Abstract
The neurotransmitter γ-aminobutyric acid (GABA) is an extracellular signaling molecule in the brain and in pancreatic islets. Here, we demonstrate that GABA regulates cytokine secretion from human peripheral blood mononuclear cells (PBMCs) and CD4+ T cells. In anti-CD3 stimulated PBMCs, GABA (100 nM) inhibited release of 47 cytokines in cells from patients with type 1 diabetes (T1D), but only 16 cytokines in cells from nondiabetic (ND) individuals. CD4+ T cells from ND individuals were grouped into responder or non-responder T cells according to effects of GABA (100 nM, 500 nM) on the cell proliferation. In the responder T cells, GABA decreased proliferation, and inhibited secretion of 37 cytokines in a concentration-dependent manner. In the non-responder T cells, GABA modulated release of 8 cytokines. GABA concentrations in plasma from T1D patients and ND individuals were correlated with 10 cytokines where 7 were increased in plasma of T1D patients. GABA inhibited secretion of 5 of these cytokines from both T1D PBMCs and ND responder T cells. The results identify GABA as a potent regulator of both Th1- and Th2-type cytokine secretion from human PBMCs and CD4+ T cells where GABA generally decreases the secretion.
Keywords: PBMCs, Immune cells, Proliferation, Cytokine, GABAA receptor, Diabetes, T1D, Autoimmune disease, T cell
Highlights
-
•
GABA regulates cytokine secretion in anti-CD3-stimulated peripheral blood mononuclear cells (PBMCs) and CD4+ T cells.
-
•
GABA inhibits secretion of 47 cytokines in PBMCs from type 1 diabetes patients.
-
•
GABA regulates secretion of pro- and anti-inflammatory cytokines in a concentration-dependent manner.
GABA is a signal molecule in the brain, blood and pancreatic islets where it is secreted by the insulin-producing β cells. GABA has many roles in human islets including optimizing function and survival of β cells. Bhandage et al. now show that GABA is a potent regulator of secretion of both pro- and anti-inflammatory cytokines in stimulated immune cells. In type 1 diabetes the β-cell mass is diminished and thus the protective effect of GABA in the islets although not in blood. Targeting GABA signaling in diabetes mellitus is likely to be a part of the solution when curing diabetes.
1. Introduction
Neurotransmitter signaling is normally associated with the nervous system but compiling evidence suggests that many neurotransmitters exert modifying influence on cells of the immune system (Levite, 2008; Jin et al., 2011b; Barragan et al., 2015; Levite et al., 2001; Franco et al., 2007). The amino acid γ-aminobutyric acid (GABA) is best known as the main inhibitory neurotransmitter in the brain, but it is also present in the pancreatic islets and in blood (Baekkeskov et al., 1990; Petty and Sherman, 1984; Farrant and Nusser, 2005; Li et al., 2015). In the brain, GABA is made in neurons from the amino acid glutamate by the enzyme glutamic acid decarboxylase (GAD) that is present in two isoforms, GAD65 and 67 (Bu et al., 1992). GAD is also found in the insulin-secreting β cells in the pancreatic islets, where GAD65 is one of the main autoantigen in T1D in humans (Kanaani et al., 2015, Bu et al., 1992, Baekkeskov et al., 1990). Interestingly, some immune cells may also produce and release GABA (Fuks et al., 2012; Bhat et al., 2010). Where GABA in blood comes from is still being explored, but the recently discovered drainage system of the brain, the glymphatic system (Plog and Nedergaard, 2018), identifies the brain, in addition to peripheral organs, as a potential source for GABA in blood. In the pancreatic islets, GABA is an auto- and paracrine signaling molecule activating GABA receptors on the endocrine cells and, perhaps, also on immune cells that may enter the islets (Birnir and Korpi, 2007; Kanaani et al., 2015; Caicedo, 2013; Bhandage et al., 2015). Similarly, in blood, immune cells may be regulated by GABA (Bhandage et al., 2015; Bjurstom et al., 2008; Tian et al., 2004; Tian et al., 1999). In T1D, the β cell mass declines and, thereby, also the local source for GABA in the pancreatic islets (Fiorina, 2013, Tian et al., 2013).
GABA activates two types of receptors in the plasma membrane of cells; the GABAA receptors, that are Cl− ion channels opened by GABA, and the G-protein-coupled GABAB receptor (Marshall et al., 1999; Olsen and Sieghart, 2008; Olsen and Sieghart, 2009). The GABAA receptors are pentameric, homo- or heteromeric, receptors formed from 19 known subunit isoforms (α1–6, β1–3, γ1–3, δ, ε θ, π, ρ1–3) (Olsen and Sieghart, 2009). In contrast, the GABAB receptor is normally formed as a dimer of the two isoforms identified to date (Marshall et al., 1999; Gassmann et al., 2004). GABA receptors are expressed in immune cells, but their ability to influence the functional phenotype, i.e. proliferation, migration or cytokine secretion, of the cells is still relatively unexplored (Barragan et al., 2015; Jin et al., 2011b). Here, we studied effects of GABA on human PBMCs and T cells from ND individuals and T1D patients. The results revealed that GABA in a concentration-dependent manner regulates release of a variety of cytokines from PBMCs and CD4+ T cells. In PBMCs from T1D patients, GABA significantly altered secretion of both pro- and anti-inflammatory types of cytokines.
2. Materials and Methods
2.1. Study Individuals and Ethical Permits
The study was approved by Regional Ethical Review Board in Uppsala, and the reported investigations were carried out in accordance with the principles of the Declaration of Helsinki as revised in 2000. The study includes 30 healthy controls and 64 T1D patients. All participants signed a written consent form before entering the study. The participants were recruited at Uppsala University Hospital. Demographic characteristics of the participants are summarized in Table S1. All the participants were screened for islet autoantibodies (GAD and islet antigen-2, IA2), which were not present in any of the healthy controls. None of the healthy controls had a first degree relative diagnosed with T1D. None of the participants was ill from, or had recently recovered from, an infectious disease. All blood samples were collected in the morning after an overnight fasting under standardized conditions. Routine lab parameters were analyzed at the Central Clinical Chemistry Laboratory, Uppsala University Hospital. The venous blood samples were collected in EDTA tubes and processed for further experimentation.
2.2. Plasma, PBMCs and T Cell Isolation
Plasma, PBMCs and T cells were isolated from freshly derived blood samples and CD4+ T cells from buffy coats as previously described (Bhandage et al., 2015; Bhandage et al., 2017). The plasma was isolated by centrifugation at 3600 rpm for 10 min at 4 °C directly after collection of blood, and immediately frozen at −80 °C.
The blood samples or buffy coats were diluted in 1:1 ratio in MACS buffer (Miltenyi Biotec, Madrid, Spain), and layered on Ficoll-paque plus (Sigma-Aldrich, Hamburg, Germany). Briefly, the samples were then subjected to density gradient centrifugation at 400g for 30 min at room temperature. The PBMCs were carefully withdrawn and washed twice in MACS buffer. A portion of PBMCs was saved in RNAlater (Sigma-Aldrich) at −80 °C for mRNA extraction for qPCR, and other portions were used for either proliferation experiments or isolation of T cells using human CD3 MicroBeads and human CD4+ T Cell Isolation Kits (Miltenyi Biotec). The CD3+ T cells were used for RNA sequencing, and the CD4+ T cells were used for proliferation and electrophysiological patch-clamp experiments.
2.3. Total RNA Isolation, Real-Time Quantitative Reverse Transcription PCR and Western Blot Analysis
Total RNAs were extracted with RNA/DNA/Protein Purification Plus Kit (Norgen Biotek, Ontario, Canada). The real-time qPCR method has been described previously (Schmittgen and Livak, 2008, Bhandage et al., 2015, Kreth et al., 2010, Ledderose et al., 2011, Bhandage et al., 2017). The extracted total RNA was quantified using Nanodrop (Nanodrop Technologies, Thermo Scientific, Inc., Wilmington, DE, USA). Then, 1.5 μg RNA was treated with 0.6 U DNase I (Roche, Basel, Switzerland) for 30 min at 37 °C to degrade genomic DNA in the sample, and then with 8 mM EDTA for 10 min at 75 °C for inactivation of DNase I enzyme. The cDNA was then synthesized using Superscript IV reverse transcriptase (Invitrogen, Stockholm, Sweden) in a 20 μl reaction mixture using standard protocol provided by manufacturer. To confirm efficient degradation of genomic DNA by DNase I treatment, we performed reverse transcriptase negative reaction which did not yield any amplification in real-time PCR, confirming the absence of genomic DNA contamination. The gene-specific primer pairs are listed in Table S2. The real-time qPCR amplification was performed on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) in a standard 10 μl reaction with an initial denaturation step of 5 min at 95 °C, followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 1 min, followed by melting curve analysis.
