Abstract
DNA damage–induced NF-κB activation and the secretion of inflammatory cytokines play crucial roles in carcinogenesis and cellular senescence. However, the underlying mechanisms, especially the initial sensors and transducers connecting the nuclear DNA damage signal with cytoplasmic NF-κB activation remain incompletely understood. Here, we report that TRAF-interacting protein with forkhead-associated domain (TIFA), an established NF-κB activator in the cytosol, unexpectedly exhibited nuclear translocation and accumulation on damaged chromatin following genotoxic stress. Accordingly, we also found that DNA damage–induced transcriptional activation and the resulting secretion of classic NF-κB targets, including interleukin (IL)-6 and IL-8, was greatly enhanced in TIFA-overexpressing cells compared with control cells. Mechanistically, DNA damage–induced TIFA phosphorylation at threonine 9 (pThr-9), and this phosphorylation event, involving the pThr-binding forkhead-associated domain, was crucial for its enrichment on damaged chromatin and subsequent NF-κB activation. Moreover, in conjunction with its partner protein, the E3 ligase TNF receptor–associated factor 2 (TRAF2), TIFA relayed the DNA damage signals by stimulating ubiquitination of NF-κB essential modulator (NEMO), whose sumoylation, phosphorylation, and ubiquitination were critical for NF-κB's response to DNA damage. Consistently, TRAF2 knockdown suppressed TIFA overexpression–enhanced NEMO ubiquitination under genotoxic stress, and a unphosphorylatable Thr-9–mutated TIFA variant had only minor effects on NEMO poly-ubiquitination. Finally, in agreement with the model of DNA damage–associated secretory senescence barrier against carcinogenesis, ectopic TIFA expression limited proliferation of multiple myeloma cancer cells. In conclusion our results indicate that TIFA functions as a key transducer in DNA damage–induced NF-κB activation.
Keywords: DNA damage, NF-κB transcription factor, ubiquitylation (ubiquitination), carcinogenesis, TNF receptor associated factor (TRAF), NEMO, TIFA, TRAF-interacting protein with forkhead-associated domain
Introduction
Genomic instability and associated DNA damage response (DDR)3 are common hallmarks in cancer and aging (1). An emerging theme in DDR-induced cellular phenotypic change is the dramatically increased secretion of a myriad inflammatory factors, including cytokines, chemokines, and interferons, which contribute to cancer development and senescence progression in autocrine, paracrine, or endocrine fashions via their collaborations with DDR (1). Such secretome alternations are manifested in the conditions with oncogene overexpression or tumor suppressor inactivation, and hence are designated as “senescence-associated secretory phenotype” in the senescence barrier model of malignancy (2).
The NF-κB family transcription factors are master regulators for transcriptional activation of secretory factors, especially during cellular response to the genotoxic stress (3). Despite the diversity of upstream stimuli, the NF-κB cascade shares a common activation scheme consisting of phosphorylation, ubiquitination, and degradation of IκB (inhibitors of NF-κB) proteins, which result in the nuclear translocation of NF-κB with masked nuclear localization signal of IκB exposed (4). A plethora of physical and chemical stresses engage specific receptors and intracellular adaptors to transduce signals, and they generally converge on activation of the IκB-kinase complex (IKK), which is composed by the catalytic subunit (IKKα or IKKβ) and the regulatory subunit (IKKγ, also known as NEMO) (4). TRAF (TNF receptor–associated factor) family proteins, represented by TRAF2 and TRAF6 with a N-terminal RING finger domain, are key intermediates in many NF-κB signaling pathways, employing their E3 ligase activity to synthesize a regulatory lysine 63-linked poly-ubiquitin chain on target proteins (4, 5). The K63-linked poly-ubiquitin chain, critical for the assembly of TAK1-TAB2/TAB3 and IKK complexes, is directly bound by ubiquitin recognizing modules from TABs or NEMO, and the TAK1-TAB2/TAB3 complex could subsequently trigger IKK phosphorylation and activation (5).
The “inside-out” transduction of DNA damage signals obliges nucleocytoplasmic shuttling mechanisms, and NEMO exploits its post-translational modifications to meet this requirement and hence takes center stage in DNA damage–induced NF-κB activation (3, 6). Following genotoxic stress, NEMO in the nucleus is sequentially modified with sumoylation, phosphorylation, and ubiquitination (7). Sumoylation of WT NEMO promotes its nuclear localization, whereas the nonsumoylatable NEMO is almost exclusively retained in the cytoplasm and deficient for NF-κB activation following DNA damage (3, 7). The necessity of NEMO phosphorylation was demonstrated by the failure of the serine 85 to alanine mutant of NEMO for mono-ubiquitination and subsequent nuclear transport (8). NEMO mono-ubiquitination occurs on the same lysine (277 and 309) as sumoylation, but functionally counteracts with the nuclear localization preference caused by sumoylation, thereby contributing to the propagation of DNA damage signals outside of the nucleus and to the ultimate IKK activation taking place in cytoplasm (3, 7).
In early two-hybrid screening with TRAF2 or TRAF6 used as bait, TRAF-interacting protein with forkhead-associated domain (TIFA) was first identified as a TRAF-interacting protein (9, 10). It contains a characteristic forkhead-associated (FHA) domain, which specifically binds to phosphorylated threonine (11). The FHA domain is intimately linked to DNA damage–repair pathways due to the prevalence of phosphorylation events in multiprotein complex assembly following genotoxic stress, and such a significance could be demonstrated by several FHA-containing proteins, such as MDC1, NBS1, and CHK1 in DDR and cell-cycle checkpoint activation (12). Along with the critical function of TRAF in the NF-κB cascade, TIFA was reported to participate in the canonical NF-κB signaling pathway by promoting oligomerization and ubiquitination of TRAF proteins (13). Interestingly, under tumor necrosis factor α (TNFα) stimulation, TIFA is phosphorylated at threonine 9, which could be recognized by its FHA domain. The resultant intermolecular binding between FHA and threonine phosphorylation leads to TIFA oligomerization and TIFA-mediated NF-κB activation (14).
Here, with our long-term interest in the interface between carcinogenesis and aging (15–21), we focused on the molecular mechanisms of DNA damage–induced NF-κB activation and secretion in this study. We identified TIFA as a novel regulator for this pathway and delineated the biochemical mechanisms underlying the TIFA-TRAF2 complex-mediated NF-κB activation following genotoxic stress.
Results
Enrichment of TIFA on chromatin following DNA damage
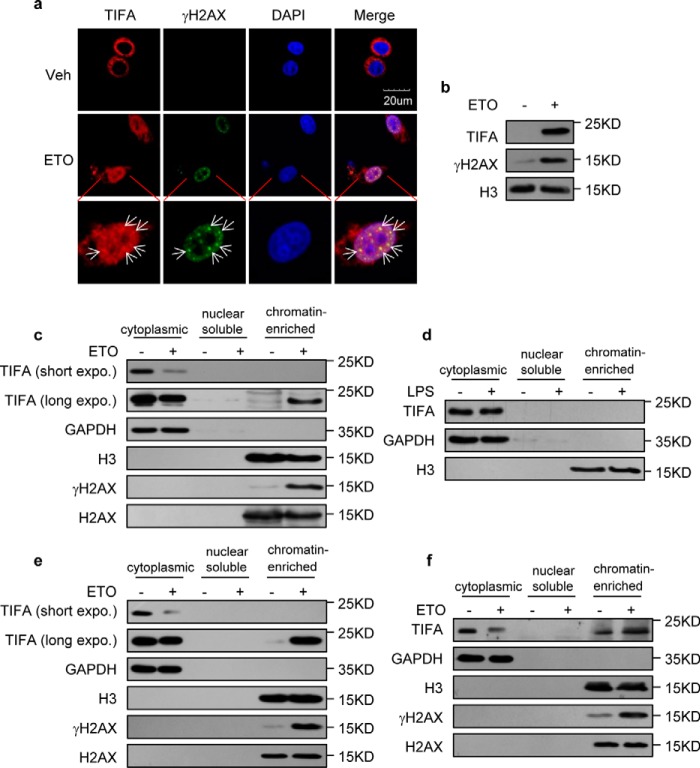
In an attempt to identify novel adaptors of DNA damage–induced NF-κB activation, we screened a panel of candidates from known NF-κB activators. We first monitored their localization following DNA damage, reasoning that initial sensors of genotoxic stress should be enriched in nucleus. Microscopic examination using FLAG-fused proteins revealed an established NF-κB regulator, TIFA, which was reported to be cytosolic under IL-1 and TNF stimulation (9), and showed significant nuclear translocations and partial co-localization with γH2AX following etoposide (ETO) treatment (Fig. 1a). This observation was independently supported with biochemical fractionation experiments using chromatins isolated from HeLa cells, as the loading of FLAG-tagged TIFA onto chromatin in ETO-treated cells was evident along with DNA damage–induced γH2AX enrichment compared with control cells (Fig. 1b). Another fractionation approach with nuclear lysis buffer containing 150 mm KOAc (22) further confirmed that TIFA could be loaded onto damaged chromatin, with a corresponding decrease in cytoplasm (Fig. 1c). Moreover, TIFA enrichment on chromatin was not observed following LPS treatment, indicating translocation of TIFA is specific to DNA damage treatment (Fig. 1d).
