ABSTRACT
RAD51-associated protein 1 (RAD51AP1) is a member of the multiprotein complexes postulated to carry out RAD51-mediated homologous recombination and DNA repair in mammalian cells. In the present study, we showed that hepatitis C virus (HCV) NS5A directly bound RAD51AP1 and increased the protein level of RAD51AP1 through modulation of the ubiquitin-proteasome pathway. We also demonstrated that RAD51AP1 protein levels were increased in the liver tissues of HCV-infected patients and NS5A-transgenic mice. Importantly, NS5A impaired DNA repair by disrupting the RAD51/RAD51AP1/UAF1 complex and rendered HCV-infected cells more sensitive to DNA damage. Silencing of RAD51AP1 expression resulted in a decrease of viral propagation. We further demonstrated that RAD51AP1 was involved in the assembly step of the HCV life cycle by protecting viral RNA. These data suggest that HCV exploits RAD51AP1 to promote viral propagation and thus that host DNA repair is compromised in HCV-infected cells. Overall, our findings provide mechanistic insight into the pathogenesis of HCV infection.
IMPORTANCE Chronic infection with HCV is the leading cause of hepatocellular carcinoma (HCC). However, the molecular mechanisms underlying HCV-induced HCC are not fully understood. Here we demonstrate that the HCV NS5A protein physically interacts with RAD51AP1 and increases the RAD51AP1 protein level through modulation of the ubiquitin-proteasome pathway. HCV coopts host RAD51AP1 to protect viral RNA at an assembly step of the HCV life cycle. Note that the RAD51 protein accumulates in the cytoplasm of HCV-infected cells, and thus the RAD51/RAD51AP1/UAF1-mediated DNA damage repair system in the nucleus is compromised in HCV-infected cells. Our data may provide new insight into the molecular mechanisms of HCV-induced pathogenesis.
KEYWORDS: NS5A, RAD51AP1, RAD51, DNA repair, homologous recombination
INTRODUCTION
Overwhelming lines of epidemiological evidence have indicated that chronic infection with hepatitis C virus (HCV) causes the development of hepatocellular carcinoma (HCC). HCV infection enhances mutation frequencies of cellular genes, causing double-stranded DNA breaks (DSBs) and thus leading to chromosomal instability and mutation accumulation (1). Many viral proteins are directly involved in cellular pathogenesis. It has been reported that HCV proteins, including core, E1, and NS3, produce reactive oxygen species (ROS) contributing to DSBs (2). Nonstructural protein 5A (NS5A) of HCV also modulates various cellular signaling pathways involved in cell transformation, differentiation, and tumorigenesis. The oncogenic role of NS5A is known to be mediated by upregulation of prosurvival genes though p53 and NF-κB pathways (3). NS5A activates the phosphatidylinositol 3-kinase (PI3K)/Akt/mTOR pathway and perturbs mitogen-activated protein kinase (MAPK) signaling pathways (4, 5). In animal models, HCV NS5A induces a range of hepatic pathologies, including extraordinary steatosis and HCC in transgenic mice, and all nude mice expressing NS5A develop tumors (6, 7). NS5A is an RNA-binding phosphoprotein that is essential for both viral replication and viral particle assembly (8).
Homologous recombination repair (HRR) is an error-free pathway for the repair of DSBs, whereas nonhomologous end joining is an error-prone repair pathway for the broken DNA ends (9). HRR can repair a variety of DNA lesions, including DSBs, single-strand DNA breaks, and interstrand cross-links. RAD51 recombinase plays a central role in homologous recombination (HR) in eukaryotes. RAD51 facilitates the formation of a heteroduplex DNA, the D-loop, which connects the joint molecules between the damaged DNA and the undamaged repair template and mediates homologous pairing (10).
RAD51-associated protein 1 (RAD51AP1) is a vertebrate-specific protein that binds to RAD51 and stimulates D-loop formation, thereby enhancing RAD51 recombinase activity. Thus, RAD51AP1 seems to act downstream of the presynaptic stage of HRR (11, 12). RAD51AP1 is a structure-specific DNA binding protein that is also capable of binding RNA (13). Moreover, RAD51AP1 acts as a bridge between USP1-associated factor 1 (UAF1; a deubiquitinating enzyme [DUB] binding protein) and RAD51. UAF1 synergizes with RAD51AP1 to promote assembly of the synaptic complex, a critical nucleoprotein intermediate in HR (14). Therefore, cells expressing a UAF1-binding-defective mutant of RAD51AP1 display reduced HR efficiency and increased chromosomal aberrations in response to DNA interstrand cross-linking (ICL)-inducing agents (15). RAD51AP1 is specifically required not only for the cellular response to ICL-inducing agents, including mitomycin and cisplatin, but also for the response to other DSB-inducing agents, including ionizing radiation and camptothecin (CPT) (16). Indeed, RAD51AP1 is highly expressed in aggressive lymphomas and carcinomas (12). UAF1, a WD40 repeat-containing protein, was first identified as a stoichiometric binding partner of ubiquitin carboxyl-terminal hydrolase 1 (USP1). UAF1 regulates both the enzyme activity and the stability of USP1 by forming a deubiquitinating complex (17). UAF1-USP1 is involved in FANCD2 and PCNA monoubiquitination (18). UAF1 mediates the interaction between RAD51AP1 and USP1, and depletions of USP1 and UAF1 lead to decreased stability of RAD51AP1. Therefore, the UAF1-USP1 complex promotes HRR via deubiquitination of the FANCD2-RAD51AP1 interaction (15).
In the present study, we demonstrated that NS5A directly interacted with RAD51AP1 and enhanced RAD51AP1 protein stability through the ubiquitin-proteasome pathway. NS5A also abrogated the protein interplay between RAD51AP1 and UAF1, and thereby the RAD51/RAD51AP1/UAF1 trimeric complex was disrupted in HCV-infected cells. HCV infection impaired DNA repair and increased hypersensitivity to DNA damaging reagents. Silencing of RAD51AP1 expression resulted in a decrease of viral propagation. We further showed that RAD51AP1 is involved in the assembly step of the HCV life cycle by protecting HCV RNA binding to NS5A. Collectively, these data show that RAD51AP1 is a novel host factor involved in both HCV propagation and HCV-mediated cellular pathogenesis.
RESULTS
RAD51AP1 is a host factor interacting with NS5A.
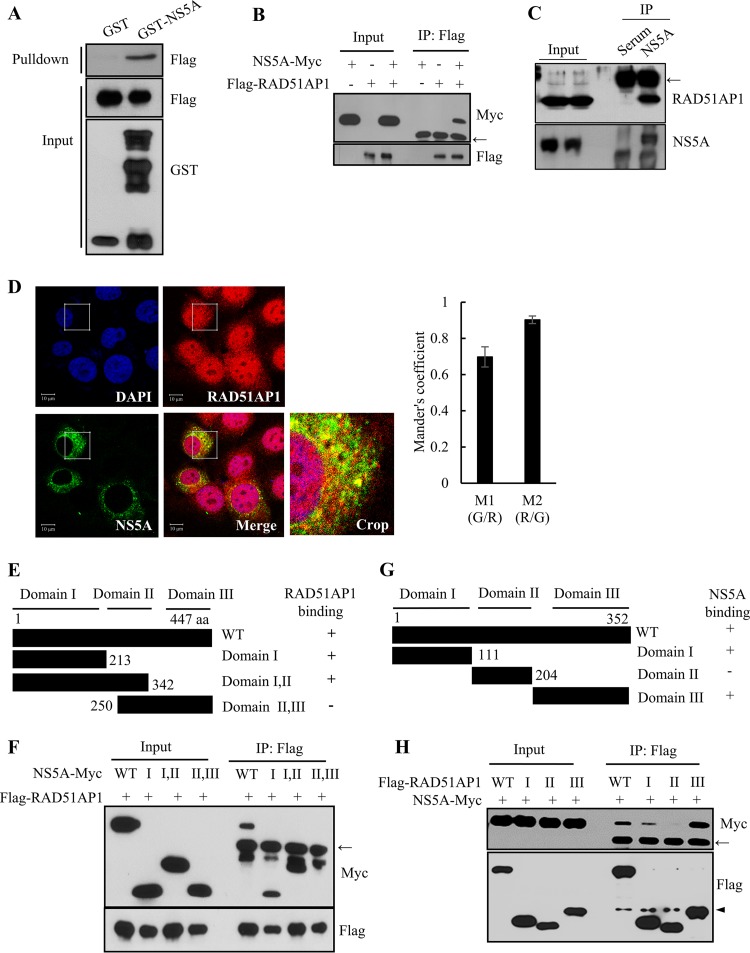
We previously performed protein microarray assays and identified RAD51AP1 as an HCV NS5A interaction partner (19). RAD51AP1 was identified as one of 90 cellular proteins. We first verified the protein array data by performing an in vitro glutathione S-transferase (GST) pulldown assay. As shown in Fig. 1A, NS5A selectively interacted with RAD51AP1. By employing a coimmunoprecipitation assay, we further confirmed that NS5A specifically interacted with RAD51AP1 (Fig. 1B). Next, we investigated whether the NS5A protein interacted with endogenous RAD51AP1. Cell lysates harvested 4 days after HCV RNA electroporation were immunoprecipitated with either control mouse IgG or an anti-RAD51AP1 antibody, and bound proteins were analyzed by immunoblotting with an anti-NS5A antibody. As shown in Fig. 1C, HCV NS5A interacted with endogenous RAD51AP1. These data also suggest that RAD51AP1 may colocalize with NS5A in HCV-infected cells. Indeed, RAD51AP1 was localized mainly in the nucleus but also in the cytoplasm and partially colocalized with NS5A in the cytoplasm, as shown by yellow fluorescence in the merged image in Fig. 1D. Collectively, these data suggest that RAD51AP1 specifically interacts with NS5A both in vitro and in vivo. To dissect the region in NS5A responsible for RAD51AP1 binding, the interactions between RAD51AP1 and various deletion mutants of NS5A (Fig. 1E) were determined by a transfection-based coimmunoprecipitation assay. As shown in Fig. 1F, RAD51AP1 interacted with domain I of NS5A. We then determined the region in RAD51AP1 for NS5A binding. We constructed various truncation mutants of RAD51AP1 (Fig. 1G) based on the results of a previous report (20). We showed that NS5A interacted with domains I and III but not with domain II, indicating that NS5A interacted with RAD51AP1 through two binding sites (Fig. 1H). In fact, domain I of RAD51AP1 is responsible for DNA binding, and domain III of RAD51AP1 has been shown to associate with RAD51 and enhances the RAD51-mediated D-loop reaction to facilitate the function of RAD51AP1 in HR (20).
