Abstract
Direct interactions between proteins are essential for the regulation of their functions in biological pathways. Targeting the complex network of protein–protein interactions (PPIs) has now been widely recognized as an attractive means to therapeutically intervene in disease states. Even though this is a challenging endeavor and PPIs have long been regarded as “undruggable” targets, the last two decades have seen an increasing number of successful examples of PPI modulators, resulting in growing interest in this field. PPI modulation requires novel approaches and the integrated efforts of multiple disciplines to be a fruitful strategy. This perspective focuses on the hub-protein 14-3-3, which has several hundred identified protein interaction partners, and is therefore involved in a wide range of cellular processes and diseases. Here, we aim to provide an integrated overview of the approaches explored for the modulation of 14-3-3 PPIs and review the examples resulting from these efforts in both inhibiting and stabilizing specific 14-3-3 protein complexes by small molecules, peptide mimetics, and natural products.
Introduction
Protein–Protein Interactions (PPIs)
Protein–protein interactions (PPIs) are important in almost all biological processes. Most proteins do not function as single isolated entities but rather are engaged in a dynamic physical network with other proteins in the biomolecular context of a cell and its environment, often as part of a multiprotein complex. This makes the interactions of proteins as important as the biochemical activity of the protein itself. To understand the biological role of a protein, it is of great importance to understand and manipulate its underlying PPI network. An excellent example of this can be found in cancer biology, where the oncogenic kinase B-Raf can activate or inhibit the MAPK pathway by mechanisms that involve changes in the interactions of B-Raf with other members of the Raf kinase family.1−3
The “druggable genome” has been initially estimated to comprise approximately 1,500 single protein targets.4 Although this is still many more than the 266 human protein targets addressed by currently approved drugs,5 intentionally targeting PPIs significantly enlarges this number. The targeting of PPIs will be particularly valuable for diseases that cannot be addressed via “conventional” targets such as enzymes, receptors, or ion channels. By considering PPIs occurring in the human body, this situation can undoubtedly be improved given the size of the so-called protein–protein “interactome” with estimates lying between 130,0006 and 650,0007 protein complexes. Successfully addressing PPIs will vastly expand our opportunities for pharmacological intervention, especially by exploiting natural products.8 However, our understanding of biological mechanisms, and thus also which PPIs are relevant to disease, is still rudimentary. No further evidence of this is needed other than to reflect on the fact that the highest attrition rate during the drug-discovery process occurs during phase II clinical trials when it also becomes more costly.9 This attrition all too often arises because the desired biological effect is not observed with a given lead candidate. The availability of a good chemical probe, in contrast to genetic methods, uniquely allows temporary and titratable knockdown of a protein of interest, permitting its “druggability” and relevance to disease to be evaluated.10−12 Such probes can drive fundamental biology; for instance, publications on BRD4 (bromodomain) and hDM2 (ubiquitin ligase) have increased dramatically since the discovery of the PPI inhibitors bromodomain inhibitor (JQ1)13 and Nutlin.14
The issue of “druggability” is not unique to PPIs. However, their extensive regulatory role in biological mechanisms dictates that high-quality tool compounds modulating PPIs are urgently required as probes of healthy/disease biology and to provide starting points for drug discovery. Here, PPIs present a further challenge in that the interacting surfaces are larger, flatter, and generally deficient in the “binding-pockets” that define conventional ligandable15,16 proteins,9 although identification of hot-spots17 permits a binding site to be defined. This challenge is sufficiently daunting that, until recently, PPIs were considered too challenging to modulate using small-molecules and amenable only to modulation using biologics.18 However, the emergence of PPIs as small-molecule targets has now been conclusively demonstrated by the Nutlin series (Roche)14 and Navitoclax (Abbott).22 Although traditional approaches (e.g., high-throughput screening, fragment-based drug discovery, and computer-aided ligand design) are recognized as having limitations in terms of the identification of hit matter,19,20 the development of design-based approaches, e.g., based on foldamers, is encouraging.21 A number of strategic approaches to modulation can be envisioned comprising competitive (or orthosteric) inhibition, allosteric inhibition, and stabilization (Figure 1A–C) with general progress in this area summarized in numerous well-cited reviews.16,19,20,23−27 In terms of intervention within a pathway, the biological effect might be complex in that competitive inhibition of a PPI might result in stabilization of a PPI elsewhere within the pathway. Similarly, allostery affects not only the activation state of a given protein but the entire pathway in which it is embedded.28 For instance, the GTPase activity of Ras is modulated through its PPI with SOS at a remote site, and modulation of this interaction affects downstream PPI-mediated kinase activity within the entire pathway.29
Figure 1.
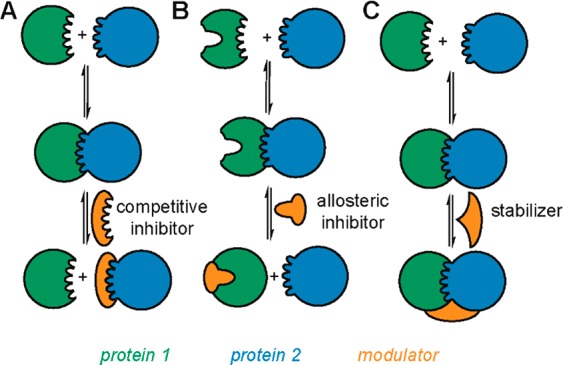
Schematic depicting different strategies for modulation of PPIs: competitive (orthosteric) inhibition (A), allosteric inhibition (B), and stabilization (C).
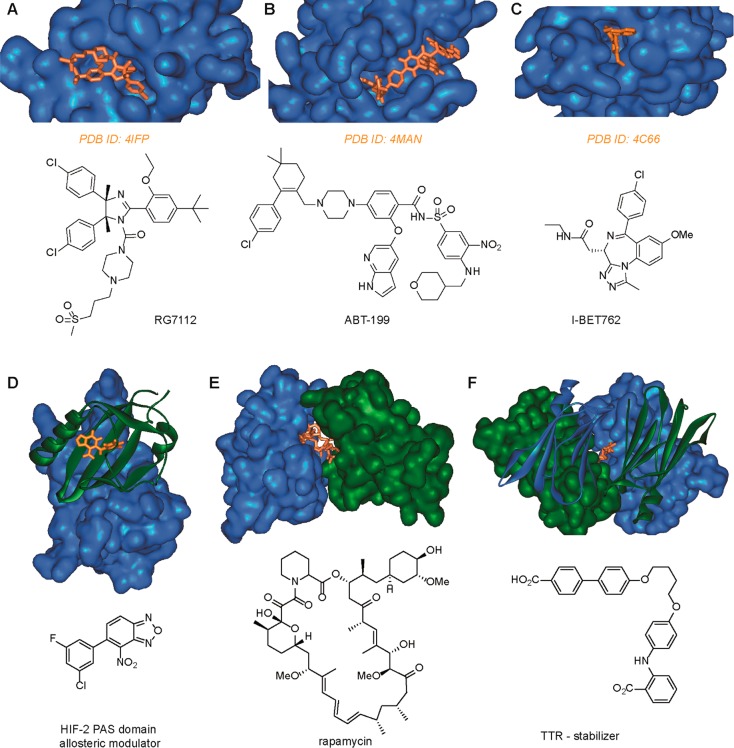
In terms of ligand approaches, competitive, allosteric, and stabilization modulation of PPIs are extremely different. Competitive inhibition is reasonably well established, and a number of inhibitors have been identified using conventional drug-discovery and design approaches. Prominent examples include a Nutlin follow-up from Roche (RG7112),30 Abbott (ABT-199),31 and GSK (I-BET762),32 which all entered clinical trials (Figure 2A–C). Challenges associated with competitive inhibition center on achieving sufficiently potent and selective recognition of either protein surface for inhibition to occur and the concomitant liability that might be introduced in terms of inhibiting all PPIs of the target protein. The biological response is proportional to the quality of the competitor. Allosteric inhibition may be more challenging to achieve by “design” and more likely to be identified by chance; however, allosteric inhibitors offer increased selectivity and self-limiting activity and, where PPIs are concerned, are much more likely to have the Lipinski properties33 characteristic of traditional small molecule drugs. A number of natural products have been identified to act through allosteric effects such as Taxol, which stabilizes tubulin so as to retard its polymerization.34 Drug-discovery and chemical biology programs have also delivered allosteric modulators. For instance, allosteric inhibitors of HIF-2 complex formation have been identified, which act through recognition of the PAS-B domain of the HIF-2α subunit (Figure 2D).35 Such compounds have been used to validate HIF-2 as a viable cancer target in renal cell cancer.36,37 Stabilization is less well established; however, it features prominently among natural products Brefeldin A,38 Forskolin,39 and Rapamycin40 (among others), which all act through stabilization of a PPI (Figure 2E and F). In addition, Tafamidis, one of the few PPI modulators to successfully reach the clinic, stabilizes the PPI transthyretin, which normally exists as a functional tetramer and aggregates in neurodegenerative diseases such as transthyretin amyloidosis. Small molecules such as Tafamidis that recognize and stabilize the tetrameric complex have been shown to kinetically retard aggregation and thus amyloid fibril formation.41,42 It should be noted that stabilizers of PPIs should also exhibit self-limiting biological response and greater selectivity because they also rely on ternary complex formation.
Figure 2.
Representative examples of competitive, allosteric, and stabilizing PPI ligands (X-ray structure (above), chemical structure (below)). (A) p53/hDM2 inhibitor RG7112 (PDB ID: 4IPF).43 (B) BH3/Bcl-2 inhibitor ABT-199 (PDB ID: 4MAN).31 (C) Fragment of BRD4/Histone inhibitor I-BET762 (PDB ID: 4C66).32 (D) HIF-2 PAS domain allosteric modulator (PDB ID: 4GHI).35 (E) FKBP12/Rapamycin/FRAP stabilizer complex (PDB ID: 1FAP).40 (F) Transthyretin stabilizer (PDB ID: 2FLM).42
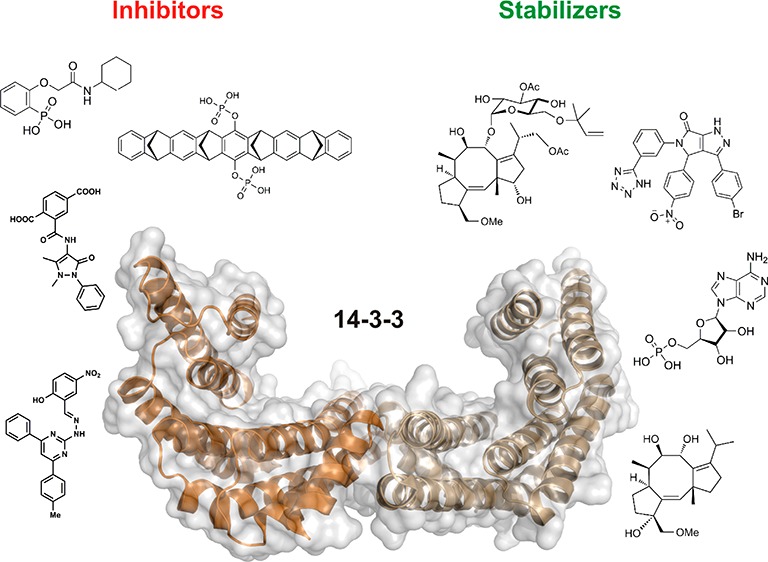
Despite these advances, PPI modulation remains a largely unsolved problem with inhibitors against only a few targets in current clinical trials.20 Progress is hampered by low success rates in identifying high-quality starting points for drug discovery19 and by a poor understanding of which PPIs may be targeted by small molecules.16 Improved ligand-discovery approaches and better conceptual understanding might well arise from the study of certain privileged protein classes. With several hundred identified protein interaction partners in eukaryotic cells, the family of the so-called 14-3-3 proteins is an especially interesting case for small-molecule PPI modulation. This protein family represents an outstanding testing ground for new conceptual approaches to PPI modulation and the elaboration of novel therapeutic approaches. PPIs of 14-3-3 proteins play key roles in numerous disease-relevant biological pathways and offer clear opportunities in terms of inhibition and stabilization. This perspective will highlight the state of the art in both areas with examples from diverse disease pathways.
14-3-3 Proteins
14-3-3 proteins are eukaryotic adaptor proteins involved in many cellular processes such as cell-cycle control, signal transduction, protein trafficking, and apoptosis.44 By binding to other proteins, 14-3-3 can assist in protein folding, protein localization, and stimulation or inhibition of other PPIs.45 Seven different mammalian 14-3-3 isoforms exist (α/β, γ, σ, δ/ζ, η, ε, and τ), which are highly conserved throughout species and mainly exist as dimers. Each monomer consists of nine alpha helices forming an amphipathic groove that can bind to (mostly phosphorylated) protein partners (Figure 3).46 Among the several hundred 14-3-3 interaction partners described so far, there are many disease-relevant proteins involved in key cellular processes, like the Raf kinases,47,48 cell-cycle phosphatase Cdc,49,50 transcriptional modulator YAP51,52 and tumor suppressor p53.53,54 This widespread involvement in human disease makes 14-3-3 proteins a highly interesting case for the development of technology to modulate their PPIs in a specific and efficient manner. Because both inhibition and stabilization of 14-3-3 PPIs have been shown with small molecules, the possibilities for novel pharmacological intervention by addressing this protein class are substantial. In this context, stabilization of 14-3-3 PPIs is an especially promising approach because the problem of specificity might be solved more easily than with inhibitors. This is due to the relatively high variability between the respective PPI interfaces. This variability might allow for the development of compounds that specifically bind to unique composite pockets at the PPI interfaces. In this way, not only can tool compounds be developed for the study of the underlying biology in, e.g., cancer, neurodegeneration, metabolic diseases, infection, and cystic fibrosis, but the approach can also be exploited in terms of drug discovery. Here, control of subcellular localization, enzymatic activity, and biological half-life can be envisioned as modes by which 14-3-3 PPI modulators could act, e.g., on transcription factors (YAP, c-Jun, MLF1, FOXOs), enzymes shuttling between cytoplasm and nucleus (Cdc25 phosphatases, HDACs), or kinases (B, C-Raf, LRRK2).
Figure 3.
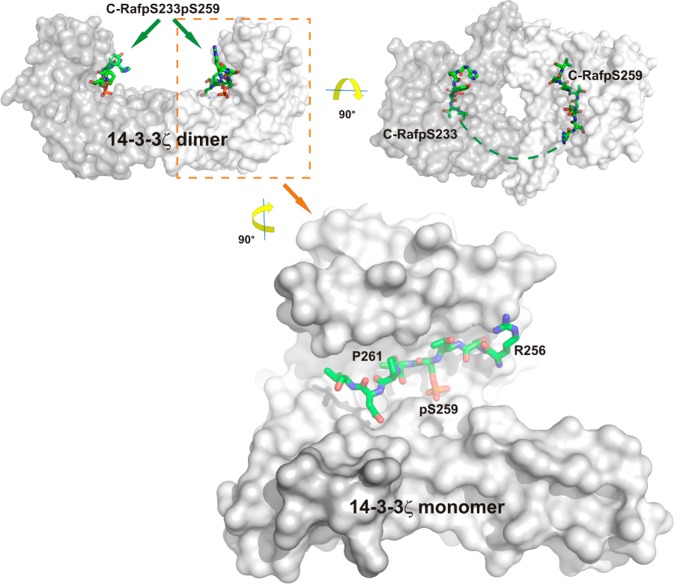
14-3-3 structure and binding of partner protein peptides exemplified by the 14-3-3ζ/C-Raf complex (PDB ID: 4FJ3).61 Top: the physiological 14-3-3 dimer can accommodate two phosphorylated peptide motifs. In the case of C-Raf, two of these motifs (pSer233 and pSer259) are located in the N-terminal region of this protein kinase. When synthesized as a diphospho peptide (C-RafpS233pS259) and crystallized with 14-3-3ζ dimer, a significant proportion of the peptide does not engage an intimate contact with 14-3-3 and is thus not visible in the X-ray crystal structure (right dimer: green dotted line). Bottom: C-RafpS259 site accommodated in the groove of a 14-3-3ζ monomer.
In recent years, a growing number of crystal structures of 14-3-3 in complex with different binding partner motifs have been published, for example, the cystic fibrosis ion channel CFTR,55 the small heat shock protein HSPB6,56 phosducin,57 and the Parkinson’s disease-related kinase LRRK2.58 As dimeric species that dock onto pairs of specific phosphorylated serine- or threonine-containing motifs, 14-3-3 proteins are endowed with special signaling, mechanical, and evolutionary properties. Although there are a few cases where a 14-3-3 dimer interacts simultaneously with phosphorylated sites in two different targets, in most documented cases a single 14-3-3 dimer binds to two phosphorylated sites that lie in tandem in the same target protein. This means that a 14-3-3 dimer can act as a signaling integrator when two binding sites on a target are phosphorylated by different kinases. The mechanical effect of 14-3-3 will depend on the location of the two docking sites. For example, these paired sites may straddle a domain or motif whose function is masked by the 14-3-3, or 14-3-3 binding to a disordered region can force a disorder-to-order transition that creates a new functional site in the target.59,60
14-3-3 binding sites lie within motifs that are phosphorylated by basophilic protein kinases such as PKB/Akt, p90RSK, PKA, and AMPK.57 This means that 14-3-3 affinity capture and quantitative mass spectrometry procedures can be used to identify targets of, for example, regulation by insulin, growth factors, energy stress, and adrenalin that activate these respective kinases. In this way, new 14-3-3-based mechanisms have been identified to explain how insulin and growth factors regulate synchronized shifts in glucose uptake, glycolysis, mTORC1 signaling, protein translation, and other regulatory events that promote cell growth and proliferation.62−64 Their roles as mediators of growth factor and nutrient signaling pathways are consistent with further findings that connect 14-3-3 proteins to a variety of human diseases. In addition to their participation in diverse cancers,44 they have been associated with the development of neurodegenerative diseases65 and virulence of human pathogenic organisms.66,67
The role of 14-3-3 proteins in parasitic organisms has only recently emerged. The rising interest in this field is justified by the limited panel of effective drugs currently available to treat parasite infections, the relevant side effects associated with these compounds, and the growing number of treatment-refractory cases.68 A survey of the recent literature has highlighted a number of reports showing the role of, and in a few cases the structural features of, 14-3-3 from parasites, which are briefly reviewed here.