Figure 1.
Enrichment of TIFA on chromatin following DNA damage. a, confocal microscopic examination of TIFA and γH2AX in HeLa cells transfected with FLAG-TIFA were treated with vehicle (Veh) or ETO. 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize the nucleus. Scale bar represents 20 μm. b, chromatin fractions were isolated from the HeLa cells expressing FLAG-TIFA in the absence or presence of ETO. These fractions were then subjected to Western blotting with the indicated antibodies. c, chromatin fractions were isolated using nuclear lysis buffer containing 150 mm KOAc from HeLa cells expressing FLAG-TIFA in the absence or presence of ETO. The purified chromatin fraction and subcellular fractions were then probed with the indicated antibodies. d, chromatin fractions were isolated using nuclear lysis buffer containing 150 mm KOAc from HeLa cells expressing FLAG-TIFA in the absence or presence of LPS. The subcellular fractions were then probed with the indicated antibodies. e, chromatin fractions were isolated using nuclear lysis buffer containing 150 mm KOAc from U266 cells stably expressing TIFA. The subcellular fractions were then probed with the indicated antibodies. f, chromatin fractions were isolated using nuclear lysis buffer containing 150 mm KOAc from RPMI-8226 cells. The subcellular fractions were then probed with indicated antibodies.
To better understand the function of TIFA, we generated an antibody against TIFA with high specificity (Fig. S1). We then consolidated DNA damage–induced TIFA enrichment on chromatin using this antibody and a pair of multiple myeloma cell lines (see below). The U266 cell lines with low endogenous TIFA expression were stably integrated with TIFA expression cassettes and the chromatin fractionation results confirmed the concomitant enrichment of TIFA and γH2AX when treated with ETO (Fig. 1e). Importantly, although RPMI-8226 cells bearing high endogenous TIFA expression showed chromatin-bound TIFA in the resting state correlating with their higher level of spontaneous DNA damages, ETO treatment caused increased TIFA loading onto chromatin, implying DNA damage–induced TIFA dynamics in in vivo settings (Fig. 1f).
TIFA potentiates DNA damage–induced NF-κB activation and secretion
Although the implications of TIFA in canonical NF-κB pathways have been studied, there have been no reports on the role of TIFA in DNA damage–induced NF-κB activation, to our knowledge. Given the novel observations on nuclear translocation and chromatin accumulation of TIFA following DDR, we then examined NF-κB activation in DNA-damaged conditions with a gain-of-function model for TIFA.
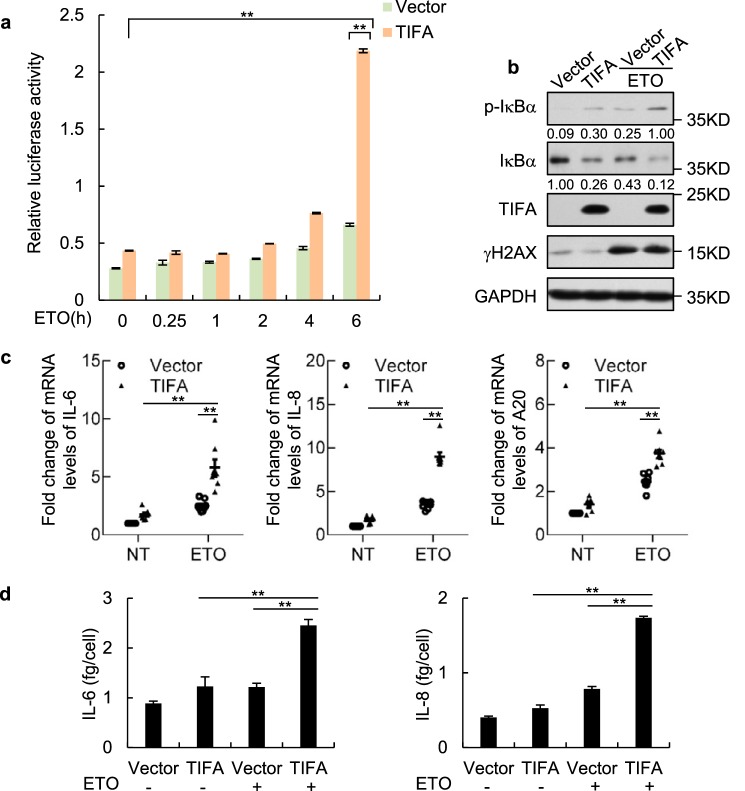
We first performed a luciferase reporter assay using the reporter construct with a classic NF-κB binding motif. In this experiment, the reporter was strongly activated in HeLa cells stably transfected with TIFA expression vector 6 h after ETO treatment, but such activation was barely detected in the control cells (Fig. 2a). Importantly, the critical event in NF-κB activation indicated by IκBα phosphorylation could only be observed in TIFA-transfected cells following ETO treatment (Fig. 2b). We then measured the mRNA expression level changes for three classic NF-κB targets, including IL-6, IL-8, and A20, following DNA damage and TIFA overexpression. Using quantitative RT-PCR, we found all the three genes were up-regulated after ETO treatment in the TIFA-overexpressed HeLa cells but not in their control cells (Fig. 2c). Quantitative assessment of IL-6 and IL-8 secretion levels using enzyme-linked immunosorbent assays (ELISA) further supported TIFA-promoted NF-κB activation following DDR, as the secretion of these two cytokines in TIFA-overexpressed cells relative to control cells was significantly increased when the cells were exposed to ETO (Fig. 2d).
Figure 2.
TIFA potentiates DNA damage–induced NF-κB activation and secretion. a, NF-κB luciferase reporter was transfected to HeLa cells stably expressing FLAG-TIFA or control cells. Renilla vector was also transfected simultaneously and served as transfection control. After treatment of cells with ETO at the indicated times, the cells were harvested for luciferase activity assay. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). b, the HeLa cells were transfected with control or FLAG-TIFA expression vectors. After 2 days, the cells were further treated with vehicle or ETO for 6 h. The cell lysate was then harvested for Western blot analysis with the indicated antibodies. p-IκBα indicates the antibody against IκBα phosphorylation on serine 32 and serine 36. c, the total mRNA was prepared from cells described in b and the mRNA levels of indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from eight independent experiments. **, p < 0.01 (Student's t test). d, the HeLa cells were transfected with control or FLAG-TIFA expression vectors and further treated with or without ETO. The secretory levels of interleukin (IL)-6 and IL-8 were then measured by ELISA. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test).