Protein extraction from PBMC samples was performed using RNA/DNA/Protein Purification Plus Kit (Norgen Biotek, Ontario, Canada). Protein amounts were quantified using the RC DCTM protein assay kit (Bio-Rad, USA) in Multiskan MS plate reader (Labsystems, Vantaa, Finland), and the concentration was calculated by plotting standard curve. Protein samples (60 μg) were subjected to SDS-PAGE using 10% polyacrylamide gels and transferred to PVDF membranes (Thermofisher Scientific, Stockholm, Sweden). The membranes were blocked with 5% non-fat milk powder in Tris buffered saline containing 0.1% Tween (TBS-T) for 1 h and incubated overnight at 4 °C with primary antibodies against NKCC1 (1:2000; Cell Signaling Technology, Cat No. 8351, USA), GABAAR ρ2 (1:500; Abcam, Cat No. ab83223, Cambridge, UK) and GAPDH (1:3000; Merck Millipore, Cat No. ABS16, USA). After 3 washings with TBS-T, the membranes were further incubated with horseradish peroxidase-conjugated secondary antibody (1:3000; Cell Signaling Technology, Cat No. 7074) for 2 h and then the immunoreactive protein bands were visualized by enhanced chemiluminescence (ECL) detection kit (Thermofisher Scientific, Stockholm, Sweden).
2.4. Determination of GABA Concentration
Plasma samples were thawed, and the level of GABA was measured using an ELISA kit (LDN Labor Diagnostika Nord, Nordhorn, Germany) as per manufacturer's guidelines (Fuks et al., 2012; Abu Shmais et al., 2012; El-Ansary et al., 2011; Lee et al., 2011). Briefly, the plasma samples and standards provided in the kit were extracted on extraction plate, derivatized using equalizing reagent and subjected to standard competitive ELISA in GABA coated microtiter strips. The absorbance of the solution in the wells was read at 450 nm within 10 min using a Multiskan MS plate reader (Labsystems, Vantaa, Finland). We used 620 nm as a reference wavelength. The outcome of the assay, optical density values, were used to plot the standard curve for each run, which was then used to interpolate the GABA concentration of the samples. The readout obtained by the GABA standards in the kit was compared to and agreed with the standards in the quality control (QC) report from the company (Fig. S1).
2.5. Electrophysiology
GABA-activated currents were recorded by the patch-clamp technique as previously described (Bjurstom et al., 2008; Jin et al., 2011a). The extracellular recording solution contained (in mM): 145 NaCl, 3 KCl, 1 CsCl, 1 CaCl2, 1 MgCl2, 10 glucose and 10 TES; the pH was adjusted to 7.4 with NaOH. To record in the whole-cell configuration, the pipette solution contained (in mM): 136 CsCl, 20 KCl, 1 MgCl2, 3 MgATP and 10 TES; pH was adjusted to 7.3 with CsOH. The pipette solution for the cell-attached configuration contained (in mM): 69 NaCl, 5 KCl, 75 CsCl, 1 CaCl2, 1 MgCl2 and 10 TES; pH was adjusted to 7.4 with NaOH. Saclofen (a GABAB receptor antagonist, 200 μM) and GABA (100 nM) were used in the experiments. The pipette potential (Vp) was −80 mV (hyperpolarizing) in the whole-cell configuration and −60 mV (depolarizing) in the cell-attached configuration.
2.6. Proliferation Assay
The proliferation of freshly isolated human PBMCs or CD4+ T cells was evaluated with MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) assay (Ring et al., 2012). Cells were suspended in complete medium (RPMI 1640 supplemented with 2 mM glutamine, 25 mM HEPES, 10% heat inactivated fetal bovine serum, 100 U/ml penicillin, 10 mg/ml streptomycin, 5 mM β-mercaptoethanol) in a concentration 1 million cells per milliliter. The assay was performed in 96-well plates in duplicates or triplicates, where each well was pre-coated with 3 μg/ml anti-CD3 antibody for 3–5 h at 37 °C. Each well was loaded with 100,000 cells. Drugs were added to the wells at the relevant concentrations. The plate was incubated for 68 h at 37 °C (95% O2, 5% CO2) and then, a media-soluble tetrazolium dye MTT was added to a final concentration of 1 mM after which the plate was incubated for additional 4 h. The plate was then centrifuged at 2000 RPM for 10 min to pellet the insoluble purple formazan crystals. The supernatant culture media was collected, stored at −80 °C and used for analysis of cytokines using the multiplex proximity extension assay (PEA). The formazan crystal pellet was dissolved in DMSO and the plate was read within 10 min using a Multiskan MS plate reader (Labsystems) at 550 nm. The optical density value was used as the proliferation index value. Drugs were purchased from Sigma-Aldrich or Tocris (Bristol, UK).
2.7. Multiplex PEA for Cytokine Measurements
Plasma samples, and culture media samples that were collected from plate wells at the end of the proliferation assay, were analyzed by multiplex PEA with an Olink Inflammation I96X96 panel, targeting 92 proteins related to inflammation (Olink Proteomics, Uppsala, Sweden) as previously described (Edvinsson et al., 2017; Assarsson et al., 2014; Larsson et al., 2015; Larssen et al., 2017). Briefly, 1 μl of sample (plasma samples or cell culture media samples) or negative control was mixed with 3 μl probe solution containing a set of 92 paires DNA-oligonucleotide-conjugated antibodies. Upon recognition of a target protein by a pair of probes, the DNA oligonucletodies on the antibodies are brought in proximity and hybridize to each other, follwed by enzymatic DNA polymerization to form a new DNA molecule. The newly formed DNA molecule is then amplified and quantified using a microfluidic real-time qPCR, BioMarkTM HD (Fluidigm, South San Francisco, CA, USA). The generated quantification cycle (Cq) values are normalized against spiked-in controls to convert Cq values to Normalized Protein eXpression (NPX) value on log2 scale. NPX is an arbitrary unit, which is positively correlated to protein concentration. These NPX data were then converted to linear data, using the formula 2NPX, prior to further statistical analysis. Limit of detetion (LOD) for each protein was defined as three standard deviations above the background. Proteins with levels below LOD were excluded from further data analysis.
2.8. Total RNA Isolation and T Cell RNA Sequencing
Total RNA was extracted from T cells using Direct-zol™ RNA MicroPrep (Zymo Research, Irvine, CA, USA) according to manufacturer's recommendation. cDNA libraries were prepared according to Smart-seq2 protocol (Picelli et al., 2013). For Illumina sequencing libraries, 2 ng of cDNA was fragmented, amplified (Picelli et al., 2014), pooled and sequenced on Illumina HiSeq 2500. Single-end 43 bp reads were generated and mapped to human reference genome GRCh38 by employing STAR (version 2.4.1) with parameter outSAMstrandField intronMotif (Dobin et al., 2013). Reads per kilobase transcript per million mapped reads (RPKM) from RefSeq gene annotations were calculated using RPKM for genes (Ramskold et al., 2009). The uniquely mapped reads were considered for the downstream analyses.
2.9. Statistical Analysis
Statistical analysis and data mining were performed using Statistica 12 (StatSoft Scandinavia, Uppsala, Sweden), GraphPad Prism 7 (La Jolla, CA, USA) and edgeR bioconductor package. The statistical tests were performed after omitting outliers identified by the Tukey test. The differences between groups were assessed by nonparametric Kruskal–Wallis ANOVA on ranks with Dunn's post hoc test. The contingency of sex equality was accessed by Fisher's exact test. Comparison of demographic data between the two groups was based on a non-parametric Mann-Whitney test for non-normally distributed data and a two-tailed Student's t-test for normally distributed. Normality of data was assessed by D'Agostino & Pearson omnibus normality test. The correlation between inflammatory cytokines and demographic factors was assessed using non-parametric Spearman rank correlation. The significance level was set to p < 0.05.
3. Results
Demographic data for the ND individuals (n = 30) and the individuals with T1D (n = 64) that participated in the study is presented in Table S1. As expected the individuals with T1D had higher levels of fasting glucose and HbA1c (Table S1). In addition, the individuals with T1D were, on the average, slightly older and had a higher BMI (Table S1). The creatinine levels did not differ between the two groups but the glomerular filtration rate was higher in the diabetes group (Table S1). Islet autoantibodies (GAD and IA2) were not detected in any of the healthy individuals.
