Abstract
The East Asian subtropics mostly occupied by evergreen broad-leaved forests (EBLFs), is one of the global diversity centers for evergreen oaks. Evergreen oaks are keystone canopy trees in EBLFs with important ecosystem function and crucial significance for regional biodiversity conservation. However, the species composition and diversity of Asian evergreen oaks are poorly understood. Here, we test whether the four chloroplast markers atpI-atpH, matK, psbA-trnH, and ycf1, can discriminate the two evergreen oak sections in Asia – Cyclobalanopsis and Ilex. Two hundred and seventy-two individuals representing 57 species were scanned and 17 species from other oaks sections were included for phylogenetic reconstruction. The genetic diversity of the Quercus sections was also compared. Overall, we found that universal chloroplast DNA (cpDNA) barcoding markers could resolve two clades in Quercus, i.e., subgenus Cerris (Old World Clade) and subgenus Quercus (New World Clade). The chloroplast markers distinguished the main sections, with few exceptions. Each cpDNA region showed no barcoding gap and none of them provided good resolution at the species level. The best species resolution (27.78%) was obtained when three or four markers were combined and analyzed using BLAST. The high conservation of the cpDNA and complicated evolutionary patterns, due to incomplete lineage sorting, interspecific hybridization and introgressions may hinder the ability of cpDNA markers to discriminate different species. When comparing diversification pattern across Quercus sections (Cyclobalanopsis, Ilex, Cerris, Quercus, and Protobalanus), we found that section Ilex was the most genetically diverse, and section Cyclobalanopsis was lower genetically diverse. This diversification pattern may have resulted from the interplay of the Eurasia Cenozoic tectonic movements, climate changes and different niches of their ancestral lineages.
Keywords: cpDNA, DNA barcoding, genetic diversity, Quercus, section Cyclobalanopsis, section Ilex
Introduction
Understanding the biodiversity of ecosystems is critical for revealing the biome assembly and its function in different ecosystems (Hebert et al., 2003; Lahaye et al., 2008; Pitman et al., 2008). Taxonomy based on morphological features has enriched our understanding of biodiversity for over 300 years, and provide an inventory of global biodiversity. However, the morphology based taxonomy system has shortcomings (Hebert et al., 2003; Vilgalys, 2003; Gonzalez et al., 2009; Goldstein and DeSalle, 2011). For example, the key diagnostic traits may be under selection or subjected to parallel evolution providing false information on the identity of the taxa. Complementing techniques such as DNA sequences under neutral evolution have become an efficient way to identify species (Hollingsworth, 2007; CBOL Plant Working Group, 2009; Valentini et al., 2009; Hollingsworth et al., 2016). DNA barcoding has become instrumental in plant research for identification of cryptic species (Bickford et al., 2007; Miwa et al., 2009; Liu et al., 2011), biodiversity assessments (Lahaye et al., 2008; Gonzalez et al., 2009; von Cräutlein et al., 2011; Yan et al., 2015), community phylogeny (Kress et al., 2009, 2010), conservation biology (Laiou et al., 2013; Veldman et al., 2014; Liu et al., 2015; Shapcott et al., 2015), invasive biology (Armstrong and Ball, 2005; Bleeker et al., 2008; Newmaster and Ragupathy, 2009; Van De Wiel et al., 2009), and disease and pest management (Ball and Armstrong, 2006; Lee et al., 2012; Montecchio and Faccoli, 2014; Shapcott et al., 2015). At present, the resolution of several candidate DNA barcodes has been tested in many plant groups (Clerc-Blain et al., 2010; Gao et al., 2010; Liu et al., 2011; Yu et al., 2011; Wang D.Y. et al., 2017). However, there is a challenge for DNA barcoding to discriminate closely related species, especially for those with shallow phylogeny and complex evolutionary history (Newmaster et al., 2008; Chen et al., 2015; Yan et al., 2015), such as species-rich genera, Quercus.
The genus Quercus (Fagaceae) has ca. 400–500 species, which are widely distributed in the warm temperate forests of the Northern Hemisphere (Camus, 1936–1954; Nixon, 1993). Many species of the genus are the canopy or dominant trees in regional EBLFs and play an important role in the function and service of ecosystem (Fang and Yoda, 1991; Nixon, 2006; Petit et al., 2013). The species identification and taxonomy in oaks are notoriously difficult (Manos et al., 1999; Simeone et al., 2013; Hubert et al., 2014), especially for Asian evergreen oaks that are composed of two species-rich sections (Cyclobalanopsis and Ilex) (Denk and Grimm, 2010; Denk et al., 2017). Species of section Cyclobalanopsis are the dominant trees in Asian (sub)tropical EBLFs with ca. 90–120 species (Denk and Grimm, 2010; Denk et al., 2017). Species of section Ilex are widely distributed in Eurasian low-middle latitude habitats comprising ca. 35–36 species (Denk and Grimm, 2010; Simeone et al., 2016). Unfortunately, the wide range of EBLFs in East Asia is now greatly diminished as a result of intensified human activities (Cao and Zhang, 1997; Tang, 2010). At least 1/3 oak species in China are now endangered or threatened due to the severe habitats loss and human activities (Deng et al., 2013a). Therefore, understanding the species composition of East Asian evergreen oak forests is important for future conservation efforts. Although the Flora of China and other regional floristic works had been finished for decades (e.g., Huang et al., 1999; Phengklai et al., 2008), the oak taxonomy and their systematic placement are still not well resolved, in part due to the high level of intra- and inter-species genetic variation. Even the fine anatomy features of the East Asian evergreen oaks, such as leaf epidermal features (Deng et al., 2014, 2017b), pollen morphology (Denk and Grimm, 2009; Deng et al., 2013b; Denk and Tekleva, 2014), and wood anatomy (Zhao et al., 2007) had been comprehensively studied in recent years. But these works do not provide useful diagnostic features to clarify species identity, and suggest paraphyletic evolution of taxonomical traits. Moreover, the interspecies gene flow has blunted genetic integrity and reduced the ability to distinguish some East Asian evergreen oaks (Tamaki and Okada, 2014; An et al., 2017).
Chloroplast DNA sequences have been applied for phylogenetic analysis and DNA barcoding of species of section Ilex in the Mediterranean (Piredda et al., 2011; Simeone et al., 2013) and China (Yang et al., 2017), providing insights into the evolutionary history of oak taxa. However, these studies included limited taxa and geographic regions (Europe or small region of China), therefore did not provide enough information on DNA barcoding for evergreen oaks. For the species-rich section Cyclobalanopsis in East Asia, little is known about evolution of chloroplast genome and phylogeny. So far, only the intergenic region of the chloroplast trnT-trnL was applied to distinguish six species of section Cyclobalanopsis distributed in Japan, and proved to have moderate efficiency (Ohyama et al., 2001). Recent studies using cpDNA show that the species of section Cyclobalanopsis, e.g., Q. glauca (Xu et al., 2015), Q. schottkyana (Jiang et al., 2016), Q. arbutifolia (Xu et al., 2016), and Q. kerrii (Jiang et al., 2018), contain high variable regions to infer population history, but the resolution of cpDNA markers in discriminating section Cyclobalanopsis species is still unknown.
Evergreen broad-leaved forests potentially cover a wide zone of monsoon-dominated regions in East Asia (Fang and Yoda, 1991). This region, as a global biodiversity hotspot, has the world’s richest flora harboring remarkable array of endemic, relic and endangered species (Myers et al., 2000; Yang et al., 2004; López-Pujol et al., 2006), especially with many endemic and endangered oak species (Menitsky, 1984; Luo and Zhou, 2000; Deng et al., 2013a). Effective DNA barcodes for species identification, conservation and resource utilization are extremely necessary in East Asian evergreen oaks. A further DNA barcoding study with comprehensive sampling on species population from the main distribution ranges is needed. In addition, comparing of the genetic diversity patterns across the main lineages in Quercus can provide more information to infer the driving forces which might contribute to their divergence. Furthermore, the knowledge on biodiversity inventory in East Asian subtropics is far from enough, and new species are continuously being identified in recent years (e.g., Xiao et al., 2015; Liang et al., 2016; Jiang et al., 2017; Wang C.W. et al., 2017). DNA barcoding of keystone species in this region can provide crucial insight into the mechanism of plant community assembly and evolutionary trajectory of the East Asian regional biota.
In this study, we comprehensively collected East Asian evergreen oaks aiming to address the following issues: (1) to reveal the discrimination ability of cpDNA barcode markers in East Asian evergreen oaks; (2) to compare the genetic diversity of the main sections in Quercus; (3) to explore the possible applications of cpDNA markers into the taxonomic and phylogenetic approaches. Our study provides insights into the species identity of Asian oaks and important information for biogeographic and population genetics of these unique oaks.
Materials and Methods
Ethics Statement
Sampling of oak species and other Fagaceae plants were granted and supported by National Forestry Bureau of China and Local National Nature Reserves.
Plant Materials
One hundred and forty-seven individuals belonging to 29 species of Quercus section Cyclobalanopsis from East Asia and 125 individuals of the 28 species of section Ilex from East Asia were included in our studies. The sampling range covered the key distribution range of East Asian evergreen oaks in China, Japan, Vietnam, and Nepal. Additionally, individuals of five species of section Ilex from the Mediterranean were analyzed. The final dataset also included four species of section Cerris (18 individuals, from Eurasia), seven species of section Quercus (11 individuals, from North America and Eurasia), five species of the American section Lobatae (7 individuals), and one species (1 individual) of section Protobalanus from western North America. One individual of Lithocarpus henryi was used as outgroup to root the tree of genus Quercus. Sequences of eight species (16 individuals) were downloaded from GenBank. Detailed information of samples used in this study is listed in Supplementary Table S1.
Healthy leaves of each individual were collected and dried instantly in silica gel for DNA extraction. Voucher specimens of each individual were deposited in the Herbarium of the Shanghai Chenshan Botanical Garden (CSH).