Significance of the phosphorylation event in TIFA-mediated NF-κB activation
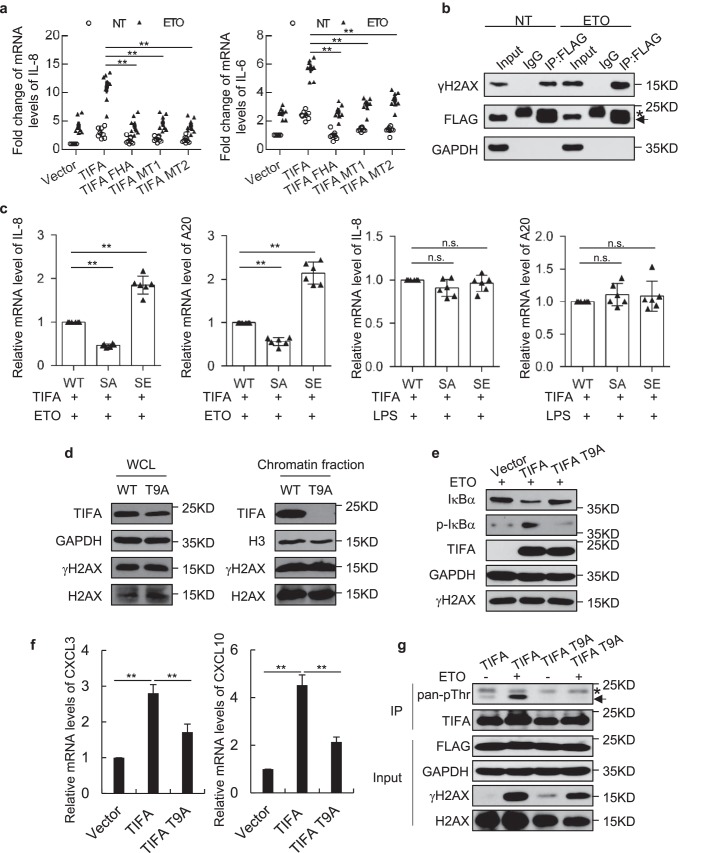
With the fact that TIFA could be accumulated on damaged DNA (Fig. 1) and the significance of FHA domain in DDR signaling pathways, it would be interesting to test the function of the FHA domain on TIFA-mediated NF-κB activation. Indeed, two groups of point mutations in the conserved residues of the FHA domain (MT1, R51A/K88A/N89A or MT2, G50E/S66A) (10, 14) abolished TIFA-mediated transcriptional activation of IL-6 and IL-8 following ETO treatment. On the other hand, the sole FHA domain of TIFA also failed to induce IL-6 and IL-8 transcription (Fig. 3a).
Figure 3.
The significance of the phosphorylation event in TIFA-mediated NF-κB activation. a, the vectors expressing full-length TIFA, the FHA domain (TIFA FHA), the R51A/K88A/N89A (MT1), or the G50E/S66A (MT2) mutants of full-length TIFA were transfected in HeLa cells with NT or treatment of ETO. The mRNA levels of the indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from eight independent experiments. **, p < 0.01 (Student's t test). b, the lysates of TIFA stably expressed HeLa cells with NT or treatment of ETO were subjected to immunoprecipitation using FLAG antibody and probed with the indicated antibodies. Before harvesting cells, cells were treated with formaldehyde for 10 min. c, FLAG-TIFA was co-transfected with WT H2AX (WT), nonphosphorylatable mutant S139A of H2AX (SA), or phosphorylation mimicking mutant S139E of H2AX (SE) in HeLa cells. Cells described were treated with ETO or LPS and the mRNA levels of the indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from six independent experiments. **, p < 0.01 (Student's t test). d, whole cell lysates and chromatin fractions from HeLa cells expressing TIFA or T9A mutant upon damage treatment were subjected to Western blot analysis probed with the indicated antibodies. e, whole cell lysates from HeLa cells expressing vector, TIFA, and T9A mutant upon damage treatment were subjected to Western blot analysis probed with indicated antibodies. f, the total mRNA was prepared from cells described in e and the mRNA levels of the indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). g, cells were transfected with FLAG-tagged TIFA or T9A mutant as indicated with or without DNA damage treatment. Cellular extracts were prepared and immunoprecipitation was performed with FLAG antibody. IgG light chain is indicated with *. IP, immunoprecipitation; n.s., nonspecific.
To understand the molecular basis for DNA damage–induced chromatin accumulation of TIFA, we first performed a co-immunoprecipitation assay to test the physical association between TIFA and γH2AX, given the pivotal role of γH2AX in marking DNA damage and orchestrating numerous signaling pathways in DDR. The results indicated that the interaction between TIFA and γH2AX was increased upon ETO treatment, whereas the immunoprecipitation efficiency using FLAG antibody was comparable in control versus damaged conditions (Fig. 3b). This point was further supported by the epistasis test using cells co-transfected with TIFA and H2AX mutants (S139A or S139E). Quantitative RT-PCR results suggested that co-transfection of the phosphorylation mimicking mutant S139E of H2AX showed greater activation of IL-8 and A20, whereas the nonphosphorylatable mutant S139A of H2AX suppressed TIFA's effect (Fig. 3c). These functional interactions could only be observed under DNA-damaged conditions but not in LPS-treated cells, implying the specific dependence of TIFA on H2AX in DNA damage–induced NF-κB activation.
The direct intermolecular association between the FHA domain and the phosphothreonine (pThr-9) (14, 23) prompted us to test whether pThr-9 was implicated in TIFA activation following genotoxic stress. Surprisingly, the single point mutation at Thr-9 (TIFA-T9A) was sufficient to abolish its enrichment on damaged chromatin (Fig. 3d). Consistent with this result, TIFA–T9A showed minimal effect on DNA damaged–induced NF-κB activation evidenced by decreased IκBα phosphorylation, sustained total IκBα protein levels (Fig. 3e), and blunted up-regulation of NF-κB targets (Fig. 3f) in TIFA–T9A–transfected cells compared with the WT TIFA–transfected cells. Examination of the phosphorylation state of TIFA suggested that DNA damage could effectively induce its phosphorylation (Fig. 3g). Importantly, such a phosphorylation event could not be detected by use of TIFA–T9A, indicating that DNA damage–elicited TIFA phosphorylation occurred at Thr-9 (Fig. 3g). These data collectively suggested that TIFA engages phosphorylation-triggered intermolecular FHA binding to propagate DNA damage signals to NF-κB.
