Abstract
Activating transcription factor 4 (ATF4) is a stress-induced transcription factor that is frequently upregulated in cancer cells. ATF4 controls the expression of a wide range of adaptive genes that allow cells to endure periods of stress, such as hypoxia or amino acid limitation. However, under persistent stress conditions, ATF4 promotes the induction of apoptosis. Recent advances point to a role for post-translational modifications (PTMs) and epigenetic mechanisms in balancing these pro- and anti-survival effects of ATF4. We review here how PTMs and epigenetic modifiers associated with ATF4 may be exploited by cancer cells to cope with cellular stress conditions that are intrinsically associated with tumor growth. Identification of mechanisms that modulate ATF4-mediated transcription and its effects on cellular metabolism may uncover new targets for cancer treatment.
Introduction
Cancer cells inevitably encounter stress during tumor development. Excessive proliferation raises the demand on protein synthesis until it eventually exceeds the protein-folding capacity of the endoplasmic reticulum (ER). This results in the accumulation of misfolded proteins, leading to a phenomenon known as ER stress [1]. As the tumor increases in size, cells in the tumor core are challenged by limited levels of oxygen, glucose, and amino acids, each of which triggers metabolic changes that tune anabolic and catabolic pathways towards the accumulation of biomass (Box 1). In turn, these changes can induce oxidative stress [2]. Therapeutic interventions can also contribute to a stressed state [3].
Box 1. Craving for Amino Acids.
ATF4 is one of the master regulators of the cellular stress response that promotes adaptation of cells to a limited availability of nutrients. It is important to realize that the rapid proliferation of tumor cells and the required accumulation of biomass that precedes each cell division make these cells extremely dependent on a continuous flow of nutrients. Poorly perfused tumors, where exogenous supply of oxygen and nutrients may be limited, must rely on alternative anabolic and catabolic pathways. However, even in well-perfused tumors, available nutrients are fed into anabolic pathways, rather than into oxidative phosphorylation, to meet the increasing demands for glycolysis-derived anabolic intermediates. While this rerouting of carbohydrate metabolism by tumor cells, known as the Warburg effect, has been long recognized, more recently the addiction of tumor cells to amino acids has received greater attention. This high demand is not limited to the essential amino acids, as sufficient uptake of non-essential amino acids, including glutamine and serine, from the extracellular environment is also necessary for sustained survival and proliferation of tumor cells. Glutamine, the second most consumed nutrient, is not only used as a building block for proteins but also serves as a carbon and nitrogen donor for amino acid synthesis and nucleotide biogenesis [101]. Similarly, high consumption of glycine and serine is necessary to increase the flux through the one-carbon and methionine cycles [102,103]. Specific metabolic addictions of some tumor cells have been recognized, as have genetic mutations that promote tumor development, including activation of PI3K [104], KRAS [105], and MYC [106], and more recently deletion of IKZF1 [107], all of which impinge on metabolic control. The importance of non-essential amino acids is underscored by the fact that dietary restriction or pharmacological interventions interfering with the availability of these nutrients is lethal to tumor cells. For example, asparaginase, a bacterially derived enzyme that depletes the serum of asparagine and glutamine, is universally used in the treatment of childhood acute lymphoblastic leukemia. While sustained depletion is catastrophic for the leukemic cells, this treatment is usually accompanied by manageable adverse effects, indicating that non-transformed cells effectively adapt to the limited availability of asparagine and glutamine [108]. Similarly, it was shown that dietary restriction of serine and glycine, or interventions affecting their metabolism, reduced tumor growth in vitro and in vivo [103]. While research and clinical practice clearly show therapeutic benefits of targeting amino acid availability, tumor cells can rapidly respond to this nutrient stress by activating adaptive pathways that start with the induced expression of ATF4. A detailed understanding of these adaptive mechanisms may aid in the selection of therapeutic targets that can prevent or overcome resistance to these metabolic therapies.
A tumor can only expand if cancer cells successfully adapt to these changes. Both normal and cancer cells rely on a combination of signaling cascades and transcriptional programs that, depending on the nature, type, and severity of the stress encountered, activate the correct set of adaptive genes. Prominent examples of such stress-induced pathways are the unfolded protein response (UPR) activated by ER stress [4], the cellular response to low oxygen levels [5], and the amino acid response (AAR) activated by amino acid deprivation [6]. Interestingly, each of these pathways share a common downstream effector protein, ATF4. This transcription factor is the master regulator of the cellular response to stress. Most of its target genes are involved in various salvage pathways that promote cell survival, but others prepare the cell for apoptosis. This review outlines the pro- and anti-survival functions of ATF4, describes molecular mechanisms that allow context-dependent activation of different ATF4 targets, and discusses how these mechanisms may be exploited by tumor cells.
ATF4-Mediated Control of Life–Death Decisions
ATF4 at the Center of Stress Signaling
Phosphorylation of eukaryotic initiation factor 2 on Ser51 of its α subunit (eIF2α) is a key event in many stress signaling pathways [7]. While this modification suppresses global protein synthesis, translation of selected mRNAs, including the transcription factor ATF4, is enhanced [8]. EIF2α phosphorylation is catalyzed by one of four different stress-induced kinases: (i) protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), a component of the ER stress response; (ii) general control non-derepressible 2 (GCN2), which signals amino acid limitation and UV exposure; (iii) protein kinase RNA-activated (PKR), that is activated in response to viral infections; and (iv) heme-regulated inhibitor (HRI), that is activated by heme deprivation and oxidative stress [7]. Hypoxia also induces phosphorylation of eIF2α and the subsequent upregulation of ATF4 by an indirect mechanism which involves activation of the UPR and PERK [9]. In addition, hypoxia regulates ATF4 stability and transcriptional activity via its oxygen-dependent degradation domain [10]. Thus, ER stress, amino acid limitation, hypoxia, and oxidative stress all activate ATF4 via both phospho (p-)eIF2α-dependent and –independent mechanisms.