DNA Extraction, PCR, and Sequencing Protocols
Genomic DNA was extracted using the modified CTAB method (Doyle and Doyle, 1987). Four cpDNA regions, the psbA-trnH intergenic spacer, a part of the matK gene, the atpI-atpH intergenic spacer and a portion of the ycf1 region were amplified using PCR and bidirectionally sequenced for analyses. PCR reactions were performed according to previously described method (Xu et al., 2015). The primer sequences used to amplify these regions are summarized in Supplementary Table S2. The sequences obtained in this study have been uploaded to GenBank (accession numbers: MH058100-MH059477).
Sequencher 4.01 (Gene Codes Corp., Ann Arbor, MI, United States) was used to assemble and edit sequences. The DNA sequences were aligned using the package Muscle (Edgar, 2004) implemented in MEGA 7.0.211 (Kumar et al., 2016) with subsequent manual adjustment.
DNA Barcoding Analyses
DNA polymorphisms were examined using DnaSP 5.10 (Librado and Rozas, 2009). To evaluate species discrimination success, four widely used methods, genetic distance-based, similarity-based, tree-based and diagnostic method, were applied to the four cpDNA markers and all their possible combinations.
Genetic distance-based method: p-distances of the four plastid regions were calculated in MEGA 7.0.21 (Kumar et al., 2016) (see footnote 1). We measured three parameters to determine intraspecific variation (Meyer and Paulay, 2005; Lahaye et al., 2008). First, we calculated average intraspecific difference between all samples collected. Second, we measured theta (𝜃), which means the average p-distance within each species. Theta eliminates biases resulted by unequal sampling between species. Lastly, we calculated average coalescent depth, which is the maximum intraspecific distance within each species. Average interspecific distance and smallest interspecific distance were used to characterize interspecific divergence (Meier et al., 2008). The distribution of intraspecific versus interspecific variability was compared using DNA barcoding gaps. Differentiations between the intraspecific and interspecific p-distance for each candidate barcode were also compared using the Wilcoxon signed rank tests in R 3.2.3 (R Core Team, 2015). To explore potential species groups, the obtained p-distance matrices for each barcode candidate and all combinations were analyzed using the Automatic Barcode Gap Discovery (ABGD) under default setting (Puillandre et al., 2012). If conspecific individuals were partitioned into the same group without sequences from other species, the species was considered as successfully delimited.
Similarity-based method: Sequences of the four candidate barcodes and all possible combinations were built as 15 local reference databases using NCBI-blast-2.6.0+ (Camacho et al., 2009; Tao, 2010). Each barcode sequence was then queried using the blastn command against its local reference database. When all individuals of a species had a top hit to conspecific individuals, species discrimination was considered successful.
Tree-based method: Bayesian trees were constructed using MrBayes 3.2.6 (Ronquist et al., 2012). Model of substitution was selected by Modeltest 3.7 (Posada and Crandall, 1998) based on the akaike information criterion (AIC). Two parallel Markov Chain Monte Carlo (MCMC) runs were performed for 20 million generations. The trees were sampled every 1,000 generations and inspected in Tracer v1.62. The first 15% trees were discarded as burn-in. Species discrimination was considered as successful only when all the conspecific individuals formed a monophyletic clade.
Diagnostic method: BLOG 2.0 (Barcoding with LOGic) was used for species identification under a Logic Mining method, which identifies the species in terms of location of key diagnostic nucleotides from the training sequences to classify species (Weitschek et al., 2013). Each candidate barcode and combination were tested with a single file input, with 80% of the sequences as the training data, and 20% as the test data. A species was considered as successfully delimited if conspecific individuals were correctly identified in both training and test datasets.
Phylogeographical and Genetic Diversity Analysis
Haplotypes were extracted by DnaSP 5.10 (Librado and Rozas, 2009). To measure the level of genetic variation, variable sites, average pairwise differences per base pair between sequence (nucleotide diversity) (Nei and Li, 1979) and haplotype diversity (Hd) were calculated using DnaSP 5.10 (Librado and Rozas, 2009). The genetic distances (p-distances) of each section were calculated in MEGA 7.0.21 (Kumar et al., 2016) (see footnote 1) for further comparison of genetic diversity. A principal coordinate analysis (PCoA) was performed using GenAlEx 6.5 (Peakall and Smouse, 2012) to illustrate both the similarity between different sections and the genetic diversity of each section.
Results
PCR Success and Sequence Characteristics
The four chloroplast candidate barcodes showed high amplification success rates (100%) at the species level. At individual level, matK, psbA-trnH, and ycf1 had high success rates (92.2–100%) (Table 1), while the atpI-atpH had the lowest success rate, potentially due to the long PCR product that was being amplified.
Table 1.
Characteristics of the four cpDNA markers used in this study.
| Vs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Barcode | N | Ns | PCR | L | Number | % | Indel | Pi | Hn | Hd |
| atpI-atpH | 345 | 72 | 92.2 | 1,187 | 128 | 10.78 | 99 | 0.00448 | 77 | 0.851 |
| matK | 374 | 74 | 100 | 691 | 64 | 9.26 | 3 | 0.00467 | 64 | 0.819 |
| psbA-trnH | 357 | 74 | 95.5 | 554 | 64 | 11.55 | 26 | 0.02012 | 68 | 0.905 |
| ycf1 | 363 | 74 | 97.1 | 796 | 90 | 11.31 | 43 | 0.00954 | 66 | 0.827 |
N, number of samples; Ns, number of species; PCR success (%); L, aligned length; Vs, variable sites; Pi, nucleotide diversity; Hn, number of haplotypes; Hd, haplotype diversity.
A total of 1,439 sequences were available for further analysis, of which 1,375 were newly generated in this study. Among the four cpDNA markers, the aligned lengths ranged from 554 bp for the matK to 1,187 bp for the atpI-atpH. The atpI-atpH region showed the highest number of variable sites, while the matK and psbA-trnH had the lowest variable sites (Table 1). The atpI-atpH region also had the highest number of indels (99), followed by ycf1 (43), psbA-trnH (26) and matK (3). Among the four chloroplast loci, psbA-trnH was the highest haplotype diversity, followed by atpI-atpH, ycf1 and matK (in descending order of Hd).
Genetic Divergence Within and Between Species
PsbA-trnH exhibited the highest level of intra-species variation, followed by ycf1, atpI-atpH and matK. When comparing the interspecific genetic divergence among the four candidate barcodes, psbA-trnH region exhibited the highest interspecific divergence, followed by ycf1, atpI-atpH, and matK (Table 1). This analysis demonstrated that psbA-trnH sequences provide the most suitable DNA barcodes.
DNA Barcoding Gap Assessment
The distribution of intraspecific and interspecific variation of four single markers lacked a distinct gap. Among the single locus, psbA-trnH had the highest variation between the distribution range of interspecific and intraspecific distances (Figure 1). Wilcoxon rank sum tests show that the intraspecific distances were always significantly lower than the interspecific distances (Table 2). However, the maximum intraspecific distances of four loci were higher than the minimum interspecific distance (Table 2).
FIGURE 1.
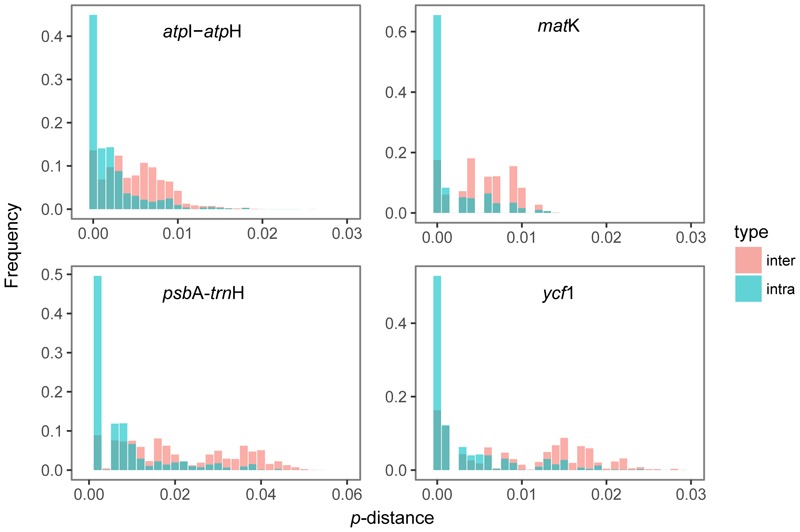
Relative distribution of intra- and inter-specific divergence of the four cpDNA markers.
Table 2.
Comparison of inter- and intraspecific p-distance of the four chloroplast markers and Wilcoxon rank sum test between inter- and intraspecific p-distance.
| Barcode | Intraspecific distance (mean) | Theta (mean) | Coalescent depth (mean) | Interspecific distance (mean) | Wilcoxon rank sum test |
|
|---|---|---|---|---|---|---|
| W statistic | p-value | |||||
| atpI-atpH | 0–0.0180 (0.0020) | 0–0.0097 (0.0023) | 0–0.0180 (0.0046) | 0–0.0260 (0.0048) | 15488000 | <0.001 |
| matK | 0–0.0130 (0.0017) | 0–0.0100 (0.0022) | 0–0.0130 (0.0039) | 0–0.0140 (0.0052) | 20437000 | <0.001 |
| psbA-trnH | 0–0.0430 (0.0069) | 0–0.0380 (0.0082) | 0–0.0430 (0.0145) | 0–0.0530 (0.0201) | 14029000 | <0.001 |
| ycf1 | 0–0.0240 (0.0030) | 0–0.0153 (0.0036) | 0–0.0240 (0.0072) | 0–0.0300 (0.0095) | 18544000 | <0.001 |
Species Identification Efficiency of Chloroplast Loci
Rates of species resolution depended on the analytic methods used (Figure 2 and Supplementary Table S3). Overall, BLAST provided the highest species discrimination rates (6.76–27.78%) among single marker or combined markers, followed by distance-based method (6.76–21.62%) and tree-based (0–20.27%) method. BLOG provided the lowest species resolution (1.35–12.16%) overall (Figure 2 and Supplementary Table S3). The diagnostic nucleotide of the highest species identification identified by BLOG is listed in Supplementary Table S4.
FIGURE 2.
