Dear Editor,
Neuroendocrine tumors (NETs) comprise a heterogeneous spectrum of neoplasms originating from neuroendocrine cells in various organs — most commonly in the endocrine glands and the gastrointestinal tract.1 The molecular and etiological features of NETs arising from different organs are still far from clarified. Therefore, systematic analysis of genomic alternations and their contribution to core pathways in NETs is urgently needed for the development of novel diagnostic, therapeutic strategies and personalized management of patients. Here, we investigated somatic mutations across 21 NET types through pan-cancer analysis and identified 86 candidate driver genes. Further analysis of druggability and panel sequencing of these genes provide potential diagnostic and therapeutic targets for NETs.
To investigate the landscape of common and specific somatic mutations in NETs, we collected mutation data of 1,103 tumors (1,034 published and 69 new whole-exome sequencing data) from 38 research projects (Supplementary information, Table S1). We performed whole-exome sequencing on tumor-normal pairs of 38 insulinoma (INS), 20 Cushing’s disease (CD) induced by corticotroph pituitary adenoma and 11 pheochromocytoma (PCC) (Supplementary information, Tables S2–5, Figure S1). Using these data, we compiled somatic mutation data across 21 NET types. The data set consisted of five types of adrenocortical tumors: aldosterone-producing adenomas (APA), cortisol-producing adrenocortical adenomas (ACA), ACTH-independent macronodular adrenocortical hyperplasia (AIMAH), adrenocortical carcinomas (ACC) and adrenocortical oncocytoma; seven types of pituitary tumors: growth hormone-secreting pituitary adenomas, gonadotropins including follicle-stimulating hormone and luteinizing hormone pituitary adenomas, prolactin pituitary adenomas, thyrotropin-stimulating hormone pituitary adenomas, CD induced by corticotroph pituitary adenoma, plurihormonal pituitary adenoma and nonfunctioning pituitary adenomas; two types of pancreatic tumors: non-functional NETs (PNETs) and INS; medullary thyroid cancer (MTC), parathyroid adenomas and parathyroid carcinomas (PTC); pulmonary carcinoids (PC); PCC and paraganglioma (PCC/PGL); small intestine NETs (SINET) and neuroblastoma (NB) (Supplementary information, Table S6).
The data from 21 NET types were re-analyzed and annotated to obtain a uniform set of somatic mutations (Supplementary information, Tables S7 and 8). Malignant NETs have a larger number of non-silent mutations and a higher mutation frequency than benign NETs (P = 7.33 × 10−8; Supplementary information, Figures S2 and 3). Mutation spectrum across 21 NET types reveals that the C−>T transversion is the predominant substitution, consistent with findings in other cancer types2 (Supplementary information, Figure S4 and Table S9). To comprehensively identify the significantly mutated genes (SMGs) with a statistically higher mutation rate in NETs, we performed systematic and stepwise analysis using the MuSiC suite.2 The results of SMG analysis are associated with the background mutation rates (BMRs) of tumor types and BMRs between benign and malignant tumors are significantly different (Supplementary information, Figure S5). Therefore, MuSiC analyses of 21 NETs were separately conducted in the combined benign set, combined malignant set, combined organ set (adrenal, gastrointestinal, pituitary and thyroid) and individual tumor types. The resulting SMGs were further filtered by gene mutation frequency, deleterious mutation rate and gene expression (Supplementary information, Figure S6). We reliably identified a total of 86 candidate driver genes in NETs, including 80 SMGs and 6 known cancer genes3–6 (Supplementary information, Figure S7 and Table S10).
Of the 86 candidate driver genes, 34 are novel SMGs in NETs and 52 have been reported in previous studies of specific NET types. The mutations of 86 genes showed common and specific distribution in NETs. Novel type-specific SMGs were identified in less frequently mutated genes, such as DNMT3A in benign NETs and AHNAK, COL1A1, SF3B1 and ZNF292 in malignant NETs (Fig. 1a and Supplementary information, Figure S8). Our data reveal that MEN1 is the most common SMGs in NETs (8 out of 21 types). Notably, mutations of three novel SMGs and six known candidate driver genes are identified in as least five NET types, indicating that more common driver genes emerge across both benign and malignant NETs (Supplementary information, Figure S9).