FIG 1.
HCV NS5A interacts with RAD51AP1. (A) HEK293T cells were transfected with Flag-tagged RAD51AP1, and total cell lysates harvested 24 h after transfection were incubated with either GST or GST-NS5A. After pulldown by GST beads, bound protein was detected by use of an anti-Flag antibody. (B) HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged RAD51AP1. Thirty-six hours later, total cell lysates were immunoprecipitated (IP) with an anti-Flag antibody. Bound protein was detected by use of an anti-Myc antibody. (C) Huh7.5 cells were electroporated with 10 μg of Jc1 RNA. Cell lysates harvested 4 days after electroporation were immunoprecipitated with either control IgG or an anti-RAD51AP1 antibody. Bound protein was immunoblotted with an anti-NS5A antibody. (D) Huh7.5 cells were infected with Jc1 for 4 h. At 48 h postinfection, cells were fixed in 4% paraformaldehyde, and immunofluorescence staining was performed to detect endogenous RAD51AP1 (red), NS5A (green), and nuclei (blue). Dual staining showed colocalization of endogenous RAD51AP1 and NS5A in the cytoplasm as yellow fluorescence in the merged image. Colocalization of RAD51AP1 and NS5A in the cytoplasm was quantified by using the Manders overlap coefficient. More than 10 cells were applied to ImageJ for quantification of the overlap coefficient, and error bars indicate the standard deviations of the means. (E) Schematic illustration of both wild-type and mutant forms of the NS5A expression plasmid. (F) RAD51AP1 interacts with domain I of NS5A. HEK293T cells were cotransfected with Flag-tagged RAD51AP1 and Myc-tagged NS5A. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with an anti-Myc antibody. (G) Schematic illustration of both wild-type and mutant RAD51AP1 proteins. (H) NS5A interacts with both domains I and III of RAD51AP1. HEK293T cells were cotransfected with Myc-tagged NS5A and Flag-tagged RAD51AP1. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with an anti-Myc antibody. Arrows indicate the position of the heavy chain, and the arrowhead denotes the light chain of IgG.
The RAD51AP1 protein level is increased in Jc1-infected cells.
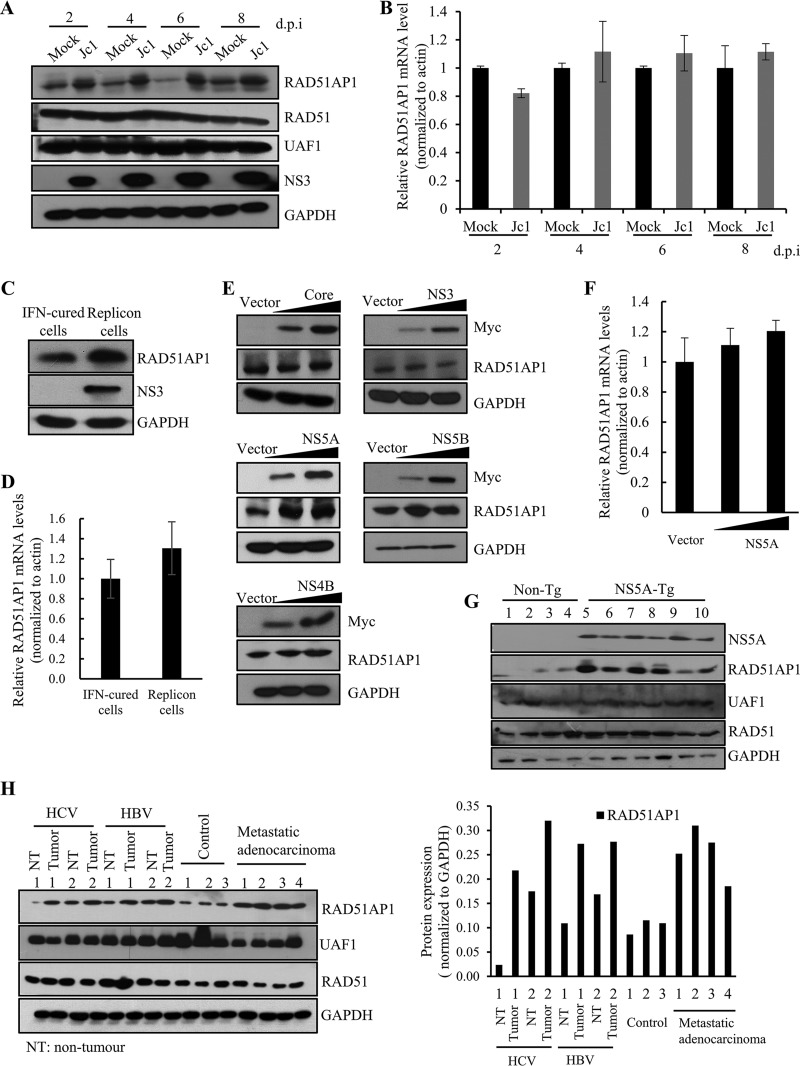
RAD51AP1 expression is upregulated in several primary human tumors, including HCC, acute myeloid leukemia with complex karyotypic abnormalities, and aggressive mantle cell lymphoma (16, 21, 22). We also showed that the RAD51AP1 protein level was markedly increased in HCV-infected cells compared to that in mock-infected cells (Fig. 2A). Note that the protein levels of both RAD51 and UAF1 remained unchanged. Interestingly, the RAD51AP1 mRNA level was not altered during the course of HCV infection (Fig. 2B), suggesting that RAD51AP1 might undergo posttranscriptional modification. We further verified that the RAD51AP1 protein level (Fig. 2C), but not its mRNA level (Fig. 2D), was elevated in Huh7 cells harboring an HCV subgenomic replicon (genotype 1b) compared to that in interferon (IFN)-cured cells. These data indicated that the nonstructural protein of HCV was responsible for the increased RAD51AP1 expression level in HCV-replicating cells. Strikingly, the RAD51AP1 protein level was increased only by NS5A, not by other HCV proteins (Fig. 2E). Here again we showed that no discernible change in RAD51AP1 mRNA level was detected by ectopic expression of NS5A (Fig. 2F). These data suggest that there may be a specific posttranslational regulation of RAD51AP1 by NS5A in HCV-infected cells. We next determined the RAD51AP1 levels in the liver tissues of NS5A-transgenic mice (6). Figure 2G shows that RAD51AP1 protein levels were remarkably increased in the liver tissues of NS5A-transgenic mice compared to those in the nontransgenic counterparts, whereas both RAD51 and UAF1 levels remained unchanged. We further showed that protein levels of RAD51AP1 were markedly increased in HCC tissues compared to those in noncancerous liver tissues from patients with HCV infection (Fig. 2H, left panel). Note that RAD51AP1 levels, but not RAD51 and UAF1 levels, were higher in tumor tissues than in their nontumor counterparts, irrespective of the viral infection (Fig. 2H, right panel). These data indicate that RAD51AP1 protein levels were elevated in all tumor tissues and thus may represent a novel tumor marker.
FIG 2.
HCV infection increases the RAD51AP1 protein expression level through NS5A. (A) Huh7.5 cells were either mock infected or infected with Jc1. Total cell lysates harvested at various time points were immunoblotted with the indicated antibodies. d.p.i, days postinfection. (B) RAD51AP1 mRNA levels for the set of experiments for panel A were analyzed by qRT-PCR. (C) Total cell lysates harvested from either IFN-cured Huh7 cells or Huh7 cells harboring an HCV subgenomic replicon (genotype 1b) were immunoblotted with the indicated antibodies. (D) RAD51AP1 mRNA levels in IFN-cured cells and HCV replicon cells were determined by qRT-PCR. (E) Huh7 cells were transiently transfected with either vector or increasing amounts of Myc-tagged core, NS3, NS4B, NS5A, or NS5B plasmid. At 36 h posttransfection, protein levels were immunoblotted by use of the indicated antibodies. (F) Huh7 cells were transiently transfected with either vector or increasing amounts of Myc-tagged NS5A plasmid. RAD51AP1 mRNA levels were analyzed by qRT-PCR. Experiments were performed in triplicate. (G) Liver tissues of either nontransgenic (n = 4) or NS5A-transgenic (n = 5) mice were homogenized and immunoblotted with the indicated antibodies. (H) (Left) Human liver tissues isolated from either control or various patients were homogenized and immunoblotted with the indicated antibodies. (Right) RAD51AP1 expression levels were quantified after normalization to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level.
NS5A stabilizes RAD51AP1 by modulating the ubiquitin-proteasome pathway.
