantic salmon (Salmo salar) infected with the salmon louse (Lepeophtheirus salmonis) and in whole-body louse homogenate. J Parasitol. 2000;86:1199–1205. doi: 10.1645/0022-3395(2000)086[1199:COPITS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Fast MD, Burka JF, Johnson SC, Ross NW. Enzymes released from Lepeophtheirus salmonis in response to mucus from different salmonids. J Parasitol. 2003;89:7–13. doi: 10.1645/0022-3395(2003)089[0007:ERFLSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Ross NW, Firth KJ, Wang A, Burka JF, Johnson SC. Changes in hydrolytic enzyme activities of naive Atlantic salmon Salmo salar skin mucus due to infection with the salmon louse Lepeophtheirus salmonis and cortisol implantation. Dis Aquat Org. 2000;41:43–51. doi: 10.3354/dao041043. [DOI] [PubMed] [Google Scholar]
- 15.Perkins PS, Haley D, Rosenblatt R. Proteolytic enzymes in the blood-feeding parasitic copepod, Phrixocephalus cincinnatus. J Parasitol. 1997;83:6–12. doi: 10.2307/3284309. [DOI] [PubMed] [Google Scholar]
- 16.Kvamme BO, Skern R, Frost P, Nilsen F. Molecular characterisation of five trypsin-like peptidase transcripts from the salmon louse (Lepeophtheirus salmonis) intestine. Int J Parasitol. 2004;34:823–832. doi: 10.1016/j.ijpara.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman W. The influence of various factors on fluid secretion by in vitro salivary glands of ixodid ticks. J Exp Biol. 1976;64:727–742. doi: 10.1242/jeb.64.3.727. [DOI] [PubMed] [Google Scholar]
- 18.Sauer JR, McSwain JL, Bowman AS, Essenberg RC. Tick salivary gland physiology. Annu Rev Entomol. 1995;40:245–267. doi: 10.1146/annurev.en.40.010195.001333. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira CJ, Anatriello E, de Miranda-Santos IK, Francischetti IM, Sá-Nunes A, Ferreira BR, et al. Proteome of Rhipicephalus sanguineus tick saliva induced by the secretagogues pilocarpine and dopamine. Ticks Tick Borne Dis. 2013;4:469–477. doi: 10.1016/j.ttbdis.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichner C, Frost P, Dysvik B, Jonassen I, Kristiansen B, Nilsen F. Salmon louse (Lepeophtheirus salmonis) transcriptomes during post molting maturation and egg production, revealed using EST-sequencing and microarray analysis. BMC Genomics. 2008;9:126. doi: 10.1186/1471-2164-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasuike M, Leong J, Jantzen SG, von Schalburg KR, Nilsen F, Jones SR, et al. Genomic resources for sea lice: analysis of ESTs and mitochondrial genomes. Mar Biotechnol (NY) 2012;14:155–166. doi: 10.1007/s10126-011-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallardo-Escárate C, Valenzuela-Muñoz V, Nuñez-Acuña G. RNA-Seq analysis using de novo transcriptome assembly as a reference for the salmon louse Caligus rogercresseyi. PLoS One. 2014;9:e92239. doi: 10.1371/journal.pone.0092239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–5. [DOI] [PMC free article] [PubMed]
- 24.Batycka M, Inglis NF, Cook K, Adam A, Fraser-Pitt D, Smith DG, et al. Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increases throughput in proteomic analysis. Rapid Commun Mass Spectrom. 2006;20:2074–2080. doi: 10.1002/rcm.2563. [DOI] [PubMed] [Google Scholar]
- 25.Taylor GK, Goodlett DR. Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3420. doi: 10.1002/rcm.2225. [DOI] [PubMed] [Google Scholar]
- 26.Thiele H, Glandorf J, Hufnagel P. Bioinformatics strategies in life sciences: from data processing and data warehousing to biological knowledge extraction. J Integr Bioinform. 2010;7:141. doi: 10.1515/jib-2010-141. [DOI] [PubMed] [Google Scholar]