A Dual Role for ATF4
Why are p-eIF2α and ATF4 expression common to many seemingly unrelated types of stress? The global suppression of translation caused by p-eIF2α helps to conserve nutrients, reducing equivalents, and ER chaperones, thereby providing temporary relief from metabolic, oxidative, and ER stress, respectively. Long-term survival subsequently requires translationally upregulated transcription factors such as ATF4 to use this ‘window of opportunity’ to promote the expression of adaptive genes. Indeed, ATF4 controls genes involved in amino acid transport and metabolism, protection from oxidative stress, and protein homeostasis [11,12]. However, ATF4 can also induce apoptosis, cell-cycle arrest, and senescence [13–19]. Despite the risk of sensitizing to apoptosis, tumor cells frequently exploit ATF4 to reduce the stress resulting from rapid proliferation and nutrient limitation inside a growing tumor mass. There are ample examples of cell-intrinsic metabolic adaptations that require ATF4 activation to alleviate stress [20–25]. Furthermore, ATF4 and its target genes have been linked to angiogenesis and metastasis [26–30]. These effects, where ATF4 expression is advantageous for tumor cells, are in sharp contrast to the tumor-suppressive effects that have also been linked to ATF4 function [31–33]. However, the benefits of ATF4 expression may diminish when conditions change, because in some cases expression of ATF4 was found to sensitize tumor cells to therapy-induced cell death [19,32,34–37]. These observations raise the question of how ATF4-expressing tumor cells benefit from its pro-survival effects while at the same time avoiding ATF4-mediated cell death.
Balancing Autophagy and Protein Synthesis To Maintain Viability
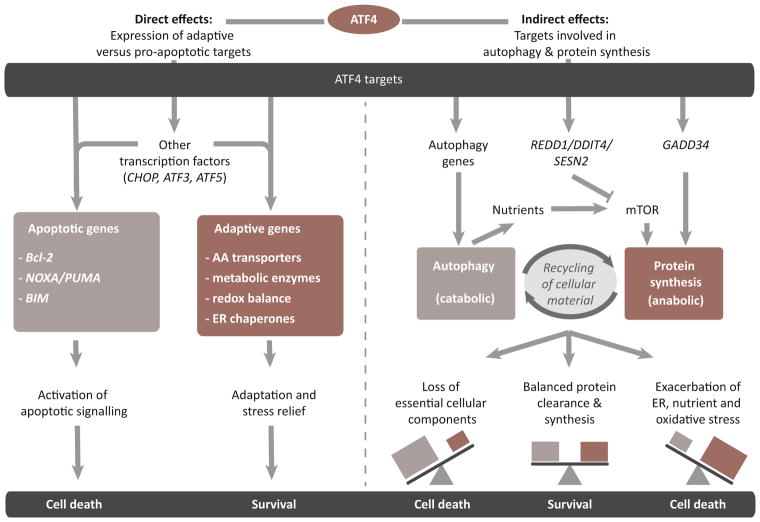
The transcription factor ATF4 directly binds to and controls the expression of target genes implicated in either adaptive responses or the induction of apoptosis (Figure 1 and Box 2). However, recent studies support an indirect influence of ATF4 on cell viability through modulation of the balance between anabolic and catabolic processes (Figure 1). ATF4 activates a set of targets involved in autophagy [38–41], but also targets that upregulate protein synthesis. For example, ATF4 induces GADD34, leading to a negative feedback loop involving dephosphorylation of eIF2α and restoration of translation [12]. ATF4 signaling also intersects with the mammalian target of rapamycin (mTOR) pathway, which senses amino acid availability and activates protein synthesis [42]. ATF4 enhances mTOR activity both by increasing amino acid availability via autophagy and by upregulating adaptive genes [42,43]. Conversely, ATF4 induces the expression of the mTOR repressors SESN2, DDIT4, and REDD1 [37,44].
Figure 1.
Activating Transcription Factor 4 (ATF4) Control of Life–Death Decisions Under Stress. ATF4 and its targets determine cell survival by specific mechanisms. (Left) ATF4 directly controls the transcription of several adaptive and pro-apoptotic genes. It also induces other transcription factors that cooperate with ATF4 to control the expression of these targets. (Right) ATF4 influences survival indirectly through modulation of autophagy and protein synthesis. Hyperactivation of either process by ATF4 can trigger cell death indirectly via downstream pathways. AA, amino acids; ER, endoplasmic reticulum.
Box 2. ATF4-Mediated Control of Transcription.
ATF4 is a member of the basic leucine-zipper (bZIP) family of transcription factors which homo- or heterodimerize with each other. Even though ATF4 homodimers can be formed in vitro, they are unstable even when bound to DNA. Instead, ATF4 is thought to function as a heterodimer, which it forms with members of the ATF, FOS/JUN, and CCAAT enhancer-binding protein (C/EBP) bZIP transcription factor subfamilies [52,109–111]. ATF4 and its binding partners bind to DNA sequences called cAMP responsive elements (CREs) or C/EBP-ATF response elements (CAREs) to control target gene expression [109,110]. Through the expression of its targets, ATF4 exerts control over life–death decisions under stress (Figure 1). By inducing the expression of amino acid transporters, metabolic enzymes, enzymes involved in redox homeostasis, and ER chaperones, ATF4 contributes to stress relief and survival [11,12]. However, the transcriptional program activated by ATF4 also downregulates the anti-apoptotic BCL2 protein and upregulates pro-apoptotic signaling through the proteins BIM, NOXA, and PUMA (reviewed in [49,54]). The outcome of ATF4 activation is therefore highly context-dependent.
The effects of these changes on cell viability are complex. Autophagy can mitigate stress by replenishing essential amino acids and clearing protein aggregates that cause ER stress [43,45]. Emerging evidence supports a role for ATF4-mediated autophagy as a survival mechanism in cancer cells [27,39,40,45,46]. However, autophagy can also induce cell death through the catastrophic breakdown of cellular constituents [47]. Similarly, recovery of protein synthesis through GADD34 or mTOR increased cell death in several mouse embryonic fibroblast (MEF) models in which it exacerbated ER, oxidative, and amino acid stress [15,42,44,48], but promoted survival in human breast cancer cells [37].
The pro- or anti-survival effects of both autophagy and protein synthesis are likely to depend on the balance between anabolism and catabolism in the cell. The threshold where autophagy starts to become damaging is context-dependent because it is influenced by both the amount of expendable protein present in the cell and the ability of the cell to compensate for the loss of essential material with newly synthesized proteins. Thus, proliferating cells, including cancer cells, may have a higher tolerance for autophagy because of a high level of protein synthesis and ER load [45].