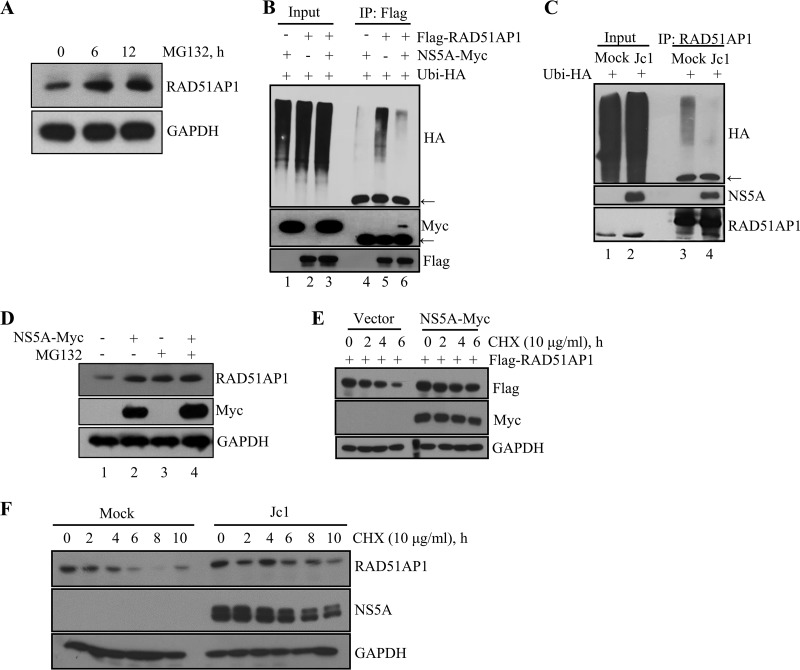
NS5A modulates transcriptional activities of numerous host genes, including the β-catechin, cyclin D1, cdk4, and epidermal growth factor receptor (EGFR) genes, and regulates ubiquitination of Pim kinase and deubiquitination of OUTD7B (19, 23–25). Moreover, a recent proteomic study suggested a possible ubiquitination of RAD51AP1 at residue K156 (26). We therefore speculated that ubiquitination of RAD51AP1 might be regulated by NS5A. As shown in Fig. 3A, the RAD51AP1 protein underwent processing by the proteasome pathway, and protein levels of RAD51AP1 were markedly increased in the presence of MG132. As we postulated, ectopic expression of NS5A resulted in a remarkable decrease in the ubiquitination level of RAD51AP1 (Fig. 3B, lane 6). We further verified that ubiquitination of endogenous RAD51AP1 was markedly suppressed in HCV-infected cells compared to that in mock-infected cells (Fig. 3C, lane 4). Moreover, ectopic expression of NS5A increased the RAD51AP1 level (Fig. 3D, lane 2). Similarly, the RAD51AP1 protein level was increased in the presence of MG132 (Fig. 3D, lane 3). However, ectopic expression of NS5A exerted no additive effect on the RAD51AP1 protein level in MG132-treated cells (Fig. 3D, lane 4). All these data suggest that NS5A stabilizes RAD51AP1 through modulation of the proteasome pathway. Since NS5A interacted with RAD51AP1 and ubiquitination of RAD51AP1 was reduced by NS5A, we postulated that NS5A might regulate RAD51AP1 protein stability. Figure 3E shows that the level of RAD51AP1 was gradually decreased in cycloheximide (CHX)-treated vector control cells, whereas the RAD51AP1 protein level remained relatively stable in the presence of NS5A. We further confirmed that the endogenous RAD51AP1 level remained relatively stable in Jc1-infected cells compared to that in mock-infected cells (Fig. 3F). Collectively, these data clearly show that NS5A protected RAD51AP1 from proteasome-dependent degradation.
FIG 3.
HCV NS5A protects RAD51AP1 from ubiquitin-dependent proteasomal degradation. (A) Huh7 cells were treated with 20 μM MG132 for the indicated time points, and protein levels were determined by immunoblot analysis with the indicated antibodies. (B) HEK293T cells were cotransfected with the indicated combinations of plasmids. At 36 h posttransfection, total cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with an anti-HA antibody. Arrows indicate the position of the heavy chain. (C) Huh7 cells that were either mock infected or infected with Jc1 for 48 h were transfected with HA-tagged ubiquitin. At 36 h posttransfection, total cell lysates were immunoprecipitated with an anti-RAD51AP1 antibody, and bound proteins were immunoblotted with an anti-HA antibody. (D) Huh7 cells were transfected with either vector or a Myc-tagged NS5A expression plasmid. At 36 h posttransfection, cells were left untreated or treated with 20 μM MG132 for 6 h, and protein levels were determined by immunoblot analysis with the indicated antibodies. (E) HEK293 cells were cotransfected with the indicated combinations of plasmids. At 30 h posttransfection, cells were treated with 10 μg/ml of CHX for the indicated time points, and protein levels were determined by immunoblot analysis with the indicated antibodies. (F) Huh7 cells that were either mock infected or infected with Jc1 for 48 h were treated with 10 μg/ml of CHX for the indicated time points, and protein levels were determined by immunoblot analysis with the indicated antibodies.
NS5A abrogates protein interaction between RAD51AP1 and UAF1.
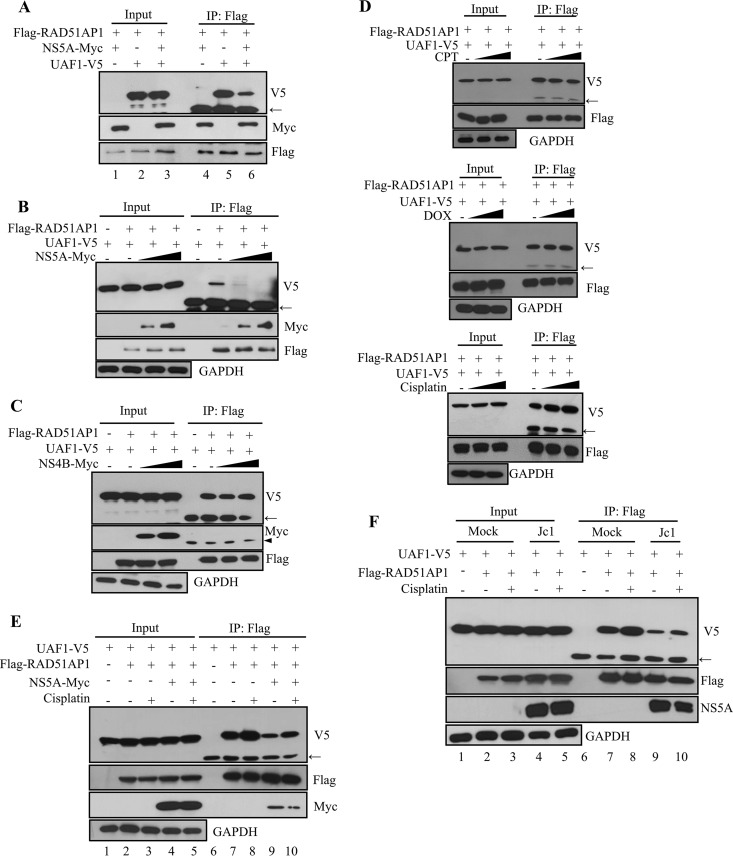
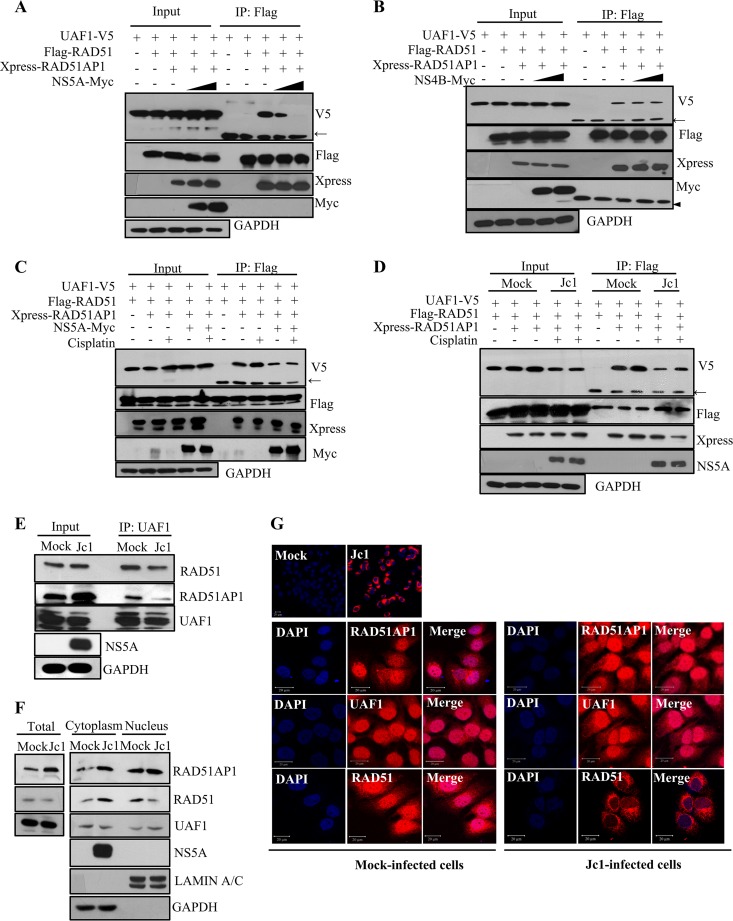
RAD51AP1 serves as a bridging factor that recruits UAF1 to RAD51. Moreover, RAD51AP1-UAF1 interaction is important for RAD51-mediated D-loop formation in the DNA strand invasion step of HRR (14). We first assessed the effect of NS5A on RAD51AP1-UAF1 interaction. For this purpose, HEK293T cells were cotransfected with Flag-tagged RAD51AP1, V5-tagged UAF1, and Myc-tagged NS5A. At 48 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and coprecipitated proteins were detected by immunoblotting with either an anti-V5 antibody or an anti-Myc antibody. As shown in Fig. 4A, both UAF1 and NS5A were coprecipitated with RAD51AP1. Note that the RAD51AP1-UAF1 association was prominently decreased in the presence of NS5A (lane 6). We verified that NS5A impaired RAD51AP1-UAF1 dimer formation in a dose-dependent manner (Fig. 4B). We showed that NS4B was unable to disrupt the RAD51AP1-UAF1 association (Fig. 4C), indicating that NS5A specifically inhibited dimer formation of RAD51AP1-UAF1. The dimeric complex of RAD51AP1 and UAF1 is critically important for HRR in response to DNA ICL-inducing agents. We therefore asked whether the RAD51AP1-UAF1 complex was sensitive to DNA damaging agents by using cisplatin, doxorubicin (DOX), and camptothecin (CPT). The effects of the chemicals on DNA damage and HRR were assessed by immunofluorescence assay with RAD51 nuclear foci. It is well known that HRR activity is monitored by RAD51 and that the steady-state level of RAD51 in the nucleus is a hallmark of the cellular response to DNA damage (27). RAD51 was redistributed to the nucleus by all of our DNA damaging agents (data not shown). With the functional doses of these exogenous DNA damaging agents, we showed that neither CPT (Fig. 4D, top panel) nor DOX (Fig. 4D, middle panel) displayed an effect on dimeric formation of RAD51AP1-UAF1. On the other hand, cisplatin increased the level of the RAD51AP1-UAF1 complex (Fig. 4D, bottom panel). We showed that protein levels of RAD51AP1, UAF1, and RAD51 remained unchanged in response to these DNA damaging agents in Huh7 cells (data not shown). Therefore, we analyzed the effect of cisplatin on RAD51AP1-UAF1 interaction in the context of HCV replication. We first investigated the effect of NS5A on RAD51AP1-UAF1 complex formation. As shown in Fig. 4E, the RAD51AP1-UAF1 complex level was increased by cisplatin (lane 8). Importantly, NS5A attenuated the effect of cisplatin on RAD51AP1-UAF1 association (Fig. 4E, lane 10). We further confirmed that cisplatin-mediated RAD51AP1-UAF1 complex formation was disrupted in HCV-infected cells (Fig. 4F, lane 10). Taken together, these data suggest that NS5A may disturb RAD51AP1-UAF1 binding to disrupt DNA repair in HCV-infected cells.