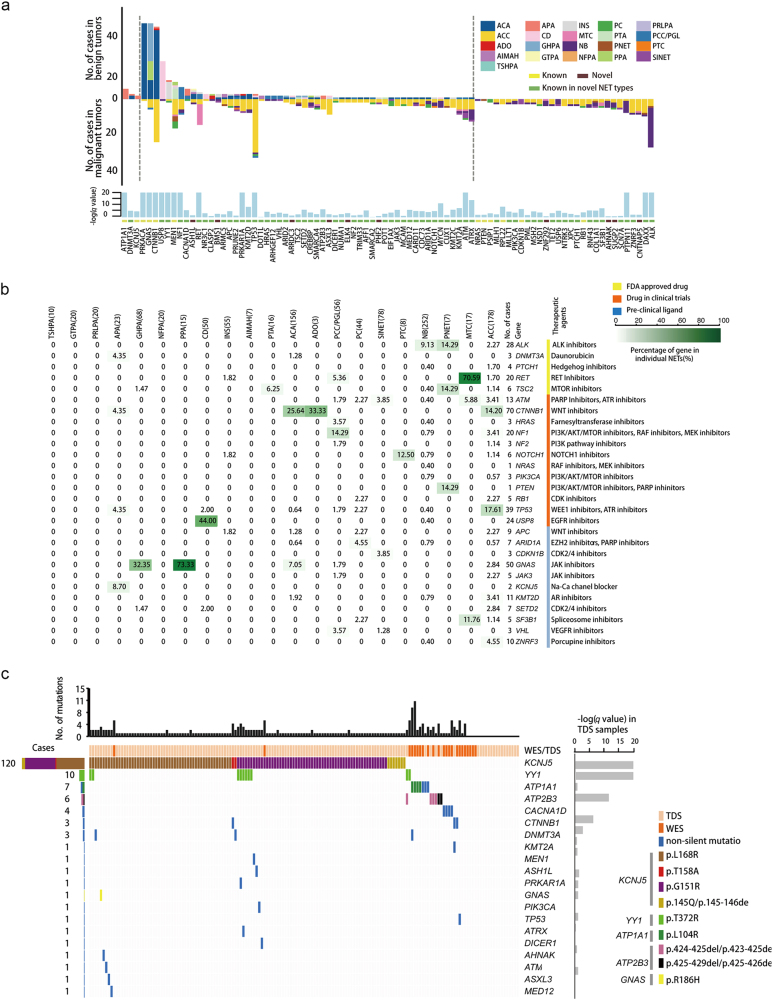
Fig. 1.
The 86 candidate driver genes identified in 21 NETs. a Distribution of candidate driver genes identified in benign and malignant tumors. Height of each colored bar represents the number of mutated cases to types. The bottom bar plot shows the most significant q-value among combined and individual sets calculated by MuSiC. b Druggable cancer gene distributions in NETs. Percentages of samples mutated in individual tumor types are shown. The types of therapeutic agents are as follows: yellow, FDA-approved drug; orange, drug in clinical trials; blue, pre-clinical ligand. The higher the frequency of each gene in 21 NETs, the deeper the color. NETs with 19 specific mutations in ALK, RET, PTCH1, DNMT3A and TSC2 might have potential to benefit from treatment of FDA-approved drugs. Mutations in 12 additional genes, such as HRAS, USP8 and CTNNB1, may present target opportunities in current clinical trials. Another 11 genes might be targeted by the agents under investigations at pre-clinical stages. c Landscape of candidate driver genes in APAs. The number of non-silent mutations in 163 APA tumors on target regions is shown on the top. The colored bars in the bottom panel indicate the mutations and mutation types of candidate driver genes in APAs; each row represents a candidate driver gene (listed on the right). The significant level of candidate driver genes is shown on the right of gene list. The number of mutated samples for each gene is listed on the left. The annotation of colored bars is shown on the right side. Collectively, 137 out of 159 APAs (86.1%) have at least one non-synonymous mutation in the 86 candidate driver genes
To comprehensively understand the mechanistic classification and further illustrate the cellular processes involved in NETs, we performed gene ontology and hierarchical clustering analysis. The 86 genes were classified into 20 categories of cellular processes2 (Supplementary information, Figure S10). Clustering analysis showed that in addition to MEN1, RET and GNAS, mutations in transcription factor genes (YY1, CTNNB1, NF1) and mutations in genome integrity genes (ATM, ATRX, TP53) are critical for clustering of multiple NET types, suggesting the importance of these SMGs in molecular classification of NETs (Supplementary information, Figure S11). Clustering of the 21 types of NET and 13 other cancers with the 86 candidate driver genes and previously described cancer genes showed that the majority of NETs, except ACC, are distinctive from common malignant tumors (Supplementary information, Figure S12).
Notably, chromatin modification and remodeling genes (22 genes in the categories of histone modifiers, genome integrity and DNA methylation) are the most significant set in NETs. In silico prediction analysis showed that all the 22 genes have pathogenic or truncating mutations (131/171, 76.6% in total), supporting the functional roles of the chromatin modification and remodeling genes in NETs (Supplementary information, Figures S13 and 14, Tables S11 and 12). MEN1 is the representative mutated driver gene in diverse inherited and sporadic NET types. The variant allele frequency (VAF) of MEN1 mutations is significantly higher than the average VAF among candidate driver genes (P = 1.2 × 10−4), suggesting that loss of heterozygosity is commonly coupled with MEN1 mutations (Supplementary information, Figure S15). The hormone secretion-associated calcium signaling (Z score = 3.74 in APA) and cAMP/PKA signaling (Z score = 3.31 in ACA) are distinctively enriched pathways in adrenal NETs (Supplementary information, Tables S13 and 14). Mutations of candidate driver genes encoding transcription factors or involved in Wnt/β-catenin signaling are frequently observed in many NETs (Supplementary information, Figure S10). Moreover, many novel SMGs have been associated with cancer development and progression in recent studies. AHNAK suppresses cell growth through modulation of tumor growth factor-β/Smad pathway.7 ARRDC3 and ZNF292 could function as tumor suppressors in breast cancer and colorectal cancer, respectively.8, 9
Identification of druggable targets and precise medical treatment by existing or novel agents is the ultimate mission for pan-cancer analysis of NETs. We analyzed the therapeutically targetable driver genes in NETs and showed that 28 candidate driver genes with 201 mutation sites are potential druggable targets in NETs (Fig. 1b and Supplementary information, Tables S15 and 16). Furthermore, defined levels of potential targets in tumors revealed that NET patients with poor prognosis or high recurrence, such as MTC, ACC and PNET, may have great potential for new clinical investigations with personalized drug therapies (Supplementary information, Figure S16).