TIFA potentiates DNA damage–induced NF-κB activation in myeloma cells
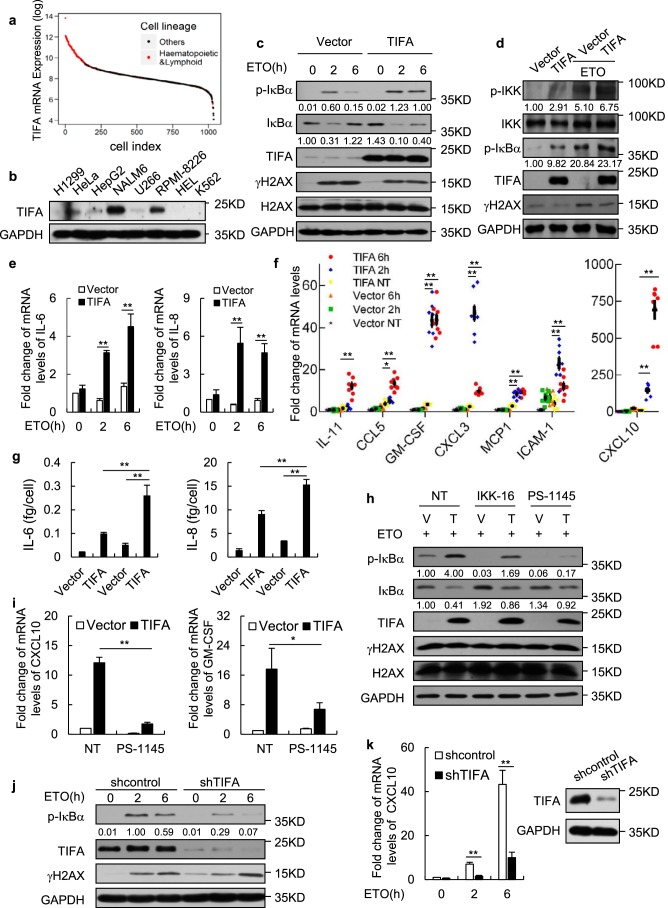
To understand the molecular function of endogenous TIFA in genotoxic stress-induced NF-κB activation, we then surveyed the Cancer Cell Line Encyclopedia (24) datasets with published mRNA profiles in over 1000 cell lines for TIFA expression patterns. We found TIFA was highly expressed in hematopoietic or lymphoid cell lines (Fig. 4a). With the antibody capable of efficiently probing endogenous TIFA proteins, we confirmed the high levels of TIFA in NALM-6 and RPMI-8226, which are both B cell-derived cancer cell lines (Fig. 4b). As a control, TIFA was lowly expressed in U266 cells. Consistent with the data in TIFA stably expressed HeLa cells, overexpression of TIFA in U266 cell lines using lentivirus caused efficient IκBα and IKK phosphorylation and significant up-regulation of IL-6 and IL-8 following ETO treatment (Fig. 4, c–e). To further explore TIFA-associated secretory changes, we screened a panel of senescence-associated secretory phenotype factors identified previously (2), and found the mRNA expression levels of IL-11, CCL5, CXCL3, CXCL10, GM-CSF, MCP1, and ICAM-1 were potently induced by DNA insults in the presence of ectopic TIFA (Fig. 4f). Consistently, ELISA results suggested that the secretion of IL-6 and IL-8 was significantly increased in TIFA–overexpressed cells relative to control cells when they were exposed to ETO (Fig. 4g).
Figure 4.
TIFA potentiates DNA damage-induced NF-κB activation in myeloma cells. a, the dataset of TIFA mRNA expression levels across ∼1000 cell lines were retrieved from the Cancer Cell Line Encyclopedia (CCLE). The sorted data were log transformed and the hematopoietic or lymphoid cells were highlighted in red. b, whole cell lysates from the indicated cell lines were subjected to Western blot analysis with anti-TIFA antibody to assess its endogenous protein levels. c, time course measurement of protein levels in U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) and treated with ETO. d, the U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) were treated with ETO as indicated. The cell lysate was then harvested for Western blot analysis with the indicated antibodies. e, the U266 cells were infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) and then treated with ETO for 2 or 6 h. The mRNA levels of the indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). f, the U266 cells were infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) and then treated with ETO for 2 or 6 h. The mRNA levels of the indicated genes were examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from eight independent experiments. *, p < 0.05; **, p < 0.01 (Student's t test). g, U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) were further treated with (last two) or without ETO (first two). The secretory levels of IL-6 and IL-8 were then measured by ELISA. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). h and i, U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) were further treated with ETO and IKK inhibitors IKK-16 or PS-1145. The whole cell lysate and mRNA from cells described above were then prepared for Western blot analysis or quantitative RT-PCR analysis. Data of quantitative RT-PCR analysis were represented as the mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01 (Student's t test). j, RPMI-8226 cells infected with shRNA control or shRNA against TIFA using lentivirus were further treated with ETO for 2 or 6 h. Western blot analysis was then performed to examine the expression levels of indicated proteins. k, the total mRNA was prepared from cells described in j and the mRNA levels of the indicated gene was examined using quantitative RT-PCR analysis. Data were represented as the mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). The right panel shows knockdown efficiency of TIFA protein. IL, interleukin.
To validate TIFA-induced NF-κB activation following DNA damage was indeed dependent on the IKK pathway, the IKKβ inhibitor IKK-16 (25) and IKKα inhibitor PS-1145 (26) were used. As shown in Fig. 4h, TIFA-induced IκBα phosphorylation and degradation of total IκBα upon genotoxic stress were impaired by either IKK-16 or PS-1145 treatment. Consistently, TIFA-induced up-regulation of CXCL10 and GM-CSF following DNA damage were severely suppressed by PS-1145 (Fig. 4i). Together, these results suggested that TIFA-induced NF-κB activation under genotoxic stress depended on IKK activation.
The necessity of TIFA for DNA damage–induced NF-κB activation was then determined using the RPMI-8226 cell line-based loss-of-function model. We transduced RPMI-8226 cells with lentiviral shRNA against TIFA or a nonsilenced control and the efficiency of TIFA depletion was validated by Western blotting (Fig. 4k, right). Importantly, both IκBα phosphorylation and up-regulation of CXCL10 were greatly suppressed upon TIFA removal (Fig. 4, j and k), supporting the essential function of endogenous TIFA in DNA damage–activated NF-κB cascade.
TIFA-TRAF2 complex promotes DNA damage–induced NEMO ubiquitination
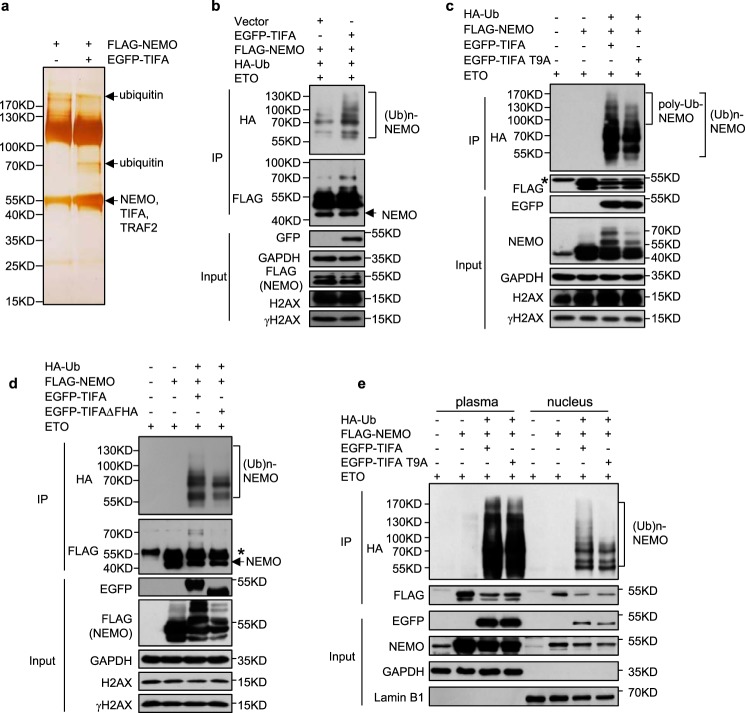
We next sought to address the molecular mechanisms underlying TIFA-promoted NF-κB activation following DDR. NEMO as a regulatory subunit of the IKK complex is the controlling nexus for genotoxic stress-induced NF-κB activation (3, 6), we therefore determined the differences of NEMO's interaction partners in TIFA-overexpressing cells versus the control cells when ETO was added, by use of affinity purification and mass spectrometry (MS) analysis (Fig. 5a). Surprisingly, overexpression of TIFA resulted in multiple additional bands in NEMO's interactome, and MS analysis revealed the presence of both ubiquitin and NEMO in these species (Fig. 5a). Given the crucial function of NEMO ubiquitination in NF-κB activation and DNA damage–response (3, 6), we then co-transfected cells with FLAG-tagged NEMO and HA-tagged ubiquitin to examine TIFA-enhanced NEMO ubiquitination following genotoxic stress. Immunoprecipitation and Western blot analysis confirmed that TIFA could prominently enhance NEMO ubiquitination as indicated by multiple HA-linked NEMO bands under DNA-damaged conditions (Fig. 5b).