FIG 4.
HCV NS5A disrupts the dimeric complex of RAD51AP1 and UAF1. (A) HEK293T cells were cotransfected with the indicated combinations of plasmids. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with anti-V5 and anti-Myc antibodies. (B) HEK293T cells were cotransfected with Flag-tagged RAD51AP1 and V5-tagged UAF1 and with increasing amounts of Myc-tagged NS5A. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with anti-V5 and anti-Myc antibodies. Immunoprecipitation efficiency was verified by immunoblot analysis using an anti-Flag antibody. (C) HEK293T cells were cotransfected with Flag-tagged RAD51AP1 and V5-tagged UAF1 and with increasing amounts of Myc-tagged NS4B. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with anti-V5 and anti-Myc antibodies. (D) HEK293T cells were cotransfected with Flag-tagged RAD51AP1 and V5-tagged UAF1. At 24 h posttransfection, cells were either left untreated (vehicle) or treated with the indicated DNA damaging agents. Sixteen hours after treatment, total cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with an anti-V5 antibody. (E) HEK293T cells were cotransfected with V5-tagged UAF1 and Flag-tagged RAD51AP1 in the absence or presence of a Myc-tagged NS5A expression plasmid. At 24 h posttransfection, cells were left untreated or treated with 25 μM cisplatin. Sixteen hours after treatment, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with anti-V5 and anti-Myc antibodies. (F) Huh7 cells were cotransfected with V5-tagged UAF1 and Flag-tagged RAD51AP1. At 24 h posttransfection, cells were either mock infected or infected with Jc1 for 4 h, followed by treatment with either vehicle or 25 μM cisplatin for 16 h. Cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with anti-V5 and anti-NS5A antibodies. Arrows indicate the position of the heavy chain, and arrowheads denote the light chain of IgG.
HCV disturbs the trimeric complex of RAD51, RAD51AP1, and UAF1 and accumulates RAD51 in the cytoplasm.
Given that NS5A interfered with protein interaction between RAD51AP1 and UAF1, it is also possible that NS5A might disrupt UAF1 binding with RAD51, since UAF1 interacts with RAD51 via RAD51AP1 (14). To investigate this possibility, HEK293T cells were cotransfected with V5-tagged UAF1, Flag-tagged RAD51, and Xpress-tagged RAD51AP1 in the absence or presence of Myc-tagged NS5A. As shown in Fig. 5A, the RAD51/RAD51AP1/UAF1 complex was disrupted by NS5A in a dose-dependent manner. We further demonstrated that this trimeric complex formation was not disrupted by NS4B (Fig. 5B), indicating that NS5A specifically inhibited trimeric complex formation. We further verified that the increase of RAD51/RAD51AP1/UAF1 complex formation by cisplatin was disrupted by both ectopic expression of NS5A (Fig. 5C) and infection with Jc1 (Fig. 5D). We finally confirmed that the endogenous formation of the RAD51/RAD51AP1/UAF1 complex was markedly decreased in Jc1-infected cells compared to that in mock-infected cells (Fig. 5E). Next, we investigated the subcellular localization of the RAD51, RAD51AP1, and UAF1 proteins in both mock- and Jc1-infected cells. As expected, the RAD51AP1 protein level was increased in Jc1-infected cells compared to that in mock-infected cells (Fig. 5F), whereas protein levels of both UAF1 and RAD51 were not altered by HCV infection. Note that the cytoplasmic RAD51 level was increased in Jc1-infected cells compared to that in mock-infected cells. There were no alterations in the subcellular localization of RAD51AP1 and UAF1. Using immunofluorescence analysis, we demonstrated that RAD51AP1, UAF1, and RAD51 were localized mainly in the nucleus, with partial localization in the cytoplasm, in mock-infected cells (Fig. 5G). Importantly, the majority of RAD51 accumulated in the cytoplasm in HCV-infected cells (Fig. 5G). Since the RAD51/RAD51AP1/UAF1 complex as well as the nuclear subcellular localization of RAD51 is critical for the HRR, these data suggest that DNA damage repair may be defective in HCV-infected cells.
FIG 5.
HCV disturbs the trimeric complex of RAD51, RAD51AP1, and UAF1 via NS5A and accumulates RAD51 in the cytoplasm. (A) HEK293T cells were cotransfected with V5-tagged UAF1 and either vector, Flag-tagged RAD51, Xpress-tagged RAD51AP1, or increasing amounts of Myc-tagged NS5A. At 36 h posttransfection, total cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with the indicated antibodies. (B) HEK293T cells were cotransfected with V5-tagged UAF1 and either vector, Flag-tagged RAD51, Xpress-tagged RAD51AP1, or increasing amounts of Myc-tagged NS4B. At 36 h posttransfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with the indicated antibodies. (C) Huh7 cells were cotransfected with the indicated combinations of plasmids. At 24 h posttransfection, cells were treated with 25 μM cisplatin. At 16 h posttreatment, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with the indicated antibodies. (D) Huh7 cells were cotransfected with the indicated combinations of plasmids. At 24 h posttransfection, cells were either mock infected or infected with Jc1 for 4 h, followed by treatment with 25 μM cisplatin. At 16 h posttreatment, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were immunoblotted with the indicated antibodies. (E) Huh7.5 cells were either mock infected or infected with Jc1. Cell lysates harvested at 6 days postinfection were immunoprecipitated with an anti-UAF1 antibody. Bound proteins were immunoblotted with anti-RAD51 and anti-RAD51AP1 antibodies. (F) HCV induces cytoplasmic distribution of RAD51 protein. Huh7.5 cells were either mock infected or infected with Jc1. At 6 days postinfection, both cytosolic and nuclear fractions were immunoblotted with the indicated antibodies. (G) Huh7.5 cells were either mock infected or infected with Jc1. If more than 90% of cells were infected with Jc1 (upper panel), immunofluorescence staining was performed. Arrows indicate the position of the heavy chain, and the arrowhead denotes the light chain of IgG.
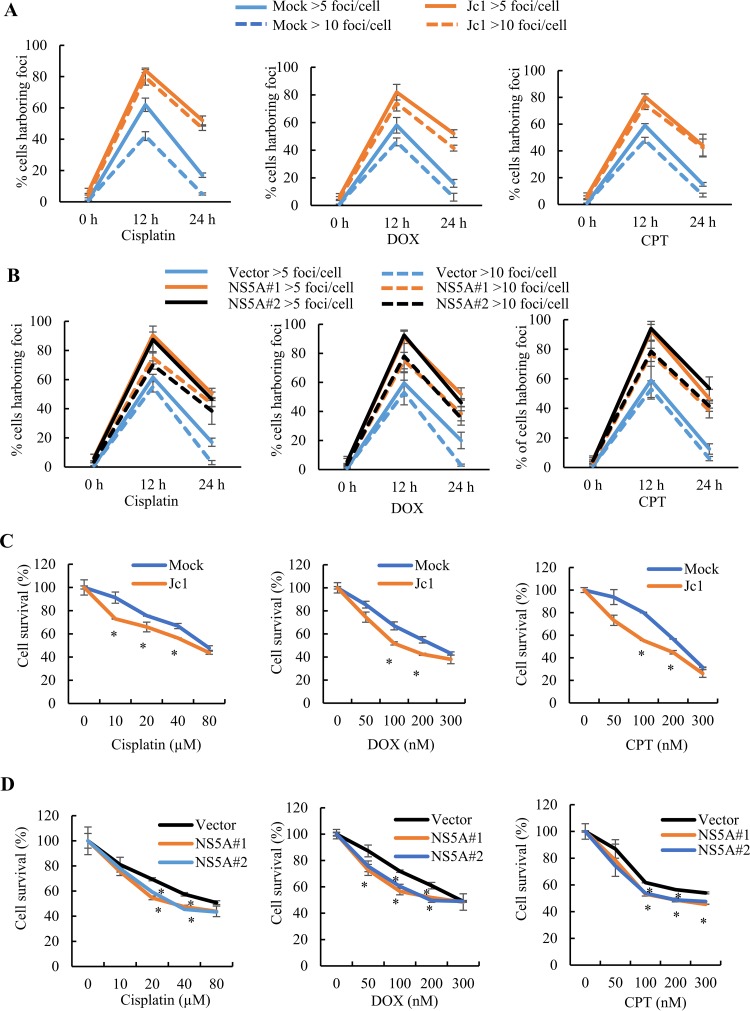
HCV NS5A enhances sensitivity to DNA damaging agents and impairs DNA repair.