Plasmodium falciparum and Plasmodium knowlesi are two species of protozoan parasites that can cause severe malaria infection in humans.69 In P. falciparum and P. knowlesi, the single isoform of 14-3-3 was shown to act as a chaperone only in specific life stages of the parasite.68 In Plasmodium berghei, the host skeletal protein dematin is translocated from the erythrocyte membrane within the parasite, where it interacts with the Plasmodium 14-3-3, thus influencing the remodeling of the erythrocytic cytoskeleton and modulating the host erythrocyte invasion.70
Eimeria tenella is a coccidian parasite that causes a serious intestinal disease in chickens. Although human infection by E. tenella has not been reported yet, this parasite has a significant economic impact with an estimated cost to the poultry industry of around $2.4 billion per annum worldwide, thus justifying the in depth study of its lifecycle and infection mechanisms.71 In E. tenella, a single isoform of 14-3-3 seems to be involved in the regulation of the mannitol pathway. In particular, the binding of 14-3-3 to the mannitol-1-phosphate dehydrogenase (M1PDH) was shown to inactivate the enzyme as soon as mannitol biosynthesis is complete. From a drug discovery perspective, it is important to note that this pathway is missing in higher eukaryotes, thus representing an attractive target for the development of selective drugs.68
Toxoplasma gondii is a protozoan parasite that causes a disease known as toxoplasmosis, a generally asymptomatic infection. Despite this, the parasite is known to cause severe congenital infection in humans and animals. The sexual reproduction of this parasite occurs in the intestine of definitive hosts (cats) while asexual multiplication takes place in various hosts, including humans.72 14-3-3 proteins have been detected in the asexual form of the parasite, namely the tachyzoite stage, that is virulent in humans.73 Moreover, it was demonstrated that, in this stage, 14-3-3 proteins from T. gondii induce hypermotility in infected host cells.74
Alveolar echinococcosis (AE) is a rare parasitic disorder that occurs after ingestion of eggs of Echinococcus multilocularis. AE is a tumorlike chronic disease, which can be fatal if left untreated.75 In E. multilocularis, 14-3-3 proteins have been reported to be implicated in the tumor-like growth process.76 Furthermore, it has been hypothesized that overexpressed 14-3-3 proteins may be involved in the promotion and/or maintenance of the progressive growth capacity of E. multilocularis larvae.77
Schistosoma mansoni is one of the major intestinal parasites that can cause schistosomiasis, the most widespread parasitic disease after malaria. This parasite encodes four 14-3-3 isoforms that have roles in host immunity, parasite development, and survival.78
Trichinella spiralis is a nematode parasite that is responsible for the development of trichinellosis, which is an important foodborne parasitic disease worldwide. The infection in humans is generally acquired by eating raw or inadequately cooked meat that contains encysted larvae of T. spiralis. 14-3-3 proteins from this parasite were shown to play a crucial role in the early stages of the infection by maintaining the host–parasite relationship.79
Cryptosporidium parvum is a parasite responsible for cryptosporidiosis, a diarrheal disease that affect humans and animals especially in developing countries.80 This infection is mainly caused by the ingestion of contaminated water, and an estimated 748,000 cryptosporidiosis cases occur annually.81 Unfortunately, only a limited number of drugs can be used to treat infections by C. parvum and most of them have low efficacy and an unknown mechanism of action. The three isoforms of 14-3-3 found in C. parvum (Cp14ε, Cp14a, and Cp14b) were crystallized in 2011, and two of them showed some interesting features that are unique among 14-3-3 proteins.80 In particular, in the isoform Cp14a, the substrate pocket is much more open compared to the classical folding of 14-3-3, leading to the hypothesis that this isoform can accommodate large substrates. The Cp14b isoform was able to bind a phosphorylated copy of the last six amino acid residues of its own C-terminus. This binding is much stronger when the C-terminus is truncated, thus suggesting a competition between C-terminus and its phosphorylated mimic.80 Even if other studies are necessary to understand the role of 14-3-3 in the C. parvum life cycle, these unique features described above can be exploited to develop novel strategies for cryptosporidiosis treatment.
Giardia duodenalis is a protozoan parasite that causes giardiasis by colonizing the upper portion of the small intestine in mammals. Giardiasis is the most common gastrointestinal diarrheal illness worldwide, and more than 200 million symptomatic human cases are reported annually.82,83 In this parasite, the single 14-3-3 isoform (g14-3-3) is essential for the development of cysts (the infective stage).84
Three crystallographic structures of G. duodenalis are available, which makes this protein amenable to study through structure-based computational methods. The crystal structure of g14-3-3 in the apo form revealed an unusual “open” conformation,85 whereas computational studies (supported by crystallographic evidence) proved that the post-translational modification (phosphorylation) on Thr214 of g14-3-3 induces a conformational rearrangement that leads to the “closed” and stable g14-3-3 conformation.84 This form corresponds to the peptide-bound g14-3-3 structure.
In summary, all these findings demonstrate that 14-3-3 proteins have substantial involvement in parasites’ life cycles. Even if the molecular basis for the role of 14-3-3 in pathogenicity is not well understood, we can speculate that this protein family could represent an alternative and promising axis to treat parasite infections. Despite rising interest in the role of 14-3-3 in parasites, significant advancement in basic research is required. Specifically, the development of a 14-3-3 modulator could be essential for unravelling the contribution of 14-3-3 proteins to parasite growth and survival in the host.
In general, the ubiquity of 14-3-3 protein involvement in numerous human diseases has sparked interest in their use as novel targets for drug discovery.44,49,65,86 The fact that 14-3-3 proteins serve purely as adapter proteins means that active compounds against 14-3-3 alone will impact several binding partners and thus likely give rise to unwanted pharmacology. However, if the drugs target the complex formation between 14-3-3 and a protein partner, either by inhibition or stabilization, then intrinsic specificity should be possible.24
Structural Biology of 14-3-3 PPIs
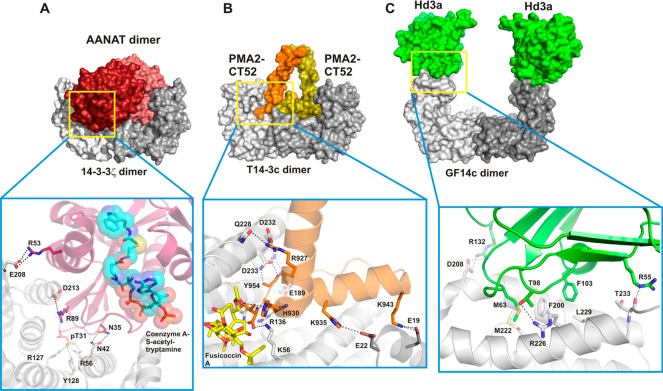
Most PDB entries of 14-3-3 crystal structures represent binary complexes of 14-3-3 with a peptide mimic of the phosphorylated binding site of the PPI partner protein. Only in a limited number of cases has crystallization of 14-3-3 with a larger part of the partner protein been possible. One reason for the difficulty in obtaining full-length structures of these complexes is the fact that the partner proteins themselves are often multidomain proteins and thus challenging to crystallize. A second reason is that the 14-3-3 binding sequences are prevalently localized in disordered regions of their target proteins and only undergo a disorder-to-order transition when binding to 14-3-3. In this transition, it is common for only the directly neighboring parts of the phosphorylated anchor residues to be involved. Thus, large parts of the partner protein regions remain disordered, which is a disadvantage for crystal growth. A commonly adopted alternative strategy therefore centers on using synthetic peptides comprising around 10–40 amino acid residues to mimic the partner protein binding motif. It is of course vital that the activity of any stabilizer or inhibitor found using the simplified 14-3-3/partner-protein-peptide system is also shown in the context of more physiologically relevant partner protein constructs. So far, however, crystallography of 14-3-3 in complex with synthetic peptides has been a useful system to obtain structural data of 14-3-3 modulators. These synthetic peptides typically bind in the 14-3-3 amphipathic binding groove like the 14-3-3ζ/C-Raf complex shown in Figure 3. Additionally, many structures have been reported where a small molecule is also bound to form a ternary complex. Historically, 14-3-3 binding sequences have been categorized in different motifs. Mode I and II interaction partners were defined to require an arginine at position −3 with respect to the phosphorylated serine or threonine residue and a proline residue at position +2 (more specifically; (I) RSX(pS/T)XP or (II) RX(F/Y)X(pS)XP).46 Mode III motifs were later defined as C-terminal sequences, where the phosphorylated serine or threonine is the penultimate residue of the binding partner.87 However, as recent reviews of the known 14-3-3 interactome have illustrated, 14-3-3 binding motifs can deviate from these well-defined motifs.60,88 Here, we will focus on examples where a larger domain of the partner protein is crystallized with 14-3-3: AANAT, PMA2, Hd3a (Figure 4), and more recently for HSPB6 (Figure 5).56,89,90 This set of crystal structures conveniently shows the variety of binding modes possible with 14-3-3 and illustrates their relevance in the identification of important and distinct interaction interfaces.
Figure 4.
Crystal structures of 14-3-3 complexes with larger partner protein constructs. (A) 14-3-3ζ/AANAT (PDB ID: 1IB1),89 (B) T14-3c/PMA2-CT52 (PDB ID: 2O98),108 and (C) GF14c/Hd3a (PDB ID: 3AXY).107 Upper row: surface representation of the complex. Lower row: details of the protein–protein complex interfaces.
Figure 5.
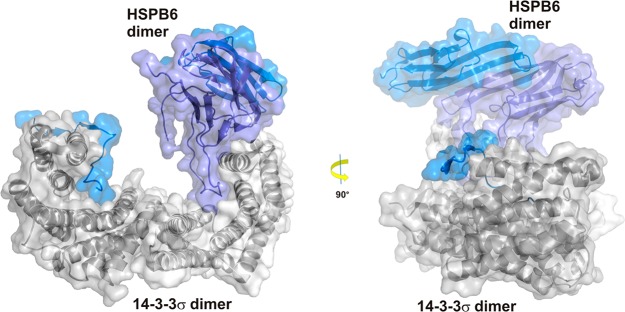
Complex between 14 and 3-3σ and HSPB6. Both proteins bind in a 2:2 stoichiometry but in contrast to the examples displayed in Figure 4 interact in an asymmetric fashion with the ACD dimer of HSPB6 binding to one 14-3-3 monomer and both N-terminal domains in the phospho-accepting grooves of 14-3-3 (PDB ID: 5LTW).56
14-3-3ζ/AANAT
Serotonin N-acetyltransferase (arylalkylamine N-acetyltransferase, AANAT) catalyzes the transfer of acetyl from acetyl-coenzyme A to serotonin, thus producing N-acetylserotonin, which is the precursor of melatonin. Melatonin levels are believed to govern the vertebrate daily rhythm with high levels occurring at night thus providing a hormonal analog signal of environmental lighting, which can be used to optimize circadian physiology and possibly form the basis of treatment in sleeping disorders.91,92 In 2001, the crystal structure of 14-3-3ζ in complex with AANAT was published by the group of Dyda showing that binding to 14-3-3 activates the enzyme by significantly increasing its affinity for its substrates serotonin and acetyl-coenzyme A.89 In the crystal structure, two AANAT monomers (residues 18–196) bind to the central channel of a 14-3-3 dimer (Figure 4A). In addition to the phosphorylation-dependent interaction of the N-terminus of AANAT, which is accommodated in an extended conformation by the amphipathic groove of the 14-3-3 monomer, the well-structured C-terminal part of the enzyme makes extensive contacts with the inner wall of the 14-3-3 channel.
T14-3c/PMA2-CT52 Complex
The plant plasma membrane H+-ATPase (PMA) generates both a chemical proton as well as an electrical gradient (membrane potential) across the plasma membrane.93 Because almost all transport mechanisms at the plant plasma membrane are energetically dependent on the PMA-generated electrochemical proton gradient, PMA plays a central role in plant physiology as the powerhouse for plant growth.94−97 One of the most important regulatory events for PMA is phosphorylation of the penultimate C-terminal threonine residue followed by binding of 14-3-3 proteins, which activates PMA.98,99 This activation is strongly increased by the natural product fusicoccin,100,101 which fills a gap in the interface of 14-3-3 with the regulatory C-terminus (CT) of PMA.102,103 The structure of the last 52 amino acid residues of the C-terminus of the PMA isoform 2 (PMA2-CT52) from tobacco (Nicotiana plumbaginifolia) in complex with Tobacco 14-3-3 isoform c (T14-3c) showed two PMA C-termini bound to one 14-3-3 dimer (Figure 4B). The C-terminal 30 amino acid residues bind as an elongated peptide and a short helix in the amphipathic groove of each 14-3-3 monomer, whereas the N-terminal 22 amino acid residues of PMA2-CT52 form a helix that perpendicularly leaves the 14-3-3 binding channel.
14-3-3/Hd3a Complex
Approximately 80 years ago, a substance was proposed to be synthesized in the leaves of flowering plants and transported to the shoot to induce flowering.104 This substance was named “florigen”, but its molecular identity was uncovered by showing that “florigen” is encoded by the highly conserved plant gene FLOWERING LOCUS T (FT),105 whose product is a mobile protein.106 In 2011, it was shown that the FT protein from rice (Hd3a) binds to 14-3-3 proteins in the apical cells of shoots to form a complex that migrates into the nucleus where it interacts with the basic leucine zipper (bZIP) transcription factor FD.107 Interestingly, the authors cocrystallized the complex between FT (Hd3a) and a rice 14-3-3 protein (GF14c) and used crystals of the binary complex to soak a short phosphopeptide derived from FD (OsFD1) and obtain the ternary complex of GF14c/Hd3a/OsFD1. In the crystal structure, the entire construct of Hd3a (residues 6–170) is visible, making it the second-largest 14-3-3 partner protein after AANAT (residues 18–196) that has been cocrystallized with 14-3-3. Two Hd3a molecules bind to one 14-3-3 dimer and occupy an unusual position that is not part of the central binding channel as seen with all other 14-3-3 ligands including AANAT. Rather, Hd3a binds to the “upper” edges of the horseshoe-like 14-3-3 dimer (Figure 4C). This site is close to the additional Cdc25C binding site predicted by a mutation study of human 14-3-3σ that lies outside of the central phosphopeptide binding channel.90
14-3-3σ/HSPB6 Complex
In early 2017, Sluchanko, Strelkov, and co-workers reported a 14-3-3 assembly with the full-length HSPB6 dimer (Figure 5).56 The authors emphasize this is the first crystal structure of a human small heat shock protein (HSP) in its functional state. The small HSPs comprise a family of ten ATP-independent chaperones with molecular masses in the range of 17–23 kDa.109 HSPB6 (also known as HSP20) is involved in smooth muscle relaxation and cardio protection and was identified as a binding partner for 14-3-3 upon phosphorylation of Ser16.109,110 HSPB6 dimerizes via its highly conserved α-Crystallin domain (ACD) that forms a β-sandwich, whereas both the N-terminal domain and C-terminal extension (NTD and CTE) that flank this region are highly unstructured. The interaction motif for 14-3-3 consists of a classical RRApSAP pattern located in the NTD.56
The authors cocrystallized 14-3-3 with two HSPB6-derived phosphopeptides (residues 13–20 and 11–23) containing the phosphorylated Ser16 before solving the structure of the full-length pHSPB6 complex (residues 1–149) with 14-3-3σ to a resolution of 4.5 Å. The asymmetric unit was found to contain three heterotetrameric complexes.56 Both intrinsically disordered pHSPB6 N-terminal domains interact with the 14-3-3 dimer grooves identical to the cocrystal structure with the phosphopeptides. Remarkably, the ACD dimer of HSPB6 was found to dock onto one 14-3-3 unit, resulting in an asymmetric overall complex (Figure 5), which is in contrast to the structures described above. The interaction surface spans ∼400 Å2 and contains an essential salt bridge between Arg224 (14-3-3) and Glu86 (HSPB6). The positioning of the flexible parts of the NTDs not covered in the electron density were further characterized in solution by small-angle X-ray scattering, confirming the 2:2 stoichiometry of the complex. The authors state that by fully sequestering 14-3-3 binding sites, the chaperone HSPB6 blocks interactions with other partner proteins, thus acting as a phospho-switching 14-3-3 regulator.
Inhibitors of 14-3-3 PPIs
R18 Peptide
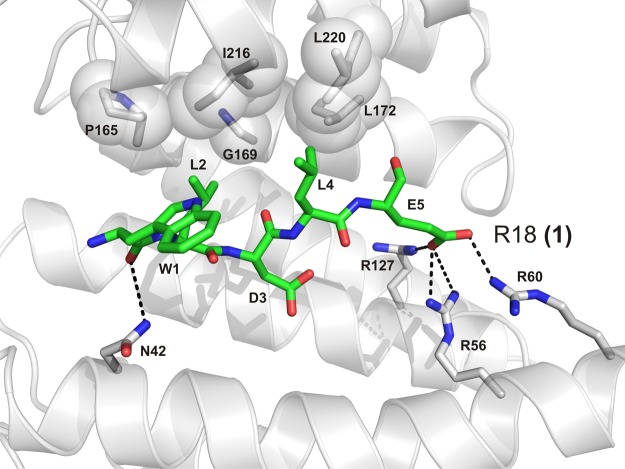
The first reported 14-3-3 PPI inhibitor, peptide R18 (1) (20 amino acid residues), was identified from a phage display by the Fu Laboratory.111 In the 14-3-3 complex crystal structure, the central sequence (WLDLE) can be seen in the amphipathic binding groove of 14-3-3.112 This structure revealed that, in addition to salt-bridge interactions between the carboxyl group of the glutamic acid side chain and three arginines of 14-3-3, there is an extensive hydrophobic contact surface between the two leucine residues of 1 and a number of 14-3-3 residues (Figure 6). In this way, 1 efficiently exploits the amphipathic character of the 14-3-3 binding channel to compete for both phosphorylation-dependent and -independent 14-3-3 PPIs. In later studies, Fu et al. showed that expression of a longer peptide of 64 amino acid residues with two such inhibiting sequences resulted in apoptosis, sensitized cancer cells for the antineoplastic drug cisplatin, and suppressed tumor growth in mice.113,114 These breakthrough studies with 1 proved the principal feasibility and efficacy of inhibiting 14-3-3 PPIs.
Figure 6.
Binding of peptide 1 (green sticks) to 14-3-3ζ (white cartoon). Residues from 14-3-3 important for interaction with 1 are shown as sticks. Polar interactions are depicted as black dotted lines, and hydrophobic contact surfaces from 14-3-3 are displayed as semitransparent spheres (PDB ID: 1A38).112
ExoS Macrocyclic Peptide
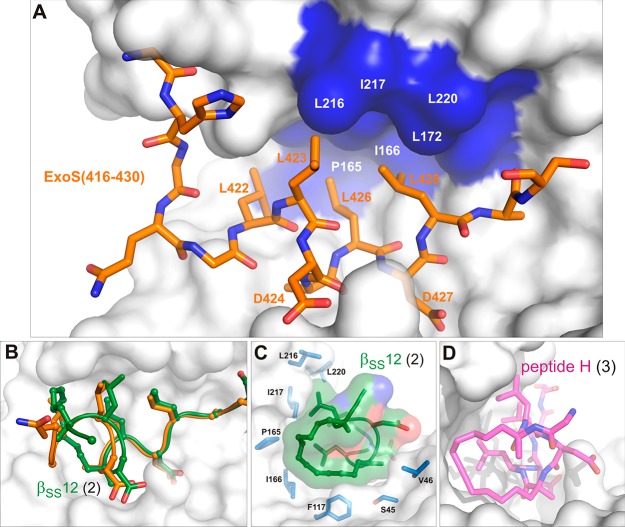
The groups of Ottmann and Grossmann recently developed a strategy for the macrocyclization of bioactive peptides with an irregular secondary structure and showed that macrocyclic molecules derived from peptides containing 14-3-3 binding motifs can efficiently inhibit the interaction between 14-3-3ζ and their binding partners.115 As a proof of concept, they prepared macrocyclic peptides targeting the interaction between 14-3-3ζ and the virulence factor of the pathogenic bacterium Pseudomonas aeruginosa Exoenzyme S (ExoS). These inhibitors were prepared from the ExoS peptide stretch that binds to 14-3-3 in an irregular and mostly extended conformation (Figure 7A, sequence Q420GLLDALDLAS430) by replacing two hydrophobic residues crucially involved in 14-3-3 binding with non-natural amino acid residues cross-linked by a (CH2)n chain. The most efficient inhibition was obtained for the βSS12 (2) inhibitor in which residues Leu422 and Ala425 were replaced with S-configured non-natural amino acid residues cross-linked with the chain containing 12 methylene groups (Figure 7B and C).115 This macrocyclic inhibitor binds to 14-3-3ζ with ∼30-fold higher affinity compared to that of the unmodified peptide (a Kd value of 41 nM vs 1.14 μM). The structural analysis revealed that the hydrophobic cross-link is involved in interactions with nonpolar residues within the ligand binding groove of 14-3-3ζ, whereas the conformation of the backbone is very similar to that of the unmodified peptide. Further biophysical analyses suggested that the improved binding affinity resulting from the incorporation of the cross-link comes from the significantly decreased conformational flexibility of the macrocyclic molecule. Because all 14-3-3 binding motifs structurally characterized so far adopt irregular and extended conformations within the 14-3-3 ligand-binding grooves, this approach should be applicable to the majority of 14-3-3 binding interactions. Figure 7D shows the crystal structure of an ExoS-derived peptide H (3), which has been obtained by a ring-closing alkyne metathesis used here for the first time for the stabilization of an irregular peptide secondary structure.116
Figure 7.