Figure 5.
TIFA promotes DNA damage–induced NEMO ubiquitination. a, FLAG-NEMO was co-transfected with control vector or EGFP-TIFA to HeLa cells. Cellular extracts were immunopurified with FLAG M2 resin and eluted with FLAG peptide. Eluted proteins were resolved by SDS-PAGE and silver stained. The bands were retrieved and analyzed by MS. b, HeLa cells were co-transfected with FLAG-NEMO, HA-Ub, and EGFP-TIFA as indicated. All the cells were treated with ETO before cellular extracts were prepared. The immunoprecipitation was performed with FLAG antibody and the immunoprecipitated (IP) proteins were examined with the indicated antibody. c, HeLa cells were transfected with plasmids expressing WT TIFA or its T9A mutant (TIFA T9A) together with the other indicated constructions. The immunoprecipitation assay was performed as in b. IgG heavy chain is indicated with *. d, HeLa cells were transfected with plasmids expressing WT TIFA or its FHA domain-deleted mutant (TIFAΔFHA) together with other indicated constructions. The immunoprecipitation assay was performed as in b. IgG heavy chain is indicated with *. e, HeLa cells were transfected with plasmids expressing WT TIFA or its T9A mutant (TIFA–T9A) together with other indicated constructions. The cells were harvested and then plasma and nucleus was isolated. The immunoprecipitation (IP) assay was performed.
Because the phosphorylation event was the key switch for TIFA-mediated NF-κB activation, we then tested the effect of the TIFA–T9A mutant on NEMO ubiquitination, which failed to accumulate on damaged chromatin (Fig. 3). In agreement, this mutant suppressed DNA damage–elicited NEMO ubiquitination, especially its poly-ubiquitination forms at higher molecular weight, compared with the WT TIFA (Fig. 5c), highlighting the significance of chromatin-bound TIFA in NF-κB activation. This point was further supported by the marginal induction of NEMO ubiquitination by FHA domain-deleted TIFA following ETO treatment (Fig. 5d), in light of the critical involvement of the FHA domain to recognize pThr-9 for TIFA oligomerization in NF-κB activation (13, 14, 23). Interestingly, with the use of the nuclear lysate prepared from ETO-treated cells, we found the cells transfected with the TIFA–T9A mutant showed decreased DNA damage–elicited NEMO ubiquitination, especially its poly-ubiquitination forms at higher molecular weight, compared with the result from WT TIFA-transfected cells, whereas the ubiquitination pattern of cytoplasmic NEMO was nearly unaffected (Fig. 5e). Given the critical function of NEMO ubiquitination for DNA damage–induced NF-κB activation, this result suggested that the nucleus translocation of TIFA for its enrichment on damaged chromatin and the possible intermolecular association between FHA and pThr-9 for TIFA oligomerization around DNA-damaged sites could enhance the efficiency of NEMO ubiquitination upon genotoxic stress.
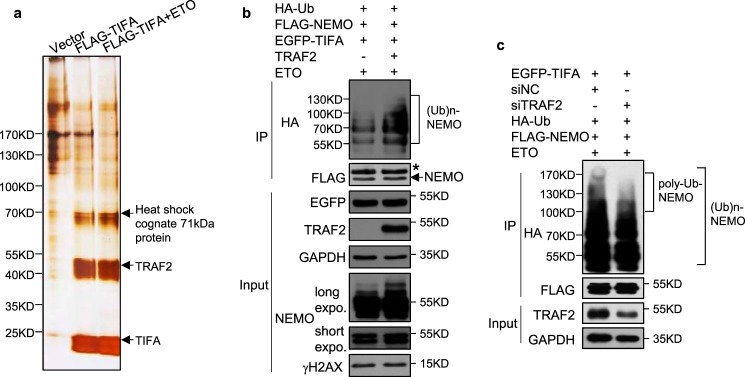
Furthermore, TIFA-containing protein complexes were affinity purified from extracts of HeLa cells stably expressing FLAG-tagged TIFA under control or DNA-damage conditions. These protein complexes were then resolved on SDS-PAGE and silver stained (Fig. 6a). As reported in other canonical NF-κB pathways, MS analysis identified TRAF2 as the major interacting partner for TIFA following ETO treatment (Fig. 6a). TRAF2 could act as a RING finger-type E3 ubiquitin ligase (27), we thus speculated that TRAF2 might be involved in NEMO ubiquitination in TIFA-mediated NF-κB activation responding to genotoxic stress. Indeed, poly-ubiquitination of NEMO was enhanced when TIFA and TRAF2 were overexpressed simultaneously in ETO-treated cells (Fig. 6b). Importantly, epistasis analysis indicated that TIFA-enhanced NEMO poly-ubiquitination at high molecular weight (indicated by brackets in Fig. 6c) was severely inhibited when endogenous TRAF2 was depleted, implying the necessity of TRAF2 in this pathway.
Figure 6.
The critical function of TRAF2 in TIFA-mediated NEMO ubiquitination. a, the extracts from cells stably expressing control vector or FLAG-TIFA in the absence or presence of ETO were immunoprecipitated (IP) with FLAG M2 resin and eluted with FLAG peptide. Silver stain and mass spectrometric analysis were performed as described in the legend to Fig. 5a. b, the HeLa cells were co-transfected with FLAG-NEMO, HA-Ub, EGFP-TIFA, and TRAF2 as indicated. All cells were treated with ETO before cellular extracts were prepared. The immunoprecipitation was performed with FLAG antibody and the immunoprecipitated proteins were examined with HA antibody. c, the HeLa cells transfected with the indicated plasmids were further co-transfected with siRNA control or siRNA targeting TRAF2. All cells were treated with ETO before cellular extracts were prepared. The immunoprecipitation was performed as in b.
TIFA overexpression is correlated with decreased cancer cell proliferation
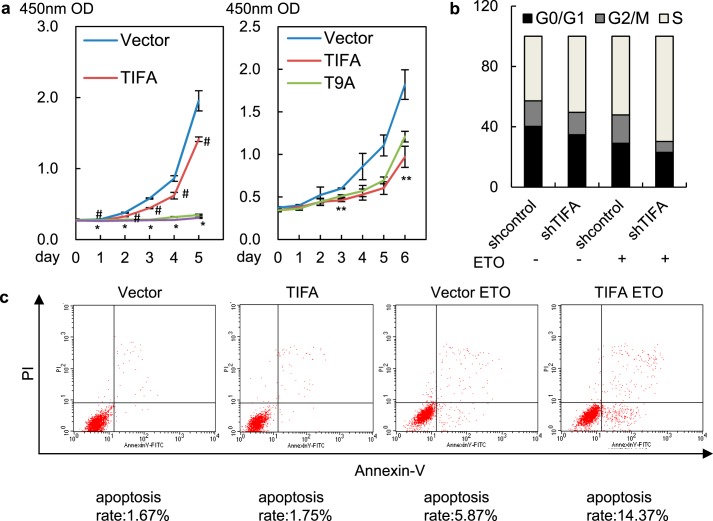
As stated earlier, DNA damage–induced NF-κB activation constitutes a secretory senescence barrier against tumorigenesis. To test whether TIFA-mediated NF-κB activation could be translated into a physiologically relevant response in multiple myeloma cells, we first examined the effect of TIFA on cancer cell proliferation and growth. The results in TIFA-negative U266 cells showed that overexpression of TIFA was correlated with decreased cancer cell proliferation in both ETO-treated and nontreated cells, suggesting minute but inherent genomic instability in cancer cells could take advantage of exogenous TIFA to defeat unlimited proliferation (Fig. 7a, left). To connect TIFA-associated phenotypical changes to its translocation to DNA lesions, we compared cell proliferation of WT- to T9A-mutated–TIFA overexpressed cancer cells (Fig. 7a, right). The data indicated that the suppression of cancer cell proliferation by TIFA was impaired by its T9A mutation, implying that TIFA-associated phenotypical changes correlated with its ability to accumulate on damaged chromatin. Flow cytometry analysis further revealed that cancer cell cycle progression was significantly accelerated in TIFA-depleted RPMI-8226 cells as indicated by the decreased G0/G1 phase cells when TIFA was depleted (Fig. 7b). Moreover, the apoptosis analysis suggested that U266 cells transduced with lentiviral TIFA showed an increased apoptosis rate compared with cells transduced with empty vector, especially when ETO was added (Fig. 7c).