HCV infection caused subcellular relocalization of RAD51, and HCV NS5A disrupted the RAD51/RAD51AP1/UAF1 complex. We therefore investigated whether DNA damage repair was impaired by HCV infection. Histone H2AX phosphorylation on Ser-139 (γH2AX) is a sensitive marker of DSBs and can be analyzed semiquantitatively by immunofluorescence microscopy (28). The rate of disappearance of γH2AX foci reflects the DNA repair ability in the cells (29). Therefore, we analyzed the extent of γH2AX foci in mock- and HCV-infected cells in the absence or presence of cisplatin, DOX, and CPT. The number of cells harboring either >5 or >10 foci was counted in each experiment. Consistent with a previous report (30), few γH2AX foci were detected for both mock- and Jc1-infected cells in the absence of DNA damaging agents (data not shown). The percentage of cells harboring >5 γH2AX foci among chemically treated cells was elevated from ∼60% for mock-infected cells to ∼80% for HCV-infected cells (Fig. 6A). It is noteworthy that the percentage of cells harboring >10 γH2AX foci was significantly higher for HCV-infected cells than for mock-infected cells. Twenty-four hours after chemical treatments, the rate of loss of foci for HCV-infected cells was considerably lower than that for mock-infected cells (Fig. 6A). These data support previous reports that HCV impairs DNA repair activity (31, 32). Next, we investigated the possible involvement of NS5A in γH2AX focus formation. Two clones of both stably vector-expressing cells (vector-stable cells) and stably NS5A-expressing cells (NS5A-stable cells) were analyzed. We showed that the percentage of cells harboring >5 to >10 γH2AX foci among chemically treated cells was significantly increased for NS5A-stable cells compared to that for vector-stable cells (Fig. 6B). Next, we determined the cellular sensitivity to DNA damaging agents for both mock-infected and Jc1-infected cells. We demonstrated that both mock- and Jc1-infected cells showed increased sensitivity to escalating doses of cisplatin, DOX, and CPT (Fig. 6C). Most importantly, NS5A-stable cells exhibited lower cell survival (Fig. 6D), indicating that the repair of DSBs and DNA cross-links was compromised in HCV-infected cells. Collectively, these data indicate that NS5A impairs DNA damage repair by interrupting RAD51/RAD51AP1/UAF1 complex formation and rendering HCV-infected cells more sensitive to DNA damage.
FIG 6.
HCV exerts persistence of γH2AX foci in response to DNA damaging agents and impairs DNA damage repair activity. (A) The percentages of cells harboring more than 5 or 10 γH2AX foci/cell in mock- and Jc1-infected cells were determined. (B) The percentages of cells harboring more than 5 or 10 γH2AX foci/cell in vector-stable or two different NS5A-stable cells were determined. (C) Huh7.5 cells were either mock infected or infected with Jc1 and then treated with the indicated doses of DNA damaging agents for 24 h. Cell survival was measured by WST assay. (D) Vector-stable or NS5A-stable cells were treated with the indicated doses of DNA damaging agents. At 24 h posttreatment, cell survival was measured by WST assay. Experiments were performed in triplicate. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
RAD51AP1 is required for HCV propagation.
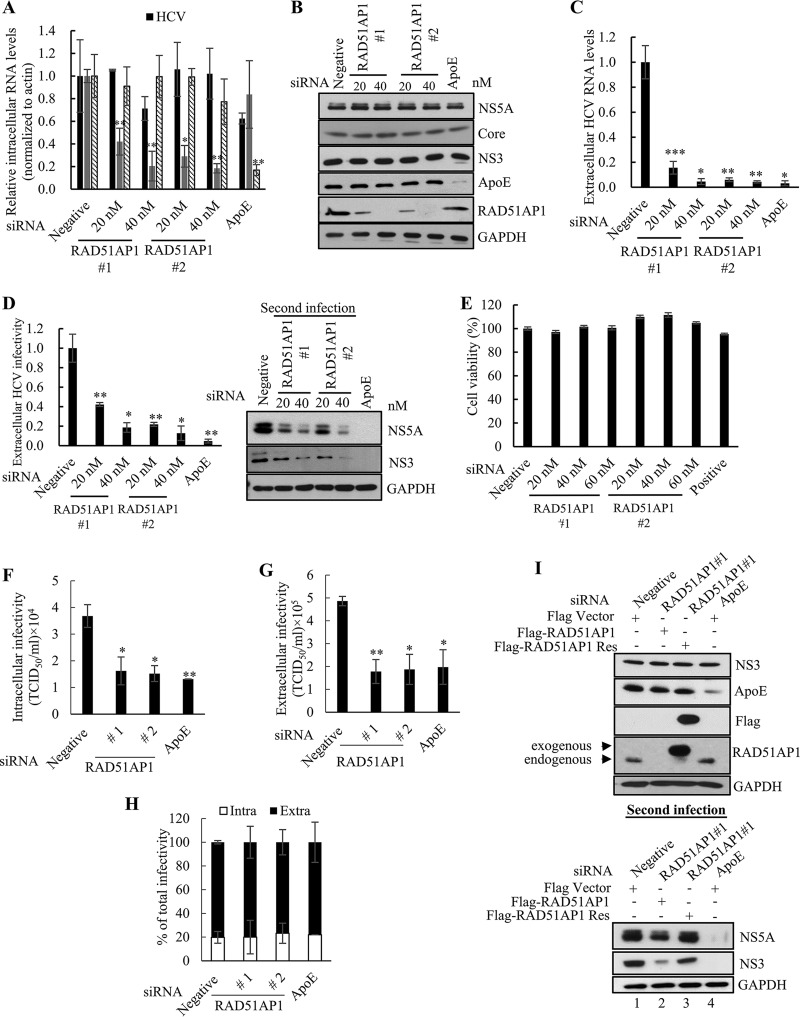
We next investigated the functional involvement of RAD51AP1 in the HCV life cycle. Silencing of RAD51AP1 had little effect on the intracellular HCV RNA level (Fig. 7A) or HCV protein level (Fig. 7B). However, extracellular HCV RNA levels were significantly reduced in RAD51AP1-silenced cells (Fig. 7C). To further investigate the role of RAD51AP1 in HCV propagation, naive Huh7.5 cells were infected with Jc1 harvested from culture supernatant of the first infection, and then both RNA and protein levels of HCV were analyzed. As shown in Fig. 7D, viral infectivity in the second infection was significantly decreased by knockdown of RAD51AP1. To determine intracellular HCV infectivity, Jc1-infected cells were disrupted by repeated cycles of freezing and thawing, and then naive Huh7.5 cells were infected with Jc1 in cellular lysates. As shown in Fig. 7F, intracellular infectivity was significantly decreased by knockdown of RAD51AP1. Consistently, extracellular infectivity was significantly decreased by knockdown of RAD51AP1 (Fig. 7G). However, the proportions of intracellular and extracellular infectivities relative to the total were not altered in RAD51AP1-silenced cells (Fig. 7H). These data indicate that RAD51AP1 might be involved in virus assembly. We further demonstrated that treatment with the same concentrations of small interfering RNAs (siRNAs) displayed no cytotoxicity (Fig. 7E). To rule out the off-target effect of a RAD51AP1 siRNA, we generated a siRNA-resistant RAD51AP1 mutant. As shown in Fig. 7I, exogenous expression of the siRNA-resistant RAD51AP1 mutant, but not that of wild-type RAD51AP1, restored the HCV protein expression level (bottom panel, lane 3). Conversely, overexpression of RAD51AP1 resulted in a significant increase in the HCV protein levels in cells with secondary infection (Fig. 7J).
FIG 7.