Structural characterization of the 14-3-3ζ/Exo S interface. (A) Wild-type ExoS (orange sticks) bound to 14-3-3ζ (white and blue surface). ExoS establishes an extensive hydrophobic contact interface with 14-3-3 with its four leucine residues (Leu422, Leu423, Leu426, Leu428) binding to a hydrophobic patch (blue surface) in the 14-3-3 channel (PDB ID: 2O02).67 (B) Structural superimposition of wild-type ExoS (orange cartoon and sticks) and the 12-carbon-linker cyclic peptide 2 (green cartoon and sticks) derived from ExoS (PDB ID: 4N84).115 (C) The 12-carbon linker of 2 engages a semicircular, hydrophobic ring in 14-3-3 (white, semitransparent surface and blue sticks; PDB ID: 4N84).115 (D) Further optimization of the constrained peptide derived from ExoS using an alkyne-cross-link in 3 (PDB ID: 5J31).116
Tau Epitope
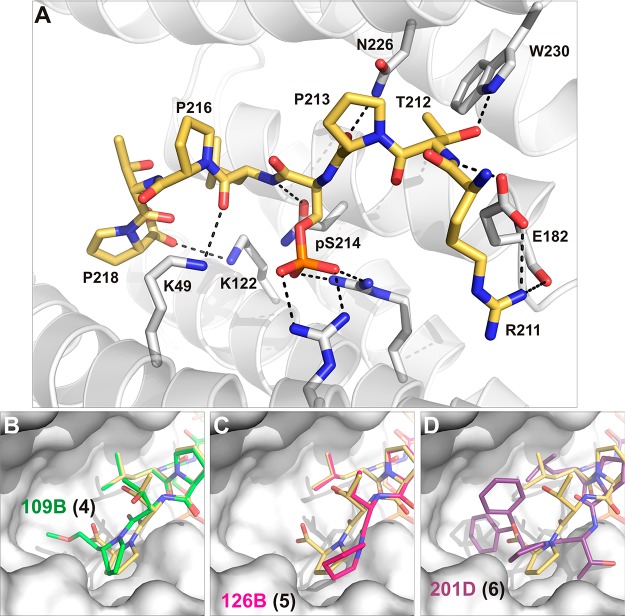
On a cellular level, the most striking pathological hallmark of Alzheimer’s disease (AD) is the occurrence of protein deposits like neurofibrillary tangles (NFTs) and amyloid plaques. NFTs are composed of hyperphosphorylated Tau displaying paired helical filaments. NFTs have been found to contain substantial amounts of 14-3-3 proteins, implicating them in the pathophysiology of AD.117 Furthermore, 14-3-3 proteins have been found to directly bind to Tau in solution via the phosphorylated residues Ser214 and Ser324 as important determinants of binding.118,119 Previously, we solved the crystal structure of 14-3-3 in complex with synthetic peptides comprising the phosphorylation sites pSer214 and pSer324.120 The sequence surrounding pSer214 (211RTPpSLPTP218) is especially interesting with three proline residues as notable structural features (Figure 8A). In particular, Pro218 occupies a position that is not used by most of the other structurally elucidated 14-3-3 recognition motifs. This observation inspired the rational design of peptide-based inhibitors using this position (Pro218) in the Tau peptide for chemical modifications that would result in peptides displaying increased affinity to 14-3-3.
Figure 8.
Targeting the 14-3-3σ/TaupS214 interface with modified peptides. (A) Wild-type TaupS214 (golden sticks) bound to 14-3-3σ (white surface and white sticks). Residues from 14-3-3σ important for binding are shown as labeled sticks; polar contacts are depicted as black dotted lines (PDB ID: 4FL5).121 (B) Structural superimposition of wild-type TaupS214 (golden sticks) and the modified Tau-peptide 4 (green sticks, PDB ID: 4Y32) binding to 14-3-3σ (white surface).121 (C) Structural superimposition of wild-type TaupS214 (golden sticks) and the modified Tau-peptide hybrid 5 (magenta sticks, PDB ID: 4Y5I) binding to 14-3-3σ (white surface).121 (D) Structural superimposition of wild-type TaupS214 (golden sticks) and the modified Tau-peptide hybrid 6 (purple sticks, PDB ID: 4Y5I) binding to 14-3-3σ (white surface).121
This concept was recently demonstrated by the groups of Ottmann, Milroy, and Landrieu who designed a potent inhibitor of the 14-3-3/Tau interaction as guided by cocrystal structures of the protein–stabilizer and protein–inhibitor complexes.121 The superposition of cocrystal structures of 14-3-3 complexes with Fusicoccin A (stabilizer) and Tau epitope (inhibitor, sequence RTPpSLPTP) showed that the C-terminal Pro218 residue of the Tau epitope and the A ring of Fusicoccin A overlap. This suggested that the poor binding affinity of this phosphopeptide for 14-3-3 could be improved by extending its C-terminus with a hydrophobic group to target the highly conserved hydrophobic pocket within the amphiphilic groove of 14-3-3, which led to 14-3-3 inhibiting modified peptides 109B (4), 126B (5), and 201D (6) (Figure 8B–D).121 Indeed, the chimeric inhibitor containing the sterically bulky and conformationally rigid benzhydryl pyrrolidine moiety at the C-terminus of the Tau epitope bound to 14-3-3 with 3 orders of magnitude higher binding affinity compared to that of the unmodified phosphopeptide. Consequent NMR spectroscopic studies on 14-3-3ζ and full-length Tau confirmed that this chimeric compound inhibits the binding of 14-3-3ζ to phosphorylated full-length Tau by disrupting its interaction with the phospho-epitope sites located within the C-terminal part of Tau.
2-5, Prodrug 15, and 19a
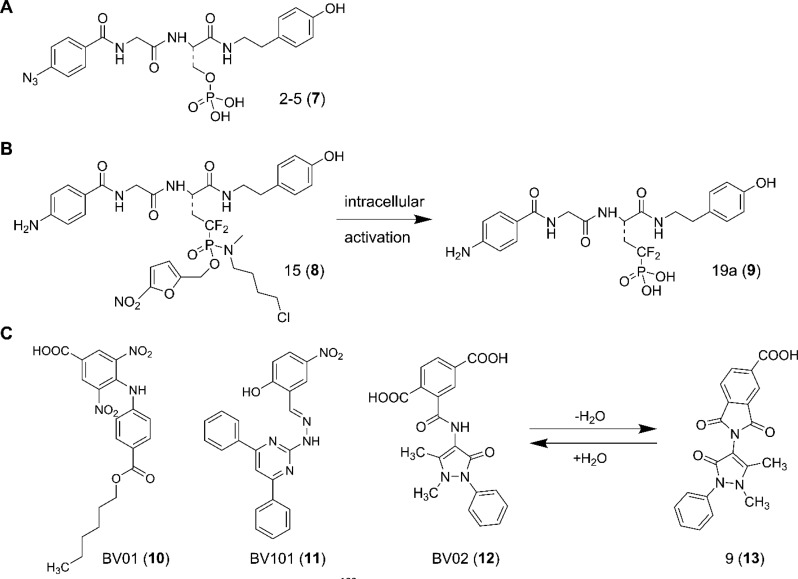
Shao et al. reported in 2010 a 14-3-3 PPI inhibitor called 2-5 (7) that was found using the small-molecule microarray (SMM) technique.122 The library was based on an optimal 14-3-3 binding amino acid sequence (RFRpSYPP), where they coupled 50 diverse amines to the C-terminus of the N-terminal peptide (RFRpS) or 243 diverse acid building blocks to the N-terminus of the C-terminal peptide (pSYSPP). These hybrids were spotted on a glass slide, and binding of fluorescently labeled GST-14-3-3 was measured. Five hits were found with Kd values between 0.6 and 1.03 μM, of which three featured a substitution of the N-terminal peptide part and two of the C-terminal peptide part. By combining these fragments into the six possible nonpeptidic molecules, compounds were found with IC50 values between 2.6 and 3.6 μM (fluorescence polarization (FP) assay), including 7 (Figure 9A).
Figure 9.
(A) Chemical structure of compound 7.122 (B) Membrane permeable prodrug 8 is converted to active component 9 by intracellular metabolic transformation.123 (C) Chemical structure of 14-3-3 PPI inhibitors 10–13 identified by the group of Botta. The reversible hydration pathway converts 12 to 13 and vice versa.124,126,127
Phospho-serine mimetic prodrug 15 (8) based on 7 was reported that showed potent 14-3-3 inhibitory activity in cells.123 The basic idea of the group of Borch was to transform bioactive peptides with phospho-serine groups, which suffer from poor membrane permeability and hydrolysis by phosphatases, into stable and cell permeable molecules with druglike properties. The phosphate to serine bridging oxygen group of 7 was replaced by a difluoromethylene linker with the aim of preventing phosphatase cleavage under physiological conditions. This also enables the retention of an appropriate pKa to keep the phosphate mimetic in a doubly charged state to maintain all essential interactions. For enabling membrane permeability, the two charges of the phosphonate group were temporarily neutralized by derivatization of the two phosphonate oxygens with nitro-furfuryl and 4-chloro-N-methylbutan-1-amine groups (Figure 9B). Upon entry into the cell, the nitrofurfuryl group was cleaved off by enzymatic reduction and spontaneous expulsion to deliver a phosphoamidate anion. Subsequent intracellular cyclization of an intermediate followed by spontaneous hydrolysis yields the biologically active phosphonate product 19a (9).123 Compound 8 inhibits viability of DG75 leukemia cells with an IC50 value of 5 μM and induces apoptosis in the same concentration range, whereas free phosphonate 9 does not show any significant inhibition at concentrations up to 100 μM.
For confirming the relevance of 14-3-3 proteins for the cellular effects of compound 8, a cellular assay for measuring 14-3-3-mediated inhibition of FOXO transcription factors was used. FOXO3A is a member of the Forkhead family of transcription factors that is inactivated by Akt1 phosphorylation and subsequent 14-3-3 binding. FOXO3a-dependent reporter gene activation was abolished by cotransfection with Akt1 in DG75 cells. The repressed FOXO3a activity was recovered in a dose-dependent manner by addition of compound 8 in concordance with reduced retention of phosphorylated FOXO3a by 14-3-3 in the cytoplasm. Active metabolite 9 was able to inhibit the interaction between immobilized 14-3-3τ and phosphorylated FOXO3a at physiologically relevant concentrations in lysates of DG75 leukemia cells transfected with FOXO3a and Akt1, whereas prodrug 8 was inactive.
BV01, BV02, BV101, and 9
The group of Botta reported the identification of small-molecule inhibitors of 14-3-3σ PPIs by using structure-based pharmacophore modeling, virtual screening, and molecular docking simulations with library design and organic synthesis. They started by in silico screening of 200,000 compounds from the ASINEX chemical collection, of which 14 compounds were eventually selected and tested in cellular and biochemical assays. This resulted in the identification of BV02 (12) as a lead inhibitor of the interaction between 14-3-3σ and cAbl in chronic myelogenous leukemia (CML) (Figure 9C).124 Indeed, 12 was able to inhibit 14-3-3/c-Abl interaction and promote c-Abl nuclear translocation at low micromolar concentration in Ba/F3 cells expressing the wild-type Bcr-Abl as well as its Imatinib-resistant T315I mutation.125 Accordingly, 12 represented a useful starting point for the development of an alternative treatment of CML, particularly for the Imatinib-resistant forms.
Following the discovery of 12, two additional 14-3-3 PPI inhibitors have been reported. These molecules, namely BV01 (10) and BV101 (11) (Figure 9C), were also initially discovered by an in silico approach,126 and promoted c-Abl nuclear translocation in Ba/F3 cells expressing the WT and the Imatinib-resistant T315I-mutated Bcr-Abl constructs. Furthermore, the interaction of 10 with 14-3-3σ was supported by transfer NOE experiments.126 In 2014, the same group published the discovery of compound 9 (13, Figure 9C), a phthalimide derivative of 12 that is able to promote c-Abl nuclear translocation as well as to sensitize multidrug-resistant (MDR) cancer stem cells.127 This discovery was facilitated by in silico docking of a virtual library of 12 and 10 analogues to a 14-3-3σ crystal structure using a well-established computational protocol.124 The most promising molecules were synthesized and submitted to biological tests. Most notably, molecule 13 was found to promote c-Abl nuclear translocation at 25 μM and has been shown to decrease the IC50 of doxorubicin by increasing its accumulation in MDR cancer cells at 10 μM concentration.
Subsequently, it was discovered that 13 was the product of spontaneous dehydration of 12 under aqueous conditions (Figure 9C) and was, in fact, the bioactive form of 12, as shown by a detailed NMR spectroscopy study.128 The direct interaction between 13 and recombinant 14-3-3σ was demonstrated by NOESY experiments, thus corroborating the mechanism of action of 13 at the molecular level. Indeed, in the presence of 14-3-3σ, the conversion of 13 back into 12 was slowed down, indicating that temperature and pH are not the only variables that influence the compounds interconversion. This observation provides important information for the appropriate setup of biological and biochemical experiments.
HSP20 Compound 85070
The phosphorylated form of HSP20 (phospho-HSP20) interacts with 14-3-3 proteins, the complex playing a regulating role on the actin depolymerizing protein cofilin. Phospho-HSP20 competition with phospho-cofilin for binding to 14-3-3 proteins frees phospho-cofilin, resulting in its dephosphorylation and subsequent depolymerization of the actin cytoskeleton. Free phosho-HSP20 is additionally able to directly destabilize the cytoskeleton. The phospho-HSP20/14-3-3 interaction could be a critical step in cofilin-mediated disruption of actin stress fibers and hence smooth muscle relaxation. Small molecules targeting the phospho-HSP20/14-3-3 interaction could thus lead to new therapeutic compounds to treat constriction of the airways in asthma. By screening a 58,019-compound library obtained from ChemDiv and ChemBridge (SanDiego CA) by high-throughput polarization assay, researchers at Prolexys Pharmaceuticals and the Johns Hopkins Bloomberg School of Public Health identified 268 modulators.129 These primary screen hits show at least 20% reduction of the polarization emission in the assay that was set up using a FAM-labeled 8-mer phosphopeptide derived from HSP20 to test for full-length 14-3-3 γ isoform (247 amino acid residues) binding. Compounds belonging to the scaffold PRLX24905 (US patent 20090136561) were further analyzed by FP for their concentration-dependent activity of inhibition. Structurally related scaffolds show a range of activity from no inhibition to 50 μM IC50 for compound 85070 (14, structure not disclosed).129 These compounds were additionally evaluated in cell-based assays. Compound 14 was the most efficient in causing, in a dose-dependent manner, a decrease in cell stiffness, decrease in contractile force in ASM cells, and an attenuation of active force development of intact tissue ex vivo. For the time being, the mechanism of action of compound 14 and the basis of these functional effects are not known.
FOBISIN101
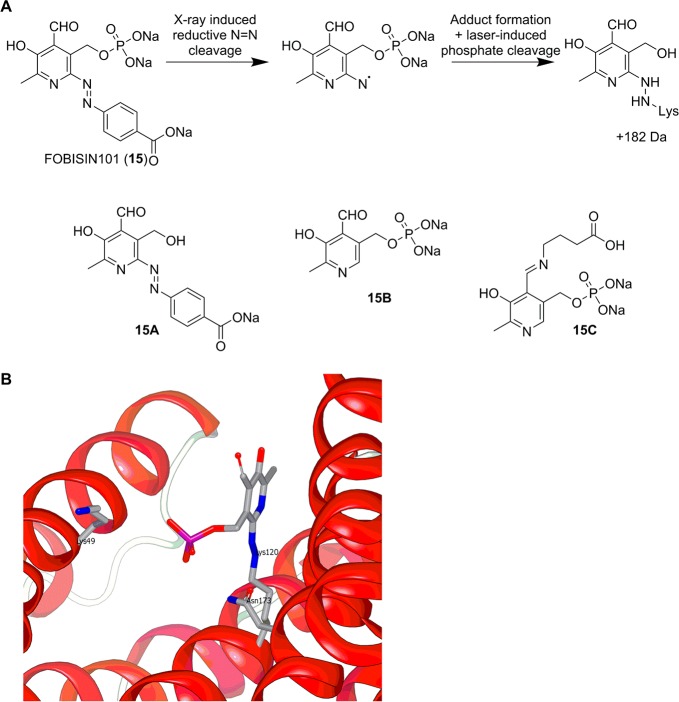
In 2011, the group of Fu reported the first covalent 14-3-3 inhibitor FOBISIN101 (15, FOurteen-three-three BInding Small molecule INhibitor).130 Compound 15 (Figure 10A), initially reported in 1998 as the P2X receptor antagonist MRS-2159,131 was identified by screening of the Sigma-Aldrich LOPAC library in a FP-based binding assay based on 14-3-3γ and a phosphorylated Raf-1 peptide. Affinity chromatography was used to demonstrate the ability of 15 to inhibit the binding of 14-3-3γ to two known 14-3-3γ binding partners, Raf-1 and p-PRAS40, from COS-7 cell lysates. Compound 15 was shown to be a pan-14-3-3 inhibitor and inhibited the binding of PRAS40 to 14-3-3ζ and 14-3-3γ with similar potencies (9.3 and 16.4 μM, respectively). Compound 15 also inhibited the 14-3-3-dependent activation of nonphosphorylated 14-3-3 client protein ExoS in a functional assay, suggesting that 15 inhibits the binding of both phosphorylated and nonphosphorylated client proteins. A limited SAR study of analogues of 15 was carried out, employing an ELISA assay based on 14-3-3ζ and Raf-1. Compounds 15A and 15B (Figure 10A) both exhibited almost complete loss of potency, indicating that the phosphate and phenyldiazene moieties were required for binding to 14-3-3ζ.
Figure 10.
(A) The proposed mechanism of adduct formation between 15 and 14-3-3ζ and the chemical structure of various derivatives of 15 (15A–C). (B) Complex structure of covalent adduct formed between 15 and 14-3-3ζ upon X-ray irradiation (PDB ID: 3RDH).130
A structure of the complex of 15 and 14-3-3ζ was obtained by X-ray crystallography, indicating the formation of an unexpected covalent adduct with the protein (Figure 10B). This was explained by X-ray-induced cleavage of the N=N bond and reaction of the radical thus formed with the side-chain terminal nitrogen atom of Lys120. The phosphate moiety of 15 was shown to interact with Lys49 (a key residue for recognition of phosphorylated client proteins) and Asn173.
A color change (from orange to colorless) of the crystals upon X-ray exposure was attributed to the loss of the conjugated aromatic system caused by reductive photo cleavage of the N=N bond. MALDI-MS also showed that X-ray exposure was required for formation of this covalent adduct as only irradiated samples exhibited the 183 Da increase in molecular weight corresponding to the 15 fragment observed in the crystal structure. However, X-ray activation is not required for 15 to bind to 14-3-3ζ, as it was able to inhibit binding of a Raf-1 peptide to 14-3-3ζ in an ELISA-based assay. This suggests that the covalent bond may be a radiation-induced crystallographic artifact rather than the functional binding mode of 15 in the ELISA assay.