Figure 7.
TIFA overexpression is correlated with decreased cancer cell proliferation. a, growth curves of U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or control (pHBLV) in the absence or presence of ETO were determined by CCK-8 (left). Growth curves of U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA), TIFA-T9A (pHBLV-TIFA-T9A), or control (pHBLV) in the presence of ETO were determined by CCK-8 (right). Data were represented as the mean ± S.D. from three independent experiments. #, p < 0.05, TIFA versus vector. *, p < 0.05, TIFA versus vector in the presence of ETO. **, p < 0.05, TIFA versus TIFA-T9A. b, the RPMI-8226 cells infected with lentivirus expressing shRNA control or shRNA against TIFA were further treated with vehicle or ETO. The flow cytometry was then used to evaluate the effect of TIFA on cell cycle progression. c, the U266 cells infected with lentivirus expressing TIFA (pHBLV-TIFA) or the control (pHBLV) were further treated with vehicle or ETO. The cells were then subjected to apoptosis analysis using PI and the Annexin-V double staining method. A portion of apoptotic cells (the upper right and bottom right quadrants) were indicated.
Discussion
Mounting evidence suggested the DNA damage–induced NF-κB activation and secretory phenotypes played crucial roles in both carcinogenesis and senescence, and the identification of TIFA as a novel regulator in this pathway added a new bridge linking DNA damage sensing, and IKK complex assembly occurred within two separate compartments. This connection is biochemically composed by the TIFA/TRAF2/NEMO trio and initiated by TIFA's accumulation on damaged chromatin. Regarding the subcellular localization change of TIFA following genotoxic stress, we proved the significance of TIFA's FHA domain in this process. Our biochemical data suggested that TIFA could interact with γH2AX-containing nucleosomes; however, in vitro pulldown did not support the direct association between TIFA and the C-terminal peptide of H2AX phosphorylated at serine 139 (data not shown). This observation was consistent with the binding preference of the FHA domain for phosphorylated threonine over phosphorylated serine (11, 12, 28). Indeed, we found threonine 9 of TIFA was phosphorylated in response to DNA insults, and the FHA–pThr interaction was crucial for TIFA-mediated NF-κB activation in genotoxic conditions as its roles in other inflammatory pathways (14, 23, 29). Benefits of chromatin enrichment of TIFA for NF-κB activation could be easily grasped with the fact that oligomerization is a prevailing mechanism for ubiquitination-based efficient assembly of IKK complex (4, 5). Even in the canonical inflammation signaling pathways, intermolecular binding between FHA and pThr-9 was the key event for TIFA oligomerization and downstream TRAFs oligomerization and IκB phosphorylation (13, 14, 23). On the other hand, the magnitude of DNA damages could be translated into the amounts of active centers for TIFA association, thus more DNA damage would cause more TIFA concentration and oligomerization to engage stronger NEMO ubiquitination. Because K63-linked poly-ubiquitination was the key signaling molecule catalyzed by TRAF family E3s (5), the interesting junction that UBC13, as the key E2-conjugating enzyme for K63-linked ubiquitin-chain formation, was also involved in DNA damage–response (30–32), suggested that the great availability of UBC13 surrounding DNA-damaged sites would facilitate nuclear TIFA/TRAF2-mediated NEMO poly-ubiquitination.
Another interesting finding from this study was the common Thr-9 phosphorylation events for both canonical inflammation and DNA damage–signaling pathways, despite that different kinases might be involved (29, 33). Although the whole picture for the differences of these cascades was currently unknown, nucleus translocation and chromatin enrichment of TIFA could only be observed in DNA-damaged conditions, but not in inflammatory cascades, suggested that additional regulations exist to account for DNA damage–induced subcellular translocation.
The DNA damage–induced secretory phenotype is a crucial component in the senescence barrier model for carcinogenesis, and our data suggested that TIFA-mediated NF-κB activation and secretion responding to genotoxic stress played an inhibitory role for proliferation of multiple myeloma cells. Of note, overexpression of TIFA dramatically augmented transcription of CXCL10, which could attenuate cell proliferation in the presence of IL-6 and eliminate precancerous cells by stimulating immune responses in vivo (34, 35). Hence, the identification of the TIFA-TRAF2 complex as novel molecular targets with regulatory roles in DNA damage–elicited signaling pathways would benefit the development of an intervention strategy for carcinogenesis.
Experimental procedures
Cell culture
HeLa and HEK293T cell lines were maintained by our laboratory. RPMI-8226 cell lines were purchased from China Infrastructure of Cell Line Resources (Beijing, China). U266 cell lines were kindly provided by Dr. Qing Ge (Peking University Health Science Center, China). HeLa and HEK293T cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. RPMI-8226 and U266 cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Etoposide (ETO, 40 μm) was purchased from Sigma (E1383). IKK-16 and PS-1145 were purchased from Selleck.
Western blotting and antibodies
Western blot analysis was performed according to procedures previously described (19). Western blot analysis was generally performed 3–4 times, with a representative blot shown in the figures. The anti-TIFA antibody was raised in a rabbit immunized with synthetic C-terminal peptide (CSSQSSSPTEMDENES) from human TIFA protein. The whole serum was collected after six rounds of immunization, and the final antibody was recovered through affinity purification by use of TIFA C-terminal peptide-conjugated resin. Other antibodies used in this study were anti-FLAG (Sigma, F3165), anti-GAPDH (Santa Cruz, sc-47724), anti-Tubulin (Santa Cruz, sc-8035), anti-GFP (Santa Cruz, sc-9996), anti-NEMO (Santa Cruz, sc-8330), anti-TRAF2 (Cell Signaling Technology Inc., 4724), anti-Phospho-IκBα (Ser-32/36) (Cell Signaling Technology Inc., 9246), anti-IκBα (Cell Signaling Technology Inc., 9242), anti-Phospho-IKKα/β (Ser-176/180) (Cell Signaling Technology Inc., 2697), anti-HA (Cell Signaling Technology Inc., 3724), anti-H3 (Abgent, AM8433), anti-γH2AX (Cell Signaling Technology Inc., 9718; Millipore, 05–636), and anti-phosphothreonine (Cell Signaling Technology Inc., 9381).
Plasmids, siRNAs, and lentiviral transfections
The template DNA of TIFA was a gift from Dr. Ming-Daw Tsai. Human TIFA was cloned into the pcDNA3.1MycHis vector, EGFP-N1 vector, or pHBLV vector, respectively. FLAG-tagged TIFA was subcloned into pcDNA3.1MycHis vector. The FHA domain mutation and T9A mutation of TIFA were generated by PCR and cloned into pcDNA3.1MycHis vector. pNF-κB-luc, TRAF2, and HA-tagged ubiquitin were maintained by our laboratory. The template DNA of NEMO was kindly provided by Dr. Tom Gilmore and FLAG-tagged NEMO was subcloned into pcDNA3.1MycHis vector.
siRNAs against TRAF2 and TIFA were purchased from Genepharm (Shanghai, China). The sequence of TRAF2 siRNA oligonucleotides was 5′-AGAGGCCAGUCAACGACAU-3′. The sequence of TIFA siRNA oligonucleotides was 5′-GGCCGAAAUUCCAACAUCU-3′. Cells transfected with plasmids using Lipofectamine 2000 (Invitrogen) were collected after 48 h of incubation. Cells transfected with RNA oligonucleotides using Lipofectamine RNAiMAX (Invitrogen) were collected after 72 h of incubation.