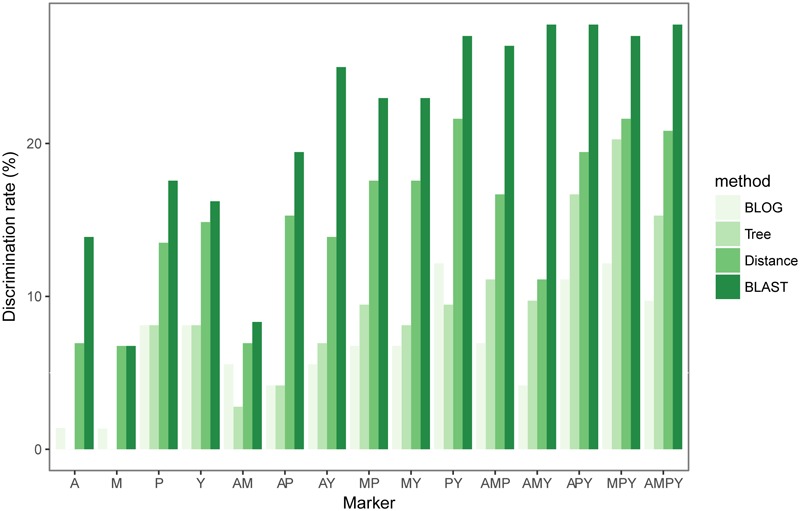
Species resolution of the four cpDNA markers and all combinations based on different methods.
Among the four single markers, psbA-trnH region showed the highest species discrimination rate for all methods except the distance-based method. When distance-based method was used, ycf1 exhibited the highest species discrimination rate (Figure 2 and Supplementary Table S3). In general, combinations with more loci could increase the probability that a species is identified correctly. Using three different combinations (atpI-atpH + matK + psbA-trnH, atpI-atpH + matK + ycf1 and atpI-atpH + matK + psbA-trnH + ycf1) with BLAST, they all generated the highest species identification rate (27.78%) (Figure 2 and Supplementary Table S3).
Phylogenetic Relationship
Subgenus Quercus, also known as the New World Clade that includes sections Lobatae, Protobalanus, and Quercus, formed a monophyletic clade (posterior probability = 1.00) (Figure 3). Section Lobatae was the only monophyletic clade (posterior probability = 1.00) inferred in this study. Two lineages were resolved in section Quercus, one with Eurasian species and the other one with the two American species (Figure 3). Section Protobalanus (Q. chrysolepis) was the sister group to the American lineage of section Quercus (Figure 3). However, we did not find strong support for the monophyletic status of subgenus Cerris (the Old World Clade). Species in section Cerris almost formed an monophyletic lineage, except for two individuals. The Mediterranean Q. suber mixed with species of section Ilex and an individual of East Asian Q. variabilis nested in a sister lineage to a section Ilex-Cyclobalanopsis mixed lineage (Figure 3). We also inferred one main clade and 11 small clades in section Cyclobalanopsis. However, we did not find strong support for the monophyletic status of section Cyclobalanopsis (Figure 3). Species within the 11 small Cyclobalanopsis clades were mixed with section Ilex species. In turn, the cpDNA based phylogenetic reconstruction inferred that section Ilex was polyphyletic and its evolutionary history appeared rather complicated compared to the other sections in Quercus (Figure 3).
FIGURE 3.
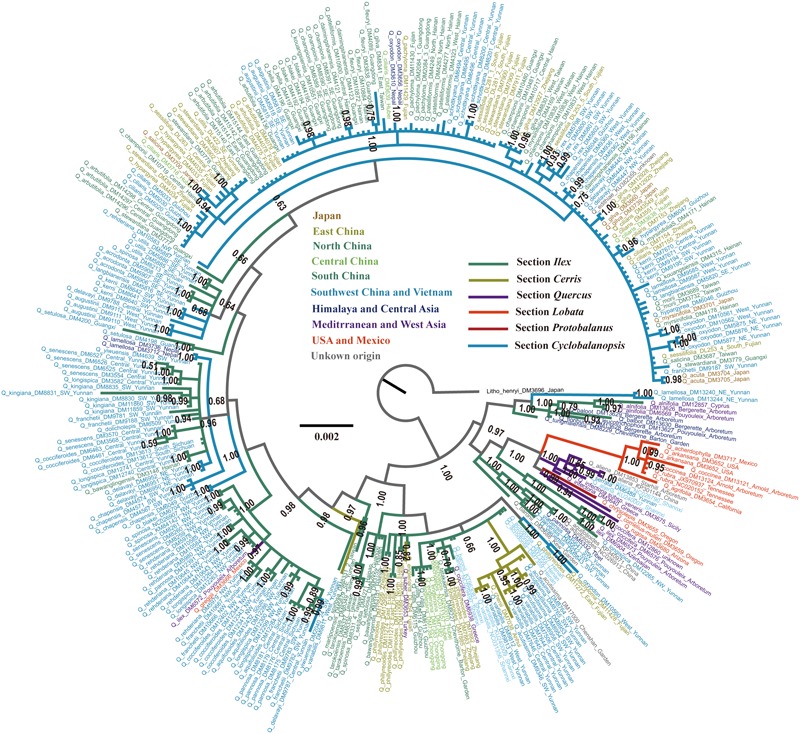
BI phylogram of Quercus inferred using combined cpDNA sequences. The tree was rooted using Lithocarpus henryi as the outgroup. Numbers close to the nodes are posterior probability values. Branch colors indicate different sections and colors of individuals indicate their localities with detail indication in the center of this figure.
Similar to the phylogenetic reconstruction results, PCoA using genetic distance among all the six sections (Figure 4) showed that section Cerris is closely to a group of East Asian subtropical species, including Q. acrodonta, Q. baronii, Q. phillyreoides, Q. dolicholepis, and Q. engleriana. Species of section Quercus from Oregon showed close similarity to section Lobatae. The major clade of section Cyclobalanopsis was relatively isolated, with only a few samples from Yunnan, SW China, are closely related to the individuals of section Ilex from the same region.
FIGURE 4.
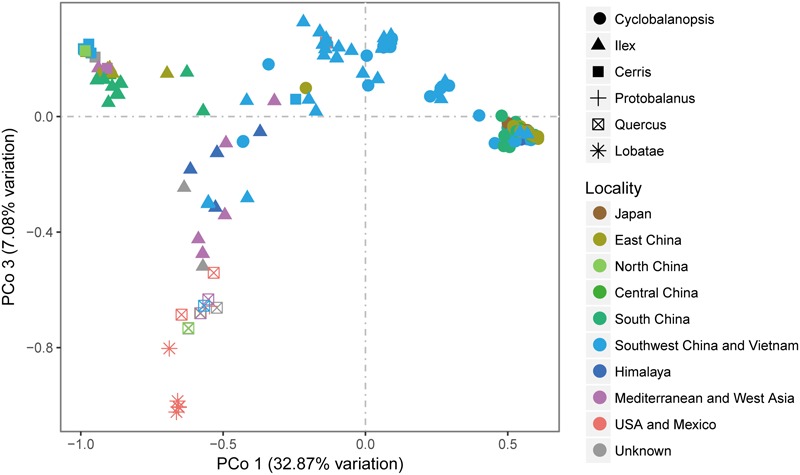
PCoA analysis of Quercus species based on the genetic distance obtained from combined cpDNA sequences.
Genetic Diversity Pattern
Section Ilex showed the highest diversity values for all the parameters (Table 3 and Figure 5), and formed three clusters in different quadrants on the PCoA plot (Figure 4). This suggests that section Ilex was the most diverse section in Quercus. Section Quercus was the second diverse section, with the second highest p-distance and haplotype diversity even only having a small number of samples (Table 3 and Figure 5). However, section Cyclobalanopsis with both high number of species and population sampling, yielded low genetic diversity levels, as the p-distance concentrated on low values (Figure 5) and the majority of individuals formed a small cluster in the PCoA analysis (Figure 5). Sections Cerris and Lobatae exhibited low genetic diversity as well (Table 3 and Figure 5). Nonetheless, we could not rule out the possibility that the low diversity might be caused by limited sampling, as we only covered four species and 18 individuals of section Cerris and five species (seven individuals) of section Lobatae.
Table 3.
Genetic diversity obtained from the combined cpDNA sequences.
| Dataset (section) | N | Ns | p-distance | Vs |
Pi | Hn | Hd | Indel | |
|---|---|---|---|---|---|---|---|---|---|
| Number | % | ||||||||
| Cyclobalanopsis | 147 | 29 | 0–0.019 (0.0020 ± 0.0031) | 134 | 4.19 | 0.00205 | 54 | 0.837 | 55 |
| Ilex | 125 | 28 | 0–0.018 (0.0090 ± 0.0041) | 220 | 6.88 | 0.00867 | 77 | 0.989 | 70 |
| Cerris | 18 | 4 | 0–0.011 (0.0025 ± 0.0026) | 51 | 1.59 | 0.00257 | 9 | 0.895 | 32 |
| Quercus | 11 | 7 | 0–0.013 (0.0059 ± 0.0045) | 67 | 2.10 | 0.00588 | 9 | 0.964 | 25 |
| Lobatae | 7 | 5 | 0–0.006 (0.0017 ± 0.0015) | 17 | 0.53 | 0.00867 | 4 | 0.714 | 4 |
| Protobalanus | 1 | 1 | – | – | – | – | – | – | – |
N, number of samples; Ns, number of species; p-distance [min. – max. (mean ± SD)]; Vs, variable sites; Pi, nucleotide diversity; Hn, number of haplotypes; Hd, haplotype diversity.
FIGURE 5.