A similar line of reasoning can be applied to the effects of protein synthesis. Recovery can be beneficial by allowing the translation of ATF4 adaptive targets [12], whereas excessive or premature stimulation of protein synthesis exacerbates stress by increasing protein load in the ER [15].
Selection of ATF4 Targets via Heterodimerization
The abovementioned effects of autophagy and protein synthesis on cell survival illustrate how expression of the same ATF4 target genes can differentially affect normal and cancer cells. Similarly, it has been suggested that cancer cells might be more resistant to pro-apoptotic ATF4 targets because they already have defects in apoptotic signaling downstream of ATF4 [4]. Although it is likely that the global alteration of cellular signaling and metabolism in cancer cells influences responses downstream of ATF4, this context-dependency may also be determined by regulation at the level of ATF4 itself.
How does ATF4 switch between adaptive and pro-apoptotic gene expression? This switch has been attributed to the formation of different ATF4 heterodimers that control specific targets and follow distinct kinetics of expression [6,24,49,50]. Some binding partners of ATF4, for example C/EBPβ and C/EBPγ, were suggested to signal adaptation [51,52], whereas dimerization with C/EBP homologous protein (CHOP) was associated with the switch to pro-apoptotic signaling [15,41,53]. Indeed, CHOP was found to regulate mediators of apoptosis such as BCL2 and BIM, and studies using overexpression or knockdown have demonstrated a key role for CHOP in stress-induced apoptosis in several cell models (reviewed in [50,54,55]). Moreover, the miR-211 microRNA can prevent apoptosis induced by ER stress in MEFs by blocking CHOP expression, and is upregulated in both mammary carcinoma and B cell lymphoma compared to normal tissue [56]. By contrast, others have reported that ATF4 can induce apoptosis in cancer cell lines independently of CHOP, and even noted an unexpected increase in cell death after CHOP knockdown [33,36]. These results suggest a pro-survival function for CHOP and demonstrate that transcriptional regulation of ATF4 targets is more complex than previously thought. Even though heterodimerization certainly adds flexibility to ATF4 target expression, it cannot be the only explanation for a switch from adaptive to pro-apoptotic signaling, because the same heterodimer can apparently have different effects on survival depending on the context. As discussed below, other possible mechanisms include ATF4 regulation through PTMs and epigenetics.
Post-Translational Control of ATF4
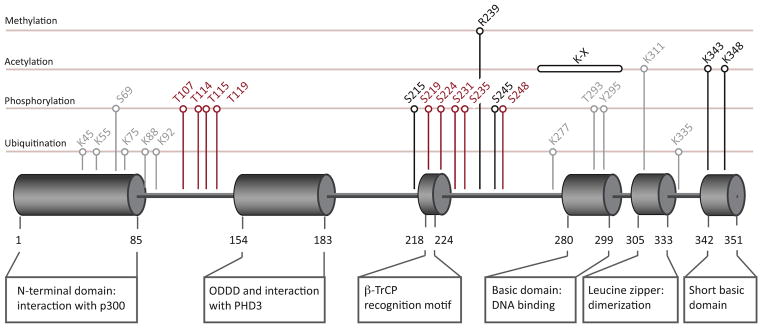
ATF4 is extensively regulated at the post-translational level (Table 1 and Figure 2). Some modifications are known to affect ATF4 stability or transcriptional activity, whereas the function of other modifications remains unknown. This section provides an overview of the known ATF4 PTMs and their potential roles in regulating ATF4 activity and/or target-specificity.
Table 1.
PTMs of ATF4 and Their Functiona
| Modification | Residue(s) | Evidence | Biological function | Refs |
|---|---|---|---|---|
| Phosphorylation | Ser69 | Screening (n = 3) | Unknown | [57] |
| Thr107 Thr114 Thr115 Thr119 |
Validated | Protein destabilization and transcriptional inactivation of NOXA and PUMA | [70] | |
| Thr213 Ser231 Ser235 |
Predicted | Interaction with βTrCP and protein degradation | [13] | |
| Ser248 | Screening (n = 2) | Interaction with βTrCP and protein degradation | [13,57] | |
| Ser219 Ser224 |
Validated | Interaction with βTrCP and protein degradation | [13,59, 60,113] | |
| Ser215 | Validated | Protein destabilization; transcriptional activation of CHOP and ATF3 | [71] | |
| Ser245 | Validated | Transcriptional activation in osteoblasts, chondrocytes, prostate cancer, breast cancer, and lung cancer | [65–69] | |
| Thr293 Tyr295 |
Screening (n = 1) | Unknown | [57,77] | |
| Methylation | Arg239 | Validated | Transcriptional activation of pro-apoptotic targets | [64] |
| Ubiquitination | Lys45 Lys55 Lys75 Lys88 Lys277 Lys335 |
Screening (n = 1) | Unknown | [57,58] |
| Lys92 | Predicted | Unknown | [57] | |
| Acetylation | Lys343 Lys348 LysX (between 270 and 300) |
Validated | Transcriptional activation, possibly by strengthening DNA binding | [57,76] |
| Lys311 | Validated | Unknown | [61] |
Residue numbers listed refer to the human ATF4 amino acid sequence (UniProtKB entry P18848). Evidence: ‘predicted’ is based on homology with mouse ATF4, ‘screening’ by mass spectrometry (n = number of independent datasets), or ‘validated’ by a targeted approach.
Figure 2.
Location of Functional Domains and Post-Translational Modifications (PTMs) in Activating Transcription Factor 4 (ATF4). Residue numbers refer to the human amino acid sequence (UniProtKB entry P18848). ATF4 domain structure was adapted from [112]. PTMs were mapped using information from the PhosphoSite database [57] or literature as indicated in Table 1. Colors indicate the effect of the modification: black, activating; red, inactivating/destabilizing; grey, unknown.