- 27.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustafa A, MacWilliams C, Fernandez N, Matchett K, Conboy GA, Burka JF. Effects of sea lice (Lepeophtheirus salmonis Kroyer, 1837) infestation on macrophage functions in Atlantic salmon (Salmo salar L.) Fish Shellfish Immunol. 2000;10:47–59. doi: 10.1006/fsim.1999.0229. [DOI] [PubMed] [Google Scholar]
- 29.Guo X, Booth CJ, Paley MA, Wang X, DePonte K, Fikrig E, et al. Inhibition of neutrophil function by two tick salivary proteins. Infect Immun. 2009;77:2320–2329. doi: 10.1128/IAI.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MW, Hutchinson AT, Dalton JP, Donnelly S. Peroxiredoxin: a central player in immune modulation. Parasite Immunol. 2010;32:305–313. doi: 10.1111/j.1365-3024.2010.01201.x. [DOI] [PubMed] [Google Scholar]
- 31.Werr M, Cramer J, Ilg T. Identification and characterization of two arginine kinases from the parasitic insect Ctenocephalides felis. Insect Biochem Mol Biol. 2009;39:634–645. doi: 10.1016/j.ibmb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Khora SS. Seafood-associated shellfish allergy: a comprehensive review. Immunol Investig. 2016;45:504–530. doi: 10.1080/08820139.2016.1180301. [DOI] [PubMed] [Google Scholar]
- 33.Xing P, Yu H, Li M, Xiao X, Jiang C, Mo L, et al. Characterization of arginine kinase, a novel allergen of Dermatophagoides farinae (Der f 20) Am J Transl Res. 2015;7:2815–2823. [PMC free article] [PubMed] [Google Scholar]
- 34.Friedland N, Liou HL, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Natl Acad Sci USA. 2003;100:2512–7. [DOI] [PMC free article] [PubMed]
- 35.Horácková J, Rudenko N, Golovchenko M, Grubhoffer L. Der-p2 (Dermatophagoides pteronyssinus) allergen-like protein from the hard tick Ixodes ricinus - a novel member of ML (MD-2-related lipid-recognition) domain protein family. Parasitology. 2010;137:1139–1149. doi: 10.1017/S0031182009992083. [DOI] [PubMed] [Google Scholar]
- 36.Boulard C. Degradation of bovine C3 by serine proteases from parasites Hypoderma lineatum (Diptera, Oestridae) Vet Immunol Immunopathol. 1989;20:387–398. doi: 10.1016/0165-2427(89)90083-4. [DOI] [PubMed] [Google Scholar]
- 37.Franta Z, Sojka D, Frantova H, Dvorak J, Horn M, Srba J, et al. IrCL1 - the haemoglobinolytic cathepsin L of the hard tick, Ixodes ricinus. Int J Parasitol. 2011;41:1253–1262. doi: 10.1016/j.ijpara.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Prowse RK, Chaplin P, Robinson HC, Spithill TW. Fasciola hepatica cathepsin L suppresses sheep lymphocyte proliferation in vitro and modulates surface CD4 expression on human and ovine T cells. Parasite Immunol. 2002;24:57–66. doi: 10.1046/j.0141-9838.2001.00438.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. E/S products common to both adult female and pre-adult stages. Data includes accession number, protein name, Proteinscape scores, no. of matched peptides, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 32 kb)
Table S2. E/S products unique to adult female stage. Data includes accession number, protein name, Proteinscape scores, no. of matched peptides, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 48 kb)
Table S3. E/S products unique to pre-adult stage. Data includes accession number, protein name, Proteinscape scores, no. of peptides matched, sequence coverage, signal peptide prediction and peptide sequence. (XLSX 18 kb)
Data Availability Statement
The datasets which support the conclusions are included within the article and its additional files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009391.