A similar pyridoxal phosphate derivative 15C (Figure 10A) was reported by Ottmann and co-workers as a 14-3-3 protein-binding ligand.132 In contrast to 15, no cleavage of the N=N bond was observed in the 1.8 Å resolution crystal structure of 15C complexed to 14-3-3σ. Instead, transimination with the terminal side chain amino moiety of Lys122 (which corresponds to 14-3-3ζ Lys120) led to a covalent adduct with the N=N bond and phenyldiazene moiety intact. The phosphate moiety of 15C adopted a similar position to that observed for 15 bound to 14-3-3ζ as reported by Fu. They then obtained a structure of 15 with 14-3-3σ, which in their hands also exhibited imine formation with the aldehyde. It also exhibited with an intact diazene moiety; however, the electron density indicated a high degree of ligand flexibility in this region when compared with the hydrazine adduct observed by Fu et al. Ottmann and co-workers also demonstrated attachment of up to four intact 15 molecules to the 14-3-3σ protein using ESI-MS with no observed N=N bond cleavage. The differences in the observed mechanisms of covalent complex formation of 15 were attributed to differences in X-ray wavelength and MS conditions used.133 Further work is needed to fully elucidate the exact mechanism of the diazene cleavage and subsequent covalent adduct formation observed for 15.
UTKO1
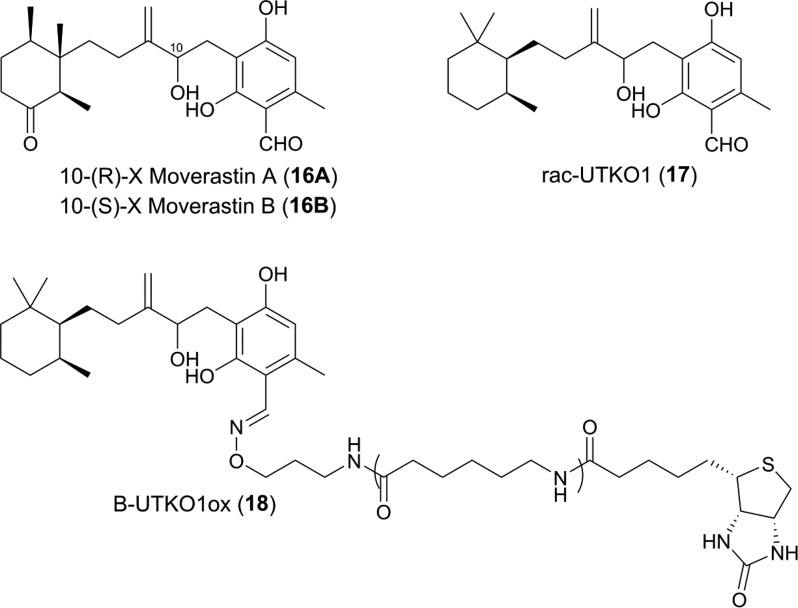
In 2005, the group of Imoto reported the discovery of the Aspergillus-derived natural product Moverastin as inhibitors of cancer cell migration by the screening of microbial extracts.134 Natural Moverastin is produced as a diastereomeric mixture of secondary alcohols, and separation led to the isolation of the pure diastereoisomers Moverastin A (16A) and B (16B) (Figure 11).134 The Moverastins are members of the cylindrol family of natural products, which are known inhibitors of farnesyl transferase (FT).135 Structurally, the Moverastins differ from other cylindrols in that they possess a methylidene moiety rather than a trisubstituted alkene.
Figure 11.
Structures of 16A, 16B, 17, and biotinylated probe molecule 18.134,136,137
HRas is a common human oncogene, and HRAS protein plays a key role in cancer cell migration. HRAS activity is dependent on posttranslational prenylation (farnesylation) by FT, which leads to activation of HRAS by membrane localization. Inhibition of FT therefore leads to a reduction of HRAS farnesylation and inhibition of membrane localization, subsequently leading to a reduction in cell migration and tumor metastasis. Compounds 16A and 16B were shown to inhibit FT in vitro with similar potency and also shown to decrease the nuclear localization of Ras in a tumor cell line.134
In 2011, the same group published a series of Moverastin derivatives with enhanced inhibitory activity against the migration of human oesophogeal tumor cells.136 The most potent compound identified, rac-UTKO1 (17, Figure 11) had 3.5-fold increased potency on inhibition of cell migration (IC50 1.98 μM) compared to 16; however, 17 was shown not to inhibit FT (IC50 > 100 μM), and therefore, another target was likely responsible for its activity.136 Biotinylated UTKO1 derivative B-UTKO1ox (18, Figure 11) was used as a probe in a series of experiments to identify the molecular mode of action of 17.134,137 Coprecipitation experiments with 18 and unlabeled 17 identified two 14-3-3 isoforms (ε and ζ) as potential 17-binding proteins of which 14-3-3ζ was considered to be the most likely candidate for the biological activity due to previous reports of its involvement in cell migration. The direct binding of 17 to 14-3-3ζ was confirmed by a competition pulldown experiment using a GST-tagged 14-3-3ζ protein. All seven mammalian 14-3-3 isoforms were then tested for binding to 17, and 14-3-3ζ was confirmed as the isoform with the strongest binding affinity. Compound 17 was also shown to bind to the C-terminal domain of 14-3-3ζ. This C-terminal domain is the most variable region within the 14-3-3 family, and this was proposed as an explanation for the selective binding of 17 to 14-3-3ζ. siRNA silencing of 14-3-3ζ led to suppression of lamellopodia formation (which is key to cell migration) in a tumor cell line thus increasing confidence that the observed effect of 17 is due to binding to 14-3-3ζ.
Additional pulldown experiments with GST-tagged 14-3-3ζ were performed to identify the relevant 14-3-3ζ partner proteins with two proteins (Tiam1 and βPix) being identified as promising candidates.134 siRNA knockdown experiments showed only Tiam1 to have an effect on cell migration and inhibition of the binding of Tiam1 to 14-3-3ζ by 17 was confirmed both in cell lysates and cultured cells. Interestingly, neither the stability nor cellular localization of Tiam1 was affected by 17, suggesting that a conformational change upon binding to 14-3-3ζ is required for activation of Tiam1 and that this is inhibited by 17.
Phosphonate-type Inhibitors of 14-3-3
Starting from the identified RFRpSYPP binding motif of an inhibitory peptide of 14-3-3, Wu et al. derived cell-permeable small molecule PPI inhibitor 7 that contains the phosphorylated central serine.122 The group of Ottmann went a step further and identified a small molecule 14-3-3 inhibitor by means of virtual screening.138 An implemented set of filters on a ZINC library of more than 8 million small molecules yielded 512 diverse compounds that incorporate one phosphate or phosphonate group and obey the Lipinski’s rule of five. Their virtual docking into a high-resolution crystal structure of 14-3-3σ (PDB ID: 3P1N) and follow-up analysis led to the synthesis and cocrystallization of 11 14-3-3 inhibitors (e.g., compound B2 (19, Figure 12)).138 A detailed computational analysis of the binding mode of eight of these molecules was performed by the group of Wang et al.139 They showed that the hydrophilic residues (Arg56, Arg129, and Tyr130 of 14-3-3σ) at the bottom of the binding pocket form seven stable hydrogen bonds with the phosphate group. In addition, two residues (Leu174 and Val178) in contact with a moiety accommodating the phosphate group contribute large van der Waals energies, and residue Leu126 provides large electrostatic energies. This is in agreement with the statement that the phosphate has the strongest pharmacophoric properties. There are three unfavorable interactions with residues (Asp126, Glu133, and Glu182) for inhibitor binding to protein. The averaged free energies for these three residues in the eight compounds are 0.93, 1.03, and 0.97 kcal/mol. Because the aspartic acid and glutamic acid residues have negative charges, they repel the phosphate group and attract the residues with positive charge in the binding pocket. By contrast, several residues surround the second hydrophobic moiety of the inhibitors, whereas there are only weak interactions between this part of the inhibitor and protein residues.
Figure 12.
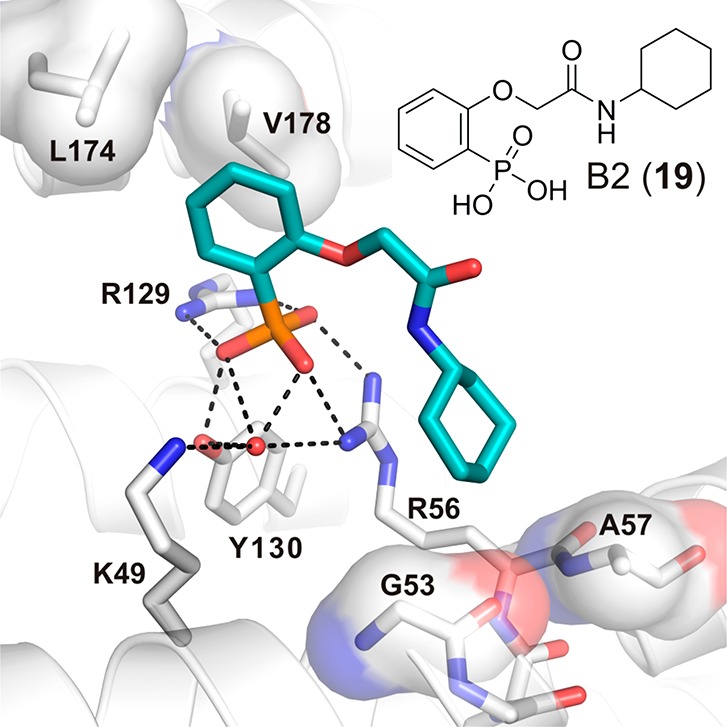
Binding of phosphonate inhibitor 19 (cyan sticks) to 14-3-3σ (white cartoon, sticks and surface). Residues from 14-3-3σ important for accommodation of 19 are shown as sticks; polar interactions are depicted as dotted black lines, and the semitransparent surface represents hydrophobic contacts (PDB ID: 4DHT).138
Molecular Tweezers
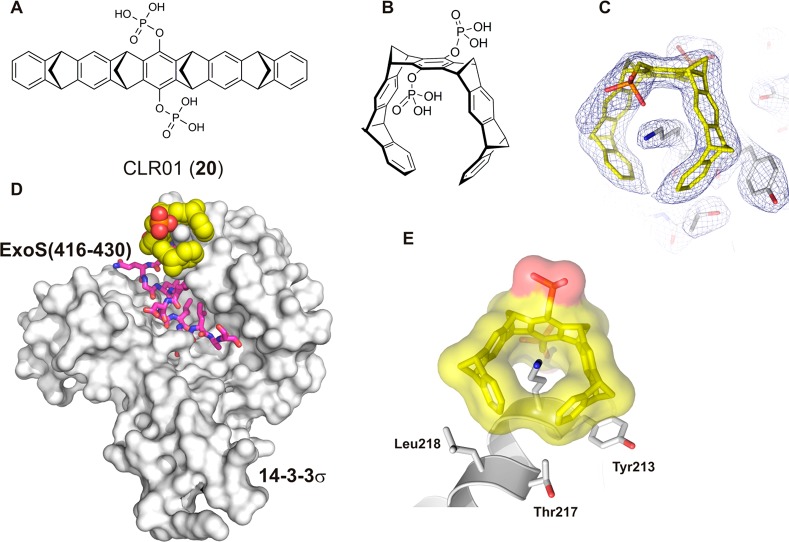
Bier et al. published in 2013 another class of phosphate-containing inhibitors called molecular tweezers (Figure 13A and B).140 With crystal structures, they showed that this molecule binds around residue Lys214 that is positioned at the edge of the amphipathic binding groove of 14-3-3σ (Figure 13C–E). By binding to this position, it can interfere with the function of 14-3-3 as an adapter protein, inhibiting, for example, the binding of phosphorylated C-Raf and unphosphorylated ExoS (Figure 13) to 14-3-3σ. Surprisingly, the crystal structure of 14-3-3 in complex with the molecular tweezer CLR01 (20) revealed only one molecule binding to 14-3-3σ.140 This was unexpected because 14-3-3σ displays 17 surface-exposed lysine residues potentially able to bind this supramolecular ligand. Closer inspection of the environment of Lys214 and extensive modeling of the 14-3-3σ/20 interaction identified a set of structural requirements for the efficient interaction of the tweezer with lysine residues. In particular, the arrangement of a predominantly hydrophobic interaction surface formed by Tyr213, Thr217, and Leu218 seems to be beneficial for a more stable accommodation of the tweezer molecule. These structural findings can help in the design of more specific molecular tweezers, an approach currently followed in the groups of Schrader and Ottmann.
Figure 13.
Chemical structure of molecular tweezer 20 (A) and a 3D view of the molecule conformation it adopts for protein recognition (B). (C) Binding of molecular tweezer 20 (yellow sticks) to Lys214 of 14-3-3σ (white sticks) and the electron density (blue mesh, 2FO-FC, contoured at 1.0 σ). (D) Superimposition of the binding of molecular tweezer 20 (yellow spheres) and the ExoS peptide (416–430, purple sticks) to 14-3-3σ (white surface). (E) Molecular tweezer 20 (yellow sticks and surface) binding to Lys214 of 14-3-3σ (white cartoon and sticks) (PDB IDs: 4HQW and 4HRU).140
Stabilizers of 14-3-3 PPIs
Fusicoccanes
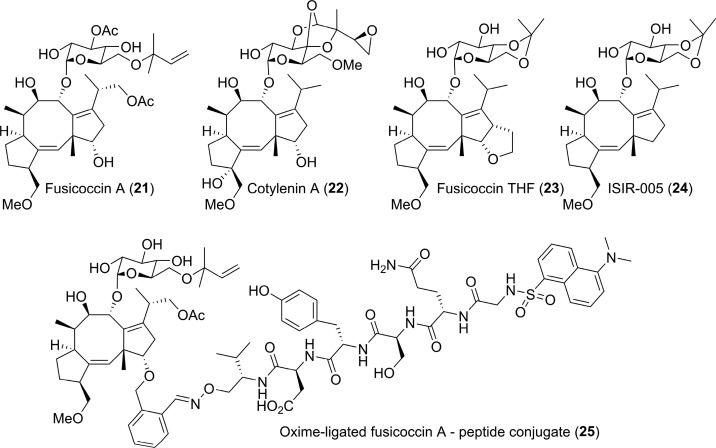
Fusicoccin A (21) is a diterpene glycoside produced by the phytopathogenic fungus Phomopsis amygdali (formerly Fusicoccum amygdali) that was initially described in the mid-1960s to be a wilt-inducing toxin.141 It was, however, not until 1994 that the molecular target was identified as the binary complex between the regulatory domain of the plasma membrane H+-ATPase (PMA) and 14-3-3 adapter proteins, which 21 stabilizes by acting like a “molecular glue”.142 Since then, 21 and the related natural product cotylenin A (22)145 and semisynthetic (e.g., Fusicoccin THF (23),150 ISIR-005 (24)151) fusicoccane analogues have proven to be valuable tool compounds to study the “molecular glue” model for stabilizing 14-3-3 binary structures (Figure 14).
Figure 14.
Fusicoccane analogues, natural (21, and 22) or semi-synthetic (23, 24, and 25) that act as “molecular glue” model for stabilizing 14-3-3 binary structures.142,145,150−152
Compound 21 itself has now been shown to stabilize 14-3-3 complexation with a number of medicinally relevant partner proteins in humans. For example, 21 promotes platelet adhesion to von Willebrand factor by stabilizing the 14-3-3 interaction with the C-terminus of the human protein glycoprotein (GP)Ibα.143 Compound 21 also stabilizes the 14-3-3 interaction with the C-terminus of the F-domain of estrogen receptor α (ERα) and thus inhibits ERα-dependent transcription.144 However, 21 stabilization is not limited to 14-3-3 partners bearing C-terminal (or “mode III”) 14-3-3 binding motifs. Although the physiological potency is relatively weak, 21 has recently been shown to stabilize the 14-3-3 interaction with the cystic fibrosis transmembrane conductance regulator (CFTR), thus promoting trafficking of CFTR to the plasma membrane.55
Compound 22 is another natural product produced by a fungus (Cladosporium sp. 501-7W) that acts as a bioactive substance against plants.145 Years after its discovery as a cytokinin-like substance, this natural product was reported to induce differentiation in human acute myeloid leukemia in both cell culture and mouse models.146,147 Interestingly, anticancer properties were found by combining 22 with other agents such as vincristine.148 The crystal structure of 22 bound to a complex of 14-3-3 with the N-terminal binding motifs of the protein kinase C-Raf published in 2013 gave important structural insight into how 22 can mediate its antitumor activity.149
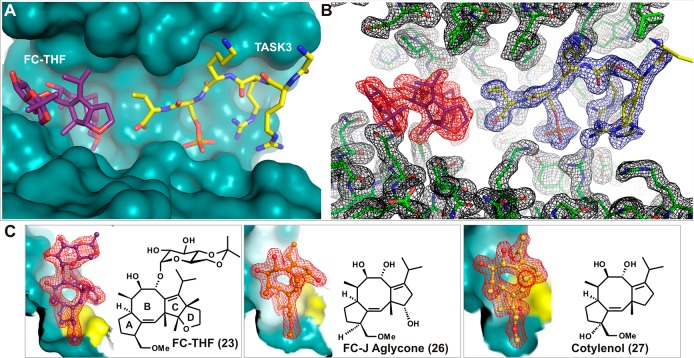
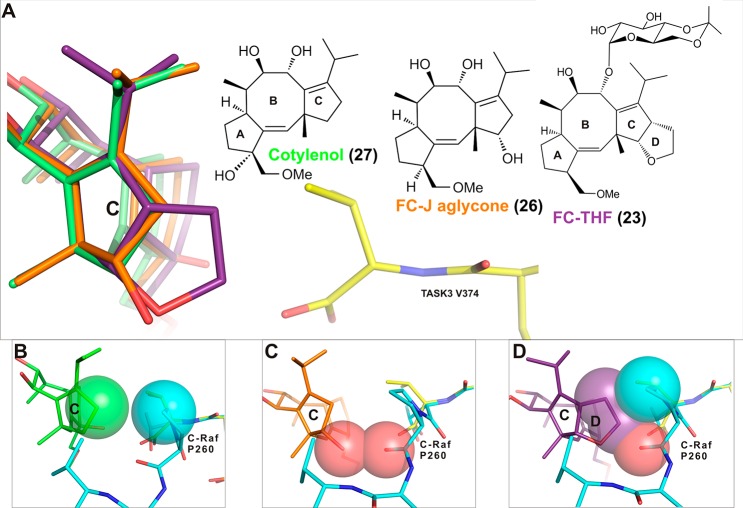
The 5-8-5-fused ring system of the fusicoccane scaffold is highly complex, and thus, investigating structural variation in the search for selectivity or enhanced potency is challenging. Nevertheless, structure-based design and semisynthesis have enabled the discovery of potent analogues. For example, semisynthetic derivative 23 (Figure 15A–C) was designed as a “mode III”-specific stabilizer and resulted in a 20-fold stabilization of the interaction between 14-3-3 and the potassium channel TASK3.150 In this study, a number of other fusicoccanes like Fusicoccin J aglycone (26)150 and cotylenol (27)150 (the aglycone of cotylenin) were tested (Figure 15C). The respective crystal structures of the complexes of the different fusicoccanes with 14-3-3 and the TASK3 peptide revealed the structural basis for their “mode III” preference (26, 22) or “mode III” specificity (23, Figure 16A). With a C12-dehydroxy fusicoccane like 22, concomitant binding of a “mode I” or “mode II” 14-3-3 partner like C-Raf and consequently its stabilization is possible (Figure 16B). However, hydroxylation of C12, which is present in 21 and 26, introduces a steric conflict with the proline carbonyl oxygen at position +2 C-terminal in the 14-3-3 binding motif of C-Raf (Figure 16C). Whereas this steric conflict might not exclude every “mode I” or “mode II” interaction with 14-3-3, the additional ring system in 23 makes this molecule a specific “mode III” stabilizer whose binding to a 14-3-3/partner protein interface should be significantly hampered with any 14-3-3 interaction motif that extends beyond the +1 position (Figure 16D). Importantly, the utility of 23 could be demonstrated in Xenopus oocytes, which have been transfected to express human TASK3. Here, adding 10 μM 23 to the culturing solution results in a 45% increase of TASK3 expression in the plasma membrane.150
Figure 15.