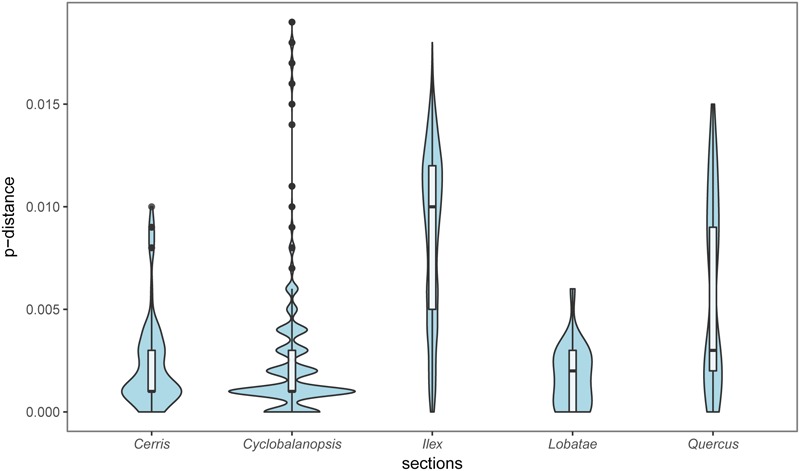
Genetic divergence of the five sections of genus Quercus based on the combined cpDNA sequences.
Discussion
The Application of cpDNA Markers on Oak Taxonomy and Systematics
We were able to successfully amplify and sequence the four cpDNA markers at high success rates (over 90%). There were differences in discrimination ability among each cpDNA marker. PsbA-trnH and ycf1 had the highest discrimination ability whereas matK had the lowest. The chloroplast region psbA-trnH had the highest discriminatory power in other plant groups and community phylogeny (Kress et al., 2010; Hollingsworth et al., 2011). Three different combinations (atpI-atpH + matK + psbA-trnH, atpI-atpH + matK + ycf1 and atpI-atpH + matK + psbA-trnH + ycf1) generated the highest species identification rate (27.78%) when used BLAST. Unfortunately, the overall identification success rates are not ideal and even the highest species resolution was below 30%. Meanwhile, none of the four chloroplast regions showed a barcode gap, which is an essential evaluation index for DNA barcodes (Hebert et al., 2003; Meier et al., 2006, 2008). The poor species resolution of universal chloroplast markers has been reported in East Asian oaks (Ohyama et al., 2001; Dong et al., 2015; Yang et al., 2017) and Euro-Mediterranean oaks (Piredda et al., 2011; Simeone et al., 2013), suggesting that these cpDNAs may not be ideal for resolving species identity in closely related oaks.
However, the universal cpDNA markers can provide information on resolving main infrageneric groups in oaks. First, two large clades of oaks, subgenera Cerris and Quercus could be distinguished when using the concatenated marker. Second, section Cerris is almost recognizable among the East Asian oaks, because it formed a monophyletic lineage with only two exceptional individuals falling into different groups. Third, most individuals of section Cyclobalanopsis (84.4%) formed a large lineage that was easily distinguished from section Ilex. Few individuals of section Ilex showed close relationship with few species in section Cyclobalanopsis from SW China. In addition to the concentric rings and scales on the cupule wall, section Cyclobalanopsis and Ilex have distinct morphological traits, such as leaf architecture structures (Zhou et al., 1995; Luo and Zhou, 2002), leaf blade shape and margin teeth (Menitsky, 1984; Huang et al., 1999) that can easily be used to distinguish the two sections.
Chloroplast markers are effective barcodes in discriminating many plant species, such as Myristicaceae (Newmaster et al., 2008), Pedicularis (Yu et al., 2011), Asteraceae (Gao et al., 2010), Carex and Kobresia (Clerc-Blain et al., 2010). However, they have limitations when discriminating closely related species, which is quite common, not only in oaks but also in other plant taxa, such as Salix (Percy et al., 2014), Rhododendron (Yan et al., 2015), Viburnum (Clement and Donoghue, 2012), and Curcuma (Chen et al., 2015). In this study, various cpDNA haplotypes exist within a species and some of these haplotypes are shared interspecifically. Recent phylogeographic studies of the section Ilex from the Himalayas (Feng et al., 2016; Meng et al., 2017) and the pan-Mediterranean (Simeone et al., 2016; Vitelli et al., 2017) also showed similar scenario. Moreover, even using the complete plastid genome sequences for phylogenetic reconstruction, the monophyletic status of species was still rare in section Quercus (Pham et al., 2017). The chloroplast genome may not be an efficient marker for fine-scale DNA barcoding nor for inferring systematics of closely related oak species (Pham et al., 2017). Therefore, either subsets of chloroplast barcode sequences, or the complete chloroplast genome are unlikely to provide confident identification of the source species. All these facts restrict the application of cpDNA sequences on species identification in closely related oaks.
Aside from cpDNA markers, ITS (incl. ITS1, 5.8S, ITS2) has been used as a universal DNA barcode marker for species-level phylogenetic studies (Kress et al., 2005; Hughes et al., 2006). The nrDNA ITS and 5S-IGS have been applied to infer oak phylogeny, which roughly illustrate the phylogeny skeleton of the main lineages in Quercus, however, the deep node phylogeny among sections are not resolved (Manos et al., 1999; Denk and Grimm, 2010). Indeed, orthologous sequences of nrDNA has better resolution than cpDNA at low taxonomical level to infer the phylogeny of oaks due to its biparental inheritance (Simeone et al., 2013). Nonetheless, nrDNA in oaks has paralogous copies and pseudogenes (Mayol and Rosselló, 2001; Bellarosa et al., 2005; Ma, 2006). Incorporating ITS paralogs in plant evolutionary studies is very risky because it can distort the phylogenetic signal (Mayol and Rosselló, 2001; Ma and Zhou, 2005; Feliner and Rosselló, 2007). To avoid incorporating ITS paralogs, PCR products need to be cloned into a vector for sequencing, which is time consuming and costly. Therefore, routine applicability of the ITS for barcoding is difficult.
Even single copy nuclear genes or a small batch of nuclear sequences may be not ideal barcoding makers for precise species identification in oaks, due to limited numbers of informative SNPs (Oh and Manos, 2008; Hubert et al., 2014). Recently, high-throughput markers, such as RAD-seq (Hipp et al., 2014; Cavender-Bares et al., 2015; Deng et al., 2017a,c) have demonstrated their power to successfully resolve the oak phylogeny with rich species and complex evolutionary history. Genome skimming also has great advantages for extending the plant barcode (Li et al., 2015; Coissac et al., 2016; Hollingsworth et al., 2016), allowing for the near-complete assembly of high-copy plastids, mitochondria and ribosomal DNA and possibly a fragmented nuclear genome assembly (Hollingsworth et al., 2016). These advanced technologies offer a promising future for DNA barcoding in oaks and probably other taxonomical difficult groups.
Performance of Different Barcoding Analysis Methods
The different performances of the four methods may reflect the analytical theories implemented in each method. BLAST had the best performance, providing the highest species resolution for all single markers and combinations. BLAST calculates the similarity of sequences, and assume that individuals from a species will be more similar to those from other species (van Velzen et al., 2012). This method also yields higher identification rates in other barcoding studies in Quercus, Curcuma, Rhododendron, and bryophytes (Hassel et al., 2013; Chen et al., 2015; Yan et al., 2015; Yang et al., 2017). The distance-based method (ABGD) automatically finds the distance where the barcode gap is located to partition the data set into candidate species, and can be used even when the intra- and inter-species distances overlap (Puillandre et al., 2012). This explained though intra- and inter-species distances overlap in our dataset, which adds difficulty to discriminate species. Nevertheless, ABGD still provided a relatively high species discrimination rate. Conversely, the tree-based methods and diagnostic methods generate low species identification rate. The tree-based approach is depended on monophyletic status of species. However, we only found few species were resolved as monophyletic clades and successfully identified. The poor resolution of tree-based method is in concordance with other studies (Meier et al., 2006; Little and Stevenson, 2007; Virgilio et al., 2010; Little, 2011). It is worth noting that BLOG has been reported to generate the highest sequence identification success in oaks and other plants (van Velzen et al., 2012; Weitschek et al., 2013; Yang et al., 2017). Actually, in our study BLOG also provides the highest individual identification success (36.23%), but it demonstrated the lowest species identification success, which requires correct identification of all individuals of a species in both training and test sequences. BLOG identifies potential diagnostic nucleotide positions from training sequences and assigns species using logic formulas based on the species-specific (diagnostic) codes (Weitschek et al., 2013). However, the diagnostic codes based on training sequences may not always cover the total variance of all sequences. Meanwhile, the total sequences are divided into training and test datasets in BLOG, as a result there are fewer samples (only one or two) in each dataset. Fewer samples may increase the discrimination rate and lead to a superficially high success rate. Hence, the high success rate produced by BLOG should be checked carefully, whether a species is correctly identified in both training and test dataset.
The Factors That Might Contributed to the Low Resolution of cpDNA in East Asian Evergreen Oaks
Conservation of the Chloroplast Genome
The poor species resolution of the four universal markers might be caused by multiple factors. Low mutation rates of chloroplast genome might lead to the poor resolution among different species. Actually, cpDNA has a low mutation rate, and much lower comparing to nrDNA (Lynch, 1997; Drouin et al., 2008). In theory, taxa with shorter generation time is likely to accumulate more mutations than long-lived species (Martin and Palumbi, 1993; Gaut et al., 1996). For woody plants with long reproductive cycle, their cpDNA mutation rates are much lower than the herbaceous species (Gaut et al., 1996). Generally, oaks have long generation time and oak seedlings need 3–45 years to produce their first acorns (Ducousso et al., 1993; Guyette et al., 2004; Kormanik et al., 2004). Consistent with the theory, the cpDNA substitution rates of oaks (0.19–0.96 × 10-9 s/s/y) (Chen et al., 2012; Xu et al., 2015, 2016; Feng et al., unpublished) are very low, below the average values reported for non-coding regions in other angiosperm lineages (1.2–1.7 × 10-9 s/s/y) (Graur and Li, 2000). Similarly, the low mutation rate of chloroplast genome is also reported in other woody species (Qi et al., 2012; Guo et al., 2014). The low substitution rate of cpDNA of oaks resulted in fewer mutations between different individuals, which partly explains the low species resolution of cpDNA in oaks. However, the low resolution may also be due to selective sweeps in chloroplast genome and past genetic drift (Ellstrand and Elam, 1993; Muir and Filatov, 2007; Percy et al., 2014). Because of the lower effective population size compared to nuclear genome, chloroplast genome may be greatly affected by genetic drift, which reduces the genetic diversity (Birky, 1988; Ellstrand and Elam, 1993; Schaal et al., 1998). As the result, chloroplast genome demonstrates low genetic diversity.