3.1. Cytokines in Plasma From ND and T1D Individuals
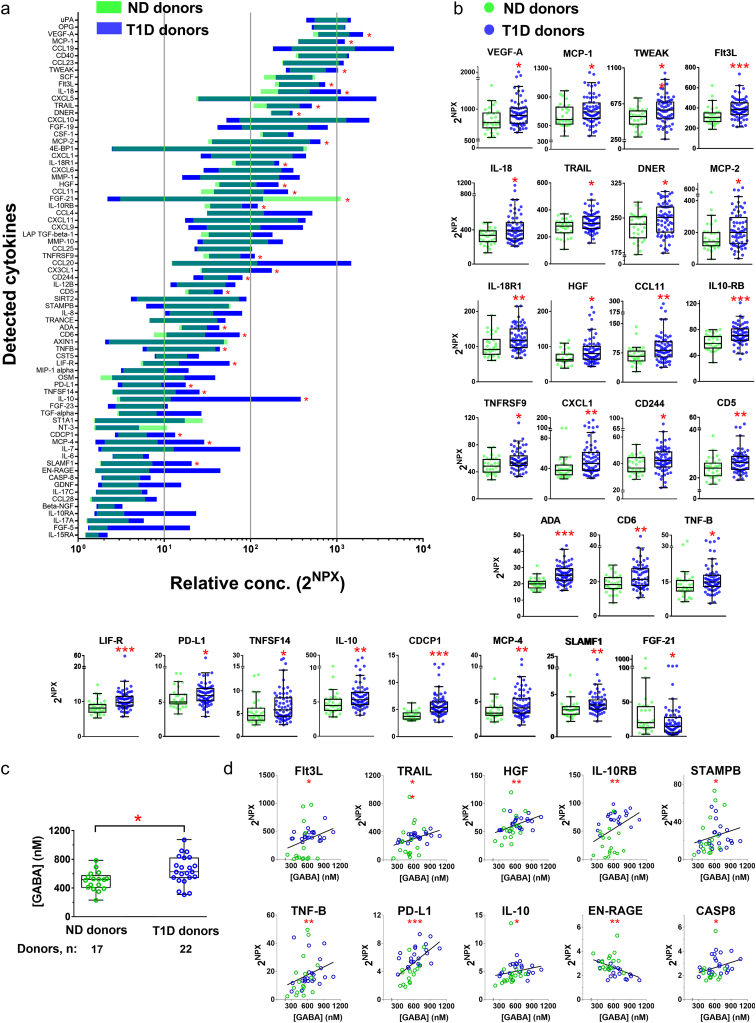
Immune cells secrete a large number of small proteins, collectively termed cytokines, which may have a protective function or act as pro-inflammatory molecules. We investigated whether the types of cytokines in plasma differed between ND individuals and T1D patients. We used the multiplex PEA to measure the blood levels of a panel of 92 cytokines that are most commonly associated with inflammation (http://www.olink.com/products/inflammation/#). The assay that uses paired cytokine-specific antibodies for the different cytokines allows comparison of the levels of the same cytokine in samples from e.g. ND individuals and T1D patients. However, the assay format does not support comparison of the absolute levels of one cytokine to another as the affinities of the antibodies for their cognate targets may vary. As illustrated in Fig. 1a, 73 out of 92 analyzed cytokines were detected in plasma from the donors, of which 26 cytokines were significantly up-regulated and only one cytokine, FGF-21 down-regulated in plasma from T1D patients as compared to ND individuals (Fig. 1b).
Fig. 1.
Cytokines in plasma from ND and T1D individuals and identification of those that correlate with plasma GABA concentration. (a) Screening of 92 inflammatory cytokines in plasma samples from ND (n = 30) and T1D individuals (n = 64) by Olink Multiplex PEA inflammation panel I detects expression of 73 cytokines. Data is presented by 2NPX (Normalized Protein Expression) values as floating bars (minimum to maximum) arranged in descending order of mean expression level of cytokines. (b) Inflammatory cytokines with significant change in the expression levels in the plasma of T1D as compared to ND individuals. Data is shown as box and whiskers overlapped with scatter dot plot. (c) Quantification of GABA levels in plasma samples from ND and T1D individuals. (d) Correlation between GABA levels and cytokine levels in plasma samples from ND individuals and T1D individuals. Only cytokines with significant correlation are shown. R values are given in Table S3. *p < 0.05, **p < 0. 01, ***p < 0.001.
We then examined if the neurotransmitter GABA varied in concentration in plasma between the ND and T1D individuals (Fig. 1c). The GABA concentration range was similar for the two groups but there was a trend for increased plasma GABA concentration in T1D patients resulting in a significantly higher average concentration in the T1D group (ND: 501 ± 32 nM; T1D: 649 ± 42 nM; p < 0.05). No correlation was observed for GABA concentrations with age or disease duration. In contrast, when the concentration of GABA was correlated with cytokines detected in plasma, levels of 10 cytokines were significantly correlated (p < 0.05) with the plasma GABA concentration (Fig. 1d; Table S3).
3.2. GABA Inhibits Proliferation of PBMCs From T1D Patients and Responder CD4+ T Cells
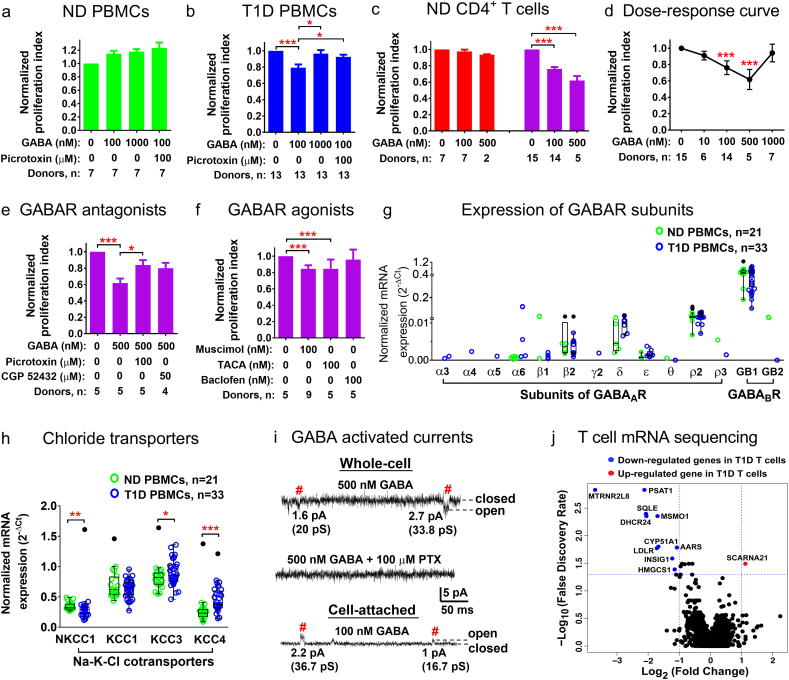
To further examine the effects of GABA on the immune cells, we stimulated PBMCs and CD4+ T cells with anti-CD3 antibody to induce proliferation of CD3+ positive T cells. We then examined effects on proliferation of GABA and the GABAA antagonist, picrotoxin. In PBMCs from ND individuals, GABA did not inhibit proliferation of the cells (Fig. 2a). In contrast, cell proliferation of PBMCs from T1D patients was inhibited in the presence of 100 nM GABA by approximately 20%, while GABA at concentration of 1 μM did not have any inhibitory effect on the cell proliferation. The inhibition was reversed by picrotoxin (Fig. 2b). We have previously demonstrated that GABA may inhibit T cells proliferation (Bjurstom et al., 2008) and, thus, examined whether CD4+ T cells from ND individuals varied in their sensitivity to GABA. The results revealed that CD4+ T cells from the ND individuals could be grouped into responders (n = 15) and non-responders (n = 7) depending on whether or not GABA affected the proliferation of the cells (Fig. 2c). We examined further if GABA, in a dose-dependent manner, could regulate proliferation of the responder CD4+ T cells (Fig. 2d). Application of different GABA concentrations demonstrated increased inhibition of proliferation from 1 to 500 nM GABA with a maximum inhibition of approximately 40% by 500 nM GABA, whereas no inhibition of proliferation of the cells was recorded in 1 μM GABA (Fig. 2d). This is in line with the fact that saturating GABA concentrations cause desensitization and thus, non-functionality of GABAA receptors. The open channel blocker picrotoxin partially reversed the 500 nM GABA-induced inhibition (Fig. 2e) and the GABAA agonists muscimol (100 nM) and TACA (100 nM) inhibited the proliferation similar to GABA (Fig. 2f), whereas the GABAB antagonist CGP52432 (50 μM) and the GABAB agonist baclofen (100 nM) were ineffective (Fig. 2e, f). Together the results highlight that immune cells and in particular CD4+ T cells can be divided into subgroups depending on whether or not their proliferation is regulated by GABA. The greater inhibition observed for the PBMCs from the T1D patients as compared to ND individuals is consistent with the presence of expanded T cells clones that respond to GABA in T1D (Tong et al., 2016).