Semisynthetic derivatives 23, 26, and 27 stabilize the interaction between 14-3-3σ and TASK3 peptide. (A) Semisynthetic derivative 23 (purple sticks) and the C-terminus of TASK3 peptide (yellow sticks) in the binding groove of 14-3-3σ (cyan surface). (B) Electron density (red, blue, and black mesh, 2FO-FC, contoured at 1.0 σ) around 23 (purple sticks), C-terminus of TASK3 peptide (yellow sticks), and 14-3-3σ (green sticks). (C) Comparison of 23 (purple sticks) with 26 (orange sticks) and 27 (yellow sticks) in the binding pocket formed by 14-3-3σ (cyan surface) and TASK3 peptide (yellow surface) (PDB IDs: 3SMN, 3SMM, and 3SP5).150
Figure 16.
Comparison among 27, 26, and 23 in the stabilization of a “mode III” binder (TASK3) and “mode I/II” binder (C-Raf). (A) Overlay of 27 (green sticks), 26 (orange sticks), and 23 (purple stick) in the 14-3-3σ/TASK3 peptide (yellow sticks) complex. (B) The C-ring of 27 does not clash with “mode I/II” C-Raf peptide. (C) The hydroxylation of C12 in 26 clashes with the carbonyl oxygen of C-Raf P260. (D) The additional ring D of 23 clashes with both the carbonyl oxygen and the side group of C-Raf P260 (PDB IDs: 3SP5, 3SMM, 3SMN, and 4IEA).149,150
More recently, C-12 dehydroxy derivative 24 (Figure 14) was shown to be an analogue well-suited for the stabilization of 14-3-3 interactions with partner proteins bearing internal (or “mode I or II”) binding motifs.151 Biophysical and cellular experiments showed 24 to stabilize the 14-3-3 – Gab2 PPI by a factor of 5.3 and the crystal structure of the ternary complex provided further structural insight.151
The potential of fusicoccane semisynthetics is not limited to the search for potent stabilizers. Ohkanda, Kato, and co-workers elegantly demonstrated the power of intracellular oxime ligation to generate a fusicoccane–peptide hybrid (25) that induced cell death, presumably through inhibition of 14-3-3 PPIs (Figure 14).152 Interestingly, 25 that does not contain a phosphorylated residue was shown to bind 14-3-3 with a Kd value of 0.37 μM, stronger even than the interaction with a PMA2-derived phosphopeptide (Kd = 1.24 μM).152 This example not only further highlights the potential for modified peptide inhibitors of 14-3-3 PPIs (i.e., based on ExoS and Tau as discussed above) but also the importance of expanding the fusicoccane toolbox. Perhaps the emergence of new synthetic approaches to sesterterpenes153 can be coupled with previous total synthesis efforts154 to achieve this aim. Or perhaps ever greater understanding of the structure and function of fusicoccadiene synthase155 will provide the necessary starting points for diverse synthetic strategies.
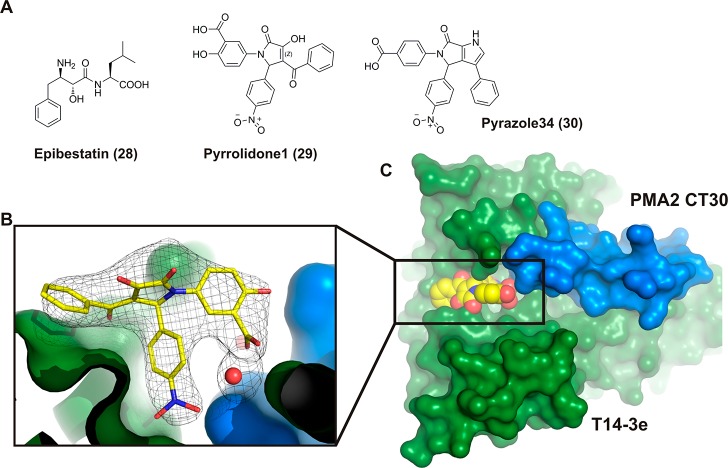
Epibestatin, Pyrrolidone1, and Pyrazole34
In 2010, the results of the first high-throughput screening for 14-3-3 PPI stabilizers were published.156 Out of a library of 37,000 small molecules, two compounds were found by a surface-based format monitoring the binding of GFP-14-3-3 to surface-immobilized PMA2-CT52: Epibestatin (28)156 and Pyrrolidone1 (29)156 (Figure 17). The crystal structures of these two compounds in the 14-3-3/PMA2 complexes revealed two distinctive binding pockets in the 14-3-3/PMA2 protein–protein interface. Two years later, Pyrazole34 (30) was published.157 This was based on optimization of the template of 29 in which the pyrrolidone scaffold had been converted into a more rigid pyrazole ring.157
Figure 17.
Compound 29 and derivative 30 stabilize the interaction between 14-3-3 and PMA2. (A) Chemical structures of 28–30. (B) Compound 29 (yellow spheres) in the binding groove of T14-3-3e (green surface) having contact with PMA2 CT30 (blue surface). (C) Close-up of the T14-3e/PMA2/29 (green surface/blue surface/yellow sticks) interaction showing the electron density of 29 (gray mesh, 2FO-FC, contoured at 1.0 σ; PDB ID: 3M51).156,157
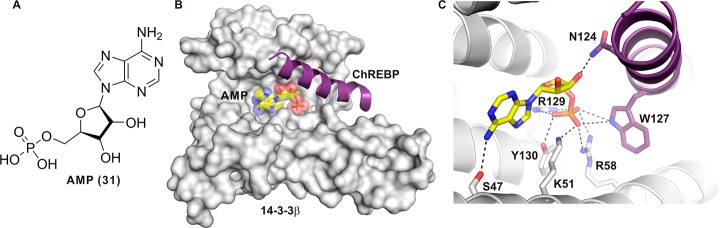
AMP/ChREBP
In 2016, the group of Uyeda published the PPI-stabilizing effect of AMP (adenosine monophosphate, Figure 18A) (31) toward the complex of 14-3-3 and the carbohydrate-response element-binding protein (ChREBP).158 ChREBP is a glucose-responsive transcription factor that is implicated in the regulation of fat storage in the liver by facilitating the conversion of carbohydrate to fat.159,160 The N-terminal region of ChREBP binds to 14-3-3 proteins and importin, which regulates subcellular localization in response to changing glucose levels. Low glucose leads to phosphorylation of Ser196 of ChREBP by PKA followed by complexation with 14-3-3 and cytoplasmic sequestration.161 In addition to the phosphorylation-dependent regulation of the 14-3-3/ChREBP interaction, a number of metabolites have been shown to influence this PPI, among them β-hydroxybutyrate (β-HB) and acetoacetate (AcAc). Both β-HB and AcAc have been shown to stabilize 14-3-3 binding to ChREBP,162 a finding that was later extended to 31 including a convincing structural biology explanation for this activity.158 A very interesting feature of the interaction between 14-3-3 and ChREBP is that it also employs a phosphorylation-independent binding mechanism. Here, the α2 helix of ChREBP (residues 117–137) binds to the central channel of 14-3-3β, engages both polar and hydrophobic interactions, and is partly dependent on the presence of a sulfate ion in the phosphate-accepting pocket of 14-3-3.163 Using the same crystallization conditions but adding 100 mM 31 during complexation of 14-3-3β and ChREBP yielded crystals showing that 31 occupies the same place where, in the phosphorylation-dependent 14-3-3 complexes, the phosphorylated serine or threonine residues can be found (Figure 18B). The phosphate group binds to Lys51, Arg58, Arg129, and Tyr130 residues, which are also used to accommodate the phosphorylated motifs of 14-3-3 partner proteins. In addition, the adenine ring of 31 interacts with Ser47 of 14-3-3, and maybe most importantly for its PPI stabilizing activity, the phosphate establishes polar contacts with Trp127 and Arg128 of ChREBP. Finally, Asn124 of ChREBP forms a contact with one of the hydroxyl groups of AMP’s sugar ring. Compound 31 is thus a direct orthosteric PPI interface stabilizer (Figure 18C).
Figure 18.
Crystal structure of the 14-3-3β/ChREBP/31 (AMP) complex. (A) Overview of ChREBP (purple cartoon) and 31 (yellow sticks and semitransparent spheres) bound to a monomer of 14-3-3β (solid white surface). (B) Detailed view of the contacts between 31 (yellow sticks), ChREBP (purple cartoon and sticks), and 14-3-3β (white cartoon and sticks). Polar contacts are depicted as black dotted lines (PDB ID: 5F74).158
Conclusions and Future Directions
In this perspective, we demonstrated that 14-3-3 proteins are highly relevant targets in drug discovery and provide a valuable tool in chemical biology. By presenting an overview of the wide range of 14-3-3 PPI modulators currently published, we illustrate the increasing evidence for the potential to modulate the activity of key proteins in various physiological processes, including Tau, p53, HSP20, and LRRK2, by targeting their distinct interaction with 14-3-3. Importantly, in addition to structural insights on the molecular mechanisms of the different small molecules, in a number of cases the influence on biological pathways has been studied, resulting in promising findings in cell-based model systems relevant to treating a variety of diseases (e.g., Alzheimer’s disease, various cancers, asthma, and cystic fibrosis). Bearing in mind that the modulation of only a small percentage of the several hundred identified 14-3-3 PPIs has been investigated so far, we believe that this is just the start of the opportunity that 14-3-3 PPI modulation can offer. However, this wide range of opportunities that 14-3-3 interaction partners hold also provides focus to where the biggest obstacles in the field will lie.
The great number of similar binding modes of 14-3-3 PPI partners makes it difficult for small molecule modulation to achieve specificity for one partner over the others. This is most strikingly illustrated by 14-3-3 PPI inhibitors, where binding of a competing molecule in the 14-3-3 binding groove will cause inhibition of binding of most other 14-3-3 binding partners, potentially leading to many side effects. For circumventing the nonspecific targeting of the phospho-binding pocket of 14-3-3 proteins, two approaches can be envisioned for future efforts toward more selective 14-3-3 PPI modulation. First, although based on a somewhat limited set of 14-3-3 crystal structures in complex with larger domain partner proteins, it is becoming clear that 14-3-3 itself is a relatively rigid molecule that allows for the docking of flexible partner proteins onto different surfaces of its dimer. By studying the “hot spots” responsible for the binding of the two partner proteins in these binary structures, we observe that there is a considerable variety of potentially distinct druggable pockets. In this regard, we very recently found that fragments from an NMR-based screen can bind to secondary binding sites outside the central phosphopeptide-accepting binding channel.164 The identified pockets are located on the upper rim of the 14-3-3 dimer, which is less conserved than the central channel and is used for binding of 14-3-3 to AANAT and FT. Second, the general approach of stabilization of 14-3-3 PPIs is suggested to be likely more beneficial compared to inhibition. This has already been illustrated by several examples where small molecules have been identified that make contacts with both 14-3-3 and the PPI partner. In this manner, they act as molecular glue and thereby provide better opportunities for selectivity. The semisynthesis of the Fusicoccane family provides a great case study for selectivity in the 14-3-3 PPI stabilization; the hydroxylation of C12 promotes a preference for stabilization for C-terminal “mode III” binders over “mode I/II” binders. Furthermore, molecules like 30 may be a good starting point to obtain selectivity over other 14-3-3 PPI partners by expanding the molecule and gaining more contact with the desired 14-3-3 binding partner. To reach the full potential in the field of 14-3-3 PPI modulation, we need to think “out-of-the-binding groove” and explore the rest of the protein. This should be possible by using a combination of high-throughput screening, fragment-based approaches, and rational design, and success will lead to the realization of small molecule inhibition and stabilization of 14-3-3 PPIs as a viable option in drug discovery.
Acknowledgments
This work was funded by The Netherlands Organization for Scientific Research (NWO) via Gravity program 024.001.035 and VICI grant 016.150.366 and by the Deutsche Forschungsgemeinschaft (DFG) via Collaborative Research Centre 1093.
Glossary
Abbreviations Used
- PPI
protein–protein interaction
- AANAT
arylalkylamine N-acetyltransferase
- PMA
plasma membrane H+-ATPase
- FT
flowering locus T
- HSP
heat shock protein
- ACD
α-Crystallin domain
- NTD
N-terminal domain
- CTE
C-terminal extension
- ExoS
Exoenzyme S
- NFT
neurofibrillary tangle
- SMM
small molecule microarray
- CML
chronic myelogenous leukemia
- FP
fluorescence polarization
- MDR
multidrug resistant
- FT
farnesyl transferase
- ERα
estrogen receptor α
- CFTR
cystic fibrosis transmembrane conductance regulator
- ChREBP
carbohydrate-response element-binding protein
- β-HB
β-hydroxybutyrate
Biographies
Loes M. Stevers received her B.S. (2010) and M.S. (2013) in Biomedical Engineering at Eindhoven University of Technology, The Netherlands. During her M.S., she worked on supramolecular membrane protein dimerization in living cells. Additionally, she did an internship in in vitro mapping of interactions between human SNX-BAR proteins at the Institute of Molecular Bioscience, University of Queensland, Brisbane, Australia. She started her Ph.D. research in Chemical Biology at Eindhoven University of Technology in 2013, focusing on characterization and modulation of multivalent 14-3-3 protein–protein interactions.
Eline Sijbesma obtained her M.Sc. degree with honors (cum laude) in Biomedical Engineering at the Eindhoven Univeristy of Technology. To complete her Master’s, she joined the lab of Michelle Arkin, Ph.D. in the Small Molecule Discovery Center at UCSF to initiate a fragment-based drug discovery project that explores protein–protein interactions. At the end of 2015, she returned to the Eindhoven University of Technology and started her Ph.D. research in the group of Prof. Luc Brunsveld in Chemical Biology under the supervision of Dr. Christian Ottmann. The project focuses on 14-3-3 proteins and nuclear receptors and combines chemical biology, drug discovery, and medicinal chemistry aided by structural insights from X-ray crystallography to study the potential and molecular mechanism of PPI stabilization by small molecules.
Maurizio Botta is a Full Professor of Medicinal Chemistry of the University of Siena. He obtained a degree with laude in Chemistry at the University of Rome in 1974. In December 1979, he obtained his Ph.D. at the University of New Brunswick under the supervision of Prof. K. Wiesner. He has been a researcher in Organic Chemistry at the University of Rome from 1981 to 1987. From January 2008, he is an Adjunct Professor at the Temple University College of Science and Technology in Philadelphia (USA). From November 2009 to December 2012, he has been Dean of the Faculty of Pharmacy at the University of Siena. He is author of more than 450 papers, 10 publications on volumes, 26 patents, and more than 240 proceedings at congresses.
Carol MacKintosh is Professor of Molecular Signaling in the University of Dundee, where she is also Head of Postgraduate Studies in the School of Life Sciences. Her laboratory is working to define how large sets of 14-3-3–phosphoprotein interactions coordinate diverse responses of human cells and tissues to nutrients, insulin, and growth factors. Following their discovery that the human 14-3-3-interactome is highly enriched in Ohnologues, members of protein families that were generated by the two rounds of whole genome duplication at the origin of the vertebrate animals, Carol’s group has developed a special interest in understanding how the 14-3-3-interactome has contributed to the evolution of vertebrate complexity, variety, and polygenic disorders such as diabetes, cancer, and neurological syndromes.
Tomas Obsil received his Ph.D. in Physical Chemistry from the Charles University, Prague, Czech Republic in 1998 after which he performed a postdoctoral stay at the National Institutes of Health (Bethesda, USA). In 2002, he returned to Charles University, where he obtained habilitation in 2007. Since 2014, he is a Professor of Physical Chemistry in the Faculty of Science, Charles University. His current research focuses on investigating the structural basis of 14-3-3 protein-mediated regulation of various signaling proteins.
Isabelle Landrieu received her Ph.D. in Biochemical Engineering (1997) from Liège University (Be) prior to postdoctoral studies at Ghent University (Be) in the department of Plant System Biology lead by Professor D. Inzé. I.L. is currently a CNRS (French National Research Centre) Research Director at Lille University (Fr) and a P.I. in the laboratory of excellence DISTALZ, which includes several French groups involved in Alzheimer’s disease research.
Ylenia Cau is a postdoctoral fellow at the University of Siena in the department of Biotechnology, Chemistry, and Pharmacy in the research group of Professor Maurizio Botta. She graduated with laude in Pharmaceutical Chemistry at the University of Siena in November 2012 under the supervision of Professor Maurizio Botta. In February 2016, she obtained her Ph.D in “Chemical and Pharmaceutical Sciences” at the University of Siena in the same research group with a thesis entitled “In silico identification and optimization of 14-3-3 inhibitors through molecular modelling and computational methods”. She is author of six papers.
Andy J. Wilson joined the University of Leeds in 2004 and was promoted to full professor in 2012. He currently serves as Deputy Director of the Astbury Centre. He completed a Ph.D. at Warwick University supervised by Prof. David Leigh FRS before postdoctoral research with Prof. Andrew Hamilton FRS at Yale University and with Prof. E. Meijer and Prof. Rint Sijbesma at Technische Universiteit Eindhoven. His research is concerned with (a) modulating protein–protein interactions, (b) developing fundamental approaches and building blocks for self-assembly, and (c) mechanistic studies of self-assembly using photo-cross-linking. Andy was recognized through the Royal Society of Chemistry (RSC) Bob Hay Lectureship (2012) and the RSC Norman Heatley Award (2016).
Anna Karawajczyk obtained her Ph.D. in chemistry in 2007 from the Leiden Institute of Chemistry, Leiden University (NL). Afterward, she worked as a postdoctoral fellow at the Centre for Molecular and Biomolecular Informatics, Computational Drug Discovery, Radboud University Medical Centre, Nijmegen and at the Molecular Design & Informatics; Schering-Plough Corporation, Oss. From 2009 to 2013, she worked as a computational chemist in a Medicinal Chemistry group of Lead Pharma BV. At the present time, she is a Principal Scientist in the Medicinal Chemistry Department of Taros Chemicals GmbH & KG and is working in the field of cancer, antibacterials, and fungicides.
Jan Eickhoff studied biochemistry at the University Bayreuth and Imperial College in London followed by Ph.D. studies at the Max Planck Institute of Biochemistry in Martinsried. He started his industrial career in 2001 at Axxima Pharmaceuticals, and GPC Biotech, before he moved to Dortmund in 2008 to build up the screening and sample management department of the Lead Discovery Center GmbH (LDC). Furthermore, he was involved in conceptual planning and implementation of the compound management and screening center of the Max Planck Society (COMAS). In addition to his role as Head of Assay Development and Screening at the LDC. From 2013 to 2015, he was holding a role as managing director of the Hit Discovery Constance GmbH in Constance, Germany.