Complicated cpDNA Evolutionary History Involved Introgression and Incomplete Lineage Sorting
Chloroplast is maternally inherited in oaks (Dumolin et al., 1995). As a result, cpDNA is easy to transfer during the hybridization process, especially with asymmetric gene flow in the parental populations. Therefore, the cpDNA phylogeny only reflects the maternal evolutionary history. Oaks are notorious for interspecific hybridization, especially among the species in the same section (Curtu et al., 2007; Neophytou et al., 2011b; Moran et al., 2012; Leroy et al., 2017; McVay et al., 2017). Natural hybridization in East Asian evergreen oaks has been revealed and well demonstrated among sympatric species. In section Cyclobalanopsis, introgression between sympatric species (Q. sessilifolia and Q. acuta distributed in Korea and Japan; Q. austrochinchinensis and Q. kerrii distributed in Southwest China) were fully revealed using morphological traits and molecular markers (Tamaki and Okada, 2014; Song et al., 2015; An et al., 2017). The morphological intermediates have been frequently reported in section Ilex (Huang et al., 1999; Neophytou et al., 2007) and several hybridization zones with a series of morphological transitions have been discovered according to our field observation, indicating frequent interspecies hybridization. Meanwhile, the inconsistent evolutionary patterns between cpDNA and nrDNA were commonly detected in regional phylogeographic and population genetic analysis of section Ilex, which also implied introgression occurs in the Mediterranean and East Asia (Neophytou et al., 2011a,b; Meng et al., 2017; Vitelli et al., 2017). We found that the evolutionary history of the chloroplast genome is net-like rather than dichotomous, which hinders the discriminatory ability to infer the species tree using these maternal markers, especially in a frequent-hybridized taxa (Hollingsworth et al., 2011; Coissac et al., 2016). Moreover, driven by the peculiar paleogeographical histories of the studied regions, plastid genome may reveal geographic differentiation or distribution range of the ancestral lineage instead of the true species phylogeny (Petit et al., 2002; Gugger and Cavender-Bares, 2013; Simeone et al., 2016; Pham et al., 2017; Vitelli et al., 2017). Eurasian evergreen oaks also have strong cpDNA genetic differentiation across different geographical regions instead of species taxonomy. As we found in section Ilex, individuals from Mediterranean-Himalaya, SW China and East China formed independent clusters, respectively. It is likely that active tectonic movements induced uplift of SE Himalaya fringes and adjacent region, which blocked the regional seed-mediated gene flow and resulted in the strong cpDNA geographic differentiation in these oaks.
Another explanation for haplotype sharing between species which located across long distance is the incomplete lineage sorting of the ancestral lineages (Piredda et al., 2011; Simeone et al., 2013). In addition, occasional introgression following the long-distance dispersal events might result in this pattern as well (Lumaret et al., 2002; Toumi and Lumaret, 2010). Therefore, the low cpDNA mutation rates, strong asymmetric interspecies gene flow and incomplete lineage sorting of ancestral polymorphism may have contributed to the low resolution of cpDNA for species delimitation for East Asian evergreen oaks.
Different Genetic Diversity Patterns in Asian Evergreen Lineages
Why Is the cpDNA Phylogeny Non-monophyletic?
Within the subgenus Cerris, our data shows that section Ilex is closely related to both sections Cyclobalanopsis and Cerris. A similar unresolved complex phylogenic relationship within subgenus Cerris was also found in other studies using cpDNA markers (Manos et al., 1999; Simeone et al., 2016). The chloroplast capture events within a genus are mostly due to hybridization (Fehrer et al., 2007; Acosta and Premoli, 2010). However, the hybridization between the extant sections in oaks is extremely rare in the wild (Costello et al., 2011). The non-monophyly of section Ilex plastomes and its close relationship with sections Cyclobalanopsis and Cerris may reflect an ancient introgression or incomplete lineage sorting of the chloroplast genome in the ancestor lineages of subgenus Cerris. In the future, this issue should be subjected to the further study using high-throughput molecular marker (e.g., GBS, RAD-seq) to resolve the species phylogeny. In addition, it needs to recruit multiple species populations of the three sections to reveal the ancient and/or contemporary gene flow among the lineages.
Why the Genetic Diversity in Sections of Quercus Is Different?
Theoretically, the deciduous lineages which are usually fast growing species, may have higher genetic variation, because they have shorter sexual maturation time comparing to evergreen lineages (Zuidema et al., 2009). Generally, it takes at least five or more years for seedlings of evergreen oaks to produce first acorns. But for the deciduous species, this cycle only needs 2–3 years (Wang and Gao, 2005; Wang et al., 2011), such as in Q. serrata, Q. fabri, and Q. aliena. However, the genetic variation patterns in oak sections may not match this theory well, as our study revealed that sections Ilex and Quercus have higher genetic variation, and other sections have lower variation. This result is slightly different to previous reports that sections Ilex, Lobatae and Quercus were the most variable lineages, and section Cerris was the least variable (Simeone et al., 2016). Currently, it is too early to conclude the overall genetic diversity patterns of the genus Quercus, because of the uneven sampling of the main sections.
However, in the well-sampled Asian evergreen sections Cyclobalanopsis and Ilex, we found different patterns of genetic diversity. Trees of section Ilex grow at semi-arid habitats with slow biomass accumulation ratio and long sexual maturation cycle (Mediavilla and Escudero, 2003). All these biological traits of species of section Ilex tend to cause low cpDNA genetic diversity level. However, it was the most diverse lineage based on our study. Interestingly, section Cyclobalanopsis with high number of species and great morphological variation showed low genetic diversity. The different biogeographical histories and adaptabilities to the environment in the two sections may have driven their genetic diversity patterns. Section Cyclobalanopsis was an early derived (at the early Oligocene) section of subgenus Cerris, but their lineage diversification did not occur until the early Miocene and the highest diversification ratio achieved at the mid-late Miocene (Deng et al., 2017a). Instead, section Ilex was later derived at the early-middle Oligocene, but it shows a stepwise diversification pattern since the middle Oligocene onward (Deng et al., 2017c). Therefore, most species in section Cyclobalanopsis are probably younger than those in section Ilex. With longer evolutionary time, more cpDNA mutations may have accumulated, which may explain why the genetic diversity level of section Ilex is much higher than that of section Cyclobalanopsis. Moreover, section Ilex can grow in a wide geographic range with diversified habitats, aside from the prominent geographic barriers, the local adaptation to different environments may have also played a role in boosting the genetic diversity of section Ilex (Du et al., 2016; Feng et al., 2016; Meng et al., 2017; Feng et al., unpublished). In contrast, trees in section Cyclobalanopsis live mainly in warm and humid subtropical habitats (Huang et al., 1999; Deng et al., 2017a). As dominant trees, their large effective population size and the similar habitats may make them less influenced by topographic and climate changes. Shorter divergence time, long-term environment stability and strong gene flow among regional populations, may have altogether resulted in the low genetic diversity of the cpDNA in section Cyclobalanopsis.
Conclusion and Perspective
This study reveals that the cpDNA markers have limited efficiency to identify the Asian evergreen oaks, but these cpDNA markers are still informative to infer the species placement to the main sections of Quercus. The different genetic diversity patterns between the two evergreen oak sections Ilex and Cyclobalanopsis may have been shaped by their spatio-temporal histories and different adaptabilities to environments. The different evolutionary pattern of oaks revealed by the chloroplast and nuclear genomes is most likely due to the historical introgression in the ancestral lineages and recent or on-going gene flow between the closely related species. All these factors reduce the efficiency of cpDNA as species level barcodes. Even single copy nuclear genes or a small batch of nuclear sequences may not be ideal barcoding makers for precise species identification in oaks, due to limited number of informative SNPs. The advanced technologies, such as high-throughput markers (e.g., RAD-seq and GBS) and genome skimming offer a promising future for DNA barcoding in oaks and other taxonomic difficult groups.
Author Contributions
MD conceived and designed the experiments. YX, RL, and MY performed the experiments. MY and JS analyzed the data. MD was responsible for field collections and specimen identification. MY and MD wrote and revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Xiaolong Jiang, Yigang Song, and Quanjian Li for their assistance with the sampling.