Fig. 2.
GABA activation of GABAA receptors inhibits proliferation of PBMCs and responder CD4+ T cells from T1D and ND individuals. Effects of GABA and GABAA receptors antagonist, picrotoxin, on proliferation of PBMCs from (a) ND individuals and (b) T1D individuals. (c) Effect of GABA on proliferation of CD4+ T cells from ND individuals identifying GABA non-responder (orange) and GABA responder (magenta) populations of CD4+ T cells. (d) GABA dose-dependent inhibition of proliferation of responder CD4+ T cells. (e) Effects of GABAA (picrotoxin, muscimol, TACA) and GABAB (CGP 52432, baclofen) receptor antagonists and (f) agonists on proliferation of responder CD4+ T cells. (g) Expression of GABAARs and GABABR subunits and (h) chloride transporters in PBMCs from ND individuals and T1D patients. Data are presented as normalized mRNA expression (2-ΔCt). (i) GABA-activated single-channel current measurements from CD4+ T cells from ND individuals. In the whole-cell and cell-attached configurations single-channel currents were activated by 500 and 100 nM GABA application, respectively, and inhibited by picrotoxin (PTX, 100 μM). # marks single-channel events with typical current amplitudes at the given Vp. (j) A volcano plot for T cell mRNA sequencing expression data as log2 (fold change in T1D T cells compared to ND T cells) against –log10 (false discovery rate). The p values are shown in Table S5. *p < 0.05, **p < 0. 01, ***p < 0.001.
GABA can potentially activate GABAA and GABAB receptors in the immune cells (Tian et al., 2004; Bjurstom et al., 2008; Bhandage et al., 2015). We, therefore, measured the expression level of the GABAA and GABAB receptor subunits in PBMCs. The most prominent GABAA receptor subunit was the ρ2 that was similarly expressed and present in most samples from both ND and T1D individuals (Fig. 2g, Table S4). Protein expression of ρ2 was confirmed by western blot analysis (Fig. S2). Of the two GABAB receptor subunits, only the GABABR1 was commonly expressed. Our results confirmed previously reported expression of GABA receptors in PBMCs (Bhandage et al., 2015). We further found that the mRNA expression levels of auxiliary proteins of GABAA receptors gephyrin and GABARAP were similar but the GABA transporters GAT3, BGT1 and the enzyme GABA-T plus the insulin receptor were significantly increased, whereas the expression level of radixin decreased in PBMCs from T1D individuals (Fig. S3, Table S4). Interestingly, expression of Cl− transporters was altered in T1D (Fig. 2h). The transporter that moves Cl− into the cell, NKCC1, was significantly down-regulated, whereas the transporters that move Cl− out of the cells, KCC3 and KCC4, were up-regulated in PBMCs from T1D patients. Protein expression of NKCC1 was confirmed by western blot analysis (Fig. S2). Since the effects of GABAA receptors are related to the Cl− equilibrium potential in the cells, any changes in intracellular chloride will have consequences for GABAA signaling. We further investigated if GABA in submicromolar (100 or 500 nM) concentrations activated single-channel currents in the T cells. GABA activated GABAA single-channel currents in the cells (n = 54), which were blocked by picrotoxin. The GABAA receptors conductance ranged from 9 to 45 pS (Fig. 2i).
3.3. Cholesterol Biosynthesis Gene Levels Are Regulated in T Cells From T1D Patients
We applied RNA-seq to examine the transcriptome of isolated CD3+ T cells from ND individuals and T1D patients (Fig. 2j). A total of 16,684 genes were identified after passing the quality control and deposited at GEO database (GSE111876). To reduce the bias of genes with low counts and only expressed in a few samples, we used the dataset with 8669 genes that are expressed in >1/3 of the samples (RPKM>5) in both T1D patients and ND individuals. Eleven genes (edgeR p < 0.05, FDR < 0.05) were differentially expressed by more than two-fold between T1D and ND samples, of which only 1 gene (SCARNA21) was up-regulated and 10 genes were down-regulated in samples from T1D patients (Fig. 2j). Among the down-regulated genes, 5 genes (SQLE, MSMO1, DHCR24, CYP51A1, HMGCS1) are involved in the cholesterol biosynthesis pathway and 1 gene (INSIG1) encodes an endoplasmic reticulum protein that regulates cholesterol biosynthesis (REACTOME, http://reactome.org) (Table S5). Interestingly, the expression levels of MSMO1 and CYP51A1 were significantly correlated with the plasma GABA concentration (p < 0.05) (Fig. S4). Furthermore, the expression of MSMO1 was also significantly correlated with BMI, fasting glucose and HbA1c levels (p < 0.05) (Fig. S4).
3.4. GABA Regulates Release of Pro- and Anti-inflammatory Cytokines From PBMCs
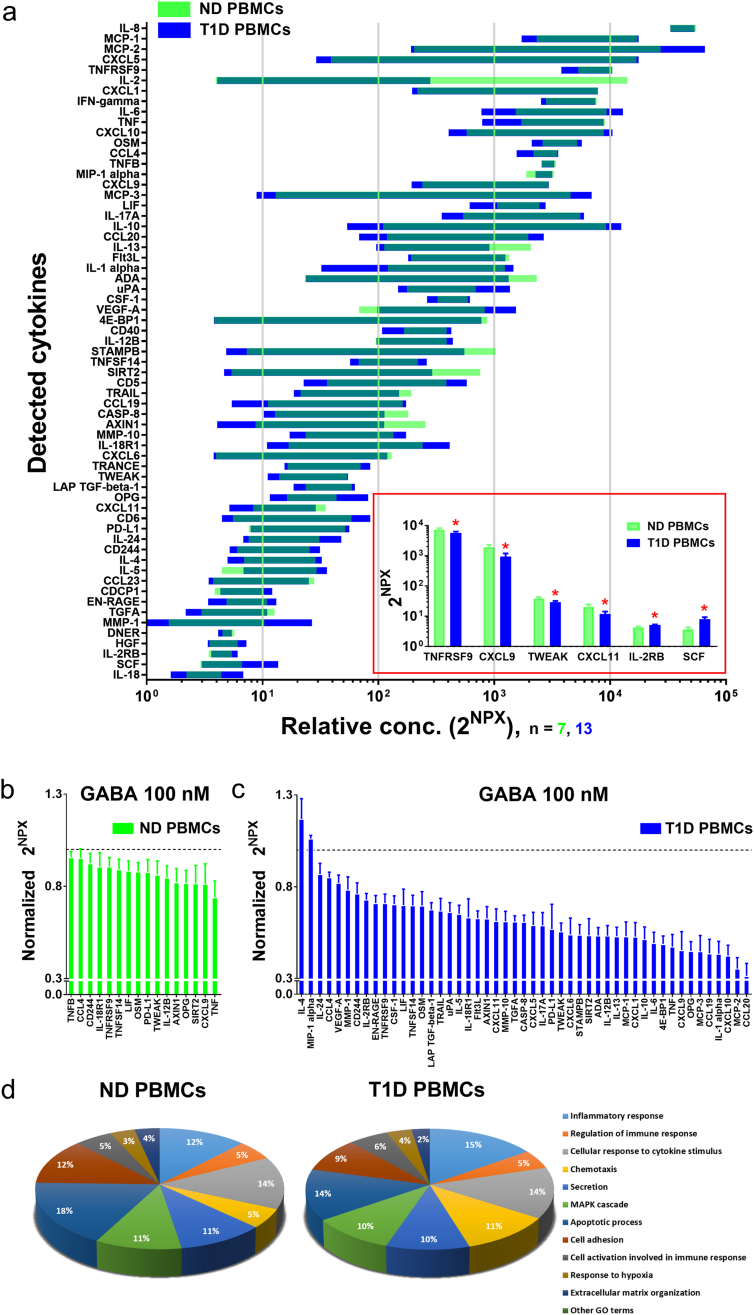
It is possible that GABA signaling regulates what cytokines are released from the immune cells. We, therefore, examined the culture media from the anti-CD3 stimulated PBMCs using the inflammatory related protein panel described above to study which of the 92 cytokines are released by the cells and whether GABA affects secretion of specific cytokines. Fig. 3a shows the levels of the different cytokines detected in the culture media harboring proliferating cells from ND individuals and T1D patients. In the absence of GABA, a difference in the secretion level was observed for six cytokines (Fig. 3a insert). Somewhat surprisingly, the results shown in Fig. 3b and c and also Table S6 and S7 demonstrate that GABA regulated the release of many pro- and anti-inflammatory cytokines. In the culture media from stimulated PBMCs, GABA (100 nM) significantly inhibited release of 16 cytokines from ND individuals (n = 7; Fig. 3b) and 47 cytokines from T1D patients (n = 13; Fig. 3c), respectively, and, additionally, increased the release of two cytokines from T1D patients (Fig. 3c). Cytokines released by stimulated PBMCs, which were affected by GABA can be grouped according to their function (Fig. 3d). The group of cytokines with the largest difference between ND and T1D PBMCs was the one associated with chemotaxis. The results are in agreement with that GABA regulates release of cytokines from PBMCs in both health and in disease.