Lentivirus used for knockdown of TIFA in RPMI-8226 cells was purchased from Genepharm (Shanghai, China). According to the manufacturer's instructions, stable transfection was conducted. Briefly, cells with optimal confluence were infected with lentivirus twice in the presence of 5 μg/ml of Polybrene and selected for the stable cell line with puromycin.
RNA isolation and primers for quantitative RT-PCR
Total RNA was isolated using an RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher) following the manufacturer's protocol. The relative quantification was calculated using the ΔΔCt method and normalized to the vector control group with no treatment (NT). The primers used for quantitative RT-PCR were: IL-6, forward 5′-TACCCCCAGGAGAAGATTCC-3′, reverse 5′-TTTTCTGCCAGTGCCTCTTT-3′; IL-8, forward 5′-TAGCAAAATTGAGGCCAAGG-3′, reverse 5′-AAACCAAGGCACAGTGGAAC-3′; A20, forward 5′-AATCCGAGCTGTTCCACTTG-3′, reverse 5′-TGGACGGGGATTTCTATCAC-3′; IL-11, forward 5′-ACATGAACTGTGTTTGCCGC-3′, reverse 5′-AGCTGGGAATTTGTCCCTCAG-3′; CCL5, forward 5′-CTGCTGCTTTGCCTACATTG-3′, reverse 5′-ACACACTTGGCGGTTCTTTC-3′; GM-CSF, forward 5′-ATGTGAATGCCATCCAGGAG-3′, reverse 5′-AGGGCAGTGCTGCTTGTAGT-3′; CXCL3, forward 5′-GCAGGGAATTCACCTCAAGA-3′, reverse 5′-GGTGCTCCCCTTGTTCAGTA-3′; MCP1, forward 5′-CCCCAGTCACCTGCTGTTAT-3′, reverse 5′-TGGAATCCTGAACCCACTTC-3′; ICAM-1, forward 5′-GGCTGGAGCTGTTTGAGAAC-3′, reverse 5′-ACTGTGGGGTTCAACCTCTG-3′; CXCL10, forward 5′-CCACGTGTTGAGATCATTGC-3′, reverse 5′-CTTGATGGCCTTCGATTCTG-3′; GAPDH, forward 5′-CGACCACTTTGTCAAGCTCA-3′, reverse 5′-AGGGGTCTACATGGCAACTG-3′.
Luciferase activity assay
All plasmids used in luciferase activity assay were transfected using Lipofectamine 2000 (Invitrogen). Cells were collected after 24 h transfection and cell lysates were prepared with the Dual Luciferase reporter assay kit (Promega) following the manufacturer's instructions. Reporter plasmid was transfected together with Renilla plasmid for normalizing of the transfection.
Chromatin fractionation
Chromatin fractionation was performed as previously described with modifications (36). In brief, about 5 × 107 cells were collected and washed with ice-cold PBS. Then cell pellets were resuspended in 500 μl of buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.05% Triton X-100, PMSF, and phosphatase inhibitor mixture) followed by incubation on ice for 5 min. Then samples were centrifuged at 14,000 rpm for 5 min at 4 °C. The pellets were washed once with 500 μl of buffer A at 14,000 rpm for 5 min. Then the pellets were resuspended in 500 μl of buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, PMSF, and phosphatase inhibitor mixture) followed by incubation on ice for 10 min. Then samples were centrifuged at 2,000 rpm for 5 min at 4 °C. The pellets were washed once with 500 μl of buffer B at 14,000 rpm for 1 min. Finally, the final chromatin fraction was collected. For Western blot analysis, the chromatin pellets were resuspended in 200 μl of 2× SDS sample buffer and sonicated for 15 s. Chromatin fractionation using nuclear lysis buffer containing 150 mm KOAc was performed as previously described with little modification (22). Cells were incubated in cytoplasmic lysis buffer (10 mm Tris-HCl, pH 7.9, 0.34 m sucrose, 3 mm CaCl2, 2 mm MgOAc, 0.1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40 and protease inhibitors) for 10 min on ice. Then the cell lysis were centrifuged at 3,500 × g for 15 min. Supernatants were collected. Nuclei pellets were further lysed in nuclear lysis buffer (20 mm HEPES, pH 7.9, 3 mm EDTA, 10% glycerol, 150 mm KOAc, 1.5 mm MgCl2, 1 mm DTT, 0.1% Nonidet P-40 and protease inhibitors) for 10 min on ice. Then the cell lysis was centrifuged at 10,000 × g for 30 min. Supernatants were collected. The chromatin-enriched pellet was resuspended in 200 μl of 2× SDS sample buffer and sonicated for 15 s.
Growth curve assay
The growth curve was determined using Cell Counting Kit-8 (CCK-8) (Vazyme) and performed according to the manufacturer's protocol. In brief, cells were seeded in 96-well plates at a density of 2× 103 per well. 10 μl of CCK-8 reagent was added to each well and incubated for 2 h. Following shaking, the absorbance at 450 nm was measured. The absorbance was measured at 0, 1, 2, 3, 4, and 5 days after plating.
Flow cytometry assay
For cell cycle analysis, suspended cells were collected by centrifugation and then washed with cold PBS. Then cells were fixed with 70% ethanol overnight at 4 °C and RNase A (Sigma) was added at 37 °C for 30 min for RNA digestion. After which, propidium iodide (PI) (Millipore) was used for staining. A BD Bioscience FACS flow cytometer was used for analysis of DNA content. For apoptosis analysis, samples were prepared according to the manufacturer's instructions (KeyGEN BioTECH). Briefly, cells were stained with Annexin V and PI, after which, a BD Bioscience FACS flow cytometer was used for analysis of apoptosis.