Jeremy Davis is Director of Hit Discovery and Enabling Technologies at UCB, based in Slough. He obtained his Ph.D. in Organic Chemistry from Southampton University under the guidance of Professor Richard Whitby before joining Celltech as a medicinal chemist in 1993. During this time, Jeremy has worked on a diverse range of protein targets across immunology, inflammation, and oncology disease areas always with a keen interest in structure-based drug design. His current role as Director of Hit Discovery is to generate small molecule starting points across UCB’s target portfolio. In addition, he leads a technology group focused on developing advanced NMR and Mass Spectrometry applications to understand the structure and function of protein–ligand complexes.
Michael Hann completed his Ph.D. in organic chemistry in 1980 and has worked in Pharma R&D initially as a medicinal chemist and then as a computational chemist. He joined Glaxo in 1986 and was responsible for helping initially build and then lead the Computational Chemistry department. More recently, he led the biophysics and protein crystallography activities including developing fragments theory and practice in lead identification. His current role is in looking at new ways to enhance our discovery approaches, reducing attrition, and promoting scientific excellence across the GSK R&D sites. A particular current interest is in understanding drug distribution at cellular and subcellular resolution. Michael is a GSK Senior Fellow and an Adjunct Professor in the Chemistry department at Imperial College London.
Gavin O’Mahony is a Principal Scientist in the Cardiovascular and Metabolic Diseases Medicinal Chemistry department of the Innovative Medicines and Early Development Biotech Unit at AstraZeneca Gothenburg, Sweden. After B.Sc. and Ph.D. studies in chemistry at Queen’s University Belfast, Northern Ireland, he carried out two years of postdoctoral work in medicinal and nucleoside chemistry at the University of Gothenburg, Sweden. In 2005, he started working at AstraZeneca as a Senior Research Scientist in the Cardiovascular and Gastrointestinal Lead Generation department. He has worked on many targets including the mineralocorticoid receptors GPR40 and PPARγ. His research interests currently focus on lead generation within the diabetes field, nuclear hormone receptor medicinal chemistry, as well as novel approaches to the small molecule modulation of transcription factor activity.
Richard G. Doveston graduated from the University of Leicester in 2008 with a first class M.Chem. degree in Pharmaceutical chemistry (including a year in industry). He went on to complete a Ph.D. in the group of Prof. Richard J. K. Taylor at the University of York and in 2012 took up a postdoctoral position with Prof. Adam Nelson and Prof. Steve Marsden at the University of Leeds to work in the area of lead-oriented synthesis. In 2015, Richard moved to work in the Chemical Biology Group at the TU/e and was awarded a Marie Curie Fellowship in 2016. His research, carried out under Prof. Luc Brunsveld and Dr. Christian Ottmann, is focused on the discovery and evaluation of novel bioactive small molecules.
Luc Brunsveld is Professor of Chemical Biology at Eindhoven University of Technology after having previously worked at the Organon Research Laboratories (now MSD), the Max Planck Institute of Molecular Physiology and the Chemical Genomics Centre of the Max Planck Society. His research interests lie at the interfaces of drug discovery, protein assembly, and supramolecular chemistry. Research topics he likes to dive into together with group members and colleagues include nuclear receptors, protein–protein interactions, synthetic signaling systems, and supramolecular protein assemblies. Luc is recipient of the golden medal of the Royal Netherlands Chemical Society and his passions include teaching and research across disciplines.
Christian Ottmann, Ph.D., is Associate Professor for Molecular Cell and Structural Biology at Eindhoven University of Technology, The Netherlands. He works on small molecule modulation of protein–protein interactions (PPIs) with a special focus on stabilization of 14-3-3 adapter protein PPIs. He is involved in early drug discovery projects with the pharmaceutical industry and is coordinator of the FP7 Industry-Academia Partnership and Pathways (IAPP) 14-3-3STABS and the Horizon2020 ETN Targeted Stabilization of Protein–Protein Interactions (TASPPI). Before taking up his current position in Eindhoven, he was a group leader at the Chemical Genomics Centre (CGC) of the Max Planck Society in Dortmund, Germany. He obtained his Ph.D. in 2003 from the University of Tübingen with Prof. Claudia Oecking.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.7b00574.
14-3-3 crystal structures deposited in the PDB (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hatzivassiliou G.; Song K.; Yen I.; Brandhuber B. J.; Anderson D. J.; Alvarado R.; Ludlam M. J.; Stokoe D.; Gloor S. L.; Vigers G.; Morales T.; Aliagas I.; Liu B.; Sideris S.; Hoeflich K. P.; Jaiswal B. S.; Seshagiri S.; Koeppen H.; Belvin M.; Friedman L. S.; Malek S. RAF Inhibitors Prime Wild-Type RAF to Activate the MAPK Pathway and Enhance Growth. Nature 2010, 464 (7287), 431–435. 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Poulikakos P. I.; Persaud Y.; Janakiraman M.; Kong X.; Ng C.; Moriceau G.; Shi H.; Atefi M.; Titz B.; Gabay M. T.; Salton M.; Dahlman K. B.; Tadi M.; Wargo J. A.; Flaherty K. T.; Kelley M. C.; Misteli T.; Chapman P. B.; Sosman J. A.; Graeber T. G.; Ribas A.; Lo R. S.; Rosen N.; Solit D. B. RAF Inhibitor Resistance Is Mediated by Dimerization of Aberrantly Spliced BRAF(V600E). Nature 2011, 480 (7377), 387–390. 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn S. J.; Milagre C.; Whittaker S.; Nourry A.; Niculescu-Duvas I.; Dhomen N.; Hussain J.; Reis-Filho J. S.; Springer C. J.; Pritchard C.; Marais R. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell 2010, 140 (2), 209–221. 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L.; Groom C. R. The Druggable Genome. Nat. Rev. Drug Discovery 2002, 1 (9), 727–730. 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Overington J. P.; Al-Lazikani B.; Hopkins A. L. How Many Drug Targets Are There?. Nat. Rev. Drug Discovery 2006, 5 (12), 993–996. 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Venkatesan K.; Rual J.-F.; Vazquez A.; Stelzl U.; Lemmens I.; Hirozane-Kishikawa T.; Hao T.; Zenkner M.; Xin X.; Goh K.-I.; Yildirim M. A.; Simonis N.; Heinzmann K.; Gebreab F.; Sahalie J. M.; Cevik S.; Simon C.; de Smet A.-S.; Dann E.; Smolyar A.; Vinayagam A.; Yu H.; Szeto D.; Borick H.; Dricot A.; Klitgord N.; Murray R. R.; Lin C.; Lalowski M.; Timm J.; Rau K.; Boone C.; Braun P.; Cusick M. E.; Roth F. P.; Hill D. E.; Tavernier J.; Wanker E. E.; Barabási A.-L.; Vidal M. An Empirical Framework for Binary Interactome Mapping. Nat. Methods 2009, 6 (1), 83–90. 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M. P. H.; Thorne T.; de Silva E.; Stewart R.; An H. J.; Lappe M.; Wiuf C. Estimating the Size of the Human Interactome. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6959–6964. 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C.; van der Hoorn R. A. L.; Kaiser M. The Impact of Plant–pathogen Studies on Medicinal Drug Discovery. Chem. Soc. Rev. 2012, 41 (8), 3168. 10.1039/c2cs15301g. [DOI] [PubMed] [Google Scholar]
- Waring M. J.; Arrowsmith J.; Leach A. R.; Leeson P. D.; Mandrell S.; Owen R. M.; Pairaudeau G.; Pennie W. D.; Pickett S. D.; Wang J.; Wallace O.; Weir A. An Analysis of the Attrition of Drug Candidates from Four Major Pharmaceutical Companies. Nat. Rev. Drug Discovery 2015, 14 (7), 475–486. 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- Arrowsmith C. H.; Audia J. E.; Austin C.; Baell J.; Bennett J.; Blagg J.; Bountra C.; Brennan P. E.; Brown P. J.; Bunnage M. E.; Buser-Doepner C.; Campbell R. M.; Carter A. J.; Cohen P.; Copeland R. A.; Cravatt B.; Dahlin J. L.; Dhanak D.; Edwards A. M.; Frye S. V.; Gray N.; Grimshaw C. E.; Hepworth D.; Howe T.; Huber K. V. M; Jin J.; Knapp S.; Kotz J. D.; Kruger R. G.; Lowe D.; Mader M. M.; Marsden B.; Mueller-Fahrnow A.; Muller S.; O’Hagan R. C.; Overington J. P.; Owen D. R.; Rosenberg S. H.; Roth B.; Ross R.; Schapira M.; Schreiber S. L.; Shoichet B.; Sundstrom M.; Superti-Furga G.; Taunton J.; Toledo-Sherman L.; Walpole C.; Walters M. A.; Willson T. M.; Workman P.; Young R. N.; Zuercher W. J. The Promise and Peril of Chemical Probes. Nat. Chem. Biol. 2015, 11 (8), 536–541. 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye S. V. The Art of the Chemical Probe. Nat. Chem. Biol. 2010, 6 (3), 159–161. 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- Edwards A. M.; Isserlin R.; Bader G. D.; Frye S. V.; Willson T. M.; Yu F. H. Too Many Roads Not Taken. Nature 2011, 470 (7333), 163–165. 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P.; Qi J.; Picaud S.; Shen Y.; Smith W. B.; Fedorov O.; Morse E. M.; Keates T.; Hickman T. T.; Felletar I.; Philpott M.; Munro S.; McKeown M. R.; Wang Y.; Christie A. L.; West N.; Cameron M. J.; Schwartz B.; Heightman T. D.; La Thangue N.; French C. a; Wiest O.; Kung A. L.; Knapp S.; Bradner J. E. Selective Inhibition of BET Bromodomains. Nature 2010, 468 (7327), 1067–1073. 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T.; Vu B. T.; Craves B.; Carvajal D.; Podlaski F.; Filipovic Z.; Kong N.; Kammlott U.; Lukacs C.; Klein C.; Fotouhi N.; Liu E. A. In Vivo Activation of the p53 Pathway by Small-MoleculeAntagonists of MDM2. Science 2004, 303 (5659), 844–848. 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Edfeldt F. N. B.; Folmer R. H. A.; Breeze A. L. Fragment Screening to Predict Druggability (Ligandability) and Lead Discovery Success. Drug Discovery Today 2011, 16 (7–8), 284–287. 10.1016/j.drudis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Surade S.; Blundell T. L. Structural Biology and Drug Discovery of Difficult Targets: The Limits of Ligandability. Chem. Biol. 2012, 19 (1), 42–50. 10.1016/j.chembiol.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Clackson T.; Wells J. A. A Hot Spot of Binding Energy in a Hormone-Receptor Interface. Science 1995, 267 (5196), 383–386. 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Biologic Drugs Set to Top 2012 Sales. Nat. Med. 2012, 18 (5), 636. 10.1038/nm0512-636a. [DOI] [PubMed] [Google Scholar]
- Azzarito V.; Long K.; Murphy N. S.; Wilson A. J. Inhibition of α-Helix-Mediated Protein-Protein Interactions Using Designed Molecules. Nat. Chem. 2013, 5 (3), 161–173. 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- Arkin M. R.; Tang Y.; Wells J. A. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality. Chem. Biol. 2014, 21 (9), 1102–1114. 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R.; Frolov A. I.; Knerr L.; Drury W. J.; Valeur E. Therapeutic Potential of Foldamers: From Chemical Biology Tools to Drug Candidates?. J. Med. Chem. 2016, 59 (21), 9599–9621. 10.1021/acs.jmedchem.6b00376. [DOI] [PubMed] [Google Scholar]
- Wilson W. H.; O’Connor O. A.; Czuczman M. S.; LaCasce A. S.; Gerecitano J. F.; Leonard J. P.; Tulpule A.; Dunleavy K.; Xiong H.; Chiu Y. L.; Cui Y.; Busman T.; Elmore S. W.; Rosenberg S. H.; Krivoshik A. P.; Enschede S. H.; Humerickhouse R. A. Navitoclax, a Targeted High-Affinity Inhibitor of BCL-2, in Lymphoid Malignancies: A Phase 1 Dose-Escalation Study of Safety, Pharmacokinetics, Pharmacodynamics, and Antitumour Activity. Lancet Oncol. 2010, 11 (12), 1149–1159. 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milroy L. G.; Grossmann T. N.; Hennig S.; Brunsveld L.; Ottmann C. Modulators of Protein-Protein Interactions. Chem. Rev. 2014, 114 (9), 4695–4748. 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- Thiel P.; Kaiser M.; Ottmann C. Small-Molecule Stabilization of Protein-Protein Interactions: An Underestimated Concept in Drug Discovery?. Angew. Chem., Int. Ed. 2012, 51 (9), 2012–2018. 10.1002/anie.201107616. [DOI] [PubMed] [Google Scholar]
- Thompson A. D.; Dugan A.; Gestwicki J. E.; Mapp A. K. Fine-Tuning Multiprotein Complexes Using Small Molecules. ACS Chem. Biol. 2012, 7 (8), 1311–1320. 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelay-Gimeno M.; Glas A.; Koch O.; Grossmann T. N. Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem., Int. Ed. 2015, 54 (31), 8896–8927. 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeluri M.; Chamakuri S.; Dasari B.; Guduru S. K. R.; Jimmidi R.; Jogula S.; Arya P. Small Molecule Modulators of Protein-Protein Interactions: Selected Case Studies. Chem. Rev. 2014, 114 (9), 4640–4694. 10.1021/cr4004049. [DOI] [PubMed] [Google Scholar]
- Nussinov R.; Tsai C.-J. Allostery in Disease and in Drug Discovery. Cell 2013, 153 (2), 293–305. 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Samatar A. A.; Poulikakos P. I. Targeting RAS–ERK Signalling in Cancer: Promises and Challenges. Nat. Rev. Drug Discovery 2014, 13 (12), 928–942. 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- Ray-Coquard I.; Blay J. Y.; Italiano A.; Le Cesne A.; Penel N.; Zhi J.; Heil F.; Rueger R.; Graves B.; Ding M.; Geho D.; Middleton S. A.; Vassilev L. T.; Nichols G. L.; Bui B. N. Effect of the MDM2 Antagonist RG7112 on the P53 Pathway in Patients with MDM2-Amplified, Well-Differentiated or Dedifferentiated Liposarcoma: An Exploratory Proof-of-Mechanism Study. Lancet Oncol. 2012, 13 (11), 1133–1140. 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- Souers A. J.; Leverson J. D.; Boghaert E. R.; Ackler S. L.; Catron N. D.; Chen J.; Dayton B. D.; Ding H.; Enschede S. H.; Fairbrother W. J.; Huang D. C. S.; Hymowitz S. G.; Jin S.; Khaw S. L.; Kovar P. J.; Lam L. T.; Lee J.; Maecker H. L.; Marsh K. C.; Mason K. D.; Mitten M. J.; Nimmer P. M.; Oleksijew A.; Park C. H.; Park C.-M.; Phillips D. C.; Roberts A. W.; Sampath D.; Seymour J. F.; Smith M. L.; Sullivan G. M.; Tahir S. K.; Tse C.; Wendt M. D.; Xiao Y.; Xue J. C.; Zhang H.; Humerickhouse R. a; Rosenberg S. H.; Elmore S. W. ABT-199, a Potent and Selective BCL-2 Inhibitor, Achieves Antitumor Activity While Sparing Platelets. Nat. Med. 2013, 19 (2), 202–208. 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Mirguet O.; Gosmini R.; Toum J.; Clément C. A.; Barnathan M.; Brusq J. M.; Mordaunt J. E.; Grimes R. M.; Crowe M.; Pineau O.; Ajakane M.; Daugan A.; Jeffrey P.; Cutler L.; Haynes A. C.; Smithers N. N.; Chung C. W.; Bamborough P.; Uings I. J.; Lewis A.; Witherington J.; Parr N.; Prinjha R. K.; Nicodème E. Discovery of Epigenetic Regulator I-bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains. J. Med. Chem. 2013, 56 (19), 7501–7515. 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Setting. Adv. Drug Delivery Rev. 1997, 23, 3–25. 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Nogales E.; Wolf S. G.; Khan I. A.; Ludueña R. F.; Downing K. H. Structure of Tubulin at 6.5 A and Location of the Taxol-Binding Site. Nature 1995, 375 (6530), 424–427. 10.1038/375424a0. [DOI] [PubMed] [Google Scholar]
- Scheuermann T. H.; Li Q.; Ma H.-W.; Key J.; Zhang L.; Chen R.; Garcia J. a; Naidoo J.; Longgood J.; Frantz D. E.; Tambar U. K.; Gardner K. H.; Bruick R. K. Allosteric Inhibition of Hypoxia Inducible Factor-2 with Small Molecules. Nat. Chem. Biol. 2013, 9 (4), 271–276. 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.; Du X.; Rizzi J. P.; Liberzon E.; Chakraborty A. A.; Gao W.; Carvo I.; Signoretti S.; Bruick R.; Josey J. A.; Wallace E. M.; Kaelin W. G. Jr On-Target Efficacy of a HIF2α Antagonist in Preclinical Kidney Cancer Models. Nature 2016, 539 (7627), 107–111. 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Hill H.; Christie A.; Kim M. S.; Holloman E.; Pavia-Jimenez A.; Homayoun F.; Ma Y.; Patel N.; Yell P.; Hao G.; Yousuf Q.; Joyce A.; Pedrosa I.; Geiger H.; Zhang H.; Chang J.; Gardner K. H.; Bruick R. K.; Reeves C.; Hwang T. H.; Courtney K.; Frenkel E.; Sun X.; Zojwalla N.; Wong T.; Rizzi J. P.; Wallace E. M.; Josey J. A.; Xie Y.; Xie X.-J.; Kapur P.; McKay R. M.; Brugarolas J. Targeting Renal Cell Carcinoma with a HIF-2 Antagonist. Nature 2016, 539 (7627), 112–117. 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L.; Guibert B.; Cherfils J. Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor. Nature 2003, 426 (6966), 525–530. 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- Tesmer J. J.; Sunahara R. K.; Johnson R. A.; Gosslin G.; Gilman A. G.; Sprang S. R.; Gosselin G.; Gilman A. G.; Sprang S. R. Two-Metal-Ion Catalysis in Adenylyl Cyclase. Science 1999, 285 (5428), 756–760. 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- Choi J.; Chen J.; Schreiber S. L. S.; Clardyt J.; Clardy J. Structure of the FKBP12-Rapamycin Complex Interacting with the Binding Domain of Human FRAP. Science 1996, 273, 239–242. 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Johnson S. M.; Wiseman R. L.; Sekijima Y.; Green N. S.; Adamski-Werner S. L.; Kelly J. W. Native State Kinetic Stabilization as a Strategy to Ameliorate Protein Misfolding Diseases: A Focus on the Transthyretin Amyloidoses. Acc. Chem. Res. 2005, 38 (12), 911–921. 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- Green N. S.; Palaninathan S. K.; Sacchettini J. C.; Kelly J. W. Synthesis and Characterization of Potent Bivalent Amyloidosis Inhibitors That Bind Prior to Transthyretin Tetramerization. J. Am. Chem. Soc. 2003, 125 (44), 13404–13414. 10.1021/ja030294z. [DOI] [PubMed] [Google Scholar]