Fig. 3.
Identification of cytokines released into the culture media and effects of GABA treatment on the PBMCs cytokine secretion. (a) Screening of 92 cytokines in media from PBMCs from ND individuals or from T1D individuals, by Olink Multiplex PEA inflammation panel I, revealed expression of 63 cytokines. No GABA was added to the media. Data are represented by 2NPX values as floating bars (minimum to maximum) arranged in descending order of mean expression level of cytokines. Insert show cytokines with significant change in the expression levels in the media of PBMCs from ND individuals as compared with T1D individuals. Data are plotted as a bar graph with mean ± SEM. (b-c) Expression of cytokines that are significantly affected by GABA 100 nM treatment of PBMCs from (b) ND individuals and from (c) T1D individuals. Data are represented by 2NPX values normalized to controls as a bar graph with mean ± SEM. Mean values with SEM and p values are shown in Tables S6 and S7. (d) Classification based on the cellular functions of cytokines that were significantly altered by GABA 100 nM treatment of PBMCs from ND individuals (16 cytokines) and from T1D individuals (49 cytokines).
3.5. GABA Regulates Release of Pro- and Anti-inflammatory Cytokines From CD4+ T Cells
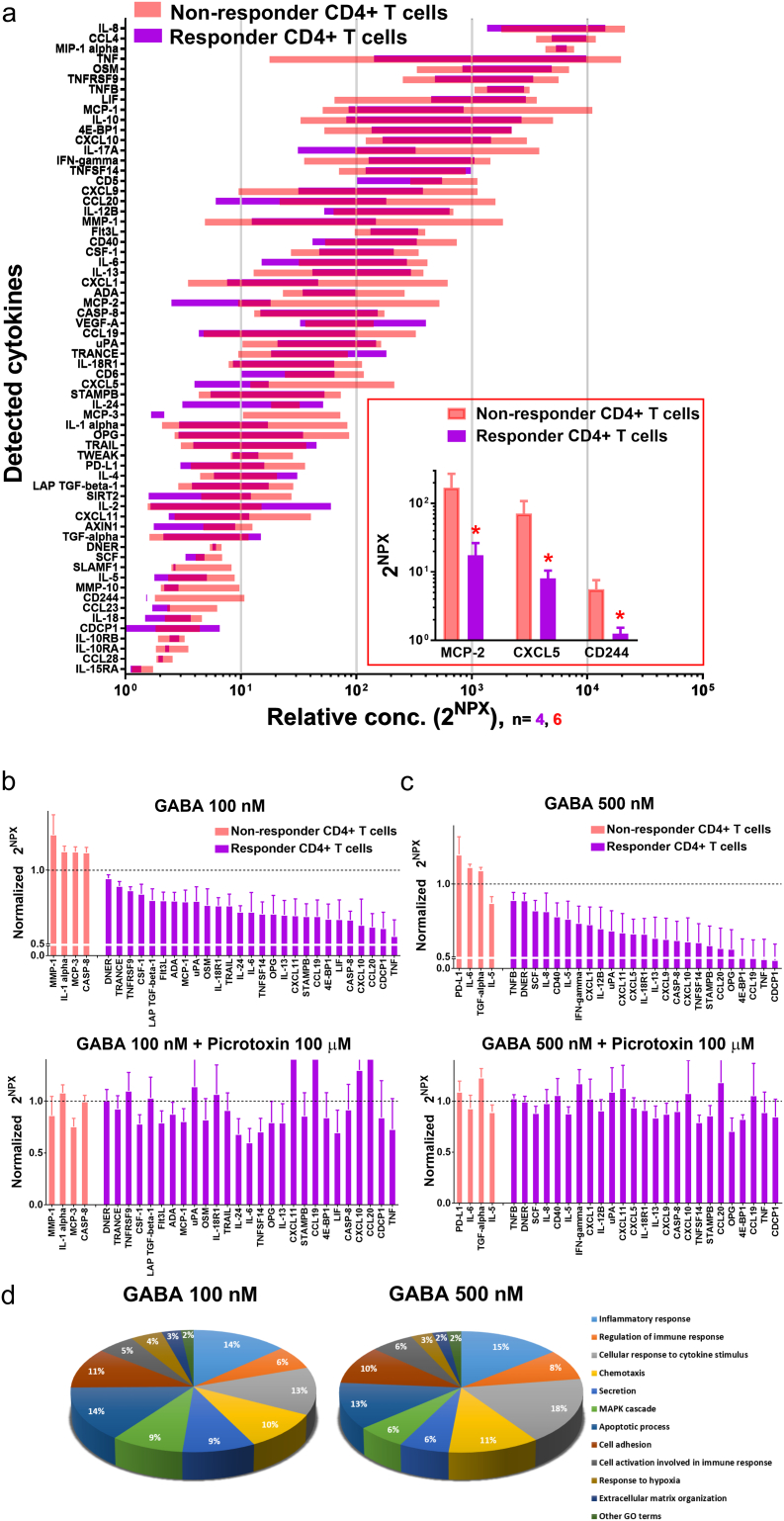
ND individuals could be divided into two groups based on whether or not their stimulated CD4+ cells responded to GABA in the proliferation assay (see Fig. 2c). We termed the two groups of stimulated CD4+ cells, responder (n = 15) and non-responder (n = 7) T cells. Stimulated T cells normally release more and higher levels of cytokines than resting T cells (Fig. S5). We examined if CD4+ T cells from responders and non-responders differentially secreted cytokines and then, if the release in the two groups were affected by GABA. Fig. 4a shows that upon stimulation, cells from both groups released a number of cytokines and to similar levels. Only levels of three cytokines were significantly different between the two groups (Fig. 4a insert). In contrast regulation of cytokine release by GABA was far more prominent in the responder as compared to the non-responder T cells. In non-responder T cells, GABA at 100 and 500 nM concentration regulated secretion of only four cytokines at each concentration (Fig. 4b, c; Table S8). This is in contrast to the responder T cells where GABA significantly inhibited release of many cytokines and, interestingly, the different concentrations of GABA inhibited release of somewhat different cytokines (Fig. 4b, c; Table S9). In the presence of 100 nM GABA, release of 27 cytokines were significantly decreased as compared to 25 cytokines in the presence of 500 nM GABA. Picrotoxin reversed the effects of GABA. Of these cytokines, secretion of 15 cytokines, including both Th1- and Th2-type cytokines e.g. TNF-α and IL-13, were inhibited by both 100 and 500 nM GABA (Fig. 5b). In the presence of 100 nM GABA, another 12 distinct cytokines, including the Th2-type IL-6 and IL-24 cytokines, were specifically inhibited. Inhibition of the Th1-type cytokines INF-γ and TNF-β plus the Th2-type cytokine IL-5 was observed only when GABA was present at 500 nM concentration. Fig. 4d shows that the proportion of cytokines associated with chemotaxis remained similar to what was determined for PBMCs from T1D patients. However, when the GABA concentration was increased from 100 to 500 nM, the proportion of cytokines associated with secretion and MAPK decreased, whereas those associated with cellular response to cytokine stimulus and regulation of immune response increased. The results demonstrate that GABA in a concentration-dependent manner regulates cytokine secretion from CD4+ T cells.
Fig. 4.