ATF4 Ubiquitination, Stability, and Degradation
High-throughput screening using mass spectrometry has revealed multiple lysine residues in ATF4 that are subject to ubiquitination [57,58]. Although none of these sites have been experimentally validated beyond the initial screen, polyubiquitination is known to play an important role in regulating ATF4 stability post-translationally [59]. Nuclear ATF4 is recognized by βTrCP, an F-box protein associated with the SCF E3-ubiquitin ligase. This interaction depends on casein kinase-dependent phosphorylation of Ser219 in the βTrCP recognition motif (Figure 2) of ATF4, and ultimately leads to ATF4 ubiquitination and proteasomal degradation [13,59]. Additional accumulation of negative charge in the region surrounding the βTrCP recognition motif, via phosphorylation of Thr213, Ser224, Ser231, Ser235, and Ser248, further promotes the interaction with βTrCP and consequent protein destabilization [13,60] (Figure 2). Conversely, binding of the hypoxia-inducible protein PHD3 stabilizes the ATF4 protein, potentially by catalyzing proline hydroxylation within the oxygen-dependent degradation domain (ODDD, Figure 2) of ATF4 [10]. Another study reported a role for histone acetyltransferase p300 in promoting ATF4 stability that was independent of its acetyltransferase activity [61]. The site of interaction with p300 was mapped to the N-terminal domain of ATF4, exactly in the region where all but two of the ubiquitinated lysines are located (Figure 2). It is possible that p300 binding promotes ATF4 stability indirectly by blocking polyubiquitination of lysines at its binding site, which would imply that ubiquitination of these residues is involved in targeting ATF4 to the proteasome.
The sites, functional significance, and types of ubiquitination that occur on ATF4 remain incompletely understood. The most common form of ubiquitination is the Lys48-linked polymer that targets proteins for proteasomal degradation. However, several other forms of polyubiquitination exist that regulate other aspects of protein function, such as subcellular localization or the interaction with other proteins. Interestingly, cellular stress promotes the interaction between the bZIP domain of ATF4 and the ABRO1–BRISC deubiquitinating complex [62]. This complex has a cytoprotective function that relies on the removal of Lys63-linked ubiquitin chains from target proteins and requires the presence of ATF4. The authors concluded that the interaction with ATF4 was important for targeting the ABRO1–BRISC complex to the nucleus, where it could de-ubiquitinate other targets. Although the relevant substrates remain to be identified, the finding that the nuclear ABRO1–BRISC complex controls P53 stability is of interest [63]. Furthermore, given the fact that ATF4 itself can contribute to survival under stress, it is tempting to speculate that ABRO1–BRISC might exert its cytoprotective effects by modulating ATF4 function via de-ubiquitination. While it remains to be established whether ATF4 itself is subject to Lys63-linked ubiquitination, it is worth noting that only two of the ubiquitinated lysines on ATF4 reported to date are located away from the p300 interaction domain, and both are in close vicinity to the bZIP domain that binds to ABRO1–BRISC (Figure 2).
Selective ATF4 Target Expression by PTM-Mediated Regulation of ATF4 Transcriptional Activity
In addition to regulating ATF4 stability, PTMs can directly affect ATF4 activity. We recently described that methylation of Arg239 in ATF4 by the methyltransferase PRMT1 enhanced its transcriptional activity. This interaction between ATF4 and PRMT1 was mediated by the tumor suppressor BTG1. Intriguingly, loss of Btg1, and thus the ATF4–PRMT1 interaction, suppressed a specific subset of ATF4 target genes associated with apoptosis, demonstrating that PTMs can shift the balance between pro- and anti-survival effects of ATF4 in tumor cells [64]. Although that study was the first to show an effect of ATF4 modification status on the expression of pro-apoptotic targets in particular, other PTMs on ATF4 may have a similar target-specific effect. For example, RSK2-mediated phosphorylation of ATF4 at Ser245 activates transcription from the osteocalcin gene in osteoblasts and chondrocytes [65,66]. Although the effect of this phosphorylation on ATF4 activity was not measured for other genes, p-Ser245 ATF4 was shown to increase VEGF expression in prostate cancer cells [67]. Because VEGF is a known target of ATF4 [30], it seems likely that this upregulation occurs through transcription. Furthermore, elevated levels of p-Ser245 ATF4 were detected in both breast and lung cancer tissue [68,69]. By promoting this modification, cancer cells may benefit from specific activation of pro-survival targets. Similarly, multisite phosphorylation at threonine residues 107, 114, 115, and 119 was shown by Bagheri-Yarmand et al. [61] to reduce ATF4 occupancy and activity at the promoters of the pro-apoptotic targets NOXA and PUMA. This phosphorylation is mediated by the receptor tyrosine kinase RET, which is hyperactivated in various types of cancer [70]. The authors suggested that RET contributes to cancer cell viability via direct repression of ATF4 at the promoters of NOXA and PUMA, supported by the observation of a direct interaction of RET with the NOXA promoter [70]. This result suggests that the repression of ATF4 activity occurs at a subset of target genes that interact with RET. It will be interesting to see whether this mechanism is specific to pro-apoptotic genes. In addition, ATF4 is phosphorylated at Ser215 by the casein kinase 2 (CK2). Although a non-phosphorylatable mutant was more stable than wild-type ATF4, it was less active at the promoters of the ATF4 targets ATF3 and CHOP, suggesting an activating function of this modification [71]. Interestingly, CK2 also phosphorylates several dimerization partners of ATF4. CK2-mediated phosphorylation of C/EBPδ increases its transcriptional activity [72], whereas its phosphorylation of CHOP has an inhibitory effect [73]. By phosphorylating ATF4 and its binding partners, CK2 may shift the balance towards a specific subset of transcriptionally active heterodimers and target genes. Indeed, CK2 is known to mediate life–death decisions, and inhibition of CK2 has been shown to induce apoptosis in a CHOP-dependent mechanism [74]. Overexpression of CK2 may be one of the mechanisms by which cancer cells repress the pro-apoptotic functions of CHOP.