- Tovar C.; Graves B.; Packman K.; Filipovic Z.; Xia B. H. M.; Tardell C.; Garrido R.; Lee E.; Kolinsky K.; To K. H.; Linn M.; Podlaski F.; Wovkulich P.; Vu B.; Vassilev L. T. MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models. Cancer Res. 2013, 73 (8), 2587–2597. 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- Hermeking H.; Benzinger A. 14-3-3 Proteins in Cell Cycle Regulation. Semin. Cancer Biol. 2006, 16 (3), 183–192. 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Aitken A. 14-3-3 Proteins: A Historic Overview. Semin. Cancer Biol. 2006, 16 (3), 162–172. 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Yaffe M. B.; Rittinger K.; Volinia S.; Caron P. R.; Aitken A.; Leffers H.; Gamblin S. J.; Smerdon S. J.; Cantley L. C. The Structural Basis for 14-3-3:phosphopeptide Binding Specificity. Cell 1997, 91 (7), 961–971. 10.1016/S0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Freed E.; Symons M.; Macdonald S. G.; McCormick F.; Ruggieri R. Binding of 14-3-3 Proteins to the Protein Kinase Raf and Effects on Its Activation. Science 1994, 265 (5179), 1713–1716. 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- Molzan M.; Schumacher B.; Ottmann C.; Baljuls A.; Polzien L.; Weyand M.; Thiel P.; Rose R.; Rose M.; Kuhenne P.; Kaiser M.; Rapp U. R.; Kuhlmann J.; Ottmann C. Impaired Binding of 14-3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling. Mol. Cell. Biol. 2010, 30 (19), 4698–4711. 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R. K.; Du X.; Berndt M. C. The 14-3-3zeta-GPIb-IX-V Complex as an Antiplatelet Target. Drug News Perspect. 2007, 20 (5), 285–292. 10.1358/dnp.2007.20.5.1120215. [DOI] [PubMed] [Google Scholar]
- Conklin D. S. 14-3-3 Proteins Associate with cdc25 Phosphatases. Proc. Natl. Acad. Sci. U. S. A. 1995, 92 (17), 7892–7896. 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A.; Kaneko K. J.; Shu H.; Zhao Y.; DePamphilis M. L. TEAD/TEF Transcription Factors Utilize the Activation Domain of YAP65, a Src/Yes-Associated Protein Localized in the Cytoplasm. Genes Dev. 2001, 15 (10), 1229–1241. 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B.; Skwarczynska M.; Rose R.; Ottmann C. Structure of a 14-3-3σ-YAP Phosphopeptide Complex at 1.15 Å Resolution. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2010, 66 (9), 978–984. 10.1107/S1744309110025479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S.; Sade R. S.; Townsley F. M.; Fersht A. R. Mechanistic Differences in the Transcriptional Activation of p53 by 14-3-3 Isoforms. Nucleic Acids Res. 2010, 38 (3), 893–906. 10.1093/nar/gkp1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B.; Mondry J.; Thiel P.; Weyand M.; Ottmann C. Structure of the p53 C-Terminus Bound to 14-3-3: Implications for Stabilization of the p53 Tetramer. FEBS Lett. 2010, 584 (8), 1443–1448. 10.1016/j.febslet.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Stevers L. M.; Lam C. V.; Leysen S. F. R.; Meijer F. A.; van Scheppingen D. S.; de Vries R. M. J. M.; Carlile G. W.; Milroy L. G.; Thomas D. Y.; Brunsveld L.; Ottmann C. Characterization and Small-Molecule Stabilization of the Multisite Tandem Binding between 14-3-3 and the R Domain of CFTR. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (9), E1152–E1161. 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluchanko N. N.; Beelen S.; Kulikova A. A.; Weeks S. D.; Antson A. A.; Gusev N. B.; Strelkov S. V. Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator. Structure 2017, 25 (2), 305–316. 10.1016/j.str.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacirova M.; Kosek D.; Kadek A.; Man P.; Vecer J.; Herman P.; Obsilova V.; Obsil T. Structural Characterization of Phosducin and Its Complex with the 14-3-3 Protein. J. Biol. Chem. 2015, 290 (26), 16246–16260. 10.1074/jbc.M115.636563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevers L. M.; de Vries R. M. J. M.; Doveston R. G.; Milroy L. G.; Brunsveld L.; Ottmann C. Structural Interface between LRRK2 and 14-3-3 Protein. Biochem. J. 2017, 474 (7), 1273–1287. 10.1042/BCJ20161078. [DOI] [PubMed] [Google Scholar]
- Johnson C.; Tinti M.; Wood N. T.; Campbell D. G.; Toth R.; Dubois F.; Geraghty K. M.; Wong B. H. C.; Brown L. J.; Tyler J.; Gernez A.; Chen S.; Synowsky S.; MacKintosh C. Visualization and Biochemical Analyses of the Emerging Mammalian 14-3-3-Phosphoproteome. Mol. Cell. Proteomics 2011, 10 (10), M110.005751. 10.1074/mcp.M110.005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.; Crowther S.; Stafford M. J.; Campbell D. G.; Toth R.; MacKintosh C. Bioinformatic and Experimental Survey of 14-3-3-Binding Sites. Biochem. J. 2010, 427 (1), 69–78. 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzan M.; Ottmann C. Synergistic Binding of the Phosphorylated S233- and S259-Binding Sites of C-RAF to One 14-3-3ζ Dimer. J. Mol. Biol. 2012, 423 (4), 486–495. 10.1016/j.jmb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Larance M.; Rowland A. F.; Hoehn K. L.; Humphreys D. T.; Preiss T.; Guilhaus M.; James D. E. Global Phosphoproteomics Identifies a Major Role for AKT and 14-3-3 in Regulating EDC3. Mol. Cell. Proteomics 2010, 9 (4), 682–694. 10.1074/mcp.M900435-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Synowsky S.; Tinti M.; MacKintosh C. The Capture of Phosphoproteins by 14-3-3 Proteins Mediates Actions of Insulin. Trends Endocrinol. Metab. 2011, 22 (11), 429–436. 10.1016/j.tem.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Chen S.; Murphy J.; Toth R.; Campbell D. G.; Morrice N. a; Mackintosh C. Complementary Regulation of TBC1D1 and AS160 by Growth Factors, Insulin and AMPK Activators. Biochem. J. 2008, 409 (2), 449–459. 10.1042/BJ20071114. [DOI] [PubMed] [Google Scholar]
- Berg D.; Holzmann C.; Riess O. 14-3-3 Proteins in the Nervous System. Nat. Rev. Neurosci. 2003, 4 (9), 752–762. 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- Fu H.; Coburn J.; Collier R. J. The Eukaryotic Host Factor That Activates Exoenzyme S of Pseudomonas Aeruginosa Is a Member of the 14-3-3 Protein Family. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (6), 2320–2324. 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C.; Yasmin L.; Weyand M.; Veesenmeyer J. L.; Diaz M. H.; Palmer R. H.; Francis M. S.; Hauser A. R.; Wittinghofer A.; Hallberg B. Phosphorylation-Independent Interaction between 14-3-3 and Exoenzyme S: From Structure to Pathogenesis. EMBO J. 2007, 26 (3), 902–913. 10.1038/sj.emboj.7601530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siles-Lucas M. D. M.; Gottstein B. The 14-3-3 Protein: A Key Molecule in Parasites as in Other Organisms. Trends Parasitol. 2003, 19 (12), 575–581. 10.1016/j.pt.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Auburn S.; Barry A. E. Dissecting Malaria Biology and Epidemiology Using Population Genetics and Genomics. Int. J. Parasitol. 2016, 47 (2–3), 77–85. 10.1016/j.ijpara.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Lalle M.; Currà C.; Ciccarone F.; Pace T.; Cecchetti S.; Fantozzi L.; Ay B.; Breton C. B.; Ponzi M. Dematin, a Component of the Erythrocyte Membrane Skeleton, Is Internalized by the Malaria Parasite and Associates with Plasmodium 14-3-3. J. Biol. Chem. 2011, 286 (2), 1227–1236. 10.1074/jbc.M110.194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal K.; Bromley E.; Oakes R.; Prieto J. H.; Sanderson S. J.; Kurian D.; Hunt L.; Yates J. R.; Wastling J. M.; Sinden R. E.; Tomley F. M. Proteomic Comparison of Four Eimeria Tenella Life-Cycle Stages: Unsporulated Oocyst, Sporulated Oocyst, Sporozoite and Second-Generation Merozoite. Proteomics 2009, 9 (19), 4566–4576. 10.1002/pmic.200900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.; Dubey J. P. Toxoplasma Gondii: Transmission, Diagnosis, and Prevention. Clin. Microbiol. Infect. 2002, 8 (10), 634–640. 10.1046/j.1469-0691.2002.00485.x. [DOI] [PubMed] [Google Scholar]
- Assossou O.; Besson F.; Rouault J. P.; Persat F.; Brisson C.; Duret L.; Ferrandiz J.; Mayençon M.; Peyron F.; Picot S. Subcellular Localization of 14-3-3 Proteins in Toxoplasma Gondii Tachyzoites and Evidence for a Lipid Raft-Associated Form. FEMS Microbiol. Lett. 2003, 224 (2), 161–168. 10.1016/S0378-1097(03)00479-8. [DOI] [PubMed] [Google Scholar]
- Weidner J. M.; Kanatani S.; Uchtenhagen H.; Varas-Godoy M.; Schulte T.; Engelberg K.; Gubbels M. J.; Sun H. S.; Harrison R. E.; Achour A.; Barragan A. Migratory Activation of Parasitized Dendritic Cells by the Protozoan Toxoplasma Gondii 14-3-3 Protein. Cell. Microbiol. 2016, 18 (11), 1537–1550. 10.1111/cmi.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulakci M.; Kartal M. G.; Yilmaz S.; Yilmaz E.; Yilmaz R.; Sahin D.; Asik M.; Erol O. B. Multimodality Imaging in Diagnosis and Management of Alveolar Echinococcosis: An Update. Diagn. Interv. Radiol. 2016, 22 (3), 247–256. 10.5152/dir.2015.15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siles-Lucas M.; Felleisen R. S. J.; Hemphill A.; Wilson W.; Gottstein B. Stage-Specific Expression of the 14-3-3 Gene in Echinococcus Multilocularis. Mol. Biochem. Parasitol. 1998, 91 (2), 281–293. 10.1016/S0166-6851(97)00208-9. [DOI] [PubMed] [Google Scholar]
- Siles-Lucas M.; Nunes C. P.; Zaha A. Comparative Analysis of the 14-3-3 Gene and Its Expression in Echinococcus Granulosus and Echinococcus Multilocularis Metacestodes. Parasitology 2001, 122 (Pt 3), 281–287. 10.1017/S0031182001007405. [DOI] [PubMed] [Google Scholar]
- McGonigle S.; Pearce E. J. 14-3-3 Proteins in Schistosoma Mansoni; Identification of a Second Epsilon Isoform. Int. J. Parasitol. 2002, 32 (6), 685–693. 10.1016/S0020-7519(01)00323-X. [DOI] [PubMed] [Google Scholar]
- Yang J.; Pan W.; Sun X.; Zhao X.; Yuan G.; Sun Q.; Huang J.; Zhu X. Immunoproteomic Profile of Trichinella Spiralis Adult Worm Proteins Recognized by Early Infection Sera. Parasites Vectors 2015, 8 (1), 20. 10.1186/s13071-015-0641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokx S. J.; Wernimont A. K.; Dong A.; Wasney G. A.; Lin Y. H.; Lew J.; Vedadi M.; Lee W. H.; Hui R. Characterization of 14-3-3 Proteins from Cryptosporidium Parvum. PLoS One 2011, 6 (8), e14827. 10.1371/journal.pone.0014827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E.; Hoekstra R. M.; Angulo F. J.; Tauxe R. V.; Widdowson M. A.; Roy S. L.; Jones J. L.; Griffin P. M. Foodborne Illness Acquired in the United States-Major Pathogens. Emerging Infect. Dis. 2011, 17 (1), 7–15. 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliez M. C. M.; Buret A. G. Extra-Intestinal and Long Term Consequences of Giardia Duodenalis Infections. World J. Gastroenterol. 2013, 19 (47), 8974–8985. 10.3748/wjg.v19.i47.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo A. A.; Almirall P.; Robertson L. J.; Franco R. M. B.; Hanevik K.; Mørch K.; Cimerman S. Giardiasis: The Ever-Present Threat of a Neglected Disease. Infect. Disord.: Drug Targets 2010, 10 (5), 329–348. 10.2174/187152610793180821. [DOI] [PubMed] [Google Scholar]
- Cau Y.; Fiorillo A.; Mori M.; Ilari A.; Botta M.; Lalle M. Molecular Dynamics Simulations and Structural Analysis of Giardia Duodenalis 14-3-3 Protein-Protein Interactions. J. Chem. Inf. Model. 2015, 55 (12), 2611–2622. 10.1021/acs.jcim.5b00452. [DOI] [PubMed] [Google Scholar]
- Fiorillo A.; Di Marino D.; Bertuccini L.; Via A.; Pozio E.; Camerini S.; Ilari A.; Lalle M. The Crystal Structure of Giardia Duodenalis 14-3-3 in the Apo Form: When Protein Post-Translational Modifications Make the Difference. PLoS One 2014, 9 (3), e92902. 10.1371/journal.pone.0092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K. The 14-3-3 Proteins: Integrators of Diverse Signaling Cues That Impact Cell Fate and Cancer Development. Trends Cell Biol. 2009, 19 (1), 16–23. 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coblitz B.; Wu M.; Shikano S.; Li M. C-Terminal Binding: An Expanded Repertoire and Function of 14-3-3 Proteins. FEBS Lett. 2006, 580 (6), 1531–1535. 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Tinti M.; Madeira F.; Murugesan G.; Hoxhaj G.; Toth R.; MacKintosh C. ANIA: ANnotation and Integrated Analysis of the 14-3-3 Interactome. Database 2014, bat085, bat085. 10.1093/database/bat085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsil T.; Ghirlando R.; Klein D. C.; Ganguly S.; Dyda F. Crystal Structure of the 14-3-3zeta:serotonin N-Acetyltransferase Complex. a Role for Scaffolding in Enzyme Regulation. Cell 2001, 105 (2), 257–267. 10.1016/S0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Wilker E. W.; Grant R. A.; Artim S. C.; Yaffe M. B. A Structural Basis for 14-3-3sigma Functional Specificity. J. Biol. Chem. 2005, 280 (19), 18891–18898. 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- Arendt J. Melatonin in Humans: It’s about Time. J. Neuroendocrinol. 2005, 17 (8), 537–538. 10.1111/j.1365-2826.2005.01333.x. [DOI] [PubMed] [Google Scholar]
- Ooms S.; Ju Y.-E. Treatment of Sleep Disorders in Dementia. Curr. Treat. Options Neurol. 2016, 18 (9), 40. 10.1007/s11940-016-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J.; Pedersen J. T.; Fuglsang A. T.; Palmgren M. Plasma Membrane H+-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9 (3), 323–337. 10.1016/j.molp.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Palmgren M. G. PLANT PLASMA MEMBRANE H+-ATPases: Powerhouses for Nutrient Uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 817–845. 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- Arango M.; Gévaudant F.; Oufattole M.; Boutry M. The Plasma Membrane Proton Pump ATPase: The Significance of Gene Subfamilies. Planta 2003, 216 (3), 355–365. [DOI] [PubMed] [Google Scholar]
- Sondergaard T. E.; Schulz A.; Palmgren M. G. Energization of Transport Processes in Plants. Roles of the Plasma Membrane H+-ATPase. Plant Physiol. 2004, 136 (1), 2475–2482. 10.1104/pp.104.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G.; Boutry M. The Plant Plasma Membrane Proton Pump ATPase: A Highly Regulated P-Type ATPase with Multiple Physiological Roles. Pfluegers Arch. 2009, 457 (3), 645–655. 10.1007/s00424-008-0457-x. [DOI] [PubMed] [Google Scholar]
- Jahn T.; Fuglsang A. T.; Olsson A.; Brüntrup I. M.; Collinge D. B.; Volkmann D.; Sommarin M.; Palmgren M. G.; Larsson C. The 14-3-3 Protein Interacts Directly with the C-Terminal Region of the Plant Plasma Membrane H(+)-ATPase. Plant Cell 1997, 9 (10), 1805–1814. 10.2307/3870526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M.; Morsomme P.; Boutry M.; Oecking C. Complementation of the Saccharomyces Cerevisiae Plasma Membrane H+-ATPase by a Plant H+-ATPase Generates a Highly Abundant Fusicoccin Binding Site. J. Biol. Chem. 1998, 273 (45), 30018–30023. 10.1074/jbc.273.45.30018. [DOI] [PubMed] [Google Scholar]
- Olsson A.; Svennelid F.; Ek B.; Sommarin M.; Larsson C. A. Phosphothreonine Residue at the C-Terminal End of the Plasma Membrane H+-ATPase Is Protected by Fusicoccin-Induced 14-3-3 Binding. Plant Physiol. 1998, 118 (2), 551–555. 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F.; Olsson A.; Piotrowski M.; Rosenquist M.; Ottman C.; Larsson C.; Oecking C.; Sommarin M. Phosphorylation of Thr-948 at the C Terminus of the Plasma Membrane H(+)-ATPase Creates a Binding Site for the Regulatory 14-3-3 Protein. Plant Cell 1999, 11 (12), 2379–2391. 10.2307/3870962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele M.; Jelich-Ottmann C.; Wittinghofer A.; Oecking C. Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex. EMBO J. 2003, 22 (5), 987–994. 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C.; Marco S.; Jaspert N.; Marcon C.; Schauer N.; Weyand M.; Vandermeeren C.; Duby G.; Boutry M.; Wittinghofer A.; Rigaud J.-L.; Oecking C. Structure of a 14-3-3 Coordinated Hexamer of the Plant Plasma Membrane H+-ATPase by Combining X-Ray Crystallography and Electron Cryomicroscopy. Mol. Cell 2007, 25 (3), 427–440. 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Chailakhyan M. K. About the Mechanism of the Photoperiodic Response (in Russian). Dokl Akad Nauk SSSR 1936, 1, 85–89. [Google Scholar]