Identification of cytokines released into the culture media and effects of GABA treatment on CD4+ T cells cytokine secretion (a) Screening of 92 cytokines in culture media from GABA non-responder and from GABA responder CD4+ T cells, by Olink Multiplex PEA inflammation panel I, revealed expression of 64 cytokines. No GABA was added to the media. Data are represented by 2NPX values as floating bars (minimum to maximum) arranged in descending order of mean expression level of cytokines. Insert show cytokines with significant change in the expression levels in the media of GABA non-responder and GABA responder CD4+ T cells. Data are plotted as a bar graph with mean ± SEM. (b-c) Cytokines with significant change in the expression levels after (b) 100 nM GABA or (c) 500 nM GABA treatment of non-responder and responder CD4+ T cells, and then in the presence of GABA plus 100 μM picrotoxin (lower panel b, c). Mean and SEM for CXCL11, CCL19 and CCL20 are 1.55 ± 0.51, 1.72 ± 0.74, 1.43 ± 0.51, respectively. Data are represented by 2NPX values normalized to controls as a bar graph with mean ± SEM. Mean values with SEM and p values are shown in Tables S8, S9. (d) Classification based on the cellular functions of cytokines that were significantly altered by GABA 100 nM (27 cytokines) and GABA 500 nM (25 cytokines) treatment of responder CD4+ T cells from ND individuals.
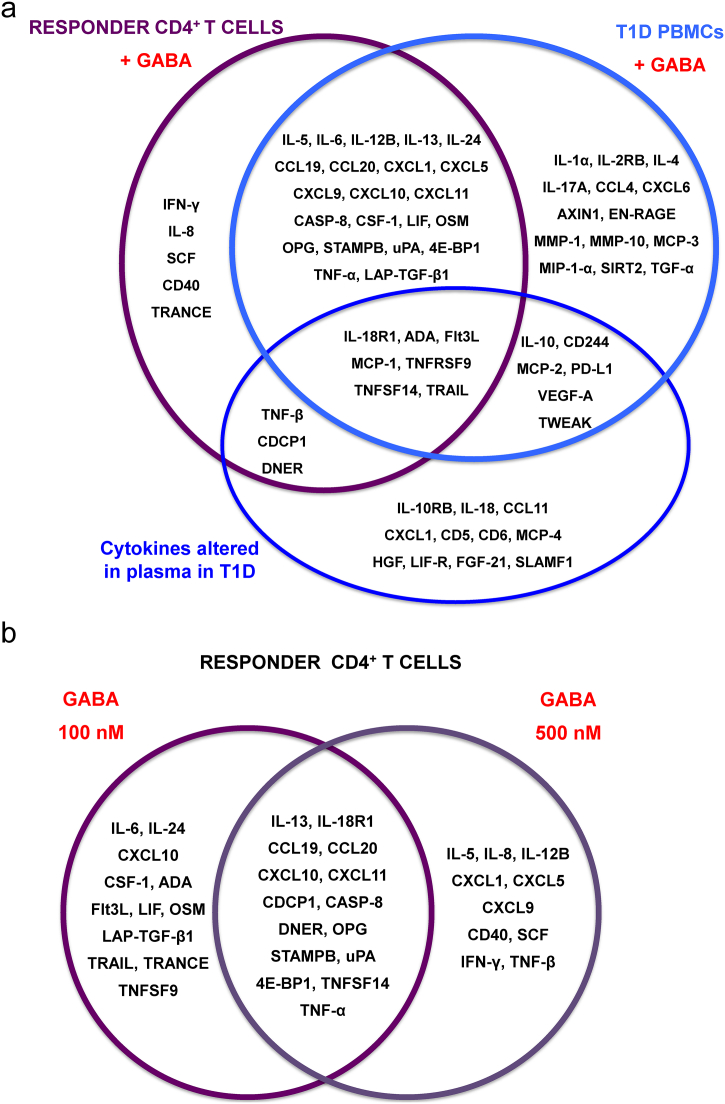
Fig. 5.
GABA and T1D regulate secretion of cytokines. (a) Purple circle: responder CD4+ T cell cytokines regulated by GABA (100 nM, 500 nM). Light blue circle: T1D PBMC cytokines regulated by GABA (100 nM). Dark blue circle: cytokines significantly altered in plasma from T1D patients as compared to ND individuals. (b) The cytokines regulated by 100 nM or 500 nM GABA or both concentrations.
4. Discussion
The results identify GABA as a potent regulator of cytokine secretion from human PBMCs and CD4+ T cells. GABA altered proliferation and cytokine secretion in a concentration-dependent manner and decreased the release of most of the cytokines. Importantly, immunomodulatory submicromolar GABA concentrations are normally present in plasma of both ND individuals and T1D patients.
We and others have shown that immune cells express the genes, proteins and functional GABAA receptors and also the genes encoding the chloride transporters that regulate the intracellular chloride concentration in the cells (Tian et al., 2004, Bjurstom et al., 2008, Bhat et al., 2010, Bhandage et al., 2015, Alam et al., 2006, Mendu et al., 2012, Jin et al., 2011b). The most prominent GABAA subunit in the PBMCs is the ρ2 subunit that can form homomeric or heteromeric receptors (Pan et al., 2006, Wang et al., 1994, Johnston et al., 2010, Bhandage et al., 2015). Interestingly, subtypes of GABAA receptors in the brain rarely contain the ρ2 subunit (Olsen and Sieghart, 2009) making the ρ2-containing GABAA receptors in the immune cells, potentially, a rather specific target for drug-development aimed at GABA signaling in immune cells.
In T1D, the mass of the pancreatic islet β cells is greatly diminished (Baekkeskov et al., 2017, Baekkeskov et al., 1990), highlighting that GABA in blood must be maintained by another mechanism than secretion from the pancreatic islets. An important source supplying GABA to blood might be the brain, by the way of the newly discovered mechanism of volume-transmission of small molecules from the brain, via the glymphatic system, to the periphery (Plog and Nedergaard, 2018). In pancreatic islets where the β cells are intact and secrete GABA, as in ND individuals, the islet interstitial GABA concentrations can be expected to fall within the GABA immunomodulatory range. In contrast, GABA immunosuppression in pancreatic islets of T1D patients is likely to decrease as the disease progresses and the β cells disappear.
This study reveals that GABA regulates secretion of a far greater number of cytokines than was previously known. In plasma of T1D patients, levels of 26 cytokines were increased and of those, 16 were inhibited by GABA in the cell assays (Fig. 1, Fig. 5). Moreover, of the 10 cytokines in plasma that correlated with the GABA plasma concentration, 7 cytokines were significantly increased in T1D patients and 5 of those (Flt3L, TRAIL, TNF-β, PD-L1, IL-10) were inhibited by GABA in the cell assays (Figs. 1d and 5). The few studies that previously have examined effects of GABA on cytokine secretion in immune cells, have only identified a limited number of cytokines e.g. IL-1β (Bhat et al., 2010), IL-2 (Tian et al., 1999), IFNγ (Tian et al., 2004), TNF-α (Duthey et al., 2010) and IL-6, IL-12 (Reyes-Garcia et al., 2007). Our study revealed that about three times more cytokines were inhibited by GABA in stimulated PBMCs from T1D individuals (47 cytokines) as compared to stimulated PBMCs from ND individuals (16 cytokines). We and others have previously shown that GABA can regulate proliferation of immune cells (Tian et al., 1999; Bjurstom et al., 2008; Jin et al., 2011b; Dionisio et al., 2011; Mendu et al., 2011; Tian et al., 2004). In this study, we used this effect of GABA to divide the stimulated CD4+ T cell samples from ND donors into non-responder and responder groups in terms of proliferation and then, examined how these groups differed in cytokine secretion. GABA effectively decreased proliferation and secretion of cytokines only in the responder group. Here GABA decreased secretion of 37 cytokines in a concentration-dependent manner. Of the inhibited cytokines from T1D stimulated PBMCs and the responder cells, 29 cytokines were common to both cell populations (Fig. 5a). Although the GABA-induced immunosuppression was somewhat similar for the responder and T1D cell populations, it clearly differed for a number of specific cytokines (Fig. 5a). Furthermore, GABA did not decrease proliferation of stimulated ND PBMCs nor proliferation or cytokine secretion in the non-responder T cell population. The dramatic effect GABA had on stimulated T1D PBMCs and responder T cell but not on non-responder T cells, implies that GABA is a central molecule determining cytokine secretion in primed, activated cells. It is possible that GABA has a homeostatic role in the immune system acting as a “brake” but not a “full stop”.