Abbreviations
- cpDNA
chloroplast DNA
- EBLF
evergreen broad-leaved forest
Funding. This work was supported by grants from the Shanghai Municipal Administration of Forestation and City Appearances (G162405, G172406, and G162404), Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01), National Natural Science Foundation of China (31270267), Science and Technology Basic Work (S&T Basic Work) (2013FY112100), and International Partnership Program of Chinese Academy of Sciences (151111KYSB20170021).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00569/full#supplementary-material
References
- Acosta M. C., Premoli A. C. (2010). Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). 54 235–242. 10.1016/j.ympev.2009.08.008 [DOI] [PubMed] [Google Scholar]
- An M., Deng M., Zheng S. S., Jiang X. L., Song Y. G. (2017). Introgression threatens the genetic diversity of Quercus austrocochinchinensis (Fagaceae), an endangered oak: a case inferred by molecular markers. 8:229. 10.3389/fpls.2017.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K., Ball S. (2005). DNA barcodes for biosecurity: invasive species identification. 360 1813–1823. 10.1098/rstb.2005.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S. L., Armstrong K. F. (2006). DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). 36 337–350. 10.1098/rstb.2005.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellarosa R., Simeone M. C., Papini A., Schirone B. (2005). Utility of ITS sequence data for phylogenetic reconstruction of Italian Quercus spp. 34 355–370. 10.1016/j.ympev.2004.10014 [DOI] [PubMed] [Google Scholar]
- Bickford D., Lohman D. J., Sodhi N. S., Ng P. K., Meier R., Winker K., et al. (2007). Cryptic species as a window on diversity and conservation. 22 148–155. 10.1016/j.tree.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Birky C. W. (1988). “Evolution and variation in plant chloroplast and mitochondrial genomes,” in eds Gottlieb L. D., Jain S. K. (Dordrecht: Springer; ) 23–53. [Google Scholar]
- Bleeker W., Klausmeyer S., Peintinger M., Dienst M. (2008). DNA sequences identify invasive alien Cardamine at Lake Constance. 141 692–698. 10.1016/j.biocon.2007.12.015 [DOI] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus A. (1936–1954). Paris: P. Lechevalier. [Google Scholar]
- Cao M., Zhang J. (1997). Tree species diversity of tropical forest vegetation in Xishuangbanna, SW China. 7 995–1006. 10.15517/rbt.v59i13212 21513205 [DOI] [Google Scholar]
- Cavender-Bares J., González-Rodríguez A., Eaton D. A., Hipp A. A., Beulke A., Manos P. S. (2015). Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. 24 3668–3687. 10.1111/mec.13269 [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group (2009). A DNA barcode for land plants. 106 12794–12797. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. M., Zhang X. X., Kang H. H., Sun X., Yin S., Du H. E., et al. (2012). Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple glacial refugia and mainland-migrated island populations. 7:e47268. 10.1371/journal.pone.0047268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhao J., Erickson D. L., Xia N., Kress W. J. (2015). Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. 15 337–348. 10.1111/1755-0998.12319 [DOI] [PubMed] [Google Scholar]
- Clement W. L., Donoghue M. J. (2012). Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. 12:73. 10.1186/1471-2148-12-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc-Blain J. L., Starr J. R., Bull R. D., Saarela J. M. (2010). A regional approach to plant DNA barcoding provides high species resolution of sedges (Carex and Kobresia, Cyperaceae) in the Canadian Arctic Archipelago. 10 69–91. 10.1111/j.1755-0998.2009.02725.x [DOI] [PubMed] [Google Scholar]
- Coissac E., Hollingsworth P. M., Lavergne S., Taberlet P. (2016). From barcodes to genomes: extending the concept of DNA barcoding. 25 1423–1428. 10.1111/mec.13549 [DOI] [PubMed] [Google Scholar]
- Costello L. R., Hagen B. W., Katherine S. J. (2011). Richmond, VA: California University of California. [Google Scholar]
- Curtu A. L., Gailing O., Finkeldey R. (2007). Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. 7:218. 10.1186/1471-2148-7-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Hipp A., Song Y. G., Li Q. S., Coombes A., Cotton A. (2014). Leaf epidermal features of Quercus subgenus Cyclobalanopsis (Fagaceae) and their systematic significance. 176 224–259. 10.1111/boj.12207 [DOI] [Google Scholar]
- Deng M., Jiang X. L., Hipp A. L., Manos P. S., Hahn M. (2017a). Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. 119 170–181. 10.1016/j.ympev.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Deng M., Jiang X. L., Song Y. G., Coombes A., Yang X. R., Xiong Y. S., et al. (2017b). Leaf epidermal features of Quercus Group Ilex (Fagaceae) and their application to species identification. 237 10–36. 10.1016/j.revpalbo.2016.11.006 [DOI] [Google Scholar]
- Deng M., Jiang X. L., Su T., Hipp A., Zhou Z. K. (2017c). “Young dispersal from East Asia through the Himalya to the Mediterranean of the Tethys disjunction lineage-Biogeography of Quercus section Ilex (Fagaceae),” in Shanghai. [Google Scholar]
- Deng M., Li Q. S., Yang S. T., Xu J., Song Y. G., Li Q. J. (2013a). “Endangered Oaks in China and the conservation challenges,” in Guangzhou. [Google Scholar]
- Deng M., Song Y. G., Li Q. J., Li Q. S. (2013b). Pollen morphology of Quercus subg. Cyclobalanopsis (Fagaceae) and its systematic implication. 33 368–375. 10.5586/asbp.2012.005 [DOI] [Google Scholar]
- Denk T., Grimm G. W. (2009). Significance of pollen characteristics for infageric classification and phylogeny in Quercus (Fagaceae). 170 926–940. [Google Scholar]
- Denk T., Grimm G. W. (2010). The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. 59 351–366. [Google Scholar]
- Denk T., Grimm G. W., Manos P. S., Deng M., Hipp A. L. (2017). “An updated infrageneric classification of the oaks: review of previous taxonomic schemes and synthesis of evolutionary patterns,” in eds Gil-Pelegrín E., Peguero-Pina J. J., Sancho-Knapik D. (Berlin: Springer; ) 13–38. [Google Scholar]
- Denk T., Tekleva M. V. (2014). Pollen morphology and ultrastructure of Quercus with focus on Group Ilex (=Quercus subgenus Heterobalanus (Oerst.) Menitsky): implications for oak systematics and evolution. 53 255–282. 10.1080/00173134.2014918647 [DOI] [Google Scholar]
- Dong W., Xu C., Li C., Sun J., Zuo Y., Shi S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. 5:8348. 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. 19 11–15. [Google Scholar]
- Drouin G., Daoud H., Xia J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. 49 827–831. 10.1016/j.ympev.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Du F. K., Hou M., Wang W., Mao K., Hampe A. (2016). Phylogeography of Quercus aquifolioides provides novel insights into the Neogene history of a major global hotspot of plant diversity in south-west China. 44 294–307. 10.1111/jbi.12836 [DOI] [Google Scholar]
- Ducousso A., Michaud H., Lumaret R. (1993). Reproduction and gene flow in the genus Quercus L. 50(Suppl.) 91s–106s. 10.1051/forest:19930708 [DOI] [Google Scholar]
- Dumolin S., Demesure B., Petit R. (1995). Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. 91 1253–1256. 10.1007/BF00220937 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand N. C., Elam D. R. (1993). Population genetic consequences of small population size: implications for plant conservation. 24 217–242. 10.1146/annurev.es.24.110193.001245 [DOI] [Google Scholar]
- Fang J. Y., Yoda K. (1991). Climate and vegetation in China V. Effect of climatic factors on the upper limit of distribution of evergreen broadleaf forest. 6 113–125. 10.1007/bf02353874 [DOI] [Google Scholar]
- Fehrer J., Gemeinholzer B., Chrtek J., Bräutigam S. (2007). Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). 42 347–361. 10.1016/j.ympev.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Feliner G. N., Rosselló J. A. (2007). Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. 44 911–919. 10.1016/j.ympev.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Feng L., Zheng Q. J., Qian Z. Q., Yang J., Zhang Y. P., Li Z. H., et al. (2016). Genetic structure and evolutionary history of three Alpine Sclerophyllous oaks in East Himalaya-Hengduan Mountains and adjacent regions. 7:1688. 10.3389/fgls.2016.01688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Yao H., Song J. Y., Zhu Y. J., Liu C., Chen S. L. (2010). Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. 10:324. 10.1186/1471-2148-10-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Morton B. R., McCaig B. C., Clegg M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. 93 10274–10279. 10.1073/pnas.93.19.10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein P. Z., DeSalle R. (2011). Integrating DNA barcode data and taxonomic practice: determination, discovery, and description. 33 135–147. 10.1002/bies.201000036 [DOI] [PubMed] [Google Scholar]
- Gonzalez M. A., Baraloto C., Engel J., Mori S. A., Pétronelli P., Riéra B., et al. (2009). Identification of Amazonian trees with DNA barcodes. 4:e7483. 10.137/journal.pone.0007483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D., Li W. H. (2000). 2nd Edn. Sunderland, MA: Sinauer Associates Inc. [Google Scholar]
- Gugger P. F., Cavender-Bares J. (2013). Molecular and morphological support for a Florida origin of the Cuban oak. 40 632–645. 10.1111/j.1365-2699.2011.02610.x [DOI] [Google Scholar]
- Guo X. D., Wang H. F., Bao L., Wang T. M., Bai W. N., Ye J. W., et al. (2014). Evolutionary history of a widespread tree species Acer mono in East Asia. 4 4332–4345. 10.1002/ece3.1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyette R. P., Muzika R.-M., Kabrick J., Stambaugh M. C. (2004). Gen. Tech. Rep. SRS-73. Asheville, NC: Department of Agriculture; 138–142. [Google Scholar]
- Hassel K., Segreto R., Ekrem T. (2013). Restricted variation in plant barcoding markers limits identification in closely related bryophyte species. 13 1047–1057. 10.1111/1755-0998.12074 [DOI] [PubMed] [Google Scholar]