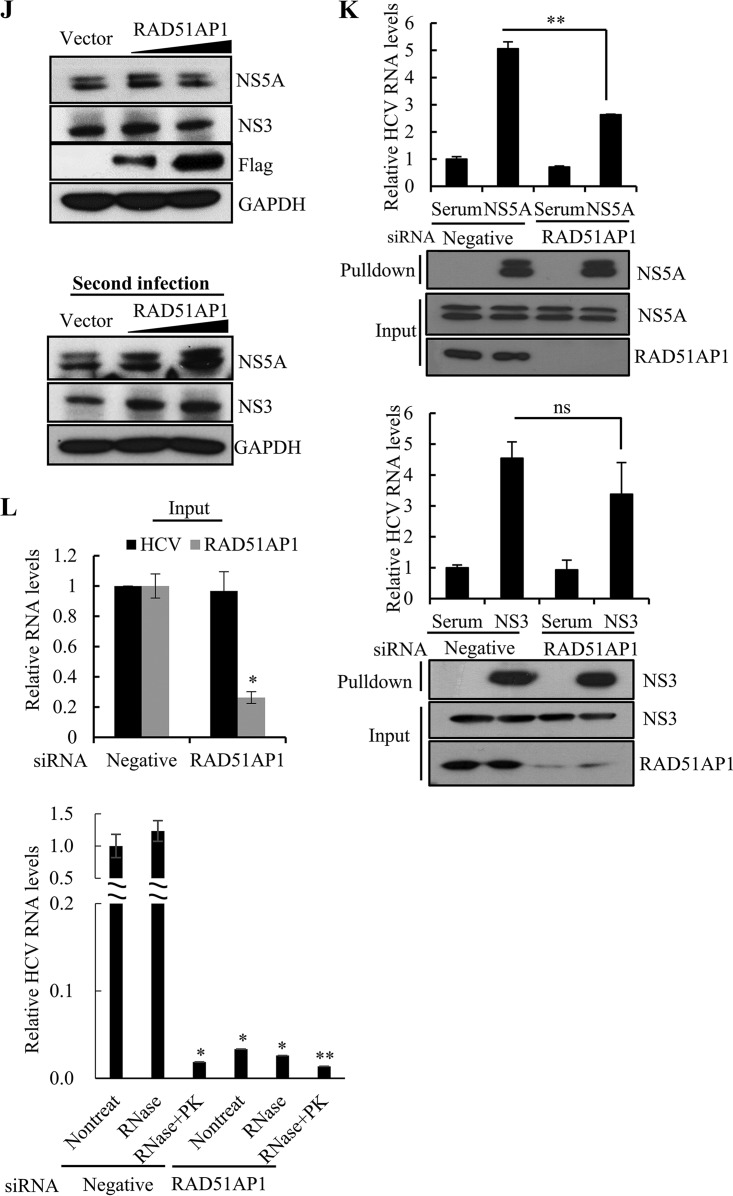
RAD51AP1 is required for the assembly step of the HCV life cycle. (A to D) Huh7.5 cells infected with Jc1 were transfected with the indicated siRNAs. At 48 h posttransfection, intracellular RNA levels (A), protein levels (B), and extracellular HCV RNA levels (C) were analyzed by qRT-PCR. (D) Naive Huh7.5 cells were infected with HCV-containing culture supernatant harvested from the cells for panel A, and viral infectivity was determined by measuring intracellular HCV RNA (left) and protein (right) levels. (E) Huh7.5 cells were transfected with the indicated siRNAs. At 96 h posttransfection, cell viability was determined by WST assay. (F) Huh7.5 cells infected with Jc1 were transfected with the indicated siRNAs. At 48 h posttransfection, cells were lysed by repetitive cycles of freezing and thawing. Intracellular infectivity was determined by limiting-dilution assay (TCID50). (G) Huh7.5 cells infected with Jc1 were transfected with the indicated siRNAs. At 48 h posttransfection, supernatants were collected and extracellular infectivity was determined by limiting-dilution assay (TCID50). (H) Percentages of intracellular and extracellular infectivity relative to the total were determined. (I) (Top) Huh7.5 cells infected with Jc1 were transfected with the indicated siRNAs. At 24 h post-siRNA transfection, cells were further transfected with either wild-type or siRNA-resistant RAD51AP1 plasmid. At 48 h post-plasmid transfection, cell lysates were immunoblotted using the indicated antibodies. (Bottom) Naive Huh7.5 cells were infected with HCV-containing culture supernatant harvested from the cells described for the top panel. At 48 h postinfection, protein expression was analyzed by immunoblot analysis using the indicated antibodies. (J) (Top) Huh7.5 cells infected with Jc1 were transfected with either empty vector or Flag-tagged RAD51AP1 plasmid. At 2 days postinfection, cell lysates were subjected to immunoblot analysis using the indicated antibodies. (Bottom) Naive Huh7.5 cells were infected with Jc1-containing culture supernatant harvested from the cells described for the top panel. At 48 h postinfection, protein expression was analyzed by immunoblot analysis using the indicated antibodies. (K) Huh7.5 cells were infected with Jc1. At 72 h postinfection, cells were transfected with either negative-control or RAD51AP1 siRNA. At 48 h posttransfection, total cell lysates were immunoprecipitated with either NS5A antibody (top panels) or NS3 antibody (bottom panels). Bound HCV RNAs were analyzed by qRT-PCR. (L) Huh7.5 cells infected with Jc1 were further transfected with the indicated siRNAs. At 48 h posttransfection, the cells were lysed by three repeated cycles of freezing and thawing and subjected to RNase protection assay. As a positive control, samples were treated with proteinase K (PK) prior to RNase treatment to remove protecting capsids. (Top) Input RNA amounts of both HCV and RAD51AP1 before RNase protection assay were quantified by qRT-PCR. (Bottom) The amounts of residual HCV RNA were analyzed by qRT-PCR. Experiments were performed in triplicate. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not significant).
Since both RAD51AP1 and NS5A bind RNA, we asked whether RAD51AP1 may modulate RNA binding to NS5A in HCV-infected cells. We showed that HCV RNA levels binding to NS5A were significantly decreased in RAD51AP1 knockdown cells (Fig. 7K, upper panel). The NS5A protein level was not altered in RAD51AP1-silenced cells, suggesting that RAD51AP1 might mediate HCV RNA-NS5A interaction. HCV NS3 is also an RNA binding protein. However, we showed that the amount of HCV RNA coprecipitated with NS3 was not significantly reduced in RAD51AP1 knockdown cells (Fig. 7K, bottom panel). To further investigate whether RAD51AP1 was involved in RNA protection, Huh7.5 cells infected with Jc1 were transfected with either negative-control or RAD51AP1-specific siRNAs. Total cell lysates harvested 48 h after transfection were either treated with RNase alone or treated with proteinase K prior to RNase treatment, and residual RNAs were quantified by quantitative reverse transcription-PCR (qRT-PCR). As shown in Fig. 7L, the majority of HCV RNAs were protected from RNase digestion in negative-control-siRNA-treated cells. Meanwhile, the majority of RNAs were degraded in the presence of RNase if samples were pretreated with proteinase K. Strikingly, no HCV RNAs were detected in RAD51AP1-silenced cells, regardless of RNase treatment. These results suggest that the viral genome may be protected by RAD51AP1 to promote viral assembly.
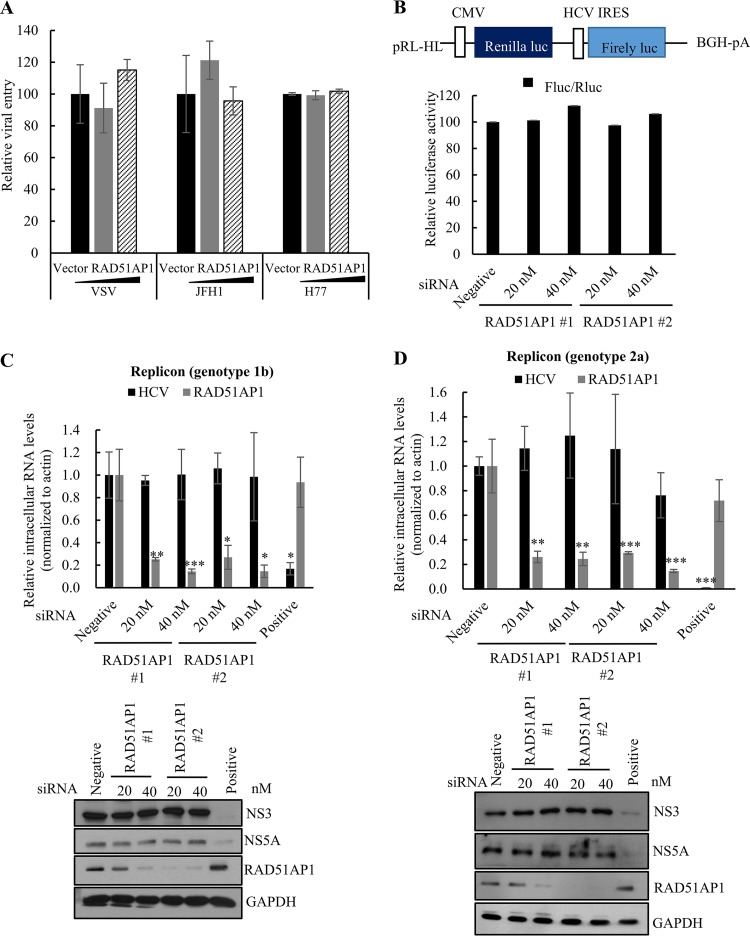
To corroborate the functional involvement of RAD51AP1 in late stages of HCV propagation, we investigated the possible role of RAD51AP1 in the early and replication steps of the HCV life cycle. We verified that RAD51AP1 was not involved in entry (Fig. 8A), internal ribosome entry site (IRES)-mediated translation (Fig. 8B), or replication (Fig. 8C and D) steps of the HCV life cycle. Collectively, these data suggest that HCV exploits host RAD51AP1 for its own propagation.
FIG 8.
RAD51AP1 is not required for the entry, translation, and replication steps of the HCV life cycle. (A) Huh7 cells were transfected with either vector or increasing amounts of Flag-tagged RAD51AP1 plasmids. At 48 h posttransfection, cells were infected with HCV pseudoparticles (HCVpp) derived from either genotype 1a (H77) or genotype 2a (JFH1). At 48 h postinfection, viral entry was determined by measuring luciferase activity. (B) (Top) Schematic diagram of the pRL-HL plasmid. (Bottom) Huh7 cells were transfected with the indicated siRNAs. At 48 h posttransfection, cells were cotransfected with the pRL-HL dual-reporter plasmid and the pCH110 β-galactosidase plasmid. At 48 h posttransfection, relative luciferase activities were determined. BGH-pA, bovine growth hormone polyadenylation signal sequence. (C and D) Huh7 cells harboring an HCV subgenomic replicon derived from genotype 1b (C) or Huh6 cells harboring an HCV subgenomic replicon derived from genotype 2a (D) were transfected with the indicated siRNAs. At 72 h post-siRNA transfection, RNA (top panels) and protein (bottom panels) levels were analyzed by qRT-PCR and immunoblot assay, respectively. Experiments were performed in triplicate. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
By employing protein array screening, we previously identified ∼90 cellular proteins as NS5A interaction partners. Among these, we delineated the roles of RAD51AP1 in HCV-infected cells. We confirmed that NS5A physically interacted with RAD51AP1 and stabilized RAD51AP1 by interrupting the ubiquitin-proteasome pathway. It was previously reported that RAD51AP1 interacts with USP1 through UAF1 and that the stability of RAD51AP1 is promoted by the USP1-UAF1 complex (15). However, the mechanisms for how NS5A competes with UAF1 and stabilizes RAD51AP1 need further study.
HCV infection directly causes DSBs by many mechanisms. HCV can stimulate the production of NO via core and NS3 and can induce ROS through core, E1, and NS3 (33). HCV core protein inhibits the formation of the DNA damage sensor protein complex Mre11-Rad50-NBS1 as well as ATM activation. Core also inhibits nonhomologous end joining and oxidatively damaged DNA repair (32). NS3/4A translocates ATM from the nucleus to the cytoplasm and delays the dephosphorylation of γH2AX, leading to the persistence of γH2AX foci (33). We also demonstrated that HCV impaired the HRR by disrupting the formation of the RAD51/RAD51AP1/UAF1 complex and translocating RAD51 to the cytoplasm (Fig. 5). This is why γH2AX foci persisted in HCV-infected cells and HCV-infected cells exhibited high sensitivity to DNA damaging agents. It was previously reported that γH2AX is elevated in cells expressing either HCV NS2 or NS3/4A (33, 34). In the present study, we showed that NS5A delayed dephosphorylation of γH2AX and impaired DNA damage repair. It is noteworthy that γH2AX foci were increased 8% in NS5A-stable cells compared to those in vector-stable cells, even in the absence of DNA damaging agents. Upon chemical treatment, NS5A not only increased the number of γH2AX foci but also increased the size and intensity of the foci. These data indicated that NS5A induced DSBs in hepatocytes.