GABA inhibited cytokines involved in chemotaxis in stimulated T1D PBMCs more than in ND PBMCs cells. When the GABA concentration was increased from 100 to 500 nM for the stimulated responder T cells, the prominence of inhibited cytokines associated with secretion and MAPK was decreased. In contrast, inhibition of cytokines that affect either the cellular response to cytokine stimulus or regulate the immune response increased in 500 nM GABA. The specific profile of cytokines regulated by GABA indicates that the 100 nM GABA response tended to modulated levels of Th2-type cytokines, whereas the 500 nM GABA inhibited both Th1- and Th2-type cytokine release (Fig. 5a, b). The results are consistent with a concentration-dependent immunomodulatory effect of GABA.
Interestingly, of the 10 genes down-regulated more than two-fold in the CD3+ T cells from T1D individuals, six were associated with cholesterol biosynthesis (Fig. 2j) and included MSMO1 and CYP51A1 that correlated negatively with the plasma GABA concentrations (Fig. S4). MSMO1 further correlated negatively with HbA1c, fasting glucose and BMI raising the question of its potential suitability as a biomarker in T1D. Non-random distribution of proteins in the cell membranes are dependent on e.g. cholesterol and the actin cytoskeleton (Goyette and Gaus, 2017). The T cell receptor resides in lipid rafts that are microdomains within the plasma membrane and where cholesterol is an essential component and contributes to membrane fluidity and signal transduction (Kidani and Bensinger, 2016; Hubler and Kennedy, 2016). The change in cholesterol biosynthesis genes observed in the T1D T cells are likely to have an impact on signaling processes associated with signaling complexes located in the cell membrane.
The general inhibitory effect of GABA on the release of cytokines from the immune cells was enlightening. It lends further support to the immunomodulatory function of GABA (Jin et al., 2011b; Barragan et al., 2015; Fiorina, 2013). It also highlights the importance of extracellular GABA in the blood and in tissues such as the pancreatic islets and the brain. Clearly, future studies are required to identify and characterize the various subpopulations of immune cells with respect to their response to GABA and further determine how GABA, in a concentration-dependent manner, manages to differentially regulate secretion of cytokines from the cells.
Funding Sources
This work was supported by the Swedish Research Council Grant to B.B. (Dnr 2015-02417), The Swedish Diabetes Foundation, The Swedish Child Diabetes Foundation, the strategic grant consortium Excellence of Diabetes Research in Sweden (EXODIAB), Family Ernfors Foundation and Uppsala University. Thurings Foundation to A.K·B., and the European Union, Seventh Framework Program, Gastric Glyco Explorer Initial Training Network: Grant No 316929 to M.K.-M.
Conflict of Interest
B. Birnir has filed a patent application based on the findings described in this manuscript. All others authors (AKB, ZJ, SVK, QS, YP, QD, DE, POC, MKM) declare no conflicts of interests.
Author Contributions
B.B. and A.K.B. designed the study. A.K.B executed most experiments. Z.J., S.V.K. participated in electrophysiological experiments, Q.S. in PEA experiments, Y.P., Q.D., Z.J. performed RNAseq experiments. A.K.B., Z.J., S.V.K., Q.S., Y.P., Q.D., M.K.M. and B.B. analyzed data. D.E. and P.O.C. collected blood samples and demographic data from study participants. B.B. and A.K.B. wrote the article that was then critically reviewed by M.K.M., Z.J., S.V.K. and then the other co-authors. We thank Karin Nygren for technical assistance and Priya Gohel for help with the western blot. B.B. is the guarantor of this work and, as such, has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.03.019.
Appendix A. Supplementary data
Supplementary material
References
- Abu Shmais G.A., Al-Ayadhi L.Y., Al-Dbass A.M., El-Ansary A.K. Mechanism of nitrogen metabolism-related parameters and enzyme activities in the pathophysiology of autism. J. Neurodev. Disord. 2012;4:4. doi: 10.1186/1866-1955-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Laughton D.L., Walding A., Wolstenholme A.J. Human peripheral blood mononuclear cells express GABAA receptor subunits. Mol. Immunol. 2006;43:1432–1442. doi: 10.1016/j.molimm.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Assarsson E., Lundberg M., Holmquist G., Bjorkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., Andersson A.C., Lindstedt P., Stenvang J., Gullberg M., Fredriksson S. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;e95192:9. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekkeskov S., Aanstoot H.J., Christgau S., Reetz A., Solimena M., Cascalho M., Folli F., Richter-Olesen H., de Camilli P. Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature. 1990;347:151–156. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- Baekkeskov S., Hubbell J.A., Phelps E.A. Bioengineering strategies for inducing tolerance in autoimmune diabetes. Adv. Drug Deliv. Rev. 2017;114:256–265. doi: 10.1016/j.addr.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Barragan A., Weidner J.M., Jin Z., Korpi E.R., Birnir B. GABAergic signalling in the immune system. Acta Physiol (Oxford) 2015;213:819–827. doi: 10.1111/apha.12467. [DOI] [PubMed] [Google Scholar]
- Bhandage A.K., Hellgren C., Jin Z., Olafsson E.B., Sundstrom-Poromaa I., Birnir B. Expression of GABA receptors subunits in peripheral blood mononuclear cells is gender dependent, altered in pregnancy and modified by mental health. Acta Physiol (Oxford) 2015;213:575–585. doi: 10.1111/apha.12440. [DOI] [PubMed] [Google Scholar]
- Bhandage A.K., Jin Z., Hellgren C., Korol S.V., Nowak K., Williamsson L., Sundstrom-Poromaa I., Birnir B. AMPA, NMDA and kainate glutamate receptor subunits are expressed in human peripheral blood mononuclear cells (PBMCs) where the expression of GluK4 is altered by pregnancy and GluN2D by depression in pregnant women. J. Neuroimmunol. 2017;305:51–58. doi: 10.1016/j.jneuroim.2017.01.013. [DOI] [PubMed] [Google Scholar]
- Bhat R., Axtell R., Mitra A., Miranda M., Lock C., Tsien R.W., Steinman L. Inhibitory role for GABA in autoimmune inflammation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2580–2585. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnir B., Korpi E.R. The impact of sub-cellular location and intracellular neuronal proteins on properties of GABAA receptors. Curr. Pharm. Des. 2007;13:3169–3177. doi: 10.2174/138161207782341330. [DOI] [PubMed] [Google Scholar]
- Bjurstom H., Wang J., Ericsson I., Bengtsson M., Liu Y., Kumar-Mendu S., Issazadeh-Navikas S., Birnir B. GABA, a natural immunomodulator of T lymphocytes. J. Neuroimmunol. 2008;205:44–50. doi: 10.1016/j.jneuroim.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Bu D.F., Erlander M.G., Hitz B.C., Tillakaratne N.J., Kaufman D.L., Wagner-Mcpherson C.B., Evans G.A., Tobin A.J. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc. Natl. Acad. Sci. U S A. 1992;89:2115–2119. doi: 10.1073/pnas.89.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo A. Paracrine and autocrine interactions in the human islet: more than meets the eye. Semin. Cell Dev. Biol. 2013;24:11–21. doi: 10.1016/j.semcdb.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio L., Jose De Rosa M., Bouzat C., Esandi Mdel C. An intrinsic GABAergic system in human lymphocytes. Neuropharmacology. 2011;60:513–519. doi: 10.1016/j.neuropharm.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthey B., Hubner A., Diehl S., Boehncke S., Pfeffer J., Boehncke W.H. Anti-inflammatory effects of the GABAB receptor agonist baclofen in allergic contact dermatitis. Exp. Dermatol. 2010;19:661–666. doi: 10.1111/j.1600-0625.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- Edvinsson A., Brann E., Hellgren C., Freyhult E., White R., Kamali-Moghaddam M., Olivier J., Bergquist J., Bostrom A.E., Schioth H.B., Skalkidou A., Cunningham J.L., Sundstrom-Poromaa I. Lower inflammatory markers in women with antenatal depression brings the M1/M2 balance into focus from a new direction. Psychoneuroendocrinology. 2017;80:15–25. doi: 10.1016/j.psyneuen.2017.02.027. [DOI] [PubMed] [Google Scholar]
- El-Ansary A.K., Bacha A.B., Ayahdi L.Y. Relationship between chronic lead toxicity and plasma neurotransmitters in autistic patients from Saudi Arabia. Clin. Biochem. 2011;44:1116–1120. doi: 10.1016/j.clinbiochem.2011.06.982. [DOI] [PubMed] [Google Scholar]