- Hebert P. D., Cywinska A., Ball S. L. (2003). Biological identifications through DNA barcodes. 270 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp A. L., Eaton D. A., Cavender-Bares J., Fitzek E., Nipper R., Manos P. S. (2014). A framework phylogeny of the American oak clade based on sequenced RAD data. 9:e93975. 10.1371/journal.pone.0093975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth P. M. (2007). DNA barcoding: potential users. 3 44–47. 10.1186/1746-5354-3-2-44 [DOI] [Google Scholar]
- Hollingsworth P. M., Graham S. W., Little D. P. (2011). Choosing and using a plant DNA barcode. 6:e19254. 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth P. M., Li D. Z., van der Bank M., Twyford A. D. (2016). Telling plant species apart with DNA: from barcodes to genomes. 371:20150338. 10.1098/rstb.2015.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Zhang Y., Bartholomew B. (1999). “Fagaceae,” in eds Wu Z., Raven P. (Beijing: Science Press; ) 314–400. [Google Scholar]
- Hubert F., Grimm G. W., Jousselin E., Berry V., Franc A., Kremer A. (2014). Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. 12 405–423. 10.1080/14772000.2014.941037 [DOI] [Google Scholar]
- Hughes C. E., Eastwood R. J., Bailey C. D. (2006). From famine to feast? Selecting nuclear DNA sequence loci for plant species-level phylogeny reconstruction. 361 211–225. 10.1098/rstb.2005.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Xu K., Fan Q., Peng H. (2017). A new species of Ilex (Aquifoliaceae) from Jiangxi Province, China, based on morphological and molecular data. 298 147–157. 10.11646/phytotaxa.298.2.4 [DOI] [Google Scholar]
- Jiang X. L., An M., Zheng S. S., Deng M., Su Z. H. (2018). Geographical isolation and environmental heterogeneity contribute to the spatial genetic patterns of Quercus kerrii (Fagaceae). 120 219–233. 10.1038/s41437-017-0012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. L., Deng M., Li Y. (2016). Evolutionary history of subtropical evergreen broad-leaved forest in Yunnan Plateau and adjacent areas: an insight from Quercus schottkyana (Fagaceae). 12:104 10.1007/s11295-016-1063-2 [DOI] [Google Scholar]
- Kormanik P. P., Sung S.-J. S., Kormanik T., Tibbs T., Zarnoch S. J. (2004). Gen. Tech. Rep. SRS 71. Asheville, NC: Department of Agriculture Forest Service; 555–558. [Google Scholar]
- Kress W. J., Erickson D. L., Jones F. A., Swenson N. G., Perez R., Sanjur O., et al. (2009). Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. 106 18621–18626. 10.1073/pnas.0909820106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress W. J., Erickson D. L., Swenson N. G., Thompson J., Uriarte M., Zimmerman J. K. (2010). Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. 5:e15409. 10.1371/journal.pone.0015409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress W. J., Wurdack K. J., Zimmer E. A., Weigt L. A., Janzen D. H. (2005). Use of DNA barcodes to identify flowering plants. 102 8369–8374. 10.1073/pnas.0503123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye R., van der Bank M., Bogarin D., Warner J., Pupulin F., Gigot G., et al. (2008). DNA barcoding the floras of biodiversity hotspots. 105 2923–2928. 10.1073/pnas.0709936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiou A., Mandolini L. A., Piredda R., Bellarosa R., Simeone M. C. (2013). DNA barcoding as a complementary tool for conservation and valorisation of forest resources. 365 197–213. 10.3897/zookeys.365.5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Koh S. H., Choi W. I., Jung C. S., Kim I. K., Byun B. K., et al. (2012). Barcoding forest insect pests in South Korea: constructing a basic endemic species dataset. 15 363–368. 10.1016/j.aspen.2012.01.008 [DOI] [Google Scholar]
- Leroy T., Roux C., Villate L., Bodénès C., Romiguier J., Paiva J. A., et al. (2017). Extensive recent secondary contacts between four European white oak species. 214 865–878. 10.1111/nph.14413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. W., Yang Y., Henry R. J., Rossetto M., Wang Y. T., Chen S. L. (2015). Plant DNA barcoding: from gene to genome. 90 157–166. 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- Liang Y. Y., Liu J., Huang Y. S., Lin C. R. (2016). Aspidistra erythrocephala sp. nov. (Asparagaceae) from Guangxi, China. 247 295–298. 10.1146/phytotaxa.247.4.9 [DOI] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. 25 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Little D. P. (2011). DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. 6:e20552 10.1371/journal.pone.002052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D. P., Stevenson D. W. (2007). A comparison of algorithms for the identification of specimens using DNA barcodes: examples from gymnosperms. 23 1–21. 10.1111/j.1096-0031.2006.00126.x [DOI] [PubMed] [Google Scholar]
- Liu J., Moller M., Gao L. M., Zhang D. Q., Li D. Z. (2011). DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. 11 89–100. 10.1111/j.1755-0998.2010.02907.x [DOI] [PubMed] [Google Scholar]
- Liu J., Yan H. F., Newmaster S. G., Pei N., Ragupathy S., Ge X. J. (2015). The use of DNA barcoding as a tool for the conservation biogeography of subtropical forests in China. 21 188–199. 10.1111/ddi.12276 [DOI] [Google Scholar]
- López-Pujol J., Zhang F. M., Ge S. (2006). Plant biodiversity in China: richly varied, endangered, and in need of conservation. 15 3983–4026. 10.1007/s10531-005-3015-2 [DOI] [Google Scholar]
- Lumaret R., Mir C., Michaud H., Raynal V. (2002). Phylogeographical variation of chloroplast DNA in holm oak (Quercus ilex L.). 11 2327–2336. 10.1046/j.1365-294X.2002.01611.x [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhou Z. (2000). Phytogeography of Quercus subg. Cyclobalanopsis. 23 1–16. 10.3969/j.issn.2095-0845.2001.01.001 29114171 [DOI] [Google Scholar]
- Luo Y., Zhou Z. K. (2002). Leaf architecture in Quercus subgenus Cyclobalanopsis (Fagaceae) from China. 140 283–295. 10.1046/j.1095-8339.2002.00097.x [DOI] [Google Scholar]
- Lynch M. (1997). Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. 14 914–925. 10.1093/oxfordjournals.molbev.a025834 [DOI] [PubMed] [Google Scholar]
- Ma C. L. (2006). Ph.D. dissertation, Kunming Institute of Botany; Kunming. [Google Scholar]
- Ma C. L., Zhou Z. K. (2005). Effect of ITS pseudogene on the phylogenetic study of Quercus (Fagaceae) and its revelation on the plant molecular phylogenetics. 28 127–132. 10.3969/j.issn.2095-0845.2006.02.007 [DOI] [Google Scholar]
- Manos P. S., Doyle J. J., Nixon K. C. (1999). Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). 12 333–349. 10.1006/mpev.1999.0614 [DOI] [PubMed] [Google Scholar]
- Martin A. P., Palumbi S. R. (1993). Body size, metabolic rate, generation time, and the molecular clock. 90 4087–4091. 10.1073/pnas.90.9.4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayol M., Rosselló J. A. (2001). Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. 19 167–176. 10.1006/mpev.2001.0934 [DOI] [PubMed] [Google Scholar]
- McVay J. D., Hipp A. L., Manos P. S. (2017). A genetic legacy of introgression confounds phylogeny and biogeography in oaks. 284:20170300. 10.1098/rspb.2017.0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediavilla S., Escudero A. (2003). Relative growth rate of leaf biomass and leaf nitrogen content in several Mediterranean woody species. 168 321–332. 10.1093/aob/mcl284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R., Shiyang K., Vaidya G., Ng P. K. (2006). DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. 55 715–728. 10.1080/10635150600969864 [DOI] [PubMed] [Google Scholar]
- Meier R., Zhang G., Ali F. (2008). The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. 57 809–813. 10.1080/10635150802406343 [DOI] [PubMed] [Google Scholar]
- Meng H. H., Su T., Gao X. Y., Li J., Jiang X. L., Sun H., et al. (2017). Warm-cold colonization: response of oaks to uplift of the Himalaya-Hengduan Mountains. 26 3276–3294. 10.1111/mec.14092 [DOI] [PubMed] [Google Scholar]
- Menitsky L. L. (1984). St. Petersburg: Leningosed Sciences. [Google Scholar]
- Meyer C. P., Paulay G. (2005). DNA barcoding: error rates based on comprehensive sampling. 3:e422. 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Odrzykoski I. J., Matsui A., Hasegawa M., Akiyama H., Jia Y., et al. (2009). Adaptive evolution of rbcL in Conocephalum (Hepaticae, bryophytes). 441 169–175. 10.1016/j.gene.2008.11.020 [DOI] [PubMed] [Google Scholar]
- Montecchio L., Faccoli M. (2014). First record of thousand cankers disease Geosmithia morbida and walnut twig beetle Pityophthorus juglandis on Juglans nigra in Europe. 98 696–696. 10.1094/PDIS-10-13-1027-PDN [DOI] [PubMed] [Google Scholar]
- Moran E. V., Willis J., Clark J. S. (2012). Genetic evidence for hybridization in red oaks (Quercus sect. Lobatae, Fagaceae). 99 92–100. 10.3732/ajb.1100023 [DOI] [PubMed] [Google Scholar]
- Muir G., Filatov D. (2007). A selective sweep in the chloroplast DNA of dioecious Silene (Section Elisanthe). 177 1239–1247. 10.1534/genetics.107.071969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N., Mittermeier R. A., Mittermeier C. G., Da Fonseca G. A., Kent J. (2000). Biodiversity hotspots for conservation priorities. 403 853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. 76 5269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou C., Aravanopoulos F. A., Fink S., Dounavi A. (2011a). Interfertile oaks in an island environment. II. Limited hybridization between Quercus alnifolia Poech and Q. coccifera L. in a mixed stand. 130 623–635. 10.1007/s10342-010-0454-4 [DOI] [Google Scholar]
- Neophytou C., Dounavi A., Fink S., Aravanopoulos F. A. (2011b). Interfertile oaks in an island environment: I. High nuclear genetic differentiation and high degree of chloroplast DNA sharing between Q. alnifolia and Q. coccifera in Cyprus. A multipopulation study. 130 543–555. 10.1007/s10342-010-0454-4 [DOI] [Google Scholar]
- Neophytou C. H., Palli G., Dounavi A., Aravanopoulos F. A. (2007). Morphological differentiation and hybridization between Quercus alnifolia Poech and Quercus coccifera l. (Fagaceae) in Cyprus. 56 271–277. 10.1515/sg-2007-0038 [DOI] [Google Scholar]
- Newmaster S., Fazekas A., Steeves R., Janovec J. (2008). Testing candidate plant barcode regions in the Myristicaceae. 8 480–490. 10.1111/j.1471-8286.2007.02002.x [DOI] [PubMed] [Google Scholar]
- Newmaster S. G., Ragupathy S. (2009). Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae. Fabaceae). 9(Suppl. 1) 172–180. 10.1111/j.1755-0998.2009.02642.x [DOI] [PubMed] [Google Scholar]