Author contributions
Y. Z. conceptualization; Y. Z. and J. F. resources; J. F., D. H., F. Y., N. X., Q. L., and X. S. data curation; J. F. formal analysis; J. F. validation; J. F. and D. H. investigation; J. F., X. Z., and G. L. methodology; Y. Z. and J. F. writing-original draft; Y. Z. and T. T. supervision; Y. Z. and T. T. funding acquisition; Y. Z. writing-review and editing.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 81771495, 81300254, and 81372164 and National Basic Research Programs of China Grant 2013CB530801. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- DDR
- DNA damage response
- TIFA
- TRAF-interacting protein with forkhead-associated domain
- TRAF2
- TNF receptor–associated factor 2
- IL
- interleukin
- NEMO
- NF-κB essential modulator
- IκB
- inhibitors of NF-κB
- TNF
- tumor necrosis factor
- IKK
- IκB-kinase complex
- ETO
- etoposide
- LPS
- lipopolysaccharide
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- HA
- hemagglutinin
- EGFP
- enhanced green fluorescent protein
- PI
- propidium iodide
- NT
- no treatment
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., and Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coppe J. P., Patil C. K., Rodier F., Sun Y., Munoz D. P., Goldstein J., Nelson P. S., Desprez P. Y., and Campisi J. (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLos Biol. 6, 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCool K. W., and Miyamoto S. (2012) DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol. Rev. 246, 311–326 10.1111/j.1600-065X.2012.01101.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayden M. S., and Ghosh S. (2008) Shared principles in NF-κB signaling. Cell 132, 344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 5. Chen Z. J. (2005) Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 10.1038/ncb0805-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyamoto S. (2011) Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 21, 116–130 10.1038/cr.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang T. T., Wuerzberger-Davis S. M., Wu Z. H., and Miyamoto S. (2003) Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell 115, 565–576 10.1016/S0092-8674(03)00895-X [DOI] [PubMed] [Google Scholar]
- 8. Wu Z. H., Shi Y., Tibbetts R. S., and Miyamoto S. (2006) Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science 311, 1141–1146 10.1126/science.1121513 [DOI] [PubMed] [Google Scholar]
- 9. Kanamori M., Suzuki H., Saito R., Muramatsu M., and Hayashizaki Y. (2002) T2BP, a novel TRAF2 binding protein, can activate NF-κB and AP-1 without TNF stimulation. Biochem. Biophys. Res. Commun. 290, 1108–1113 10.1006/bbrc.2001.6315 [DOI] [PubMed] [Google Scholar]
- 10. Takatsuna H., Kato H., Gohda J., Akiyama T., Moriya A., Okamoto Y., Yamagata Y., Otsuka M., Umezawa K., Semba K., and Inoue J. (2003) Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J. Biol. Chem. 278, 12144–12150 10.1074/jbc.M300720200 [DOI] [PubMed] [Google Scholar]
- 11. Pennell S., Westcott S., Ortiz-Lombardia M., Patel D., Li J., Nott T. J., Mohammed D., Buxton R. S., Yaffe M. B., Verma C., and Smerdon S. J. (2010) Structural and functional analysis of phosphothreonine-dependent FHA domain interactions. Structure 18, 1587–1595 10.1016/j.str.2010.09.014 [DOI] [PubMed] [Google Scholar]
- 12. Reinhardt H. C., and Yaffe M. B. (2013) Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14, 563–580 10.1038/nrm3640 [DOI] [PubMed] [Google Scholar]
- 13. Ea C. K., Sun L., Inoue J., and Chen Z. J. (2004) TIFA activates IκB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. U.S.A. 101, 15318–15323 10.1073/pnas.0404132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang C. C., Weng J. H., Wei T. Y., Wu P. Y., Hsu P. H., Chen Y. H., Wang S. C., Qin D., Hung C. C., Chen S. T., Wang A. H., Shyy J. Y., and Tsai M. D. (2012) Intermolecular binding between TIFA-FHA and TIFA-pT mediates tumor necrosis factor α stimulation and NF-κB activation. Mol. Cell. Biol. 32, 2664–2673 10.1128/MCB.00438-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Q., Zhang Y., Fu J., Han L., Xue L., Lv C., Wang P., Li G., and Tong T. (2013) FOXA1 mediates p16(INK4a) activation during cellular senescence. EMBO J. 32, 858–873 10.1038/emboj.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Zhang D., Li Q., Liang J., Sun L., Yi X., Chen Z., Yan R., Xie G., Li W., Liu S., Xu B., Li L., Yang J., He L., and Shang Y. (2016) Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat. Genet. 48, 1003–1013 10.1038/ng.3635 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Yang X., Gui B., Xie G., Zhang D., Shang Y., and Liang J. (2011) Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J. Biol. Chem. 286, 42414–42425 10.1074/jbc.M111.271064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang W., Wu J., Zhang Z., and Tong T. (2001) Characterization of regulatory elements on the promoter region of p16(INK4a) that contribute to overexpression of p16 in senescent fibroblasts. J. Biol. Chem. 276, 48655–48661 10.1074/jbc.M108278200 [DOI] [PubMed] [Google Scholar]
- 19. Cao X., Xue L., Han L., Ma L., Chen T., and Tong T. (2011) WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J. Biol. Chem. 286, 33447–33456 10.1074/jbc.M111.225565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen T., Xue L., Niu J., Ma L., Li N., Cao X., Li Q., Wang M., Zhao W., Li G., Wang J., and Tong T. (2012) The retinoblastoma protein selectively represses E2F1 targets via a TAAC DNA element during cellular senescence. J. Biol. Chem. 287, 37540–37551 10.1074/jbc.M111.260679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., and Tong T. (2014) FOXA1 antagonizes EZH2-mediated CDKN2A repression in carcinogenesis. Biochem. Biophys. Res. Commun. 453, 172–178 10.1016/j.bbrc.2014.09.092 [DOI] [PubMed] [Google Scholar]
- 22. Zhao Q., Wang Q. E., Ray A., Wani G., Han C., Milum K., and Wani A. A. (2009) Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem. 284, 30424–30432 10.1074/jbc.M109.044982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weng J. H., Hsieh Y. C., Huang C. C., Wei T. Y., Lim L. H., Chen Y. H., Ho M. R., Wang I., Huang K. F., Chen C. J., and Tsai M. D. (2015) Uncovering the mechanism of forkhead-associated domain-mediated TIFA oligomerization that plays a central role in immune responses. Biochemistry 54, 6219–6229 10.1021/acs.biochem.5b00500 [DOI] [PubMed] [Google Scholar]
- 24. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duckworth C., Zhang L., Carroll S. L., Ethier S. P., and Cheung H. W. (2016) Overexpression of GAB2 in ovarian cancer cells promotes tumor growth and angiogenesis by upregulating chemokine expression. Oncogene 35, 4036–4047 10.1038/onc.2015.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hideshima T., Chauhan D., Richardson P., Mitsiades C., Mitsiades N., Hayashi T., Munshi N., Dang L., Castro A., Palombella V., Adams J., and Anderson K. C. (2002) NF-κB as a therapeutic target in multiple myeloma. J. Biol. Chem. 277, 16639–16647 10.1074/jbc.M200360200 [DOI] [PubMed] [Google Scholar]
- 27. Pineda G., Ea C. K., and Chen Z. J. (2007) Ubiquitination and TRAF signaling. Adv. Exp. Med. Biol. 597, 80–92 10.1007/978-0-387-70630-6_7 [DOI] [PubMed] [Google Scholar]
- 28. Durocher D., Taylor I. A., Sarbassova D., Haire L. F., Westcott S. L., Jackson S. P., Smerdon S. J., and Yaffe M. B. (2000) The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6, 1169–1182 10.1016/S1097-2765(00)00114-3 [DOI] [PubMed] [Google Scholar]
- 29. Lin T. Y., Wei T. W., Li S., Wang S. C., He M., Martin M., Zhang J., Shentu T. P., Xiao H., Kang J., Wang K. C., Chen Z., Chien S., Tsai M. D., and Shyy J. Y. (2016) TIFA as a crucial mediator for NLRP3 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 113, 15078–15083 10.1073/pnas.1618773114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Messick T. E., and Greenberg R. A. (2009) The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187, 319–326 10.1083/jcb.200908074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang B., and Elledge S. J. (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brusky J., Zhu Y., and Xiao W. (2000) UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37, 168–174 10.1007/s002940050515 [DOI] [PubMed] [Google Scholar]
- 33. Wei T. W., Wu P. Y., Wu T. J., Hou H. A., Chou W. C., Teng C. J., Lin C. R., Chen J. M., Lin T. Y., Su H. C., Huang C. F., Yu C. R., Hsu S. L., Tien H. F., and Tsai M. D. (2017) Aurora A and NF-κB survival pathway drive chemoresistance in acute myeloid leukemia via the TRAF-interacting protein TIFA. Cancer Res. 77, 494–508 10.1158/0008-5472.CAN-16-1004 [DOI] [PubMed] [Google Scholar]
- 34. Brownell J., and Polyak S. J. (2013) Molecular pathways: hepatitis C virus, CXCL10, and the inflammatory road to liver cancer. Clin. Cancer Res. 19, 1347–1352 10.1158/1078-0432.CCR-12-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barash U., Zohar Y., Wildbaum G., Beider K., Nagler A., Karin N., Ilan N., and Vlodavsky I. (2014) Heparanase enhances myeloma progression via CXCL10 downregulation. Leukemia 28, 2178–2187 10.1038/leu.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li D. Q., Nair S. S., Ohshiro K., Kumar A., Nair V. S., Pakala S. B., Reddy S. D., Gajula R. P., Eswaran J., Aravind L., and Kumar R. (2012) MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2, 1657–1669 10.1016/j.celrep.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.