A Potential Role for ATF4 Acetylation in Controlling Heterodimerization and DNA Binding
A study by Lassot et al., investigating ATF4 stabilization by the acetyltransferase p300, revealed that p300 acetylates ATF4 at Lys311. As discussed previously, a stabilizing effect of p300 on ATF4 was independent of its enzymatic activity [61], and the function of acetylation at Lys311 remains unknown. However, it is noteworthy that this residue is located in the bZIP domain of ATF4 that controls heterodimerization (Figure 2). Phosphorylation in the bZIP domain of the related transcription factor C/EBPβ has been shown to regulate its affinity for binding partners and thereby heterodimerization specificity [75]. It remains to be determined whether ATF4 heterodimerization is likewise regulated by the modification of residues (such as Lys311) in its bZIP domain.
Gachon et al. detected p300-dependent acetylation of multiple lysines in the two basic regions of ATF4 (Figure 2). Mutation of these residues impaired the transcription of a reporter construct containing an ATF4-binding CRE element, whereas transcription from a viral promoter with a Tax responsive element (TxRE) (also bound by ATF4) was not affected [76]. The authors hypothesized that ATF4 acetylation stabilizes its interaction with DNA at the CRE element. This effect is redundant at TxRE promoters, where stabilization is provided by the viral protein Tax instead. Although the TxRE promoter is absent in normal cells, this study provides proof of principle that modifications in the ATF4 DNA-binding domain can differentially affect target promoters. The effect of stabilization with DNA may depend on the exact sequence and the presence of other proteins that interact with ATF4. This process would provide another mechanism by which PTMs can influence ATF4 target specificity.
The PhosphoSite database contains additional residues on ATF4 where phosphorylation has been detected in screens, but these have not been experimentally validated [57,77]. Ser69, Thr293, and Tyr295 each fall into this category of phosphorylation sites with unknown function (Figure 2 and Table 1). With the advance of high-throughput screens, it will be interesting to see what other PTMs will be detected in the future. Although such a screen alone is not sufficient to identify which PTMs control ATF4 function or target specificity, it can provide a starting point for more detailed research into the mechanisms that influence stress responses in cancer cells.
The Emerging Role of Epigenetic Modifications in Controlling ATF4 Target Gene Expression
Target-Specific Requirements for Chromatin Modification
Another potential mechanism of modulating ATF4-mediated transcription is through epigenetic control. Several studies have shown that transcriptional activation of ATF4 targets requires the recruitment of enzymes that modify chromatin structure (Table 2). For example, inhibition of deacetylase activity via HDAC inhibitors was able to rescue transcription from the Atf3, Jmjd3, and Chop genes in ATF4 knockout cells, but not from the Asns and Cat1 genes [78]. This illustrates that the recruitment of histone acetyltransferase (HAT) activity to target promoters is an important part of ATF4 function, but also highlights that the requirement for epigenetic modifiers differs between targets and illustrates flexibility in ATF4-induced transcription. Similarly, Gjymishka et al. showed that, even though both amino acid deprivation and ER stress were able to recruit ATF4 and C/EBPβ to the CARE of the SNAT2 gene, only the AAR increased transcription from this gene because the UPR failed to induce histone acetylation. By contrast, the ASNS gene was transcribed in response to both stressors. When the UPR and the AAR were induced simultaneously, the UPR actively repressed the induction of SNAT2 by the AAR, suggesting that the UPR pathway either promoted the formation of repressive chromatin marks at the SNAT2 promoter or prevented the recruitment of a required HAT activity. Interestingly, the UPR could induce transcription from the SNAT2 promoter when it was placed in a reporter plasmid, further highlighting the importance of the chromatin structure in situ [79]. A study by Bruhat et al. reported that the recruitment of phosphorylated ATF2 (p-ATF2) to the promoter of the CHOP gene was essential for acetylation of histones H2 and H4 and for ATF4-mediated transcription following amino acid deprivation. The ATF3 locus was regulated in the same manner, whereas transcription from the ASNS promoter required acetylation of histone H3 and a form of H4 acetylation that was independent of p-ATF2 [80].
Table 2.
Histone Modifications Required for ATF4 Target Gene Expression
| Modification | Enzyme | Model | Target gene specificity | Ref. |
|---|---|---|---|---|
| H4 acetylation | Unknown | Histidine deprivation in MEFs | Required for Jmjd3, Chop, and Atf3, but not for Asns and Cat1 | [78] |
| H3 acetylation | Unknown | Histidine deprivation or ER stress (thapsigargin) in human HepG2 cells | Required for SNAT2, but not for ASNS | [79] |
| H2B and H4 acetylation | p-ATF2 | Amino acid deprivation in MEFs | Required for Chop and Atf3, but not for Asns | [80] |
| H3K9 demethylation | KDM4C | Serine deprivation in the human cancer cell lines HeLa and BE2C (neuroblastoma) | Required for the expression of most amino acid responsive genes | [81] |
These findings highlight the importance of chromatin modifiers in adding target gene specificity to ATF4-mediated responses. Interestingly, Zhao et al. recently reported a requirement for the histone H3 Lys9 (H3K9) demethylase KDM4C in ATF4-mediated control of amino acid transport and metabolism after serine deprivation [81]. In contrast to the examples given above, this dependency was not target-specific because KDM4C was required for transcription of all tested amino acid-responsive genes. Furthermore, genes upregulated by KDM4C were enriched for UPR targets, suggesting that the requirement for KDM4C may be a mechanism of regulating ATF4-dependent transcription in general rather than activating specific target genes. High KDM4C expression occurs in many cancers as well as in mouse embryonic stem cells, and may be a mechanism for rapidly proliferating cells to activate ATF4 signaling [81].
DNA Methylation as a Potential Mechanism Influencing Heterodimer DNA-Binding Specificity
Methylation of DNA CpG motifs alters the DNA binding specificity of transcription factor complexes [82,83]. Both CRE elements (5′-TGACGTCA-3′) and some CAREs (consensus 5′-TGATGX1AAX2-3′, where X1 = C) contain a central GC motif and may be subject to methylation [6,24]. Interestingly, the C/EBPβ–ATF4 heterodimer gained the ability to bind the non-canonical sequence 5′-CGATGCAA-3′ when the first cytosine was methylated. By contrast, ATF4 homodimers show decreased affinity for methylated DNA [82,84]. Furthermore, methyl-binding proteins may compete with the bZIP heterodimers for binding to methylated promoters [85]. Differential DNA methylation could therefore be another way in which ATF4 and its binding partners are directed to subsets of target genes depending on cell type, metabolic context, stage of development, or disease.