Consistent with a previous report (15), RAD51AP1-UAF1 interaction was not altered by CPT and DOX. However, cisplatin promoted RAD51AP1-UAF1 interaction. We reasoned that these three chemicals have their own inherent functional mechanisms. CPT inhibits DNA topoisomerase I, causing positive supercoils in advance of replication forks and replication-associated DSBs (35); DOX inhibits DNA topoisomerase II, leading to DSBs (36); and cisplatin induces intrastrand and interstrand cross-links in DNA (37). Likewise, the protein level of RAD51AP1 remains stable in response to a variety of DNA damaging agents, including DOX, mitomycin C, UV light, and gamma irradiation, whereas the protein level of RAD51AP1 increases upon exposure to hydroxyurea (16). Moreover, RAD51AP1 is localized mainly in the nucleus in Ramos and Granta-519 cells, whereas both RAD51AP1 and RAD51 are localized to the nucleus and the cytoplasm in Huh7 cells (16).
The role of USP1 in the RAD51/RAD51AP1/UAF1-dependent synaptic complex has been controversial. Although RAD51AP1 interacts with USP1, USP1 plays no role in the D-loop reaction (14). Meanwhile, Cukras et al. reported that USP1 is involved in D-loop formation via RAD51 and RAD51AP1 (15). In the present study, we were unable to dissect the function of USP1 in HCV-infected cells. Nevertheless, we demonstrated the critical role of RAD51/RAD51AP1/UAF1 interaction in HRR of HCV-infected cells. Further studies are needed to determine if USP1 is involved in RAD51/RAD51AP1/UAF1-mediated HRR in HCV-infected cells.
NS5A is a multifunctional protein that functions as a key regulator of HCV RNA replication and viral assembly. HCV RNA replication occurs at endoplasmic reticulum (ER)-derived modified membranes. NS5A binds to viral RNA and colocalizes with the HCV core protein in close proximity to lipid droplets (LDs) on the virion assembly site. Therefore, NS5A may function as a transport vehicle that brings newly synthesized viral RNA from replication sites to LDs for encapsidation. Domain I of NS5A appears to play an important role in both active RNA replication and mobilization of replication complexes toward LDs for assembly. Note that RAD51AP1 interacted with NS5A through domain I and orchestrated NS5A-HCV RNA interaction. Since viral RNAs are highly susceptible to RNase attack, HCV coopts host RAD51AP1 to protect viral RNA from RNase degradation. We showed that HCV RNA levels were significantly decreased in RAD51AP1-silenced cells, suggesting that RAD51AP1 may be required for the early step in the assembly of infectious HCV particles. All these data indicate that HCV exploits host RAD51AP1 to facilitate viral propagation and thus that HRR activity is compromised in HCV-infected cells (Fig. 9). Collectively, our data provide unprecedented insights into the molecular mechanisms of HCV-induced hepatocarcinogenesis.
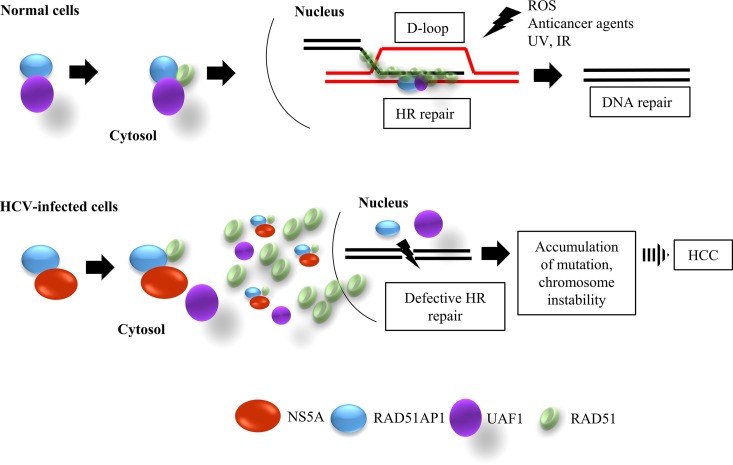
FIG 9.
Proposed model for defective DNA repair mechanism in HCV-infected cells. In normal cells, UAF1 forms a complex with RAD51AP1 and cooperates with RAD51 to promote the assembly of the synaptic complex, a critical nucleoprotein intermediate in HR repair. RAD51/RAD51AP1/UAF1 is indispensable for genome maintenance and against the DNA damage caused by exogenous agents. In HCV-infected cells, NS5A interacts with RAD51AP1, and thus RAD51AP1-UAF1 interaction is inhibited. This in turn disturbs the trimeric complex of RAD51, RAD51AP1, and UAF1 and leads to the accumulation of RAD51 in the cytoplasm. Consequently, DNA repair is compromised in HCV-infected cells. Defective DNA repair results in an increased mutation frequency and a consequent high incidence of HCC. IR, ionizing radiation.
MATERIALS AND METHODS
Cell culture.
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin in 5% CO2 at 37°C. Huh7 cells harboring a subgenomic replicon derived from genotype 1b or Huh6 cells harboring a subgenomic replicon derived from genotype 2a were grown as reported previously (38).
Chemicals.
Cisplatin was purchased from EMD Millipore. Both camptothecin and doxorubicin were purchased from Sigma-Aldrich.
RNA interference.
siRNAs targeting RAD51AP1 (RAD51AP1 1 [sense, 5′-GCA GTG TAG CCA GTG ATT A-3′; and antisense, 5′-TAA TCA CTG GCT ACA CTG C-3′] and RAD51AP1 2 [sense, 5′-TGA ACA ATC TCC GGA AAG A-3′; and antisense, 5′-TCT TTC CGG AGA TTG TTC A-3′]), siRNA targeting ApoE (sense, 5′-CTC TCA AGT GGT TAC CTG T-3′; and antisense, 5′-ACA GGT AAC CAC TTG AGA G-3′), and a universal negative-control siRNA were purchased from Bioneer. siRNA transfection was performed using the Lipofectamine RNAiMax reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A positive, HCV-specific siRNA targeted the 5′ nontranslated region (NTR) of Jc1.
Quantification of RNA.
Quantitative real-time PCR (qRT-PCR) experiments were carried out by using an iQ5 multicolor real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) as reported previously (19).
GST pulldown assay.
The glutathione S-transferase (GST)–NS5A fusion protein was expressed in Escherichia coli BL21 and purified with glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions. HEK293T cells were transfected with the Flag-tagged RAD51AP1 plasmid. At 24 h posttransfection, cells were harvested in lysis buffer. The cell lysate was centrifuged at 13,500 rpm for 15 min, and the protein concentration was determined by the Bradford assay (Bio-Rad). For the in vitro binding assay, Flag-tagged RAD51AP1 was incubated with either GST or the GST-NS5A fusion protein for 2 h at 4°C in cell lysis buffer. The samples were washed four times in lysis buffer, and bound protein was detected by immunoblot assay.
WST assay.
Cells seeded into 12-well plates were transfected with either 20 nM negative-control siRNA or RAD51AP1-specific siRNAs. Cell viability was determined by using 30 μl of water-soluble tetrazolium salt (WST) in each well, with the plate incubated for 1 h at 37°C. After plate shaking for 1 min, the aqueous layer in each well was transferred to a 96-well plate, and the absorbance at 450 nm was measured as reported previously (19).
Protein stability assay.
Huh7.5 cells were transiently transfected with the indicated plasmids. At 24 h posttransfection, cells were treated with 10 μg/ml cycloheximide (Sigma-Aldrich) for the indicated times. The resulting cell lysates were immunoblotted with the indicated antibodies to monitor protein turnover.
RNase digestion assay.
Cells were scraped into 200 μl proteinase K buffer (50 mM Tris-HCl, pH 8.0, 10 mM CaCl2, 1 mM dithiothreitol [DTT]) and subjected to five cycles of freezing and thawing. Crude cell lysates were either (i) left untreated, (ii) treated with RNase A (50 μg/ml) for 4 h at room temperature, or (iii) pretreated with proteinase K (5 μg/ml) for 1 h on ice. The reaction was stopped by the addition of 10 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail prior to RNase A digestion. Subsequently, total RNAs were extracted using RiboEx (GeneAll) according to the manufacturer's instructions, and HCV RNAs were quantified by qRT-PCR.
γH2AX focus formation assay.
Either mock- or Jc1-infected cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 10 min at 37°C. After three washes with PBS, fixed cells were blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature. The cells were then stained with an anti-phospho-histone-H2AX antibody (Trevigen Inc.) overnight at 4°C. After three washes with PBS, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG for 1 h at room temperature. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. After three washes with PBS, cells were analyzed using a Zeiss LSM 700 laser confocal microscopy system (Carl Zeiss, Inc., Thornwood, NY). The number of phosphorylated H2AX foci in 200 cells for each sample was counted manually.
Patient and mouse tissues.
Human liver tissue specimens were obtained from the Liver Cancer Specimen Bank at Yonsei University in Seoul, South Korea. All patients participating in this study gave informed consent before surgery, and the use of human tissue for this research was authorized by the Institutional Review Board of the College of Medicine at Yonsei University (39). Transgenic mice were generated as we previously reported (6). Mice were maintained in accordance with the guidelines of the Institutional Animal Care and Use Committee, Korea Research Institute of Bioscience and Biotechnology (KRIBB) (Daejeon, South Korea). Briefly, liver tissues were homogenized in RIPA buffer (50 mM Tris-HCl [pH 7.5], 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1 mM Na3VO4, and 1 mM PMSF) by use of a homogenizer. The homogenates were centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was collected, and equal amounts of protein were subjected to SDS-PAGE followed by immunoblot analysis using the indicated antibodies.
Plasmids and DNA transfection.