- Nixon K. (2006). “Globe and neotropical distribution and diversity of oak (genus Quercus) and oak forests,” in ed. Kappella M. (Berlin: Springer; ) 3–13. [Google Scholar]
- Nixon K. C. (1993). Infrageneric classification of Quercus (Fagaceae) and typification of sectional names. 50(Suppl. 1) 25s–34s. 10.1051/forest:19930701 [DOI] [Google Scholar]
- Oh S.-H., Manos P. S. (2008). Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. 57 434–451. [Google Scholar]
- Ohyama M., Baba K. I., Itoh T. (2001). Wood identification of Japanese Cyclobalanopsis species (Fagaceae) based on DNA polymorphism of the intergenic spacer between trnT and trnL 5’ exon. 47 81–86. 10.1007/bf00780554 [DOI] [Google Scholar]
- Peakall R., Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. 28 2537–2539. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy D. M., Argus G. W., Cronk Q. C., Fazekas A. J., Kesanakurti P. R., Burgess K. S., et al. (2014). Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep? 23 4737–4756. 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
- Petit R. J., Carlson J., Curtu A. L., Loustau M. L., Plomion C., González-Rodríguez A., et al. (2013). Fagaceae trees as models to integrate ecology, evolution and genomics. 197 369–371. 10.1111/nph.12089 [DOI] [PubMed] [Google Scholar]
- Petit R. J., Csaikl U. M., Bordács S., Burg K., Coart E., Cottrell J., et al. (2002). Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. 156 5–26. 10.1016/S0378-1127(01)00645-4 [DOI] [Google Scholar]
- Pham K. K., Hipp A. L., Manos P. S., Cronn R. C. (2017). A time and a place for everything: phylogenetic history and geography as joint predictors of oak plastome phylogeny. 60 720–732. 10.1139/gen-2016-0191 [DOI] [PubMed] [Google Scholar]
- Phengklai C., Santisuk T., Larsen K. (2008). “Fagaceae,” in eds Santisuk T., Larsen K. (Bangkok: The Forest Herbarium; ) 179–410. [Google Scholar]
- Piredda R., Simeone M. C., Attimonelli M., Bellarosa R., Schirone B. (2011). Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. 11 72–83. 10.1111/j.1755-0998.2010.02900.x [DOI] [PubMed] [Google Scholar]
- Pitman N. C., Mogollón H., Dávila N., Ríos M., García-Villacorta R., Guevara J., et al. (2008). Tree community change across 700 km of lowland Amazonian forest from the Andean foothills to Brazil. 40 525–535. 10.1111/j.1744-7429.2008.00424.x [DOI] [Google Scholar]
- Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. 14 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Puillandre N., Lambert A., Brouillet S., Achaz G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. 21 1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- Qi X. S., Chen C., Comes H. P., Sakaguchi S., Liu Y. H., Tanaka N., et al. (2012). Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). 196 617–630. 10.1111/j.1469-8137.2012.04242.x [DOI] [PubMed] [Google Scholar]
- R Core Team (2015). Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal B., Hayworth D., Olsen K. M., Rauscher J., Smith W. (1998). Phylogeographic studies in plants: problems and prospects. 7 465–474. [Google Scholar]
- Shapcott A., Forster P. I., Guymer G. P., McDonald W. J., Faith D. P., Erickson D., et al. (2015). Mapping biodiversity and setting conservation priorities for SE Queensland’s rainforests using DNA barcoding. 10:e0122164. 10.1371/journal.pone.o122164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone M. C., Grimm G. W., Papini A., Vessella F., Cardoni S., Tordoni E., et al. (2016). Plastome data reveal multiple geographic origins of Quercus Group Ilex. 4:e1897. 10.7287/peerj.preprints.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone M. C., Piredda R., Papini A., Vessella F., Schirone B. (2013). Application of plastid and nuclear markers to DNA barcoding of Euro-Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. 172 478–499. 10.1111/boj.12059 [DOI] [Google Scholar]
- Song Y., Deng M., Hipp A. L., Li Q. (2015). Leaf morphological evidence of natural hybridization between two oak species (Quercus austrocochinchinensis and Q. kerrii) and its implications for conservation management. 134 139–151. 10.1007/s10342-014-0839-x [DOI] [Google Scholar]
- Tamaki I., Okada M. (2014). Genetic admixing of two evergreen oaks, Quercus acuta and Q. sessilifolia (subgenus Cyclobalanopsis), is the result of interspecific introgressive hybridization. 10 989–999. 10.1007/s11295-014-0737-x [DOI] [Google Scholar]
- Tang C. Q. (2010). Subtropical montane evergreen broad-leaved forests of Yunnan, China: diversity, succession dynamics, human influence. 4 22–32. 10.1007/s11707-009-0057-x [DOI] [Google Scholar]
- Tao T. (2010). Bethesda, MD: National Center for Biotechnology Information. [Google Scholar]
- Toumi L., Lumaret R. (2010). Genetic variation and evolutionary history of holly oak: a circum-Mediterranean species-complex [Quercus coccifera L./Q. calliprinos (Webb) Holmboe, Fagaceae]. 290 159–171. 10.1007/s00606-010-0358-2 [DOI] [Google Scholar]
- Valentini A., Pompanon F., Taberlet P. (2009). DNA barcoding for ecologists. 24 110–117. 10.1016/j.tree.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Van De Wiel C., Van Der Schoot J., Van Valkenburg J., Duistermaat H., Smulders M. (2009). DNA barcoding discriminates the noxious invasive plant species, floating pennywort (Hydrocotyle ranunculoides Lf), from non-invasive relatives. 9 1086–1091. 10.1111/j.1755-0998.2009.02547.x [DOI] [PubMed] [Google Scholar]
- van Velzen R., Weitschek E., Felici G., Bakker F. T. (2012). DNA barcoding of recently diverged species: relative performance of matching methods. 7:e30490. 10.1371/journal.pone.0030490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman S., Otieno J., Gravendeel B., van Andel T., de Boer H. (2014). “Conservation of endangered wild harvested medicinal plants: use of DNA barcoding,” in ed. Gurib-Fakim A. (Chichester: John Wiley & Sons; ) 81–88. 10.1002/9781118460566.ch6 [DOI] [Google Scholar]
- Vilgalys R. (2003). Taxonomic misidentification in public DNA databases. 160 4–5. 10.1046/j.1469-8137.2003.00894.x [DOI] [PubMed] [Google Scholar]
- Virgilio M., Backeljau T., Nevado B., De Meyer M. (2010). Comparative performances of DNA barcoding across insect orders. 11:206. 10.1186/1471-2105-11-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli M., Vessella F., Cardoni S., Pollegioni P., Denk T., Grimm G. W., et al. (2017). Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). 13:3 10.1007/s11295-016-1086-8 [DOI] [Google Scholar]
- von Cräutlein M., Korpelainen H., Pietiläinen M., Rikkinen J. (2011). DNA barcoding: a tool for improved taxon identification and detection of species diversity. 20 373–389. 10.1007/s10531-010-9964-0 [DOI] [Google Scholar]
- Wang C. W., Yang B. Y., Jin X. H. (2017). Herminium motuoensis sp. nov. (Orchidaceae, Orchidoideae), a new species from Tibet, China. 329 197–200. 10.11646/phytotaxa.329.2.14 [DOI] [Google Scholar]
- Wang D. Y., Wang Q., Wang Y. L., Xiang X. G., Huang L. Q., Jin X. H. (2017). Evaluation of DNA barcodes in Codonopsis (Campanulaceae) and in some large angiosperm plant genera. 12:e0170286. 10.1371/journal.pone.0170286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. X., Liang S. C., Wei F., Li J. W. (2011). Spatial Pattern of Quercus fabri population in Karst Hills of Kaili in Guizhou. 39 9639–9642. 10.3969/j.issn.0517-6611.2011.16.068 [DOI] [Google Scholar]
- Wang Z. L., Gao X. M. (2005). The regeneration of Quercus aliena var. acuteserrata: acorn status, seedling pool and size structure. 25 986–993. 10.4028/www.scientific.net/amm.52-54.949 [DOI] [Google Scholar]
- Weitschek E., Velzen R., Felici G., Bertolazzi P. (2013). BLOG 2.0: a software system for character-based species classification with DNA Barcode sequences. What it does, how to use it. 13 1043–1046. 10.1111/1755-0998.12073 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Li C., Hsieh T. Y., Tian D. K., Zhou J. J., Zhang D. G., et al. (2015). Eutrema bulbiferum (Brassicaceae), a new species with bulbils from Hunan, China. 219 233–242. 10.11646/phytotaxa.219.3.3 [DOI] [Google Scholar]
- Xu J., Deng M., Jiang X. L., Westwood M., Song Y. G., Turkington R. (2015). Phylogeography of Quercus glauca (Fagaceae), a dominant tree of East Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. 11:805 10.1007/s11295-014-0805-2 [DOI] [Google Scholar]
- Xu J., Jiang X. L., Deng M., Westwood M., Song Y. G., Zheng S. S. (2016). Conservation genetics of rare trees restricted to subtropical montane cloud forests in southern China: a case study from Quercus arbutifolia (Fagaceae). 12:90 10.1007/s11295-016-1048-1 [DOI] [Google Scholar]
- Yan L. J., Liu J., Moller M., Zhang L., Zhang X. M., Li D. Z., et al. (2015). DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. 15 932–944. 10.1111/1755-0998.12353 [DOI] [PubMed] [Google Scholar]
- Yang J., Vázquez L., Chen X., Li H., Zhang H., Liu Z., et al. (2017). Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. 8:816. 10.3389/fpls.2017.00816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. M., Tian K., Hao J. M., Pei S. J., Yang Y. X. (2004). Biodiversity and biodiversity conservation in Yunnan, China. 13 813–826. 10.1023/b:bioc.0000014464.80847.02 [DOI] [Google Scholar]
- Yu W. B., Huang P. H., Ree R. H., Liu M. L., Li D. Z., Wang H. (2011). DNA barcoding of Pedicularis L.(Orobanchaceae): evaluating four universal barcode loci in a large and hemiparasitic genus. 49 425–437. 10.1111/j.1759-6831.2011.00154.x [DOI] [Google Scholar]
- Zhao J. F., Feng D. J., Lei Y. F. (2007). Wood study on Quercus and Cyclobalanopsis of Fagaceae in Shaanxi Province. 35 196–202. 10.3321/j.issn:1671-9387.2007.10.038 [DOI] [Google Scholar]
- Zhou Z. K., Wilkinson H., Wu C. Y. (1995). Taxonomical and evolutionary implications of the leaf anatomy and architecture of Quercus L. subgenus Quercus from China. 7 1–34. 10.1046/j.1095-8339.2002.00097.x [DOI] [Google Scholar]
- Zuidema P. A., Brienen R. J., During H. J., Güneralp B. (2009). Do persistently fast-growing juveniles contribute disproportionately to population growth? A new analysis tool for matrix models and its application to rainforest trees. 174 709–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