Concluding Remarks and Future Perspectives
Cellular stress is increasingly recognized as a major factor in cancer development. In fact, it is tightly linked to several of the ‘hallmarks of cancer’ that are intrinsic to tumor cells [86]. The activation of oncogenes and the high proliferation rate that follows both increase the demand on the protein synthesis machinery and thereby contribute to ER stress [87–89]. This is exacerbated further by other cancer hallmarks such as metabolic reprogramming, which can disrupt ER homeostasis by causing redox imbalances and genomic instability, resulting in an increased mutation rate and protein misfolding [89]. However, mounting evidence supports a model in which stress is not merely a result of malignant transformation but also actively promotes tumor development. For example, the importance of hypoxic signaling in metabolic reprogramming and angiogenesis has long been established [5], and the UPR has likewise been linked to various aspects of cancer biology including transformation, drug resistance, immunosuppression, angiogenesis, and metastasis [89–91]. A similar role is now emerging for ATF4. The conditions that promote ATF4 expression are not limited to the challenges of rapidly proliferating cells within a growing tumor mass. During treatment, tumor cells must cope with therapy-induced stress. Because many anticancer drugs act through the induction of metabolic, oxidative, or ER stress, it is not surprising that ATF4 mediates resistance to proteasome inhibitors and chemotherapeutic agents in a variety of cancers [38,92–96]. The potential of ATF4 in promoting growth is further illustrated by its importance during normal development. ATF4 has a key role in maintaining the viability of highly secretory cells such as osteoblasts [65] and plasma cells [97] by mitigating the ER stress caused by high rates of protein synthesis. Moreover, recent studies have uncovered a role for ATF4 in determining stem cell fate. Induction of the AAR modulates the differentiation of mouse embryonic stem cells to favor the formation of the endodermal lineage, a process that was further enhanced in ATF4-deficient cells [98]. Activation of the PERK–ATF4 arm of the UPR was found to induce differentiation of intestinal stem cells to transit amplifying cells [99]. ER stress accompanied by ATF4 activation induced apoptosis in hematopoietic stem cells, whereas progenitor cells exhibited lower levels of ATF4 expression favoring survival [100]. These studies demonstrate a key role for ATF4 in promoting the survival of cells with high secretory activity or proliferation rate, which may also be relevant to cancer development. That ATF4 is overexpressed in many cancers and its potential contribution to tumor cell behavior, including proliferation, adaptation to microenvironmental stress, drug resistance and metastasis, make ATF4 and downstream pathways interesting targets for cancer treatment. Nevertheless, the ambiguous role of ATF4 in promoting both cell survival and apoptosis makes targeting ATF4 a risky approach. Drugs that could ‘switch’ ATF4 between adaptive to pro-apoptotic targets hold the potential to abolish these pro-tumorigenic features and actively promote tumor cell death. Although there are strong indications that specific enzymes modulate ATF4 activity in a target-specific manner, their importance in balancing the pro- and anti-survival functions of ATF4 is currently difficult to assess. Most research investigating the effect of PTMs or histone modifications on ATF4 activity has focused on a limited number of ATF4 target genes. Studies using genome-wide analysis, such as RNA-sequencing and chromatin immunoprecipitation (ChIP)-sequencing, will help to determine whether these factors are indeed responsible for making the choice between adaptive and pro-apoptotic expression profiles (see Outstanding Questions). Nevertheless, given that the enzymes involved in controlling ATF4 action may be susceptible to drug modulation, the study of PTMs and epigenetic modifiers associated with ATF4 is a promising avenue of research.
Outstanding Questions.
How do tumor cells selectively exploit the ATF4 pro-survival benefits at the expense of its apoptosis-inducing effects?
How important are PTMs or epigenetic regulators in modulating ATF4 function?
How do oncogenes and tumor suppressors impinge upon post-translational control or epigenetic regulation of ATF4?
Can we block the pro-survival effects or reactivate the pro-apoptotic effects of ATF4 in tumor cells to enhance response to therapy without affecting the stem cell compartment?
Trends.
Mammalian cells utilize sophisticated mechanisms to respond to unfavorable conditions such as nutrient limitation. The transcription factor ATF4 plays a central role in this response. Although activation of ATF4 predominantly serves to promote survival under stress, it can also induce apoptosis.
ATF4 signaling supports many normal biological processes, such as the maintenance of stem and progenitor cells or immune regulation, but these functions can be hijacked by cancer cells to sustain rapid tumor growth and survive the hostile tumor microenvironment.
How cancer cells selectively exploit the pro-survival effects of ATF4 could relate to changes in PTMs of ATF4 or in ATF4 association with epigenetic modifiers that are specifically altered in cancer cells. Deeper insight into these tumor-associated mechanisms may lead the way to improving therapy responses in cancer patients.
Acknowledgments
The authors apologize for not citing other relevant publications owing to space limitations. I.M.N.W is supported by a PhD grant from the Radboud university medical center. L.T.v.d.M. and F.N.v.L. are supported by the Dutch Cancer Society (grant 10072) and Children Cancer Free (KiKa, grant 134). M.S.K. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (grant CA203565).