Total RNAs were isolated from Huh7 cells by using RiboEx (GeneAll), and full-length RAD51AP1 was amplified from cDNA synthesized by use of a cDNA synthesis kit (Toyobo) according to the manufacturer's instructions. PCR products were inserted into the corresponding enzyme sites of the plasmid pCMV10-3X Flag (Sigma-Aldrich). RAD51AP1 was subcloned into either the plasmid pEF6/V5-HisB (Invitrogen) or pcDNA 4-HisA (Invitrogen). pEF6B Myc-tagged wild-type and mutant NS5A constructs were described previously (38). All DNA transfections were performed by using polyethyleneimine reagent (Sigma-Aldrich) as we described previously (38).
Immunoprecipitation.
HEK293T cells were cotransfected with Flag-tagged RAD51AP1 and Myc-tagged NS5A plasmids. Total amounts of DNA were adjusted by adding an empty vector. At 48 h posttransfection, cell lysates were centrifuged at 13,500 rpm for 15 min. The supernatant was incubated with the indicated antibody at 4°C overnight. The samples were further incubated with 30 μl of protein A beads (Sigma-Aldrich) for 1 h. The beads were washed five times in washing buffer, and then bound protein was detected by immunoblot assay as we described previously (38).
Immunofluorescence assay.
Huh7.5 cells grown on cover slides were fixed in 4% paraformaldehyde in PBS for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 10 min at 37°C. After three washes with PBS, fixed cells were blocked with 1% BSA in PBS for 1 h at room temperature. The cells were then incubated with the indicated antibody and rabbit anti-NS5A overnight at 4°C. After three washes with PBS, cells were incubated with either fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG or tetramethylrhodamine isothiocyanate (TRITC)-conjugated donkey anti-rabbit IgG for 1 h at room temperature. Cells were counterstained with DAPI to label nuclei. After three washes with PBS, cells were analyzed using a Zeiss LSM 700 laser confocal microscopy system (Carl Zeiss, Inc., Thornwood, NY).
TCID50 assay.
A 50% tissue culture infective dose (TCID50) assay was performed to determine the infectious titer of cell culture-produced HCV. Briefly, 6.4 × 103 Huh7.5 cells/well were seeded into collagen-coated 96-well plates and incubated overnight. Cells were infected with 5-fold serial dilutions of the HCV-containing supernatants and were further cultured for 96 h. HCV-infected cells were fixed in ice-cold methanol at −20°C for 20 min. Endogenous peroxidase was quenched with 3% H2O2 in PBS for 5 min. Cells were treated with blocking buffer, followed by incubation with an anti-NS5A polyclonal antibody (1:1,000) at 4°C overnight. Cells were further incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:200) for 30 min at room temperature. Bound peroxidase was developed by use of diaminobenzidine (DAB) substrate (DAB+; Dako, USA) for 10 min. HCV NS5A-positive wells were counted under a light microscope to determine the TCID50, as reported previously (40).
Statistical analysis.
Data are presented as means ± standard deviations (SD). Student's t test was used for statistical analysis. The asterisks in the figures indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not significant).
ACKNOWLEDGMENTS
We thank Ralf Bartenschlager (University of Heidelberg) for the Jc1 construct and Charles Rice (The Rockefeller University) for Huh7.5 cells.
We have no conflicts of interest to report.
This work, including the efforts of Soon B. Hwang and Yun-Sook Lim, was funded by the Ministry of Science, ICT and Future Planning (MSIP) (grants 2016R010886 and 2017R1C1B2004989).
REFERENCES
- 1.Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, Lai MM. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci U S A 101:4262–4267. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machida K, Cheng KT, Lai CK, Jeng KS, Sung VM, Lai MM. 2006. Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J Virol 14:7199–7207. doi: 10.1128/JVI.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang YF, He B, Li NP, Ma J, Gong GZ, Zhang M. 2011. The oncogenic role of NS5A of hepatitis C virus is mediated by up-regulation of survivin gene expression in the hepatocellular cell through p53 and NF-κB pathways. Cell Biol Int 12:1225–1232. doi: 10.1042/CBI20110102. [DOI] [PubMed] [Google Scholar]
- 4.He Y, Nakao H, Tan SL, Polyak SJ, Neddermann P, Vijaysri S, Jacobs BL, Katze MG. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J Virol 76:9207–9217. doi: 10.1128/JVI.76.18.9207-9217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Street A, Macdonald A, Crowder K, Harris M. 2004. The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J Biol Chem 279:12232–12241. doi: 10.1074/jbc.M312245200. [DOI] [PubMed] [Google Scholar]
- 6.Wang AG, Lee DS, Moon HB, Kim JM, Cho KH, Choi SH, Ha HL, Han YH, Kim DG, Hwang SB, Yu DY. 2009. Non-structural 5A protein of hepatitis C virus induces a range of liver pathology in transgenic mice. J Pathol 219:253–262. doi: 10.1002/path.2592. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh AK, Steele R, Meyer K. 1999. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol 80:1179–1183. doi: 10.1099/0022-1317-80-5-1179. [DOI] [PubMed] [Google Scholar]
- 8.Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96:727–738. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 9.Richardson C. 2005. Rad51, genome instability, and tumorigenesis. Cancer Lett 218:127–139. doi: 10.1016/j.canlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Gupta RC, Bazemore LR, Golub EI, Radding CM. 1997. Activities of human recombination protein Rad51. Proc Natl Acad Sci U S A 94:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberq B, Sung P, Schild D. 2007. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell 28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. 2007. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell 28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM. 1997. A novel nucleic acid-binding protein that interacts with human Rad51 recombinase. Nucleic Acids Res 25:4946–4953. doi: 10.1093/nar/25.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang F, Longerich S, Miller A, Tang C, Buzovetsky O, Xiong Y, Maranon DG, Wiese C, Kupfer GM, Sung P. 2016. Promotion of RAD51-mediated homologous DNA pairing by the RAD51AP1-UAF1 complex. Cell Rep 15:2118–2126. doi: 10.1016/j.celrep.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cukras S, Lee E, Palumbo E, Benavidez P, Moldovan GL, Kee Y. 2016. The USP1-UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair. Cell Cycle 15:2636–2646. doi: 10.1080/15384101.2016.1209613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henson SE, Tsai SC, Malone CS, Soghomonian SV, Ouyang Y, Wall R, Marahrens Y, Teitell MA. 2006. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutat Res 601:113–124. doi: 10.1016/j.mrfmmm.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Park C, Min S, Park EM, Lim YS, Kang S, Suzuki T, Shin EC, Hwang SB. 2015. Pim kinase interacts with nonstructural 5A protein and regulates hepatitis C virus entry. J Virol 89:10073–10086. doi: 10.1128/JVI.01707-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai MS, Leung SG, Schild D, Wiese C, Sung P. 2012. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem 287:12343–12347. doi: 10.1074/jbc.C112.352161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. 2004. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol 10:509–513. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoch C, Kern W, Kohlmann A, Hiddemann W, Schnittger S, Haferlach T. 2005. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosomes Cancer 43:227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 23.Milward A, Mankouri J, Harris M. 2010. Hepatitis C virus NS5A protein interacts with beta-catenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. J Gen Virol 91:373–381. doi: 10.1099/vir.0.015305-0. [DOI] [PubMed] [Google Scholar]
- 24.Igloi Z, Kazlauskas A, Saksela K, Macdonald A, Mankouri J, Harris M. 2015. Hepatitis C virus NS5A protein blocks epidermal growth factor receptor degradation via a proline motif-dependent interaction. J Gen Virol 96:2133–2144. doi: 10.1099/vir.0.000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sianipar IR, Matsui C, Minami N, Gan X, Deng L, Hotta H, Shoji I. 2015. Physical and functional interaction between hepatitis C virus NS5A protein and ovarian tumor protein deubiquitinase 7B. Microbiol Immunol 59:466–476. doi: 10.1111/1348-0421.12278. [DOI] [PubMed] [Google Scholar]
- 26.Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gyqi SP, Elledge SJ. 2015. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell 59:867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gildemeister OS, Sage JM, Knight KL. 2009. Cellular redistribution of Rad51 in response to DNA damage: novel role of Rad51C. J Biol Chem 284:31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothkamm EP, Lobrich M. 2003. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc Natl Acad Sci U S A 100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taneja N, Davis M, Choy JS, Beckett MA, Singh R, Kron SJ, Weichselbaum RR. 2004. Histone H2AX phosphorylation as a predictor of radiosensitivity and target radiotherapy. J Biol Chem 279:2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 30.Buscemi G, Perego P, Carenini N, Nakanishi M, Chessa L, Chen J, Khanna K, Delia D. 2004. Activation of ATM and Chk2 kinases in relation to the amount of DNA strand breaks. Oncogene 23:7691–7700. doi: 10.1038/sj.onc.1207986. [DOI] [PubMed] [Google Scholar]
- 31.Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L, Ou JH, Lai MM. 2010. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol 185:6985–6998. doi: 10.4049/jimmunol.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machida K, Cheng KT, Sung VM, Lee KJ, Levine AM, Lai MM. 2004. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J Virol 78:8835–8843. doi: 10.1128/JVI.78.16.8835-8843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. 2008. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology 370:295–309. doi: 10.1016/j.virol.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 34.Bittar C, Shrivastava S, Bhanja Chowdhury J, Rahal P, Ray RB. 2013. Hepatitis C virus NS2 protein inhibits DNA damage pathway by sequestering p53 to the cytoplasm. PLoS One 8:e62581. doi: 10.1371/journal.pone.0062581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koster DA, Palle K, Bot ESM, Bjornsti MA, Dekker NH. 2007. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 36.Ganapathi RN, Ganapathi MK. 2013. Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol 4:89. doi: 10.3389/fphar.2013.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noll DM, Mason TM, Miller PS. 2006. Formation and repair of interstrand cross-links in DNA. Chem Rev 106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran GVQ, Luong TTD, Park EM, Kim JW, Choi JW, Park C, Lim YS, Hwang SB. 2016. Nonstructural 5A protein of hepatitis C virus regulates soluble resistance-related calcium-binding protein activity for viral propagation. J Virol 90:2794–2805. doi: 10.1128/JVI.02493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. 2009. Nonstructural 5A protein activates β-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol 51:853–864. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 40.Reed LJ, Muench HA. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]