Gene sequencing panels for ultra-deep and high-coverage targeted sequencing can effectively evaluate cancer gene alterations. To detect somatic mutations in NETs, we designed a sequencing panel consisting of 61 candidate driver genes (lacking 25 SMGs due to data update) and 118 additional genes potentially related to NETs, and applied targeted high-coverage sequencing (depth ≥ 300×) to 140 pairs of APAs (Supplementary information, Figure S17, Tables S17 and 18). We identified five known and two novel recurrently mutated genes in APAs (Fig. 1c and Supplementary information, Table S19). KCNJ5 was mutated in 80.7% (113/140) of the panel-sequenced APA cohorts. KCNJ5 contains four known hotspot mutations and a novel E145del mutation. One novel in-frame deletion (p.425–429del) was detected in the Ca2+ ATPase gene ATP2B3. Activating mutations of CTNNB1 were identified in tumors from two female patients. We identified 10 novel SMGs in APAs, including recurrently mutated gene YY1 and DNMT3A (Fig. 1c; Supplementary information, Figure S18 and Table S20). Activating hotspot YY1T372R mutations, which enhance cell proliferation and hormone production in INS,10 were found in 6.3% (10/159) of APAs. Recurrent DNMT3A mutations are identified in APAs and ACAs. Moreover, single SMG mutations in APAs include activating GNASR201H mutation, inactivating PRKAR1A mutation and stop-gain MEN1 mutation. These results showed that NET panel sequencing could be an efficient and powerful approach.
Highly frequent and dominant activating hotspot mutations in oncogenes, which are particularly valuable for developing targeted drug therapies, are salient features of multiple NET types. We identified 36 hotspot mutations in 19 SMGs in 21 NETs (Supplementary information, Figure S19, Tables S21 and 22). These activating hotspot mutations could be the most promising choice for the development of druggable targets and personalized medicine for NETs.
In summary, we systematically analyzed somatic mutations across 21 types of NETs and identified 86 candidate driver genes. Mutation features and mutated genes are distinct in benign and malignant NETs; some candidate driver genes are highly specific in one tumor type, while others are common across NETs. Further sequencing of APAs using a customized NET-specific panel efficiently identified somatic mutations in candidate driver genes. Taken together, our findings may advance genomics-based diagnosis and personalized targeting therapies of NETs.
Materials and Methods are available in Supplementary information, Data S1.
Electronic supplementary material
Acknowledgements
We thank Dr. Huanming Yang, Dr. Qiang Gao, Mr. Xuanlin Huang and Ms. Peina Du at BGI-Shenzhen for the helpful discussion. This work is supported by the National Natural Science Foundation of China (81522032 to Y.C. and 81530020 to G.N.), the National Key Research and Development Program (2016YFC0905001 and 2017YFC0909703 to Y.C.), the Shanghai Rising-Star Program (15QA1402900 to Y.C.) and the National Human Genome Research Institute (U01HG006517 to L.D.).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally to this work: Yanan Cao, Weiwei Zhou, Lin Li, Jiaqian Wang, and Zhibo Gao.
Materials and Methods are available in Supplementary information, Data S1. Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0019-5.
Contributor Information
Yanan Cao, Email: caoyanan@vip.sina.com.
Li Ding, Email: lding@genome.wustl.edu.
Guang Ning, Email: guangning@medmail.com.cn.
References
- 1.Kunz PL. Carcinoid and neuroendocrine tumors: building on success. J. Clin. Oncol. 2015;33:1855–63. doi: 10.1200/JCO.2014.60.2532. [DOI] [PubMed] [Google Scholar]
- 2.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013;45:279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Cuesta L, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat. Commun. 2014;5:3518. doi: 10.1038/ncomms4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee IH, et al. Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene. 2014;33:4675–84. doi: 10.1038/onc.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draheim KM, et al. ARRDC3 suppresses breast cancer progression by negatively regulating integrin β4. Oncogene. 2010;29:5032–47. doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda H, et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 2015;47:142–50. doi: 10.1038/ng.3175. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, et al. Whole exome sequencing of insulinoma reveals recurrent T372R mutations in YY1. Nat. Commun. 2013;4:2810. doi: 10.1038/ncomms3810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.