References
- 1.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagelkerke A, et al. The unfolded protein response as a target for cancer therapy. Biochim Biophys Acta. 2014;1846:277–284. doi: 10.1016/j.bbcan.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 5.Harris AL. Hypoxia – a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 6.Kilberg MS, et al. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wek RC, et al. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 8.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 9.Blais JD, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köditz J, et al. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110:3610–3617. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 11.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 12.Novoa I, et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank CL, et al. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J Biol Chem. 2010;285:33324–33337. doi: 10.1074/jbc.M110.140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin HK, et al. Skp2 targeting suppresses tumorigenesis by Arf–p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiramatsu N, et al. Translational and posttranslational regulation of XIAP by eIF2alpha and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol Biol Cell. 2014;25:1411–1420. doi: 10.1091/mbc.E13-11-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto H, et al. Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open. 2013;2:1084–1090. doi: 10.1242/bio.20135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohoka N, et al. TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizawa J, et al. ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci Signal. 2016;9:ra17. doi: 10.1126/scisignal.aac4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao Y, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971. doi: 10.1038/ncomms11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Geldermalsen M, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren P, et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J Pathol. 2015;235:90–100. doi: 10.1002/path.4429. [DOI] [PubMed] [Google Scholar]
- 23.Hart LS, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122:4621–4634. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ameri K, et al. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood. 2004;103:1876–1882. doi: 10.1182/blood-2003-06-1859. [DOI] [PubMed] [Google Scholar]
- 25.Bi M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H, et al. Crosstalk between ATF4 and MTA1/HDAC1 promotes osteosarcoma progression. Oncotarget. 2016;7:7329–7342. doi: 10.18632/oncotarget.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey S, et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest. 2015;125:2592–2608. doi: 10.1172/JCI78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagelkerke A, et al. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mujcic H, et al. Hypoxic activation of the unfolded protein response (UPR) induces expression of the metastasis-associated gene LAMP3. Radiother Oncol. 2009;92:450–459. doi: 10.1016/j.radonc.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, et al. The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res. 2012;72:5396–5406. doi: 10.1158/0008-5472.CAN-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagheri-Yarmand R, et al. ATF4 targets RET for degradation and is a candidate tumor suppressor gene in medullary thyroid cancer. J Clin Endocrinol Metab. 2017;102:933–941. doi: 10.1210/jc.2016-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qing G, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armstrong JL, et al. Regulation of endoplasmic reticulum stress-induced cell death by ATF4 in neuroectodermal tumor cells. J Biol Chem. 2010;285:6091–6100. doi: 10.1074/jbc.M109.014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zong Y, et al. Up-regulated ATF4 expression increases cell sensitivity to apoptosis in response to radiation. Cell Physiol Biochem. 2017;41:784–794. doi: 10.1159/000458742. [DOI] [PubMed] [Google Scholar]
- 35.Dilshara MG, et al. Glutamine deprivation sensitizes human breast cancer MDA-MB-231 cells to TRIAL-mediated apoptosis. Biochem Biophys Res Commun. 2017;485:440–445. doi: 10.1016/j.bbrc.2017.02.059. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, et al. Anacardic acid induces cell apoptosis associated with induction of ATF4-dependent endoplasmic reticulum stress. Toxicol Lett. 2014;228:170–178. doi: 10.1016/j.toxlet.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Wang S, et al. ATF4 gene network mediates cellular response to the anticancer PAD inhibitor YW3-56 in triple-negative breast cancer cells. Mol Cancer Ther. 2015;14:877–888. doi: 10.1158/1535-7163.MCT-14-1093-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milani M, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 39.Rouschop KM, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B’chir W, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto H, et al. Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open. 2013;2:1084–1090. doi: 10.1242/bio.20135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan BJ, et al. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. J Biol Chem. 2014;289:12593–12611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, Long YC. Autophagy modulates amino acid signaling network in myotubes: differential effects on mTORC1 pathway and the integrated stress response. FASEB J. 2015;29:394–407. doi: 10.1096/fj.14-252841. [DOI] [PubMed] [Google Scholar]
- 44.Dennis MD, et al. Regulated in DNA damage and development 1 (REDD1) promotes cell survival during serum deprivation by sustaining repression of signaling through the mechanistic target of rapamycin in complex 1 (mTORC1) Cell Signal. 2013;25:2709–2716. doi: 10.1016/j.cellsig.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding WX, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 46.Pike LR, et al. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 47.Denton D, et al. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87–95. doi: 10.1038/cdd.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang YJ, et al. The TSC1 and TSC2 tumor suppressors are required for proper ER stress response and protect cells from ER stress-induced apoptosis. Cell Death Differ. 2011;18:133–144. doi: 10.1038/cdd.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 50.Kilberg MS, et al. The transcription factor network associated with the amino acid response in mammalian cells. Adv Nutr. 2012;3:295–306. doi: 10.3945/an.112.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halterman MW, et al. Loss of c/EBP-beta activity promotes the adaptive to apoptotic switch in hypoxic cortical neurons. Mol Cell Neurosci. 2008;38:125–137. doi: 10.1016/j.mcn.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huggins CJ, et al. C/EBPgamma is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol Cell Biol. 2015;36:693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teske BF, et al. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szegezdi E, et al. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozpedek W, et al. The role of the PERK/eIF2alpha/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med. 2016;16:533–544. doi: 10.2174/1566524016666160523143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chitnis NS, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48:353–364. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lassot I, et al. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pons J, et al. Transfer-NMR and docking studies identify the binding of the peptide derived from activating transcription factor 4 to protein ubiquitin ligase beta-TrCP. Competition STD-NMR with beta-catenin. Biochemistry. 2008;47:14–29. doi: 10.1021/bi7014212. [DOI] [PubMed] [Google Scholar]