- Kobayashi Y.; Weigel D. Move on Up, It’s Time for Change--Mobile Signals Controlling Photoperiod-Dependent Flowering. Genes Dev. 2007, 21 (19), 2371–2384. 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Tamaki S.; Matsuo S.; Wong H. L.; Yokoi S.; Shimamoto K. Hd3a Protein Is a Mobile Flowering Signal in Rice. Science 2007, 316 (5827), 1033–1036. 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K.; Ohki I.; Tsuji H.; Furuita K.; Hayashi K.; Yanase T.; Yamaguchi M.; Nakashima C.; Purwestri Y. A.; Tamaki S.; Ogaki Y.; Shimada C.; Nakagawa A.; Kojima C.; Shimamoto K. 14-3-3 Proteins Act as Intracellular Receptors for Rice Hd3a Florigen. Nature 2011, 476 (7360), 332–335. 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Ottmann C.; Marco S.; Jaspert N.; Marcon C.; Schauer N.; Weyand M.; Vandermeeren C.; Duby G.; Boutry M.; Wittinghofer A.; Rigaud J. L.; Oecking C. Structure of a 14-3-3 Coordinated Hexamer of the Plant Plasma Membrane H+-ATPase by Combining X-Ray Crystallography and Electron Cryomicroscopy. Mol. Cell 2007, 25 (3), 427–440. 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Bakthisaran R.; Tangirala R.; Rao C. M. Small Heat Shock Proteins: Role in Cellular Functions and Pathology. Biochim. Biophys. Acta, Proteins Proteomics 2015, 1854 (4), 291–319. 10.1016/j.bbapap.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Chernik I. S.; Seit-Nebi A. S.; Marston S. B.; Gusev N. B. Small Heat Shock Protein Hsp20 (HspB6) as a Partner of 14-3-3gamma. Mol. Cell. Biochem. 2007, 295 (1–2), 9–17. 10.1007/s11010-006-9266-8. [DOI] [PubMed] [Google Scholar]
- Wang B.; Yang H.; Liu Y. C.; Jelinek T.; Zhang L.; Ruoslahti E.; Fu H. Isolation of High-Affinity Peptide Antagonists of 14-3-3 Proteins by Phage Display. Biochemistry 1999, 38 (38), 12499–12504. 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- Petosa C.; Masters S. C.; Bankston L. A.; Pohl J.; Wang B.; Fu H.; Liddington R. C. 14-3-3ζ Binds a Phosphorylated Raf Peptide and an Unphosphorylated Peptide via Its Conserved Amphiphatic Groove. J. Biol. Chem. 1998, 273 (26), 16305–16310. 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- Masters S. C.; Fu H. 14-3-3 Proteins Mediate an Essential Anti-Apoptotic Signal. J. Biol. Chem. 2001, 276 (48), 45193–45200. 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- Cao W.; Yang X.; Zhou J.; Teng Z.; Cao L.; Zhang X.; Fei Z. Targeting 14-3-3 Protein, Difopein Induces Apoptosis of Human Glioma Cells and Suppresses Tumor Growth in Mice. Apoptosis 2010, 15 (2), 230–241. 10.1007/s10495-009-0437-4. [DOI] [PubMed] [Google Scholar]
- Glas A.; Bier D.; Hahne G.; Rademacher C.; Ottmann C.; Grossmann T. N. Constrained Peptides with Target-Adapted Cross-Links as Inhibitors of a Pathogenic Protein-Protein Interaction. Angew. Chem., Int. Ed. 2014, 53 (9), 2489–2493. 10.1002/anie.201310082. [DOI] [PubMed] [Google Scholar]
- Cromm P. M.; Wallraven K.; Glas A.; Bier D.; Fürstner A.; Ottmann C.; Grossmann T. N. Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis. ChemBioChem 2016, 17 (20), 1915–1919. 10.1002/cbic.201600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layfield R.; Fergusson J.; Aitken A.; Lowe J.; Landon M.; Mayer R. J. Neurofibrillary Tangles of Alzheimer’s Disease Brains Contain 14-3-3 Proteins. Neurosci. Lett. 1996, 209 (1), 57–60. 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- Sadik G.; Tanaka T.; Kato K.; Yamamori H.; Nessa B. N.; Morihara T.; Takeda M. Phosphorylation of Tau at Ser214 Mediates Its Interaction with 14-3-3 Protein: Implications for the Mechanism of Tau Aggregation. J. Neurochem. 2009, 108 (1), 33–43. 10.1111/j.1471-4159.2008.05716.x. [DOI] [PubMed] [Google Scholar]
- Sluchanko N. N.; Seit-Nebi A. S.; Gusev N. B. Phosphorylation of More than One Site Is Required for Tight Interaction of Human Tau Protein with 14-3-3ζ. FEBS Lett. 2009, 583 (17), 2739–2742. 10.1016/j.febslet.2009.07.043. [DOI] [PubMed] [Google Scholar]
- Joo Y.; Schumacher B.; Landrieu I.; Bartel M.; Smet-Nocca C.; Jang A.; Choi H. S.; Jeon N. L.; Chang K. A.; Kim H. S.; Ottmann C.; Suh Y. H. Involvement of 14-3-3 in Tubulin Instability and Impaired Axon Development Is Mediated by Tau. FASEB J. 2015, 29 (10), 4133–4144. 10.1096/fj.14-265009. [DOI] [PubMed] [Google Scholar]
- Milroy L. G.; Bartel M.; Henen M. A.; Leysen S.; Adriaans J. M. C.; Brunsveld L.; Landrieu I.; Ottmann C. Stabilizer-Guided Inhibition of Protein-Protein Interactions. Angew. Chem., Int. Ed. 2015, 54 (52), 15720–15724. 10.1002/anie.201507976. [DOI] [PubMed] [Google Scholar]
- Wu H.; Ge J.; Yao S. Q. Microarray-Assisted High-Throughput Identification of a Cell-Permeable Small-Molecule Binder of 14-3-3 Proteins. Angew. Chem., Int. Ed. 2010, 49 (37), 6528–6532. 10.1002/anie.201003257. [DOI] [PubMed] [Google Scholar]
- Arrendale A.; Kim K.; Choi J. Y.; Li W.; Geahlen R. L.; Borch R. F. Synthesis of a Phosphoserine Mimetic Prodrug with Potent 14-3-3 Protein Inhibitory Activity. Chem. Biol. 2012, 19 (6), 764–771. 10.1016/j.chembiol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi V.; Mancini M.; Manetti F.; Petta S.; Santucci M. A.; Botta M. Identification of the First Non-Peptidic Small Molecule Inhibitor of the c-Abl/14-3-3 Protein-Protein Interactions Able to Drive Sensitive and Imatinib-Resistant Leukemia Cells to Apoptosis. Bioorg. Med. Chem. Lett. 2010, 20 (20), 6133–6137. 10.1016/j.bmcl.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Mancini M.; Corradi V.; Petta S.; Barbieri E.; Manetti F.; Botta M.; Santucci M. A. A New Nonpeptidic Inhibitor of 14-3-3 Induces Apoptotic Cell Death in Chronic Myeloid Leukemia Sensitive or Resistant to Imatinib. J. Pharmacol. Exp. Ther. 2011, 336 (3), 596–604. 10.1124/jpet.110.172536. [DOI] [PubMed] [Google Scholar]
- Corradi V.; Mancini M.; Santucci M. A.; Carlomagno T.; Sanfelice D.; Mori M.; Vignaroli G.; Falchi F.; Manetti F.; Radi M.; Botta M. Computational Techniques Are Valuable Tools for the Discovery of Protein-Protein Interaction Inhibitors: The 14-3-3σ Case. Bioorg. Med. Chem. Lett. 2011, 21 (22), 6867–6871. 10.1016/j.bmcl.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Mori M.; Vignaroli G.; Cau Y.; Dinic̈ J.; Hill R.; Rossi M.; Colecchia D.; Pešic̈ M.; Link W.; Chiariello M.; Ottmann C.; Botta M. Discovery of 14-3-3 Protein-Protein Interaction Inhibitors That Sensitize Multidrug-Resistant Cancer Cells to Doxorubicin and the Akt Inhibitor GSK690693. ChemMedChem 2014, 9 (5), 973–983. 10.1002/cmdc.201400044. [DOI] [PubMed] [Google Scholar]
- Valensin D.; Cau Y.; Calandro P.; Vignaroli G.; Dello Iacono L.; Chiariello M.; Mori M.; Botta M. Molecular Insights to the Bioactive Form of BV02, a Reference Inhibitor of 14-3-3σ Protein-Protein Interactions. Bioorg. Med. Chem. Lett. 2016, 26 (3), 894–898. 10.1016/j.bmcl.2015.12.066. [DOI] [PubMed] [Google Scholar]
- An S. S.; Askovich P. S.; Zarembinski T. I.; Ahn K.; Peltier J. M.; von Rechenberg M.; Sahasrabudhe S.; Fredberg J. J. A Novel Small Molecule Target in Human Airway Smooth Muscle for Potential Treatment of Obstructive Lung Diseases: A Staged High-Throughput Biophysical Screening. Respir. Res. 2011, 12 (1), 8. 10.1186/1465-9921-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Du Y.; Horton J. R. R.; Upadhyay A. K. K.; Lou B.; Bai Y.; Zhang X.; Du L.; Li M.; Wang B.; et al. Discovery and Structural Characterization of a Small Molecule 14-3-3 Protein-Protein Interaction Inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (39), 16212–16216. 10.1073/pnas.1100012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-C.; Camaioni E.; Ziganshin A. U.; Ji X.-D.; King B. F.; Wildman S. S.; Rychkov A.; Yoburn J.; Kim H.; Mohanram A.; Harden T. K.; Boyer J. L.; Burnstock G.; Jacobson K. A. Synthesis and Structure-Activity Relationships of Pyridoxal-6-Arylazo-5′-phosphate and Phosphonate Derivatives as P2 Receptor Antagonists. Drug Dev. Res. 1998, 45 (2), 52–66. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röglin L.; Thiel P.; Kohlbacher O.; Ottmann C. Covalent Attachment of Pyridoxal-Phosphate Derivatives to 14-3-3 Proteins. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (18), E1051–3. author reply E1054. 10.1073/pnas.1116592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A. K.; Horton J. R.; Du Y.; Bai Y.; Cheng X.; Fu H. Reply to Roglin et Al.: Synchrotron Radiation-Induced Covalent Modification of 14-3-3 by Diazene Compounds Containing Pyridoxal Phosphate. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (18), E1054–E1054. 10.1073/pnas.1203341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto Y.; Watanabe H.; Uchida K.; Matsumura K.; Nakae K.; Tashiro E.; Shindo K.; Kitahara T.; Imoto M. Chemistry and Biology of Moverastins, Inhibitors of Cancer Cell Migration, Produced by Aspergillus. Chem. Biol. 2005, 12 (12), 1337–1347. 10.1016/j.chembiol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Singh S. B.; Ball R. G.; Bills G. F.; Cascales C.; Gibbs J. B.; Goetz M. A.; Hoogsteen K.; Jenkins R. G.; Liesch J. M.; Lingham R. B.; Silverman K. C.; Zink D. L. Chemistry and Biology of Cylindrols: Novel Inhibitors of Ras Farnesyl-Protein Transferase from Cylindrocarpon Lucidum. J. Org. Chem. 1996, 61 (22), 7727–7737. 10.1021/jo961074p. [DOI] [PubMed] [Google Scholar]
- Sawada M.; Kubo S. I.; Matsumura K.; Takemoto Y.; Kobayashi H.; Tashiro E.; Kitahara T.; Watanabe H.; Imoto M. Synthesis and Anti-Migrative Evaluation of Moverastin Derivatives. Bioorg. Med. Chem. Lett. 2011, 21 (5), 1385–1389. 10.1016/j.bmcl.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Tashiro E.; Imoto M. Screening and Target Identification of Bioactive Compounds That Modulate Cell Migration and Autophagy. Bioorg. Med. Chem. 2016, 24 (15), 3283–3290. 10.1016/j.bmc.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Thiel P.; Röglin L.; Meissner N.; Hennig S.; Kohlbacher O.; Ottmann C. Virtual Screening and Experimental Validation Reveal Novel Small-Molecule Inhibitors of 14-3-3 Protein–protein Interactions. Chem. Commun. 2013, 49 (76), 8468–8470. 10.1039/c3cc44612c. [DOI] [PubMed] [Google Scholar]
- Hu G.; Cao Z.; Xu S.; Wang W.; Wang J. Revealing the Binding Modes and the Unbinding of 14-3-3σ Proteins and Inhibitors by Computational Methods. Sci. Rep. 2015, 5, 16481. 10.1038/srep16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier D.; Rose R.; Bravo-Rodriguez K.; Bartel M.; Ramirez-Anguita J. M.; Dutt S.; Wilch C.; Klärner F.-G.; Sanchez-Garcia E.; Schrader T.; Ottmann C. Molecular Tweezers Modulate 14-3-3 Protein-Protein Interactions. Nat. Chem. 2013, 5 (3), 234–239. 10.1038/nchem.1570. [DOI] [PubMed] [Google Scholar]
- Ballio A.; Chain E. B.; de Leo P.; Erlanger B. F.; Mauri M.; Tonolo A. Fusicoccin: A New Wilting Toxin Produced by Fusicoccum Amygdali Del. Nature 1964, 203, 297–297. 10.1038/203297a0. [DOI] [Google Scholar]
- Oecking C.; Eckerskorn C.; Weiler E. W. The Fusicoccin Receptor of Plants Is a Member of the 14-3-3 Superfamily of Eukaryotic Regulatory Proteins. FEBS Lett. 1994, 352 (2), 163–166. 10.1016/0014-5793(94)00949-X. [DOI] [PubMed] [Google Scholar]
- Camoni L.; Di Lucente C.; Visconti S.; Aducci P. The Phytotoxin Fusicoccin Promotes Platelet Aggregation via 14-3-3-Glycoprotein Ib-IX-V Interaction. Biochem. J. 2011, 436 (2), 429–436. 10.1042/BJ20102037. [DOI] [PubMed] [Google Scholar]
- De Vries-van Leeuwen I. J.; da Costa Pereira D.; Flach K. D.; Piersma S. R.; Haase C.; Bier D.; Yalcin Z.; Michalides R.; Feenstra K. A.; Jiménez C. R.; de Greef T. F. A.; Brunsveld L.; Ottmann C.; Zwart W.; de Boer A. H. Interaction of 14-3-3 Proteins with the Estrogen Receptor Alpha F Domain Provides a Drug Target Interface. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (22), 8894–8899. 10.1073/pnas.1220809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T.; Tojyo T.; Munakata K. Isolation of a New Plant Growth Substance with Cytokinin-like Activity. Nature 1970, 227, 379. 10.1038/227379a0. [DOI] [PubMed] [Google Scholar]
- Honma Y.; Ishii Y.; Yamamoto-Yamaguchi Y.; Sassa T.; Asahi K. I. Cotylenin A, a Differentiation-Inducing Agent, and IFN-Alpha Cooperatively Induce Apoptosis and Have an Antitumor Effect on Human Non-Small Cell Lung Carcinoma Cells in Nude Mice. Cancer Res. 2003, 63 (13), 3659–3666. [PubMed] [Google Scholar]
- Yamada K.; Honma Y.; Asahi K.-I.; Sassa T.; Hino K.-I.; Tomoyasu S. Differentiation of Human Acute Myeloid Leukaemia Cells in Primary Culture in Response to Cotylenin A, a Plant Growth Regulator. Br. J. Haematol. 2001, 114 (4), 814–821. 10.1046/j.1365-2141.2001.03029.x. [DOI] [PubMed] [Google Scholar]
- Takahashi T.; Honma Y.; Miyake T.; Adachi K.; Takami S.; Okada M.; Kumanomidou S.; Ikejiri F.; Jo Y.; Onishi C.; Kawakami K.; Moriyama I.; Inoue M.; Tanaka J.; Suzumiya J. Synergistic Combination Therapy with Cotylenin A and Vincristine in Multiple Myeloma Models. Int. J. Oncol. 2015, 46 (4), 1801–1809. 10.3892/ijo.2015.2882. [DOI] [PubMed] [Google Scholar]
- Molzan M.; Kasper S.; Röglin L.; Skwarczynska M.; Sassa T.; Inoue T.; Breitenbuecher F.; Ohkanda J.; Kato N.; Schuler M.; Ottmann C. Stabilization of Physical RAF/14-3-3 Interaction by Cotylenin A as Treatment Strategy for RAS Mutant Cancers. ACS Chem. Biol. 2013, 8 (9), 1869–1875. 10.1021/cb4003464. [DOI] [PubMed] [Google Scholar]
- Anders C.; Higuchi Y.; Koschinsky K.; Bartel M.; Schumacher B.; Thiel P.; Nitta H.; Preisig-Müller R.; Schlichthörl G.; Renigunta V.; Ohkanda J.; Daut J.; Kato N.; Ottmann C. A Semisynthetic Fusicoccane Stabilizes a Protein-Protein Interaction and Enhances the Expression of K+ Channels at the Cell Surface. Chem. Biol. 2013, 20 (4), 583–593. 10.1016/j.chembiol.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Bier D.; Bartel M.; Sies K.; Halbach S.; Higuchi Y.; Haranosono Y.; Brummer T.; Kato N.; Ottmann C. Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface. ChemMedChem 2016, 11 (8), 911–918. 10.1002/cmdc.201500484. [DOI] [PubMed] [Google Scholar]
- Parvatkar P.; Kato N.; Uesugi M.; Sato S. I.; Ohkanda J. Intracellular Generation of a Diterpene-Peptide Conjugate That Inhibits 14-3-3-Mediated Interactions. J. Am. Chem. Soc. 2015, 137 (50), 15624–15627. 10.1021/jacs.5b09817. [DOI] [PubMed] [Google Scholar]
- Brill Z. G.; Grover H. K.; Maimone T. J. Enantioselective Synthesis of an Ophiobolin Sesterterpene via a Programmed Radical Cascade. Science 2016, 352 (6289), 1078–1082. 10.1126/science.aaf6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A.; Hedberg C.; Waldmann H. Enantioselective Synthesis of the C10-C20 Fragment of Fusicoccin A. J. Org. Chem. 2011, 76 (16), 6694–6702. 10.1021/jo201020v. [DOI] [PubMed] [Google Scholar]
- Chen M.; Chou W. K. W.; Toyomasu T.; Cane D. E.; Christianson D. W. Structure and Function of Fusicoccadiene Synthase, a Hexameric Bifunctional Diterpene Synthase. ACS Chem. Biol. 2016, 11 (4), 889–899. 10.1021/acschembio.5b00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R.; Erdmann S.; Bovens S.; Wolf A.; Rose M.; Hennig S.; Waldmann H.; Ottmann C. Identification and Structure of Small-Molecule Stabilizers of 14-3-3 Protein-Protein Interactions. Angew. Chem., Int. Ed. 2010, 49 (24), 4129–4132. 10.1002/anie.200907203. [DOI] [PubMed] [Google Scholar]
- Richter A.; Rose R.; Hedberg C.; Waldmann H.; Ottmann C. An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction. Chem. - Eur. J. 2012, 18 (21), 6520–6527. 10.1002/chem.201103761. [DOI] [PubMed] [Google Scholar]
- Sato S.; Jung H.; Nakagawa T.; Pawlosky R.; Takeshima T.; Lee W. R.; Sakiyama H.; Laxman S.; Wynn R. M.; Tu B. P.; MacMillan J. B.; De Brabander J. K.; Veech R. L.; Uyeda K. Metabolite Regulation of Nuclear Localization of Carbohydrate-Response Element-Binding Protein (ChREBP): Role of Amp as an Allosteric Inhibitor. J. Biol. Chem. 2016, 291 (20), 10515–10527. 10.1074/jbc.M115.708982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S.; Iizuka K.; Miller B. C.; Uyeda K. Carbohydrate Response Element Binding Protein Directly Promotes Lipogenic Enzyme Gene Transcription. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (44), 15597–15602. 10.1073/pnas.0405238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K.; Bruick R. K.; Liang G.; Horton J. D.; Uyeda K. Deficiency of Carbohydrate Response Element-Binding Protein (ChREBP) Reduces Lipogenesis as Well as Glycolysis. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (19), 7281–7286. 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama H.; Wynn R. M.; Lee W. R.; Fukasawa M.; Mizuguchi H.; Gardner K. H.; Repa J. J.; Uyeda K. Regulation of Nuclear Import/export of Carbohydrate Response Element-Binding Protein (ChREBP): Interaction of An α-Helix of ChREBP with the 14-3-3 Proteins and Regulation by Phosphorylation. J. Biol. Chem. 2008, 283 (36), 24899–24908. 10.1074/jbc.M804308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T.; Ge Q.; Pawlosky R.; Wynn R. M.; Veech R. L.; Uyeda K. Metabolite Regulation of Nucleo-Cytosolic Trafficking of Carbohydrate Response Element-Binding Protein (ChREBP): Role of Ketone Bodies. J. Biol. Chem. 2013, 288 (39), 28358–28367. 10.1074/jbc.M113.498550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q.; Huang N.; Wynn R. M.; Li Y.; Du X.; Miller B.; Zhang H.; Uyeda K. Structural Characterization of a Unique Interface between Carbohydrate Response Element-Binding Protein (ChREBP) and 14-3-3β Protein. J. Biol. Chem. 2012, 287 (50), 41914–41921. 10.1074/jbc.M112.418855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbesma E.; Skora L.; Leysen S.; Brunsveld L.; Koch U.; Nussbaumer P.; Jahnke W.; Ottmann C. Identification of Two Secondary Ligand binding sites in 14-3-3 Proteins Using Fragment Screening. Biochemistry 2017, 56 (30), 3972–3982. 10.1021/acs.biochem.7b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.