- Farrant M., Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABAA receptors. Nat. Rev. Neurosci. 2005;6:215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- Fiorina P. GABAergic system in beta-cells: from autoimmunity target to regeneration tool. Diabetes. 2013;62:3674–3676. doi: 10.2337/db13-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R., Pacheco R., Lluis C., Ahern G.P., O'connell P.J. The emergence of neurotransmitters as immune modulators. Trends Immunol. 2007;28:400–407. doi: 10.1016/j.it.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Fuks J.M., Arrighi R.B., Weidner J.M., Kumar Mendu S., Jin Z., Wallin R.P., Rethi B., Birnir B., Barragan A. GABAergic signaling is linked to a hypermigratory phenotype in dendritic cells infected by Toxoplasma gondii. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann M., Shaban H., Vigot R., Sansig G., Haller C., Barbieri S., Humeau Y., Schuler V., Muller M., Kinzel B., Klebs K., Schmutz M., Froestl W., Heid J., Kelly P.H., Gentry C., Jaton A.L., van der Putten H., Mombereau C., Lecourtier L., Mosbacher J., Cryan J.F., Fritschy J.M., Luthi A., Kaupmann K., Bettler B. Redistribution of GABAB1 protein and atypical GABAB responses in GABAB2-deficient mice. J. Neurosci. 2004;24:6086–6097. doi: 10.1523/JNEUROSCI.5635-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyette J., Gaus K. Mechanisms of protein nanoscale clustering. Curr. Opin. Cell Biol. 2017;44:86–92. doi: 10.1016/j.ceb.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Hubler M.J., Kennedy A.J. Role of lipids in the metabolism and activation of immune cells. J. Nutr. Biochem. 2016;34:1–7. doi: 10.1016/j.jnutbio.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Jin Y., Birnir B. GABA-activated single-channel and tonic currents in rat brain slices. J. Vis. Exp. 2011;53 doi: 10.3791/2858. pii 2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Mendu S.K., Birnir B. GABA is an effective immunomodulatory molecule. Amino Acids. 2011;45:87–94. doi: 10.1007/s00726-011-1193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G.A., Chebib M., Hanrahan J.R., Mewett K.N. Neurochemicals for the investigation of GABAC receptors. Neurochem. Res. 2010;35:1970–1977. doi: 10.1007/s11064-010-0271-7. [DOI] [PubMed] [Google Scholar]
- Kanaani J., Cianciaruso C., Phelps E.A., Pasquier M., Brioudes E., Billestrup N., Baekkeskov S. Compartmentalization of GABA synthesis by GAD67 differs between pancreatic beta cells and neurons. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani Y., Bensinger S.J. Modulating cholesterol homeostasis to build a better T cell. Cell Metab. 2016;23:963–964. doi: 10.1016/j.cmet.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Kreth S., Heyn J., Grau S., Kretzschmar H.A., Egensperger R., Kreth F.W. Identification of valid endogenous control genes for determining gene expression in human glioma. Neuro-Oncology. 2010;12:570–579. doi: 10.1093/neuonc/nop072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larssen P., Wik L., Czarnewski P., Eldh M., Lof L., Ronquist K.G., Dubois L., Freyhult E., Gallant C.J., Oelrich J., Larsson A., Ronquist G., Villablanca E.J., Landegren U., Gabrielsson S., Kamali-Moghaddam M. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol. Cell. Proteomics. 2017;16:502–511. doi: 10.1074/mcp.M116.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A., Carlsson L., Gordh T., Lind A.L., Thulin M., Kamali-Moghaddam M. The effects of age and gender on plasma levels of 63 cytokines. J. Immunol. Methods. 2015;425:58–61. doi: 10.1016/j.jim.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Ledderose C., Heyn J., Limbeck E., Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res. Notes. 2011;4:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Schwab C., Mcgeer P.L. Astrocytes are GABAergic cells that modulate microglial activity. Glia. 2011;59:152–165. doi: 10.1002/glia.21087. [DOI] [PubMed] [Google Scholar]
- Levite M. Neurotransmitters activate T-cells and elicit crucial functions via neurotransmitter receptors. Curr. Opin. Pharmacol. 2008;8:460–471. doi: 10.1016/j.coph.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Levite M., Chowers Y., Ganor Y., Besser M., Hershkovits R., Cahalon L. Dopamine interacts directly with its D3 and D2 receptors on normal human T cells, and activates beta1 integrin function. Eur. J. Immunol. 2001;31:3504–3512. doi: 10.1002/1521-4141(200112)31:12<3504::aid-immu3504>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang Z., Liu X., Wang Y., Mao F., Mao J., Lu X., Jiang D., Wan Y., Lv J.Y., Cao G., Zhang J., Zhao N., Atkinson M., Greiner D.L., Prud'homme G.J., Jiao Z., Li Y., Wang Q. Study of GABA in healthy volunteers: pharmacokinetics and pharmacodynamics. Front. Pharmacol. 2015;6:260. doi: 10.3389/fphar.2015.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall F.H., Jones K.A., Kaupmann K., Bettler B. GABAB receptors - the first 7TM heterodimers. Trends Pharmacol. Sci. 1999;20:396–399. doi: 10.1016/s0165-6147(99)01383-8. [DOI] [PubMed] [Google Scholar]
- Mendu S.K., Akesson L., Jin Z., Edlund A., Cilio C., Lernmark A., Birnir B. Increased GABAA channel subunits expression in CD8+ but not in CD4+ T cells in BB rats developing diabetes compared to their congenic littermates. Mol. Immunol. 2011;48:399–407. doi: 10.1016/j.molimm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Mendu S.K., Bhandage A., Jin Z., Birnir B. Different subtypes of GABAA receptors are expressed in human, mouse and rat T lymphocytes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R.W., Sieghart W. International Union of Pharmacology. LXX. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol. Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R.W., Sieghart W. GABAA receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology. 2009;56:141–148. doi: 10.1016/j.neuropharm.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Ripps H., Qian H. Random assembly of GABA rho1 and rho2 subunits in the formation of heteromeric GABAC receptors. Cell. Mol. Neurobiol. 2006;26:289–305. doi: 10.1007/s10571-006-9001-8. [DOI] [PubMed] [Google Scholar]
- Petty F., Sherman A.D. Plasma GABA levels in psychiatric illness. J. Affect. Disord. 1984;6:131–138. doi: 10.1016/0165-0327(84)90018-1. [DOI] [PubMed] [Google Scholar]
- Picelli S., Bjorklund A.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Picelli S., Bjorklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plog B.A., Nedergaard M. The glymphatic system in central nervous system health and disease: past, present, and future. Annu. Rev. Pathol. 2018;13:379–394. doi: 10.1146/annurev-pathol-051217-111018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Garcia M.G., Hernandez-Hernandez F., Hernandez-Tellez B., Garcia-Tamayo F. GABAA receptor subunits RNA expression in mice peritoneal macrophages modulate their IL-6/IL-12 production. J. Neuroimmunol. 2007;188:64–68. doi: 10.1016/j.jneuroim.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Ring H., Mendu S.K., Shirazi-Fard S., Birnir B., Hallbook F. GABA maintains the proliferation of progenitors in the developing chick ciliary marginal zone and non-pigmented ciliary epithelium. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Tian J., Chau C., Hales T.G., Kaufman D.L. GABAA receptors mediate inhibition of T cell responses. J. Neuroimmunol. 1999;96:21–28. doi: 10.1016/s0165-5728(98)00264-1. [DOI] [PubMed] [Google Scholar]
- Tian J., Lu Y., Zhang H., Chau C.H., Dang H.N., Kaufman D.L. Gamma-aminobutyric acid inhibits T cell autoimmunity and the development of inflammatory responses in a mouse type 1 diabetes model. J. Immunol. 2004;173:5298–5304. doi: 10.4049/jimmunol.173.8.5298. [DOI] [PubMed] [Google Scholar]
- Tian J., Dang H., Chen Z., Guan A., Jin Y., Atkinson M.A., Kaufman D.L. gamma-Aminobutyric acid regulates both the survival and replication of human beta-cells. Diabetes. 2013;62:3760–3765. doi: 10.2337/db13-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Li Z., Zhang H., Xia L., Zhang M., Xu Y., Wang Z., Deem M.W., Sun X., He J. T cell repertoire diversity is decreased in type 1 diabetes patients. Genom. Proteom. Bioinform. 2016;14:338–348. doi: 10.1016/j.gpb.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.L., Guggino W.B., Cutting G.R. A novel gamma-aminobutyric acid receptor subunit (rho 2) cloned from human retina forms bicuculline-insensitive homooligomeric receptors in Xenopus oocytes. J. Neurosci. 1994;14:6524–6531. doi: 10.1523/JNEUROSCI.14-11-06524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material