- 61.Lassot I, et al. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 62.Ambivero CT, et al. ATF4 interacts with Abro1/KIAA0157 scaffold protein and participates in a cytoprotective pathway. Biochim Biophys Acta. 2012;1823:2149–2156. doi: 10.1016/j.bbamcr.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, et al. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat Commun. 2014;5:5059. doi: 10.1038/ncomms6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuniati L, et al. Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress. Oncotarget. 2016;7:3128–3143. doi: 10.18632/oncotarget.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin–Lowry syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 66.Li TF, et al. BMP-2 induces ATF4 phosphorylation in chondrocytes through a COX-2/PGE2 dependent signaling pathway. Osteoarthr Cartil. 2014;22:481–489. doi: 10.1016/j.joca.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain S, et al. Prostaglandin E2 regulates tumor angiogenesis in prostate cancer. Cancer Res. 2008;68:7750–7759. doi: 10.1158/0008-5472.CAN-07-6689. [DOI] [PubMed] [Google Scholar]
- 68.Fan CF, et al. Elevated p-CREB-2 (ser245) expression is potentially associated with carcinogenesis and development of breast carcinoma. Mol Med Rep. 2012;5:357–362. doi: 10.3892/mmr.2011.657. [DOI] [PubMed] [Google Scholar]
- 69.Fan CF, et al. Expression of a phosphorylated form of ATF4 in lung and non-small cell lung cancer tissues. Tumour Biol. 2014;35:765–771. doi: 10.1007/s13277-013-1104-5. [DOI] [PubMed] [Google Scholar]
- 70.Bagheri-Yarmand R, et al. A novel dual kinase function of the RET protooncogene negatively regulates activating transcription factor 4-mediated apoptosis. J Biol Chem. 2015;290:11749–11761. doi: 10.1074/jbc.M114.619833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ampofo E, et al. Functional interaction of protein kinase CK2 and activating transcription factor 4 (ATF4), a key player in the cellular stress response. Biochim Biophys Acta. 2013;1833:439–451. doi: 10.1016/j.bbamcr.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 72.Schwind L, et al. CK2 phosphorylation of C/EBPδ regulates its transcription factor activity. Int J Biochem Cell Biol. 2015;61:81–89. doi: 10.1016/j.biocel.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 73.Ubeda M, Habener JF. CHOP transcription factor phosphorylation by casein kinase 2 inhibits transcriptional activation. J Biol Chem. 2003;278:40514–40520. doi: 10.1074/jbc.M306404200. [DOI] [PubMed] [Google Scholar]
- 74.Intemann J, et al. ER stress signaling in ARPE-19 cells after inhibition of protein kinase CK2 by CX-4945. Cell Signal. 2014;26:1567–1575. doi: 10.1016/j.cellsig.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 75.Lee S, et al. RSK-mediated phosphorylation in the C/EBPbeta leucine zipper regulates DNA binding, dimerization, and growth arrest activity. Mol Cell Biol. 2010;30:2621–2635. doi: 10.1128/MCB.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gachon F, et al. Activation of HTLV-I transcription in the presence of Tax is independent of the acetylation of CREB-2 (ATF-4) Virology. 2002;299:271–278. doi: 10.1006/viro.2002.1501. [DOI] [PubMed] [Google Scholar]
- 77.Mertins P, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shan J, et al. ATF4-dependent regulation of the JMJD3 gene during amino acid deprivation can be rescued in Atf4-deficient cells by inhibition of deacetylation. J Biol Chem. 2012;287:36393–36403. doi: 10.1074/jbc.M112.399600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gjymishka A, et al. Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells. J Biol Chem. 2008;283:27736–27747. doi: 10.1074/jbc.M803781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bruhat A, et al. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 2007;35:1312–1321. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao E, et al. KDM4C and ATF4 cooperate in transcriptional control of amino acid metabolism. Cell Rep. 2016;14:506–519. doi: 10.1016/j.celrep.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mann IK, et al. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013;23:988–997. doi: 10.1101/gr.146654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sayeed SK, et al. C/EBPβ (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim Biophys Acta. 2015;1849:583–589. doi: 10.1016/j.bbagrm.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kribelbauer JF, et al. Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 2017;19:2383–2395. doi: 10.1016/j.celrep.2017.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ren Y, et al. Methylation of the asparagine synthetase promoter in human leukemic cell lines is associated with a specific methyl binding protein. Oncogene. 2004;23:3953–3961. doi: 10.1038/sj.onc.1207498. [DOI] [PubMed] [Google Scholar]
- 86.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 87.Maurel M, et al. Controlling the unfolded protein response-mediated life and death decisions in cancer. Semin Cancer Biol. 2015;33:57–66. doi: 10.1016/j.semcancer.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 88.Dejeans N, et al. Novel roles of the unfolded protein response in the control of tumor development and aggressiveness. Semin Cancer Biol. 2015;33:67–73. doi: 10.1016/j.semcancer.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 89.Urra H, et al. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer. 2016;2:252–262. doi: 10.1016/j.trecan.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Chevet E, et al. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- 91.Dufey E, et al. ER proteostasis addiction in cancer biology: novel concepts. Semin Cancer Biol. 2015;33:40–47. doi: 10.1016/j.semcancer.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 92.Igarashi T, et al. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene. 2007;26:4749–4760. doi: 10.1038/sj.onc.1210289. [DOI] [PubMed] [Google Scholar]
- 93.Hu J, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119:826–837. doi: 10.1182/blood-2011-07-366492. [DOI] [PubMed] [Google Scholar]
- 94.Zhu H, et al. Activating transcription factor 4 confers a multidrug resistance phenotype to gastric cancer cells through transactivation of SIRT1 expression. PLoS One. 2012;7:e31431. doi: 10.1371/journal.pone.0031431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu H, et al. Activating transcription factor 4 mediates a multidrug resistance phenotype of esophageal squamous cell carcinoma cells through transactivation of STAT3 expression. Cancer Lett. 2014;354:142–152. doi: 10.1016/j.canlet.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 96.Ye P, et al. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol Cell Biol. 2014;34:3421–3434. doi: 10.1128/MCB.00221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goldfinger M, et al. Protein synthesis in plasma cells is regulated by crosstalk between endoplasmic reticulum stress and mTOR signaling. Eur J Immunol. 2011;41:491–502. doi: 10.1002/eji.201040677. [DOI] [PubMed] [Google Scholar]
- 98.Shan J, et al. Activation of the amino acid response modulates lineage specification during differentiation of murine embryonic stem cells. Am J Physiol Endocrinol Metab. 2013;305:E325–E335. doi: 10.1152/ajpendo.00136.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heijmans J, et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 2013;3:1128–1139. doi: 10.1016/j.celrep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 100.van Galen P, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 101.Hosios AM, et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maddocks OD, et al. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maddocks OD, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Engelman JA, et al. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 105.Kawada K, et al. Targeting metabolic reprogramming in KRAS-driven cancers. Int J Clin Oncol. 2017 doi: 10.1007/s10147-017-1156-4. [DOI] [PubMed] [Google Scholar]
- 106.Stine ZE, et al. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan LN, et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature. 2017;542:479–483. doi: 10.1038/nature21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pieters R, et al. L-asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer. 2011;117:238–249. doi: 10.1002/cncr.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vallejo M, et al. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad Sci U S A. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci U S A. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 113.Wang A, et al. Ribosomal protein RPL41 induces rapid degradation of ATF4, a transcription factor critical for tumour cell survival in stress. J Pathol. 2011;225:285–292. doi: 10.1002/path.2918. [DOI] [PubMed] [Google Scholar]